Abstract
Mutations in Parkin or PINK1 are the most common cause of recessive familial parkinsonism. Recent studies suggest that PINK1 and Parkin form a mitochondria quality control pathway that identifies dysfunctional mitochondria, isolates them from the mitochondrial network, and promotes their degradation by autophagy. In this pathway the mitochondrial kinase PINK1 senses mitochondrial fidelity and recruits Parkin selectively to mitochondria that lose membrane potential. Parkin, an E3 ligase, subsequently ubiquitinates outer mitochondrial membrane proteins, notably the mitofusins and Miro, and induces autophagic elimination of the impaired organelles. Here we review the recent rapid progress in understanding the molecular mechanisms of PINK1- and Parkin-mediated mitophagy and the identification of Parkin substrates suggesting how mitochondrial fission and trafficking are involved. We also discuss how defects in mitophagy may be linked to Parkinson's disease.
PINK1 senses mitochondrial fidelity and recruits Parkin to dysfunctional mitochondria. Parkin then ubiquitinates membrane proteins of the dysfunctional organelle and induces mitophagy.
Parkinson's disease (PD) is the second most common neurodegenerative disorder and is characterized by cardinal motor symptoms: slowness of movement, rigidity, rest tremor, and postural instability (Ropper et al. 2009). Although these symptoms initially respond to drugs that modulate dopamine metabolism or surgeries that alter basal ganglia circuitry, the disease eventually progresses. With a modest exception (Olanow et al. 2009), no therapy has been shown to alter the disease course.
The pathogenesis of sporadic Parkinson's disease is likely complex involving altered metabolism of the protein α-synuclein, lysosomal dysfunction, and a dysregulated inflammatory response (reviewed in Shulman et al. 2011). Several lines of evidence also point to mitochondrial dysfunction as a central player in the pathogenesis of PD. Complex I dysfunction is associated with sporadic PD and is sufficient to induce parkinsonism (reviewed in Schapira 2008). The inhibitors of complex I, MPTP (Langston et al. 1983) and rotenone (Betarbet et al. 2000), replicate the symptoms of PD, and rotenone recapitulates key pathognomonic features of PD, such as the α-synuclein-rich inclusion bodies (Betarbet et al. 2000). The cause of mitochondrial dysfunction in sporadic PD is not entirely clear, but laser capture microdissection of substantia nigra neurons from patients with PD reveal a higher burden of mitochondrial DNA deletions relative to age-matched controls (Bender et al. 2006). That such deletions are sufficient to cause parkinsonism is suggested by the occurrence of parkinsonism in patients with rare mutations in their mtDNA replication machinery (e.g., the catalytic subunit of the mtDNA polymerase POLG [Luoma et al. 2004] or the mtDNA helicase Twinkle [Baloh et al. 2007]). The defective mtDNA replicative machinery generates high levels of mtDNA deletions throughout the body that are qualitatively similar to those observed in the substantia nigra in patients with sporadic PD (Reeve et al. 2008). Thus, mitochondrial dysfunction is both associated with sporadic PD and sufficient to cause the parkinsonian syndrome.
As is discussed in this review, recent insights from certain genetic forms of PD—resulting from mutations in Parkin or PINK1—support the model that mitochondrial damage is a central driver of PD pathogenesis. Additionally, they provide a rationale for targeting mitochondrial quality control pathways in patients with PD.
GENETIC STUDIES SUPPORT A COMMON PINK1/PARKIN PATHWAY
Mutations in Parkin or PINK1 are the leading cause of recessive parkinsonism (Hardy 2010). The parkinsonian syndrome caused by loss of Parkin (Kitada et al. 1998) or PINK1 (Valente et al. 2004) function is clinically indistinguishable from idiopathic Parkinson's disease at its outset, although it tends to present before the age of 40 and to progress more slowly. Additionally, only some patients with Parkin mutations develop the α-synuclein-rich inclusions, known as Lewy bodies and Lewy neurites, which are pathognomonic for idiopathic Parkinson's disease (Pramstaller et al. 2005).
The clinical similarity between patients with Parkin or PINK1 mutations suggests that they might function in a common pathway. Supporting this notion, deletion of either dParkin or dPink1 in Drosophila melanogaster leads to a similar (and relatively unique) phenotype with the triad of muscle degeneration, disrupted spermatogenesis, and loss of dopaminergic neurons within a specific cluster (Greene et al. 2003; Whitworth et al. 2005; Clark et al. 2006; Park et al. 2006; Yang et al. 2006). Moreover, overexpression of dParkin in the dPink1 null Drosophila and not the converse partially compensates for this phenotype, indicating that dPink1 is upstream of dParkin in the same pathway (Clark et al. 2006; Park et al. 2006; Yang et al. 2006).
The function of the dPink1/dParkin pathway at least in Drosophila is suggested by ultrastructural examination of the tissues before their degeneration. Mitochondria in the flight muscles become swollen and fragmented early in the course of the degeneration, suggesting that a mitochondrial defect may be the primary event (Greene et al. 2003; Whitworth et al. 2005; Clark et al. 2006; Park et al. 2006; Yang et al. 2006). Overexpression of components of the glutathione-dependent detoxification pathway partially prevents the mitochondrial damage, implying that mitochondria in Drosophila lacking the dPink1/dParkin pathway are less able to cope with oxidative stress (Whitworth et al. 2005). The apparent damage to mitochondria in the flight muscles correlates with a severe bioenergetic deficit as well as increased oxidative damage of the surrounding tissue, suggesting a mechanism for the subsequent apoptosis of the muscle fibers (Greene et al. 2003; Whitworth et al. 2005; Clark et al. 2006; Park et al. 2006; Yang et al. 2006).
Studies in mice are largely consistent with the notion of a common Pink1/Parkin pathway important for maintaining mitochondrial integrity. Loss of Parkin or Pink1 causes one phenotype similar to, albeit milder than, that in man, in which nigrostriatal dysfunction correlates with mitochondrial dysfunction in the striatum (Palacino et al. 2004; Gautier et al. 2008; Kitada et al. 2009). Interestingly, PINK1 knock out (KO) mice also display pronounced cardiac hypertrophy (Billia et al. 2011) and Parkin KO mice display less subcutaneous fat, are resistant to high-fat-diet-induced weight gain (Kim et al. 2011a), and may be predisposed to hepatocellular carcinomas (Fujiwara et al. 2008). It remains to be determined if these non-neuronal phenotypes are shared between PINK1 and Parkin deficient mice.
Although Parkin and PINK1 may have independent functions in some cellular pathways, parsimony would suggest that they function together in pathways relevant to PD pathogenesis. Additionally, as is discussed in more depth below, the difference in phenotype between insects and mammals suggests that, although features (including substrates) of PINK1/Parkin pathway are shared among insects and mammals, likely there are differences in the physiologic functions they serve.
PINK1 SENSES MITOCHONDRIAL STRESS
PINK1 is a serine/threonine protein kinase targeted to mitochondria (Valente et al. 2004; Silvestri et al. 2005; Zhou et al. 2008). Under basal conditions, mammalian Parkin is an E3 ubiquitin ligase residing (likely inert) in the cytosol. Upon mitochondrial damage, however, Parkin is activated and recruited selectively to bioenergetically impaired mitochondria (Narendra et al. 2008) by an elegant mechanism in which PINK1 functions as a sort of molecular scout reporting the internal state of individual mitochondria to Parkin in the cytosol (Geisler et al. 2010b; Matsuda et al. 2010; Narendra et al. 2010b; Vives-Bauza et al. 2010) (Fig. 1).
