Abstract
The regulation of translation provides a mechanism to control not only the abundance of proteins, but also the precise time and subcellular location that they are synthesized. Much of what is known concerning the molecular basis for translational control has been gleaned from experiments (e.g., luciferase assays and polysome analysis) that measure average changes in the protein synthesis of a population of cells, however, mechanistic insights can be obscured in ensemble measurements. The development of fluorescent microscopy techniques and reagents has allowed translation to be studied within its cellular context. Here we highlight recent methodologies that can be used to study global changes in protein synthesis or regulation of specific mRNAs in single cells. Imaging of translation has provided direct evidence for local translation of mRNAs at synapses in neurons and will become an important tool for studying translational control.
Fluorescent microscopy techniques and reagents allow translation to be studied in its cellular context—even at the level of individual mRNA molecules.
A recent genome-wide study of mRNA and protein abundance in mammalian cells determined that cellular levels of proteins are predominantly controlled at the level of translation (Schwanhausser et al. 2011). Although methods have been developed that allow transcription to be quantified at the single molecule level in living cells, similar measurements of translation in eukaryotic cells have not yet been possible. Only in bacteria has it been possible to measure the mRNA and protein levels of a gene with single molecule sensitivity in single cells (Taniguchi et al. 2010). Here we highlight what has been learned about the behavior of single mRNA molecules in gene expression and the current methodologies for imaging translation in single cells. We conclude by discussing what could be performed to allow the translation of single mRNA molecules to be monitored in living cells.
SINGLE MOLECULE IMAGING OF mRNA IN GENE EXPRESSION
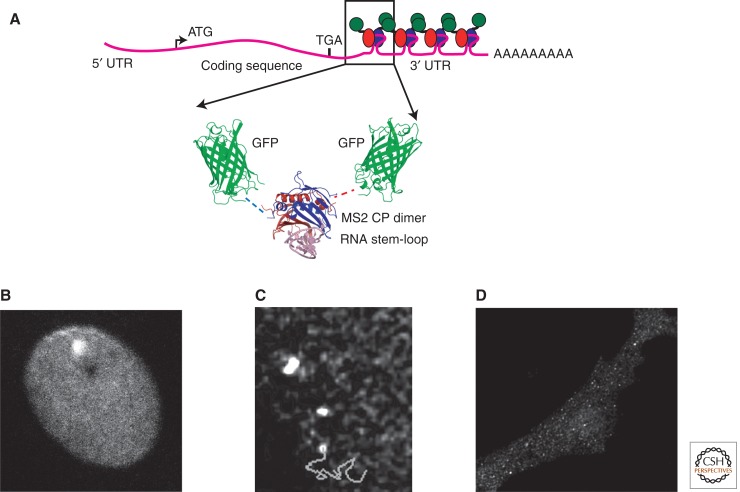
The development of imaging technologies capable of quantifying and tracking single RNA molecules has allowed gene expression to be characterized in living cells. The most common method for imaging single molecules of mRNA in living cells uses a unique RNA stem-loop sequence that is specifically recognized by the MS2 bacteriophage coat protein (MCP) (Valegard et al. 1994). By constructing a reporter mRNA that contains multiple copies of the MS2 operator RNA stem-loop and coexpressing a chimeric MS2 coat protein fusion with green fluorescent protein (GFP), single RNA molecules can be detected above the fluorescent background of the unbound MCP-GFP molecules (Fig. 1A) (Bertrand et al. 1998; Beach et al. 1999). Because the MCP binds to its cognate RNA stem-loop with high affinity, this interaction allows stable labeling of the reporter RNA in cells (Lowary and Uhlenbeck 1987; Lim and Peabody 1994). Although labeling of RNAs with the MS2 systems requires the addition of multiple RNA stem-loops and MCP-GFP molecules, this increase in size and mass does not significantly perturb RNA metabolism as evidenced by the creation of a transgenic mouse that has an insertion of the MS2 stem-loops in the endogenous β-actin gene, which has no effect on viability (Lionnet et al. 2011). This system has been used to study many aspects of gene expression ranging from transcription to RNA localization in the cytoplasm in a variety of experimental systems.
Figure 1.
MS2 system for fluorescent labeling of mRNAs in living cells. (A) Schematic of a reporter mRNA that contains multiple copies of the MS2 stem-loop (boxed) that bind to the MS2 coat protein fused to GFP (arrows). The binding of many MCP-GFP molecules to a single transcript allows the RNA to be observed in living cells. (B) Transcription in the nucleus detected by binding of MCP-GFP to nascent mRNAs at the transcription site. (From Darzacq et al. 2007; reprinted, with permission, from the author.) (C) Tracking of single mRNAs (trajectory shown as line) in the nucleoplasm. (From Shav-tal et al. 2004; reprinted, with permission, from The American Association for the Advancement of Science © 2004.) (D) Single mRNAs detected in the cytoplasm. (From Fusco et al. 2003; reprinted, with permission, from Elsevier © 2003.)
