Abstract
The conditions in densely populated Bangladesh favor picornavirus transmission, resulting in a high rate of infection in the human population. Data suggest that nonhuman primates (NHP) may play a role in the maintenance and transmission of diverse picornaviruses in Bangladesh. At the Dhaka Zoo, multiple NHP species are caged in close proximity. Their proximity to other species and to humans, both zoo workers and visitors, provides the potential for cross-species transmission. To investigate possible interspecies and intraspecies transmission of picornaviruses among NHP, we collected fecal specimens from nine NHP taxa at the Dhaka Zoo at three time points, August 2007, January 2008, and June 2008. Specimens were screened using real-time PCR for the genera Enterovirus, Parechovirus, and Sapelovirus, and positive samples were typed by VP1 sequencing. Fifty-two picornaviruses comprising 10 distinct serotypes were detected in 83 fecal samples. Four of these serotypes, simian virus 19 (SV19), baboon enterovirus (BaEV), enterovirus 112 (EV112), and EV115, have been solely associated with infection in NHP. EV112, EV115, and SV19 accounted for 88% of all picornaviruses detected. Over 80% of samples from cages housing rhesus macaques, olive baboons, or hamadryas baboons were positive for a picornavirus, while no picornaviruses were detected in samples from capped langurs or vervet monkeys. In contrast to our findings among synanthropic NHP in Bangladesh where 100% of the picornaviruses detected were of human serotypes, in the zoo population, only 15% of picornaviruses detected in NHP were of human origin. Specific serotypes tended to persist over time, suggesting either persistent infection of individuals or cycles of reinfection.
INTRODUCTION
Picornaviruses are ubiquitous viruses that commonly infect humans, as well as a wide range of other animal species (1). They cause a broad spectrum of disease, from minor febrile illness to myocarditis, meningitis, encephalitis, and paralysis. Much of the early research characterizing the biological and pathogenic properties of human picornaviruses was performed in the mid-20th century, using nonhuman primates (NHP), most commonly rhesus macaques, which had been imported from South Asia as biomedical models. Recently, renewed interest has focused on NHP-to-human transmission of enteroviruses (EV), stemming from the finding that some enterovirus serotypes associated with disease in humans closely resemble serotypes found in NHP from South Asia (2). It is important to determine whether NHP could be reservoirs for picornaviruses that cause severe disease in human populations.
Picornaviruses are small viruses with a single-stranded RNA genome (3). In humans, infection occurs through the fecal-oral route. After an incubation period of 4 to 7 days, clinical illness, which is mild in the overwhelming majority of cases, may develop (4). Enteric picornaviruses are typically shed in stool for 4 to 8 weeks postinfection (5). Prolonged shedding of the virus in asymptomatic individuals occurs rarely. Individuals who recover from infection are thought to have developed long-lasting immunity to the specific picornavirus serotype that had infected them.
Recent data suggest that, at least in captive populations of NHP, enteroviral serotypes may persist for years. This is based on a study done at the Yerkes National Primate Research Center, Atlanta, GA, on fecal samples from 56 NHP with diarrhea, which detected a high prevalence of known simian and previously undescribed enteroviruses but no known human enteroviruses (6). It is likely that either these persistent serotypes circulate within a colony and evolve over time or multiple strains of each type were introduced to the colony at different times. Perhaps individual NHP can maintain infection over long periods or can be reinfected with a given serotype, or both.
The current study focuses on picornaviruses that were detected in NHP at the Dhaka Zoo. This sample population was part of a larger project designed to characterize the picornavirus pathogen landscape among NHP in Bangladesh during the period 2007 to 2008. Here we present data characterizing the prevalence of picornaviruses in a captive population comprising multiple species of NHP, surveyed at multiple time points, focusing on whether the picornaviruses resemble known human serotypes or serotypes characteristic of NHP. This study complements our research for this project, characterizing enteroviruses found in free-ranging NHP in Bangladesh during the same period (7).
MATERIALS AND METHODS
Study site and sample collection.
A total of 83 fecal specimens, representing nine NHP taxa, were collected at the Dhaka Zoo. Dhaka Zoo, located 16 kilometers from the center of Dhaka, was established in 1974 and houses, in very basic conditions, fauna native to the region and exotic species from around the world. Tens of thousands of local visitors arrive each day. It is common to observe visitors reaching out to touch and offer food to the animals, particularly the NHP.
