Abstract
During S phase, not only does DNA have to be replicated, but also newly synthesized DNA molecules have to be connected with each other. This sister chromatid cohesion is essential for the biorientation of chromosomes on the mitotic or meiotic spindle, and is thus an essential prerequisite for chromosome segregation. Cohesion is mediated by cohesin complexes that are thought to embrace sister chromatids as large rings. Cohesin binds to DNA dynamically before DNA replication and is converted into a stably DNA-bound form during replication. This conversion requires acetylation of cohesin, which in vertebrates leads to recruitment of sororin. Sororin antagonizes Wapl, a protein that is able to release cohesin from DNA, presumably by opening the cohesin ring. Inhibition of Wapl by sororin therefore “locks” cohesin rings on DNA and allows them to maintain cohesion for long periods of time in mammalian oocytes, possibly for months or even years.
Cohesin complexes may embrace sister chromatids as large rings that can be opened by Wapl. Sororin inhibits Wapl, locking the rings and maintaining cohesion for long periods of time (possibly years in mammalian oocytes).
DNA replication during the synthesis (S) phase generates identical DNA molecules, which, in their chromatinized form, are called sister chromatids. The pairs of sister chromatids remain united as part of one chromosome during the subsequent gap (G2) phase and during early mitosis, in prophase, prometaphase, and metaphase. During these stages of mitosis chromosomes condense, in most eukaryotes the nuclear envelope breaks down, and in all species chromosomes are ultimately attached to both poles of the mitotic spindle. Only once this biorientation has been achieved for all chromosomes, the sister chromatids are separated from each other in anaphase and transported toward opposite spindle poles of the mother cell, enabling its subsequent division into two genetically identical daughter cells.
This series of events critically depends on the fact that sister chromatids remain physically connected with each other from S phase until metaphase. This physical connection, called sister chromatid cohesion, opposes the pulling forces that are generated by microtubules that attach to kinetochores and thereby enables the biorientation of chromosomes on the mitotic spindle (Tanaka et al. 2000b). Without cohesion, sister chromatids could therefore not be segregated symmetrically between the forming daughter cells, resulting in aneuploidy. For the same reasons, cohesion is essential for chromosome segregation in meiosis I and meiosis II. Cohesion defects in human oocytes can lead to aneuploidy, which is thought to be the major cause of spontaneous abortion, because only a few types of aneuploidy are compatible with viability, such as trisomy 21 (Down syndrome), trisomy 18 (Edwards syndrome), and trisomy 13 (Patau syndrome) (Hunt and Hassold 2010). Studying the mechanisms of cohesion is therefore essential for understanding how the genome is passed properly from one cell generation to the next.
In addition, sister chromatid cohesion facilitates the repair of DNA double-strand breaks in cells that have replicated their DNA, where such breaks can be repaired by a homologous recombination mechanism that uses the undamaged sister chromatid as a template (for review, see Watrin et al. 2006). Furthermore, mutations in the proteins that are required for sister chromatid cohesion can cause defects in chromatin structure and gene regulation, and can in rare cases lead to congenital developmental disorders, called Cornelia de Lange syndrome, Roberts/SC Phocomelia syndrome, and Warsaw Breakage syndrome (for review, see Mannini et al. 2010).
SISTER CHROMATID COHESION IS MEDIATED BY COHESIN
Sister chromatid cohesion depends on a multi-subunit protein complex called cohesin (Fig. 1). Three of cohesin’s subunits—Smc1, Smc3, and Scc1/Rad21/Mcd1—form an unusual ring-shaped structure (Anderson et al. 2002; Haering et al. 2002). Smc1 and Smc3 are members of the “structural maintenance of chromosomes” (SMC) protein family. Members of this family are also found in other DNA-binding protein complexes, such as condensins, the Smc5/6 complex, and in bacterial SMC complexes. Like other SMC proteins, Smc1 and Smc3 are highly elongated molecules almost 50 nm in length that contain a central rod-shaped domain that is composed of anti-parallel coiled coils. Smc1 and Smc3 bind directly to each other at one of their ends via a “hinge” domain. At the other end of their coiled-coil domains, Smc1 and Smc3 form a nucleotide-binding domain (NBD). These NBDs form two ATP-binding sites, where one molecule of Mg2+-ATP is simultaneously bound to Walker A and Walker B motifs in the NBD of one SMC subunit and to an ABC signature motif in the other SMC subunit (Haering et al. 2004). The NBDs of Smc1 and Smc3 are connected by the kleisin (“bridging”) subunit Scc1, with Scc1’s amino terminus being bound to Smc3 and its carboxy-terminal Winged helix domain being bound to Smc1 (Haering et al. 2002). These interactions between Smc1, Smc3, and Scc1 lead to the formation of a tripartite ring with an inner diameter of 30–40 nm.
Figure 1.
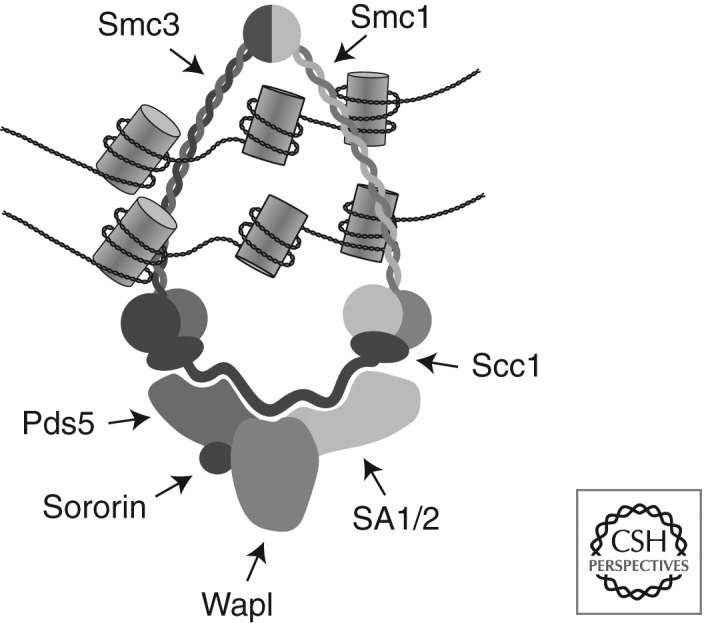
The cohesin complex. A schematic illustration of how cohesin subunits might be arranged in vertebrate cohesin complexes that mediate sister chromatid cohesion by embracing the two sister chromatid strands.
The integrity of this ring structure is essential for its ability to mediate sister chromatid cohesion (Gruber et al. 2003), and experimental observations using yeast mini-chromosomes indicate that cohesin mediates cohesion by topologically embracing the two sister chromatids (Ivanov and Nasmyth 2005; Haering et al. 2008). It has therefore been proposed that cohesin mediates cohesion by embracing both sister chromatids as a single ring (for review, see Nasmyth 2011). Other models propose that only a single DNA molecule can be embraced by a cohesin ring, and that two cohesin rings have to interact to connect two sister chromatids (Huang et al. 2005; Zhang et al. 2008b).
A fourth cohesin subunit, called Scc3 in yeast and stromal antigen (SA/STAG) in higher eukaryotes, binds directly to Scc1 (Haering et al. 2002). Scc3 is also essential for sister chromatid cohesion (Michaelis et al. 1997; Nishiyama et al. 2010). In vertebrate somatic cells, cohesin complexes contain either one of two different SA/STAG proteins, called SA1 and SA2 (Losada et al. 2000; Sumara et al. 2000). In most organisms, Scc1 and Scc3/SA interact with several other proteins called Wapl/Rad61, Pds5, and Sororin (Table 1). These proteins are believed to determine how stably the cohesin ring interacts with DNA, and if these interactions are able to mediate cohesion (see below).
Table 1.