Quantifying transcription in real time in living cells has revealed that the expression of individual genes is stochastic and has allowed direct measurement of the kinetics of the process. In bacteria, the counting of individual mRNAs in single cells showed that transcriptional bursting, infrequent transcription events that produce many transcripts within a short period of time, was responsible for the variability in gene expression (Golding and Cox 2004; Golding et al. 2005). Similarly, in the eukaryote Dictyostelium, transcription of an endogenous developmental gene, dscA, was observed to be expressed in discrete bursts of gene activity (Chubb et al. 2006). It has also been possible to measure the speed of the RNA polymerase II in mammalian cells and it was found to elongate at 4.3 kb min−1. Surprisingly, gene expression was found to be inefficient, with only 1% of polymerases that contact the gene resulting in transcription of an mRNA (Fig. 1B) (Darzacq et al. 2007).
Once a transcript leaves the site of its transcription, its movements can be tracked within the nucleoplasm. Rapid time-lapse imaging and single particle tracking revealed that the diffusion of individual mRNAs is governed by simple diffusion and that transcripts can be partially corralled by the chromatin domains within the nucleoplasm (Fig. 1C) (Shav-Tal et al. 2004). Transcripts diffuse within the nucleoplasm for times ranging from 5 to 40 minutes; however, once a transcript engages a nuclear pore, export into the cytoplasm is fast (less than 0.2 sec) (Grunwald and Singer 2010; Mor et al. 2010). Interestingly, transport through the nuclear pore takes only 5–20 min and docking on the nuclear side and release into the cytoplasm both take approximately 80 min, indicating that translocation through the nuclear pore is not the rate-limiting step (Grunwald and Singer 2010).
In contrast to what was observed in the nucleus, once a transcript is in the cytoplasm, it can undergo rapid directional movements indicating active transport by molecular motors along the cytoskeleton. The MS2 system was first employed to characterize the localization of ASH1 mRNA to the bud tip in Saccharomyces cerevisiae (Bertrand et al. 1998). Time-lapse imaging of ASH1 mRNA tagged with MCP-GFP showed that it was transported at speeds of 0.20–0.44 µm sec−1 by Myo4p, a type V myosin motor, along the actin cytoskeleton. Characterization of mRNA movements in mammalian cells indicated that transcripts can undergo static, corralled, diffusive and directed movements in the cytoplasm, and individual transcripts frequently switch between these different modes of movement (Fig. 1D) (Fusco et al. 2003). The β-actin mRNA zipcode, a cis-acting element that is responsible for the localization of this transcript, was found to increase both the frequency and length of directed movements. In the Drosophila melanogaster oocyte, the posterior localization of oskar mRNA was found to be achieved by a biased random walk with a 14% excess of posterior directed movements by kinesin along weakly polarized microtubules (Zimyanin et al. 2008).
Characterization of mRNAs in the cytoplasm has also provided indirect evidence for how translation may affect these movements. In rat pheochromocytoma cells (PC12), a reporter mRNA that labeled peripherin mRNA with MCP-YFP (yellow fluorescent protein) indicated that RNA particles were motile with an average speed of 0.42 µm sec−1 and that often, when particles became stationary, the peripherin protein could subsequently be detected (Chang et al. 2006). This led to the hypothesis that motile mRNAs are translationally repressed. Further evidence for this model was obtained by tracking ARC mRNA with MCP-GFP in cultured rat neurons. ARC mRNA was observed to undergo rapid bidirectional movements while in transit, but would often reverse direction and decrease in velocity before becoming stationary suggesting that local signals in the dendrite regulated ARCs mRNA movement and determined the location for its translation (Dynes and Steward 2007). Although these experiments have provided hints as to how protein synthesis is regulated, more direct methods are required to study the translation of mRNAs in the cytoplasm.
GLOBAL MEASUREMENTS OF PROTEIN SYNTHESIS IN SINGLE CELLS
To distinguish newly synthesized proteins from ones that have been previously translated, cells can be incubated with radioisotopically labeled amino acids (35S-methionine) that are then incorporated into nascent polypeptides. On cell lysis, the fraction of the proteome that has been synthesized after the addition of the radiolabeled amino acid can be measured by autoradiography. Although these types of experiments have been tremendously informative for deducing the mechanisms by which factors exert their translational control, they cannot be performed on intact cells and, consequently, information concerning the spatial regulation of protein synthesis within the cell is lost. Several fluorescent microscopy approaches have been developed that allow global changes in translation to be measured in single cells.
