Abstract
Effective vaccination programs have dramatically reduced the number of measles-related deaths globally. Although all the available data suggest that measles eradication is biologically feasible, a structural and biochemical basis for the single serotype nature of measles virus (MV) remains to be provided. The hemagglutinin (H) protein, which binds to two discrete proteinaceous receptors, is the major neutralizing target. Monoclonal antibodies (MAbs) recognizing distinct epitopes on the H protein were characterized using recombinant MVs encoding the H gene from different MV genotypes. The effects of various mutations on neutralization by MAbs and virus fitness were also analyzed, identifying the location of five epitopes on the H protein structure. Our data in the present study demonstrated that the H protein of MV possesses at least two conserved effective neutralizing epitopes. One, which is a previously recognized epitope, is located near the receptor-binding site (RBS), and thus MAbs that recognize this epitope blocked the receptor binding of the H protein, whereas the other epitope is located at the position distant from the RBS. Thus, a MAb that recognizes this epitope did not inhibit the receptor binding of the H protein, rather interfered with the hemagglutinin-fusion (H-F) interaction. This epitope was suggested to play a key role for formation of a higher order of an H-F protein oligomeric structure. Our data also identified one nonconserved effective neutralizing epitope. The epitope has been masked by an N-linked sugar modification in some genotype MV strains. These data would contribute to our understanding of the antigenicity of MV and support the global elimination program of measles.
INTRODUCTION
Measles virus (MV) used to be a major cause of death in children. The WHO estimated that ∼4% of deaths in children under 5 years of age were caused by measles during the period from 2000 to 2003 (1). However, effective vaccination programs have eliminated measles in the Western Hemisphere, and several other countries are approaching measles elimination (2). In the last decade, the number of measles-related deaths was reduced by more than 90% in all WHO regions, except for Southeast Asia (2). This year, the Measles and Rubella Initiative (http://www.measlesinitiative.org/) developed the Global Measles and Rubella Strategic Plan 2012–2020, which aims to reduce global measles mortality by at least 95% compared with 2000 by the end of 2015 and to achieve measles elimination in at least five WHO regions by the end of 2020. One of the factors favoring measles eradication is the monotypic nature of MV. All the available data suggest that measles eradication is biologically feasible (3, 4). However, a biochemical, molecular, and virological basis for the monotypic nature of MV remains to be provided.
MV is an enveloped virus in the genus Morbillivirus of the family Paramyxoviridae and possesses two types of glycoprotein spikes, the hemagglutinin (H) and fusion (F) proteins, on the viral envelope. The H protein is responsible for binding to cellular receptors on the target host cells. The signaling lymphocyte activation molecule (SLAM) expressed on immune system cells and nectin4 expressed at adherens junctions in epithelia function as the principal receptors for MV (5–8). Binding of the H protein to a receptor triggers F protein-mediated membrane fusion between the virus envelope and the host cell plasma membrane. Although neutralizing Abs directed against each of the viral envelope glycoproteins are elicited, H protein-specific Abs mainly account for the protection against MV infection (9–11). All measles vaccines consist of live attenuated MV strains isolated about a half a century ago. Currently, 24 genotypes are recognized for MV, and all vaccine strains belong to the same single genotype (genotype A) (12). To date, measles vaccines have been effective, despite differences in the endemic genotypes present in different countries or regions. Consequently, based on these observations, there is no evidence to suggest that MV undergoes a major antigenic drift. Nevertheless, several studies have suggested that currently circulating MV strains show antigenic variations, which could potentially affect the efficacy of vaccination (4, 13–17). Many amino acid residues have been documented to constitute a portion of an epitope. The data show that the H protein has several neutralizing epitopes (NEs), which may locate at the receptor-binding site (RBS) or a region interacting with the F protein. A list of amino acids or regions, which may constitute an epitope, and Abs, which recognize these epitopes, has been provided by Bouche et al. (10). Recently, Hashiguchi et al. determined a crystal structure of the head domain of the H protein in complexes with the V domain of SLAM (18). The head domain of the H protein is formed with six β-sheets arranged in a six-bladed propeller fold (19). SLAM binds to a β-sheet using the side of the propeller fold structure (18). The H protein head forms a homodimer, which is further assembled into a tetrameric structure by forming a dimer of dimers (18). These data allowed us to conduct a fine characterization of epitopes on the H protein. In the present study, we identified the location of several neutralizing epitopes on the MV H protein structure, and characterized these epitopes, providing a molecular basis for the sustainability of the monotypic nature of MV.
MATERIALS AND METHODS
Cells.
II-18 (20) and B95a (21) cells were maintained in RPMI medium (Invitrogen) supplemented with 7.5% fetal calf serum (FCS). BHK/T7-9 cells constitutively expressing T7 RNA polymerase (22) were maintained in E-MEM (Invitrogen) supplemented with 10% tryptose phosphate broth and 5% FCS. Vero and Vero/hSLAM cells (Vero cells constitutively expressing human SLAM) (23) were maintained in DMEM (GIBCO) supplemented with 7.5% FCS.
MAbs.
