Abstract
Germ cell development creates totipotency through genetic as well as epigenetic regulation of the genome function. Primordial germ cells (PGCs) are the first germ cell population established during development and are immediate precursors for both the oocytes and spermatogonia. We here summarize recent findings regarding the mechanism of PGC development in mice. We focus on the transcriptional and signaling mechanism for PGC specification, potential pluripotency, and epigenetic reprogramming in PGCs and strategies for the reconstitution of germ cell development using pluripotent stem cells in culture. Continued studies on germ cell development may lead to the generation of totipotency in vitro, which should have a profound influence on biological science as well as on medicine.
A key function of the germline is to establish totipotency. Understanding the transcriptional and signaling mechanisms behind primordial germ cell development may lead to generating totipotency in vitro.
1. INTRODUCTION
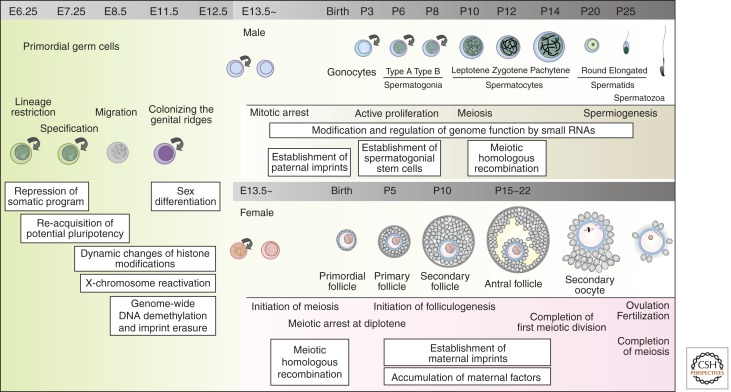
In most multicellular organisms, including mammals, germ cells are the origin of new organisms and ensure the perpetuation of the genetic and epigenetic information across the generations. Accordingly, they prepare for totipotency during their ontogeny through genetic and epigenetic regulations of their genome function (Fig. 1).
Figure 1.
A schematic representation of germ cell development in mice. A brief outline of germ cell development in mice is shown schematically. Key events associated with each stage of germ cell development are also shown. (Left) PGC development (from E6.25 to ∼E12.5); (upper right) male germ cell development (from ∼E13.5); (lower right) female germ cell development (from ∼E13.5).
There are several key properties/critical themes commonly associated with germ cell development in animals. Generally, the germ cell lineage is set aside from the somatic lineages early in development, either through localized maternal determinants (most typically referred to as germ plasm) or induction from pluripotent embryonic cells (Box 1). In many animals, primordial germ cells (PGCs) are the first germline cell population (Saitou and Yamaji 2010), which colonize the developing gonads by active migration (Richardson and Lehmann 2010). In the gonads, PGCs initiate differentiation either toward a spermatogenic (male) or an oogenic (female) pathway. The spermatogenic pathway most typically involves the establishment of spermatogonial stem cells (or male germline stem cells [GSCs]) with an enormous mitotic potential, whereas it appears that female GSCs in adults are rare across the phylogenetic spectrum (Spradling et al. 2011): Caenorhabditis elegans, Drosophila, and medaka fish (Nakamura et al. 2010) seem to be among the rare examples of female GSCs in adults. In other animals, including mammals, the oogenesis is dependent on PGCs set aside in development, although some cells in the adult ovary may retain germline potential (White et al. 2012).
BOX 1. PREFORMATION VERSUS EPIGENESIS IN GERM CELL SPECIFICATION.
There are two distinct mechanisms for the specification of the germ cell lineage: One is through the inheritance of localized determinants, often called the germ plasm, in the egg (pre-formation), and the other is through the induction from pluripotent embryonic cells (epigenesis) (Extavour and Akam 2003). The former mode is seen in many model organisms, including C. elegans, D. melanogaster, Danio rerio, and Xenopus laevis, whereas the latter mode is seen typically in mammals. Evolutionarily, the latter epigenesis mode appears to be more prevalent and ancestral to the metazoa, whereas the pre-formation mode appears to have arisen independently at multiple times, probably because of its better ability to secure the germline at the very onset of development (Extavour and Akam 2003).
Although the mechanisms for the segregation of the germ cell fate are divergent, some of the properties that the specified PGCs are endowed with appear to be conserved. One such property is the repression in PGCs of the somatic differentiation program robustly activated in surrounding somatic cells. In C. elegans and D. melanogaster, this is achieved through transient global repression of the RNA polymerase II (Pol II) activity by PIE-1 and the polar granule component, respectively (Nakamura et al. 1996; Seydoux et al. 1996). PIE-1, bearing two CCCH RNA-binding domains but showing no similarity to known transcriptional repressors, inhibits the transcriptional initiation and elongation activity of RNAPII by distinct mechanisms (Ghosh and Seydoux 2008). As a mechanism to inhibit transcriptional elongation, PIE-1 appears to inhibit the positive transcriptional elongation factor b (P-TEFb), which promotes the transcriptional elongation activity of RNAPII by phosphorylating the Ser-2 of the carboxy-terminal domain (CTD) of RNAPII (Zhang et al. 2003). On the other hand, the polar granule component, which is a short, 71-amino-acid protein conserved only among Drosophila species, inhibits the recruitment of P-TEFb to transcriptional sites (Hanyu-Nakamura et al. 2008). In contrast, in mice, germ cell specification requires active transcription, but the transcriptional repressor BLIMP1 specifically shuts off gene expression for a somatic mesodermal program (Kurimoto et al. 2008) (see main text).
In this article, we focus on recent advances in the study of PGCs in mice on the mechanisms of PGC specification, potential pluripotency, and genome-wide epigenetic reprogramming in PGCs, as well as the induction of PGC fate in culture. Related themes, such as the properties of germline stem cells and small RNA-based genome defense mechanisms, have been reviewed elsewhere (Malone and Hannon 2009; Saito and Siomi 2010; Spradling et al. 2011).
2. GERM CELL DEVELOPMENT IN MICE
In the mouse, PGCs first become identifiable as a cluster of approximately 40 cells at the base of the incipient allantois at around embryonic day 7.25 (∼E7.25) (Figs. 1 and 2) (Chiquoine 1954; Ginsburg et al. 1990). They migrate to the developing hindgut endoderm at ∼E7.75, into the mesentery at ∼E9.5, and colonize the genital ridges at ∼E10.5 (Tam and Snow 1981; Molyneaux et al. 2001; Seki et al. 2007; Richardson and Lehmann 2010). A key event that occurs in PGCs during this proliferative phase both in the male and female is epigenetic reprogramming—most notably, a genome-wide DNA demethylation that includes the erasure of genomic imprinting (Fig. 1) (Saitou et al. 2012).
Figure 2.
A schematic representation of PGC development in mice. A brief outline of PGC specification, migration, and colonization of the gonads in mice is shown schematically. (Green) Blimp1- and Prdm14-positive cells that appear in the most proximal epiblasts. (Lower middle panel) A view of the transverse section of the position indicated by a red bar in the lower left panel. Epi, epiblast; (AVE, anterior visceral endoderm; ExE, extra-embryonic ectoderm; PGCs, primordial germ cells; EM, embryonic mesoderm; ExM, extra-embryonic mesoderm; Hg, hindgut; DA, dorsal aorta; NT, neural tube; NoC, notochord; IM, intermediate mesoderm; NeC, nephrogenic cord.
In the female XX embryo, the PGCs continue to proliferate until ∼E13.5 (when they reach around 25,000 in number) and subsequently enter into the prophase I of meiotic divisions (Hilscher et al. 1974; Speed 1982). Subsequently, they are arrested at the diplotene stage of prophase I of meiosis. Upon hormonal stimulation, oocytes complete the first meiotic division with concomitant extrusion of the first polar body. At the fertilization with a haploid spermatozoon, the oocyte completes the second meiotic division and extrudes the second polar body. In contrast to those in the female, XY PGCs enter into mitotic arrest upon entry into the genital ridges, and stay quiescent in the G0/G1 phase of the cell cycle for the remaining embryonic period (Hilscher et al. 1974; Western et al. 2008). Around day 5 postpartum (P5), many of them resume active proliferation, while some are recruited as spermatogonial stem cells (SSCs) (Fig. 1) (Russell 1990; Yoshida 2010; Spradling et al. 2011). In culture, germline stem cells (GSCs) bearing the abilities for long-term proliferation and for spermatogenesis upon transplantation into testes are established in the presence of GDNF (glial cell–derived neurotrophic factor), most readily from neonatal testes (Kanatsu-Shinohara et al. 2003). Thus, compared with the very limited size of the oocyte pools, the spermatozoa can be supplied essentially infinitely by the SSC system.