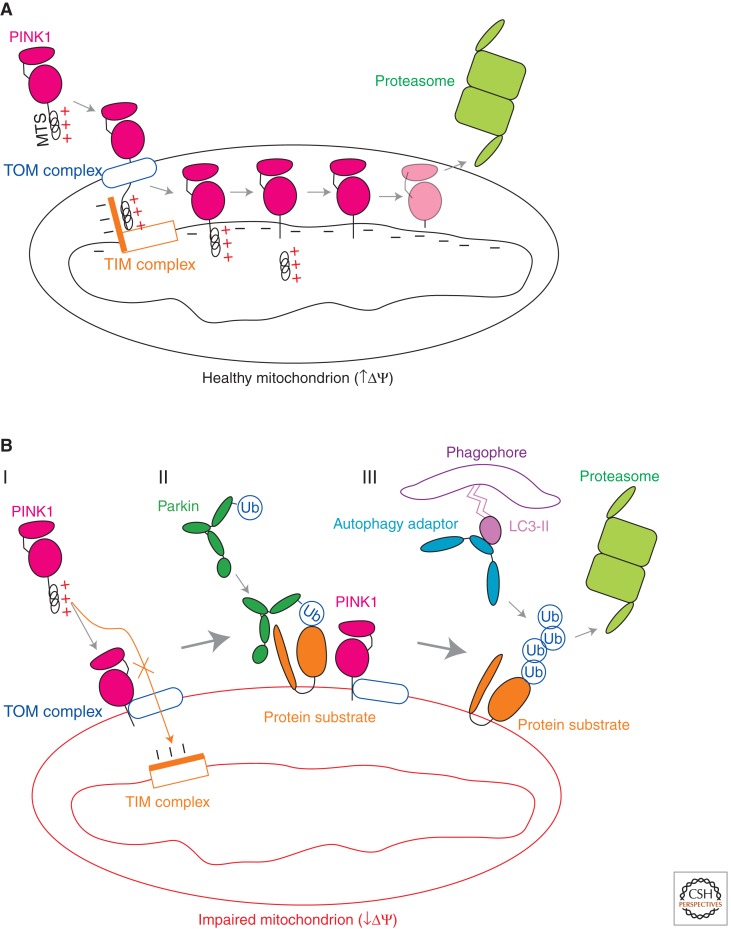
Figure 1.
Regulation of the PINK1-Parkin mitochondrial quality control pathway by inner mitochondrial membrane potential. (A) In healthy mitochondria, PINK1 is imported to the inner mitochondrial membrane by the TOM (translocase of the outer membrane) and TIM23 (translocase of the inner membrane 23) complexes, along a path that depends on the voltage component of the mitochondrial inner membrane potential (ΔΨ). At the inner membrane, the mitochondrial-targeting signal (MTS) of PINK1 is cleaved by the matrix metalloprotease. PINK1 is cleaved again in its transmembrane domain by the rhomboid protease PARL. The resulting PINK1 cleavage product is unstable and is degraded in a manner that depends on proteasomal activity. (B, I) In impaired mitochondria with collapsed ΔΨ, PINK1 cannot be imported to the inner membrane along the ΔΨ-dependent TIM23 pathway. Instead, PINK1 is directed to the outer mitochondrial membrane by a cryptic signal amino terminal to its transmembrane domain, in which it associates with the TOM complex. (B, II) The accumulation of PINK1 on the outer mitochondrial membrane recruits Parkin from the cytosol and activates its ubiquitin ligase activity. Parkin ubiquitinates outer membrane proteins preferentially on the mitochondrion on which PINK1 has accumulated. (B, III) Ubiquitination of outer mitochondrial membrane proteins by Parkin leads either to their degradation by the proteasome or to the recruitment of ubiquitin-binding adaptor proteins to effect the removal of the damaged mitochondrion by autophagy.
Like most other mitochondrial proteins, PINK1 is encoded in the nuclear genome and synthesized in the cytosol before its import via the TOM/TIM complexes (Silvestri et al. 2005). And like most other mitochondrial proteins expressing a mitochondrial targeting presequence, PINK1 requires the electrical component of the inner mitochondrial membrane potential (ΔΨ) for its import into mitochondria (Silvestri et al. 2005). Two features of PINK1 processing and targeting, however, make PINK1 uniquely suited for its role in signaling mitochondrial dysfunction. First, unlike most other mitochondrial proteins, imported PINK1 is cleaved into an unstable product by PARL (Whitworth et al. 2008; Jin et al. 2010; Deas et al. 2011; Meissner et al. 2011; Shi et al. 2011), an inner mitochondrial membrane protease (McQuibban et al. 2003), and other mitochondrial proteases (Greene et al. 2012) and consequently is present at very low levels in mitochondria that are healthy enough to maintain the ΔΨ dependent import pathway (Fig. 1A). (As an indication of its low abundance, PINK1 is usually not detected in proteomics studies of mitochondrial proteins despite its relatively large size [see for instance the MitoCarta database (Pagliarini et al. 2008)].) Second, PINK1 contains a second weaker targeting signal that directs it to the outer mitochondrial membrane (Zhou et al. 2008), if the TOM/TIM import pathway is blocked by collapse of ΔΨ (Narendra et al. 2010b). Thus, PINK1 acts as a kind of scout, dispatched centrally to all the mitochondria in the cell to assess their internal state. The healthy mitochondria with a strong inner membrane potential quickly dispose of PINK1, whereas failing mitochondria, unable to import and degrade the kinase, accumulate it on their cysotolic face, in effect displaying their inner dysfunction on their outer surface. The accumulation appears to be very rapid with a several fold increase in PINK1 protein expression within minutes of ΔΨ collapse (Narendra et al. 2010b). However, an alternative model has been presented in which PARL-mediated PINK1 cleavage activates PINK1 rather than eliminates it (Whitworth et al. 2008; Deas et al. 2011). Once on the outer mitochondrial membrane, PINK1 constitutively associates with the TOM complex. This association has been suggested to mediate rapid reimport and degradation of PINK1 if mitochondrial membrane potential is restored thereby yielding a quick on/off signal of mitochondrial dysfunction (Lazarou et al. 2012).
PINK1 RECRUITS PARKIN TO MITOCHONDRIA AND ACTIVATES ITS LATENT E3 LIGASE ACTIVITY
PINK1, when exposed on the cytosolic surface, directs Parkin specifically to the ailing mitochondrion resulting in ubiquitination of several outer mitochondrial membrane proteins on that dysfunctional mitochondrion (Geisler et al. 2010b; Matsuda et al. 2010; Narendra et al. 2010b; Vives-Bauza et al. 2010) (Fig. 1B). Precisely how PINK1 activates and directs Parkin selectively to the mitochondria on which PINK1 accumulates is not resolved fully. Although there is evidence that Parkin and PINK1 directly bind each other (Parkin and PINK1 co-immunoprecipitate [Xiong et al. 2009; Sha et al. 2010]). The bulk of PINK1 and Parkin appear to reside in complexes of different sizes by size exclusion chromatography and native PAGE electrophoresis (Thomas et al. 2011; Lazarou et al. 2012). There is also mixed evidence that PINK1 directly phosphorylates Parkin (Kim et al. 2008; Narendra et al. 2010b; Sha et al. 2010; Vives-Bauza et al. 2010).
An alternative (and not mutually exclusive) hypothesis suggests that PINK1 may direct Parkin to particular substrates on the outer mitochondrial membrane by priming the substrates through phosphorylation. A recent study of one of the substrates of Parkin, Miro1, lends support for this shared substrate model. The investigators report that PINK1 phosphorylates Miro1 on a specific serine residue, S156 (Wang et al. 2011). In addition, the investigators found that if phosphorylation of S156 is blocked by mutating S156 to an alanine, Miro1 is no longer degraded in a Parkin-dependent manner. However, a recent study was unable to confirm that dMiro is phosphorylated by dPink1 in an in vitro assay or that the Miro1 S156A mutant is degraded by Parkin less efficiently than wild-type Miro1 (Liu et al. 2012). Additional studies will be required to validate whether Miro1 is a substrate of PINK1, although, as discussed below, it appears that Miro1 is a physiological substrate of Parkin.
If confirmed, the shared substrate model implies that there may be a motif that is recognized in the unphosphorylated form by PINK1 and the phosphorylated form by Parkin. It will be interesting to learn whether other Parkin substrates (in particular the mitofusins) are similarly phosphorylated by PINK1 and whether a common motif among PINK1/Parkin substrates is found. A preferred peptide substrate for a recombinant insect PINK1 has been identified in vitro, and it is notable for a proline in the 1+ position relative to the phosphorylated serine and positively charged residues in other positions: KKWIPYRRSPRRR (Woodroof et al. 2011). Of note, the context of the phosphorylated serine in this peptide is not obviously similar to that in the proposed Miro1 site, NLKNISELFYYAQK (Wang et al. 2011). This could reflect differences between the in vitro system and the cell-based system (e.g., the efficiency of phospho-transfer within the cell will likely be influenced by the local concentration of the kinase and substrate on the mitochondrial membrane as well as binding between kinase and substrate outside of the phosphorylated region.) Alternatively, the in vitro findings may cast doubt on the validity of the proposed Miro site.
