Abstract
During lytic infections, the herpes simplex virus (HSV) virion host shutoff (Vhs) endoribonuclease degrades many host and viral mRNAs. Within infected cells it cuts mRNAs at preferred sites, including some in regions of translation initiation. Vhs binds the translation initiation factors eIF4H, eIF4AI, and eIF4AII, suggesting that its mRNA degradative function is somehow linked to translation. To explore how Vhs is targeted to preferred sites, we examined the in vitro degradation of a target mRNA in rabbit reticulocyte lysates containing in vitro-translated Vhs. Vhs caused rapid degradation of mRNAs beginning with cleavages at sites in the first 250 nucleotides, including a number near the start codon and in the 5′ untranslated region. Ligation of the ends to form a circular mRNA inhibited Vhs cleavage at the same sites at which it cuts capped linear molecules. This was not due to an inability to cut any circular RNA, since Vhs cuts circular mRNAs containing an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) at the same sites as linear molecules with the IRES. Cutting linear mRNAs at preferred sites was augmented by the presence of a 5′ cap. Moreover, mutations that altered the 5′ proximal AUG abolished Vhs cleavage at nearby sites, while mutations that changed sequences surrounding the AUG to improve their match to the Kozak consensus sequence enhanced Vhs cutting near the start codon. The results indicate that mutations in an mRNA that affect its translation affect the sites at which it is cut by Vhs and suggest that Vhs is directed to its preferred cut sites during translation initiation.
INTRODUCTION
During lytic herpes simplex virus (HSV) infections, viral and cellular gene expressions are regulated at multiple steps of mRNA metabolism (1, 2), including transcription (2–8), splicing (2, 9–12), transport from the nucleus to the cytoplasm (2, 13–20), translation (21–32), and stabilization of mRNAs in the cytoplasm (33–43). Among the posttranscriptional controls are the regulation of mRNA half-lives (36–42) and translation (23, 29) by the HSV virion host shutoff (Vhs) protein (UL41). Vhs is an endoribonuclease (33, 44–47) that is a minor component of virions (48–50). At early times, copies of Vhs from infecting virions accelerate the degradation of many housekeeping and constitutively expressed cellular mRNAs (40, 42, 48, 51–55), with a concomitant downturn in the levels of those mRNAs and in the synthesis of the proteins that they encode (52, 56, 57). Following the onset of viral transcription, Vhs ensures the rapid turnover of most, if not all, viral mRNAs (36, 37, 57–60). In so doing, it helps determine viral mRNA levels and facilitates the sequential expression of different classes of viral mRNAs (38). Significantly, during animal infections, Vhs impedes the establishment of an interferon-induced antiviral state (61–67). It blocks the activation of dendritic cells (68–70) and inhibits other components of the innate and adaptive immune responses (41, 71–75). As such, it is an important determinant of HSV virulence (61–67, 76–85).
Although Vhs degrades many cellular and viral transcripts, not all mRNAs are equally sensitive. For unknown reasons, a number of host mRNAs appear to be refractory, or at least less sensitive to Vhs degradation, and actually increase in abundance following infection (43, 86–93). These include some mRNAs that contain AU-rich instability elements (AREs) (43, 86–88, 92, 94, 95), as well as others that lack these elements (43, 86). At least one ARE-containing host mRNA is stabilized through the activation of the p38 mitogen-activated protein kinase (MAPK) pathway by the HSV-1 immediate-early protein ICP27 (94, 95). HSV also has been reported to cause the Vhs-dependent stabilization of deadenylated full-length and 3′ truncated forms of some ARE-containing mRNAs (86–88, 93). Whether this is due to the direct action of Vhs on these mRNAs or is a downstream consequence of Vhs degradation of mRNAs encoding proteins that affect the half-lives of ARE-containing mRNAs remains to be determined. In addition, recent studies have suggested that the rate and overall extent of Vhs-mediated mRNA degradation may depend upon the cell type that is infected (23, 29).
For the many mRNAs that are sensitive to Vhs, a central unanswered question is how Vhs is targeted to the mRNAs and to preferred cut sites within them. In the absence of key cellular factors, Vhs lacks the cleavage specificity that it shows in vivo (33, 45–47). Thus, a preparation of Vhs from partially purified virions (47), or a purified complex of recombinant Vhs and the cellular translation factor eIF4H (33), does not distinguish mRNAs from non-mRNAs and cleaves target RNAs at many sites. In addition, a purified glutathione S-transferase (GST)-Vhs fusion protein has been reported to cleave single-stranded RNAs after C's and U's (45). In contrast, within infected cells or cytoplasmic extracts, Vhs is targeted to mRNAs, as opposed to non-mRNAs (36, 37, 40, 42, 47, 58, 96–98), and cleaves those mRNAs at preferred sites (44, 86, 93, 99–102). While the Vhs cut sites have been mapped in only a few mRNAs, for several they fall in regions of translation initiation (44, 99–102). Thus, in infected cells Vhs degrades sequences near the 5′ end of the viral thymidine kinase (TK) mRNA prior to those at the 3′ end (100), while in rabbit reticulocyte lysates, in vitro-translated Vhs cuts signal recognition particle (SRP) mRNA at sites in the 5′ quadrant of the transcript and, in particular, at several sites near the start codon (44). In addition, in vitro-translated Vhs cuts mRNAs containing an encephalomyocarditis (EMCV) internal ribosome entry site (IRES) at sites downstream from the IRES (99).
A potential clue to the mechanism of Vhs targeting comes from our observation that Vhs binds the cellular translation initiation factors eIF4H, eIF4AI, and eIF4AII (33–35). eIF4AI and 4AII are isoforms of an ATP-dependent RNA helicase that is a component of the cap-binding complex eIF4F. They play essential roles in cap-dependent initiation by unwinding mRNA secondary structure near the 5′ cap to allow the binding and subsequent scanning of the 40S ribosomal subunit (103–107). eIF4H is a helicase accessory factor that binds to and stimulates the helicase activities of eIF4AI and 4AII (104, 106, 108–113). Apparently, eIF4H is essential for Vhs activity, since small interfering RNA (siRNA)-mediated depletion of eIF4H from cells prior to infection inhibits Vhs degradation of housekeeping mRNAs (39). Furthermore, to date, every mutation in Vhs that abrogates its interaction with eIF4H also abolishes its ability to degrade constitutively expressed mRNAs, even if the mutant proteins retain endonuclease activity in vitro (33–35). Vhs also associates with the eIF4F cap-binding complex, presumably through its interaction with eIF4AI and 4AII (114).
To further investigate the mechanism by which Vhs is directed to its preferred cut sites, in this study we examined in vitro degradation of target mRNAs in rabbit reticulocyte lysates containing in vitro-translated Vhs. Vhs caused rapid degradation of pBK2 mRNA, a modified form of the HSV-1 thymidine kinase (TK) mRNA, beginning with cleavages at sites in the first 250 nucleotides, including a number near the start codon and in the 5′ untranslated region (UTR). Ligation of the ends of pBK2 mRNA to form a circle inhibited translation and inhibited Vhs cleavage at the same sites at which it cuts capped linear molecules. This was not due to an inability of Vhs to cut any circular RNA, since it cut circular mRNAs containing an EMCV IRES at the same sites at which it cuts linear molecules with the IRES. Cleavage of linear pBK2 mRNA at preferred sites was greatly augmented by the presence of a 5′ cap, while blocking ATP hydrolysis with a nonhydrolyzable analogue of ATP inhibited Vhs degradation of the mRNA. Furthermore, mutations that altered the 5′ proximal AUG abolished Vhs cleavage at nearby sites, while mutations that changed the sequence surrounding the AUG to improve its match to the Kozak consensus sequence enhanced Vhs cutting at sites near the start codon. The results indicate that mutations or alterations in a target mRNA that affect its initiation of translation can affect the sites at which it is cut by Vhs and suggest that Vhs is directed to its preferred cut sites during translation initiation.
MATERIALS AND METHODS
Plasmids.
The plasmid pKOSamp contains the Vhs open reading frame from HSV-1, strain KOS, cloned into pcDNA1.1amp (Invitrogen) downstream from a promoter recognized by T7 RNA polymerase (115, 116). It was used to produce the Vhs protein by in vitro transcription and translation.
pBK2 and pCITE-RLuc were used to produce target mRNAs for in vitro Vhs mRNA degradation assays. In vitro transcription of EcoRI-linearized pBK2 with SP6 RNA polymerase yields a 1,244-nucleotide mRNA containing the HSV-1 thymidine kinase (TK) open reading frame, preceded by a 91-nucleotide 5′ untranslated region (UTR) and terminated with a 35-nucleotide poly(A) tail encoded by the plasmid (100). It is translated by 5′ end-dependent ribosome scanning. To construct pCITE-RLuc, a 1,716-nucleotide XhoI-BamHI fragment containing the Renilla luciferase (RLuc) open reading frame was excised from pGEMLUC (Promega) and inserted between the corresponding sites of the vector pCITE-4c(+) (Invitrogen). In vitro transcription of SpeI-linearized pCITE-RLuc using T7 RNA polymerase produces an mRNA encoding RLuc translated from an EMCV IRES.
Synthesis of target mRNAs.
Target mRNAs for in vitro mRNA degradation assays were produced by in vitro transcription of EcoRI-linearized pBK2 or SpeI-linearized pCITE-RLuc using SP6 or T7 RNA polymerase, respectively. Capped internally labeled pBK2 mRNA was synthesized using an SP6 Riboprobe in vitro transcription system (Promega) in reactions that included 0.5 mM (each) rATP, rUTP, and rCTP, 50 μM rGTP, 50 μCi [α-32P]GTP, and 0.5 mM Ribo m7G cap analog (Promega). After in vitro transcription, template DNA was removed by digestion with RQ1 DNase (Promega). mRNAs were extracted twice with phenol-chloroform, precipitated from ethanol, and resuspended in a small volume of water.
To produce pBK2 mRNA with a 32P-labeled cap and a cold poly(A) tail, uncapped unlabeled pBK2 mRNA with a 35-nucleotide poly(A) tail encoded by the plasmid was synthesized using an SP6 RiboMAX in vitro transcription kit (Promega). Samples of mRNA were heated at 65°C for 8 min to melt secondary structures, quenched on ice, and labeled by the addition of a 32P-labeled cap using vaccinia virus capping enzyme from a ScriptCap m7G capping system (Epicentre) in the presence of [α-32P]GTP. After the addition of a cap, mRNAs were extracted with phenol-chloroform, precipitated from ethanol, resuspended in water, and electrophoresed through gels of 1.4% (wt/vol) agarose in Tris-borate-EDTA (TBE) buffer (117). Bands containing intact mRNAs were excised. The mRNAs were extracted using Bio-Rad Freeze 'N Squeeze gel extraction spin columns, precipitated from ethanol, and resuspended in water.
Target mRNAs with an unlabeled cap and a cold poly(A) tail were produced as described above, except that the capping reaction mixture contained unlabeled GTP and no [α-32P]GTP. These were used in experiments in which mRNA degradation products were detected by primer extension.
Uncapped mRNAs containing a 32P-labeled 5′ monophosphate were synthesized by in vitro transcription of linearized pBK2 with SP6 RNA polymerase to yield an mRNA with a 5′ triphosphate. This mRNA was dephosphorylated using calf intestine alkaline phosphatase and labeled by the addition of a 32P-5′ monophosphate using polynucleotide kinase and [γ-32P]ATP, according to procedures described for the KinaseMax 5′ end-labeling kit (Ambion).
mRNAs with an unlabeled cap and a 32P-labeled poly(A) tail were transcribed in vitro and then capped in capping reactions that lacked [α-32P]GTP. A labeled poly(A) tail was added using a poly(A) polymerase tailing kit (Epicentre) and [α-32P]ATP.
Preparation of circular mRNAs.
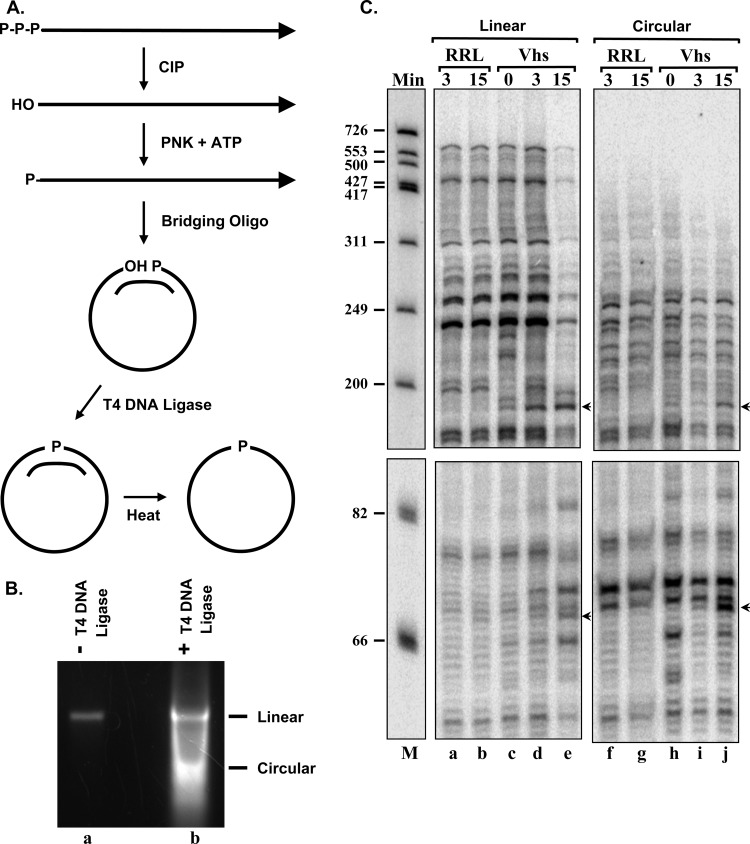
Circular forms of the pBK2 and pCITE-RLuc mRNAs were prepared using a modification of the procedure described by Chen and Sarnow (118, 119). Initially, linear mRNAs with 5′ triphosphates were prepared by in vitro transcription of linearized pBK2 and pCITE-RLuc using the SP6 and T7 RiboMAX in vitro transcription kits (Promega). The mRNAs were dephosphorylated with calf intestine alkaline phosphatase according to procedures described for the KinaseMax 5′-end-labeling kit (Ambion), after which the phosphatase was removed using phosphatase removal reagent (Ambion), and the mRNAs were precipitated from ethanol. The mRNAs were resuspended in a small volume of water and incubated with polynucleotide kinase in the presence of 1 mM ATP to add a 5′ monophosphate. The reactions were stopped by heating to 90°C for 2 min and adjusted to final concentrations of 10 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, and 100 mM NaCl. Each reaction received a 6.5 M excess of a circularization oligonucleotide that contained sequences complementary to both the extreme 5′ and 3′ ends of the mRNA and was designed to hold it in a circular configuration following annealing. The circularization oligonucleotide for pBK2 mRNA was 5′-TGCAGCCCAAGCTTGTATTCGGGCGTCGCAGATCGTCGGT-3′, while that for pCITE-RLuc mRNA was 5′-TTTTTTTTTTTCAAATTTGTCCCGCTTAATTAAGGCCAAT-3′. After addition of the oligonucleotides, the samples were heated to 90°C for 5 min and allowed to cool slowly to room temperature to anneal the oligonucleotide. The samples were incubated with T4 DNA ligase overnight at room temperature to join the 5′ and 3′ ends of the mRNAs. The mRNAs were precipitated from ethanol, resuspended in water, heated briefly to melt the circularization oligonucleotide from the mRNA, and electrophoresed through a 1.6% (wt/vol) agarose gel in TBE buffer containing 0.5 μg/ml of ethidium bromide. Included was a control lane with a small amount of mRNA that had been through the procedure described above, except for ligation, to allow for identification of the bands containing linear and circular forms of the mRNA. The bands containing the circular mRNAs were excised from the gel, after which the mRNAs were isolated using Freeze 'N Squeeze gel extraction spin columns, precipitated from ethanol, and resuspended in a small volume of water.