Subunits and regulators of cohesin in yeasts, fruit flies, and humans
| Saccharomyces cerevisiae | Schizosaccharomyces pombe | Drosophila melanogaster | Homo sapiens | |||||
|---|---|---|---|---|---|---|---|---|
| Mitosis | Meiosisa | Mitosis | Meiosisa | Mitosis | Meiosisa | Mitosis | Meiosisa | |
| SMC | Smc1 | Psm1 | Smc1 | Smc1 | Smc1b | |||
| Smc3 | Psm3 | Smc3 | Smc3 | |||||
| Kleisin | Mcd1/Scc1 | Rec8 | Rad21 | Rec8 | Rad21 | C(2)M | Scc1 | Rec8,b Rad21L |
| Kleisin-associated | Scc3 | Psc3 | Rec11 | SA/Scc3 | SNM | SA1/STAG1 | SA3/STAG3 | |
| HEAT repeat protein | SA2/STAG2 | |||||||
| Releasin | Rad61/Wpl1 | Wpl1 | Wapl | Wapl | ||||
| Pds5 | Spo76/Pds5 | Pds5 | Pds5A, Pds5B | |||||
| Sororin | Dalmatian | Sororin | ||||||
| Adherin/kollerin | Scc2 | Mis4 | Nipped-B | NIPBL/Scc2 | ||||
| Cohesin loading | Scc4 | Ssl3 | MAU-2/Scc4 | |||||
| Cohesin positioning | CTCF | CTCF | BORIS/CTCFLc | |||||
| Cohesin cleavage | Esp1 | Cut1 | Separase | Separase | ||||
| Acetyltransferase | Eco1 | Eso1 | Deco | Esco1, Esco2 | ||||
| Deacetylase | Hos1 | HDAC8 | ||||||
aExpressed specifically in meiosis and largely/partially replace mitotic subunit.
bAlso expressed in B cells.
cParalog of CTCF specifically expressed in testis.
In many organisms, different kleisin subunits and isoforms of Smc1, Scc3/SA, and Pds5 exist, some of which are differentially expressed between somatic and meiotic cells. For example, in mammalian oocytes and spermatocytes, many cohesin complexes contain Smc1β instead of Smc1 (the latter is therefore also called Smc1α), and the kleisins Rec8 or Rad21L instead of Scc1. Table 1 provides an overview regarding the different isoforms of these and other cohesin subunits and their regulators that have been identified in different species.
LOADING OF COHESIN ONTO DNA DEPENDS ON ADHERIN, COHESIN'S ATPASE ACTIVITY, AND POSSIBLY OPENING OF THE COHESIN RING
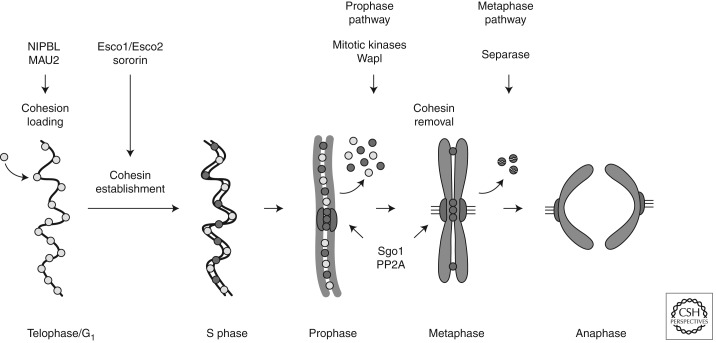
Cohesin is loaded onto DNA before DNA replication, in vertebrates already at the end of mitosis in telophase (Fig. 2). This process depends on cohesin’s ATPase activity (Arumugam et al. 2003; Weitzer et al. 2003) and on a separate complex, called adherin (Furuya et al. 1998) or kollerin (Nasmyth 2011). Because cohesin is thought to embrace DNA as a ring, it has furthermore been proposed that cohesin rings have to be opened transiently to allow entry of the DNA into the ring (Gruber et al. 2003).
Figure 2.
The cohesin cycle in vertebrate cells. A schematic illustration showing cohesin loading, cohesion establishment, and cohesin removal from chromosomes in telophase/G1 phase, S phase, and mitosis, respectively. (Small circles) Cohesin complexes; (light gray circles) cohesin complexes that are dynamically bound to DNA; (dark gray circles) “cohesive” complexes that are stably bound to DNA (Gerlich et al. 2006).
Adherin is a chromatin-bound protein complex composed of two subunits, a large protein (with a molecular mass of 316 kDa in humans), called Scc2 in budding yeast and NIPBL in mammals, and a smaller subunit, called Scc4 in yeast and MAU2 in mammals (Table 1). In Xenopus eggs, adherin is recruited to chromatin by binding to pre-replicative complexes (pre-RCs) (Gillespie and Hirano 2004; Takahashi et al. 2004), whereas in yeast, adherin is particularly enriched at centromeres but is also found on chromosome arms and near telomeres (Lengronne et al. 2004). Both pre-RC components in Xenopus and centromere-associated kinetochore proteins in yeast are also required for efficient cohesin loading (for review, see Peters et al. 2008). At least in the case of the yeast kinetochore, its role in cohesin loading might go beyond the recruitment of adherin to chromatin, because tethering Scc2 to a chromosomal locus where it is normally not present is not sufficient for recruiting cohesin to such a site (Hu et al. 2011).
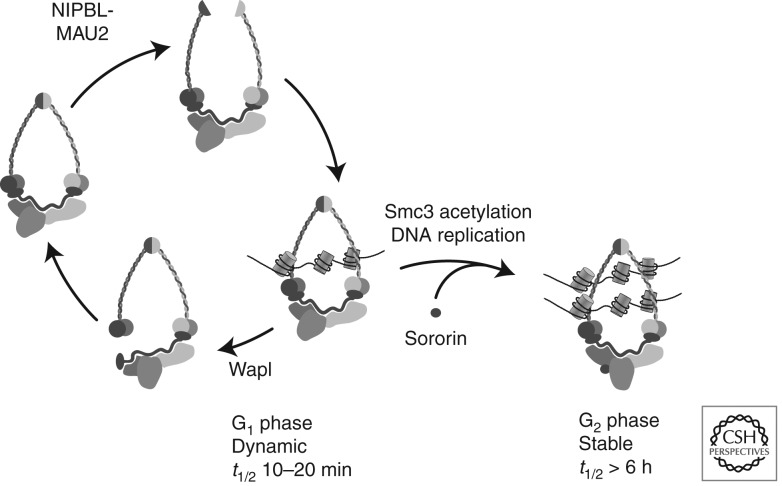
Both subunits of adherin are composed of repetitive sequence elements, and no enzymatic functions are known for them. Because loading of cohesin onto DNA depends on both adherin and on cohesin’s ATPase activity, and because the loading reaction is thought to require opening of the cohesin ring, adherin could be an allosteric regulator of cohesin’s ATPase activity, and it is conceivable that this activity would facilitate ring opening. If this were true, how might ring opening be achieved, and how might ATP hydrolysis mediate this process? Because mutation of Smc1’s and Smc3’s hinge domains, or artificially tethering these domains to each other, blocks the loading of cohesin onto DNA, it has been proposed that Smc1’s and Smc3’s hinge domains have to be separated from each other to create an “entry gate” for DNA (Fig. 3) (Gruber et al. 2006; Mishra et al. 2010; Kurze et al. 2011; Nasmyth 2011). Studies of a cohesin-related bacterial SMC complex support this view because the hinge domain of this complex is also required for DNA binding (Hirano and Hirano 2006).
Figure 3.
Dynamic and stable binding modes of cohesin to DNA. A schematic drawing illustrating how adherin-mediated separation of the hinge domains of Smc1 and Smc3 might create an entry gate for DNA into the cohesin ring, whereas a transient Wapl-mediated dissociation of Scc1 from the SMC subunits may release cohesin from DNA by generating an exit gate (Nasmyth 2011; Chan et al. 2012). Wapl’s ability to open this exit gate might be inhibited by sororin, which binds to cohesin in vertebrates following Smc3 acetylation and possibly other unidentified events during DNA replication (Nishiyama et al. 2010).