Study of ectopic targeting of PINK1 to cellular membranes has provided additional insight into Parkin recruitment by PINK1 (Lazarou et al. 2012). Ectopic targeting of PINK1 to alternate organelles can recruit Parkin to those same alternate subcellular locations. Interestingly, when cytosolic PINK1 is recruited to mitochondria or other membranes using a heterodimerization system, Parkin recruitment to the same membranes lags by tens of minutes. This suggests that PINK1 assumes a form competent for binding Parkin after it associates with membranes or that Parkin recruitment to membranes reflects an interaction with membrane components other than PINK1 (e.g., binding to its protein substrates).
Failing mitochondria that are selectively tagged by activated Parkin are altered to promote their isolation within the cell and their degradation by selective macroautophagy (hereafter, “autophagy”). Several events in cultured mammalian cells overexpressing Parkin are observed following mitochondrial uncoupling: (1) The mitochondria fragment and fail to refuse when the membrane potential is restored (Tanaka et al. 2010); (2) the spherical mitochondria form clumps that resemble bunches of grapes (Narendra et al. 2010a; Okatsu et al. 2010); (3) the clumps of mitochondria are gathered in a perinuclear localization possibly because of increased minus end transport along microtubules (Lee et al. 2010; Vives-Bauza et al. 2010); and (4) mitochondria are degraded by a selective form of autophagy (mitophagy) (Narendra et al. 2008). Within 24 h, all of the mitochondria within mammalian cells overexpressing Parkin can be eliminated by autophagy. The specific targeting of Parkin to damaged mitochondria by PINK1 together with the autophagic elimination of such mitochondria suggests that this pathway mediates mitochondrial quality control. Although this hypothesis has yet to be validated in animal models, a study of cultured cybrid cells containing a stable heteroplasmic mixture of wild-type mitochondrial DNA and mitochondrial DNA mutated in the COX1 gene supports the idea. Stable overexpression of Parkin at levels 3–6-fold above endogenous Parkin levels found in some normal tissues mediated the selective removal of the mutated mitochondrial DNA and enrichment of cytochrome oxidase activity (Suen et al. 2010). In a complementary study, mitophagy of dysfunctional mitochondria in cells homoplasmic for mtDNA mutations was found to be dependent on both Parkin expression and mTORC1 inhibition (Gilkerson et al. 2012). Consistent with a housekeeping function of Parkin, it is widely distributed throughout mitochondria-rich tissues, including brain, muscle, and liver (Suen et al. 2010).
PARKIN UBIQUITINATES MITOCHONDRIAL OUTER MEMBRANE PROTEINS FOLLOWING PINK1 RECRUITMENT
Parkin mediates directly or indirectly the ubiquitination of mitochondrial outer membrane proteins with various ubiquitin linkages (Geisler et al. 2010b; Narendra et al. 2010a; Chan et al. 2011). Some of these proteins with predominantly K48-linked ubiquitin chains are eliminated by proteosomal degradation, whereas others with K63-linked ubiquitin chains may recruit adaptor proteins from the cytosol. Changes in mitochondrial dynamics and transport appear to be mediated by elimination of specific outer mitochondrial membrane proteins, whereas mitochondrial clumping and mitophagy appear to depend on recruitment of specific adaptor proteins to ubiquitinated substrates on the outer mitochondrial membrane (as discussed briefly below and in more depth elsewhere).
Proteins that accumulate on mitochondria concurrent with Parkin translocation and ubiquitination of mitochondrial substrates include p97 and proteasome subunits consistent with an ERAD type pathway of mitochondrial outer membrane protein degradation by the proteasome (Tanaka et al. 2010; Xu et al. 2011). Other proteins such as p62 accumulate on mitochondria by binding ubiquitin chains (with a preference for binding K63-linked chains [Kirkin et al. 2009]). p62 is an adaptor protein that recruits unfolded protein aggregates to autophagosomes, and its role in mitophagy will be discussed later. Which process, proteosomal elimination versus adaptor protein recruitment, mediates autophagosome engulfment of mitochondria is still not clear.
UNBIASED IDENTIFICATION OF SUBSTRATES DEGRADED BY PARKIN
Parkin likely mediates its effect on mitochondrial dynamics, distribution, and turnover through ubiquitination of specific substrates in the outer mitochondrial membrane. These effects could be mediated either through elimination or inhibition of the normal function of the outer membrane proteins or by recruitment of adaptor proteins to the post-translationally modified OMM proteins. Particularly when Parkin is present at supraphysiologic levels many proteins on the OMM appear to be ubiquitinated in a manner that depends on activated Parkin (Chan et al. 2011; Yoshii et al. 2011). Although it is clear that Parkin can promote the ubiquitination of many OMM proteins under special circumstances (e.g., in the presence of high levels of Parkin and PINK1), which substrate is ubiquitinated most efficiently in vivo and which is of greatest physiological importance is less clear.
At least for the subset of Parkin substrates that are degraded after ubiquitination their specificity as substrates in vivo may be reflected in the ease with which they are eliminated by activated Parkin in cultured cells. To help identify the subset of substrates targeted for degradation following Parkin ubiquitination as well as adaptor proteins recruited to OMM proteins ubiquitinated by Parkin, we measured changes in membrane protein abundance using a stable isotope labeling in cell culture (SILAC) proteomics approach (DP Narendra, unpubl.). The experiment was designed to assess changes in membrane protein abundance in response to two variables: the presence or absence of Parkin and the presence or absence of mitochondrial uncoupler CCCP (which activates Parkin by increasing endogenous PINK1 protein levels). Assessing changes along these two dimensions helps to separate changes caused by activated Parkin from secondary effects of the uncoupler CCCP (Fig. 2).
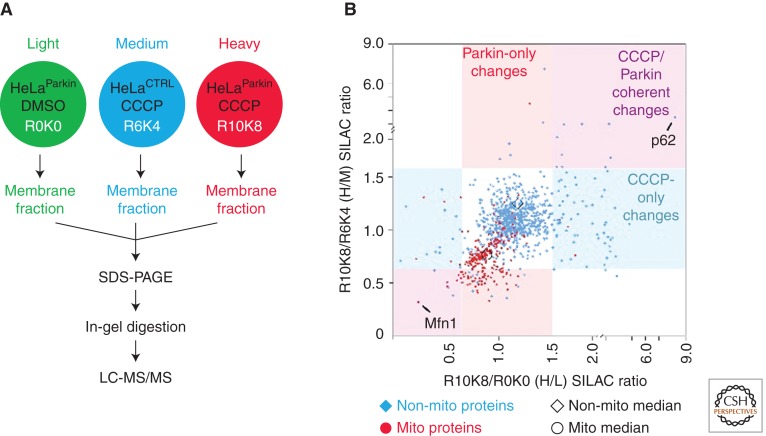
Figure 2.
Quantitative proteomics-based screen for the identification of Parkin substrates and adaptors in the PINK1/Parkin pathway. (A) Schematic depicting the experimental setup. Three cell lines were labeled with stable isotopes of arginine and lysine to give a light (L) labeled HeLa cell line stably expressing YFP-Parkin treated with vehicle (HeLaParkin); a medium (M) labeled HeLa cell line without Parkin expression treated with the mitochondrial uncoupler CCCP for 2 h (HeLaCTRL); and a heavy (H) labeled HeLaParkin cell line treated with CCCP. A crude membrane fraction was extracted from each cell line by permeabilizing the membranes with a low dose of digitonin, isolating the membranes by centrifugation, and then extracting proteins from the remaining membranes with DDM (n-dodecyl-β-maltoside). The protein extracts from each cell line were mixed in a 1:1:1 ratio, before separation on a SDS-PAGE gel, in-gel digestion, and LC-MS/MS on an Orbitrap instrument with a top 5 duty cycle. The data was analyzed in MaxQuant. (B) Scatter plot depicting H/L (x-axis) and H/M (y-axis) ratios for 992 proteins identified in the experiment. Proteins with “mitochondrial” in the protein name appear as red circles, whereas the others appear as blue diamonds. The median for proteins annotated as mitochondrial is shifted down and to the left relative to the median for other proteins consistent with the en masse degradation of mitochondrial proteins following treatment with CCCP in the presence of Parkin expression. Proteins in the upper right quadrant represent potential adaptors recruited to mitochondria. Proteins in the lower left quadrant represent potential substrates of Parkin. The protein with the largest increase along the x- and y-axes, p62, and the largest decrease, Mfn1, are labeled on the plot.