Synthesis of Vhs mRNA.
Capped Vhs mRNA, for in vitro translation of the Vhs protein, was produced by the transcription of linearized pKOSamp using a T7 RiboMAX in vitro transcription kit (Promega) in reaction mixtures containing 3 mM Ribo m7G cap analog.
In vitro Vhs mRNA degradation assays.
In vitro Vhs mRNA degradation assays were performed using a modification of the procedure described by Smiley and colleagues (44, 120). Initially, Vhs protein was produced by in vitro translation of capped in vitro-transcribed Vhs mRNA using a Flexi rabbit reticulocyte lysate system (Promega). Reaction mixtures contained 14 μg of Vhs mRNA, 33 μl of Flexi rabbit reticulocyte lysate, 20 μM all 20 amino acids, 40 units of RNasin, 1 mM magnesium acetate, 70 mM KCl, and 2 mM dithiothreitol in a total volume of 50 μl. In vitro translation reaction mixtures were incubated for 1.5 h at 30°C, after which they were chilled on ice until the start of the in vitro mRNA degradation reactions. Mock in vitro translation reactions were performed as described above, except they did not receive any Vhs mRNA.
To initiate the in vitro mRNA degradation reactions, target mRNA was added to 30 μl of either the Vhs or mock in vitro translation reactions, and the reactions were placed at 30°C. Aliquots were removed immediately (0 min) or at various times after the start of incubation, supplemented with 15 μg of tRNA, and extracted with TRIzol (Invitrogen), and the mRNAs were precipitated with isopropanol. 32P-labeled mRNAs were analyzed by electrophoresis through a 1.3% or 1.4% (wt/vol) agarose-formaldehyde gel cast in NorthernMax denaturing gel buffer (Ambion) or an 8% polyacrylamide-8 M urea sequencing gel cast in TBE buffer. The gels were dried, and the RNAs were visualized using a Storm PhosphorImager (model 840; Molecular Dynamics, Inc.). For assays involving unlabeled target mRNAs, the reaction products were analyzed by primer extension.
To compare the rates of mRNA degradation caused by different amounts of Vhs, mRNA degradation assays were performed using serial 2-fold dilutions of in vitro-translated Vhs prepared by diluting the standard Vhs in vitro translation reaction with naïve rabbit reticulocyte lysate to yield mixtures containing 1×, 0.5×, and 0.25× Vhs.
Primer extension analysis of mRNAs.
Unlabeled pBK2 and pCITE-RLuc mRNAs were analyzed by primer extension using a primer extension system (AMV reverse transcriptase; Promega) according to protocols recommended by the manufacturer. Primer extensions of pBK2 mRNA were performed using the primers pe165, pe235, and pe325. Reverse transcriptase-mediated extensions of these primers to the 5′ end of intact pBK2 mRNA yield products of 165, 235, and 325 nucleotides, respectively. The sequence of pe165 is 5′-TCGGTTGCTATGGCCGCGAGAACGC-3′, that of pe235 is 5′-CGTGGGCATTTTCTGCTCCAGGCGG-3′, and that of pe325 is 5′-GATATCGTCGCGCGAACCCAGGGCC-3′. Primer extension analysis of pCITE-RLuc mRNA was performed with the primer peCITE-RLuc, which has the sequence 5′-TCATAGCCTTATGCAGTTGC-3′. The primer anneals to a sequence from 163 to 182 nucleotides downstream from the 3′ end of the EMCV IRES. Primer extension to the 5′ end of intact pCITE-RLuc mRNA yields a product of 700 nucleotides.
Primers were 5′ end labeled by incubating 10 pmol of primer with 10 U of T4 polynucleotide kinase (Ambion) and 30 μCi of [γ-32P]ATP in a total volume of 10 μl at 37°C for 10 min. The samples were heated to 90°C for 2 min to inactivate the enzyme and diluted to 100 fmol/μl with water.
To initiate primer extension, 5 μl of mRNA was combined with 1 μl of labeled primer plus 5 μl of 2× AMV primer extension buffer (Promega). The mixtures were heated at 67°C (or a temperature near the melting point of the primer) for 20 min and then cooled at room temperature for 10 min to allow annealing of the primer. Each reaction mixture received 5 μl of 2× AMV primer extension buffer (Promega), 1.4 μl of 40 mM sodium pyrophosphate, 1.6 μl of H2O, and 1 unit of AMV reverse transcriptase. The reaction mixtures were incubated at 42°C for 30 min, after which 20 μl of gel loading buffer was added. Samples were heated at 90°C for 10 min, loaded onto a preheated 8% polyacrylamide-8 M urea gel cast in TBE buffer, and electrophoresed at a constant power of 50 W for 1.5 h on a Bio-Rad Sequi-Gen GT sequencing cell. The gels were dried, and the RNAs were visualized using a Storm PhosphorImager (model 840; Molecular Dynamics, Inc.).
Site-directed mutagenesis.
A QuikChange Lightning multisite-directed mutagenesis kit (Stratagene) was used for site-directed mutagenesis of pBK2 to generate plasmids encoding mRNAs with mutations in and around the first AUG from the 5′ cap. This procedure utilizes single-stranded mutagenic primers and thermal cycling to introduce the desired mutations into the target plasmid. Two mutant forms of pBK2 were constructed. In the first, which was designated AUG1→CCC, the 5′ proximal AUG was changed to CCC. This was done using the mutagenic primer 5′-CCTTGTAGAAGCGCGTCCCGCTTCGTACCCCTGCC-3′. The 5′ proximal AUG of pBK2 is in a suboptimal context compared to the Kozak consensus sequence of (A/G)ccAUGG, where the start codon is in bold and strongly conserved bases are capitalized (121–126). For the second mutant pBK2 allele, designated AUG1-OPT, the 5′ proximal AUG was maintained, and the surrounding nucleotides were altered to place it in an optimal Kozak context. This was done using the mutagenic primer 5′-GCGCCTTGTAGAAGCCACCATGGCTTCGTACCCC-3′. For both mutagenic primers, the nucleotides in bold are at the position of the AUG of wild-type pBK2, and the underlined nucleotides are those that have been changed. Both mutations were confirmed by sequencing the mutant plasmids.
Oligonucleotides and primers.
All oligonucleotides and primers were purchased from IDT Technologies (Coralville, IA).
RESULTS
Vhs cleavage of pBK2 mRNA.
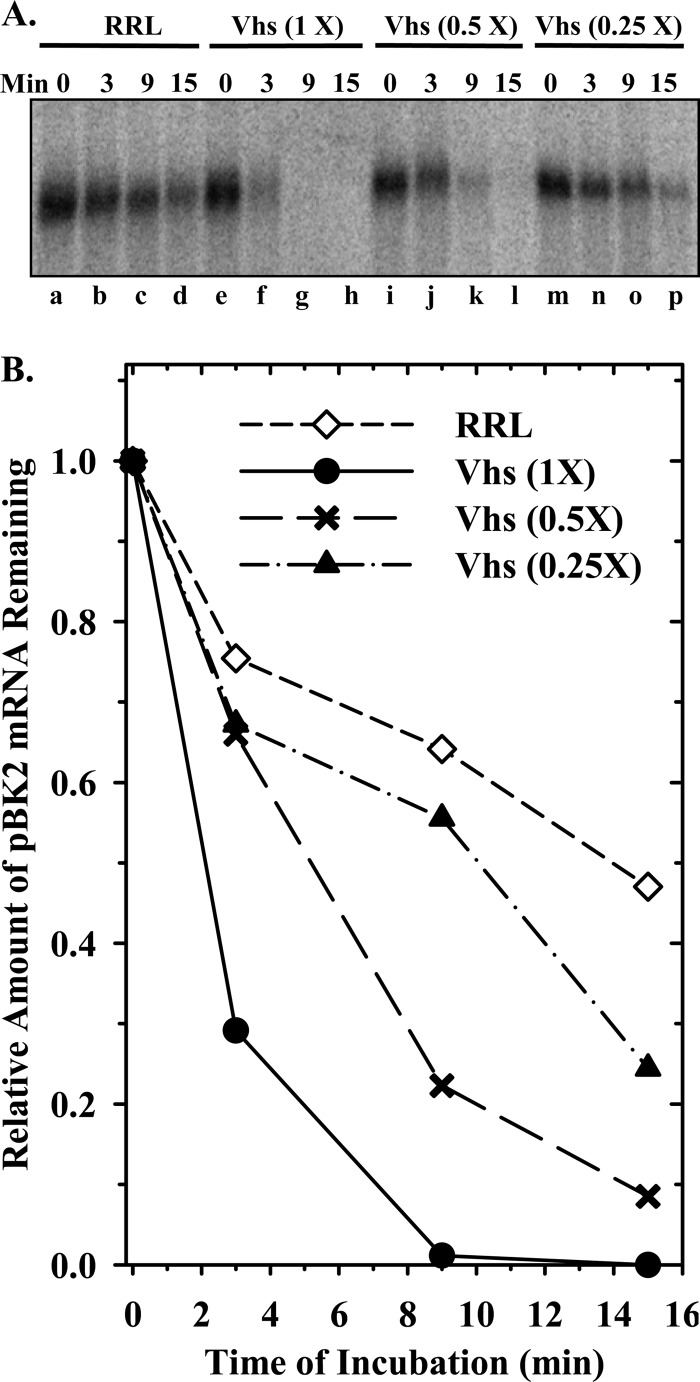
Before one can determine the mechanism by which Vhs is targeted to preferred cut sites, the locations of those sites must be mapped in specific mRNAs. To date, Vhs cut sites have been mapped in relatively few mRNAs (44, 86, 93, 99, 100, 102). Therefore, at the start of this study, we mapped the Vhs cleavage sites in pBK2 mRNA, a modified form of the HSV-1 thymidine kinase (TK) mRNA (100), using an in vitro mRNA degradation system (44, 99) in which the decay of pBK2 mRNA was followed in rabbit reticulocyte lysates containing in vitro-translated Vhs. In this system, Vhs-mediated decay is similar to that observed in intact cells (36, 37, 40, 42, 58, 97) or cytoplasmic extracts from infected cells (47, 96, 98), in that Vhs is targeted to mRNAs, as opposed to non-mRNAs, even though the purified enzyme does not distinguish one kind of RNA from another (33, 45, 46). Our initial experiments focused on preliminary characterization of the system and determination of whether the in vitro decay of pBK2 mRNA is similar to the decay of TK mRNA in infected cells, in which Vhs-mediated decay begins with the degradation of sequences near the 5′ end of the mRNA (100). Figure 1 shows the results of an experiment in which 5′ cap-labeled pBK2 mRNA was incubated for various intervals in lysates containing serial 2-fold dilutions of in vitro-translated Vhs. As expected, Vhs caused dose-dependent degradation of pBK2 mRNA, with the most rapid degradations occurring in reactions that contained the largest amounts of Vhs.
Fig 1.
Dose-dependent degradation of mRNAs by Vhs. Vhs protein was produced by in vitro translation. Once translation was complete, the reaction [Vhs (1×)] was diluted using naïve rabbit reticulocyte lysate to yield lysates containing serial 2-fold dilutions of Vhs. mRNA degradation reactions were performed using cap-labeled pBK2 target mRNA and rabbit reticulocyte lysates containing 1×, 0.5×, or 0.25× amounts of Vhs or no Vhs (RRL). (A) Aliquots were removed after 0, 3, 9, or 15 min, and the RNAs were electrophoresed through a 1.3% (wt/vol) agarose-formaldehyde gel; (B) the relative amounts of full-length mRNAs were quantified using ImageQuant software and plotted to compare the relative mRNA decay rates.
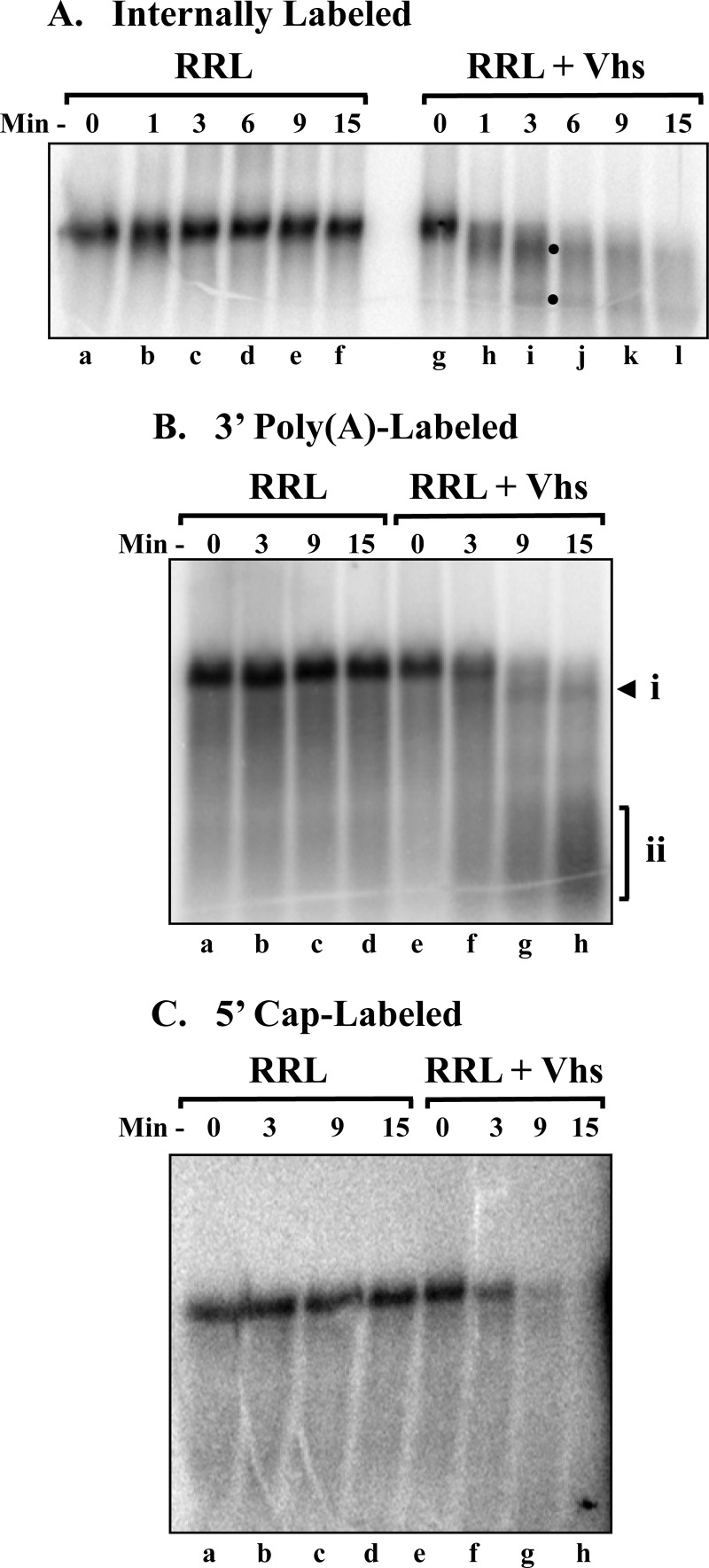
To determine whether Vhs degradation of pBK2 occurs by cleavage at multiple sites throughout the mRNA or initiates with cutting near one end or the other of the molecule, in vitro mRNA degradation reactions were performed with pBK2 mRNA that had been labeled at many internal sites by transcription in the presence of [α-32P]GTP (Fig. 2A). Vhs caused the rapid disappearance of full-length pBK2 mRNA, which was accompanied by the appearance (at the 1- and 3-min time points) of a band migrating slightly faster than the intact mRNA, as well as a faster-migrating band (Fig. 2A, lane i, closed circles). These bands became less abundant and more diffuse with increasing times, consistent with their being decay intermediates. The results are consistent with one of two possibilities: (i) Vhs cleaves pBK2 mRNA beginning near one end or the other of the molecule, or (ii) it cleaves small fragments from both ends of the transcript, leaving a large internal fragment almost the size of the intact mRNA. In either case, fragments cleaved from the ends would be too small to be seen on a 1.3% (wt/vol) agarose gel.