Fluorescent Noncanonical Amino Acid Tagging (FUNCAT)
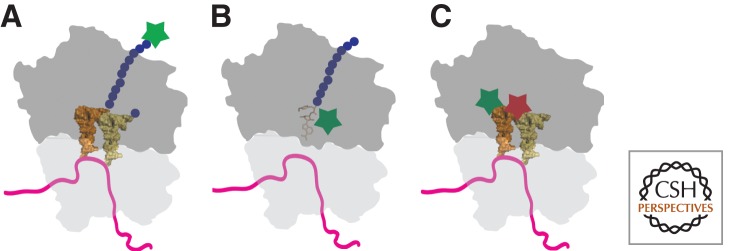
Noncanonical amino acids that can be recognized by the appropriate aminoacyl tRNA synthetase and incorporated into elongating polypeptides by the ribosome provide a tool for fluorescent labeling of newly synthesized proteins. Schuman and colleagues devised a labeling strategy based on two methionine analogs, azidohomoalanine (AHA) and homopropargylglycine (HPG) (Dieterich et al. 2010). These noncanonical amino acids can be added at methionine codons. Because they contain chemical functionality that standard amino acids to do not have, they can be specifically conjugated to fluorescent dyes (Texas Red or carboxyfluoroscein) via “click chemistry” that uses a copper-catalyzed azide-alkyne [3 + 2] cycloaddition (Fig. 2A) (Rostovtsev et al. 2002). This metabolic labeling strategy is conceptually very similar to radiolabeling with 35S-methionine; however, it allows detection of newly synthesized proteins by fluorescent microscopy. Because AHA and HPG are not as efficient substrates for methionyl-tRNA-synthetase (metRS) as methionine, it is necessary to reduce the cellular concentration of methionine by starving the cells of this particular amino acid, so that the analogs can effectively compete for charging to tRNA and incorporation by the ribosome. The AHA and HPG amino acids are added to proteins in positions normally occupied by methionines, which can result in nonuniform labeling because the accessibility of the noncanonical side chains to participate in “click chemistry” may not be equivalent for every protein.
Figure 2.
Global measurements of translation in single cells. (A) The incorporation of noncanonical amino acids using FUNCAT allows newly synthesized proteins to be fluorescently labeled (green star). Methionine analogs are incorporated into the elongating polypeptide chain that can be conjugated to fluorescent dyes. (B) Fluorescent analogs of puromycin (green star) prematurely terminate translation and allow nascent polypetides to be fluorescently labeled. (C) FRET between adjacent fluorescent tRNAs (green and red stars are FRET pairs) allows the cellular location of active translation to be observed. Only when fluorescently labeled tRNAs are bound to the ribosome in adjacent sites, is the FRET signal observable.
FUNCAT has been used to investigate the dynamics of protein synthesis in rat hippocampal neurons. Although this metabolic labeling strategy labels all newly synthesized cellular proteins after addition of AHA or HPG, the noncanonical amino acid can be locally microperfused using a small delivery pipet combined with a suction pipet to discrete subcellular regions. Using this approach FUNCAT was used to observe protein synthesis in individual dendrites that was diminished on addition of anisomycin, a translation inhibitor, and stimulated on addition of brain-derived neurotropic factor (BDNF), which induces translation-dependent enhancement of synaptic strength (Kang and Schuman 1996). The engineering of a mutant MetRS that is capable of charging the noncanonical amino acid azidonorleucine, which is not recognized by the cell's endogenous MetRS, can be used to restrict the incorporation of azidonorleucine to a defined population of cells (Ngo et al. 2009).
Fluorescent Puromycin Derivatives
Puromycin is a natural product derived from Streptomyces alboniger bacteria that causes premature termination of the elongating protein during translation. The small molecule contains a region that is structurally analogous to the 3′-terminus of aminoacylated-tRNA and can enter the ribosome at the acceptor site. When puromycin is bound in this position, its primary amine forms a peptide bond with the carboxyl terminus of the nascent polypeptide, thereby stalling the ribosome and prematurely terminating protein synthesis (Yarmolinsky and Haba 1959; Nathans 1964). By conjugating a fluorescent dye to puromycin, newly synthesized polypeptide fragments can be detected in living cells. (Fig. 2B) It has been shown that puromycin substituted at the 5′ OH has decreased efficacy as a translation inhibitor, so a deoxycytidine (dC) was used to link fluorescent dyes (Cy5 and fluorescein) to puromycin (Starck and Roberts 2002; Starck et al. 2004).
Fluorescein-dC-puromycin (F2P) has been used to measure global protein synthesis in the dendrites of rat hippocampal neurons. Addition of F2P to neuronal cultures resulted in an increase in fluorescence in both the soma and dendrites and this signal was attenuated by addition of anisomycin or unlabeled puromycin (Smith et al. 2005). When neurons were treated with a dopamine D1/D5 agonist, SKF-38393, a significant increase in fluorescence was measured in both dendrites and spines consistent with the ability of dopamine agonist to stimulate protein synthesis.
A second puromycin analog has been developed that contains a terminal alkyne group, O-propargyl-puromycin (OP-puro) (Liu et al. 2011). This substitution enables “click chemistry” to be performed on the polypeptide-OP-puro fusions allowing conjugation with fluorescent dyes in a similar manner as described for FUNCAT. Importantly, this labeling strategy can be used to fluorescently detect nascent proteins in whole animals. When OP-puro is injected intraperitoneally into mice, protein synthesis can be detected in tissue that has been fixed and stained with tetramethylrhodamine-azide. Protein synthesis was found to be highest in the crypts and at the base of intestinal villi in the small intestine, which is consistent with the function of these cells and shows the sensitivity of this method to detect differences in translation rates amongst and within different organs.