For each protein sequenced, two ratios, heavy labeled (H)/light labeled (L) and heavy labeled (H)/medium labeled (M), reflect changes in membrane protein abundance caused by CCCP treatment and Parkin expression, respectively (Tables 1 and 2). Proteins that increase in response to both Parkin expression and CCCP treatment may be adaptor proteins that are recruited to mitochondria from the cytosol following ubiquitination of outer mitochondrial membrane (OMM) proteins, whereas proteins that decrease in response to both Parkin expression and CCCP treatment may be substrates of Parkin degraded by the proteasome. Of note, this approach does not identify proteins ubiquitinated by Parkin that are not subsequently degraded in disproportion to other mitochondrial proteins (Fig. 3).
Table 1.
Proteins with coherent changes in abundance
| Protein | Localization | Unique peptides | PEP | Ratio H/L | Ratio H/M | Ratio H/L (Chan et al. 2011) |
|---|---|---|---|---|---|---|
| Proteins up-regulated | ||||||
| p62/SQSTM1 | Cyto | 4 | 3.37E-05 | 7.58 | 2.59 | 5.8 |
| Heme oxygenase 1 | ER | 3 | 5.65E-07 | 2.42 | 2.11 | NR |
| Heat shock protein β-1 | Cyto/Nuc | 9 | 0 | 2.54 | 1.68 | NR |
| Ferritin light chain | Cyto | 4 | 4.64E-32 | 1.79 | 2.07 | NR |
| Thioredoxin | 9 | 0 | 2.15 | 1.59 | 3.2 | |
| Proteins down-regulated | ||||||
| Mitofusin1 | OMM | 3 | 4.54E-09 | 0.19a | 0.28a | 0.09b |
| GDAP1 | OMM | 14 | 2.49E-08 | 0.53 | 0.34 | 0.40 |
| MitoNEET | OMM | 18 | 0 | 0.48a | 0.42 | 0.26b |
| MOSC1 | OMM | 3 | 0 | 0.47 | 0.44 | 0.39 |
| TOM70 | OMM | 5 | 0 | 0.47a | 0.48 | 0.13b |
| Additional proteins down-regulated in Chan et al. (2011) | ||||||
| Mitofusin2 | OMM | ND | ND | 0.1b | ||
| MIRO1/RHOT1 | OMM | ND | ND | 0.13b | ||
| CPT1A | OMM | 3 | 2.76E-24 | 0.73 | 0.60 | 0.23b |
| MOSC2 | OMM | ND | ND | 0.26b | ||
| GPAM | OMM | ND | ND | 0.42b | ||
| MIRO2/RHOT2 | OMM | ND | ND | 0.44b | ||
| FIS1 | OMM | 2 | 9.93E-14 | 0.59 | 0.64 | 0.55b |
| Additional substrates identified by candidate approaches | ||||||
| VDAC1 | OMM | 5 | 5.27E-29 | 0.50 | 0.68 | NR |
| Tom20 | ND | ND | NR | |||
| Bcl-2 | OMM | 3 | 1.61E-23 | 0.99 | 0.87 | NR |
Proteins with coherent changes in abundance (i.e., the H/L and H/M ratios are both high or both low) are listed. These represent proteins with a change in abundance that is dependent on both CCCP treatment and Parkin expression. ND, not detected; NR, not reported.
aRatios reaching statistical significance after correction for multiple comparisons in this study.
bRatios reaching statistical significance after correction for multiple corrections in Chan et al. (2011).
Table 2.
Proteins increased or decreased along a single dimension
| Protein | Function | Unique peptides | PEP | Ratio H/L | Ratio H/M | H/L (p-value) | H/M (p-value) |
|---|---|---|---|---|---|---|---|
| CCCP-only changes | |||||||
| Subunits of ETC complexes | |||||||
| Complex I-ESSS | Supernumary subunit of β subcomplex | 2 | 0.00055987 | 0.23986 | 1.3004 | 4.92E-07 | 0.20850717 |
| Complex I-PDSW | Supernumary subunit of β subcomplex | 4 | 1.97E-21 | 0.44766 | 1.1732 | 3.74E-05 | 0.362282139 |
| Complex IV - subunit 4 isoform 1 | Regulatory subunit downregulated by hypoxia | 9 | 0 | 0.43761 | 1.2583 | 2.33E-05 | 0.181333382 |
| Subunits of vacuolar proton pump | |||||||
| ATP6V1H | Component of vacuolar proton pump | 3 | 4.63E-06 | 2.9342 | 0.72304 | 7.85E-06 | 0.347560475 |
| ATP6B2 | Component of vacuolar proton pump | 14 | 0 | 2.6621 | 0.90669 | 7.59E-06 | 0.722666052 |
| ATP6A1 | Component of vacuolar proton pump | 18 | 0 | 2.5312 | 0.88796 | 2.34E-05 | 0.671846651 |
| ATP6G | Component of vacuolar proton pump | 3 | 3.22E-31 | 2.4512 | 0.82108 | 0.000228262 | 0.56428798 |
| ATP6E | Component of vacuolar proton pump | 5 | 9.67E-22 | 2.3293 | 0.91277 | 0.000131918 | 0.739212408 |
| ATP6M | Component of vacuolar proton pump | 2 | 0.01103 | 2.2548 | 0.70783 | 0.000895118 | 0.317310509 |
| Cholestrol metabolism | |||||||
| 2,3-epoxysqualene–lanosterolcyclase | Cholestrol synthesis | 11 | 1.11E-39 | 2.4972 | 0.77465 | 3.13E-05 | 0.38264962 |
| LDL receptor | Cell-surface receptor for LDL particles | 11 | 1.46E-34 | 2.164 | 1.0538 | 0.0016753 | 0.816681405 |
| Sterol-4-α-carboxylate 3-dehydrogenase | Cholestrol synthesis | 13 | 0 | 2.0767 | 1.0301 | 0.001109734 | 0.904184966 |
| Misc. increased | |||||||
| α-SGT | Co-chaperone of HSP70 | 4 | 3.20E-28 | 4.0283 | 1.04 | 4.91E-09 | 0.866974414 |
| Glyceraldehyde-3-phosphate dehydrogenase | Glycolysis | 14 | 0 | 2.8911 | 1.1959 | 1.06E-06 | 0.304181529 |
| Adult folate-binding protein | Folate transport | 5 | 3.01E-39 | 2.1425 | 0.58908 | 0.000640711 | 0.075974088 |
| Histone H4 | Core component of nucelosome | 6 | 0 | 2.0654 | 1.3682 | 0.001218908 | 0.064947946 |
| Misc. decreased | |||||||
| Nucleobindin-1 | Multidomain calcium binding protein | 18 | 0 | 0.27213 | 0.74534 | 8.74E-11 | 0.317310509 |
| Cysteine-rich FGF receptor | FGF binding intracellular glycoprotein | 10 | 0 | 0.34651 | 0.58858 | 9.31E-08 | 0.075513331 |
| Mesothelin | GPI-linked cell surface protein | 4 | 1.35E-13 | 0.35673 | 0.76751 | 0.000233983 | 0.441886649 |
| Retinol dehydrogenase 13 | Oxidoreductase | 4 | 3.30E-22 | 0.39441 | 0.75978 | 0.000845024 | 0.42493045 |
| Cytochrome c type heme lyase | Covalently attaches heme to cytochrome c | 13 | 0 | 0.39837 | 0.81099 | 2.91E-06 | 0.470230202 |
| Proliferating cell nuclear antigen | DNA replication/repair | 5 | 0 | 0.41676 | 1.3729 | 0.0016321 | 0.127915585 |
| Parkin-only changes | |||||||
| Increased | |||||||
| Integrin α-1 | Cell adhesion | 19 | 0 | 1.4191 | 6.3657 | 0.162165287 | 4.54E-29 |
| OPA1 | Mitochondrial fusion | 3 | 0 | 1.2757 | 3.7024 | 0.401069865 | 1.99E-10 |
| GPRC5C | G-protein signaling | 3 | 9.69E-09 | 1.4069 | 2.1807 | 0.205413008 | 0.000156889 |
| Decreased | |||||||
| Adenylate kinase isoenzyme 4, mitochondrial | ATP/AMP transphosphorylase | 3 | 0.034025 | 0.87783 | 0.29612 | 0.537881606 | 0.00048901 |
Proteins with an increase or decrease along either the x- or y-axis, indicating an increase or decrease in response to either CCCP treatment or Parkin expression but not both in the same direction, are listed.