Fig 2.
Vhs degradation of pBK2 mRNA initiates near the 5′ end. Parallel in vitro mRNA degradation reactions were performed using rabbit reticulocyte lysates that contained (RRL + Vhs) or lacked (RRL) Vhs and target pBK2 mRNAs that were internally labeled (A), 3′ poly(A) tail labeled (B), or 5′ cap labeled (C). Aliquots were removed after 0, 1, 3, 6, 9, or 15 min, and the RNAs were electrophoresed through 1.3% (wt/vol) agarose-formaldehyde gels. Bands arising from shorter-than-full-length degradation products are indicated by the solid circles to the right of lane i in panel A and by the arrowhead labeled “i” and the bracket labeled “ii” to the right of lane h in panel B.
To distinguish between these possibilities, in vitro degradation reactions were performed using pBK2 mRNAs that had been labeled posttranscriptionally by the addition of a 32P-labeled cap (Fig. 2C) or a 32P-labeled poly(A) tail (Fig. 2B). The products from reactions with poly(A)-labeled pBK2 (Fig. 2B) were similar in many respects to those produced from internally labeled mRNA. Vhs caused the rapid disappearance of full-length pBK2 mRNA, which was accompanied by the appearance of a labeled band that migrated slightly faster than the intact mRNA (Fig. 2B, lane h, arrowhead). This fragment exhibited an electrophoretic mobility similar to that of the slowest migrating of the degradation intermediates observed for internally labeled mRNA (Fig. 2A) and could have been generated only by the removal of sequences from the 5′ end of the full-length mRNA, since it had a labeled poly(A) tail. With increasing time, there was the accumulation of a broad smear of faster-migrating bands (Fig. 2B, lane h, bracket) consisting of polyadenylated molecules that were truncated to various extents from the 5′ end. In contrast, Vhs digestion of 5′ cap-labeled mRNA (Fig. 2C) resulted in the disappearance of full-length pBK2 without the production of any labeled bands that were large enough to be seen on a 1.3% agarose gel. The results indicate that Vhs degradation of pBK2 mRNA commences with cleavage near the 5′ end of the transcript.
Mapping Vhs cut sites.
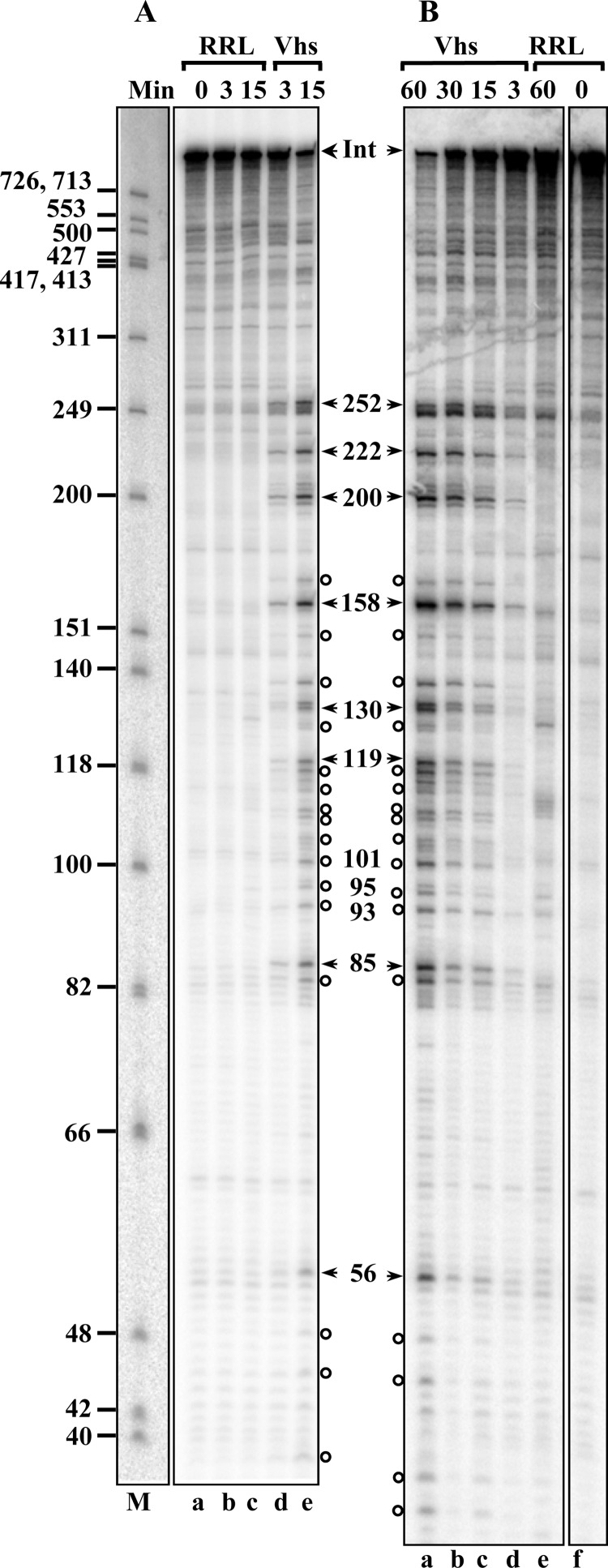
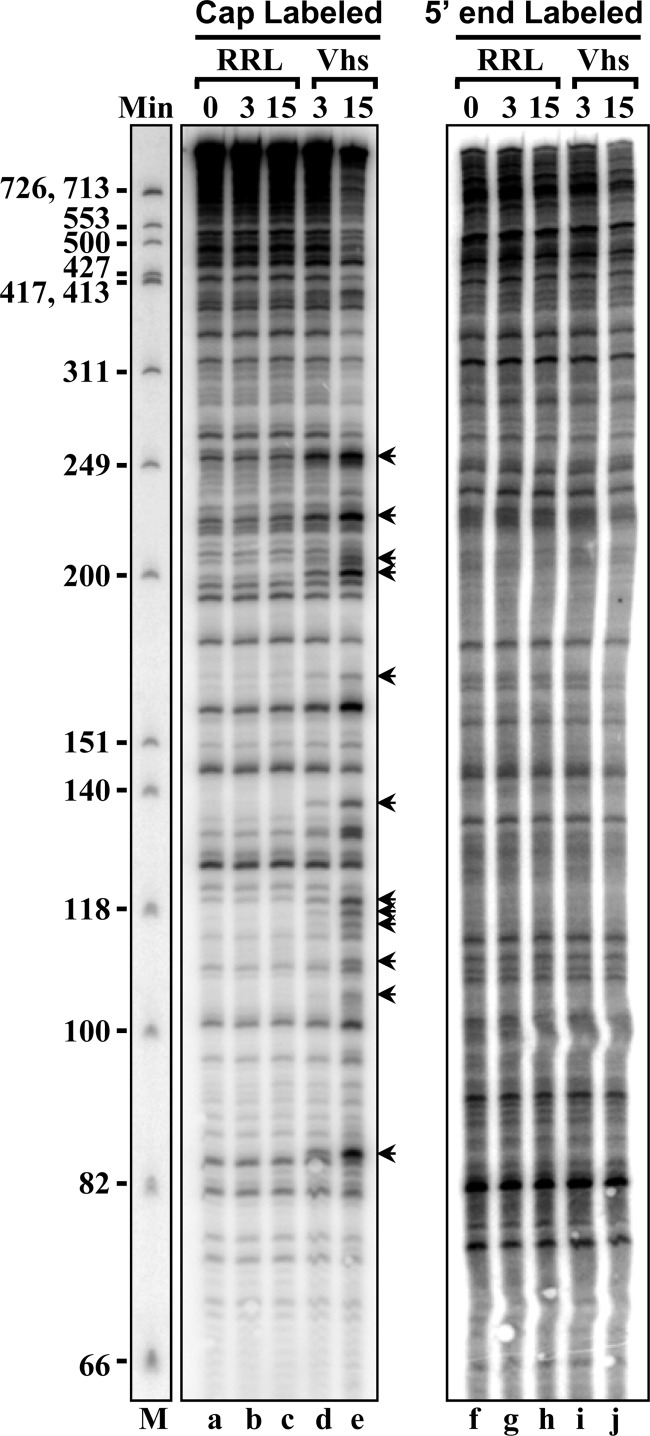
To define the cut sites more precisely, the products of Vhs digestion of cap-labeled pBK2 mRNA were electrophoresed through an 8% polyacrylamide-8 M urea sequencing gel (Fig. 3). Vhs cleaved the population of mRNAs at 8 prominent sites approximately 252, 222, 200, 158, 130, 119, 85, and 56 nucleotides downstream from the cap (Fig. 3A and B, arrows), as well as at a number of less prominent sites in the first 250 nucleotides (Fig. 3, open circles). While cleavage at these sites was easily detectable by 3 min after the start of the reaction, extending the incubation time to as long as 60 min simply led to progressive increases in the abundances of the same degradation products without the generation of prominent new bands, suggesting that the upstream products of Vhs cleavage were relatively stable, at least over a period of 60 min. In contrast, electrophoresis of the products of Vhs digestion of 3′ end-labeled pBK2 mRNA on an 8% polyacrylamide-8 M urea sequencing gel did not reveal any labeled degradation products smaller than 700 nucleotides (the upper limit of resolution on this gel), further supporting the conclusion that Vhs degradation begins with cleavage near the 5′ end of pBK2 (data not shown).
Fig 3.
Mapping Vhs cut sites in cap-labeled pBK2 mRNA. Two identical in vitro mRNA degradation reactions were performed using rabbit reticulocyte lysates that contained (Vhs) or lacked (RRL) Vhs and cap-labeled pBK2 target mRNAs. One was carried out for 15 min (A), while the other was incubated for up to 60 min (B). Aliquots were removed after 0, 3, 15, 30, or 60 min, and the RNAs were electrophoresed through 8% polyacrylamide-8 M urea sequencing gels. The lane labeled “M” in panel A contains labeled marker fragments with the sizes (in nucleotides) of the fragments indicated on the left. The most prominent Vhs degradation products are indicated by the arrowheads to the right of panel A and to the left of panel B, along with the sizes of the products in nucleotides. Less prominent degradation products are designated by open circles. The band formed by intact pBK2 mRNA is indicated by the arrowheads labeled “Int.”
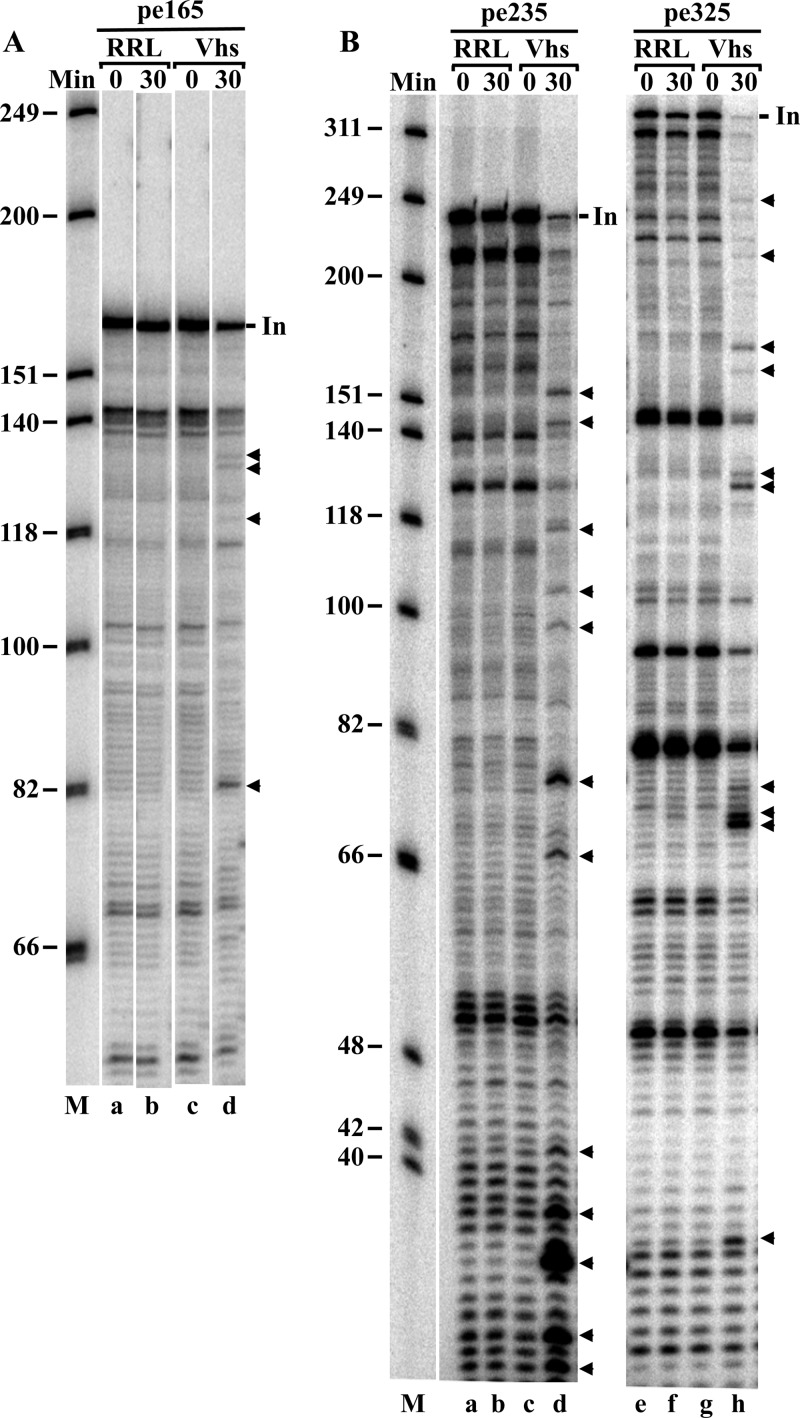
Digestion of cap-labeled mRNAs revealed only the 3′ ends of the upstream products of Vhs cleavage. To examine the 5′ ends of the downstream products, we analyzed Vhs cleavage products by primer extension using primers that anneal to pBK2 mRNA at 3 different sites downstream from the cap (Fig. 4). These primers were chosen because they should allow detection of Vhs cleavage at the sites detected and shown in Fig. 3. Reverse transcription to the end of intact pBK2 mRNA using the primers pe165, pe235, and pe325 resulted in products of 165, 235, and 325 nucleotides, respectively (Fig. 4, lines labeled “In”). Numerous shorter bands also were produced, even in the absence of Vhs, apparently due to pausing or stalling of the reverse transcriptase on the intact mRNA. Therefore, the only primer extension products that were attributed to Vhs cutting were those observed with mRNAs that had been incubated with Vhs for 30 min, but not with mRNAs from the 0-min time point, or mRNAs exposed to rabbit reticulocyte lysates lacking Vhs.