3. MECHANISM FOR PGC SPECIFICATION
3.1. Gene Expression Dynamics during PGC Specification
A single-cell analysis of gene expression of the founder population of PGCs at E7.25 identified two genes, Fragilis and Stella, that are highly and specifically, respectively, expressed in PGCs (Saitou et al. 2002). Fragilis (also known as mouse interferon-induced protein like gene-1, mil-1/interferon-induced transmembrane protein 3, Ifitm3) (Tanaka and Matsui 2002) is a member of the interferon-inducible transmembrane proteins, whereas Stella (also known as Primordial germ cell 7, Pgc7/developmental pluripotency-associated 3, Dppa3) (Sato et al. 2002) is a small, highly basic nucleo–cytoplasmic shuttling protein. Fragilis begins to show expression around the most proximal epiblast cells at ∼E6.25–E6.5, and its expression intensifies in the posterior extra-embryonic mesoderm, where AP-positive PGCs arise at ∼E7.0–E7.25. Stella begins to express specifically in Fragilis-expressing cells in the extra-embryonic mesoderm at ∼E7.0–E7.25 and continues to be expressed in migrating PGCs. The Stella-positive cells show high expression of tissue nonspecific alkaline phosphatase (Tnap), a gene for AP activity of PGCs (MacGregor et al. 1995). Cells with positivity for Stella and high levels of Fragilis repress the expression of Homeobox genes such as Hoxb1 and Hoxa1, whereas Fragilis-positive but Stella-negative cells retain Hox gene expression (Saitou et al. 2002). It has thus been proposed that the Stella-positive and Hox-negative cells are the established PGCs (Saitou et al. 2002). Gene-knockout studies, however, revealed that neither Fragilis nor Stella is essential for PGC specification (Payer et al. 2003; Lange et al. 2008). Instead, Stella has been found to be a critical factor to protect the maternal genome and paternally imprinted genes from genome-wide DNA demethylation that occurs in the zygotes (Nakamura et al. 2007), whereas Fragilis has a critical function in restricting the replication of multiple pathogenic viruses including influenza (Brass et al. 2009; Everitt et al. 2012).
Further screenings of the PGC transcriptome led to the identification of two key regulators for PGC specification (Fig. 3; See Table 1 for key genes affecting PGC specification and proliferation/survival): Blimp1 (B-lymphocyte-induced maturation protein 1, also known as PR domain-containing protein 1 [Prdm1]) and Prdm14 (PR domain-containing protein 14) (Ohinata et al. 2005; Vincent et al. 2005; Yabuta et al. 2006; Kurimoto et al. 2008; Yamaji et al. 2008). BLIMP1 and PRDM14 are evolutionarily conserved proteins. Together with Tcfap2c (also known as AP2γ) (Weber et al. 2010), these genes are found to be required for PGC specification in mammals.
Figure 3.
Transcriptional regulation of PGC specification. A potential genetic network for PGC specification is shown schematically. Arrows and lines with terminal bars indicate genetic pathways for activation and for repression, respectively, as demonstrated by in vivo experiments (Kurimoto et al. 2008; Yamaji et al. 2008). Dotted arrows and dotted lines with terminal bars indicate genetic pathways for activation and for repression, respectively, as proposed based on in vitro experiments (Covello et al. 2006; West et al. 2009; Weber et al. 2010).
Table 1.
Key genes affecting PGC specification and proliferation/survival
| Gene | Function | Expressiona | Loss-of-function phenotype in PGCs |
|---|---|---|---|
| PGC SPECIFICATION | |||
| Cytokines | |||
| Bmp4 | A BMP/GDF/MIS subfamily ligand of the TGF-β family | ICM, ExE from E5.5, ExM during gastrulation | No and reduced number of PGCs in homozygous and heterozygous mutants, respectively; loss of Blimp1 and Prdm14 expression in homozygous mutants; loss of Bmp4 in the ExM leading to abnormal PGC localization and survival (Lawson et al. 1999; Fujiwara et al. 2001; Yamaji et al. 2008; Ohinata et al. 2009) |
| Bmp8b | A BMP/GDF/MIS subfamily ligand of the TGF-β family | ExE from E5.5 | No and reduced number of PGCs in homozygous and heterozygous mutants, respectively (C57BL/6 background); severely impaired Blimp1 and Prdm14 expression in homozygous mutants (Ying et al. 2000; Ohinata et al. 2009) |
| Bmp2 | A BMP/GDF/MIS subfamily ligand of the TGF-β family | VE | Reduced numbers of PGCs both in the heterozygous and homozygous mutants (C57BL/6 background) (Ying and Zhao 2001) |
| Wnt3 | A secreted glycoprotein of the Wnt family | Posterior proximal epiblast and VE, primitive streak, mesodermal wing | Loss of Blimp1 and Prdm14 expression in homozygous mutants (Ohinata et al. 2009) |
| Signal transducers | |||
| Smad1 | A receptor Smad for the BMP/GDF/MIS subfamily | Ubiquitous | No PGCs in homozygous mutants; severely impaired Blimp1 and Prdm14 expression in homozygous mutants (Tremblay et al. 2001; Hayashi et al. 2002; Aubin et al. 2004; Yamaji et al. 2008; Ohinata et al. 2009) |
| Smad5 | A receptor Smad for the BMP/GDF/MIS subfamily | Ubiquitous | No and reduced number of PGCs in homozygous and heterozygous mutants, respectively (Chang and Matzuk 2001) |
| Smad4 | A common partner for all receptor Smads | Ubiquitous | Severely reduced number of PGCs in epiblast-specific Smad4 knockouts (Chu et al. 2004) |
| Smad2 | A receptor Smad for the TGF-β/Activin/Nodal subfamily | Ubiquitous | Abundant PGCs at E8.5 in surviving embryos; widespread Blimp1 expression in early gastrulating embryos (Ohinata et al. 2009) |
| FoxH1 | A winged-helix transcription factor | Ubiquitous | Widespread Blimp1 expression in early gastrulating embryos (Ohinata et al. 2009) |
| Receptors | |||
| Alk2 | A type I BMP receptor (serine/threonine kinase) | Primarily VE before gastrulation | No and reduced number of PGCs in homozygous and heterozygous mutants, respectively (de Sousa Lopes et al. 2004) |
| Transcription factors (TFs) | |||
| Blimp1 | A TF with PR domain and Zinc fingers | PGCs, VE, many other lineages | Impaired PGC specification; reduced number of PGCs in heterozygous mutants (Vincent et al. 2005; Ohinata et al. 2009) |
| Prdm14 | A TF with PR domain and zinc fingers | PGCs, ICM | Impaired PGC specification (Yamaji et al. 2008) |
| Tcfap2c | A basic helix–span–helix TF | All cells until the blastocyst stage, ExE, PGCs | Impaired PGC specification in epiblast-specific Tcfap2c knockouts (Weber et al. 2010) |
| Oct4 | A POU-homeobox domain TF | Ubiquitous in the embryo proper until E7.5, specific to germ cells thereafter | Impaired PGC specification; apoptosis of PGCs (Kehler et al. 2004; Okamura et al. 2008) |
| RNA-binding protein | |||
| Lin28 | An RNA-binding protein | Ubiquitous | Reduced number of PGCs by RNAi-mediated Lin28 knockdown (West et al. 2009) |
| Cell adhesion molecules | |||
| Cdh1 | A homophilic cell–cell adhesion molecule | Epiblast, PGCs, and their somatic neighbors | An inhibitory antibody blocking PGC specification (Okamura et al. 2003) |
| PGC PROLIFERATION/SURVIVAL | |||
| Cytokines | |||
| Scf | A short-chain helical cytokine | The migratory path of PGCs | Impaired proliferation/survival and migration of PGCs (McCoshen and McCallion 1975; Gu et al. 2009) |
| Cxcl12 | A chemokine ligand | Gonad and subjacent mesonephric tissue at E12.5, widespread in embryos | Impaired migration of PGCs (Ara et al. 2003; Molyneaux et al. 2003) |
| Fgf9 | An FGF family ligand | Specific to male gonads after E12.5 | Impaired survival of male PGCs after E11.5 (DiNapoli et al. 2006) |
| Bmp7 | A BMP/GDF/MIS subfamily ligand of the TGF-β family | Specific to male gonads after E12.5 | Impaired PGC proliferation during E10.5–E11.5 in males (Ross et al. 2007) |
| Wnt5a | A secreted glycoprotein of the Wnt family | Widespread, up-regulated in testicular interstitial cells after E13.5 | Impaired proliferation/survival and migration of PGCs (Chawengsaksophak et al. 2011; Laird et al. 2011) |
| Signal transducers | |||
| Pten | A lipid phosphatase | Ubiquitous | Bilateral testicular teratoma (Kimura et al. 2003) |
| Bax | A proapoptotic factor of the Bcl2 family | Ubiquitous | Enhanced PGC survival (Stallock et al. 2003; Runyan et al. 2006; Suzuki et al. 2008; Cook et al. 2009; Gu et al. 2009) |
| Receptors | |||
| Kit | A receptor tyrosine kinase | PGCs | Impaired proliferation/survival and migration of PGCs (Mintz and Russell 1957; Buehr et al. 1993) |
| Cxcr4 | A seven-transmembrane spanning G-protein-coupled receptor | PGCs | Impaired migration/survival of PGCs (Ara et al. 2003; Molyneaux et al. 2003) |
| Fgfr2-IIIb | A receptor tyrosine kinase | Widespread | Impaired PGC survival at E11.5 (Takeuchi et al. 2005) |
| Ror2 | A receptor tyrosine kinase | Widespread | Impaired proliferation/survival and migration of PGCs (Laird et al. 2011) |
| Transcription factors | |||
| Oct4 | A POU-homeobox domain TF | Ubiquitous in the embryo proper until E7.5, specific to germ cells thereafter | Impaired PGC specification; apoptosis of PGCs (Kehler et al. 2004; Okamura et al. 2008) |
| Nanog | A divergent homeodomain TF | Morula, ICM, epiblast, primitive streak, PGCs | Apoptosis of PGCs (Chambers et al. 2007; Yamaguchi et al. 2009) |
| Hif2α | A bHLH-PAS TF | Ubiquitous | Reduced number of PGCs at E8.5 and eventual loss of PGCs (Covello et al. 2006) |
| RNA-binding proteins | |||
| Nanos3 | An RNA-binding protein | PGCs | Reduced number of PGCs at E8.0 and eventual loss of PGCs (Tsuda et al. 2003; Suzuki et al. 2008) |
| Dnd1 | An RNA-binding protein | PGCs, widespread in embryos at E7.5 | Reduced number of PGCs at E8.0 and eventual loss of PGCs (Sakurai 1995; Youngren et al. 2005; Cook et al. 2009) |
| Tiar | An RNA-binding protein | Ubiquitous | Reduced number of PGCs in the genital ridges at E11.5 and eventual loss of PGCs (Beck et al. 1998) |
| Mvh | A DEAD-box RNA helicase | PGCs after E10.5 | Impaired male PGC proliferation after E11.5 (Tanaka et al. 2000) |
| Dicer | RNase | Ubiquitous | Impaired PGC proliferation (Hayashi et al. 2008) |
| miRNAs | |||
| Mir-290-295 | Posttranscriptional regulation of gene expression | ESCs, embryos before E6.5, PGCs | Impaired migration/survival of PGCs (Medeiros et al. 2011) |
| Cell adhesion molecules | |||
| Gja1 | A gap-junction protein | PGCs, widespread in embryos | Impaired colonization of PGCs (Juneja et al. 1999; Francis and Lo 2006) |
| Itgb1 | A β subunit of integrin complex | PGCs, widespread in embryos | Impaired migration of PGCs (Anderson et al. 1999) |
| Others | |||
| Hp1γ | Heterochromatin protein with binding to H3K9me | Ubiquitous | Reduced number of PGCs after E7.25 due to impaired cell cycle (Abe et al. 2011) |
| Mcm9 | A minichromosome maintenance helicase paralog | Ubiquitous | Impaired PGC proliferation (Hartford et al. 2011) |
| Ft locus | A region of mouse chromosome 8 bearing six genes | Detected in embryonic gonads | Impaired PGC proliferation (Kim et al. 2011) |
aTissue types: ESC, embryonic stem cell; ExE, extra-embryonic ectoderm; ExM, extra-embryonic mesoderm; ICM, inner cell mass; PGC, primordial germ cell; VE, visceral endoderm.