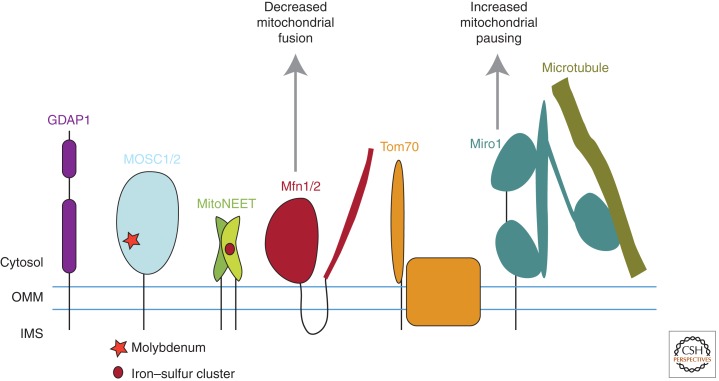
Figure 3.
Figure depicting the five mitochondrial proteins with the largest relative decrease in abundance in response to Parkin activation in unbiased quantitative proteomics experiments. They differ in the number of transmembrane domains they possess, their size, and their association with other proteins, indicating a diversity of mitochondrial substrates toward which Parkin has high activity. Three of the substrates, Mfn1, Mfn2, and Miro1, have been extensively validated as physiologic substrates of Parkin in both insects and mammals by multiple groups.
After filtering out contaminants according to published guidelines (Cox et al. 2009), a total of 992 membrane proteins were identified and quantified, roughly a quarter of which were annotated as mitochondrial in their protein name. Proteins annotated as mitochondrial exhibited a 43.8% and 30.9% decrease in abundance along both the Parkin-dependent and CCCP-dependent axes relative to all other proteins (two-tailed Student’s t-test; p-values = 3.0 × 10−47 and 2.5 × 10−29, respectively), consistent with Parkin promoting the selective degradation of uncoupled mitochondria under these conditions.
p62/SQSTM1, a cytosolic ubiquitin-binding adaptor protein, exhibited the largest increase in abundance, consistent with previous studies showing that it binds proteins ubiquitinated by Parkin (Geisler et al. 2010a; Lee et al. 2010; Narendra et al. 2010a; Okatsu et al. 2010). Its possible role as an adaptor in the PINK1/Parkin pathway is discussed further below.
The five proteins exhibiting the largest decrease in abundance along both dimensions are resident to the OMM, consistent with their potential ubiquitination and degradation by Parkin. These are Mfn1, MitoNEET, GDAP1, MOSC1, and Tom70. Mfn1, MitoNEET, and Tom70 reached statistical significance along the CCCP-treated axis, following correction for multiple comparisons, whereas the others trended toward significance. Mfn1, which also reached statistical significance along the Parkin-dependent axis, exhibited an almost twofold greater decrease at 2 h relative to the other four OMM proteins that decreased in this experiment (DP Narendra, unpubl.).
These results are largely in concordance with those from a quantitative proteomics experiment recently published by an independent group (Chan et al. 2011). Similar to our results, Chan et al. (2011) report that p62, as well as its homolog NBR1 (not identified in our experiment), increased in the membrane fraction of HeLa cells expressing exogenous Parkin following depolarization with CCCP for 2 h. Chan et al. also observed an increase in autophagy related proteins, such as LC3, consistent with ongoing mitophagy. Of note although both groups (Chan et al. 2011; DP Narendra, unpubl.) observed an increase in subunits of the vacuolar proton pump in response to CCCP treatment, this increase did not depend on the presence on Parkin in our experiment, indicating that it is because of CCCP treatment alone and is not an effect mediated by Parkin.
Additionally, five of the top ten OMM proteins Chan et al identified as significantly decreased in the membrane fraction following treatment with CCCP (namely, Mfn1, Mfn2, MitoNEET, MOSC2, and Tom70) are the same or homologs of the five proteins we identified as significantly decreased under these conditions. As in our experiment, Mfn1 (as well as its homolog Mfn2) exhibited the largest decrease. Additionally, Chan et al found that Miro1 (not identified in our experiment) exhibited a decrease of similar magnitude to Mfn1/Mfn2. In both the Chan et al. and our experiments, other OMM proteins identified as potential substrates of Parkin by candidate approaches (such as VDAC1 [Geisler et al. 2010a] and Bcl-2 [Chen et al. 2010a]) were not significantly altered in abundance following depolarization with CCCP for 2 h. The concordance of these findings suggests that Parkin exhibits considerable specificity for Mfn1/Mfn2, Miro1, and Tom70 (and possibly a limited number of other mitochondrial proteins, such as MitoNEET). Mfn1/Mfn2, Miro1, and Tom70 emerge from these unbiased proteomics screens as the most likely mitochondrial candidates for physiologic substrates of Parkin.
Detailed studies of mitofusins and Miro1 support their relative physiological importance among proposed Parkin substrates. To our knowledge, Mfn1/Mfn2 and Miro1 are the only mitochondrial substrates shown to be ubiquitinated or degraded by endogenous levels of Parkin (Poole et al. 2010; Tanaka et al. 2010; Wang et al. 2011; Liu et al. 2012). Additionally, they are the only proteins reported to be targeted by the PINK1/Parkin pathway in both insects and mammals (Gegg et al. 2010; Poole et al. 2010; Tanaka et al. 2010; Ziviani et al. 2010; Chan et al. 2011; Rakovic et al. 2011; Wang et al. 2011). Finally, Mfn1/Mfn2 and Miro1 are quantitatively modified following PINK1/Parkin activation, whereas a relatively small portion of the protein pool appears to be modified in the case of other proposed substrates, such as VDAC1. Together these findings argue that Mfn1/2 and Miro1 are physiologically relevant substrates of the PINK1/Parkin mitochondrial quality control pathway.
PINK1 AND PARKIN REGULATE MITOFUSINS TO ISOLATE IMPAIRED MITOCHONDRIA
The mitofusin orthologs (Mfn1 and Mfn2 in mammals, Fzo1 and MARF in flies, and Fzo1 in yeast) are GTPases that mediate outer mitochondrial membrane fusion (Hales and Fuller 1997). Their disruption in mice and yeast leads to a highly fragmented mitochondrial network as well as loss of mitochondrial fidelity, as reflected in decreased mtDNA copy number, an increased number of mtDNA mutations, and an accompanying bioenergetic deficit (Hermann et al. 1998; Rapaport et al. 1998; Chen et al. 2010b).
The degradation of the mitofusins by Parkin appears to help isolate failing mitochondria from the mitochondrial network by preventing their outer membrane fusion with more bioenergetically competent mitochondria. If the isolation of the dysfunctional mitochondria is inhibited (e.g., by disruption of the mitochondrial fission machinery), then mitophagy of the dysfunctional mitochondria is delayed (Tanaka et al. 2010).
Whereas the PINK1/Parkin pathway appears to regulate the fusion machinery selectively on dysfunctional mitochondria as a quality control measure, it does not appear to play an important role in the basal regulation of the fusion machinery in mammalian cells. Mitofusin levels are not substantially altered following knockdown or knockout of endogenous Parkin or PINK1 in the absence of mitochondrial stress (Gegg et al. 2010; Tanaka et al. 2010; Chan et al. 2011; Rakovic et al. 2011). Likewise, several reports have shown a minimally changed or more fragmented mitochondrial network following knockdown or elimination of Parkin or PINK1 in cultured mammalian cells (Exner et al. 2007) or in tissues analyzed ex vivo (Gautier et al. 2008).