Fig 4.
Mapping Vhs cut sites in pBK2 mRNA by primer extension. In vitro mRNA degradation reactions were performed using rabbit reticulocyte lysates that contained (Vhs) or lacked (RRL) Vhs and unlabeled pBK2 target mRNA. Aliquots were removed after 0 or 30 min, and the RNAs were analyzed by primer extension using the 5′-end-labeled primers pe165 (A), pe235 (B, lanes a through d), and pe325 (B, lanes e through h). The primer extension products were electrophoresed through 8% polyacrylamide-8 M urea sequencing gels. All of the samples were from the same experiment. Those analyzed using the primer pe165 were electrophoresed through the gel shown in panel A. Samples analyzed with the primers pe235 and pe325 were electrophoresed through a second gel shown in panel B. Bands due to prominent Vhs cleavage sites in pBK2 are indicated by arrowheads to the right of panel A, lane d, and to the right of panel B, lanes d and h. Bands due to extension of the primers to the end of intact pBK2 mRNA are indicated by the short lines labeled “In.”
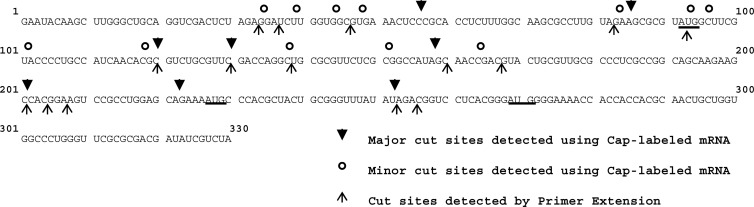
A number of cut sites were identified using each of the primers (Fig. 4, arrowheads). The locations of these sites are indicated in Fig. 5 by the upward-pointing arrows below the first 330 nucleotides of the pBK2 mRNA sequence. For comparison, the major cut sites identified in Fig. 3 by the digestion of cap-labeled mRNAs are indicated by the downward-pointing arrows above the sequence, while some of the less prominent sites are identified by open circles. Many cut sites were identified by both primer extension and digestion of cap-labeled mRNA, confirming the previous report by Smiley and colleagues that Vhs is an endonuclease rather than an exonuclease (44). Several Vhs cut sites were located near the first three AUG codons of pBK2 mRNA. The first two were surrounded by suboptimal contexts, compared to the Kozak consensus sequence (123–127), while the third AUG was in an optimal context. In vitro translation of pBK2 mRNA has been reported to yield one prominent polypeptide of the size expected for initiation from the first AUG, as well as several less prominent polypeptides (100), suggesting that translation initiates by leaky scanning, at least part of the time, in vitro.
Fig 5.
Summary of Vhs cut sites identified by analysis of Vhs cutting of 5′ cap-labeled pBK2 mRNA and primer extension analysis of unlabeled pBK2 mRNA. The sequence of the first 330 nucleotides of pBK2 mRNA is shown. Downward-pointing arrowheads above the sequence indicate the most prominent Vhs cut sites identified by Vhs cutting of 5′ cap-labeled pBK2 mRNA. Open circles above the sequence indicate less prominent Vhs cut sites identified using cap-labeled pBK2 mRNA. Upward-pointing arrows below the sequence indicate the location of cut sites identified by primer extension analysis of Vhs degradation products. The first three AUG codons (beginning at nucleotides 92, 227, and 269) of pBK2 mRNA are underlined.
Ligating the ends of pBK2 mRNA to form a circle significantly reduces Vhs cleavage at the sites where it cuts capped linear molecules.
In their classic study, Chen and Sarnow showed that circular mRNAs containing an EMCV IRES can be translated in vitro, even though a circular form of an mRNA that is normally translated by cap-dependent ribosome scanning cannot be translated in vitro (118). The locations of Vhs cut sites (Fig. 3 to 5) (44, 99, 100), together with the fact that Vhs binds eIF4H, eIF4AI, and eIF4AII (33–35), suggest that Vhs targeting occurs during the initiation of translation. If this is the case, its targeting to preferred sites in mRNAs that are translated by cap-dependent scanning would be expected to require that the mRNAs have a 5′ end, even though Vhs is an endonuclease. Alternatively, since Vhs cuts linear mRNAs downstream from an EMCV IRES (99), cleavage of mRNAs containing this IRES should not require a 5′ end. To test these predictions, we compared Vhs cutting of circular and capped linear forms of pBK2 mRNA and pCITE-RLuc mRNA, which encodes Renilla luciferase translated from an EMCV IRES.
Circular forms of pBK2 and pCITE-RLuc mRNAs were prepared by annealing linear mRNAs to bridging oligonucleotides that juxtaposed the 5′ and 3′ ends, followed by ligation with T4 DNA ligase, heating of the mRNAs to dissociate the bridging oligonucleotides, and purification of the circular mRNAs by agarose gel electrophoresis (Fig. 6A and B). Circular and capped linear forms of the mRNAs then were incubated in rabbit reticulocyte lysates that contained or lacked in vitro-translated Vhs, and mRNA cleavage was monitored by primer extension. pCITE-RLuc mRNA was analyzed using a primer that anneals to a sequence from 150 to 170 nucleotides downstream from the RLuc start codon. Reverse transcription of intact linear mRNA yielded a runoff transcript of 700 nucleotides (Fig. 6C, lanes a through e). In contrast, reverse transcription of circular pCITE-RLuc mRNA did not yield a distinct runoff transcript (Fig. 6C, lanes f through j). Instead, the reverse transcripts appeared to fade out near the top of the gel, presumably because reverse transcription of different molecules stopped at many different sites around the circle. This absence of a distinct 700-nucleotide runoff transcript was strong evidence that the pCITE-RLuc mRNA had been circularized efficiently. Primer extension revealed that Vhs cut linear pCITE-RLuc mRNA at three prominent sites (Fig. 6C, lane e, arrowheads). Two were closely spaced approximately 100 nucleotides downstream from the RLuc start codon, while the third was within the 3′ end of the IRES 20 nucleotides upstream of the start codon. Of particular relevance to this study, Vhs readily cut circular pCITE-RLuc mRNA at the same three sites as it cut capped linear molecules (Fig. 6C, compare lanes h and j to lanes d and e).
Fig 6.
Vhs cuts circular mRNAs that have an EMCV IRES. (A) Diagram of the procedure for preparing circular mRNAs. (B) Linear and circular forms of the RNA were resolved by electrophoresis through a 1.6% (wt/vol) agarose gel. Lane b contains RNAs that were taken through the circularization procedure, and lane a contains an aliquot of RNA that was taken through the same procedure, except that T4 DNA ligase was omitted from the ligation reaction. (C) In vitro RNA degradation reactions were performed using rabbit reticulocyte lysates that contained (Vhs) or lacked (RRL) Vhs and capped linear or circular pCITE-RLuc RNA. Aliquots were removed after 0, 3, or 15 min, and the RNAs were analyzed by primer extension. Bands due to Vhs cleavage sites are indicated by arrows to the right of lanes e and j.
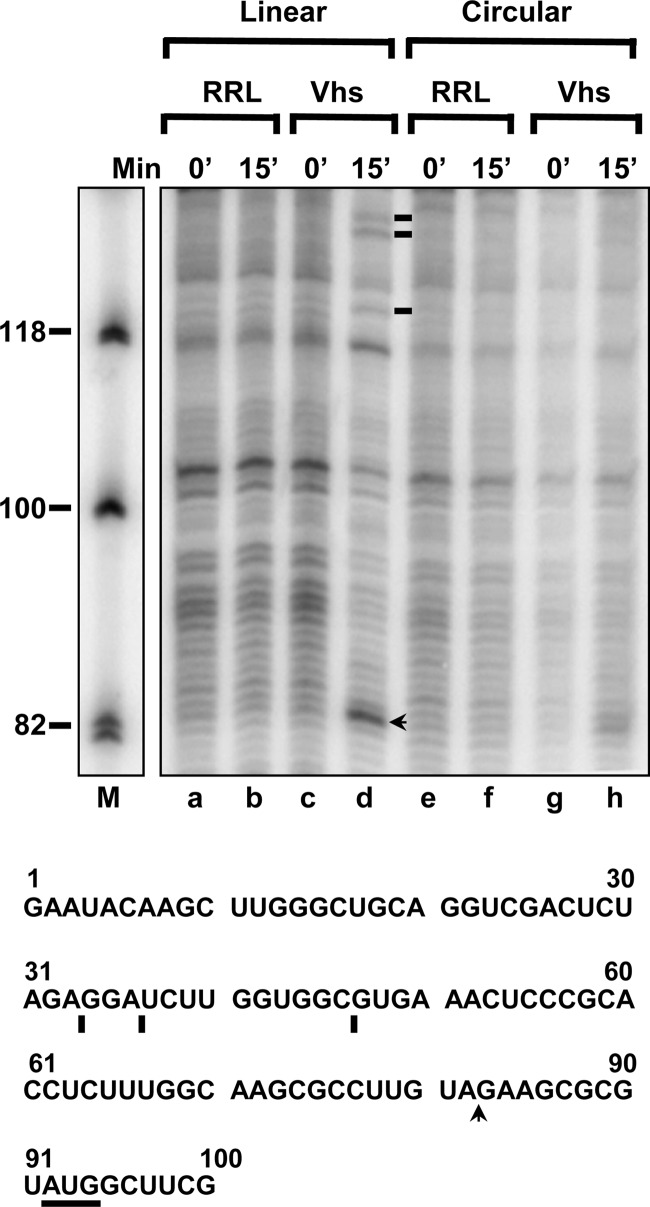
Primer extension using the primer pe165 revealed that Vhs cut capped linear pBK2 mRNA at a prominent site 9 nucleotides upstream from the start codon (Fig. 7, lane d, arrow), as well as at 3 less prominent sites farther upstream in the 5′ UTR (Fig. 7, lane d, lines). These are the same sites that are shown in Fig. 4A and summarized in Fig. 5. In contrast, Vhs cleavages of circular mRNA at these sites were undetectable or greatly reduced (Fig. 7, lane h). Thus, no Vhs cleavage was detected at the three upstream sites in the 5′ UTR, while a faint, indistinct smudge of approximately 82 nucleotides was seen for the 15-min time point of Vhs-digested circular mRNA (Fig. 7, lane h). While always faint, the intensities of these bands varied in different gels, making it unclear whether they were actually due to mRNA cleavage. Nevertheless, even if some cleavage did occur on circular molecules, it clearly was significantly reduced relative to that observed for capped linear mRNAs. The results suggest that, although Vhs readily cleaves circular mRNAs that contain an EMCV IRES at specific sites, targeted cleavage of mRNAs that are translated by ribosome scanning is greatly augmented by an mRNA 5′ end.
Fig 7.
Vhs fails to cut circular pBK2 RNA at the same sites at which it cuts capped linear molecules. Circular (lanes e through h) and capped linear (lanes a through d) forms of pBK2 RNA were incubated in rabbit reticulocyte lysates that contained (Vhs) or lacked (RRL) in vitro-translated Vhs. Aliquots were removed after 0 or 15 min, and the RNAs were analyzed by primer extension using primer pe165. Bands due to prominent Vhs cleavage sites in capped linear pBK2 are indicated by the arrow and lines to the right of lane d. The sequence of the first 100 nucleotides of pBK2 mRNA is shown below the gel, with the 5′ proximal AUG underlined, and the prominent Vhs cut sites are indicated by the arrow and lines below the sequence.
Cleavage of pBK2 mRNA at preferred sites is augmented by a 5′ cap.
Because, for most mRNAs, initiation by ribosome scanning is dependent upon a 5′ cap, it was important to determine whether the targeting of Vhs to specific cut sites requires not just a 5′ end but also the presence of a cap. To this end, 5′-end-labeled pBK2 mRNAs were prepared by the posttranscriptional addition of a 32P-labeled cap or a 32P-labeled 5′ monophosphate. These were incubated in parallel reactions that contained rabbit reticulocyte lysates with or without Vhs, and the reaction products were analyzed by electrophoresis through 8% polyacrylamide-8 M urea sequencing gels to compare 5′ proximal Vhs cleavage products. With the cap-labeled substrate (Fig. 8, lanes a through e), Vhs produced the same set of labeled fragments as those shown in Fig. 3 and 5. In striking contrast, Vhs did not cut the 5′ monophosphate-labeled mRNA at the same preferred sites to any appreciable extent (Fig. 8, lanes f through j). These results indicate that the targeting of Vhs to preferred cut sites in pBK2 mRNA is significantly enhanced by the presence of a cap.
Fig 8.
Importance of a 5′ cap for Vhs cleavage of an mRNA at preferred sites. pBK2 RNA was labeled with a 32P-labeled 5′ cap (lanes a through e) or with a 32P-labeled 5′ monophosphate (lanes f through j) as described in the text. Cap-labeled or 5′ monophosphate-labeled RNAs were incubated in rabbit reticulocyte lysates that contained (Vhs) or lacked (RRL) in vitro-translated Vhs. Aliquots were removed after 0, 3, or 15 min, and the RNAs were analyzed by electrophoresis through an 8% polyacrylamide-8 M urea sequencing gel. The most prominent sites at which Vhs cuts cap-labeled pBK2 mRNA are indicated by arrows to the right of lane e.
Mutations in and around the start codon alter the sites of Vhs cleavage.
Given the importance of a capped 5′ end and the fact that Vhs interacts with the cap-binding complex eIF4F (114), our observation that some of its preferred cut sites cluster near the first AUG of pBK2 and in the 5′ UTR suggests a possible role for scanning-dependent translation initiation in targeting Vhs. To test this idea, we examined the effect on Vhs cleavage of mutations in or surrounding the first AUG codon of pBK2 mRNA which affect ribosome scanning.