Ideas for how this entry gate might be opened have come from structural and biochemical analyses of proteins whose NBDs are related in structure to the NBDs of SMC proteins. Such NBDs are found in ATP-binding cassette (ABC) transporters and in Rad50 (Hopfner and Tainer 2003). ABC transporters are homodimeric or heterodimeric ATPases that are using cycles of ATP binding and hydrolysis to transport small molecules across membranes, whereas Rad50 is a homodimeric protein required for DNA repair that contains both NBDs and coiled-coil domains that are similar to the ones of SMC proteins. In these proteins, ATP binding leads to a transient physical interaction between the two NBDs (Hopfner et al. 2000), and in the case of ABC transporters, this “engagement” step facilitates transport of substrate molecules through the transmembrane channel (for review, see Rees et al. 2009). ATP hydrolysis reverts these conformational changes by leading to disengagement of the NBDs. Similarly to how one cycle of ATP binding and hydrolysis leads to transient opening of the transmembrane channel in ABC transporters, it is therefore possible that ATP binding and hydrolysis would lead to transient opening of an entry gate for DNA in the cohesin ring.
But how could binding of ATP to the NBDs of Smc1 and Smc3 lead to separation of the hinge domains of Smc1 and Smc3, which are almost 50 nm away from the NBDs? Atomic force microscopy and crystallographic studies have revealed that ATP binding to the signature motif of Rad50 is not only accompanied by NBD engagement but also induces a dramatic conformational change that alters the angle at which the coiled-coil domain of Rad50 emanates from the NBD (Moreno-Herrero et al. 2005; Lammens and Hopfner 2010; Lim et al. 2011; Williams et al. 2011). This conformational change is mediated by helices in Rad50, called the signature-coupling helices, which move relative to each other and thereby function as a lever that exerts force on the coiled-coil domain. It is therefore conceivable that ATP binding and hydrolysis also induce changes in the coiled-coil domains of cohesin, and thereby could affect interactions between Smc1’s and Smc3’s dimerization domains over relatively long distances.
Further support for the hypothesis that cohesin loading onto DNA is controlled by ATP hydrolysis comes from the recent identification of a cohesin mutant in the Walker B motif of Smc1 and Smc3 that might be locked in a transition state of the loading reaction (Hu et al. 2011). This mutant binds to DNA in an Scc2-dependent manner, in particular at centromeres, but is unable to associate with DNA stably and to mediate cohesion. Based on analogy with ABC transporters, it has been speculated that this mutant might be locked in a state where the NBDs are engaged but the hinge domains are physically separated, which would explain why these mutants can interact with DNA only transiently.
COHESIN IS POSITIONED AT SPECIFIC SITES IN THE GENOME
Chromatin immunoprecipitation (ChIP) experiments performed in several different organisms have revealed that cohesin can predominantly be detected at specific sites in the genome, indicating that cohesin is not distributed randomly along the length of chromosomes (for review, see Peters et al. 2008). For example, in budding yeast and fission yeast, large amounts of cohesin are found at centromeres and peri-centric regions, and smaller amounts are found in discrete cohesin attachment regions (CARs) along chromosome arms, which on average are 10 kb apart from each other. Because adherin is essential for the ability of cohesin to associate with DNA, it is generally assumed that cohesin loading occurs at adherin-binding sites. However, ChIP experiments in budding yeast and fission yeast indicate that most adherin complexes are bound to different regions of the genome from cohesin, with the exception of centromeres where both complexes are abundant (Lengronne et al. 2004; Schmidt et al. 2009; Hu et al. 2011; for a conflicting view, see Kogut et al. 2009). It has therefore been proposed that cohesin is first loaded onto DNA at adherin sites and that it is subsequently translocated from these sites to peri-centric regions or CARs (Lengronne et al. 2004). Consistent with this notion, the Walker B mutant of cohesin, which is thought to be locked in a transition state of the loading reaction, accumulates at yeast centromeres but to a much lesser degree than wild-type cohesin at peri-centric regions and CARs, as if cohesin were normally loaded predominantly at centromeres and subsequently moved from there to chromosomal arm regions (Hu et al. 2011).
How cohesin is moved from one site to the other remains unknown, but many CARs in budding and fission yeast are found in regions of convergent transcription, and it has been shown that the induction of regulatable genes leads to removal of cohesin from an intragenic position to the end of the transcription unit (Lengronne et al. 2004). This finding and biochemical experiments that suggest that cohesin rings are able to “slide off” short linearized yeast mini-chromosomes in vitro (Ivanov and Nasmyth 2005) have led to the proposal that cohesin is able to move laterally along DNA while encircling it. According to this idea, RNA polymerases or the native transcripts and RNPs produced by them would simply push cohesin out of actively transcribed regions. If this is true, the pattern of cohesin distribution that is seen along yeast chromosome arms could largely be an indirect consequence of gene activity, rather than a pattern that is specified because cohesin could only fulfill its function in particular chromosomal locations. An exception to this rule could be the centromere, where a high number of cohesin complexes are thought to resist the pulling forces that are generated by the mitotic spindle.
Because cohesin mediates sister chromatid cohesion from yeast to men, the principle mechanisms of how cohesin associates with DNA might have evolved early during the evolution of eukaryotes, or possibly even before, because DNA-binding SMC complexes related to cohesin exist in all kingdoms of life. Although this may well be the case for the initial loading process in which the cohesin rings entrap DNA, it is less clear if the subsequent positioning of cohesin in the genome occurs according to the same principles in different organisms. In contrast to the situation in yeasts, where cohesin and adherin can be found in different chromosomal locations, the two complexes largely colocalize in the Drosophila genome, where in many cases they occupy the entire length of actively transcribed genes (Misulovin et al. 2008). Yet a different situation is found in mammalian cells, where NIPBL is frequently located at the promoters of active genes, often together with mediator, a large transcriptional coactivator complex, and cohesin (Kagey et al. 2010). However, although some cohesin can be detected at the majority of NIPBL sites (Kagey et al. 2010), the opposite is not true. There are many more cohesin-binding sites detectable in ChIP experiments (estimates range up to 55,000 sites for the nonrepetitive part of mammalian genomes) (Schmidt et al. 2010) than NIPBL sites, implying that also in mammals, cohesin loading may occur at sites that are distinct from the sites where cohesin finally accumulates. But unlike in yeasts, there is presently no evidence that cohesin is moved to these positions by the transcriptional machinery.
Instead, there are numerous observations that indicate that the pattern of cohesin distribution in mammalian genomes is specified by DNA sequence elements that are recognized by CCCTC binding factor (CTCF), a protein that binds to DNA via one or several of its 11 zinc finger motifs. CTCF is found at the majority, but possibly not at all cohesin-binding sites (for review, see Merkenschlager 2010). Interestingly, partial depletion of CTCF or mutation of CTCF-binding sites reduces cohesin binding at these sites, without detectably reducing the total amount of cohesin on DNA (Parelho et al. 2008; Stedman et al. 2008; Wendt et al. 2008). The implication is that CTCF is dispensable for loading cohesin onto DNA but required for a subsequent positioning step. This situation is particularly evident at the H19-Igf2 locus. In this chromosomal location, the maternal and paternal alleles are differentially methylated, leading to different gene expression patterns on the two alleles within one and the same cell. This “imprinted” gene expression pattern depends on CTCF, which can bind to an imprinting control region (ICR) on the maternal allele but not on the paternal allele, where cytosine methylation prevents CTCF binding to the ICR. Interestingly, cohesin binds only to the ICR on the maternal allele, where CTCF is present, but not on the paternal allele, where CTCF is not bound, further supporting the hypothesis that CTCF specifies where cohesin accumulates once it has been loaded onto DNA (Rubio et al. 2008; Stedman et al. 2008; Wendt et al. 2008; Nativio et al. 2009).
Recent experiments have shown that the carboxy-terminal region of CTCF can directly bind to the SA2 subunit of mammalian cohesin (Xiao et al. 2011), suggesting that cohesin may be recruited to CTCF-binding sites through direct protein–protein interactions. But how cohesin moves from its presumed loading sites where NIPBL is present to CTCF sites remains unknown. Either cohesin can somehow be translocated along DNA, or loading would normally have to occur directly at CTCF sites. In the latter case, one would have to postulate that NIPBL is present, at least transiently, at many more CTCF sites than could be detected in ChIP experiments so far, or NIPBL might be able to promote cohesin loading in cis. In this case, an NIPBL site would have to be able to interact with multiple CTCF sites, possibly in a dynamic fashion that depends on chromatin folding.