Blimp1-positive cells first appear in a few of the most proximal epiblast cells at the posterior side of the embryo ∼E6.25 (pre-primitive streak/no streak [P/0S] stage) (Fig. 2). They increase in number and form a cluster of cells with strong AP activity, and repression of Stella and the Hox gene (Ohinata et al. 2005). Prdm14 is first expressed at ∼E6.5 (early streak [ES] stage) in Blimp1-positive cells and later in the PGCs (Yamaji et al. 2008). Although activation of Prdm14 is independent of Blimp1 (Yamaji et al. 2008), the expression of Tcfap2c at ∼E6.75 (late streak/no allantoic bud [LS/0B] stage) appears to be dependent on Blimp1 (Kurimoto et al. 2008).
All the Blimp1-positive PGC precursors initially express the Hox genes and repress Sox2 (Yabuta et al. 2006; Kurimoto et al. 2008). However, from E6.75 onward, Blimp1-positive cells begin to repress the Hox genes and express Sox2, Stella, and Nanog (Yamaguchi et al. 2005; Kurimoto et al. 2008). Blimp1-positive cells continue to express Oct4 (also known as Pou5f1) PGCs at E7.25 up-regulate and down-regulate about 500 “germ cell specification” genes and around 330 “somatic program” genes, respectively (Kurimoto et al. 2008). Although Blimp1-positive PGCs delaminate from the epiblast, they do not display typical epithelial–mesenchymal transition (Kurimoto et al. 2008).
3.2. The Functions of Blimp1, Prdm14, and Tafap2c
BLIMP1 is a transcriptional repressor bearing a PR (PRDI-BF1 and RIZ) domain at its amino terminus and five Krüppel-type zinc fingers at its carboxyl terminus (Turner et al. 1994). The PR domain is structurally similar to the SET (Suppressor of hairless, Enhancer of zeste, and Trithorax) domain, which is known to show histone methyltransferase activity (Rea et al. 2000). The PR domain of BLIMP1 has not yet been shown to exhibit enzymatic activity. BLIMP1 interacts with many distinct epigenetic regulators, including HDACs (Yu et al. 2000), Groucho (Ren et al. 1999), and G9A (Gyory et al. 2004), in a context-dependent manner (Bikoff et al. 2009).
In Blimp1-knockout embryos, AP-positive PGC-like cells appear at around the E/MB stage, but they are smaller in number, do not show a migratory phenotype, and are unable to express PGC-specific genes properly (Table 1) (Ohinata et al. 2005; Vincent et al. 2005). A single-cell cDNA microarray analysis has shown that, in the absence of Blimp1, nearly all of the somatic genes fail to be repressed, whereas approximately half of the specification genes are up-regulated normally (Kurimoto et al. 2008). Therefore, primarily, BLIMP1 is a potent transcriptional repressor responsible for the repression of the somatic genes. The BMP4-SMAD1/5 signaling pathway may mediate the BLIMP1-independent acquisition of the specification genes (Fig. 3) (see below). The precise mechanism of action of BLIMP1 in PGC specification remains to be determined.
PRDM14 also bears the PR-domain and five Krüppel-type zinc fingers. Prdm14 is transiently expressed in the ICM and later only in the germline until ∼E13.5 (Yamaji et al. 2008). Prdm14 is essential for PGC specification (Table 1). Prdm14-deficient cells fail to reactivate Sox2, although they repress the somatic mesodermal program normally. They also fail to repress GLP (also known as EHMT1), an enzyme essential for the genome-wide H3K9me2 modification (Tachibana et al. 2005), and fail to reduce and up-regulate their H3K9me2 and H3K27me3 levels, respectively, for the acquisition of pluripotency and genome-wide epigenetic reprogramming (Fig. 3). Prdm14-deficient cells cannot form embryonic germ cells (EGCs) in culture. Prdm14 may act as a sequence-specific transcriptional regulator and is essential for the pluripotency of mice and human embryonic stem (ES) cells (Tsuneyoshi et al. 2008; Chia et al. 2010; Ma et al. 2011).
TCFAP2C (AP2γ) is a basic helix–span–helix domain transcription factor. It shows maternal expression in all cells until the blastocyst stage, but its expression is restricted in ExE and PGCs from E6.75 up to E12.5–E13.5 (Auman et al. 2002; Werling and Schorle 2002; Weber et al. 2010). Epiblast-specific deletion of Tcfap2c results in the drastically reduced PGCs, which show little migration and are lost later (Table 1) (Weber et al. 2010). TCFAP2C may function downstream from BLIMP1 in PGCs (Fig. 3).
3.3. Signaling for PGC Specification
Understanding the signaling requirements for the induction of PGCs from the epiblasts is of critical importance to develop a culture system to recapitulate PGC specification in culture. An integrated view of the signaling pathways for PGC specification has been proposed based on genetic as well as in vitro culture experiments that examined the influence of signaling molecules on the induction of Blimp1 and Prdm14 expression in the epiblast (Fig. 3) (Ohinata et al. 2009).
Accordingly, bone morphogenetic protein 4 (BMP4) emitted from the extra-embryonic ectoderm (ExE) activates the expression of Blimp1 and Prdm14 in a dose-dependent manner. BMP2 expressed in the proximal VE enhances the same signaling pathway, ensuring that the highest levels of BMP signaling occur in the most proximal epiblast. These findings are consistent with the previous observations that PGC specification has been shown to depend on the dosage of BMP signaling and the level of Blimp1 induction (Lawson et al. 1999; Ohinata et al. 2005; Vincent et al. 2005), and that mutants for Bmp4, Bmp8b, Bmp2, Smad1, Smad4, Smad5, and Alk2 lack or have reduced numbers of AP-positive PGCs (Table 1) (Zhao 2003; Saitou and Yamaji 2010). The induction of Blimp1 expression in the posterior side of the embryo is explained by the inhibition of BMP signaling in the anterior epiblast by antagonist factors (e.g., LEFTY1 against NODAL, DKK1 against WNT, and Cerberus-like [CER1] against BMP, etc.) that are emanated from the anterior visceral endoderm (AVE) (Umulis et al. 2009). In Smad2 or Foxh1 mutants that fail to form AVE, Blimp1 expression becomes widespread in the epiblast. Epiblast that is striped of the AVE also express Blimp1 and Prdm14 in response to BMP4.
WNT signaling is also essential for the PGC fate (Table 1): Wnt3-knockout epiblast cells do not show Blimp1 expression in response to BMP4 (Ohinata et al. 2009). Wnt3 is expressed in the epiblast and VE from ∼E5.5 (Liu et al. 1999), and epiblast cells of embryos younger than this age do not express Blimp1 in response to BMP4 (Ohinata et al. 2009). Wnt3 may therefore prime the epiblasts to express Blimp1 in response to BMP4. Wnt3-knockout embryos express receptors (Alk3, Alk6, BmprII) and signal transducers (Smad1, Smad5) for BMP signaling (Ohinata et al. 2009), suggesting that the WNT signaling may act posttranscriptionally on BMP signaling components. In this regard, WNT signaling can stabilize SMAD1 by inhibiting GSK3-mediated phosphorylation of its linker region and prevents its degradation (Fuentealba et al. 2007; Sapkota et al. 2007).
Significantly, most, if not all, of the epiblast cells cultured under a serum-free, defined condition primarily with BMP4 show Blimp1 and Prdm14 after 36 h, and these cells go on to develop Blimp1-, Prdm14-, and Stella-positive PGC-like cells after 132 h (Ohinata et al. 2009). They not only show proper gene expression like that of the PGCs, but also show genome-wide epigenetic reprogramming, including H3K9me2 reduction, H3K27me3 up-regulation, and erasure of an imprinted gene. When transplanted into the neonatal testes of germ-cell-less W/Wv mice, these cells contribute to proper spermatogenesis, and the resultant spermatozoa contribute to fertile offspring, a bona fide demonstration that the PGC-like cells induced from the epiblasts ex vivo acquire the germ cell phenotype (Ohinata et al. 2009). These findings provide a basis for generating the germ cell lineage in mammals in vitro.