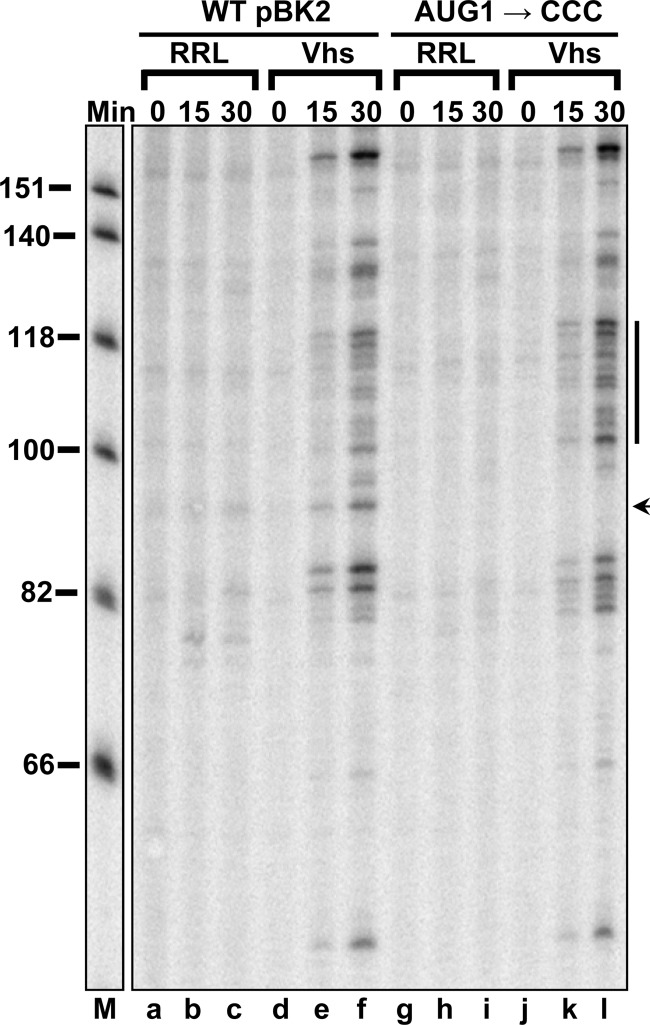
In the first set of experiments, we examined the effect on Vhs cleavage of mutations that altered the first AUG of pBK2 to CCC (AUG1→CCC). Cap-labeled mutant and wild-type pBK2 mRNAs were incubated in parallel mRNA degradation reaction mixtures containing rabbit reticulocyte lysates with or without Vhs, and the reaction products were analyzed by electrophoresis through 8% polyacrylamide-8 M urea sequencing gels to compare the Vhs cleavage sites (Fig. 9). The mutations that altered the AUG abolished Vhs cleavage at a site just upstream from the normal translational start site (Fig. 9, lane l, arrow) and moderately enhanced cleavage at a number of sites further downstream (Fig. 9, lane l, line). Thus, mutations that abolished translation initiation from the first AUG and allowed the 40S ribosomal subunit to continue scanning further downstream reduced Vhs cleavage at sites near the normal start codon and enhanced cleavage at downstream sites.
Fig 9.
Mutations that alter the 5′ proximal AUG of pBK2 mRNA abolish Vhs cleavage at nearby sites. In vitro mRNA degradation reactions were performed using wild-type (WT) pBK2 mRNA (lanes a through f) or the mutant AUG1→CCC (lanes g through l) in which the 5′ proximal AUG has been altered to CCC. Cap-labeled wild-type or mutant mRNAs were added to rabbit reticulocyte lysates that contained (Vhs) or lacked (RRL) in vitro-translated Vhs. Aliquots were removed after 0, 15, or 30 min, and the RNAs were analyzed by electrophoresis through an 8% polyacrylamide-8 M urea sequencing gel. The location of the prominent Vhs cleavage site at which cutting is abolished by the mutation is indicated by the arrow to the right of lane l, while sites at which cleavage was moderately enhanced are indicated by the line to the right of lane l.
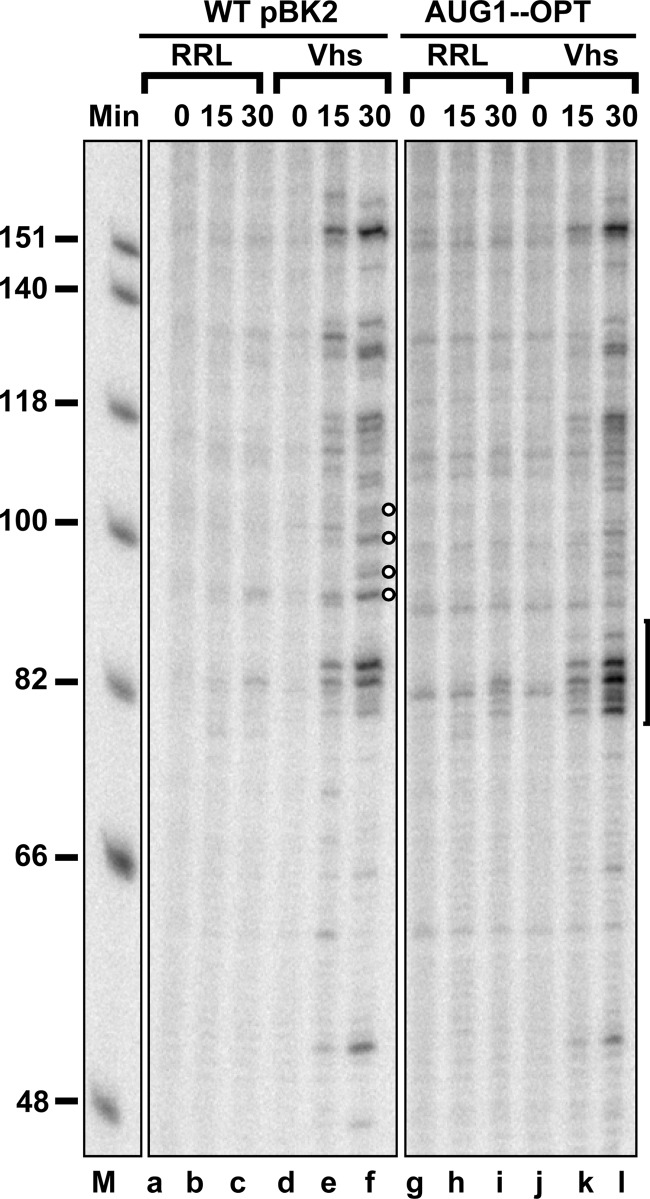
As was mentioned previously, the first AUG of wild-type pBK2 mRNA is in a suboptimal context compared to the Kozak consensus sequence (Fig. 5) (121–127), a fact that appears to allow leaky scanning to occur at least some of the time in vitro (100). Next we examined whether mutations that altered the nucleotides surrounding the first AUG of pBK2 to optimize their match to the Kozak consensus sequence would affect the efficiency of Vhs cleavage at sites near the start codon (Fig. 10). The mutant AUG1-OPT contains four point mutations that change the context of the first AUG of pBK2 mRNA so that it reads CACCAUGG, where the start codon is shown in bold and the altered residues are underlined. The result is an AUG surrounded by a context that is a perfect match to the Kozak consensus sequence. Cap-labeled wild-type pBK2 and AUG1-OPT mRNAs were incubated in rabbit reticulocyte lysates that contained or lacked Vhs, and the reaction products were analyzed by electrophoresis through an 8% polyacrylamide-8 M urea sequencing gel. The mutations that optimize the context of the first AUG caused a moderate increase in Vhs cleavage near the start codon (Fig. 10, lane l, bracket), as well as a decrease in cleavage at sites downstream from the AUG (Fig. 10, lane f, open circles). The results shown in Fig. 9 and 10 indicate that mutations in an mRNA, which affect the recognition of an AUG as a start codon, affect the efficiency of Vhs cleavage at sites surrounding the AUG.
Fig 10.
Mutations that optimize the context of the 5′ proximal AUG of pBK2 mRNA enhance Vhs cleavage at nearby sites. In vitro mRNA degradation reactions were performed using wild-type pBK2 mRNA (lanes a through f) or the mutant AUG1-OPT (lanes g through l) in which nucleotides surrounding the 5′ proximal AUG have been altered to create a match to the Kozak consensus sequence. Cap-labeled wild-type or mutant mRNAs were added to rabbit reticulocyte lysates that contained (Vhs) or lacked (RRL) in vitro-translated Vhs. Aliquots were removed after 0, 15, or 30 min, and the RNAs were analyzed by electrophoresis through an 8% polyacrylamide-8 M urea sequencing gel. The bracket to the right of lane l denotes sites at which Vhs cleavage is enhanced by the mutations that optimize the context of the 5′ proximal AUG. Sites at which Vhs cleavage is reduced are indicated by the open circles to the right of lane f.
Vhs cleavage is inhibited by a nonhydrolyzable analogue of ATP but not by cycloheximide or a nonhydrolyzable analogue of GTP.
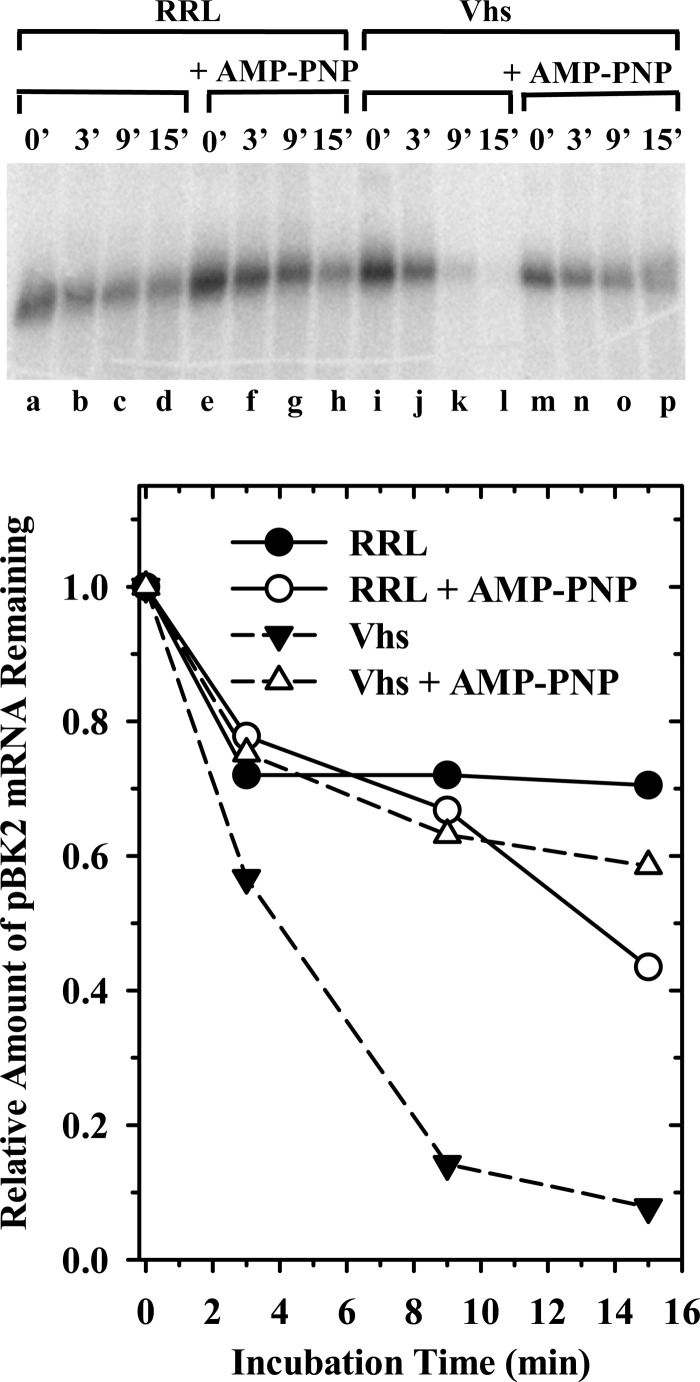
For mRNAs with even a modicum of secondary structure, ribosome scanning is dependent upon ATP hydrolysis (105, 107, 128). Therefore, to further test the apparent linkage between the selection of Vhs cut sites and translation initiation, we examined whether adenosine 5′-(β, γ-imido)triphosphate (AMP-PNP), a nonhydrolyzable analogue of ATP, would inhibit Vhs cleavage of pBK2 mRNA. The inclusion of AMP-PNP in in vitro translation reactions blocks the ATP-dependent helicase activities of eIF4AI and eIF4AII, the consequent interaction of eIF4G, eIF4H, and other translation initiation factors with the 5′ UTR, and scanning of the 40S ribosomal subunit to the start codon to form the 48S initiation complex (105, 129–131). Rabbit reticulocyte lysates that contained or lacked Vhs were supplemented with 2 mM AMP-PNP or an equal volume of water and preincubated at 30°C for 12 min. This concentration of the analogue is sufficient to block all detectable polypeptide synthesis (data not shown). Cap-labeled pBK2 mRNA was added, after which incubation was continued for various intervals, and the RNA reaction products were electrophoresed through a 1.3% (wt/vol) agarose-formaldehyde gel (Fig. 11). AMP-PNP inhibited detectable Vhs cutting of pBK2 mRNA such that in its presence, the rate of pBK2 mRNA decay was indistinguishable in the presence and absence of Vhs. These results indicate that ATP hydrolysis is essential for Vhs degradation of pBK2 mRNA, probably because it is required for the RNA helicase activities of eIF4AI and eIF4AII, and suggest that scanning-dependent initiation plays an important role in Vhs activity.
Fig 11.
Vhs degradation of pBK2 mRNA in the presence of AMP-PNP. Rabbit reticulocyte lysates containing (lanes i through p) or lacking (lanes a through h) in vitro-translated Vhs were preincubated for 12 min in the presence (lanes e through h and m through p) or absence (lanes a through d and i through l) of 2 mM AMP-PNP, after which cap-labeled pBK2 mRNA was added and incubation continued. Aliquots were removed after 0, 3, 9, or 15 min, and the RNAs were electrophoresed through a 1.3% (wt/vol) agarose-formaldehyde gel. The relative amounts of full-length mRNAs were quantified using ImageQuant software and plotted to compare the relative decay rates of pBK2 mRNA.