In Drosophila, in contrast, the dPink1/dParkin pathway does appear to play a critical role in the basal regulation of the fusion/fission machinery. Depletion of Parkin in Drosophila myocytes or cultured S2 cells leads to a substantial decrease in mitofusin ubiquitination and increase in mitofusin levels (Poole et al. 2010; Ziviani et al. 2010). Coinciding with the decrease in ubiquitinated mitofusin the mitochondrial network becomes hyperfused, indicating that the dPink1/dParkin pathway has a strong effect on the overall mitochondrial fission/fusion balance under basal conditions (Ziviani et al. 2010). Additionally, a loss of the basal regulation of mitochondrial dynamics by disruption of the dPink1/dParkin pathway appears to be at least partially responsible for the abnormal muscle and sperm phenotype observed, as evidenced by the partial amelioration of the dParkin or dPink1 null phenoytpe by genetic alterations that promote increased fission (Deng et al. 2008; Poole et al. 2008; Yang et al. 2008).
PINK1/PARKIN PATHWAY REGULATES MIRO LEVELS TO IMMOBILIZE MITOCHONDRIA
Miro1 and Miro2, homologous EF-hand-containing adaptor proteins anchored in the OMM, form a complex with Milton and kinesin to promote the transport of mitochondria toward the plus-end of microtubules (Stowers et al. 2002; Guo et al. 2005). Miro1/2 are regulated by calcium via their EF-hands to promote pausing in the presence of high calcium concentrations (Saotome et al. 2008; Macaskill et al. 2009; Wang and Schwarz 2009). The dwelling of mitochondria in pools of higher calcium may help buffer the local calcium keeping the calcium rise spatially and temporally restricted. Miro1 is also a protein component of ER-mitochondrial contact sites (Kornmann et al. 2011). These sites also help to hem in calcium transients by forming a synapse between a major site of calcium release (the ER) and a major site of calcium buffering (the mitochondria) (Rizzuto et al. 1998). In addition, these contact sites allow phospholipid exchange between the organelles, which is necessary for the synthesis of certain phospholipids (reviewed in Osman et al. 2011).
The first indication that the Parkin/PINK1 pathway may regulate the Miro1/Milton complex came from the observation that a portion of the mitochondrial PINK1 pool exists in complex with Miro1 and Milton (Weihofen et al. 2009). Subsequently, it was shown that Miro1 is degraded under conditions that activate Parkin (e.g., treatment with a mitochondrial uncoupler) and that its degradation depends on the presence of endogenous levels of Parkin and PINK1 (Wang et al. 2011; Liu et al. 2012). The degradation of Miro1 leads to arrest of the dysfunctional mitochondria in neurons, which may prevent their transport to energy-hungry areas of the cell at the expense of bioenergetically viable mitochondria (Wang et al. 2011; Liu et al. 2012). The degradation of Miro1 also appears to promote the subsequent mitophagy of the impaired mitochondria (Liu et al. 2012).
In cultured mammalian cells, activation of the Parkin/PINK1 pathway promotes net retrograde movement of mitochondria resulting in the concentration of mitochondria near the microtubule-organizing center rather than mitochondrial pausing (Vives-Bauza et al. 2010). This transport is inhibited by nocodazole, which causes microtubule depolymerization, suggesting that it occurs along the microtubular tracks (Vives-Bauza et al. 2010). Similarly, loss of Parkin causes clustering of mitochondria around the plus-end of microtubules in Drosophila follicular cells, suggesting that Parkin also promotes net mitochondrial transport toward the minus end of microtubules in this tissue (Cox and Spradling 2009). Degradation of Miro1 may also be responsible for these Parkin-induced changes in mitochondrial transport. They may also depend on HDAC6, a microtubule binding protein that is recruited to mitochondria following the activation of Parkin by PINK1 (Lee et al. 2010).
Similar to the situation with mitochondrial fission, Parkin/PINK1 appears to be more important for basal mitochondrial transport in Drosophila relative to mice. Knockdown of dParkin or dPink1 in Drosophila leads to an increase in anterograde and retrograde mitochondrial transport within neurons, presumably caused by stabilization of the Miro1/Milton/Kinesin complex (Wang et al. 2011). However, knockout of Parkin or Pink1 in mice does not appear to significantly increase the likelihood of mitochondrial transport in the absence of mitochondrial stress, consistent with a quiescent PINK1/Parkin pathway when the mitochondria are healthy (Wang et al. 2011).
The relative importance of the Pink1/Parkin pathway for the basal regulation of mitochondrial transport and mitochondria fission/fusion in insects versus mammals may help explain the disparity in phenotype severity between the two classes of organisms. Pink1/Parkin appears to play an important housekeeping function in flies by controlling levels of the mitofusins and Miro, which are produced in excess. Consistent with a basal requirement for dPink1 and dParkin, their loss leads to severe disruption of multiple tissues. In mammals, in contrast, the PINK1/Parkin pathway appears to be kept in reserve until mitochondrial damage is suffered, thus triggering its activation. Consequently, in mice and humans dysfunction appears only in the most vulnerable tissues and at a relatively late stage of life.
MECHANISM OF PARKIN-INDUCED MITOPHAGY
Although the degradation of mitofusins and Miro by Parkin may explain the effect of the PINK1/Parkin pathway on mitochondrial dynamics and transport and their degradation may promote the subsequent mitophagy (Tanaka et al. 2010; Liu et al. 2012), likely they do not fully explain how Parkin promotes mitophagy. Parkin, for instance, still induces the autophagy of depolarized mitochondria in mouse fibroblasts null for Mfn1 and Mfn2 (Tanaka et al. 2010; Chan et al. 2011).
The leading hypothesis for the mechanism of Parkin-induced mitophagy is that the stable ubiquitination of some OMM proteins by Parkin serves as a positive signal for autophagy through an interaction with adaptor proteins that mediate selective autophagy. Such as mechanism has been proposed for the selective autophagy of other organelles, such as peroxisomes, as well as some intracellular bacteria, and ribosomes (reviewed in Johansen and Lamark 2011).
To date, no Parkin substrate has been convincingly shown to be required for mitophagy. A single report proposed that VDAC1 is required for mitophagy (Geisler et al. 2010a). However, this report, which used siRNA knockdown of VDAC1, has not been reproduced. A subsequent report by our group found that Parkin-induced mitophagy occurs in mouse fibroblasts null for VDAC1 and VDAC3 (Narendra et al. 2010a). Additionally, only a small fraction of the VDAC1 pool appears to be ubiquitinated by overexpressed Parkin, and VDAC1 has not been shown to be ubiquitinated by endogenous levels of Parkin and PINK1, calling into question whether VDAC1 is a specific substrate of Parkin (Narendra et al. 2010a). This is in marked contrast to the situation with the mitofusins and Miro, in which endogenous levels of Parkin and PINK1 are sufficient to ubiquitinate most of the substrate pool (Tanaka et al. 2010; Wang et al. 2011; Liu et al. 2012).
The identification of specific Parkin substrates that are not subsequently degraded might benefit from additional quantitative proteomics studies. Using a recently described antibody, the tryptic digests of proteins can now be enriched for diglycine-modified peptides (the stigmata of ubiquitination following protein digestion with trypsin), facilitating the direct identification of ubiquitinated proteins (Xu et al. 2010; Kim et al. 2011b). These substrates may have a ubiquitin chain with a specific linkage that does not bind the proteasome subunits, explaining why they are not degraded following ubiquitination. Polyubiquitin chains with a K63 linkage, for instance, usually do not direct the substrate to the proteasome for degradation (reviewed in Winget and Mayor 2010). Parkin has been shown to induce the K63 polyubiquitination of unidentified OMM proteins, using both antibodies specific for particular diubiquitin linkages (Narendra et al. 2010a; Okatsu et al. 2010; Chan et al. 2011) and quantitative mass spectrometry based techniques (Chan et al. 2011).