4. PGCs FROM PLURIPOTENT STEM CELLS
The ability to generate PGCs from epiblast explants points to an effective strategy to produce functional PGCs from pluripotent stem cells (e.g., embryonic stem cells [ESCs], induced pluripotent stem cells [iPSCs]) in vitro by directed differentiation (Fig. 4) (Hayashi et al. 2011). The functional attributes of these in vitro–derived germline cells may be tested as for spermatogenesis potential by transplanting into neonatal testes lacking endogenous germ cells (Chuma et al. 2005), and for oogenesis potential by aggregating with gonadal somatic cells and transplanted under kidney corpuscles to allow oocyte growth and maturation (Matoba and Ogura 2011).
Figure 4.
The generation of PGC-like cells from ESCs/iPSCs in culture. A schematic representation of the generation of PGC-like cells (PGCLCs) from ESCs/iPSCs in culture (Hayashi et al. 2011). ESCs/iPSCs are induced into epiblast-like cells (EpiLCs) by the stimulation with ActivinA, bFGF, and KSR (knockout serum replacement) for 2 d. EpiLCs (about 1000 cells) are aggregated in a floating condition and are induced into PGCLCs by the stimulation with BMP4, BMP8b, SCF (stem cell factor), LIF (leukemia inhibitory factor), and EGF (epidermal growth factor). Blimp1-positive PGCLCs (green) are induced and form clusters at peripheries of the aggregate. PGCLCs are sorted by FACS (fluorescence activated cell sorting) according to their expression of Blimp1 transgene, or of surface markers, SSEA1 (stage-specific embryonic antigen 1), and Integrinβ3. PGCLCs are transplanted into neonatal testes lacking endogenous germ cells, leading to proper spermatogenesis. The ESC/iPSC-derived sperm are injected into oocytes by ICSI (intracytoplasmic sperm injection), and the resultant two-cell embryos are transferred to pseudopregnant females, resulting in the generation of fertile offspring.
Several critical milestones of germ cell development have to be recapitulated during in vitro differentiation of ESCs. First, ESCs have to be induced into epiblast-like cells (EpiLCs) by treatment with ActivinA (ACTA), bFGF, and a low concentration of KSR (Fig. 4) (Hayashi et al. 2011). The EpiLCs are a transient entity and show a global gene expression profile similar to that of the pre-gastrulating epiblasts at E5.75, but distinct from that of epiblast stem cells (EpiSCs) (Brons et al. 2007; Tesar et al. 2007; Hayashi et al. 2011): For example, EpiSCs express the key pluripotency gene Nanog at a high level, but EpiLCs and the epiblasts at E5.75 express it at a low level. EpiSCs also express endoderm markers such as Gata6, Sox17, and Cer1 at a high level, but EpiLCs and the epiblasts at E5.75 express them at a low/undetectable level (Hayashi et al. 2011). Remarkably, under conditions that induce the epiblast into PGC-like cells ex vivo, EpiLCs produce Blimp1-, Prdm14-, and Stella-positive PGC-like cells (PGCLCs), whereas EpiSCs show some Blimp1 but not Stella expression (Hayashi et al. 2011). The PGCLCs show a global gene expression profile very similar to that of PGCs at E9.5 (Hayashi et al. 2011). The PGCLCs show genome-wide epigenetic reprogramming (reduction of H3K9me2 and elevation of H3K27me3) and undergo spermatogenesis when transplanted into the testes of neonatal W/Wv mice, and the resultant sperm contribute to healthy, fertile offspring (Fig. 4) (Hayashi et al. 2011). The induction of EpiLCs and PGCLCs can be achieved in many ESC and iPSCs (Fig. 4) (Hayashi et al. 2011).
One of the key challenges for the generation of the stem cell–derived gametes in vitro is the sustenance of the proliferation of PGCLCs or PGCs to expand these germline precursor cells, in the same manner as between E9.5 and E12.5 in vivo, when their numbers increase from approximately 250 to 25,000 (Tam and Snow 1981). PGCLCs are short-lived (Hayashi et al. 2011), and PGCs in culture undergo apoptosis after several rounds of cell division (Dolci et al. 1991; Matsui et al. 1991; Pesce et al. 1993; Kawase et al. 1996) or dedifferentiate into EGCs (Matsui et al. 1992; Labosky et al. 1994).
Efficient PGCLC induction in mice depends strictly on the use of ESCs cultured under a serum-free condition with an inhibitor against MAP kinase signaling, an inhibitor against GSK3, and LIF (2i + LIF), a condition that maintains ESCs in an undifferentiated self-renewing “ground state” (Ying et al. 2008) and the derivation of EpiLCs as the immediate precursor for PGCLCs (Hayashi et al. 2011). So far, this has not been accomplished in human ESCs. That EpiSCs are functionally different from pre-gastrulating epiblast cells in germ-cell-forming capacity has significant implications for germ cell differentiation from human ESCs, which are believed to closely resemble EpiSCs (Nichols and Smith 2009). The different properties between mouse and non-rodent mammalian ESCs may be a reflection of the presence of different molecular logic in the mechanisms for early development between mouse and non-rodent mammals. It is also of note that the modes of gastrulation, during which PGCs emerge, are different between mice and non-rodent mammals, including humans (Sadler 2010; Hopf et al. 2011). It is therefore important to elucidate the process of germ cell specification in other non-rodent mammals that develop similarly to humans, before any rational approach can be devised for differentiating hESCs/iPSCs into germ cells.
5. REVEALING PLURIPOTENCY IN PGCs
PGCs have long been known as origins for teratoma/teratocarcinoma (Stevens 1964). The investigation of teratoma/teratocarcinomas and the stem cells derived from them (embryonal carcinoma cells, ECCs) eventually led to the derivation of ESCs from the ICM in culture (Evans and Kaufman 1981). The mechanism underlying cellular pluripotency has been studied most extensively using ESCs. The pluripotency is regulated and stably maintained mainly by a network of the core transcriptional regulators OCT4, SOX2, and NANOG (Niwa 2007; Young 2011). These transcription factors are expressed in the ICM and early epiblasts, but upon epiblast differentiation, although OCT4 remains expressed until relatively late, SOX2 and NANOG are down-regulated, especially in the posterior regions. As discussed above, PGCs, although essentially unipotent in vivo, reacquire the network of key genes for pluripotency upon their specification: PGCs continue to express OCT4 and regain the expression of SOX2 and NANOG (Fig. 3).
5.1. The Functions of Pluripotency Genes in PGCs
There is evidence that Oct4 is essential for the emergence of Stella-positive PGCs (Table 1) (Okamura et al. 2008). Conditional knockout of Oct4 or conditional knockdown of Nanog in migrating PGCs results in the death of PGCs by apoptosis, indicating the importance of these gene products in the maintenance of PGCs (Table 1) (Kehler et al. 2004; Chambers et al. 2007; Yamaguchi et al. 2009). Hypoxia-inducible factor 2a (HIF2α), which has a function to directly up-regulate Oct4, is also critical in PGC specification and/or survival (Fig. 3; Table 1) (Covello et al. 2006). Furthermore, LIN28, a negative regulator of let-7 microRNA (miRNA, mir) family processing and a key factor for pluripotency (Gangaraju and Lin 2009), appears to be essential for PGC specification (West et al. 2009). Because let-7 is known to inhibit Blimp1 mRNA translation by binding to its 3′ UTR (Nie et al. 2008), it is proposed that LIN28 activates Blimp1 by blocking the maturation of functional let-7 (Fig. 3; Table 1) (West et al. 2009). Mir-290-295, which is expressed in early embryos, ESCs, and PGCs and facilitates induced pluripotency (Houbaviy et al. 2003; Marson et al. 2008; Judson et al. 2009), has also been shown to be essential for PGC development (Table 1) (Medeiros et al. 2011). Thus, the genes associated with pluripotency are critical in PGC specification and proliferation/survival. Among the core pluripotency factors, SOX2 is not present in human PGCs, indicating that a different mechanism may underpin the pluripotency between the mouse and the human (Perrett et al. 2008).
Upon specification, PGCs express essential factors required for induced pluripotency, including OCT4, SOX2, NANOG, LIN28, KLF2, KLF5, and N-MYC (Fig. 3) (Takahashi and Yamanaka 2006; Yu et al. 2007; Kurimoto et al. 2008; Nakagawa et al. 2008). A key difference between PGC specification and induced pluripotency would be that the PGC fate is induced in epiblast cells that already bear key attributes of pluripotency. PGCs reacquire robust expression of SOX2, NANOG, KLF2, KLF5, and N-MYC during their specification (Kurimoto et al. 2008). Furthermore, the inactivation of the p53 pathway enhances EGC formation from PGCs (see below) (Kimura et al. 2008). p53 is a major tumor suppressor and prevents tumor formation from damaged or stressed cells by triggering apoptosis, by activating cell cycle checkpoints, or by promoting permanent cell cycle arrest (Vousden and Prives 2009; Hanahan and Weinberg 2011). Notably, the formation of pluripotent stem cells from spermatogonial stem cells and the induction of pluripotency from somatic cells by defined factors are also enhanced by the lack of p53 function (Kanatsu-Shinohara et al. 2004; Hong et al. 2009; Kawamura et al. 2009; Li et al. 2009; Marion et al. 2009; Utikal et al. 2009), indicating a common mechanism regulating the dedifferentiation of PGCs/spermatogonial stem cells into pluripotent stem cells and induced pluripotency.