Fluorescent tRNA-Derivatives
An alternative strategy for measuring protein synthesis in living cells that does not label the protein product of translation has been shown using fluorescently labeled tRNA (fl-tRNA). In this experiment, bulk uncharged tRNA from yeast was labeled with either Cy3 or Rhodamine 110 (Rho110) in the D-loop (Betteridge et al. 2007; Pan et al. 2009). Most tRNAs contain dihydroU at multiple positions within the D-loop that can be selectively reduced by treatment with NaBH4 and then conjugated to fluorescent dyes (Cerutti and Miller 1967). Modification of tRNAs in these positions does not alter their ability to bind to the ribosome and can be used to monitor translation (Pan et al. 2007).
When fl-tRNAs were transfected into Chinese hamster ovary (CHO) cells, they partially colocalized with several cellular proteins involved in tRNA charging or translation (Arg-RS, eEF1A, and rpS6) indicating that the fl-tRNAs were functional (Barhoom et al. 2011). When cells are transfected with both Cy3-tRNA and Rho110-tRNA, fluorescence resonance energy transfer (FRET) can be observed when the differentially labeled tRNAs occupy adjacent positions within the ribosome (Fig. 2C). Using this system, primary brain astrocytes that were activated with either bacterial lipopolysaccharides or interferon-γ were found to have increases in both the number of cells that had FRET signals as well as the intensity of the FRET signal per cell. These increases in FRET correlated with standard 35S metabolic labeling, indicating that this approach could provide information on sites of active translation within cells.
TRANSCRIPT-SPECIFIC MEASUREMENTS OF TRANSLATION IN LIVING CELLS
To measure changes in protein synthesis for a particular protein of interest, radioisotope labeling with 35S-methionine of all cellular proteins can be followed by immunoprecipitation with specific antibodies and standard electrophoresis. Although this approach allows the translation properties of endogenous protein to be measured, it is often of interest to know where and when specific proteins are synthesized within a cell. A number of approaches have been developed that allow the fluorescent detection of reporter constructs that have been exogenously expressed.
Fluorescent Protein Reporters
The discovery and development of fluorescent proteins has been a boon for scientists interested in visualizing intracellular gene expression. When a protein of interest is expressed as a chimeric fusion protein with a fluorescent protein, the intracellular fluorescence can report on the subcellular localization of the endogenous protein. A variety of fluorescent proteins with distinct spectral properties have been engineered (Giepmans et al. 2006; Wu et al. 2011). Recent advances have allowed fluorescent proteins to be used as translation reporters to dynamically visualize synthesis of new proteins in living cells. In neurons, the use of translation reporters has widely been used, due in large part to the cells’ pronounced polarized morphology that clearly distinguishes the cell body from the processes. The neurites provide discrete subcellular compartments away from the cell body where protein synthesis can be spatially and temporally regulated. Furthermore, the observation that synaptic plasticity requires local synthesis of new proteins at specific synapses provides a rationale for devising and advancing effective translation reporters.
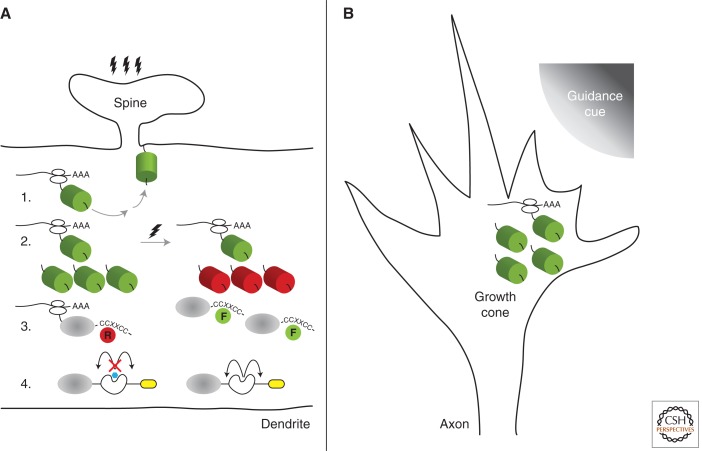
Our understanding of RNA localization and local protein synthesis has been central to the design of translation reporters. Schuman and colleagues created a dendritic translation reporter by flanking the coding sequence for GFP with the 5′ and 3′ untranslated regions (UTR) of the α subunit of Ca2+/calmodulin-dependent kinase II (αCaMKII) (Fig. 3A.1) (Aakalu et al. 2001; Sutton et al. 2004). The 3′ UTR of αCaMKII has been shown to be sufficient for dendritic localization of the mRNA in neurons. Furthermore, to address the possibility that GFP synthesized in the cell body could diffuse into the dendrite, a myristoylation tag was included before the GFP coding sequence to anchor the newly synthesized reporter proteins to the plasma membrane. The PEST sequence from ornithine decarboxylase was fused to the carboxyl terminus of GFP to reduce the half-life of the protein and enhance the temporal resolution of the reporter (Rogers et al. 1986; Corish and Tyler-Smith 1999). On treatment of hippocampal neurons with brain derived neurotrophic factor (BDNF), it was possible to visualize newly synthesized reporter fluorescence from distal regions of transected or optically isolated dendrites. Success of the dendritic translation reporter paved the way for efforts to design translation reporters targeted to different regions of the neuron. Flanagan and colleagues fused GFP with a 77-nucleotide conserved region from the 3′ UTR of the Eph2A receptor (Brittis et al. 2002). When expressed in commissural neurons of chick spinal cords, GFP fluorescence could be detected in distal axon segments. To address concerns that fast axonal transport may confound interpretation of the results, the GFP in the reporter was replaced with a fluorescent protein (Fluorescent Timer) that changes color from green to red over time (Terskikh et al. 2000). Comparison of the relative green and red fluorescence intensities from the soma and the axon growth cone gave results consistent with de novo protein synthesis within axons.