Once ubiquitinated, mitochondrial surface proteins may interact with ubiquitin-binding adaptor proteins that connect the ubiquitinated proteins with the autophagy machinery. p62 and the related NBR1 appear to act as autophagy adaptors for peroxisomes, intracellular bacteria, and protein aggregates (reviewed in Johansen and Lamark 2011). p62 has been shown to be recruited to mitochondria ubiquitinated by Parkin (Geisler et al. 2010a; Lee et al. 2010; Narendra et al. 2010a; Okatsu et al. 2010), and the quantitative proteomics studies described above show that p62 and NBR1 exhibit a larger fold increase in the membrane fraction than any other protein detected following Parkin activation (Chan et al. 2011). Although p62 mediates mitochondrial clumping, it does not appear to be necessary or sufficient for mitophagy (Narendra et al. 2010a; Okatsu et al. 2010). Parkin-induced mitophagy occurs in fibroblasts from p62 null mice to a similar extent as in wild-type cells (Narendra et al. 2010a; Okatsu et al. 2010), and recruitment of p62 to mitochondria by an OMM targeted ubiquitin fusion protein does not induce mitophagy (Narendra et al. 2010a) (but see Geisler et al. [2010a] and Lee et al. [2010] for RNA interference data suggesting that p62 is limiting for Parkin-induced mitophagy). Nonetheless, it is possible that p62 and NBR1 together are necessary and/or sufficient for Parkin-induced mitophagy. Drosophila have only one p62 ortholog, called ref(2)P. A recent report suggested that Parkin overexpression rescues mitochondrial damage induced by a misfolded matrix protein by a mechanism that requires both the autophagy gene atg1 and ref(2)P, consistent with the p62 ortholog functioning as an autophagy adaptor in this pathway (Pimenta de Castro et al. 2012). Future studies may reveal whether p62 and NBR1 together mediate autophagy in mammalian cells after OMM proteins are tagged with ubiquitin by Parkin.
A recent study showed that autophagosome formation is initiated adjacent to mitochondria following Parkin recruitment. The most upstream complex in the autophagy pathway, the ULK1–Atg13–FIP200–Atg101 complex, appears to be recruited near mitochondria (Itakura et al. 2012). Although p62/NBR1 has traditionally been thought to engage the autophagy machinery only after LC3-II has been incorporated into the mature phagophore, this may not preclude its role in the PINK1/Parkin pathway as p62/NBR1 also appears at the site of autophagosome formation early in the process: It colocalizes with the ULK1–Atg13–FIP200–Atg101 complex even in the absence of LC3 lipidation. Its subsequent interaction with LC3-II may promote efficient incorporation of the cargo (Itakura and Mizushima 2011).
CONCLUDING REMARKS
Studies published over the last six years have established a model for the PINK1/Parkin pathway of mitochondria quality control. This model is centered on three reproducible observations: (1) Parkin displays little activity toward mitochondrial proteins under basal conditions; (2) PINK1 accumulation on an impaired mitochondrion activates Parkin and constrains its activity to substrates on the impaired mitochondrion; and (3) Parkin ubiquitinates outer mitochondrial membrane proteins, most notably the mitofusins and Miro1, and induces the autophagic removal of impaired organelles. Whereas these findings are robust in cell culture in the presence of supraphysiological concentrations of Parkin, it has yet to be convincingly shown in vivo.
Given the strong genetic interaction between PINK1 and a Parkin in model systems and the similar disease caused by mutations in either PINK1 or Parkin in humans, the PINK1/Parkin mitochondrial quality control pathway is likely relevant to the pathogenesis of early onset parkinsonism.
We propose that this model may also provide a means of evaluating the relevance of novel PINK1 and Parkin substrates to the pathogenesis of early-onset parkinsonism. A zinc-finger protein renamed PARIS (parkin interacting substrate), for instance, was recently proposed to be a substrate of Parkin (Shin et al. 2011). As a putative repressor of PGC1-α, its degradation by Parkin was found to promote mitochondrial biogenesis. As with other proposed Parkin substrates, demonstrating that its degradation by Parkin depends on PINK1 would support its relevance to the pathogenesis of early-onset parkinsonism. Likewise, PINK1 has been proposed to physically or genetically interact with OMI/HTRA2 (Plun-Favreau et al. 2007), TRAP1 (Pridgeon et al. 2007), PGAM5 (Imai et al. 2010), and others. Although these interactions may reflect physiological functions of PINK1, parsimony would suggest that their relevance to the pathogenesis of early-onset parkinsonism may hinge on a genetic interaction with Parkin as well.
The unraveling of the PINK1/Parkin mitochondrial quality control pathway also holds great promise for the development of therapies aimed at sporadic Parkinson's disease as well as other disorders in which mitochondrial dysfunction drives pathogenesis. The ability of Parkin overexpression to clear mitochondria harboring deleterious mtDNA mutations shows the therapeutic potential of PINK1/Parkin upregulation for the treatment of mitochondrial disorders. The robust recruitment of fluorescent protein tagged Parkin to mitochondria on activation by PINK1 and the robust degradation of mitochondria following Parkin activation provide read-outs for high-throughput screens for activators and inhibitors of the PINK1/Parkin pathway. Whole genome siRNA and large library small compounds screens for PINK1/Parkin activators and inhibitors are currently underway by groups in government, academia, and industry. It is hoped that these efforts will uncover novel therapeutics for the treatment of Parkinson's disease as well as other mitochondrial disorders for which few disease-modifying therapies now exist.
ACKNOWLEDGMENTS
This review was supported by the intramural program at the NINDS (R.Y. and D.N.), the Medical Research Council (J.E.W. and D.N.), and NIH grant 1F30AG039185-01 (D.N.). We thank Drs. Ian Fearnley, Joe Carroll, and Shujing Ding for many helpful discussions and assistance in preparing proteomics data for Tables 1 and 2 and Figure 2 of this review.
Footnotes
Editors: Douglas C. Wallace and Richard J. Youle
Additional Perspectives on Mitochondria available at www.cshperspectives.org
REFERENCES
- Baloh RH, Salavaggione E, Milbrandt J, Pestronk A 2007. Familial parkinsonism and ophthalmoplegia from a mutation in the mitochondrial DNA helicase twinkle. Arch Neurol 64: 998–1000 [DOI] [PubMed] [Google Scholar]
- Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, et al. 2006. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet 38: 515–517 [DOI] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT 2000. Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat Neurosci 3: 1301–1306 [DOI] [PubMed] [Google Scholar]
- Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW 2011. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci 108: 9572–9577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC 2011. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet 20: 1726–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Gao F, Li B, Wang H, Xu Y, Zhu C, Wang G 2010a. Parkin mono-ubiquitinates Bcl-2 and regulates autophagy. J Biol Chem 285: 38214–38223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC 2010b. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 141: 280–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M 2006. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441: 1162–1166 [DOI] [PubMed] [Google Scholar]
- Cox RT, Spradling AC 2009. clueless, a conserved Drosophila gene required for mitochondrial subcellular localization, interacts genetically with parkin. Dis Model Mech 2: 490–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Matic I, Hilger M, Nagaraj N, Selbach M, Olsen JV, Mann M 2009. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc 4: 698–705 [DOI] [PubMed] [Google Scholar]
- Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SH, Renton AE, Harvey RJ, Whitworth AJ, Martins LM, et al. 2011. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet 20: 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M 2008. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci 105: 14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner N, Treske B, Paquet D, Holmstrom K, Schiesling C, Gispert S, Carballo-Carbajal I, Berg D, Hoepken HH, Gasser T, et al. 2007. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci 27: 12413–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Marusawa H, Wang HQ, Iwai A, Ikeuchi K, Imai Y, Kataoka A, Nukina N, Takahashi R, Chiba T 2008. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene 27: 6002–6011 [DOI] [PubMed] [Google Scholar]
- Gautier CA, Kitada T, Shen J 2008. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci 105: 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW 2010. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet 19: 4861–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W 2010a. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12: 119–131 [DOI] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Treis A, Skujat D, Weber SS, Fiesel FC, Kahle PJ, Springer W 2010b. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy 6: 871–878 [DOI] [PubMed] [Google Scholar]
- Gilkerson RW, De Vries RL, Lebot P, Wikstrom JD, Torgyekes E, Shirihai OS, Przedborski S, Schon EA 2012. Mitochondrial autophagy in cells with mtDNA mutations results from synergistic loss of transmembrane potential and mTORC1 inhibition. Hum Mol Genet 21: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ 2003. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci 100: 4078–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene AW, Grenier K, Aguileta MA, Muise S, Farazifard R, Haque ME, McBride HM, Park DS, Fon EA 2012. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep 13: 378–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE 2005. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron 47: 379–393 [DOI] [PubMed] [Google Scholar]
- Hales KG, Fuller MT 1997. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90: 121–129 [DOI] [PubMed] [Google Scholar]
- Hardy J 2010. Genetic analysis of pathways to Parkinson disease. Neuron 68: 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM 1998. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol 143: 359–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Kanao T, Sawada T, Kobayashi Y, Moriwaki Y, Ishida Y, Takeda K, Ichijo H, Lu B, Takahashi R 2010. The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet 6: e1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Mizushima N 2011. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol 192: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Koyama-Honda I, Mizushima N 2012. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J Cell Sci 125: 1488–1499 [DOI] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ 2010. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 191: 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Park J, Kim S, Song S, Kwon SK, Lee SH, Kitada T, Kim JM, Chung J 2008. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun 377: 975–980 [DOI] [PubMed] [Google Scholar]