5.2. From PGCs to EGCs
Reflecting their ability to acquire pluripotency relatively easily, PGCs have been shown to dedifferentiate into pluripotent EGCs in culture in the presence of basic FGF, SCF (stem cell factor, also known as Kit-ligand and Kitl) (provided by the Sl4-m220 feeder cells, which bear the membrane-bound form of SCF), and LIF at a ratio of ∼1% (Fig. 5) (Matsui et al. 1992; Labosky et al. 1994). PGCs convert into EGCs with the ability to contribute to chimeras within 10 d in culture, and the critical window for the requirement of bFGF is the first 24 h (Fig. 5) (Durcova-Hills et al. 2006; Durcova-Hills et al. 2008). A key effect of FGF is the repression of BLIMP1 that allows the activation of KLF4 and c-MYC (Fig. 5). LIF is required on days 2–4 to activate STAT3, a key transcription factor for pluripotency. Interestingly, trichostatin A (TSA), an inhibitor of histone deacetylases, can act as a substitute for the required bFGF. Conditional deletion of Pten or constitutive activation of AKT partly replaces the requirement of bFGF and enhances the dedifferentiation of PGCs to EGCs through the inactivation of the p53 pathway (Fig. 5) (Kimura et al. 2003, 2008).
Figure 5.
Regulatory network for EGC derivation from PGCs. Key signaling mechanisms (A) and events for the first 10 d during the derivation of EGCs from PGCs (B) are shown schematically. (A) Arrows indicate activation, whereas blunted arrows show repression. (B, upper panel) The requirement of the signaling molecules; (lower panel) gene expression dynamics.
EGCs can also be derived both in mice and rats by culturing PGCs under the 2i + LIF condition (Leitch et al. 2010), which also facilitates induced pluripotency from somatic cells (Takahashi and Yamanaka 2006; Silva et al. 2008). The ratio of EGC derivation becomes as high as ∼10% when PGCs are first stimulated by bFGF and SCF and then transferred to a 2i + LIF condition. Moreover, the culture under the simple 2i + LIF condition alone is sufficient for EGC derivation, although the efficiency decreases in the absence of bFGF (Leitch et al. 2010). It is of note that AKT signaling, which is a key for EGC derivation and a substitute for bFGF (Kimura et al. 2008), acts in part by inhibiting GSK3. Thus, the successful activation of multiple signaling pathways/underlying transcription factors converts PGCs into EGCs (Fig. 5).
6. EPIGENETIC REPROGRAMMING IN PGCs
PGCs show genome-wide DNA demethylation, which, by E13.5, leads to the erasure of genomic imprinting, reactivation of the inactivated X chromosome in females, and demethylation of the transposable/repetitive elements, resulting in the creation of an epigenetically “inert” genome (Monk et al. 1987; Kafri et al. 1992; Szabo and Mann 1995; Hajkova et al. 2002; Lee et al. 2002; Lane et al. 2003; Popp et al. 2010; Guibert et al. 2012; Saitou et al. 2012). Genome-wide methylation analyses of CpG dinucleotide sequences have revealed a significant hypomethylation of both male and female PGCs at E13.5 (Popp et al. 2010; Guibert et al. 2012). One study shows that the female PGCs bear a lower methylation level than the male PGCs (Popp et al. 2010), which may be an effect of their having two active X chromosomes because of their X-reactivation (Zvetkova et al. 2005). In PGCs, long terminal repeat (LTR) retrotransposon sequences such as intracisternal A particles (IAPs) are more resistant to demethylation, whereas genic, intergenic, and other transposon sequences are apparently globally demethylated (Popp et al. 2010; Guibert et al. 2012). The resistance of the LTR sequences to demethylation can cause trans-generational epigenetic inheritance in some cases (Daxinger and Whitelaw 2012).
Given that PGCs initially bear a genome-wide methylation level similar to that of their precursor epiblast cells (Guibert et al. 2012; Saitou et al. 2012), they should possess mechanisms for genome-wide DNA demethylation, which still remain poorly elucidated. In mammals, the mechanism of active DNA demethylation has long been elusive (Ooi and Bestor 2008; Wu and Zhang 2010; see Gehring et al. 2009 for information regarding the known mechanism of DNA demethylation involving 5-methylcytosine [5mC] demethylases in plants). Although a number of potential mechanisms have been proposed for active DNA demethylation in animals, none has yet been convincingly shown to be responsible for genome-wide DNA demethylation in PGCs (Saitou et al. 2012). For example, there is evidence that the cytosine deaminases AID and APOBEC1 can convert 5-methylcytosine (5mC) to T’s by deamination, creating T:G mismatches, which would then become targets of glycosylases such as TDG (thymine DNA glycosylase) or MBD4 (methyl-CpG binding domain protein 4) for base-excision mismatch repair (BER) (Morgan et al. 2004). Indeed, it appears that in Aid−/− PGCs, global DNA demethylation is attenuated to some degree (Popp et al. 2010). However, it is important to note that Aid and Apobec1 are expressed at very low levels in PGCs (Hajkova et al. 2010) and that both Aid−/− male and female mice are fertile, with the Aid−/− mice being maintained over many generations by intercrosses between homozygotes (Muramatsu et al. 2000), indicating that AID plays little, if any, role in germ cell development.
Recent studies have shown that the TET (ten-eleven translocation) proteins (TET1, 2, 3), which are 2-oxoglutarate and Fe(II)-dependent dioxygenases, hydroxylate 5mC into 5-hydroxymethylcytosine (5hmC) (Kriaucionis and Heintz 2009; Tahiliani et al. 2009), and successively into 5-formylcytosine (5fC) and into 5-carboxylcytosine (5caC) (He et al. 2011; Ito et al. 2011; Pfaffeneder et al. 2011). The resultant 5hmC, 5fC, and 5caC may be converted into C passively during DNA replication or removed by a BER pathway (He et al. 2011; Ito et al. 2011). The TET proteins are therefore new candidate DNA demethylases in PGCs, where Tet1 and, to a lesser extent, Tet2, but not Tet3, are expressed (Hajkova et al. 2010). Several studies have shown the roles of TET proteins, most typically the role of TET1 in ESCs (Ito et al. 2010; Ficz et al. 2011; Koh et al. 2011; Pastor et al. 2011; Song et al. 2011; Williams et al. 2011; Wu et al. 2011; Xu et al. 2011). Accordingly, one of the functions of TET1 would be to remove aberrant stochastic DNA methylation from promoters with high or intermediate CpG contents, thereby regulating DNA methylation fidelity in ESCs (Williams et al. 2012). Furthermore, significantly, TET3, which is highly expressed in the oocytes, has been shown to be involved in the active genome-wide DNA demethylation from the paternal haploid genome in the zygotes (Gu et al. 2011; Iqbal et al. 2011; Wossidlo et al. 2011). Thus, it is of particular interest and importance to explore whether the TET proteins (TET1 and TET2) are, indeed, involved in the genome-wide DNA demethylation in PGCs and, if so, whether the BER pathway would follow the TET-mediated 5mC hydroxylation (Hajkova et al. 2010). However, it has more recently been shown that Tet1-knockout ESCs are pluripotent and contribute robustly to chimeras and the Tet1-knockout mice are viable and fertile, although their litter sizes are somewhat smaller than those of the wild types (Dawlaty et al. 2011). To further examine the function of TET proteins in germ cell development and DNA demethylation in PGCs, Tet1 and Tet2 double-knockout mice need to be analyzed.
Although there is as yet no convincing evidence for an enzyme-triggered, active DNA demethylation in PGCs, there is circumstantial evidence that DNA demethylation in PGCs is mediated by a DNA repair mechanism (Hajkova et al. 2008, 2010). Accordingly, PGCs at ∼E11.5 appear to undergo a global reorganization of their chromatin state, which includes concomitant loss of linker histone H1, chromocenters (clusters of pericentric heterochromatin domains of several chromosomes) (Probst and Almouzni 2011), and various histone modification marks, along with a significant enlargement of nuclei. Evidence shows that components of the BER pathway such as XRCC1, APE1, and PARP1 are specifically concentrated, and XRCC1 is bound to chromatin in the nuclei of PGCs (Hajkova et al. 2010). It has thus been suggested that the global chromatin reorganization in PGCs is a consequence of the DNA demethylation mediated by a genome-wide BER, which results in a genome-wide histone replacement, possibly by the histone chaperone HIRA or NAP1 (nucleosome assembly protein 1) (Hajkova et al. 2008, 2010). This is a testable prediction and requires exploration by genetic experiments such as knockouts of relevant genes.
Although there is a perception that the genome-wide epigenetic reprogramming in PGCs occurs after their colonization of the gonads, at least a part of the reprogramming process starts much earlier, nearly concurrent with PGC specification, and proceeds during the PGC migration period (Seki et al. 2005, 2007; Guibert et al. 2012). The promoter DNA demethylation in migrating PGCs (E8.5–E9.5) appears to be global and influences genes of various biological functions (Guibert et al. 2012). Furthermore, PGCs reduce their genome-wide H3K9me2 from ∼E7.75, and nearly all of the PGCs show reduced H3K9me2 levels by E8.75. In contrast, they start to up-regulate their genome-wide H3K27me3 from ∼E8.25, and by E9.5 nearly all of the PGCs show up-regulation of H3K27me3. Interestingly, although PGCs at E7.25 or after E9.75 show a cell cycle distribution similar to that of ESCs, indicative of their rapid proliferation, PGCs that are migrating in the hindgut endoderm from ∼E7.75 to E8.75 show a unique cell cycle distribution, with nearly 60% of them being in the G2 phase (Seki et al. 2007). Coincident with this period, PGCs appear to shut off their RNA polymerase II–dependent global transcription (Seki et al. 2007). Therefore, the histone modification reprogramming in migrating PGCs occurs when they are in the G2 phase of the cell cycle and are transcriptionally quiescent, apparently in a progressive, cell-by-cell fashion.