Figure 3.
Strategies for visualization of newly synthesized proteins in neurons. Polarized cellular architecture of neurons provides an ideal system to study localized translation in discrete subcellular domains. In dendrites (A), a small structure called the spine receives signals and transmits the information to the cell body. During this process, localized protein synthesis can be visualized using a variety of strategies depicted in the illustration. 1. Anchored GFP: Postsynaptic stimulation leads to synthesis of myristoylated GFP (green cylinder) that becomes anchored to the membrane thereby demarcating the site of active translation. 2. Photoconvertible GFP: Conversion of the preexisting population of fluorescent proteins (cylinders) from green to red by a brief exposure to ultraviolet light allows visualization of the newly synthesized population of fluorescent proteins. 3. Biarsenical dyes: Association of cell-permeable biarsenical dyes, FlAsH (F) and ReAsH (R) to the tetracysteine motif (CCXXCC) permits detection of translation using a pulse-chase approach. 4. TimeSTAMP: A self-cleaving protease (white) cassette that carries an epitope tag (yellow) becomes detectable on addition of the protease inhibitor (blue) by immunofluorescence methods. In axon growth cones (B), the directionality of guidance cues (upper right quadrant) can lead to asymmetric translation thereby leading to rapid growth cone turning and extension. Most, if not all, of the strategies employed in dendrites can be used in growth cones and other subcellular compartments to further characterize localized GFP translation (green cylinder) in neurons.
Continued development of fluorescent proteins that are capable of undergoing photoconversion and photoswitching has provided a means to distinguish newly synthesized proteins from the preexisting population of protein. A brief exposure of these fluorescent proteins to ultraviolet light (UV) within a confined focal volume results in the photoconversion of the fluorophore to different excitation and emission characteristics and allows detection of newly synthesized protein by visualizing the recovery of the unconverted fluorescent protein (Stepanenko et al. 2008; Wu et al. 2011). Experimental strategies that use these fluorescent proteins reduce concerns of somatic contamination of the reporter protein and provide unprecedented spatial and temporal resolution.
Localization of β-actin mRNA and its translation have been shown to be important for cell migration in fibroblasts and extending axonal growth cones in neurons (Kislauskis et al. 1994; Bassell et al. 1998). To investigate the role of local β-actin mRNA translation in growth cone turning, Holt and colleagues constructed an axonal translation reporter by fusing the photoconvertible protein, Kaede, to the β-actin 3′ UTR, which contains cis-acting elements responsible for RNA localization and translational control (Leung et al. 2006). Kaede is a fluorescent protein that was isolated from the stony coral, Trachyphyllia geoffroyi, and undergoes an irreversible photoconversion (green to red) on irradiation with UV light that results in a 2000-fold increase in red-to-green fluorescent signal (Ando et al. 2002). In growth cones expressing the Kaede reporter, new translation was observed on the side of the growth cone that was closest to an external gradient of the netrin-1 guidance cue (Fig. 3B). These data indicate that local synthesis of β-actin protein is important for the attractive turning response of growth cones.
To investigate the somatodendritic presence of potassium channel Kv1.1, which has been shown to be predominantly localized to axons, Jan and colleagues designed a reporter with Kaede fused to the amino terminus of the potassium channel protein Kv1.1 and whose mRNA also contained the 3′ UTR of Kv1.1 (Raab-Graham et al. 2006). On photoconversion and rapamycin treatment, the investigators observed new Kaede-Kv1.1 expression in distal regions of the dendrites indicating that the 3′ UTR contributes to the dendritic localization of the mRNA and subsequent stimulation-dependent translation of Kv1.1 (Fig. 3A.2). Interestingly, they observed that the fluorescently labeled Kv1.1 was concentrated in stationary “hotspots” in dendrites similar to what was reported with other locally translated protein (Aakalu et al. 2001).
To directly measure the requirement for local translation in neuronal plasticity, Martin and colleagues constructed a translation reporter using the photoconvertible Dendra2 flanked by the 5′ and 3′ UTRs of sensorin (Wang et al. 2009). Dendra2, is a single point mutant (A224V) of Dendra which results in faster maturation and brighter fluorescence, that undergoes an irreversible photoconversion from green to red fluorescence on excitation with either UV or blue light (488 nm) (Gurskaya et al. 2006). Photoconversion with blue light instead of UV light reduces phototoxicity that can be induced by exposure to UV irradiation. Sensorin mRNA localizes to synapses between sensory neuron and motor neuron involved in the gill withdrawal reflex of the sea slug, Aplysia. The monosynaptic connection between a sensory neuron and a motor neuron in culture provides an elegant and yet simple system to directly test stimulus-specific responses and visualize plasticity in the synapses that express the reporter. Following photoconversion of the preexisting reporter population and serotonin stimulation, they observed local translation of the reporter in a synapse-specific and stimulus-dependent manner.