The Dhaka Zoo collection of NHP contains both species that are naturally distributed in Bangladesh (e.g., rhesus macaques, pigtail macaques, capped langurs, and hoolock gibbons) and exotic species of NHP from Africa, including olive and hamadryas baboons, mandrills, and golden mangabeys. Information about their origins or the dates on which these NHP entered the zoo was not available. However, it was known that wild-caught rhesus macaques were occasionally brought into the zoo and introduced into cages that already contained rhesus macaques.
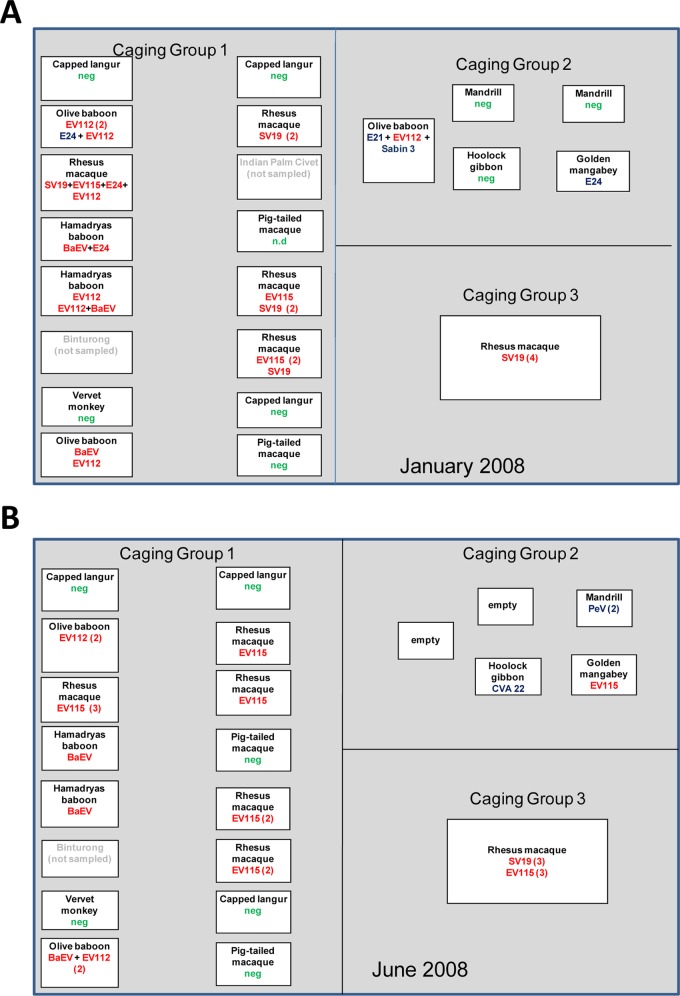
The NHP sampled at the Dhaka Zoo were housed in three groups (Fig. 1). Caging group 1 has 16 large, open-air, metal-bar cages arranged in parallel rows. Individual cages within each group are elevated on concrete platforms above ground level and separated by 3 to 10 m. The construction and arrangement of the cages in caging group 2 are similar yet smaller. This grouping is located several hundred meters from caging groups 1 and 3. The location of caging group 3 is closer to the entrance of the zoo. This is a very large, open-air cage, approximately 30 by 20 by 10 m high.
Fig 1.
Layout and occupancy of NHP cages at the Dhaka Zoo. Multiple species of NHP are housed throughout the Dhaka Zoo in three groups. Caging group 1 has 16 large, open-air, metal-bar cages arranged in parallel rows. Individual cages within each group are elevated on concrete platforms above ground level and separated by 3 to 10 m. The construction and arrangement of the cages in caging group 2 are similar yet smaller. The location of caging group 3, a very large open-air enclosure, is closer to the entrance of the zoo. Contact between zoo visitors and caged NHP is not uncommon. Fecal samples were collected in January 2008 (A) and June 2008 (B). The NHP species that was in the cage at the time of sampling is shown. The number of positive samples, if >1, is shown in parentheses. neg, picornaviruses were not detected; n.d., no samples were collected. Simian picornavirus serotypes are noted in red and human picornavirus serotypes in blue. In several fecal samples, both human and simian picornavirus serotypes were detected and are indicated by a plus sign.
Building on a study designed to characterize the picornavirus pathogen landscape among NHP in Bangladesh (7), we initiated a limited collection of NHP fecal specimens from the zoo in August 2007. This was followed up with the collection of fecal samples from all available NHP at the zoo during a single afternoon in January 2008 and in June 2008 (Table 1). January typically marks the low season for enterovirus circulation among humans in Bangladesh, and June marks the high season.