RECRUITMENT OF COHESIN TO CTCF SITES MAY DETERMINE CHROMATIN STRUCTURE
There is presently no evidence that cohesin has to accumulate at CTCF sites to be able to mediate sister chromatid cohesion, because no cohesion defects have been observed after depletion of CTCF, despite the fact that less cohesin is detected at CTCF sites under these conditions (Wendt et al. 2008). Furthermore, cohesin does not colocalize with CTCF on Drosophila chromosomes, and there is no evidence that a CTCF-related protein colocalizes with cohesin in the genome of budding yeast or fission yeast, further indicating that cohesin’s role in cohesion is independent of CTCF. However, cohesin depletion does cause defects in gene expression (for review, see Dorsett and Ström 2012), and in mammalian cells, some of these defects resemble the defects that are seen after depletion of CTCF. For example, cohesin depletion changes the levels of H19 and Igf2 transcripts in a similar way as depletion of CTCF, implying that both proteins are required for imprinted gene expression at the H19-Igf2 locus (Wendt et al. 2008; Nativio et al. 2009). Because CTCF is required for recruitment of cohesin to the ICR, whereas the opposite is not true, CTCF appears not to be sufficient for imprinted gene expression. Instead, the main function of CTCF in this process might be to recruit cohesin, and it could therefore be cohesin that affects transcription more directly than CTCF. If this were true, then cohesin positioning in mammalian cells might not be an indirect consequence of gene expression, as has been proposed to be the case in yeast, but may on the contrary control patterns of gene expression.
It is believed that cohesin controls gene expression by mediating long-range chromatin interactions, for example, by forming physical contacts between different CTCF sites that could lead to the formation of chromatin loops. This hypothesis is based on the notion that activity of gene expression depends on promoter–enhancer interactions, and that the proximity between promoters and enhancers might be controlled topologically through the formation of chromatin loops or more complex higher-order chromatin structures. For example, CTCF is thought to function as a transcriptional insulator on the maternal allele of the H19-Igf2 locus by inducing the formation of a chromatin loop, which topologically separates the H19 enhancer from the promoters of the Igf2 gene. Given that cohesin is able to mediate sister chromatid cohesion, that is, it can physically connect two DNA molecules in trans, it is conceivable that cohesin also tethers different sequence elements (CTCF sites) on the same chromosome in cis, leading to the formation of intrachromatid loops. Support for this hypothesis has come from “chromatin conformation capture” (3C) experiments (for review, see Seitan et al. 2012), but how generally important cohesin and CTCF are for regulating chromatin structure remains to be determined.
SISTER CHROMATID COHESION IS ESTABLISHED DURING DNA REPLICATION AND COINCIDES WITH STABILIZATION OF COHESIN ON DNA
Cohesin can bind to DNA both before and after DNA replication, but it can establish sister chromatid cohesion normally only during S phase (Uhlmann and Nasmyth 1998). This and other observations indicate that cohesion establishment is coupled to DNA replication. It is attractive to think that cohesion establishment must occur at or in the direct vicinity of replication forks, because such a scenario could explain how cohesion is only established between sister chromatids, and not between non-sister DNA molecules. Consistent with this possibility, numerous replication fork components are required for cohesion (Table 2), and Eco1/Ctf7, an acetyltransferase required for establishment of sister chromatid cohesion (Skibbens et al. 1999; Toth et al. 1999), is thought to travel along DNA with replication forks (Lengronne et al. 2004). Even though cohesion may normally only be established at replication forks, recent experiments in budding yeast have revealed that cohesion can also be established de novo after DNA replication if DNA has been damaged by double-strand breaks (see below). In addition, in this case, cohesion establishment depends on Eco1. However, in the absence of Eco1 and either DNA replication or DNA damage, the association of cohesin with DNA is not sufficient to establish sister chromatid cohesion. The implication is that it is not the binding of cohesin to DNA per se but an Eco1-dependent process and possibly other DNA replication or DNA damage-dependent events that make cohesin “cohesive.” An interesting hint as to what may determine the “cohesiveness” of cohesin has come from fluorescence recovery after photobleaching (FRAP) experiments in mammalian cells. These experiments revealed that cohesin binds to DNA dynamically in G1 phase with a residence time of several minutes, whereas many cohesin complexes bind to DNA much more stably after DNA replication, with a residence time of at least many hours (Gerlich et al. 2006). Thus, the ability of cohesin to mediate cohesion coincides with stabilization of cohesin on DNA, implying that stable binding of cohesin to DNA may be an inherent property of cohesive cohesin, and that this property may be required to maintain cohesion from S phase until the subsequent metaphase. Cohesin’s ability to bind to DNA for long periods of time may be particularly important in mammalian oocytes. In these cells cohesion is established during pre-meiotic S phase, which occurs before birth, and then has to be maintained until completion of meiosis, which only occurs during ovulation many months or years later (Hunt and Hassold 2010).
Table 2.
DNA replication proteins required for sister chromatid cohesion
| S. cerevisiae | S. pombe | Vertebrates | Function | References | |
|---|---|---|---|---|---|
| RfcCtf18 | Ctf18 | Ctf18 | Ctf18 | PCNA clamp loader | Hanna et al. 2001; Mayer et al. 2001; Ansbach et al. 2008; Terret et al. 2009 |
| Ctf8 | Ctf8 | Ctf8 | |||
| Dcc1 | Dcc1 | Dcc1 | |||
| RfcElg1 | Elg1 | Elg1 | Elg1 | PCNA clamp loader | Maradeo et al. 2009 |
| Chl1/ChlR1/Ddx11 | Chl1 | Chl1 | ChlR1/Ddx11 | DNA helicase | Mayer et al. 2004; Petronczki et al. 2004; Parish et al. 2006; Ansbach et al. 2008 |
| Ctf4/And1 | Ctf4 | Mcl1 | And1 | Recruitment of Polα to chromatin | Hanna et al. 2001; Williams et al. 2002; Mayer et al. 2004; Petronczki et al. 2004; Errico et al. 2009 |
| Tof1/Timeless/Tim | Tof1 | Swi1 | Tim1 | Tim1–Tipin complex stabilizes replication fork | Mercier et al. 2001; Mayer et al. 2004; Errico et al. 2009 |
| Csm3/Tipin | Csm3 | Swi3 | Tipin | Mayer et al. 2004; Ansbach et al. 2008; Errico et al. 2009 | |
| Mrc1/Claspin | Mrc1 | Mrc1 | Claspin | S-phase checkpoint mediator | Xu et al. 2004 |
| Rad27/Rad2/Fen1 | Rad27 | Rad2 | Fen1 | Maturation of Okazaki fragment | Farina et al. 2008 |
COHESIN ACETYLATION IS ESSENTIAL FOR SISTER CHROMATID COHESION
Eco1’s essential role in sister chromatid cohesion is the acetylation of cohesin’s Smc3 subunit on two lysine residues, K112 and K113 in budding yeast, and K105 and K106 in human Smc3 (Ben-Shahar et al. 2008; Unal et al. 2008; Zhang et al. 2008a; Rowland et al. 2009). Both residues are well conserved in many eukaryotes, as are orthologs of Eco1, of which there are two different ones in vertebrates, called Esco1 and Esco2 (Table 1) (Hou and Zou 2005). Eco1 orthologs are also required for sister chromatid cohesion in fission yeast (Tanaka et al. 2000a), Drosophila (Williams et al. 2003), and human cells (Hou and Zou 2005), and in the latter case, both enzymes contribute to Smc3 acetylation (Zhang et al. 2008a; Nishiyama et al. 2010). The acetylation of Smc3 can be reverted by specific enzymes. In human cells, the major enzyme responsible for Smc3 deacetylation is HDAC8 (Deardorff et al. 2012). In budding yeast, the deacetylase Hos1 is required for this process (Beckouet et al. 2010; Borges et al. 2010; Xiong et al. 2010). Hos1 deletion does not cause cohesion defects; however, it becomes crucial for cohesion either when Eco1 is inactivated (Beckouet et al. 2010) or when de novo Smc3 synthesis is suppressed (Borges et al. 2010). These observations raise the interesting possibility that cohesin may have to be acetylated during the process of cohesion establishment, whereas it may be impossible to convert previously acetylated cohesin into a cohesive state.