The mechanism and the significance of the histone modification reprogramming in migrating PGCs have been unknown. It is, however, of note that PGCs repress GLP and G9A (also known as EHMT2), key histone lysine methyltransferases for H3K9me2 (Tachibana et al. 2002, 2005), from ∼E7.75 and E9.5, respectively. The identification of the sequences from which H3K9me2 is removed and to which H3K27me3 is added should provide critical information to explore the function of these events. It would be interesting to examine whether the H3K27me3 in PGCs marks genes similar to those in ESCs (Bernstein et al. 2006; Barski et al. 2007; Mikkelsen et al. 2007; Pan et al. 2007; Zhao et al. 2007; Mohn et al. 2008). It is also critical to note that PGCs repress DNMT3A/3B and UHRF1 soon after their specification (Seki et al. 2005; Kurimoto et al. 2008) and therefore would bear limited de novo as well as maintenance methyltransferase activities during their development. The low H3K9me2, high H3K27me3, and limited DNA methyltransferase activities in PGCs may facilitate passive, rather than active, genome-wide DNA demethylation. Additionally, PGCs bear a high level of histone H2A/H4 arginine3 symmetrical dimethylation (H2A/H4R3me2s) (Ancelin et al. 2006), a modification conferred by a protein arginine methyltransferase (PRMT) (Bedford and Clarke 2009). Future studies will be needed to elucidate whether the active, passive, or both mechanisms are involved in the genome-wide DNA demethylation in PGCs, and its mechanistic linkage with histone modification reprogramming.
7. PERSPECTIVE
Investigations during the last decade have unveiled key mechanisms underlying PGC development in mice. Continuation of careful studies on the mechanism of germ cell development and the step-by-step in vitro reconstruction of the information obtained will be major initiatives for the advancement of this field in the next decade. The key function of the germline is to establish totipotency. Toward this end, PGCs first erase epigenetic information inherited from a previous generation, most remarkably as manifested by genome-wide DNA demethylation. PGCs undergo this process while maintaining the underlying potential pluripotency. Subsequently, germ cells gradually establish appropriate epigenetic modifications consistent with embryonic development in a sex-dependent manner. Moreover, in the development of oocytes, they acquire “developmental potency,” which supports the onset of embryonic development. The molecular nature of developmental potency is not well defined. The developmental potency of the oocytes is sufficient to reprogram the epigenome of any somatic cells toward totipotency, although the efficiency is evidently low (Campbell et al. 1996; Wakayama et al. 1998). The mechanism with which the oocytes reprogram somatic nuclei remains poorly understood. Future research should be aimed at understanding the nature of the totipotent epigenome as well as of the developmental potency of the oocytes. Obviously, the studies of the germ cell lineage have wider implications in stem cell biology in general. One of the key issues in stem cell biology is to explore a method to propagate the stem cell state in vitro, without altering its potential to differentiate into a certain lineage. Understanding of the epigenetic regulation of the stem cell state should provide critical information as to how the stem cell state is orchestrated. It is very much hoped that research into the germline, with an emphasis on its epigenetic regulation, will reveal a means of controlling the cells’ epigenetic states in a desirable manner.
ACKNOWLEDGMENTS
We thank the members of our laboratory for their discussion on this work. This study is supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by JST, CREST/ERATO.
Footnotes
Editors: Patrick P.L. Tam, W. James Nelson, and Janet Rossant
Additional Perspectives on Mammalian Development available at www.cshperspectives.org
REFERENCES
- Abe K, Naruse C, Kato T, Nishiuchi T, Saitou M, Asano M 2011. Loss of heterochromatin protein 1γ reduces the number of primordial germ cells via impaired cell cycle progression in mice. Biol Reprod 85: 1013–1024 [DOI] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA 2006. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol 8: 623–630 [DOI] [PubMed] [Google Scholar]
- Anderson R, Fassler R, Georges-Labouesse E, Hynes RO, Bader BL, Kreidberg JA, Schaible K, Heasman J, Wylie C 1999. Mouse primordial germ cells lacking β1 integrins enter the germline but fail to migrate normally to the gonads. Development 126: 1655–1664 [DOI] [PubMed] [Google Scholar]
- Ara T, Nakamura Y, Egawa T, Sugiyama T, Abe K, Kishimoto T, Matsui Y, Nagasawa T 2003. Impaired colonization of the gonads by primordial germ cells in mice lacking a chemokine, stromal cell-derived factor-1 (SDF-1). Proc Natl Acad Sci 100: 5319–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin J, Davy A, Soriano P 2004. In vivo convergence of BMP and MAPK signaling pathways: Impact of differential Smad1 phosphorylation on development and homeostasis. Genes Dev 18: 1482–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman HJ, Nottoli T, Lakiza O, Winger Q, Donaldson S, Williams T 2002. Transcription factor AP-2γ is essential in the extra-embryonic lineages for early postimplantation development. Development 129: 2733–2747 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Beck AR, Miller IJ, Anderson P, Streuli M 1998. RNA-binding protein TIAR is essential for primordial germ cell development. Proc Natl Acad Sci 95: 2331–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG 2009. Protein arginine methylation in mammals: Who, what, and why. Mol Cell 33: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bikoff EK, Morgan MA, Robertson EJ 2009. An expanding job description for Blimp-1/PRDM1. Curr Opin Genet Dev 19: 379–385 [DOI] [PubMed] [Google Scholar]
- Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, et al. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195 [DOI] [PubMed] [Google Scholar]
- Buehr M, McLaren A, Bartley A, Darling S 1993. Proliferation and migration of primordial germ cells in We/We mouse embryos. Dev Dyn 198: 182–189 [DOI] [PubMed] [Google Scholar]
- Campbell KH, McWhir J, Ritchie WA, Wilmut I 1996. Sheep cloned by nuclear transfer from a cultured cell line. Nature 380: 64–66 [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A 2007. Nanog safeguards pluripotency and mediates germline development. Nature 450: 1230–1234 [DOI] [PubMed] [Google Scholar]
- Chang H, Matzuk MM 2001. Smad5 is required for mouse primordial germ cell development. Mech Dev 104: 61–67 [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K, Svingen T, Ng ET, Epp T, Spiller CM, Clark C, Cooper H, Koopman P 2011. Loss of Wnt5a disrupts primordial germ cell migration and male sexual development in mice. Biol Reprod 86: 1–12 [DOI] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, et al. 2010. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468: 316–320 [DOI] [PubMed] [Google Scholar]
- Chiquoine AD 1954. The identification, origin and migration of the primordial germ cells in the mouse embryo. Anat Rec 118: 135–146 [DOI] [PubMed] [Google Scholar]
- Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ 2004. Differential requirements for Smad4 in TGFβ-dependent patterning of the early mouse embryo. Development 131: 3501–3512 [DOI] [PubMed] [Google Scholar]
- Chuma S, Kanatsu-Shinohara M, Inoue K, Ogonuki N, Miki H, Toyokuni S, Hosokawa M, Nakatsuji N, Ogura A, Shinohara T 2005. Spermatogenesis from epiblast and primordial germ cells following transplantation into postnatal mouse testis. Development 132: 117–122 [DOI] [PubMed] [Google Scholar]
- Cook MS, Coveney D, Batchvarov I, Nadeau JH, Capel B 2009. BAX-mediated cell death affects early germ cell loss and incidence of testicular teratomas in Dnd1(Ter/Ter) mice. Dev Biol 328: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, Labosky PA, Simon MC, Keith B 2006. HIF-2α regulates Oct-4: Effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev 20: 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, et al. 2011. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9: 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E 2012. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet 13: 153–162 [DOI] [PubMed] [Google Scholar]
- De Sousa Lopes SM, Roelen BA, Monteiro RM, Emmens R, Lin HY, Li E, Lawson KA, Mummery CL 2004. BMP signaling mediated by ALK2 in the visceral endoderm is necessary for the generation of primordial germ cells in the mouse embryo. Genes Dev 18: 1838–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNapoli L, Batchvarov J, Capel B 2006. FGF9 promotes survival of germ cells in the fetal testis. Development 133: 1519–1527 [DOI] [PubMed] [Google Scholar]
- Dolci S, Williams DE, Ernst MK, Resnick JL, Brannan CI, Lock LF, Lyman SD, Boswell HS, Donovan PJ 1991. Requirement for mast cell growth factor for primordial germ cell survival in culture. Nature 352: 809–811 [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G, Adams IR, Barton SC, Surani MA, McLaren A 2006. The role of exogenous fibroblast growth factor-2 on the reprogramming of primordial germ cells into pluripotent stem cells. Stem Cells 24: 1441–1449 [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G, Tang F, Doody G, Tooze R, Surani MA 2008. Reprogramming primordial germ cells into pluripotent stem cells. PLoS ONE 3: e3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156 [DOI] [PubMed] [Google Scholar]
- Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, et al. 2012. IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour CG, Akam M 2003. Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development 130: 5869–5884 [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W 2011. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473: 398–402 [DOI] [PubMed] [Google Scholar]
- Francis RJ, Lo CW 2006. Primordial germ cell deficiency in the connexin 43 knockout mouse arises from apoptosis associated with abnormal p53 activation. Development 133: 3451–3460 [DOI] [PubMed] [Google Scholar]
- Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, De Robertis EM 2007. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell 131: 980–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Dunn NR, Hogan BL 2001. Bone morphogenetic protein 4 in the extraembryonic mesoderm is required for allantois development and the localization and survival of primordial germ cells in the mouse. Proc Natl Acad Sci 98: 13739–13744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H 2009. MicroRNAs: Key regulators of stem cells. Nat Rev Mol Cell Biol 10: 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Reik W, Henikoff S 2009. DNA demethylation by DNA repair. Trends Genet 25: 82–90 [DOI] [PubMed] [Google Scholar]
- Ghosh D, Seydoux G 2008. Inhibition of transcription by the Caenorhabditis elegans germline protein PIE-1: Genetic evidence for distinct mechanisms targeting initiation and elongation. Genetics 178: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg M, Snow MH, McLaren A 1990. Primordial germ cells in the mouse embryo during gastrulation. Development 110: 521–528 [DOI] [PubMed] [Google Scholar]