Biarsenical Dyes
Although fluorescent proteins have proven to be extremely useful for measuring translation in living cells, they are limited in their ability to measure rapid changes in protein synthesis because of the time it takes for the fluorophore to mature once the protein has been translated. For many fluorescent proteins, the maturation time has been measured to be greater than 10 minutes and can take up to several hours for fluorescence to become detectable (Evdokimov et al. 2006; Kremers et al. 2006; Shaner et al. 2008). This lag in time between when the fluorescent protein has been synthesized and when it becomes observable also prevents identification of the precise location of translation because the fluorescent protein can diffuse away from the site of its synthesis. The engineering of short peptides sequences that can significantly enhance the fluorescence of dyes or small molecules provides a means to detect proteins as they are being translated.
The Tsien laboratory developed two biarsenical fluorescent dyes based on fluorescein (FlAsH) and resorufin (ReAsH) that become fluorescent when bound by a peptide that contains four cysteine residues (Griffin et al. 1998; Adams et al. 2002). The peptide sequence (FLNCCPGCCMEP) adopts a specific β-hairpin turn structure that is able to bind the biarsenical dyes with picomolar binding affinities (Madani et al. 2009). The FlAsH and ReAsH dyes are also membrane permeable, which facilitates their use in imaging translation in living cells. This labeling strategy was used to show that the AMPA receptors, GluR1 and GluR2, are locally synthesized in dendrites and localized to synapses in response to activity-dependent changes in synaptic strength (Ju et al. 2004). To observe newly synthesized proteins, the FlAsH and ReAsH dyes were used in a pulse-chase experiment, so that all preexisting GluR1 proteins were labeled with ReAsH and only proteins synthesized during the chase would be labeled by FlAsH (Fig. 3A.3). To determine the location of protein synthesis, the tetracysteine peptide sequence was added to the amino terminus of a GFP-β-actin fusion protein (Rodriguez et al. 2006). By placing the tetracysteine peptide sequence at the beginning of the reporter protein, the location of polysomes carrying multiple nascent reporter proteins that are actively being translated could be identified. Recently, a peptide that contains three copies of the tetracysteine sequence was engineered that has increased brightness compared to the original sequence, which improves the signal-to-noise in these experiments and may facilitate detection of the cellular sites of translation (Van Engelenburg et al. 2010).
TimeSTAMP
In efforts to develop novel approaches to discern newly synthesized proteins from the preexisting population, the Tsien lab devised a drug-dependent epitope tagging approach that does not use fluorescent proteins. Time-specific tagging for the age measurement of proteins, or TimeSTAMP, uses a viral protease whose activity can be controlled by a cell-permeant drug (Fig. 3A.4) (Lin et al. 2008). The crux of the idea was to design a self-cleaving cassette that can be regulated by a specific small molecule inhibitor. The cassette is composed of the NS3 protease from hepatitis C virus flanked by its cognate target sites along with an epitope tag on the opposite side of the protein of interest. In the absence of the inhibitor, BILN-2061, the protease will cleave itself, rendering the protein undetectable to antibodies against the tag. On addition of the inhibitor, the protease activity is blocked and the epitope tag then becomes available for detection. By capturing the immunofluorescence of the protein of interest and the epitope tag, it is possible to distinguish the population of newly synthesized protein during the time window of the drug administration.
The goal of TimeSTAMP was not only to visualize newly synthesized protein, but also to develop a method that circumvents some of the concerns with traditional fluorescent protein-mediated detection of new proteins. For example, phototoxicity during photoconversion or photobleaching of fluorescent proteins is a significant concern to those intending to study intracellular translation over a long period of time. Second, the time delay between synthesis and subsequent maturation of the chromophore reduces the temporal resolution of the onset of translation. In addition, limitations in detection of low copy number proteins and the inability to amplify fluorescence signal from fluorescent proteins allow only high abundance proteins as potential targets for analyses. In summary, although TimeSTAMP is not amenable to dynamic visualization of newly synthesized proteins, it provides an alternative strategy and has the potential to complement fluorescent protein-based reporter assays that are currently available. TimeSTAMP has been used to detect new protein synthesis of postsynaptic density protein (PSD)-95 and the trafficking of neuroligin (NLGN) 3 in cultured hippocampal neurons (Lin et al. 2008; De Jaco et al. 2010).