Although the importance of Smc3 acetylation for sister chromatid cohesion is well established, it is less clear how this modification enables cohesin to become cohesive. The acetylated lysine residues in Smc3 are predicted to be located close to the ATP-binding pocket in the NBD domain. Interestingly, mutations that are thought to mimic the acetylated state of Smc3 abolish a dominant-negative effect that Walker B mutations in Smc3 have on wild-type cohesin (Heidinger-Pauli et al. 2010). Because Smc3 mutations that prevent ATP binding also suppress this dominant effect, it has been proposed that Smc3 acetylation regulates the ATP-binding and hydrolysis cycle of cohesin, either by reducing ATP binding to cohesin or by preventing ATP-dependent interactions between cohesin and its loading machinery. Furthermore, cohesin is normally only acetylated when bound to chromatin. Together with the genetic interactions between acetyl-mimicking and Walker B mutations, this observation raises the possibility that Smc3 acetylation on chromatin prevents rebinding of ATP to cohesin that has entrapped DNA. According to this hypothesis, Smc3 acetylation could stabilize cohesin on DNA by blocking cycles of ATP binding and hydrolysis.
An alternative explanation for how acetylation may contribute to cohesion establishment has come from the observation that the essential function of Eco1 becomes dispensable in fission yeast and budding yeast if the cohesin-associated proteins Wapl and Pds5 are inactivated, or if specific residues in the cohesin core subunits Smc3 and Scc3 are mutated (Tanaka et al. 2001; Ben-Shahar et al. 2008; Unal et al. 2008; Zhang et al. 2008a; Rowland et al. 2009; Sutani et al. 2009). Because Wapl promotes the release of cohesin from DNA in vertebrates (see below), it has been proposed that Smc3 acetylation by Eco1 might stabilize cohesin on DNA by inactivating Wapl (Ben-Shahar et al. 2008; Unal et al. 2008). Alternatively, it has been argued that Wapl, Pds5, Smc3, and Scc3 are required for an activity that prevents cohesion establishment, and that Smc3 acetylation does not prevent the Wapl-mediated release of cohesin from DNA but instead overcomes this “anti-establishment” activity (Rowland et al. 2009). In vertebrates, the existing evidence supports the idea that Smc3 acetylation mediates cohesion by preventing dissociation of cohesin from DNA because in these species, cohesin acetylation leads to the recruitment of sororin, a protein that prevents release of cohesin from DNA (see below).
SORORIN STABILIZES COHESIN–DNA INTERACTIONS AND MEDIATES SISTER CHROMATID COHESION BY ANTAGONIZING WAPL
Sororin is a protein that has only been identified in metazoans so far, where it is essential for sister chromatid cohesion (Rankin et al. 2005) and for stabilizing cohesin on chromatin during DNA replication (Schmitz et al. 2007). Sororin only binds to cohesin once Smc3 has been acetylated. However, this modification is not sufficient for sororin recruitment because DNA replication is also necessary for this event (Lafont et al. 2010; Nishiyama et al. 2010). Remarkably, sororin’s essential function in sister chromatid cohesion becomes dispensable when Wapl is depleted from cells or Xenopus egg extracts, indicating that sororin might mediate cohesion by inhibiting Wapl (Nishiyama et al. 2010). This hypothesis could explain how sororin stabilizes cohesin on DNA (Schmitz et al. 2007), because the release of cohesin from DNA depends on Wapl (see below).
In vitro, sororin can displace Wapl from its binding partner Pds5, but there is no evidence that sororin can fully dissociate Wapl from cohesin (Nishiyama et al. 2010). It has therefore been proposed that sororin changes the topology of how Wapl and Pds5 interact with cohesin, and that these conformational changes inhibit Wapl’s ability to release cohesin from DNA. As a result, cohesin may become stably “locked” on DNA and may thus be able to mediate sister chromatid cohesion for prolonged periods of time (Fig. 3).
It remains unknown if cohesion depends on sororin in all species because related proteins have been identified in some but not all eukaryotes. Sororin exists in vertebrates and Drosophila, where it is called Dalmatian, and related proteins containing a conserved carboxy-terminal “sororin domain” have been identified in many other metazoans, including cephalochordates, echinoderms, cnidaria, placozoa, and in plants, but not yet in yeasts and worms (Nishiyama et al. 2010). It therefore remains to be understood if Smc3 acetylation regulates cohesin also in yeast and worms by recruiting a sororin-related protein or by a different mechanism.
DNA DOUBLE-STRAND BREAKS ALSO INDUCE COHESION ESTABLISHMENT
Although cohesion is normally established during DNA replication, DNA double-strand breaks also trigger de novo cohesion establishment even in postreplicative budding yeast cells in an Eco1-dependent manner (Strom et al. 2007; Unal et al. 2007). In this case, it is thought that acetylation of Scc1 by Eco1, but not of Smc3, is required for cohesion establishment (Heidinger-Pauli et al. 2009). This cohesion can be established independently of DNA replication and is induced by double-strand breaks not only on the damaged but also on undamaged chromosomes that are present in the same cells. These observations imply that a diffusible cohesion establishment factor, which is inactivated after DNA replication, is somehow reactivated in response to DNA damage. This factor is presumably Eco1 itself because Eco1 is normally degraded after S phase, but stabilized following DNA damage, and because overexpression of Eco1 is sufficient to induce cohesion establishment after DNA replication (Lyons and Morgan 2011).
COHESIN IS REMOVED FROM MITOTIC CHROMOSOMES IN TWO STEPS
Because cohesin physically connects sister chromatids, sister chromatids can only be separated from each other and transported toward opposite spindle poles in anaphase if cohesin is removed from chromosomes and cohesion is dissolved. This can be achieved by either of two mechanisms. In vertebrates, and presumably in most other metazoans, most cohesin complexes are removed from chromosome arms already in prophase and prometaphase, whereas cohesin at centromeres is protected from this “prophase pathway” and maintains cohesion until anaphase onset (Waizenegger et al. 2000). It is this differential removal of cohesin from chromosome arms and centromeres, respectively, that causes the characteristic “X-shape” (or “V-shape” in species where centromeres are close to one of the telomeres) of chromosomes in prometaphase and metaphase (Gimenez-Abian et al. 2004).
WAPL RELEASES COHESIN FROM DNA
The dissociation of cohesin from chromosomes in prophase and prometaphase depends on Wapl (Gandhi et al. 2006; Kueng et al. 2006) and on phosphorylation of cohesin’s SA2 subunit (Hauf et al. 2005) by Polo-like kinase (Plk1) and possibly other mitotic kinases, such as cyclin-dependent kinase 1 (Cdk1) and Aurora B (Losada et al. 2002; Sumara et al. 2002; Hegemann et al. 2011). This pathway of removing cohesin from mitotic chromosomes might use the same mechanism by which cohesin is released from DNA in interphase because depletion of Wapl also increases the residence time and the steady-state levels of cohesin on DNA in interphase (Kueng et al. 2006). In mitosis, this release mechanism might become activated by mitotic kinases, either by phosphorylation of SA2, which is located next to Wapl in the cohesin complex (Gandhi et al. 2006; Kueng et al. 2006), or by phosphorylation of Wapl and Pds5, which are also phosphorylated in mitosis in a Plk1-dependent manner (Hegemann et al. 2011). Because sororin antagonizes Wapl, sororin also has to be inactivated in mitosis to enable Wapl to release cohesin from DNA. In addition, sororin inactivation is mediated by phosphorylation (Nishiyama et al. 2010), which has been reported to depend on Cdk1 (Dreier et al. 2011) or Plk1 (Zhang et al. 2011). At the end of mitosis, sororin is further degraded following ubiquitylation by the ubiquitin ligase APC/CCdh1, possibly to prevent precocious formation of cohesin complexes that become too stably bound to DNA (Rankin et al. 2005). Activation of the prophase pathway of cohesin dissociation may therefore depend on mitotic phosphorylation of several proteins: sororin, SA2 (and presumably also SA1), and possibly Wapl and Pds5. In addition, adherin is also removed from chromosomes in prophase (Gillespie and Hirano 2004; Watrin et al. 2006), implying that cohesin cannot be loaded onto DNA any longer in early mitosis. Together, these events may shift the equilibrium between soluble and DNA-bound cohesin complexes so that during prophase and prometaphase most cohesin is removed from chromosome arms.