Table 1.
NHP species sampled
| Species | No. of picornaviruses detected |
Total (%) | ||
|---|---|---|---|---|
| August 2007 | January 2008 | June 2008 | ||
| Macaca mulatta, rhesus macaque | 5 | 14 | 15 | 34 (41) |
| M. nemestrina, pigtail macaque | 2 | 1 | 2 | 5 (6) |
| Trachypithecus pileatus, capped langur | 2 | 4 | 5 | 11 (13.2) |
| Hylobates hoolock, hoolock gibbon | 0 | 3 | 2 | 5 (6) |
| Papio cynocephalus, olive baboon | 0 | 6 | 4 | 10 (12.4) |
| Papio hamadryas, hamadryas baboon | 0 | 3 | 2 | 5 (6) |
| Mandrillus sphinx, mandrill | 0 | 3 | 2 | 5 (6) |
| Cercocebus chrysgaster, golden mangabey | 0 | 2 | 1 | 3 (3.6) |
| Cercopithecus aethiops, vervet | 0 | 2 | 3 | 5 (6) |
| Total | 9 | 38 | 36 | 83 |
Fecal material was collected only from freshly deposited stools. Specimens were kept cold and immediately frozen upon return from the field. Samples were stored at −20°C until shipped on dry ice to Atlanta, GA, for analysis.
Laboratory testing.
RNA was extracted directly from 10% (wt/vol) stool suspensions as described previously (8) and tested for enterovirus (EV), parechovirus (PeV), and sapelovirus by genus-specific TaqMan real-time reverse transcriptase (RT-PCR) assays, targeting the 5′ nontranslated region (5′ NTR) (9–11); assay details are described in reference 7.
Genus-specific, real-time PCR-positive specimens were confirmed by nested or seminested RT-PCR targeting a portion of the genome region encoding the VP1 capsid protein, followed by amplicon sequencing (8, 12). Virus type identity was determined by comparison of the VP1 amplicon nucleotide and deduced amino acid sequences with the VP1 sequences of all the reference strains for each virus genus by script-driven sequential pairwise comparison using the program Gap (Wisconsin Sequence Analysis Package, version 11.0; Accelrys, Inc., San Diego, CA) as described previously (8). For viruses which appeared to represent unique virus types (<75% nucleotide identity to all known types), complete VP1 sequences were determined and analyzed as described previously (13). If the complete VP1 sequences were still distinct from all known types, the sequence was forwarded to the Picornaviridae Study Group of the International Committee for the Taxonomy of Viruses for registration of a new type. VP1 nucleic acid sequences were aligned using the Pileup program (Wisconsin Package), and phylogenetic relationships were inferred by the neighbor-joining method implemented in MEGA version 4.0 (14) using the Kimura two-parameter method (15). Regions containing alignment gaps were omitted from the analysis. Support for specific tree topologies was estimated by bootstrap analysis with 1,000 pseudoreplicate data sets.
Nucleotide sequence accession numbers.
The sequences described here have been deposited in the GenBank sequence database under the accession numbers JX537975 to JX538032.
RESULTS
A total of 83 fecal samples from nine NHP species were collected and analyzed for this study (Table 1). Four of the NHP species, rhesus macaque, pigtail macaque, hoolock gibbon, and capped langur, are native to Bangladesh, but the provenance of the individual animals sampled for this study is unknown. Figure 1 shows cage locations and species represented at the January 2008 and June 2008 time points. Though the species contained in most cages remained unchanged between January 2008 and June 2008, we were unable to ascertain whether movement of individual animals had occurred between the two time points. The greatest number of samples were collected from the cages of rhesus macaques (41% of samples), followed by capped langurs (13%) and olive baboons (12%) (Table 1).