- Gu Y, Runyan C, Shoemaker A, Surani A, Wylie C 2009. Steel factor controls primordial germ cell survival and motility from the time of their specification in the allantois, and provides a continuous niche throughout their migration. Development 136: 1295–1303 [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. 2011. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477: 606–610 [DOI] [PubMed] [Google Scholar]
- Guibert S, Forne T, Weber M 2012. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 22: 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyory I, Wu J, Fejer G, Seto E, Wright KL 2004. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol 5: 299–308 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117: 15–23 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA 2008. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA 2010. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329: 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A 2008. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature 451: 730–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartford SA, Luo Y, Southard TL, Min IM, Lis JT, Schimenti JC 2011. Minichromosome maintenance helicase paralog MCM9 is dispensible for DNA replication but functions in germ-line stem cells and tumor suppression. Proc Natl Acad Sci 108: 17702–17707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Kobayashi T, Umino T, Goitsuka R, Matsui Y, Kitamura D 2002. SMAD1 signaling is critical for initial commitment of germ cell lineage from mouse epiblast. Mech Dev 118: 99–109 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K, O’Carroll D, Das PP, Tarakhovsky A, Miska EA, et al. 2008. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE 3: e1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M 2011. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146: 519–532 [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilscher B, Hilscher W, Bulthoff-Ohnolz B, Kramer U, Birke A, Pelzer H, Gauss G 1974. Kinetics of gametogenesis. I. Comparative histological and autoradiographic studies of oocytes and transitional prospermatogonia during oogenesis and prespermatogenesis. Cell Tissue Res 154: 443–470 [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S 2009. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature 460: 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf C, Viebahn C, Puschel B 2011. BMP signals and the transcriptional repressor BLIMP1 during germline segregation in the mammalian embryo. Dev Genes Evol 221: 209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA 2003. Embryonic stem cell-specific microRNAs. Dev Cell 5: 351–358 [DOI] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE 2011. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci 108: 3642–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R 2009. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 27: 459–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja SC, Barr KJ, Enders GC, Kidder GM 1999. Defects in the germ line and gonads of mice lacking connexin43. Biol Reprod 60: 1263–1270 [DOI] [PubMed] [Google Scholar]
- Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A 1992. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev 6: 705–714 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T 2003. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod 69: 612–616 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, Toyokuni S, et al. 2004. Generation of pluripotent stem cells from neonatal mouse testis. Cell 119: 1001–1012 [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase E, Shirayoshi Y, Hashimoto K, Nakatsuji N 1996. A combination of Buffalo rat liver cell-conditioned medium, forskolin and membrane-bound stem cell factor stimulates rapid proliferation of mouse primoridial germ cells in vitro similar to that in vivo. Develop Growth Differ 38: 315–322 [DOI] [PubMed] [Google Scholar]
- Kehler J, Tolkunova E, Koschorz B, Pesce M, Gentile L, Boiani M, Lomeli H, Nagy A, McLaughlin KJ, Scholer HR, et al. 2004. Oct4 is required for primordial germ cell survival. EMBO Rep 5: 1078–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Kim Y, Sakuma R, Hui CC, Ruther U, Jorgensen JS 2011. Primordial germ cell proliferation is impaired in Fused Toes mutant embryos. Dev Biol 349: 417–426 [DOI] [PubMed] [Google Scholar]
- Kimura T, Suzuki A, Fujita Y, Yomogida K, Lomeli H, Asada N, Ikeuchi M, Nagy A, Mak TW, Nakano T 2003. Conditional loss of PTEN leads to testicular teratoma and enhances embryonic germ cell production. Development 130: 1691–1700 [DOI] [PubMed] [Google Scholar]
- Kimura T, Tomooka M, Yamano N, Murayama K, Matoba S, Umehara H, Kanai Y, Nakano T 2008. AKT signaling promotes derivation of embryonic germ cells from primordial germ cells. Development 135: 869–879 [DOI] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. 2011. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8: 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Shigeta M, Yamanaka K, Saitou M 2008. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev 22: 1617–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labosky PA, Barlow DP, Hogan BL 1994. Mouse embryonic germ (EG) cell lines: Transmission through the germline and differences in the methylation imprint of insulin-like growth factor 2 receptor (Igf2r) gene compared with embryonic stem (ES) cell lines. Development 120: 3197–3204 [DOI] [PubMed] [Google Scholar]
- Laird DJ, Altshuler-Keylin S, Kissner MD, Zhou X, Anderson KV 2011. Ror2 enhances polarity and directional migration of primordial germ cells. PLoS Genet 7: e1002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W 2003. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35: 88–93 [DOI] [PubMed] [Google Scholar]
- Lange UC, Adams DJ, Lee C, Barton S, Schneider R, Bradley A, Surani MA 2008. Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Mol Cell Biol 28: 4688–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL 1999. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev 13: 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, Ogura A, Ishino F 2002. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development 129: 1807–1817 [DOI] [PubMed] [Google Scholar]
- Leitch HG, Blair K, Mansfield W, Ayetey H, Humphreys P, Nichols J, Surani MA, Smith A 2010. Embryonic germ cells from mice and rats exhibit properties consistent with a generic pluripotent ground state. Development 137: 2279–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A 1999. Requirement for Wnt3 in vertebrate axis formation. Nat Genet 22: 361–365 [DOI] [PubMed] [Google Scholar]
- Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J 2011. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat Struct Mol Biol 18: 120–127 [DOI] [PubMed] [Google Scholar]
- MacGregor GR, Zambrowicz BP, Soriano P 1995. Tissue non-specific alkaline phosphatase is expressed in both embryonic and extraembryonic lineages during mouse embryogenesis but is not required for migration of primordial germ cells. Development 121: 1487–1496 [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA 2009. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Ogura A 2011. Generation of functional oocytes and spermatids from fetal primordial germ cells after ectopic transplantation in adult mice. Biol Reprod 84: 631–638 [DOI] [PubMed] [Google Scholar]
- Matsui Y, Toksoz D, Nishikawa S, Williams D, Zsebo K, Hogan BL 1991. Effect of Steel factor and leukaemia inhibitory factor on murine primordial germ cells in culture. Nature 353: 750–752 [DOI] [PubMed] [Google Scholar]
- Matsui Y, Zsebo K, Hogan BL 1992. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell 70: 841–847 [DOI] [PubMed] [Google Scholar]
- McCoshen JA, McCallion DJ 1975. A study of the primordial germ cells during their migratory phase in Steel mutant mice. Experientia 31: 589–590 [DOI] [PubMed] [Google Scholar]
- Medeiros LA, Dennis LM, Gill ME, Houbaviy H, Markoulaki S, Fu D, White AC, Kirak O, Sharp PA, Page DC, et al. 2011. Mir-290–295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc Natl Acad Sci 108: 14163–14168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz B, Russell ES 1957. Gene-induced embryological modifications of primordial germ cells in the mouse. J Exp Zool 134: 207–237 [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D 2008. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766 [DOI] [PubMed] [Google Scholar]
- Molyneaux KA, Stallock J, Schaible K, Wylie C 2001. Time-lapse analysis of living mouse germ cell migration. Dev Biol 240: 488–498 [DOI] [PubMed] [Google Scholar]
- Molyneaux KA, Zinszner H, Kunwar PS, Schaible K, Stebler J, Sunshine MJ, O’Brien W, Raz E, Littman D, Wylie C, et al. 2003. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development 130: 4279–4286 [DOI] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S 1987. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 99: 371–382 [DOI] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK 2004. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: Implications for epigenetic reprogramming. J Biol Chem 279: 52353–52360 [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553–563 [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S 2008. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 26: 101–106 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF 1996. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science 274: 2075–2079 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. 2007. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 9: 64–71 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Kobayashi K, Nishimura T, Higashijima S, Tanaka M 2010. Identification of germline stem cells in the ovary of the teleost medaka. Science 328: 1561–1563 [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A 2009. Naive and primed pluripotent states. Cell Stem Cell 4: 487–492 [DOI] [PubMed] [Google Scholar]
- Nie K, Gomez M, Landgraf P, Garcia JF, Liu Y, Tan LH, Chadburn A, Tuschl T, Knowles DM, Tam W 2008. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: A potential pathogenetic lesion in Hodgkin lymphomas. Am J Pathol 173: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H 2007. How is pluripotency determined and maintained? Development 134: 635–646 [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, et al. 2005. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436: 207–213 [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M 2009. A signaling principle for the specification of the germ cell lineage in mice. Cell 137: 571–584 [DOI] [PubMed] [Google Scholar]
- Okamura D, Kimura T, Nakano T, Matsui Y 2003. Cadherin-mediated cell interaction regulates germ cell determination in mice. Development 130: 6423–6430 [DOI] [PubMed] [Google Scholar]
- Okamura D, Tokitake Y, Niwa H, Matsui Y 2008. Requirement of Oct3/4 function for germ cell specification. Dev Biol 317: 576–584 [DOI] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH 2008. The colorful history of active DNA demethylation. Cell 133: 1145–1148 [DOI] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA 2007. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1: 299–312 [DOI] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. 2011. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473: 394–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Saitou M, Barton SC, Thresher R, Dixon JP, Zahn D, Colledge WH, Carlton MB, Nakano T, Surani MA 2003. Stella is a maternal effect gene required for normal early development in mice. Curr Biol 13: 2110–2117 [DOI] [PubMed] [Google Scholar]