Although the majority of cohesin complexes are removed from mitotic chromosomes in vertebrate cells by Wapl, some cohesin complexes can only be removed by a different mechanism that depends on the protease separase, which cleaves the Scc1 subunit and thereby opens the cohesin ring (see below). Unlike separase, Wapl releases cohesin from DNA presumably by enabling opening of the cohesin ring without cleaving one of its subunits (Waizenegger et al. 2000; Kueng et al. 2006; Peters et al. 2008). It has been proposed that the Wapl-mediated opening of the cohesin ring occurs via an “exit gate” for DNA that is different from the proposed “entry gate” (Fig. 3) (Nasmyth 2011; Chan et al. 2012). Whereas the latter is thought to be created by separation of the hinge domains of Smc1 and Smc3 (see above), the former has been proposed to require dissociation of Scc1 from the SMC subunits (Nasmyth 2011). There is some precedence that such a conformational change could occur from structural studies of MukB, an SMC-related protein required for chromosome segregation in bacteria (Woo et al. 2009). MukB forms a homodimer whose NBDs interact with two carboxy-terminal Winged helix domains of the MukF protein, similarly to how Scc1’s carboxy-terminal Winged helix binds to Smc1 (Haering et al. 2002). Interestingly, ATP-mediated engagement of the MukB’s NBDs results in detachment of one of MukF’s Winged helix domains from MukB (Woo et al. 2009), implying that conformational changes could also lead to transient dissociation of Scc1 from cohesin’s SMC subunits.
POSSIBLE COEVOLUTION OF COHESIN ACETYLATION AND WAPL-MEDIATED COHESIN RELEASE FROM DNA
In budding yeast, it has been reported that a Plk1-dependent mechanism removes cohesin from meiotic chromosomes (Yu and Koshland 2005), but it is unknown if this pathway depends on Wapl, and in mitotic yeast cells, most if not all cohesin is cleaved by separase (Uhlmann et al. 1999). It is therefore unclear if there is a prophase pathway of cohesin dissociation in budding yeast that is related to the one in vertebrates. However, because Wapl antagonizes the function of Eco1 (see above), Wapl may also be required for releasing cohesin from DNA in budding yeast. Direct support for this notion has come from studies in fission yeast, where, as in vertebrates, Wapl is required for dissociation of cohesin from DNA (Feytout et al. 2011).
Nasmyth and Schleiffer (2004) pointed out that the genome of the simple eukaryote Encephalitozoon cuniculi lacks a gene that encodes an Eco1-related enzyme. We found 19 other species in whose genome we could not identify an Eco1-related gene (Table 3). Interestingly, in seven of these 20 species without a detectable Eco1 gene, at least one of the two lysine residues that are acetylated by Eco1 in yeast is replaced by another amino acid residue or missing. In contrast, we found only a single species (Trypanosoma brucei) with an Eco1 gene in which the KK motif in Smc3 was not conserved. More interestingly, of the seven species in whose genomes we could not identify an Eco1 gene but could find a mutated or missing KK motif in Smc3, six species have genes encoding either Pds5 or Wapl orthologs. The presence of Pds5 and Wapl therefore coincides in many cases either with the presence of Eco1, or with mutations in the lysine residues that are acetylated by Eco1 in other species. These observations raise the interesting possibility that the ability of Pds5 and/or Wapl to release cohesin from DNA may have coevolved with Smc3 acetylation by Eco1.
Table 3.
Coevolution of Smc3 acetylation by Eco1 and cohesin release by Wapl
| Organism | Smc3 | Pds5 | Wapl | Eco1 | |
|---|---|---|---|---|---|
| Species with an Eco1 ortholog | Homo sapiens | KK | + | + | + |
| Ciona intestinalis | KK | + | + | + | |
| Xenopus laevis | KK | + | + | + | |
| Arabidopsis thaliana | KK | + | + | + | |
| Saccharomyces cerevisiae | KK | + | + | + | |
| Schizosaccharomyces pombe | KK | + | + | + | |
| Drosophila melanogaster | KK | + | + | + | |
| Trypanosoma brucei TREU927 | KQ | + | + | ||
| Species without an identifiable Eco1 ortholog | Acromyrmex echinatior | KK | + | + | |
| Dictyostelium fasciculatum | NK | + | + | ||
| Gibberella zeae PH-1 | KK | + | + | ||
| Glomerella graminicola M1.001 | KK | + | + | ||
| Grosmannia clavigera kw1407 | RK | + | + | ||
| Moniliophthora perniciosa FA553 | KK | + | + | ||
| Monosiga brevicollis MX1 | HK | + | + | ||
| Trichinella spiralis | KK | + | + | ||
| Tuber melanosporum | KK | + | + | ||
| Ustilago maydis 521 | KK | + | + | ||
| Chlamydomonas reinhardtii | KK | + | |||
| Hydra magnipapillata | KK | + | |||
| Malassezia globosa CBS 7966 | TR | + | |||
| Metarhizium acridum CQMa 102 | KK | + | |||
| Trichomonas vaginalis | RK | + | |||
| Volvox carteri f. nagariensis | − | + | |||
| Cryptosporidium parvum | QK | ||||
| Aureococcus anophagefferens | KK | ||||
| Encephalitozoon cuniculi GB-M1 | KK | ||||
| Schistosoma japonicum | KK |
(KK) Lysine residues that correspond to the ones that have been shown to be acetylated by Eco1 in S. cerevisiae Smc3. In the sequence of Volvox carteri f. nagariensis Smc3, the region containing these lysine residues is missing.
Gene or protein sequences are available upon request.
SEPARASE CLEAVES COHESIN IN METAPHASE AND ENABLES SISTER CHROMATID SEPARATION IN ANAPHASE
The Wapl-dependent prophase pathway of cohesin dissociation removes most cohesin from chromosomes arms but leaves cohesin behind at centromeres, where cohesion is maintained until the onset of anaphase (Waizenegger et al. 2000). These centromeric cohesin complexes are protected from the prophase pathway by a protein called shugoshin, or Sgo1, which recruits the protein phosphatase 2A complex (PP2A) to centromeres (for review, see Gutiérrez-Caballero et al. 2012). PP2A has been proposed to protect cohesin from the prophase pathway by reverting the phosphorylation of SA2 at centromeres (Kitajima et al. 2006), but it is conceivable that PP2A also has other critical substrates whose dephosphorylation is required to maintain centromeric cohesion, such as sororin, Wapl, or Pds5. A second protein that has been shown to protect cohesin at centromeres is a protein kinase called Haspin. Haspin’s only known substrate is the core histone H3, which can be phosphorylated on threonine 5, but how Haspin contributes to protection of cohesin at centromeres remains to be understood (for review, see Higgins 2010).
Those cohesin complexes that are protected from the prophase pathway by shugoshin-PP2A can only be removed from chromosomes by separase, a protease that cleaves the Scc1 subunit of cohesin at two distinct sites and thereby opens the cohesin ring (Uhlmann et al. 1999, 2000; Waizenegger et al. 2000). This process is essential for sister chromatid separation because either expression of non-cleavable Scc1 or inactivation of separase prevents proper sister chromatid separation (for review, see Peters et al. 2008).