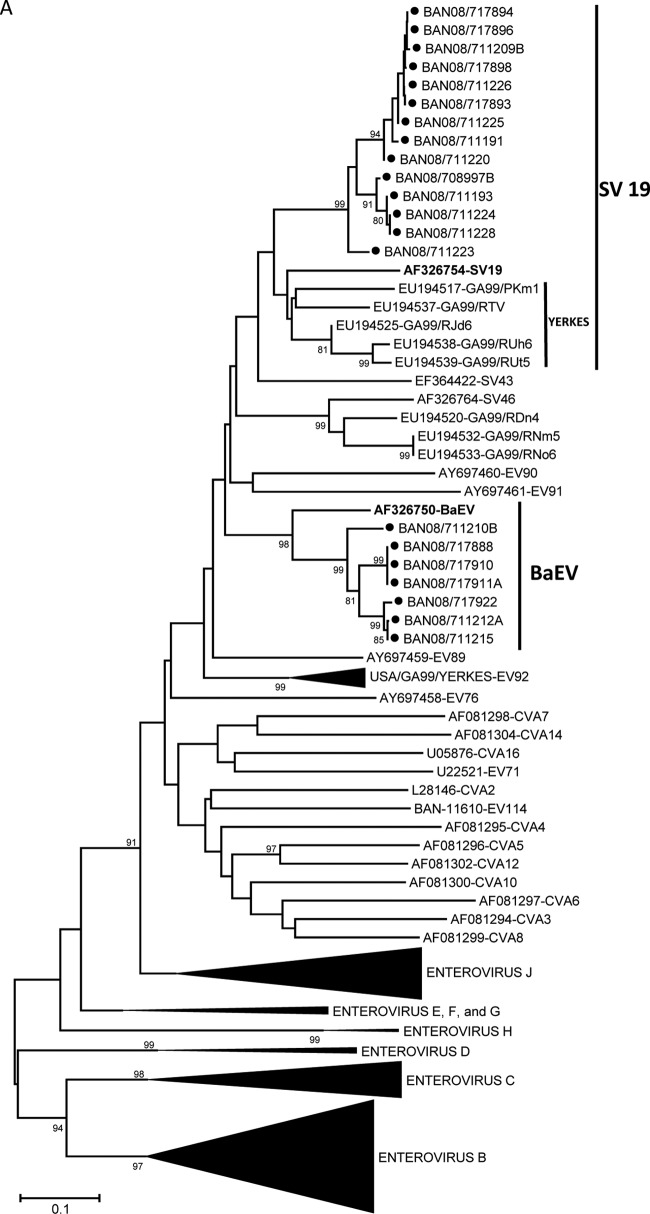
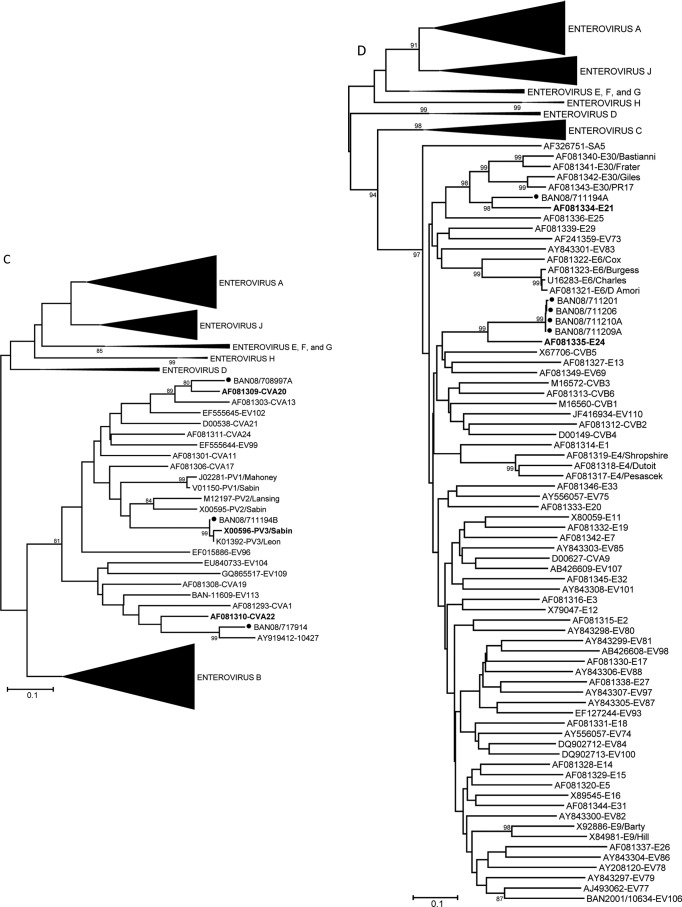
Genus-specific real-time PCR assays were used to directly screen RNA from fecal extracts for three human and simian enteric picornavirus genera (Enterovirus, Parechovirus, and Sapelovirus). Overall, picornaviruses were detected in 4 of 9 (44%) samples from August 2007, 23 of 38 (61%) samples from January 2008, and 25 of 36 (69%) samples from June 2008 (Table 2). Among these 52 fecal samples, ≥10 distinct picornavirus serotypes were detected (Table 3). Two of these serotypes, baboon enterovirus (BaEV) and simian virus 19 (SV19), have been solely associated with infection in NHP. The seven BaEV sequences were 90.2 to 100% identical to one another (92.9 to 100% amino acid identity) and 80.4 to 82.5% identical to the prototype BaEV strain (Fig. 2A). Similarly, the SV19 strains detected in 15 rhesus fecal samples were 90.1 to 100% identical to one another (91.2 to 100% amino acid identity) and 74.2 to 82.5% identical to other SV19 strains. The Bangladesh monkey SV19 sequences cluster with one another and separately from SV19 strains detected in an NHP research colony in the United States (Fig. 2A). Nineteen viruses were 93.1 to 100% identical to one another and only 70.5 to 73% identical to viruses in the SV6 group (proposed species “Enterovirus J,” forming a monophyletic cluster [Fig. 2B], suggesting they comprise a new type in EV-J) (http://www.picornastudygroup.com/proposals/2011/psg_proposals_2011.htm); these viruses have been assigned to a new type, EV115. The other 11 divergent viruses were 88.5 to 100% identical to one another and no more than 66% identical to any other serotype, forming a unique cluster of a new type, EV112, which may also be a member of EV-J (Fig. 2B). A more detailed characterization is needed to determine if EV112 should be classified as a member of EV-J or if it represents a new Enterovirus species (http://www.picornaviridae.com/enterovirus/unassigned/ev_unassigned_seq.htm). Three