FUTURE DIRECTIONS
Thus far, the methodologies used to measure translation in single cells have focused on either the process of protein synthesis or its protein output, which has made detection of single molecules challenging. Because of the critical role of translation and its regulation in gene expression, new techniques and labeling strategies will need to be developed to more precisely interrogate protein synthesis. An alternate strategy that measures translation-dependent changes to reporter mRNAs themselves may allow translation to be monitored with the single molecule resolution in living cells. Besides the well-established fluorescent labeling method based on the MS2 bacteriophage coat protein, fluorescent RNA labeling has also been shown using the PP7 bacteriophage coat protein, the λN peptide and the U1A spliceosomal protein (Brodsky and Silver 2000; Takizawa and Vale 2000; Daigle and Ellenberg 2007; Larson et al. 2011). Because all four RNA-protein complexes are orthogonal, it should be possible, in principle, to specifically label a reporter mRNA in both the coding region as well as the 3′ UTR with spectrally distinct fluorescent proteins. In such a system, an untranslated RNA would labeled with two fluorescent proteins; on translation, however, the ribosome would displace the fluorescent protein that was bound to the coding region resulting in a singly fluorescently labeled RNA. Using this type of reporter, untranslated RNAs can be distinguished from ones that have encountered the ribosome in real time in living cells. Although this experimental methodology is only designed to allow observation of the first round of translation, much of the regulation of translation takes place at the initiation step. Such a system could be used to address a variety of outstanding questions including where the pioneer round of translation takes place for mRNAs that contain premature stop codons and are targets of nonsense mediated decay, and whether localized mRNAs are translationally repressed during transport (Maquat 2004; Chang et al. 2007; Sonenberg and Hinnebusch 2009; Donnelly et al. 2010). Continued development of reagents and techniques for quantifying translation of single molecules in living cells will lead to a mechanistic understanding of the regulation of translation and, undoubtedly, a few surprises along the way.
Footnotes
Editors: John W.B. Hershey, Nahum Sonenberg, and Michael B. Mathews
Additional Perspectives on Protein Synthesis and Translational Control available at www.cshperspectives.org
REFERENCES
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM 2001. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 30: 489–502 [DOI] [PubMed] [Google Scholar]
- Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY 2002. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: Synthesis and biological applications. J Am Chem Soc 124: 6063–6076 [DOI] [PubMed] [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A 2002. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci 99: 12651–12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barhoom S, Kaur J, Cooperman BS, Smorodinsky NI, Smilansky Z, Ehrlich M, Elroy-Stein O 2011. Quantitative single cell monitoring of protein synthesis at subcellular resolution using fluorescently labeled tRNA. Nucleic Acids Res 39: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS 1998. Sorting of β-actin mRNA and protein to neurites and growth cones in culture. J Neurosci 18: 251–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach DL, Salmon ED, Bloom K 1999. Localization and anchoring of mRNA in budding yeast. Curr Biol 9: 569–578 [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM 1998. Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445 [DOI] [PubMed] [Google Scholar]
- Betteridge T, Liu H, Gamper H, Kirillov S, Cooperman BS, Hou YM 2007. Fluorescent labeling of tRNAs for dynamics experiments. RNA 13: 1594–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittis PA, Lu Q, Flanagan JG 2002. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell 110: 223–235 [DOI] [PubMed] [Google Scholar]
- Brodsky AS, Silver PA 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6: 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti P, Miller N 1967. Selective reduction of yeast transfer ribonucleic acid with sodium borohydride. J Mol Biol 26: 55–66 [DOI] [PubMed] [Google Scholar]
- Chang L, Shav-Tal Y, Trcek T, Singer RH, Goldman RD 2006. Assembling an intermediate filament network by dynamic cotranslation. J Cell Biol 172: 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF 2007. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH 2006. Transcriptional pulsing of a developmental gene. Curr Biol 16: 1018–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corish P, Tyler-Smith C 1999. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng 12: 1035–1040 [DOI] [PubMed] [Google Scholar]
- Daigle N, Ellenberg J 2007. λN-GFP: An RNA reporter system for live-cell imaging. Nat Methods 4: 633–636 [DOI] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH 2007. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol 14: 796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jaco A, Lin MZ, Dubi N, Comoletti D, Miller MT, Camp S, Ellisman M, Butko MT, Tsien RY, Taylor P 2010. Neuroligin trafficking deficiencies arising from mutations in the α/β-hydrolase fold protein family. J Biol Chem 285: 28674–28682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM 2010. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci 13: 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Fainzilber M, Twiss JL 2010. Subcellular communication through RNA transport and localized protein synthesis. Traffic 11: 1498–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynes JL, Steward O 2007. Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J Comp Neurol 500: 433–447 [DOI] [PubMed] [Google Scholar]
- Evdokimov AG, Pokross ME, Egorov NS, Zaraisky AG, Yampolsky IV, Merzlyak EM, Shkoporov AN, Sander I, Lukyanov KA, Chudakov DM 2006. Structural basis for the fast maturation of Arthropoda green fluorescent protein. EMBO Rep 7: 1006–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E 2003. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol 13: 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giepmans BN, Adams SR, Ellisman MH, Tsien RY 2006. The fluorescent toolbox for assessing protein location and function. Science 312: 217–224 [DOI] [PubMed] [Google Scholar]
- Golding I, Cox EC 2004. RNA dynamics in live Escherichia coli cells. Proc Natl Acad Sci 101: 11310–11315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC 2005. Real-time kinetics of gene activity in individual bacteria. Cell 123: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Griffin BA, Adams SR, Tsien RY 1998. Specific covalent labeling of recombinant protein molecules inside live cells. Science 281: 269–272 [DOI] [PubMed] [Google Scholar]