In budding yeast, Scc1 cleavage by separase is also a prerequisite for Smc3 deacetylation by Hos1 (Beckouet et al. 2010; Borges et al. 2010). If cohesin contains non-cleavable Scc1, Smc3 is not deacetylated at the time of anaphase onset even though cyclin B is degraded. Vice versa, Scc1 cleavage triggers Smc3 deacetylation in metaphase arrested cells in the absence of cyclin B degradation. Although Scc1 cleavage is a prerequisite for cohesin dissociation from chromatin, cleavage per se is not necessary for deacetylation in vitro because release of cohesin from DNA by nuclease digestion is sufficient for Smc3 deacetylation even if Scc1 remains intact (Borges et al. 2010). Separase may therefore enable cohesin deacetylation by simply releasing cohesin from chromatin, with which Hos1 is not associated (Borges et al. 2010).
CONCLUDING REMARKS
It is one of the most fundamental aspects of life that new cells are generated by division of older cells and that the newly formed daughter cells share most if not all properties of their mothers because the daughters inherit an identical copy of the mother cell’s genome. The two key processes that constitute this chromosome cycle are the precise duplication of the genome by DNA replication and the subsequent segregation of the two copies of the genome by chromosome segregation. Although the basic logic of this cycle was understood already during the early days of cell biology in the 19th century, it has only become obvious in the more recent past that understanding the mechanisms of sister chromatid cohesion will also be essential for understanding the eukaryotic cell division cycle. It is now well established that sister chromatid cohesion is mediated by cohesin, but exactly how cohesin performs this and possibly other functions and how these are controlled during the cell cycle and during cell differentiation remain incompletely understood. Numerous important questions will have to be answered to build a comprehensive molecular model of how cohesin mediates these functions: How and where does adherin load cohesin onto DNA? Is cohesin translocated from loading sites to its final binding sites? If so, how, and by which mechanism does CTCF recruit cohesin to these sites in vertebrates? How does Wapl release cohesin from DNA, and how is this process prevented by sororin or possibly by other mechanisms that depend on cohesin acetylation? Is cohesin converted into a cohesive form at replication forks? If so, how are cohesin acetyltransferases and sororin recruited to these forks, and exactly how do the two sister DNA strands become connected by cohesin at or in the vicinity of replication forks? What happens to cohesin when DNA or RNA polymerases or other enzymes move along DNA? Can these enzymes move through cohesin rings, or bypass them, or push them along DNA, or do they have to evict cohesin transiently from DNA? Is cohesin really able to mediate cohesion for months or years, as has been speculated to be the case in mammalian oocytes? How does cohesin’s ability to mediate cohesion relate to its role in gene regulation? Does cohesin mediate the latter function through the formation of intrachromatid loops? If yes, how is the ability of cohesin to connect DNA strands targeted to sister chromatids when cohesion is established, but to different DNA sequences in cis during the process of gene regulation? Answering these and further questions will be an exciting task for the future, and an essential prerequisite for understanding how chromosomes are organized and passed from one cell generation to the next.
ACKNOWLEDGMENTS
We thank T. Hirano and K. Shintomi for inspiring the ideas regarding the coevolution of Eco1 and Wapl, and Hannes Tkadletz for help with preparing the figures. Research in the laboratory of J.-M.P. is supported by Boehringer Ingelheim, the Vienna Science and Technology Fund (WWTF LS09-13), the Austrian Science Fund (FWF Special Research Program SFB F34, “Chromosome Dynamics,” and Wittgenstein award Z196-B20), and the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement number 241548 (MitoSys).
Footnotes
Editors: Stephen D. Bell, Marcel Mechali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
- Anderson DE, Losada A, Erickson HP, Hirano T 2002. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol 156: 419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansbach AB, Noguchi C, Klansek IW, Heidlebaugh M, Nakamura TM, Noguchi E 2008. RFCCtf18 and the Swi1–Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe. Mol Biol Cell 19: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K 2003. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr Biol 13: 1941–1953 [DOI] [PubMed] [Google Scholar]
- Beckouet F, Hu B, Roig MB, Sutani T, Komata M, Uluocak P, Katis VL, Shirahige K, Nasmyth K 2010. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol Cell 39: 689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F 2008. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321: 563–566 [DOI] [PubMed] [Google Scholar]
- Borges V, Lehane C, Lopez-Serra L, Flynn H, Skehel M, Rolef Ben-Shahar T, Uhlmann F 2010. Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol Cell 39: 677–688 [DOI] [PubMed] [Google Scholar]
- Chan KL, Roig MB, Hu B, Beckouet F, Metson J, Nasmyth K 2012. Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell 150: 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, Saitoh K, Komata M, Katou Y, Clark D, et al. 2012. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 10.1038/nature11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Strom L 2012. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr Biol 22: R240–R250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier MR, Bekier ME II, Taylor WR 2011. Regulation of sororin by Cdk1-mediated phosphorylation. J Cell Sci 124: 2976–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Cosentino C, Rivera T, Losada A, Schwob E, Hunt T, Costanzo V 2009. Tipin/Tim1/And1 protein complex promotes Pol α chromatin binding and sister chromatid cohesion. EMBO J 28: 3681–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina A, Shin JH, Kim DH, Bermudez VP, Kelman Z, Seo YS, Hurwitz J 2008. Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1. J Biol Chem 283: 20925–20936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feytout A, Vaur S, Genier S, Vazquez S, Javerzat JP 2011. Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1. Mol Cell Biol 31: 1771–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K, Takahashi K, Yanagida M 1998. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev 12: 3408–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T 2006. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol 16: 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J 2006. Live-cell imaging reveals a stable cohesin–chromatin interaction after but not before DNA replication. Curr Biol 16: 1571–1578 [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Hirano T 2004. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol 14: 1598–1603 [DOI] [PubMed] [Google Scholar]
- Gimenez-Abian JF, Sumara I, Hirota T, Hauf S, Gerlich D, de la Torre C, Ellenberg J, Peters JM 2004. Regulation of sister chromatid cohesion between chromosome arms. Curr Biol 14: 1187–1193 [DOI] [PubMed] [Google Scholar]
- Gruber S, Haering CH, Nasmyth K 2003. Chromosomal cohesin forms a ring. Cell 112: 765–777 [DOI] [PubMed] [Google Scholar]
- Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K 2006. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell 127: 523–537 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Caballero C, Cebollero LR, Pendas AM 2012. Shugoshins: From protectors of cohesion to versatile adaptors at the centromere. Trends Genet 28: 351–360 [DOI] [PubMed] [Google Scholar]
- Haering CH, Lowe J, Hochwagen A, Nasmyth K 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9: 773–788 [DOI] [PubMed] [Google Scholar]
- Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Lowe J 2004. Structure and stability of cohesin’s Smc1–kleisin interaction. Mol Cell 15: 951–964 [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K 2008. The cohesin ring concatenates sister DNA molecules. Nature 454: 297–301 [DOI] [PubMed] [Google Scholar]
- Hanna JS, Kroll ES, Lundblad V, Spencer FA 2001. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 21: 3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM 2005. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol 3: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann B, Hutchins JR, Hudecz O, Novatchkova M, Rameseder J, Sykora MM, Liu S, Mazanek M, Lenart P, Heriche JK, et al. 2011. Systematic phosphorylation analysis of human mitotic protein complexes. Sci Signal 4: rs12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Koshland D 2009. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol Cell 34: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Onn I, Koshland D 2010. Genetic evidence that the acetylation of the Smc3p subunit of cohesin modulates its ATP-bound state to promote cohesion establishment in Saccharomyces cerevisiae. Genetics 185: 1249–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JM 2010. Haspin: A newly discovered regulator of mitotic chromosome behavior. Chromosoma 119: 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Hirano T 2006. Opening closed arms: Long-distance activation of SMC ATPase by hinge–DNA interactions. Mol Cell 21: 175–186 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Tainer JA 2003. Rad50/SMC proteins and ABC transporters: Unifying concepts from high-resolution structures. Curr Opin Struct Biol 13: 249–255 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101: 789–800 [DOI] [PubMed] [Google Scholar]
- Hou F, Zou H 2005. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol Biol Cell 16: 3908–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Itoh T, Mishra A, Katoh Y, Chan KL, Upcher W, Godlee C, Roig MB, Shirahige K, Nasmyth K 2011. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr Biol 21: 12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CE, Milutinovich M, Koshland D 2005. Rings, bracelet or snaps: Fashionable alternatives for Smc complexes. Philos Trans R Soc Lond B Biol Sci 360: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt P, Hassold T 2010. Female meiosis: Coming unglued with age. Curr Biol 20: R699–R702 [DOI] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K 2005. A topological interaction between cohesin rings and a circular minichromosome. Cell 122: 849–860 [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y 2006. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441: 46–52 [DOI] [PubMed] [Google Scholar]