serotypes, EV112, EV115, and SV19, accounted for nearly 88% of all picornaviruses detected. Four of the serotypes detected in January 2008 persisted in June 2008 (Fig. 3). EV115 and SV19 were present at all three time points. Prevalence of infection was highest among the baboons, where 100% of the olive and hamadryas samples were positive. Among the rhesus macaques, 31 of 34 (91%) samples were positive for a picornavirus. In contrast, we detected no picornaviruses in samples from capped langurs or vervet monkeys.
Table 2.
Picornaviruses detected in NHP fecal samples
| Species | No. of picornaviruses detecteda |
Total no. of fecal samples in which picornavirus was detected (%) | ||
|---|---|---|---|---|
| August 2007 | January 2008 | June 2008 | ||
| Macaca mulatta, rhesus macaque | 3/5 EV115 (2), CVA20 + SV19 | 13/14 SV19 + EV115 + E24 + EV112, EV115 (3), SV19 (9) | 15/15 EV115 (11), SV19 (4) | 31/34 (91.2) |
| M. nemestrina, pigtail macaque | 1/2 EV115 + PeVb | 0/1 | 0/2 | 1/5 (40) |
| Trachypithecus pileatus, capped langur | 0/2 | 0/4 | 0/5 | 0/11 |
| Hylobates hoolock, hoolock gibbon | ND | 0/3 | 1/2 CVA22 | 1/5 (20) |
| Papio cynocephalus, olive baboon | ND | 6/6 E21 + EV112 + Sabin3, E24 + EV112, EV112 (3), BaEV | 4/4 BaEV + EV112 (2), EV112 (2) | 10/10 (100) |
| Papio hamadryas, hamadryas baboon | ND | 3/3 BaEV + E24, EV112 + BaEV, EV112 | 2/2 BaEV (2) | 5/5 (100) |
| Mandrillus sphinx, mandrill | ND | 0/3 | 2/2 PeV (2)a | 2/5 (40) |
| Cercocebus chrysgaster, golden mangabey | ND | 1/2 E24 | 1/1 EV115 | 2/3 (66) |
| Cercopithecus aethiops, vervet | ND | 0/2 | 0/3 | 0/5 |
| Total no. of positive fecal samples by sampling time point (%) | 4/9 (44) | 23/38 (61) | 25/36 (69) | 52/83 (63) |
ND, not detected. In several fecal samples, both human and simian picornavirus serotypes were detected and are indicated by a plus sign. The number of positive samples, if >1, is shown in parentheses.
Untyped PeV.
Table 3.