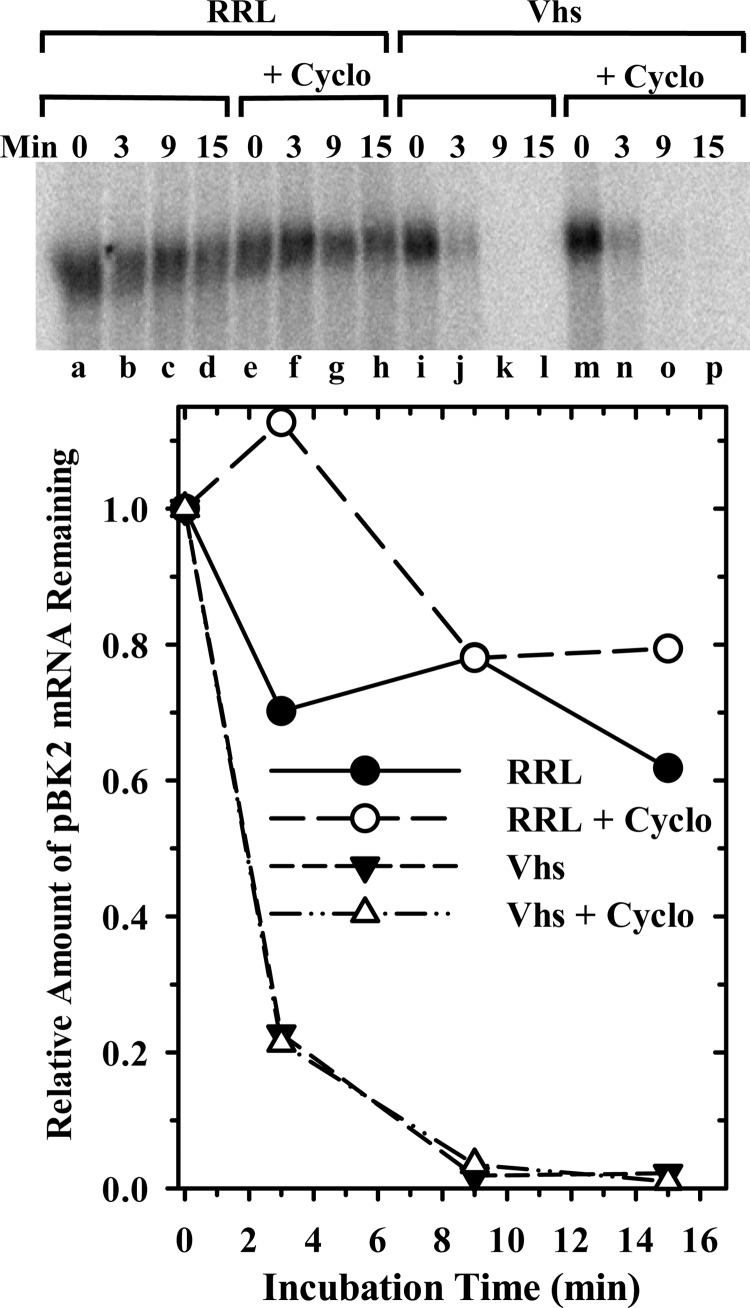
AMP-PNP blocks an early stage of translation initiation. To determine whether Vhs cleavage is dependent upon progression of the mRNA through the entire pathway of translation initiation, or on ongoing translation elongation, we examined the effect on Vhs cleavage of halting translation at different points using cycloheximide or a non-hydrolyzable analogue of GTP. Cycloheximide allows binding of initiation factors and the 40S ribosomal subunit at the cap, scanning to the start codon, and joining of the 60S subunit to form a functional 80S ribosome. However, it inhibits peptide bond formation, which prevents translation elongation (103, 132, 133). In the next set of experiments, cap-labeled pBK2 mRNA was incubated for various intervals in rabbit reticulocyte lysates that contained or lacked Vhs and contained or lacked 50 μg/ml of cycloheximide, a concentration of the drug that is sufficient to halt all detectable translation (data not shown). The reaction products were electrophoresed through a 1.3% (wt/vol) agarose-formaldehyde gel to follow the degradation of full-length cap-labeled mRNA (Fig. 12). Cycloheximide had no detectable effect on Vhs degradation of full-length pBK2 mRNA. In addition, analysis of the degradation products by electrophoresis through an 8% polyacrylamide-8 M urea sequencing gel revealed that cycloheximide had no effect upon the sites of Vhs cleavage (data not shown). Thus, Vhs cleavage is not dependent upon ongoing translation elongation.
Fig 12.
Cycloheximide has no appreciable effect upon Vhs cleavage. Rabbit reticulocyte lysates containing (Vhs) or lacking (RRL) in vitro-translated Vhs were preincubated for 12 min in the presence (lanes e through h and m through p) or absence (lanes a through d and i through l) of 50 μg/ml of cycloheximide (Cyclo), after which cap-labeled pBK2 mRNA was added and incubation continued. Aliquots were removed after 0, 3, 9, or 15 min, and the RNAs were electrophoresed through a 1.3% (wt/vol) agarose-formaldehyde gel. The relative amounts of full-length mRNAs were quantified using ImageQuant software and plotted to compare the relative decay rates of pBK2 mRNA.
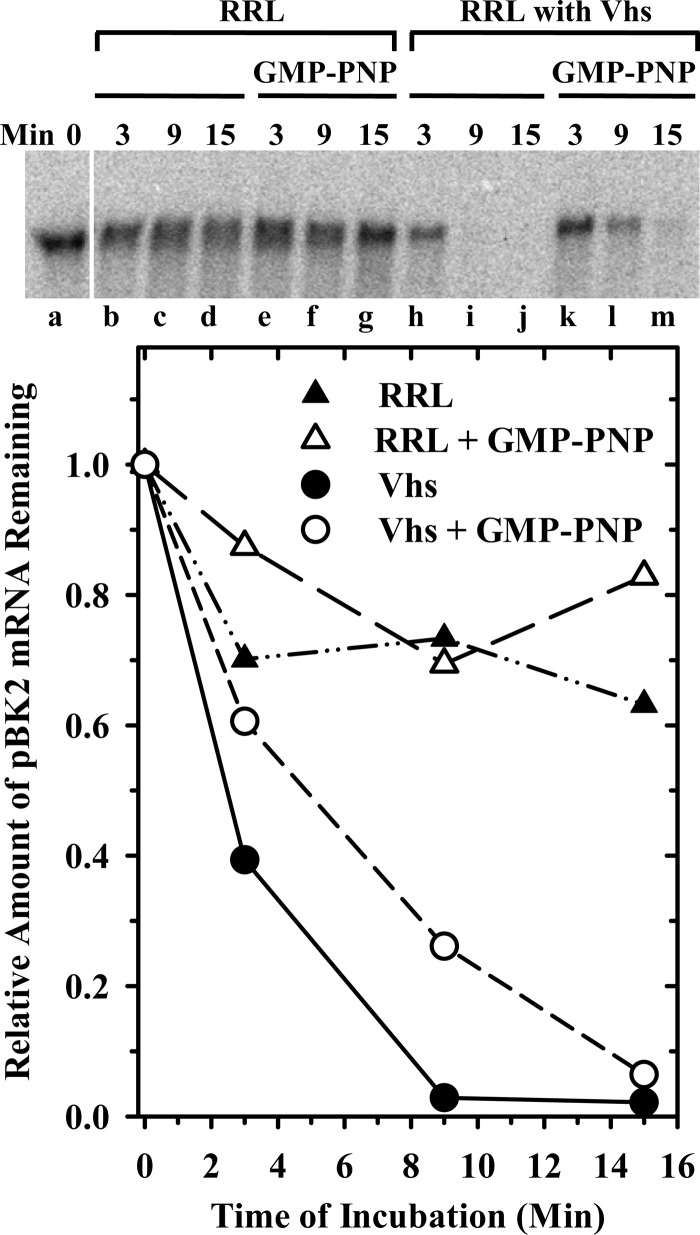
GMP-PNP is a nonhydrolyzable analogue of GTP that allows binding of initiation factors and the 40S ribosomal subunit to the mRNA, and scanning to the start codon to form the 48S complex, but blocks joining of the 60S ribosomal subunit (103, 133, 134). Rabbit reticulocyte lysates, that contained or lacked Vhs, were supplemented with 2 mM GMP-PNP or an equal volume of water and preincubated for 12 min. This concentration of the analogue is sufficient to block all detectable polypeptide synthesis (data not shown). Cap-labeled pBK2 mRNA was added, after which incubation was continued for various intervals, and the RNA reaction products were electrophoresed through a 1.3% (wt/vol) agarose-formaldehyde gel (Fig. 13). GMP-PNP had at most a modest effect on Vhs degradation of full-length pBK2 mRNA, so that by 15 min after addition of the target, more than 90% of the mRNA had been degraded. In addition, electrophoresis of aliquots of the same samples on 8% polyacrylamide-8 M urea sequencing gels showed that Vhs cleavage occurred at the same sites, regardless of whether GMP-PNP was present or absent (data not shown). Thus, the vast majority of Vhs cleavage events are not dependent upon joining of the 60S ribosomal subunit.
Fig 13.
Vhs degradation of pBK2 mRNA in the presence of GMP-PNP. Rabbit reticulocyte lysates containing (lanes h through m) or lacking (lanes a through g) in vitro-translated Vhs were preincubated for 12 min in the presence (lanes e through g and k through m) or absence (lanes a through d and h through j) of 2 mM GMP-PNP, after which cap-labeled pBK2 mRNA was added and incubation continued. Aliquots were removed after 0, 3, 9, or 15 min, and the RNAs were electrophoresed through a 1.3% (wt/vol) agarose-formaldehyde gel. The relative amounts of full-length mRNAs were quantified using ImageQuant software and plotted to compare the relative decay rates of pBK2 mRNA.
DISCUSSION
The thrust of these studies was to investigate how Vhs is targeted to preferred cut sites in mRNAs that are sensitive to Vhs degradation. Previous work has shown that Vhs interacts with the translation initiation factors eIF4H, eIF4AI, and eIF4AII (33–35, 114) and demonstrated the importance of at least eIF4H to Vhs activity (33–35, 39, 114, 135). However, none had examined what determines the sites at which Vhs cuts an mRNA or demonstrated a functional link between translation initiation and Vhs cleavage. The contribution of the present experiments is to map the sites at which Vhs cleaves a particular target mRNA and to demonstrate that mutations or alterations in the mRNA that affect its translation initiation affect the sites of Vhs cleavage. In addition, halting scanning-dependent initiation at an early step by blocking ATP hydrolysis inhibits Vhs activity. The results indicate a link between Vhs cleavage and translation initiation and suggest that Vhs reaches at least some of its preferred cut sites during translation initiation.
Studies with pBK2 mRNA provide a good illustration of the targeting of Vhs to preferred sites. Previously, we showed that a purified complex of recombinant Vhs and a GST-eIF4H fusion protein cuts pBK2 mRNA at many sites throughout the molecule (33). In contrast, the present studies show that in rabbit reticulocyte lysates, Vhs cuts the same pBK2 mRNA at a small subset of those sites, clustered within the first 250 nucleotides from the 5′ cap. This result with rabbit reticulocyte lysates is similar to that reported for Vhs cleavage of the mRNA encoding the signal recognition particle alpha subunit (SRP α) (44). For pBK2 mRNA, this targeting of Vhs to preferred sites was greatly augmented by the presence of a capped 5′ end on the mRNA. Ligating the ends of the mRNA to form a circle abolished or significantly reduced Vhs cutting at a number of sites where Vhs cuts capped linear molecules, and Vhs cutting at preferred sites was greatly reduced for linear molecules with a 5′ monophosphate compared to that for capped linear mRNAs. While these results suggest a link between Vhs cutting and translation initiation, even more persuasive data come from our observation that mutations that alter the start codon from AUG abolished Vhs cleavage at a prominent nearby site, while other mutations that optimized the context of the 5′ proximal AUG to improve its match to the Kozak consensus sequence caused a moderate increase in Vhs cleavage near the start codon and a reduction in cutting at sites farther downstream. Furthermore, our finding that AMP-PNP inhibits most, if not all, Vhs cutting suggests a prominent role for the ATP-dependent eIF4AI and eIF4AII RNA helicases, and probably ribosome scanning, in Vhs degradation of many mRNAs.
The fact that Vhs interacts with the cap-binding complex eIF4F (114) suggests a plausible mechanism for directing Vhs to cut sites close to the 5′ end of mRNAs. However, any model for Vhs targeting must also address how it is directed to cut sites farther downstream from the cap. One possibility is that, after loading onto the mRNA at the cap, Vhs reaches at least some of its cut sites by scanning with the 40S ribosomal subunit during translation initiation. Since purified Vhs lacks significant selectivity (33, 47)—it cleaves single-stranded regions of any kind of RNA downstream from C's and U's (45)—a plausible model is that preferred Vhs cut sites are determined by a combination of mRNA secondary structure and the amount of time that Vhs spends in any one area of the mRNA. Cutting at sites near the 5′ proximal AUG and in the 5′ UTR would occur in regions of single-stranded RNA where the 40S ribosomal subunit, associated factors, and Vhs pause or slow down during ribosome scanning.
If such a model is correct, an important question is why the presence of the 5′ proximal AUG does not block Vhs cleavage at downstream sites near the second and third AUGs. One possibility is that Vhs reaches these sites by leaky scanning. The first and second AUGs of pBK2 mRNA are in a suboptimal context compared to the Kozak consensus sequence. This might allow scanning of the 40S ribosomal subunit, and Vhs, to continue to the second or third AUG codons some of the time, resulting in Vhs cleavage of some mRNA molecules at these downstream sites. It is worth noting that no Vhs cleavage sites were observed downstream from the third AUG codon. Therefore, this AUG, which is in an optimal Kozak context, may block Vhs from cleaving farther downstream.
A second, equally plausible, model is one in which the selection of some Vhs cut sites is dependent upon ribosome scanning, but the Vhs endonuclease does not actually travel along the mRNA with the scanning 40S subunit. Instead, a scanning 40S subunit might remodel mRNA or messenger ribonucleoprotein structure to create structures conducive to Vhs binding and cleavage at sites downstream from the cap. Recent studies of eIF4H and eIF4AI have added credence to such a model (129, 136). Pelletier and coworkers showed that, during translation initiation, copies of eIF4H could be cross-linked to an mRNA at sites up to 52 nucleotides downstream from the cap and that this interaction was both cap and ATP dependent. They went on to propose that, following the binding of eIF4E to the cap, multiple copies of eIF4H, eIF4AI, and eIF4AII may bind the mRNA at internal sites in the 5′ UTR (129). In other experiments, they found that eIF4H stimulates the helicase activity of eIF4AI to a significantly greater extent for hairpin substrates with an internal single-stranded loop than for duplexes with a single-stranded overhang, and they suggested that this reflected higher-affinity binding of eIF4H-eIF4AI complexes to the loops of hairpins than to single-stranded tails (136). They further postulated that one role of eIF4H is to deliver eIF4AI, and perhaps other proteins, to specific structures within mRNAs. The importance of these results to the present studies is that they suggest a mechanism by which Vhs, through its binding to eIF4H, could be directed during translation initiation to sites within the 5′ UTR of an mRNA without having to scan from the 5′ cap.
Both of these models postulate a prominent role for the eIF4A helicase and ribosome scanning in directing Vhs to its preferred sites. We favor these models for pBK2 mRNA because (i) Vhs binds translation initiation factors that play a central role in ribosome scanning, (ii) mutations in and around the start codon of pBK2 mRNA, which affect ribosome scanning, also affect the sites of Vhs cleavage, and (iii) inhibition of ATP hydrolysis blocks most, if not all, Vhs degradation of pBK2 mRNA. In addition, hippuristanol, a specific inhibitor of eIF4AI and 4AII, inhibits Vhs degradation of pBK2 mRNA in rabbit reticulocyte lysates, while hippuristanol treatment or siRNA-mediated depletion of eIF4AI and 4AII from cells prior to infection inhibit Vhs degradation of beta actin mRNA in vivo (D. Agarwal, J. S. Sadek, L. A. Shiflett, and G. S. Read, unpublished data).