- Grunwald D, Singer RH 2010. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature 467: 604–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA 2006. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol 24: 461–465 [DOI] [PubMed] [Google Scholar]
- Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, Garner CC, Tsien RY, Ellisman MH, Malenka RC 2004. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci 7: 244–253 [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM 1996. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 273: 1402–1406 [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH 1994. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J Cell Biol 127: 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremers GJ, Goedhart J, van Munster EB, Gadella TW Jr 2006. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Forster radius. Biochemistry 45: 6570–6580 [DOI] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH 2011. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332: 475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE 2006. Asymmetrical β-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci 9: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim F, Peabody DS 1994. Mutations that increase the affinity of a translational repressor for RNA. Nucleic Acids Res 22: 3748–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MZ, Glenn JS, Tsien RY 2008. A drug-controllable tag for visualizing newly synthesized proteins in cells and whole animals. Proc Natl Acad Sci 105: 7744–7749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH 2011. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods 8: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xu Y, Stoleru D, Salic A 2011. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci 10.1073/pnas.1111561108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Uhlenbeck OC 1987. An RNA mutation that increases the affinity of an RNA-protein interaction. Nucleic Acids Res 15: 10483–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani F, Lind J, Damberg P, Adams SR, Tsien RY, Graslund AO 2009. Hairpin structure of a biarsenical-tetracysteine motif determined by NMR spectroscopy. J Am Chem Soc 131: 4613–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE 2004. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y 2010. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol 12: 543–552 [DOI] [PubMed] [Google Scholar]
- Nathans D 1964. Puromycin inhibition of protein synthesis: Incorporation of puromycin into peptide chains. Proc Natl Acad Sci 51: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo JT, Champion JA, Mahdavi A, Tanrikulu IC, Beatty KE, Connor RE, Yoo TH, Dieterich DC, Schuman EM, Tirrell DA 2009. Cell-selective metabolic labeling of proteins. Nat Chem Biol 5: 715–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Kirillov SV, Cooperman BS 2007. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell 25: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Qin H, Cooperman BS 2009. Synthesis and functional activity of tRNAs labeled with fluorescent hydrazides in the D-loop. RNA 15: 346–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Graham KF, Haddick PC, Jan YN, Jan LY 2006. Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science 314: 144–148 [DOI] [PubMed] [Google Scholar]
- Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J 2006. Visualization of mRNA translation in living cells. J Cell Biol 175: 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M 1986. Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science 234: 364–368 [DOI] [PubMed] [Google Scholar]
- Rostovtsev VV, Green LG, Fokin VV, Sharpless KB 2002. A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl 41: 2596–2599 [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY 2008. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods 5: 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH 2004. Dynamics of single mRNPs in nuclei of living cells. Science 304: 1797–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WB, Starck SR, Roberts RW, Schuman EM 2005. Dopaminergic stimulation of local protein synthesis enhances surface expression of GluR1 and synaptic transmission in hippocampal neurons. Neuron 45: 765–779 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Roberts RW 2002. Puromycin oligonucleotides reveal steric restrictions for ribosome entry and multiple modes of translation inhibition. RNA 8: 890–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Green HM, Alberola-Ila J, Roberts RW 2004. A general approach to detect protein expression in vivo using fluorescent puromycin conjugates. Chem Biol 11: 999–1008 [DOI] [PubMed] [Google Scholar]
- Stepanenko OV, Verkhusha VV, Kuznetsova IM, Uversky VN, Turoverov KK 2008. Fluorescent proteins as biomarkers and biosensors: Throwing color lights on molecular and cellular processes. Curr Protein Pept Sci 9: 338–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Wall NR, Aakalu GN, Schuman EM 2004. Regulation of dendritic protein synthesis by miniature synaptic events. Science 304: 1979–1983 [DOI] [PubMed] [Google Scholar]
- Takizawa PA, Vale RD 2000. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Natl Acad Sci 97: 5273–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS 2010. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329: 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, Zhao X, Lukyanov S, Matz M, Kim S, et al. 2000. “Fluorescent timer”: Protein that changes color with time. Science 290: 1585–1588 [DOI] [PubMed] [Google Scholar]
- Valegard K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L 1994. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature 371: 623–626 [DOI] [PubMed] [Google Scholar]
- Van Engelenburg SB, Nahreini T, Palmer AE 2010. FACS-based selection of tandem tetracysteine peptides with improved ReAsH brightness in live cells. Chembiochem 11: 489–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, Martin KC 2009. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science 324: 1536–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Piatkevich KD, Lionnet T, Singer RH, Verkhusha VV 2011. Modern fluorescent proteins and imaging technologies to study gene expression, nuclear localization, and dynamics. Curr Opin Cell Biol 23: 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmolinsky MB, Haba GL 1959. Inhibition by puromycin of mino acid incorporation into protein. Proc Natl Acad Sci 45: 1721–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D 2008. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 134: 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]