- Perrett RM, Turnpenny L, Eckert JJ, O’Shea M, Sonne SB, Cameron IT, Wilson DI, Rajpert-De Meyts E, Hanley NA 2008. The early human germ cell lineage does not express SOX2 during in vivo development or upon in vitro culture. Biol Reprod 78: 852–858 [DOI] [PubMed] [Google Scholar]
- Pesce M, Farrace MG, Piacentini M, Dolci S, De Felici M 1993. Stem cell factor and leukemia inhibitory factor promote primordial germ cell survival by suppressing programmed cell death (apoptosis). Development 118: 1089–1094 [DOI] [PubMed] [Google Scholar]
- Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T 2011. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl 50: 7008–7012 [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W 2010. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463: 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Almouzni G 2011. Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet 27: 177–185 [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Ren B, Chee KJ, Kim TH, Maniatis T 1999. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev 13: 125–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BE, Lehmann R 2010. Mechanisms guiding primordial germ cell migration: Strategies from different organisms. Nat Rev Mol Cell Biol 11: 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A, Munger S, Capel B 2007. Bmp7 regulates germ cell proliferation in mouse fetal gonads. Sex Dev 1: 127–137 [DOI] [PubMed] [Google Scholar]
- Runyan C, Schaible K, Molyneaux K, Wang Z, Levin L, Wylie C 2006. Steel factor controls midline cell death of primordial germ cells and is essential for their normal proliferation and migration. Development 133: 4861–4869 [DOI] [PubMed] [Google Scholar]
- Russell LD, Ettlin RA, Sinha Hikim AP, Clegg ED 1990. Histological and histopathological evaluation of the testis. Cache River Press, Clearwater, FL [Google Scholar]
- Sadler TW 2010. Langman’s medical embryology. Lippincott Williams & Wilkins, Baltimore/Philadelphia [Google Scholar]
- Saito K, Siomi MC 2010. Small RNA-mediated quiescence of transposable elements in animals. Dev Cell 19: 687–697 [DOI] [PubMed] [Google Scholar]
- Saitou M, Yamaji M 2010. Germ cell specification in mice: Signaling, transcription regulation, and epigenetic consequences. Reproduction 139: 931–942 [DOI] [PubMed] [Google Scholar]
- Saitou M, Barton SC, Surani MA 2002. A molecular programme for the specification of germ cell fate in mice. Nature 418: 293–300 [DOI] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K 2012. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 139: 15–31 [DOI] [PubMed] [Google Scholar]
- Sakurai T, Iguchi T, Moriwaki K, Noguchi M 1995. The ter mutation first causes primordial germ cell deficiency in ter/ter mouse embryos at 8 days of gestation. Develop Growth Differ 37: 293–302 [DOI] [PubMed] [Google Scholar]
- Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massague J 2007. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell 25: 441–454 [DOI] [PubMed] [Google Scholar]
- Sato M, Kimura T, Kurokawa K, Fujita Y, Abe K, Masuhara M, Yasunaga T, Ryo A, Yamamoto M, Nakano T 2002. Identification of PGC7, a new gene expressed specifically in preimplantation embryos and germ cells. Mech Dev 113: 91–94 [DOI] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y 2005. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 278: 440–458 [DOI] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M 2007. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134: 2627–2638 [DOI] [PubMed] [Google Scholar]
- Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A 1996. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature 382: 713–716 [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A 2008. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol 6: e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. 2011. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol 29: 68–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed RM 1982. Meiosis in the foetal mouse ovary. I. An analysis at the light microscope level using surface-spreading. Chromosoma 85: 427–437 [DOI] [PubMed] [Google Scholar]
- Spradling A, Fuller MT, Braun RE, Yoshida S 2011. Germline stem cells. Cold Spring Harb Perspect Biol 3: a002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallock J, Molyneaux K, Schaible K, Knudson CM, Wylie C 2003. The pro-apoptotic gene Bax is required for the death of ectopic primordial germ cells during their migration in the mouse embryo. Development 130: 6589–6597 [DOI] [PubMed] [Google Scholar]
- Stevens LC 1964. Experimental production of testicular teratomas in mice. Proc Natl Acad Sci 52: 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Tsuda M, Kiso M, Saga Y 2008. Nanos3 maintains the germ cell lineage in the mouse by suppressing both Bax-dependent and -independent apoptotic pathways. Dev Biol 318: 133–142 [DOI] [PubMed] [Google Scholar]
- Szabo PE, Mann JR 1995. Biallelic expression of imprinted genes in the mouse germ line: Implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev 9: 1857–1868 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y 2005. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 19: 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Molyneaux K, Runyan C, Schaible K, Wylie C 2005. The roles of FGF signaling in germ cell migration in the mouse. Development 132: 5399–5409 [DOI] [PubMed] [Google Scholar]
- Tam PP, Snow MH 1981. Proliferation and migration of primordial germ cells during compensatory growth in mouse embryos. J Embryol Exp Morphol 64: 133–147 [PubMed] [Google Scholar]
- Tanaka SS, Matsui Y 2002. Developmentally regulated expression of mil-1 and mil-2, mouse interferon-induced transmembrane protein like genes, during formation and differentiation of primordial germ cells. Mech Dev 119: S261–S267 [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T 2000. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev 14: 841–853 [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448: 196–199 [DOI] [PubMed] [Google Scholar]
- Tremblay KD, Dunn NR, Robertson EJ 2001. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development 128: 3609–3621 [DOI] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y 2003. Conserved role of nanos proteins in germ cell development. Science 301: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Tsuneyoshi N, Sumi T, Onda H, Nojima H, Nakatsuji N, Suemori H 2008. PRDM14 suppresses expression of differentiation marker genes in human embryonic stem cells. Biochem Biophys Res Commun 367: 899–905 [DOI] [PubMed] [Google Scholar]
- Turner CA Jr, Mack DH, Davis MM 1994. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 77: 297–306 [DOI] [PubMed] [Google Scholar]
- Umulis D, O’Connor MB, Blair SS 2009. The extracellular regulation of bone morphogenetic protein signaling. Development 136: 3715–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K 2009. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Sciammas R, Shapiro-Shalef M, Davis MM, Calame K, Bikoff EK, Robertson EJ 2005. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development 132: 1315–1325 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C 2009. Blinded by the light: The growing complexity of p53. Cell 137: 413–431 [DOI] [PubMed] [Google Scholar]
- Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R 1998. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 394: 369–374 [DOI] [PubMed] [Google Scholar]
- Weber S, Eckert D, Nettersheim D, Gillis AJ, Schafer S, Kuckenberg P, Ehlermann J, Werling U, Biermann K, Looijenga LH, et al. 2010. Critical function of AP-2γ/TCFAP2C in mouse embryonic germ cell maintenance. Biol Reprod 82: 214–223 [DOI] [PubMed] [Google Scholar]
- Werling U, Schorle H 2002. Transcription factor gene AP-2γ essential for early murine development. Mol Cell Biol 22: 3149–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, et al. 2009. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 460: 909–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western PS, Miles DC, van den Bergen JA, Burton M, Sinclair AH 2008. Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells 26: 339–347 [DOI] [PubMed] [Google Scholar]
- White YA, Woods DC, Takai Y, Ishihara O, Seki H, Tilly JL 2012. Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nat Med 18: 413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K 2011. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K 2012. DNA methylation: TET proteins—guardians of CpG islands? EMBO Rep 13: 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J 2011. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun 2: 241. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y 2010. Active DNA demethylation: Many roads lead to Rome. Nat Rev Mol Cell Biol 11: 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Eve Sun Y, Zhang Y 2011. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473: 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. 2011. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 42: 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, Saitou M 2006. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol Reprod 75: 705–716 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kimura H, Tada M, Nakatsuji N, Tada T 2005. Nanog expression in mouse germ cell development. Gene Expr Patterns 5: 639–646 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kurimoto K, Yabuta Y, Sasaki H, Nakatsuji N, Saitou M, Tada T 2009. Conditional knockdown of Nanog induces apoptotic cell death in mouse migrating primordial germ cells. Development 136: 4011–4020 [DOI] [PubMed] [Google Scholar]
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M 2008. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet 40: 1016–1022 [DOI] [PubMed] [Google Scholar]
- Ying Y, Zhao GQ 2001. Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev Biol 232: 484–492 [DOI] [PubMed] [Google Scholar]
- Ying Y, Liu XM, Marble A, Lawson KA, Zhao GQ 2000. Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol Endocrinol 14: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S 2010. Stem cells in mammalian spermatogenesis. Develop Growth Differ 52: 311–317 [DOI] [PubMed] [Google Scholar]
- Young RA 2011. Control of the embryonic stem cell state. Cell 144: 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, Lamb BT, Deng JM, Behringer RR, Capel B, et al. 2005. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 435: 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K 2000. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol 20: 2592–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zhang F, Barboric M, Blackwell TK, Peterlin BM 2003. A model of repression: CTD analogs and PIE-1 inhibit transcriptional elongation by P-TEFb. Genes Dev 17: 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GQ 2003. Consequences of knocking out BMP signaling in the mouse. Genesis 35: 43–56 [DOI] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. 2007. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1: 286–298 [DOI] [PubMed] [Google Scholar]
- Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, John R, Feil R, Brockdorff N 2005. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet 37: 1274–1279 [DOI] [PubMed] [Google Scholar]