While a version of one of these models probably is the case for pBK2 and many other mRNAs, some mRNAs may exist that require a modification of the model. Elgadi et al. have reported that in rabbit reticulocyte lysates, in vitro degradation of SRP α mRNA is not dependent upon the presence of a 5′ cap (44). While different from our result for pBK2 mRNA, this result is not necessarily inconsistent with a link between Vhs degradation and translation initiation. Uncapped mRNAs can be translated to an appreciable extent in rabbit reticulocyte lysates, particularly if they lack secondary structure near the 5′ end (128). In addition, Gunnery et al. showed that when an mRNA encoding the HIV TAT protein was engineered to be transcribed from an RNA polymerase III promoter, translation of the resulting uncapped mRNA could be detected in transfected HeLa cells and occurred by a mechanism involving ribosome scanning (137). Thus, the relative importance of a cap to Vhs-mediated degradation may vary between mRNAs (especially in vitro), depending on their secondary structures, even if for both degradation is linked to translation initiation.
Elgadi et al. also reported that some Vhs cutting of SRP α mRNA occurred in rabbit reticulocyte lysates that had been depleted of ribosomes by centrifugation (44). This does not necessarily mean that Vhs degradation of SRP α mRNA is not linked to translation initiation. Although some Vhs cleavage occurred in the ribosome-depleted extracts, it was significantly reduced relative to the control extracts, and the extent of the ribosome depletion was unclear (44). Furthermore, just as mRNAs differ in the extent to which their translation is dependent on the eIF4A helicase (138), depending upon their secondary structures, mRNAs may differ in the extent to which ribosome scanning is required to remodel them to allow Vhs cleavage at preferred sites. Some mRNAs may be able to adopt multiple secondary structures, only one of which allows Vhs cleavage at preferred sites, and ribosome scanning may drive the mRNA to adopt that structure. Such a model may explain the results of experiments in which circular pBK2 mRNA was incubated with Vhs. Vhs failed to cleave circular pBK2 at three sites at which it cuts capped linear molecules, consistent with the idea that translation initiation is required for cleavage at these sites. However, although significantly reduced, some Vhs cleavage of circles may have occurred at a fourth site. A small fraction of circular molecules may spontaneously adopt a structure that allows Vhs cleavage at this site, while at any one time most of the molecules do not. This would explain a small amount of Vhs cutting of circles at this site. However, ribosome scanning, which occurs on capped linear molecules, would cause a higher fraction of the molecules to adopt the conformation that can be cleaved, leading to significantly more Vhs cleavage of capped linear molecules at this site.
In addition, some mRNAs may exist that are cleaved by Vhs through a mechanism that is completely scanning independent. These may include certain ARE-containing mRNAs that are reportedly cleaved by Vhs at sites in the 3′ UTR (86, 87, 93, 139). Whether cleavage at these sites first involves loading Vhs onto the mRNAs by binding cap-bound eIF4F, followed by redirection to the sites in the 3′ UTR, remains to be determined. Alternatively, Vhs may completely bypass the cap and be directly targeted to sites near the ARE by an unknown mechanism.
Whether or not Vhs reaches different cut sites by different mechanisms in scanned mRNAs, several results indicate that different mechanisms target Vhs to sites in scanned mRNAs versus mRNAs that contain an EMCV IRES. First, circularization of an mRNA that is normally translated by scanning significantly reduces Vhs cleavage at the sites that are cut in capped linear molecules. In contrast, Vhs readily cuts linear and circular forms of an mRNA with an EMCV IRES at the same sites. Second, the mutant Vhs 1 contains a point mutation that changes threonine 214 to isoleucine (60,140). The mutant polypeptide retains endonuclease activity and the ability to bind eIF4AI and eIF4AII (35) but is unable to bind eIF4H (33–35). Vhs polypeptides containing the T214I mutation completely lack the ability to degrade mRNAs that are translated by scanning (36, 37, 48, 57, 58, 96) but retain the ability to cleave mRNAs containing an EMCV IRES at sites downstream from the IRES (101). Thus, Vhs requires different cellular factors for cleavage of scanned and IRES-containing mRNAs.
A number of important questions concerning the function of Vhs remain. In particular, little is known regarding what features of some mRNAs make them more or less sensitive to Vhs degradation. Despite its name, Vhs is not exclusively a host shutoff factor. Rather, it alters the balance between the transcription and degradation rates of many mRNAs, which can have many downstream effects, both negative and positive, on gene expression. Elucidating what features of mRNAs determine their different sensitivities to Vhs degradation is important to understanding virus-host interactions that are central to HSV biology. In view of our observation that Vhs reaches at least some of its cut sites during translation initiation, it will be interesting to determine whether mRNA secondary structures that modulate their efficiencies of translation may influence their sensitivities to Vhs degradation.
Finally, recent data have indicated that in some cell types, Vhs-mediated mRNA degradation is reduced and, instead, Vhs actually stimulates the translation of specific mRNAs. Thus, in HeLa and several other cell types, Vhs activates translation of the 3′ cistron of bicistronic mRNAs containing several cellular IRESs (29). Additionally, during lytic infections, it stimulates translation of several late viral mRNAs (23). Understanding how the mRNA degradative and translational stimulatory functions of Vhs are coordinated and regulated is an important avenue for future research.
ACKNOWLEDGMENTS
We thank Jouliana Sadek and our other colleagues at the University of Missouri–Kansas City, School of Biological Sciences, for many helpful discussions.
This work was supported by Public Health Services grant RO1 AI-021501 (to G.S.R.), from the National Institute of Allergy and Infectious Diseases, and by a grant from the University of Missouri Research Board.
Footnotes
Published ahead of print 17 October 2012
REFERENCES
- 1. Roizman B, Knipe DM, Whitley RJ. 2007. Herpes simplex viruses, p 2501–2601 In Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, Straus SE. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Sandri-Goldin RM. 2007. Initiation of transcription and RNA synthesis, processing and transport in HSV and VZV infected cells. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: http://www.ncbi.nlm.nih.gov/books/NBK47363/ [PubMed] [Google Scholar]
- 3. Ferenczy MW, Ranayhossaini DJ, DeLuca NA. 2011. Activities of ICP0 involved in the reversal of silencing of quiescent herpes simplex virus 1. J. Virol. 85:4993–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kristie TM. 2007. Early events pre-initiation of alphaherpes viral gene expression. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: http://www.ncbi.nlm.nih.gov/books/NBK47386/ [PubMed] [Google Scholar]
- 5. Lester JT, DeLuca NA. 2011. Herpes simplex virus 1 ICP4 forms complexes with TFIID and mediator in virus-infected cells. J. Virol. 85:5733–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sampath P, DeLuca NA. 2008. Binding of ICP4, TATA-binding protein, and RNA polymerase II to herpes simplex virus type 1 immediate-early, early, and late promoters in virus-infected cells. J. Virol. 82:2339–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zabierowski S, DeLuca NA. 2004. Differential cellular requirements for activation of herpes simplex virus type 1 early (tk) and late (gC) promoters by ICP4. J. Virol. 78:6162–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zabierowski SE, DeLuca NA. 2008. Stabilized binding of TBP to the TATA box of herpes simplex virus type 1 early (tk) and late (gC) promoters by TFIIA and ICP4. J. Virol. 82:3546–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hardy WR, Sandri-Goldin RM. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68:7790–7799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sandri-Goldin RM. 1994. Properties of an HSV-1 regulatory protein that appears to impair host cell splicing. Infect. Agents Dis. 3:59–67 [PubMed] [Google Scholar]
- 11. Sandri-Goldin RM. 1998. Interactions between a herpes simplex virus regulatory protein and cellular mRNA processing pathways. Methods 16:95–104 [DOI] [PubMed] [Google Scholar]
- 12. Sciabica KS, Dai QJ, Sandri-Goldin RM. 2003. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 22:1608–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen IH, Sciabica KS, Sandri-Goldin RM. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corbin-Lickfett KA, Rojas S, Li L, Cocco MJ, Sandri-Goldin RM. 2010. ICP27 phosphorylation site mutants display altered functional interactions with cellular export factors Aly/REF and TAP/NXF1 but are able to bind herpes simplex virus 1 RNA. J. Virol. 84:2212–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Corbin-Lickfett KA, Souki SK, Cocco MJ, Sandri-Goldin RM. 2010. Three arginine residues within the RGG box are crucial for ICP27 binding to herpes simplex virus 1 GC-rich sequences and for efficient viral RNA export. J. Virol. 84:6367–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hernandez FP, Sandri-Goldin RM. 2010. Head-to-tail intramolecular interaction of herpes simplex virus type 1 regulatory protein ICP27 is important for its interaction with cellular mRNA export receptor TAP/NXF1. mBio 1:e00268–10 doi:10.1128/mBio.00268-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson LA, Li L, Sandri-Goldin RM. 2009. The cellular RNA export receptor TAP/NXF1 is required for ICP27-mediated export of herpes simplex virus 1 RNA, but the TREX complex adaptor protein Aly/REF appears to be dispensable. J. Virol. 83:6335–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson LA, Sandri-Goldin RM. 2009. Efficient nuclear export of herpes simplex virus 1 transcripts requires both RNA binding by ICP27 and ICP27 interaction with TAP/NXF1. J. Virol. 83:1184–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sandri-Goldin RM. 2001. Nuclear export of herpes virus RNA. Curr. Top. Microbiol. Immunol. 259:2–23 [PubMed] [Google Scholar]
- 20. Sandri-Goldin RM. 2004. Viral regulation of mRNA export. J. Virol. 78:4389–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chuluunbaatar U, Mohr I. 2011. A herpesvirus kinase that masquerades as Akt: you don't have to look like Akt, to act like it. Cell Cycle 10:2064–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chuluunbaatar U, Roller R, Feldman ME, Brown S, Shokat KM, Mohr I. 2010. Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev. 24:2627–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dauber B, Pelletier J, Smiley JR. 2011. The herpes simplex virus 1 vhs protein enhances translation of viral true late mRNAs and virus production in a cell type-dependent manner. J. Virol. 85:5363–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ellison KS, Maranchuk RA, Mottet KL, Smiley JR. 2005. Control of VP16 translation by the herpes simplex virus type 1 immediate-early protein ICP27. J. Virol. 79:4120–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mohr I. 2006. Phosphorylation and dephosphorylation events that regulate viral mRNA translation. Virus Res. 119:89–99 [DOI] [PubMed] [Google Scholar]
- 26. Mulvey M, Arias C, Mohr I. 2006. Resistance of mRNA translation to acute endoplasmic reticulum stress-inducing agents in herpes simplex virus type 1-infected cells requires multiple virus-encoded functions. J. Virol. 80:7354–7363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mulvey M, Arias C, Mohr I. 2007. Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J. Virol. 81:3377–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mulvey M, Poppers J, Sternberg D, Mohr I. 2003. Regulation of eIF2alpha phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J. Virol. 77:10917–10928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saffran HA, Read GS, Smiley JR. 2010. Evidence for translational regulation by the herpes simplex virus virion host shutoff protein. J. Virol. 84:6041–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walsh D, Mohr I. 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 18:660–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walsh D, Mohr I. 2006. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev. 20:461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walsh D, Mohr I. 2011. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 9:860–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Everly DN, Jr, Feng P, Mian IS, Read GS. 2002. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J. Virol. 76:8560–8571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feng P, Everly DN, Jr., Read GS. 2001. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J. Virol. 75:10272–10280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feng P, Everly DN, Jr., Read GS. 2005. mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J. Virol. 79:9651–9664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oroskar AA, Read GS. 1987. A mutant of herpes simplex virus type 1 exhibits increased stability of immediate-early (alpha) mRNAs. J. Virol. 61:604–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oroskar AA, Read GS. 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J. Virol. 63:1897–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Read GS. 1997. Control of mRNA stability during herpes simplex virus infections, p 311–321 In Harford JB, Morris DR. (ed), mRNA metabolism and post-transcriptional gene regulation, 1st ed, vol 17 Wiley-Liss, Inc., New York, NY [Google Scholar]
- 39. Sarma N, Agarwal D, Shiflett LA, Read GS. 2008. Small interfering RNAs that deplete the cellular translation factor eIF4H impede mRNA degradation by the virion host shutoff protein of herpes simplex virus. J. Virol. 82:6600–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schek N, Bachenheimer SL. 1985. Degradation of cellular mRNAs induced by a virion-associated factor during herpes simplex virus infection of Vero cells. J. Virol. 55:601–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smiley JR. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 78:1063–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Strom T, Frenkel N. 1987. Effects of herpes simplex virus on mRNA stability. J. Virol. 61:2198–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taddeo B, Esclatine A, Roizman B. 2002. The patterns of accumulation of cellular RNAs in cells infected with a wild-type and a mutant herpes simplex virus 1 lacking the virion host shutoff gene. Proc. Natl. Acad. Sci. U. S. A. 99:17031–17036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Elgadi MM, Hayes CE, Smiley JR. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 73:7153–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taddeo B, Roizman B. 2006. The virion host shutoff protein (UL41) of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J. Virol. 80:9341–9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taddeo B, Zhang W, Roizman B. 2006. The U(L)41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc. Natl. Acad. Sci. U. S. A. 103:2827–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zelus BD, Stewart RS, Ross J. 1996. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J. Virol. 70:2411–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Read GS, Karr BM, Knight K. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 67:7149–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Read GS, Patterson M. 2007. Packaging of the virion host shutoff (Vhs) protein of herpes simplex virus: two forms of the Vhs polypeptide are associated with intranuclear B and C capsids, but only one is associated with enveloped virions. J. Virol. 81:1148–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smibert CA, Johnson DC, Smiley JR. 1992. Identification and characterization of the virion-induced host shutoff product of herpes simplex virus gene UL41. J. Gen. Virol. 73:467–470 [DOI] [PubMed] [Google Scholar]
- 51. Everett RD, Fenwick ML. 1990. Comparative DNA sequence analysis of the host shutoff genes of different strains of herpes simplex virus: type 2 strain HG52 encodes a truncated UL41 product. J. Gen. Virol. 71:1387–1390 [DOI] [PubMed] [Google Scholar]
- 52. Fenwick ML. 1984. The effects of herpesviruses on cellular macromolecular synthesis, p 359–390 In Fraenkel-Conrat H, Wagner RR. (ed), Comprehensive virology, vol 19 Plenum Publishing Corp., New York, NY [Google Scholar]
- 53. Fenwick ML, Everett RD. 1990. Inactivation of the shutoff gene (UL41) of herpes simplex virus types 1 and 2. J. Gen. Virol. 71:2961–2967 [DOI] [PubMed] [Google Scholar]
- 54. Fenwick ML, Everett RD. 1990. Transfer of UL41, the gene controlling virion-associated host cell shutoff, between different strains of herpes simplex virus. J. Gen. Virol. 71:411–418 [DOI] [PubMed] [Google Scholar]