Picornavirus serotypes detected in NHP fecal samples
| Virus | Serotype | No. of samples | Species |
|---|---|---|---|
| Simian enterovirus types | |||
| Enterovirus A | BaEVa,b | 7 | Hamadryas baboon, olive baboon |
| Enterovirus A | SV19c | 15 | Rhesus macaque |
| Enterovirus J | EV112c,d,e,f | 12 | Rhesus macaque, hamadryas baboon, olive baboon |
| Enterovirus J | EV115c,g | 19 | Rhesus macaque, pigtail macaque, golden mangabey |
| Human enterovirus types | |||
| Enterovirus B | E21d | 1 | Olive baboon |
| Enterovirus B | E24a,c,f | 4 | Olive baboon, golden mangabey, hamadryas baboon, rhesus macaque |
| Enterovirus C | CVA20h | 1 | Rhesus macaque |
| Enterovirus C | CVA22 | 1 | Hoolock gibbon |
| Enterovirus C | Sabin-3d | 1 | Olive baboon |
| Parechovirus type | |||
| Parechovirus | Unknowng | 3 | Pigtail macaque, mandrill |
Includes one sample with a mixture of BaEV and E24.
Includes three samples with a mixture of EV112 and BaEV.
Includes one sample with a mixture of EV115, SV19, EV112, and E24.
Includes one sample with a mixture of E21, EV112, and Sabin-3.
Includes one sample with a mixture of BaEV and EV112.
Includes one fecal sample with detectable EV112 and E24 viruses.
Includes one fecal sample with detectable EV115 and an untyped PeV.
Includes one fecal sample with detectable CVA20 and SV19 viruses.
Fig 2.
Phylogenetic relationships of NHP and human enteroviruses from zoo NHP at the Dhaka Zoo based on an analysis of partial VP1 nucleic acid sequences, constructed using the Kimura two-parameter method (15) and the neighbor-joining algorithm implemented in MEGA 4.0 (14). Bootstrap values ≥80% are shown. Collapsed branches are shown as black wedges. Only fully double-stranded, full-length sequences were used for tree analyses. Some short sequences, adequate for virus identification purposes but not for phylogenetic reconstruction, were omitted. Phylogenetic trees are labeled in accordance with the 2011 ICTV proposal for renaming the enterovirus species (http://www.picornaviridae.com/). Designated prototype viruses are shown in bold for viruses detected in NHP. Viruses in the species Enterovirus A (A); viruses in the proposed species “Enterovirus J” (B); viruses in the species Enterovirus C (C); and viruses in the species Enterovirus B (D). Filled circles indicate viruses detected in the present study.
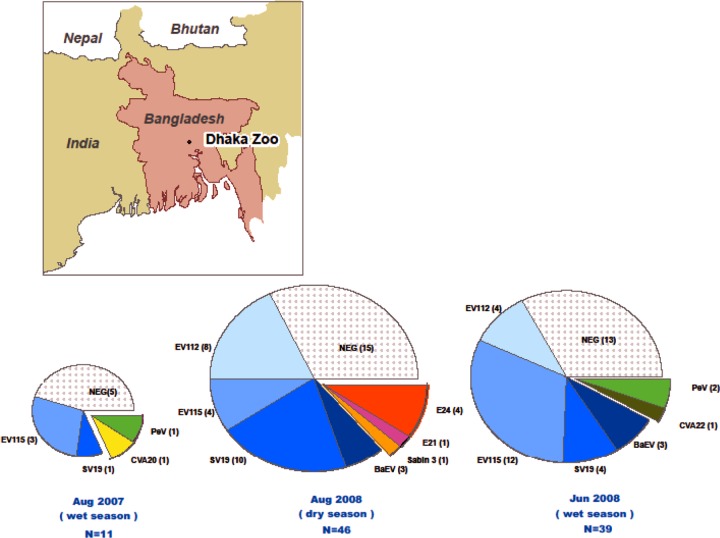
Fig 3.
Persistence of picornavirus serotypes among primates at the Dhaka Zoo. The Dhaka Zoo, located in the capital of Bangladesh (see inset map), houses multiple species of NHP. Fecal samples were collected from zoo primates in 2007 and 2008. The pie charts represent the picornavirus serotypes detected at each time point with the number of positive fecal samples shown in parentheses after each serotype. The majority of picornavirus serotypes detected (shown in the blue-shaded wedges as EV112, EV115, SV19, and BaEV) were those that have previously only been associated with infection in NHP. Human picornavirus serotypes are represented by other colored wedges. Only simian picornavirus serotypes persisted over time in these captive NHP.