- Kim KY, Stevens MV, Akter MH, Rusk SE, Huang RJ, Cohen A, Noguchi A, Springer D, Bocharov AV, Eggerman TL, et al. 2011a. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J Clin Invest 121: 3701–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. 2011b. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44: 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33: 505–516 [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392: 605–608 [DOI] [PubMed] [Google Scholar]
- Kitada T, Pisani A, Karouani M, Haburcak M, Martella G, Tscherter A, Platania P, Wu B, Pothos EN, Shen J 2009. Impaired dopamine release and synaptic plasticity in the striatum of parkin–/– mice. J Neurochem 110: 613–621 [DOI] [PubMed] [Google Scholar]
- Kornmann B, Osman C, Walter P 2011. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc Natl Acad Sci 108: 14151–14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston JW, Ballard P, Tetrud JW, Irwin I 1983. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science 219: 979–980 [DOI] [PubMed] [Google Scholar]
- Lazarou M, Jin SM, Kane LA, Youle RJ 2012. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell 22: 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP 2010. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol 189: 671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, Millan I, Shen A, Saxton W, Kanao T, et al. 2012. Parkinson's disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet 8: e1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma P, Melberg A, Rinne JO, Kaukonen JA, Nupponen NN, Chalmers RM, Oldfors A, Rautakorpi I, Peltonen L, Majamaa K, et al. 2004. Parkinsonism, premature menopause, and mitochondrial DNA polymerase γ mutations: Clinical and molecular genetic study. Lancet 364: 875–882 [DOI] [PubMed] [Google Scholar]
- Macaskill AF, Rinholm JE, Twelvetrees AE, Arancibia-Carcamo IL, Muir J, Fransson A, Aspenstrom P, Attwell D, Kittler JT 2009. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron 61: 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al. 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuibban GA, Saurya S, Freeman M 2003. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423: 537–541 [DOI] [PubMed] [Google Scholar]
- Meissner C, Lorenz H, Weihofen A, Selkoe DJ, Lemberg MK 2011. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem 117: 856–867 [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ 2010a. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6: 1090–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ 2010b. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, Kimura M, Sato S, Hattori N, Komatsu M, et al. 2010. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 15: 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olanow CW, Rascol O, Hauser R, Feigin PD, Jankovic J, Lang A, Langston W, Melamed E, Poewe W, Stocchi F, et al. 2009. A double-blind, delayed-start trial of rasagiline in Parkinson's disease. N Engl J Med 361: 1268–1278 [DOI] [PubMed] [Google Scholar]
- Osman C, Voelker DR, Langer T 2011. Making heads or tails of phospholipids in mitochondria. J Cell Biol 192: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134: 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J 2004. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem 279: 18614–18622 [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, et al. 2006. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Pimenta de Castro I, Costa AC, Lam D, Tufi R, Fedele V, Moisoi N, Dinsdale D, Deas E, Loh SH, Martins LM 2012. Genetic analysis of mitochondrial protein misfolding in Drosophila melanogaster. Cell Death Differ 19: 1308–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plun-Favreau H, Klupsch K, Moisoi N, Gandhi S, Kjaer S, Frith D, Harvey K, Deas E, Harvey RJ, McDonald N, et al. 2007. The mitochondrial protease HtrA2 is regulated by Parkinson's disease-associated kinase PINK1. Nat Cell Biol 9: 1243–1252 [DOI] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ 2008. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci 105: 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L 2010. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS ONE 5: e10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramstaller PP, Schlossmacher MG, Jacques TS, Scaravilli F, Eskelson C, Pepivani I, Hedrich K, Adel S, Gonzales-McNeal M, Hilker R, et al. 2005. Lewy body Parkinson's disease in a large pedigree with 77 Parkin mutation carriers. Ann Neurol 58: 411–422 [DOI] [PubMed] [Google Scholar]
- Pridgeon JW, Olzmann JA, Chin LS, Li L 2007. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol 5: e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovic A, Grunewald A, Kottwitz J, Bruggemann N, Pramstaller PP, Lohmann K, Klein C 2011. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS ONE 6: e16746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Brunner M, Neupert W, Westermann B 1998. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem 273: 20150–20155 [DOI] [PubMed] [Google Scholar]
- Reeve AK, Krishnan KJ, Elson JL, Morris CM, Bender A, Lightowlers RN, Turnbull DM 2008. Nature of mitochondrial DNA deletions in substantia nigra neurons. Am J Hum Genet 82: 228–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280: 1763–1766 [DOI] [PubMed] [Google Scholar]
- Ropper AH, Adams RD, Victor M, Samuels MA 2009. Adams and Victor’s principles of neurology. McGraw-Hill Medical, New York [Google Scholar]
- Saotome M, Safiulina D, Szabadkai G, Das S, Fransson A, Aspenstrom P, Rizzuto R, Hajnoczky G 2008. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc Natl Acad Sci 105: 20728–20733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH 2008. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet Neurol 7: 97–109 [DOI] [PubMed] [Google Scholar]
- Sha D, Chin LS, Li L 2010. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-κB signaling. Hum Mol Genet 19: 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, Lee JR, Grimes DA, Racacho L, Ye D, Yang H, Ross OA, Farrer M, McQuibban GA, Bulman DE 2011. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson's disease. Hum Mol Genet 20: 1966–1974 [DOI] [PubMed] [Google Scholar]
- Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM 2011. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson's disease. Cell 144: 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman JM, De Jager PL, Feany MB 2011. Parkinson's disease: Genetics and pathogenesis. Annu Rev Pathol 6: 193–222 [DOI] [PubMed] [Google Scholar]
- Silvestri L, Caputo V, Bellacchio E, Atorino L, Dallapiccola B, Valente EM, Casari G 2005. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet 14: 3477–3492 [DOI] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL 2002. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron 36: 1063–1077 [DOI] [PubMed] [Google Scholar]
- Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ 2010. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci 107: 11835–11840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ 2010. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol 191: 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KJ, McCoy MK, Blackinton J, Beilina A, van der Brug M, Sandebring A, Miller D, Maric D, Cedazo-Minguez A, Cookson MR 2011. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet 20: 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. 2004. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304: 1158–1160 [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. 2010. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci 107: 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Schwarz TL 2009. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell 136: 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL 2011. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147: 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen A, Thomas KJ, Ostaszewski BL, Cookson MR, Selkoe DJ 2009. Pink1 forms a multiprotein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking. Biochemistry 48: 2045–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth AJ, Theodore DA, Greene JC, Benes H, Wes PD, Pallanck LJ 2005. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson's disease. Proc Natl Acad Sci 102: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth AJ, Lee JR, Ho VM, Flick R, Chowdhury R, McQuibban GA 2008. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson's disease factors Pink1 and Parkin. Dis Model Mech 1: 168–174; discussion 173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winget JM, Mayor T 2010. The diversity of ubiquitin recognition: Hot spots and varied specificity. Mol Cell 38: 627–635 [DOI] [PubMed] [Google Scholar]
- Woodroof HI, Pogson JH, Begley M, Cantley LC, Deak M, Campbell DG, van Aalten DMF, Whitworth AJ, Alessi DR, Muqit MMK 2011. Discovery of catalytically active orthologues of the Parkinson's disease kinase PINK1: Analysis of substrate specificity and impact of mutations. Open Biol 1: 110012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Wang D, Chen L, Choo YS, Ma H, Tang C, Xia K, Jiang W, Ronai Z, Zhuang X, et al. 2009. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest 119: 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Paige JS, Jaffrey SR 2010. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol 28: 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Peng G, Wang Y, Fang S, Karbowski M 2011. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol Biol Cell 22: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B 2006. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci 103: 10793–10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B 2008. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci 105: 7070–7075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii SR, Kishi C, Ishihara N, Mizushima N 2011. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem 286: 19630–19640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, Dauer W, Schon EA, Przedborski S 2008. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci 105: 12022–12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ 2010. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci 107: 5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]