- 55. Fenwick ML, McMenamin MM. 1984. Early virion-associated suppression of cellular protein synthesis by herpes simplex virus is accompanied by inactivation of mRNA. J. Gen. Virol. 65:1225–1228 [DOI] [PubMed] [Google Scholar]
- 56. Fenwick ML, Clark J. 1982. Early and delayed shut-off of host protein synthesis in cells infected with herpes simplex virus. J. Gen. Virol. 61:121–125 [DOI] [PubMed] [Google Scholar]
- 57. Read GS, Frenkel N. 1983. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha (immediate early) viral polypeptides. J. Virol. 46:498–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kwong AD, Frenkel N. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. U. S. A. 84:1926–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kwong AD, Frenkel N. 1989. The herpes simplex virus virion host shutoff function. J. Virol. 63:4834–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kwong AD, Kruper JA, Frenkel N. 1988. Herpes simplex virus virion host shutoff function. J. Virol. 62:912–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Duerst RJ, Morrison LA. 2004. Herpes simplex virus 2 virion host shutoff protein interferes with type I interferon production and responsiveness. Virology 322:158–167 [DOI] [PubMed] [Google Scholar]
- 62. Korom M, Wylie KM, Morrison LA. 2008. Selective ablation of virion host shutoff protein RNase activity attenuates herpes simplex virus 2 in mice. J. Virol. 82:3642–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Murphy JA, Duerst RJ, Smith TJ, Morrison LA. 2003. Herpes simplex virus type 2 virion host shutoff protein regulates alpha/beta interferon but not adaptive immune responses during primary infection in vivo. J. Virol. 77:9337–9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pasieka TJ, Cilloniz C, Lu B, Teal TH, Proll SC, Katze MG, Leib DA. 2009. Host responses to wild-type and attenuated herpes simplex virus infection in the absence of Stat1. J. Virol. 83:2075–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pasieka TJ, Lu B, Crosby SD, Wylie KM, Morrison LA, Alexander DE, Menachery VD, Leib DA. 2008. Herpes simplex virus virion host shutoff attenuates establishment of the antiviral state. J. Virol. 82:5527–5535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pasieka TJ, Lu B, Leib DA. 2008. Enhanced pathogenesis of an attenuated herpes simplex virus for mice lacking Stat1. J. Virol. 82:6052–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wylie KM, Schrimpf JE, Morrison LA. 2009. Increased eIF2alpha phosphorylation attenuates replication of herpes simplex virus 2 vhs mutants in mouse embryonic fibroblasts and correlates with reduced accumulation of the PKR antagonist ICP34.5. J. Virol. 83:9151–9162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cotter CR, Kim WK, Nguyen ML, Yount JS, Lopez CB, Blaho JA, Moran TM. 2011. The virion host shutoff protein of herpes simplex virus 1 blocks the replication-independent activation of NF-kappaB in dendritic cells in the absence of type I interferon signaling. J. Virol. 85:12662–12672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cotter CR, Nguyen ML, Yount JS, Lopez CB, Blaho JA, Moran TM. 2010. The virion host shut-off (vhs) protein blocks a TLR-independent pathway of herpes simplex virus type 1 recognition in human and mouse dendritic cells. PLoS One 5:e8684 doi:10.1371/journal.pone.0008684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Samady L, Costigliola E, MacCormac L, McGrath Y, Cleverley S, Lilley CE, Smith J, Latchman DS, Chain B, Coffin RS. 2003. Deletion of the virion host shutoff protein (vhs) from herpes simplex virus (HSV) relieves the viral block to dendritic cell activation: potential of vhs- HSV vectors for dendritic cell-mediated immunotherapy. J. Virol. 77:3768–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Barzilai A, Zivony-Elbom I, Sarid R, Noah E, Frenkel N. 2006. The herpes simplex virus type 1 vhs-UL41 gene secures viral replication by temporarily evading apoptotic cellular response to infection: Vhs-UL41 activity might require interactions with elements of cellular mRNA degradation machinery. J. Virol. 80:505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liang L, Roizman B. 2006. Herpes simplex virus 1 precludes replenishment of the short-lived receptor of tumor necrosis factor alpha by virion host shutoff-dependent degradation of its mRNA. J. Virol. 80:7756–7759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Suzutani T, Nagamine M, Shibaki T, Ogasawara M, Yoshida I, Daikoku T, Nishiyama Y, Azuma M. 2000. The role of the UL41 gene of herpes simplex virus type 1 in evasion of non-specific host defence mechanisms during primary infection. J. Gen. Virol. 81:1763–1771 [DOI] [PubMed] [Google Scholar]
- 74. Tigges MA, Leng S, Johnson DC, Burke RL. 1996. Human herpes simplex virus (HSV)-specific CD8+ CTL clones recognize HSV-2-infected fibroblasts after treatment with IFN- gamma or when virion host shutoff functions are disabled. J. Immunol. 156:3901–3910 [PubMed] [Google Scholar]
- 75. Trgovcich J, Johnson D, Roizman B. 2002. Cell surface major histocompatibility complex class II proteins are regulated by the products of the gamma(1)34.5 and U(L)41 genes of herpes simplex virus 1. J. Virol. 76:6974–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Becker Y, Tavor E, Asher Y, Berkowiltz C, Moyal M. 1993. Effect of herpes simplex virus type-1 UL41 gene on the stability of mRNA from the cellular genes: beta-actin, fibronectin, glucose transporter-1, and docking protein, and on virus intraperitoneal pathogenicity of newborn mice. Virus Genes 7:133–143 [DOI] [PubMed] [Google Scholar]
- 77. Keadle TL, Laycock KA, Morris JL, Leib DA, Morrison LA, Pepose JS, Stuart PM. 2002. Therapeutic vaccination with vhs(-) herpes simplex virus reduces the severity of recurrent herpetic stromal keratitis in mice. J. Gen. Virol. 83:2361–2365 [DOI] [PubMed] [Google Scholar]
- 78. Keadle TL, Morrison LA, Morris JL, Pepose JS, Stuart PM. 2002. Therapeutic immunization with a virion host shutoff-defective, replication-incompetent herpes simplex virus type 1 strain limits recurrent herpetic ocular infection. J. Virol. 76:3615–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Leib DA, Harrison TE, Laslo KM, Machalek MA, Moorman NJ, Virgin HW. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189:663–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pasieka TJ, Cilloniz C, Carter VS, Rosato P, Katze MG, Leib DA. 2011. Functional genomics reveals an essential and specific role for Stat1 in protection of the central nervous system following herpes simplex virus corneal infection. J. Virol. 85:12972–12981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Smith TJ, Ackland-Berglund CE, Leib DA. 2000. Herpes simplex virus virion host shutoff (vhs) activity alters periocular disease in mice. J. Virol. 74:3598–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Smith TJ, Morrison LA, Leib DA. 2002. Pathogenesis of herpes simplex virus type 2 virion host shutoff (vhs) mutants. J. Virol. 76:2054–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Strand SS, Leib DA. 2004. Role of the VP16-binding domain of vhs in viral growth, host shutoff activity, and pathogenesis. J. Virol. 78:13562–13572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Strelow LI, Leib DA. 1995. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 69:6779–6786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Walker J, Leib DA. 1998. Protection from primary infection and establishment of latency by vaccination with a herpes simplex virus type 1 recombinant deficient in the virion host shutoff (vhs) function. Vaccine 16:1–5 [DOI] [PubMed] [Google Scholar]
- 86. Esclatine A, Taddeo B, Evans L, Roizman B. 2004. The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc. Natl. Acad. Sci.U. S. A. 101:3603–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Esclatine A, Taddeo B, Roizman B. 2004. Herpes simplex virus 1 induces cytoplasmic accumulation of TIA-1/TIAR and both synthesis and cytoplasmic accumulation of tristetraprolin, two cellular proteins that bind and destabilize AU-rich RNAs. J. Virol. 78:8582–8592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Esclatine A, Taddeo B, Roizman B. 2004. The UL41 protein of herpes simplex virus mediates selective stabilization or degradation of cellular mRNAs. Proc. Natl. Acad. Sci.U. S. A. 101:18165–18170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hobbs WE, DeLuca NA. 1999. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 73:8245–8255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Khodarev NN, Advani SJ, Gupta N, Roizman B, Weichselbaum RR. 1999. Accumulation of specific RNAs encoding transcriptional factors and stress response proteins against a background of severe depletion of cellular RNAs in cells infected with herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 96:12062–12067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ray N, Enquist LW. 2004. Transcriptional response of a common permissive cell type to infection by two diverse alphaherpesviruses. J. Virol. 78:3489–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Taddeo B, Esclatine A, Roizman B. 2004. Post-transcriptional processing of cellular RNAs in herpes simplex virus-infected cells. Biochem. Soc. Trans. 32:697–701 [DOI] [PubMed] [Google Scholar]
- 93. Taddeo B, Esclatine A, Zhang W, Roizman B. 2003. The stress-inducible immediate-early responsive gene IEX-1 is activated in cells infected with herpes simplex virus 1, but several viral mechanisms, including 3′ degradation of its RNA, preclude expression of the gene. J. Virol. 77:6178–6187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Corcoran JA, Hsu WL, Smiley JR. 2006. Herpes simplex virus ICP27 is required for virus-induced stabilization of the ARE-containing IEX-1 mRNA encoded by the human IER3 gene. J. Virol. 80:9720–9729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hsu WL, Saffran HA, Smiley JR. 2005. Herpes simplex virus infection stabilizes cellular IEX-1 mRNA. J. Virol. 79:4090–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Krikorian CR, Read GS. 1991. In vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J. Virol. 65:112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Pak AS, Everly DN, Knight K, Read GS. 1995. The virion host shutoff protein of herpes simplex virus inhibits reporter gene expression in the absence of other viral gene products. Virology 211:491–506 [DOI] [PubMed] [Google Scholar]
- 98. Sorenson CM, Hart PA, Ross J. 1991. Analysis of herpes simplex virus-induced mRNA destabilizing activity using an in vitro mRNA decay system. Nucleic Acids Res. 19:4459–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Elgadi MM, Smiley JR. 1999. Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff protein. J. Virol. 73:9222–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Karr BM, Read GS. 1999. The virion host shutoff function of herpes simplex virus degrades the 5′ end of a target mRNA before the 3′ end. Virology 264:195–204 [DOI] [PubMed] [Google Scholar]
- 101. Lu P, Saffran HA, Smiley JR. 2001. The vhs1 mutant form of herpes simplex virus virion host shutoff protein retains significant internal ribosome entry site-directed RNA cleavage activity. J. Virol. 75:1072–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Perez-Parada J, Saffran HA, Smiley JR. 2004. RNA degradation induced by the herpes simplex virus vhs protein proceeds 5′ to 3′ in vitro. J. Virol. 78:13391–13394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11:113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N. 2011. mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 12:235–245 [DOI] [PubMed] [Google Scholar]
- 105. Pestova TV, Lorsch JR, Hellen CUT. 2007. The mechanism of translation initiation in eukaryotes, p 87–128 In Mathews MB, Sonenberg N, Hershey JWB. (ed), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 106. Rogers GW, Jr, Komar AA, Merrick WC. 2002. eIF4A: the godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 72:307–331 [DOI] [PubMed] [Google Scholar]
- 107. Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. 2009. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136:447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Richter NJ, Rogers GW, Jr, Hensold JO, Merrick WC. 1999. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J. Biol. Chem. 274:35415–35424 [DOI] [PubMed] [Google Scholar]
- 110. Richter-Cook NJ, Dever TE, Hensold JO, Merrick WC. 1998. Purification and characterization of a new eukaryotic protein translation factor. Eukaryotic initiation factor 4H. J. Biol. Chem. 273:7579–7587 [DOI] [PubMed] [Google Scholar]
- 111. Rogers GW, Jr, Lima WF, Merrick WC. 2001. Further characterization of the helicase activity of eIF4A. Substrate specificity. J. Biol. Chem. 276:12598–12608 [DOI] [PubMed] [Google Scholar]
- 112. Rogers GW, Jr, Richter NJ, Lima WF, Merrick WC. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276:30914–30922 [DOI] [PubMed] [Google Scholar]
- 113. Rogers GW, Jr, Richter NJ, Merrick WC. 1999. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274:12236–12244 [DOI] [PubMed] [Google Scholar]
- 114. Page HG, Read GS. 2010. The virion host shutoff endonuclease (UL41) of herpes simplex virus interacts with the cellular cap-binding complex eIF4F. J. Virol. 84:6886–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Everly DN, Jr., Read GS. 1997. Mutational analysis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): characterization of HSV type 1 (HSV-1)/HSV-2 chimeras. J. Virol. 71:7157–7166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Everly DN, Jr., Read GS. 1999. Site-directed mutagenesis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): analysis of functional differences between HSV type 1 (HSV-1) and HSV-2 alleles. J. Virol. 73:9117–9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 118. Chen CY, Sarnow P. 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268:415–417 [DOI] [PubMed] [Google Scholar]
- 119. Chen CY, Sarnow P. 1998. Internal ribosome entry sites tests with circular mRNAs. Methods Mol. Biol. 77:355–363 [DOI] [PubMed] [Google Scholar]
- 120. Smiley JR, Elgadi MM, Saffran HA. 2001. Herpes simplex virus vhs protein. Methods Enzymol. 342:440–451 [DOI] [PubMed] [Google Scholar]
- 121. Babendure JR, Babendure JL, Ding JH, Tsien RY. 2006. Control of mammalian translation by mRNA structure near caps. RNA 12:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kozak M. 1991. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 115:887–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kozak M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867–19870 [PubMed] [Google Scholar]
- 125. Kozak M. 1992. Regulation of translation in eukaryotic systems. Annu. Rev. Cell Biol. 8:197–225 [DOI] [PubMed] [Google Scholar]
- 126. Kozak M. 1994. Features in the 5′ non-coding sequences of rabbit alpha and beta-globin mRNAs that affect translational efficiency. J. Mol. Biol. 235:95–110 [DOI] [PubMed] [Google Scholar]
- 127. Kozak M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Pestova TV, Kolupaeva VG. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16:2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lindqvist L, Imataka H, Pelletier J. 2008. Cap-dependent eukaryotic initiation factor-mRNA interactions probed by cross-linking. RNA 14:960–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Pestova TV, Borukhov SI, Hellen CU. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394:854–859 [DOI] [PubMed] [Google Scholar]
- 131. Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. 2008. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell 135:1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6:209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Wilson JE, Pestova TV, Hellen CU, Sarnow P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102:511–520 [DOI] [PubMed] [Google Scholar]
- 134. Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403:332–335 [DOI] [PubMed] [Google Scholar]
- 135. Doepker RC, Hsu WL, Saffran HA, Smiley JR. 2004. Herpes simplex virus virion host shutoff protein is stimulated by translation initiation factors eIF4B and eIF4H. J. Virol. 78:4684–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Sun Y, Atas E, Lindqvist L, Sonenberg N, Pelletier J, Meller A. 2012. The eukaryotic initiation factor eIF4H facilitates loop-binding, repetitive RNA unwinding by the eIF4A DEAD-box helicase. Nucleic Acids Res. 40:6199–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Gunnery S, Maivali U, Mathews MB. 1997. Translation of an uncapped mRNA involves scanning. J. Biol. Chem. 272:21642–21646 [DOI] [PubMed] [Google Scholar]
- 138. Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. 2001. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7:382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Taddeo B, Zhang W, Roizman B. 2009. The virion-packaged endoribonuclease of herpes simplex virus 1 cleaves mRNA in polyribosomes. Proc. Natl. Acad. Sci.U. S. A. 106:12139–12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kwong AD. 1988. The herpes simplex virus virion host shutoff and mRNA destabilization function, abstr 48. 13th Int. Herpesvirus Workshop, Irvine, CA, 7 to 13 August 1988 [Google Scholar]