Only 8 of the 64 (12.5%) enteroviruses detected in the zoo NHP were viruses that have been detected in humans. The most common human enterovirus was echovirus 24 (E24), with four detections in four NHP species at one time point (Fig. 3). Additionally there were four single EV detections (coxsackievirus A20 [CVA20], CVA22, E21, and the poliovirus type 3 (PV3) Sabin vaccine strain) (Table 3 and Fig. 2C and D). This finding is in stark contrast to what we detected among synanthropic NHP in Bangladesh, where 100% of the enteroviruses detected were also known to be circulating in the human population (7).
Three parechoviruses were detected by real-time RT-PCR, two in mandrills and one in a pigtail macaque, at two different time points (Tables 2 and 3). Parechovirus VP1 PCR was negative, so we were unable to type these viruses. Simian sapelovirus was not detected in any of the NHP fecal samples.
DISCUSSION
The majority of picornaviruses detected (53/64; 83%) in the zoo population were simian viruses, all of which were detected at more than one time point. In contrast, the 8 human EV types each appeared at only one time point. Numerous studies, mostly in the older literature, have identified simian EV in a wide range of NHP species (reviewed by reference 16). Several Asian monkey species have been shown to be experimentally infectible with coxsackie B viruses and certain other human enteroviruses (17–19), and there are a few recently documented detections of human picornaviruses in NHP (20–22).
We recently identified simian enteroviruses in rhesus and pigtail macaques in a research colony (6). The intratypic genetic diversity among the zoo viruses (up to 11.5% nucleotide sequence difference) suggests that simian EV circulate continuously among NHP in the Dhaka Zoo. In this study, the diversity among the research colony viruses suggested long-term circulation in the colony, rather than a single point source outbreak (6). Both of these captive environments apparently provide favorable conditions for continued circulation of enteric picornaviruses. It is unknown whether the viruses persistently infect individual captive NHP or the simian viruses are constantly retransmitted within the colony.
Three parechoviruses were detected in two NHP species in the Dhaka Zoo population, but none of these viruses were successfully typed. Two of the three PeV were detected at a single time point from NHP sharing a cage and are likely the same type. The pattern of high simian EV detection and low human EV type detection in the zoo NHP population may be reflected with the PeV as well. No exclusively simian PeV are currently known, but the study of PeV and reliable assays to detect them are recent developments. Synanthropic NHP had a higher incidence of PeV detection, and all of the typeable NHP PeV were of known human PeV types (7). It is somewhat unexpected that simian sapeloviruses were not detected in the zoo NHP, as they have been detected in the past in the same species and contexts as the simian enteroviruses. Unlike our assays for the other picornaviruses, which have been extensively optimized and validated (8–10, 12, 23), our sapelovirus assay is limited by the small number of available sequences from which to design primers and probes and the lack of a good collection of clinical material for assay validation (11).
Enterovirus seasonality in the human population is less pronounced in tropical and subtropical regions than in temperate climates, with peaks generally corresponding to the wet season. This pattern was mirrored in free-ranging Bangladeshi NHP that we sampled in 2007 and 2008, in which prevalence rose from 6% during the dry season to 14% during the wet season (7). In contrast, the prevalence of picornaviruses remained significantly higher among NHP at the Dhaka Zoo (mean, 62%), with no obvious seasonality. Further studies are needed to assess the role of simian picornaviruses in the health of captive NHP and their potential zoonotic transmission to humans.
ACKNOWLEDGMENTS
We thank our student assistants in the wildlife branch of the Department of Zoology at Jahangirnagar University and the management and staff of the Dhaka Zoo. We also thank the Bangladesh Forest Department for their permission and constant support of our research program. Administrative and data support were provided by R. Liszanckie and J. Johnson.
