Abstract
MAP kinases are activated within protein kinase cascades that regulate cell proliferation, differentiation, and death. In mammals, MAP kinases are grouped into three families: ERKs, JNKs, and p38/SAPKs.
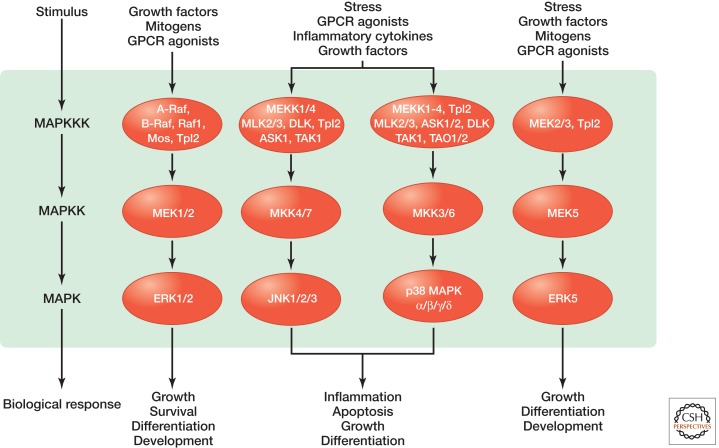
Mitogen-activated protein kinase (MAPK) modules containing three sequentially activated protein kinases are key components of a series of vital signal transduction pathways that regulate processes such as cell proliferation, cell differentiation, and cell death in eukaryotes from yeast to humans (Fig. 1) (Qi and Elion 2005; Raman et al. 2007; Keshet and Seger 2010). Each cascade is initiated by specific extracellular cues and leads to activation of a particular MAPK following the successive activation of a MAPK kinase kinase (MAPKKK) and a MAPK kinase (MAPKK) (Fig. 1). The MAPKKK is typically activated by interactions with a small GTPase and/or phosphorylation by protein kinases downstream from cell surface receptors (Cuevas et al. 2007). The MAPKKK directly phosphorylates and activates the MAPKK, which, in turn, activates the MAPK by dual phosphorylation of a conserved tripeptide TxY motif in the activation segment. Once activated, the MAPK phosphorylates diverse substrates in the cytosol and nucleus to bring about changes in protein function and gene expression that execute the appropriate biological response. MAPKs generally contain docking sites for MAPKKs and substrates, which allow high-affinity protein–protein interactions to ensure both that they are activated by a particular upstream MAPKK (Bardwell and Thorner 1996) and that they recognize specific downstream targets (Tanoue and Nishida 2003).
Figure 1.
MAPK pathways.
The MAP kinases can be grouped into three main families. In mammals, these are ERKs (extracellular-signal-regulated kinases), JNKs (Jun amino-terminal kinases), and p38/SAPKs (stress-activated protein kinases). ERK family members possess a TEY motif in the activation segment and can be subdivided into two groups: the classic ERKs that consist mainly of a kinase domain (ERK1 and ERK2) and the larger ERKs (such as ERK5) that contain a much more extended sequence carboxy-terminal to their kinase domain (Zhang and Dong 2007). The classic ERK1/2 module (Fig. 2) responds primarily to growth factors and mitogens to induce cell growth and differentiation (McKay and Morrison 2007; Shaul and Seger 2007). Important upstream regulators of this module include cell surface receptors, such as receptor tyrosine kinases (RTKs), G-protein-coupled receptors (GPCRs), and integrins, as well as the small GTPases Ras and Rap. MAPKKs for the classic ERK1/2 module are MEK1 and MEK2, and the MAPKKKs include members of the Raf family, Mos, and Tpl2.
Figure 2.
The ERK MAPK pathway.
JNK family members contain a TPY motif in the activation segment and include JNK1, JNK2, and JNK3. The JNK module (Fig. 3) is activated by environmental stresses (ionizing radiation, heat, oxidative stress, and DNA damage) and inflammatory cytokines, as well as growth factors, and signaling to the JNK module often involves the Rho family GTPases Cdc42 and Rac (Johnson and Nakamura 2007). The JNK module plays an important role in apoptosis, inflammation, cytokine production, and metabolism (Dhanasekaran and Reddy 2008; Huang et al. 2009; Rincon and Davis 2009). MAPKKs for the JNK module are MKK4 and MKK7, and the MAPKKKs include MEKK1 and MEKK4, MLK2 and MLK3, ASK1, TAK1, and Tpl2.
Figure 3.
The JNK MAPK pathway.