- Kogut I, Wang J, Guacci V, Mistry RK, Megee PC 2009. The Scc2/Scc4 cohesin loader determines the distribution of cohesin on budding yeast chromosomes. Genes Dev 23: 2345–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM 2006. Wapl controls the dynamic association of cohesin with chromatin. Cell 127: 955–967 [DOI] [PubMed] [Google Scholar]
- Kurze A, Michie KA, Dixon SE, Mishra A, Itoh T, Khalid S, Strmecki L, Shirahige K, Haering CH, Lowe J, et al. 2011. A positively charged channel within the Smc1/Smc3 hinge required for sister chromatid cohesion. EMBO J 30: 364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont AL, Song J, Rankin S 2010. Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc Natl Acad Sci 107: 20364–20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens A, Hopfner KP 2010. Structural basis for adenylate kinase activity in ABC ATPases. J Mol Biol 401: 265–273 [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HS, Kim JS, Park YB, Gwon GH, Cho Y 2011. Crystal structure of the Mre11–Rad50–ATPγS complex: Understanding the interplay between Mre11 and Rad50. Genes Dev 25: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T 2000. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol 150: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T 2002. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev 16: 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons NA, Morgan DO 2011. Cdk1-dependent destruction of Eco1 prevents cohesion establishment after S phase. Mol Cell 42: 378–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannini L, Menga S, Musio A 2010. The expanding universe of cohesin functions: A new genome stability caretaker involved in human disease and cancer. Hum Mutat 31: 623–630 [DOI] [PubMed] [Google Scholar]
- Maradeo ME, Skibbens RV 2009. The Elg1–RFC clamp-loading complex performs a role in sister chromatid cohesion. PLoS ONE 4: e4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Gygi SP, Aebersold R, Hieter P 2001. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): An alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell 7: 959–970 [DOI] [PubMed] [Google Scholar]
- Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, Newitt R, Aebersold R, Boone C, Brown GW, et al. 2004. Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell 15: 1736–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Vezon D, Bullier E, Motamayor JC, Sellier A, Lefevre F, Pelletier G, Horlow C 2001. SWITCH1 (SWI1): A novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev 15: 1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M 2010. Cohesin: A global player in chromosome biology with local ties to gene regulation. Curr Opin Genet Dev 20: 555–561 [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K 1997. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45 [DOI] [PubMed] [Google Scholar]
- Mishra A, Hu B, Kurze A, Beckouet F, Farcas AM, Dixon SE, Katou Y, Khalid S, Shirahige K, Nasmyth K 2010. Both interaction surfaces within cohesin’s hinge domain are essential for its stable chromosomal association. Curr Biol 20: 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, et al. 2008. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117: 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C 2005. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature 437: 440–443 [DOI] [PubMed] [Google Scholar]
- Nasmyth K 2011. Cohesin: A catenase with separate entry and exit gates? Nat Cell Biol 13: 1170–1177 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Schleiffer A 2004. From a single double helix to paired double helices and back. Philos Trans R Soc Lond B Biol Sci 359: 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A 2009. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet 5: e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, et al. 2010. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 143: 737–749 [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Parish JL, Rosa J, Wang X, Lahti JM, Doxsey SJ, Androphy EJ 2006. The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. J Cell Sci 119: 4857–4865 [DOI] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J 2008. The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114 [DOI] [PubMed] [Google Scholar]
- Petronczki M, Chwalla B, Siomos MF, Yokobayashi S, Helmhart W, Deutschbauer AM, Davis RW, Watanabe Y, Nasmyth K 2004. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-α-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J Cell Sci 117: 3547–3559 [DOI] [PubMed] [Google Scholar]
- Rankin S, Ayad NG, Kirschner MW 2005. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell 18: 185–200 [DOI] [PubMed] [Google Scholar]
- Rees DC, Johnson E, Lewinson O 2009. ABC transporters: The power to change. Nat Rev Mol Cell Biol 10: 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, et al. 2009. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell 33: 763–774 [DOI] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A 2008. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci 105: 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CK, Brookes N, Uhlmann F 2009. Conserved features of cohesin binding along fission yeast chromosomes. Genome Biol 10: R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT 2010. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 20: 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Watrin E, Lenart P, Mechtler K, Peters JM 2007. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr Biol 17: 630–636 [DOI] [PubMed] [Google Scholar]
- Seitan VC, Krangel MS, Merkenschlager M 2012. Cohesin, CTCF and lymphocyte antigen receptor locus rearrangement. Trends Immunol 33: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P 1999. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev 13: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C 2007. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317: 242–245 [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM 2000. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 151: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters JM 2002. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell 9: 515–525 [DOI] [PubMed] [Google Scholar]
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K 2009. Budding yeast Wpl1(Rad61)–Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol 19: 492–497 [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC 2004. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol 6: 991–996 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H 2000a. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol 20: 3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Fuchs J, Loidl J, Nasmyth K 2000b. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol 2: 492–499 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hao Z, Kai M, Okayama H 2001. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J 20: 5779–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV 2009. Cohesin acetylation speeds the replication fork. Nature 462: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev 13: 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K 1998. Cohesion between sister chromatids must be established during DNA replication. Curr Biol 8: 1095–1101 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400: 37–42 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103: 375–386 [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Koshland D 2007. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317: 245–248 [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE 2008. A molecular determinant for the establishment of sister chromatid cohesion. Science 321: 566–569 [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM 2000. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103: 399–410 [DOI] [PubMed] [Google Scholar]
- Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM 2006. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol 16: 863–874 [DOI] [PubMed] [Google Scholar]
- Weitzer S, Lehane C, Uhlmann F 2003. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr Biol 13: 1930–1940 [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Williams DR, McIntosh JR 2002. mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryot Cell 1: 758–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Garrett-Engele CM, Li Z, Williams EV, Rosenman ED, Goldberg ML 2003. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr Biol 13: 2025–2036 [DOI] [PubMed] [Google Scholar]
- Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai AS, Limbo O, Guenther G, SilDas S, Hammel M, Russell P, et al. 2011. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat Struct Mol Biol 18: 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JS, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, Joo K, Robinson H, Lee J, Park SY, et al. 2009. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell 136: 85–96 [DOI] [PubMed] [Google Scholar]
- Xiao T, Wallace J, Felsenfeld G 2011. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex, and are required for cohesin dependent insulation activity. Mol Cell Biol 31: 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong B, Lu S, Gerton JL 2010. Hos1 is a lysine deacetylase for the Smc3 subunit of cohesin. Curr Biol 20: 1660–1665 [DOI] [PubMed] [Google Scholar]
- Xu H, Boone C, Klein HL 2004. Mrc1 is required for sister chromatid cohesion to aid in recombination repair of spontaneous damage. Mol Cell Biol 24: 7082–7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HG, Koshland D 2005. Chromosome morphogenesis: Condensin-dependent cohesin removal during meiosis. Cell 123: 397–407 [DOI] [PubMed] [Google Scholar]
- Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, et al. 2008a. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell 31: 143–151 [DOI] [PubMed] [Google Scholar]
- Zhang N, Kuznetsov SG, Sharan SK, Li K, Rao PH, Pati D 2008b. A handcuff model for the cohesin complex. J Cell Biol 183: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Panigrahi AK, Mao Q, Pati D 2011. Interaction of Sororin protein with polo-like kinase 1 mediates resolution of chromosomal arm cohesion. J Biol Chem 286: 41826–41837 [DOI] [PMC free article] [PubMed] [Google Scholar]