This work was partially supported by grants NIH-NIAID R03AI064865 and NIH-NCRR P51 RR 00166.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 24 October 2012
REFERENCES
- 1. Pallansch MA, Roos R. 2006. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p 839–893 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Oberste MS, Maher K, Michele SM, Belliot G, Uddin M, Pallansch MA. 2005. Enteroviruses 76, 89, 90 and 91 represent a novel group within the species Human enterovirus A. J. Gen. Virol. 86:445–451 [DOI] [PubMed] [Google Scholar]
- 3. Racaniello VR. 2007. Picornaviridae: the viruses and their replication, p 795–838 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 4. Pallansch MA, Oberste MS. 2009. Enteroviruses and parechoviruses, p 249–282 In Specter S, Hodinka RL, Young SA. (ed), Clinical virology manual. ASM Press, Washington, DC [Google Scholar]
- 5. Alexander JP, Jr, Gary HE, Jr, Pallansch MA. 1997. Duration of poliovirus excretion and its implications for acute flaccid paralysis surveillance: a review of the literature. J. Infect. Dis. 175:S176–S182 [DOI] [PubMed] [Google Scholar]
- 6. Nix WA, Jiang B, Maher K, Strobert E, Oberste MS. 2008. Identification of enteroviruses in naturally infected captive primates. J. Clin. Microbiol. 46:2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oberste MS, Feeroz MM, Maher K, Nix WA, Engel GA, Hasan KM, Begum S, Oh G, Chowdhury AH, Pallansch MA, Jones-Engel L. 2013. Characterizing the picornavirus landscape among synanthropic nonhuman primates in Bangladesh, 2007 to 2008. J. Virol. 87:558–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nix WA, Oberste MS, Pallansch MA. 2006. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 44:2698–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kilpatrick DR, Yang C-F, Ching K, Vincent A, Iber J, Campagnoli R, Mandelbaum M, De L, Yang S-J, Nix A, Kew OM. 2009. Rapid group-, serotype-, and vaccine strain-specific identification of poliovirus isolates by real-time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. J. Clin. Microbiol. 47:1939–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nix WA, Maher K, Niklasson B, Lindberg M, Johansson S, Pallansch MA, Oberste MS. 2008. Detection of all known parechoviruses by real time-PCR. J. Clin. Microbiol. 46:2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oberste MS, Maher K, Pallansch MA. 2003. Genomic evidence that simian virus 2 and six other simian picornaviruses represent a new genus in Picornaviridae. Virology 314:283–293 [DOI] [PubMed] [Google Scholar]
- 12. Nix WA, Maher K, Pallansch MA, Oberste MS. 2010. Parechovirus typing in clinical specimens by nested or semi-nested VP1 PCR coupled with sequencing. J. Clin. Virol. 48:202–207 [DOI] [PubMed] [Google Scholar]
- 13. Oberste MS, Schnurr D, Maher K, al-Busaidy S, Pallansch MA. 2001. Molecular identification of new picornaviruses and characterization of a proposed enterovirus 73 serotype. J. Gen. Virol. 82:409–416 [DOI] [PubMed] [Google Scholar]
- 14. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 15. Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120 [DOI] [PubMed] [Google Scholar]
- 16. Kalter SS, Heberling RL. 1971. Comparative virology of primates. Bacteriol. Rev. 35:310–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lashkevich VA, Koroleva GA, Tereshkina NV, Lukashev AN, Grigor'eva LV, Titova IP. 1996. Superacute lethal liver necrosis in monkeys infected with highly pathogenic variants of enteroviruses (ECHO 11 and ECHO 19 viruses). Vopr. Virusol. 41:198–206 (In Russian.) [PubMed] [Google Scholar]
- 18. Schmidt NJ, Dennis J, Lennette EH, Ho HH, Shinomoto TT. 1965. Antibody responses of rhesus (Macaca mulatta) monkeys experimentally infected with coxsackieviruses of group B and group A, type 9. J. Immunol. 95:54–69 [PubMed] [Google Scholar]
- 19. Wenner HA, TeYong L, Kamitsuka PS. 1961. Experimental infections with coxsackie viruses. I. Studies on virulence and pathogenesis in cynomolgus monkeys. Arch. Gesamte Virusforsch. 10:426–450 [PubMed] [Google Scholar]
- 20. Harvala H, Sharp CP, Ngole EM, Delaporte E, Peeters M, Simmonds P. 2011. Detection and genetic characterization of enteroviruses circulating among wild populations of chimpanzees in Cameroon: relationship with human and simian enteroviruses. J. Virol. 85:4480–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He W, Lu H, Song D, Cheng J, Gai X, Chen Q, Gao F. 2008. Isolation and identification of coxsackievirus in Sichuan golden monkey. Chinese J. Virol. 24:312–316 [PubMed] [Google Scholar]
- 22. Shan TL, Wang CM, Cui L, Delwart E, Yuan CL, Zhao W, Guo W, Dai XQ, Yu Y, Hua XG. 2010. Human parechovirus infections in monkeys with diarrhea, China. Emerg. Infect. Dis. 16:1168–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oberste MS, Gotuzzo E, Blair PJ, Nix WA, Ksiazek TG, Comer JA, Goldsmith CA, Olson JG, Kochel T. 2009. Human encephalomyocarditis virus disease in Peru. Emerg. Infect. Dis. 15:640–646 [DOI] [PMC free article] [PubMed] [Google Scholar]