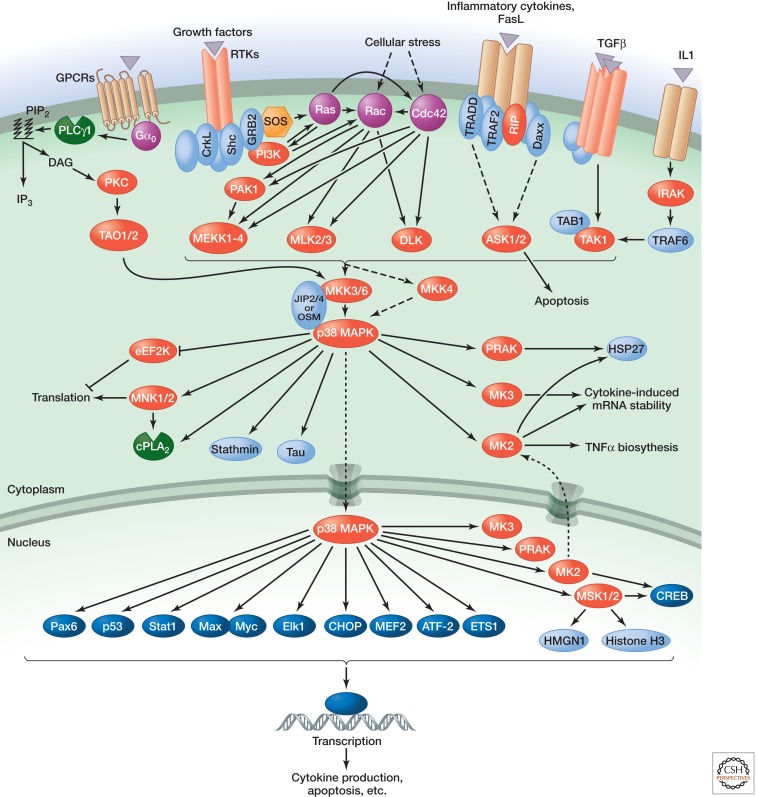
p38 family members possess a TGY motif in the activation segment and include p38α, p38β, p38γ, and p38δ. Like JNK modules, p38 modules (Fig. 4) are strongly activated by environmental stresses and inflammatory cytokines. p38 activation contributes to inflammation, apoptosis, cell differentiation, and cell cycle regulation (Cuenda and Rousseau 2007; Cuadrado and Nebreda 2010). The primary MAPKKs for p38 modules are MKK3 and MKK6, and the MAPKKKs include MLK2 and MLK3, MEKKs, ASKs, TAK1, and TAO1 and TAO2. Important substrates in p38 signaling include the downstream kinases MK2/3, PRAK, and MSK1 and MSK2, as well as various transcription factors.
Figure 4.
The p38 MAPK pathway.
For all of the MAPK modules, specific scaffold proteins (Good et al. 2011) have been identified that dock at least two of the core kinases of the module. These scaffolds contribute to MAPK signaling by increasing the local concentration of the components, providing spatial temporal regulation of cascade activation, and/or localizing the module to specific cellular sites or substrates. Scaffold proteins involved in MAPK cascade signaling include KSR and MP1 for the ERK module; JIP1, JIP2, JIP3, JIP4, and POSH for the JNK module; and JIP2, JIP4, and OSM for the p38 module (Dhanasekaran et al. 2007).
Acknowledgments
Figure 1 adapted, with permission, from Cell Signaling Technology (http://www.cellsignal.com).
Footnotes
Editors: Lewis Cantley, Tony Hunter, Richard Sever, and Jeremy Thorner
Additional Perspectives on Signal Transduction available at www.cshperspectives.org.
REFERENCES
- Bardwell L, Thorner J 1996. A conserved motif at the amino termini of MEKs might mediate high-affinity interaction with the cognate MAPKs. Trends Biochem Sci 21: 373–374 [PubMed] [Google Scholar]
- Cuadrado A, Nebreda AR 2010. Mechanisms and functions of p38 MAPK signalling. Biochem J 429: 403–417 [DOI] [PubMed] [Google Scholar]
- Cuenda A, Rousseau S 2007. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta 1773: 1358–1375 [DOI] [PubMed] [Google Scholar]
- Cuevas BD, Abell AN, Johnson GL 2007. Role of mitogen-activated protein kinase kinase kinases in signal integration. Oncogene 26: 3159–3171 [DOI] [PubMed] [Google Scholar]
- Dhanasekaran DN, Reddy EP 2008. JNK signaling in apoptosis. Oncogene 27: 6245–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran DN, Kashef K, Lee CM, Xu H, Reddy EP 2007. Scaffold proteins of MAP-kinase modules. Oncogene 26: 3185–3202 [DOI] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA 2011. Scaffold proteins: Hubs for controlling the flow of cellular information. Science 332: 680–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Shi LZ, Chi H 2009. Regulation of JNK and p38 MAPK in the immune system: Signal integration, propagation and termination. Cytokine 48: 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GL, Nakamura K 2007. The c-Jun kinase/stress-activated pathway: Regulation, function and role in human disease. Biochim Biophys Acta 1773: 1341–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet Y, Seger R 2010. The MAP kinase signaling cascades: A system of hundreds of components regulates a diverse array of physiological functions. Methods Mol Biol 661: 3–38 [DOI] [PubMed] [Google Scholar]
- McKay MM, Morrison DK 2007. Integrating signals from RTKs to ERK/MAPK. Oncogene 26: 3113–3121 [DOI] [PubMed] [Google Scholar]
- Qi M, Elion EA 2005. MAP kinase pathways. J Cell Sci 118: 3569–3572 [DOI] [PubMed] [Google Scholar]
- Raman M, Chen W, Cobb MH 2007. Differential regulation and properties of MAPKs. Oncogene 26: 3100–3112 [DOI] [PubMed] [Google Scholar]
- Rincon M, Davis RJ 2009. Regulation of the immune response by stress-activated protein kinases. Immunol Rev 228: 212–224 [DOI] [PubMed] [Google Scholar]
- Shaul YD, Seger R 2007. The MEK/ERK cascade: From signaling specificity to diverse functions. Biochim Biophys Acta 1773: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Tanoue T, Nishida E 2003. Molecular recognitions in MAP kinase cascades. Cell Signal 15: 455–462 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Dong C 2007. Regulatory mechanisms of mitogen-activated kinase signaling. Cell Mol Life Sci 64: 2771–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]