Mouse monoclonal antibodies (MAbs) (A2, A26, B5, B12, E39, E81, E103, E128, and E185) were raised against the H protein of the Toyoshima MV strain (genotype A), and some of them were reported previously (24–26). Competitive binding enzyme-linked immunosorbent assays (ELISAs) were performed as reported previously (25). Briefly, peroxidase-conjugated B5, B69, B12, A2, A26, or C149 MAb was mixed with various dilutions of an unlabeled MAb (B5, E81, E128, E185, E39, or E103) and then allowed to react with the MV antigen-coated wells for 2 h. The binding of the peroxidase-conjugated MAb to the MV antigens was detected as described previously (27).
Plasmid construction.
All full-length genome plasmids were derived from the p(+)MV323 plasmid encoding the antigenic full-length cDNA of the IC-B strain (genotype D3) (28). The p(+)MV323-Luci plasmid, which has an additional transcriptional unit for the Renilla luciferase gene, was reported previously (24). The full-length genome plasmids encoding the H gene of different genotype strains were generated by replacing the H gene region of p(+)MV323-Luci with the corresponding cDNA for the Edmonston-tag [A] (29), MVi/Massachusetts.USA/26.09[B3], MVi/New York.USA/22.09[D4], MVi/Vietnam/29.01[D5], MVi/Okinawa.JPN/37.09[D8], MVi/California.USA/5.9[D9], and MVi/Pennsylvania.USA/20.09[H1] strains. Point mutations, Q391R and Q311R, were introduced into the H gene region of p(+)MV323-Luci by site-directed mutagenesis. Similarly, G211S, E235G, Y252H, L284F, L296F, and G302R mutations were independently introduced into the H gene region of p(+)MV323-Ed/H-Luci (plasmid for the IC/Ed-H-Luci virus) (24).
Viruses.
IC323-EGFP, IC323-Luci, IC/EdH-EGFP, IC/EdH-Luci, MV323-EGFP-H-β12(N481Y), MV323-EGFP-H8, -H9, -H10, and -H11 were reported previously (24, 30–32). Other recombinant MVs were generated from the respective full-length genome plasmid as reported previously (33).
Neutralizing assay.
A suspension of recombinant MV (2,000 PFU per 75 μl) was incubated with serially diluted MAbs for 30 min at 37°C (the first dilution of each MAb was 1:10, followed by 2-fold dilutions). After incubation with the MAb, the virus solution was inoculated into culture media of confluent monolayers of II-18 (nectin4+) and B95a (SLAM+) cells. For recombinant viruses possessing the genotype A H gene (genotype A viruses; IC/EdH-Luci and IC/EdH-EGFP) and their mutants, CD46-dependent infection was blocked by a MAb against CD46 (clone M75) when the assay was performed with II-18 cells. At 2 days postinfection, the luciferase activity in the cells was measured using a Dual-Glo luciferase assay system (Promega). The neutralizing titer was indicated by the maximum dilution point, exhibiting >50% reduction of luciferase activity. The amount of Ig in each MAb solution (mouse ascites) was analyzed by an ELISA that detects the Fc region of Ig (TaKaRa). The neutralizing titers were normalized by the amount of Ig and are shown in the tables. When enhanced green fluorescent protein (EGFP)-expressing recombinant MVs were used for neutralizing assays, monolayers of II-18 and B95a cells in 24-well cluster plates were incubated with neutralized virus samples for 1 h at 37°C. After a 1-h incubation, 200 μl of RPMI medium supplemented with 7.5% fetal bovine serum (FBS) and 100 μg/ml fusion-blocking peptide (Z-D-Phe-Phe-Gly) (Peptide Institute Inc., Osaka, Japan) was added to each well to block a second round of infection by progeny viruses. At 48 h postinfection, the number of EGFP-expressing cells was counted under a fluorescence microscope. The cell number was expressed in cell infectious units (CIU). The number of CIU for each recombinant MV was also determined in the absence of the Ab and compared with that in the presence of the Ab. The number of CIU for each virus without the Ab was set to 100%.
Ab-selected escape mutants.
Recombinant MVs (IC323-EGFP and IC/EdH-EGFP) were incubated with a MAb (B5, E81, or E103) (the Ig concentrations were 0.5 to 1.4 mg/ml) for 30 min at 37°C and then propagated in B95a or II-18 cells in the presence of the MAb. At 2 days postinfection, several syncytia expressing EGFP were independently picked up, suspended in a small volume (100 μl) of culture medium, incubated with the respective MAb for 30 min at 37°C, and then cultured with fresh cells. This cycle was repeated twice, and the H gene nucleotide sequences of the selected mutants were determined as reported previously (33).
Replication kinetics.
II-18 cells on 6-well plates were infected with recombinant MVs at a multiplicity of infection (MOI) of 0.01 per cell. After various time intervals, cells were harvested with culture medium and determined for their CIU on Vero/SLAM cells (23).
H and F protein coimmunoprecipitation.
Monolayers of Vero cells on 6-well plates were transfected with 3 μg of pCG-EdmHn3xFLAG (Edmonston H protein containing an N-terminal triple Flag tag) (34) and 3 μg of pCG-EdmFc2xHA (Edmonston F protein containing a C-terminal double hemagglutinin [HA] tag) (35) in the presence of 100 μM fusion inhibitory peptide. MV H and F protein coimmunoprecipitation was performed as described previously (36). A mixture of CV1/CV4 MAbs (Millipore) served as a reference (1:1,000). MAbs B5, E81, E103, and E128 were used at a dilution of 1:2,000.
H and H-(473-477A) protein immunoprecipitation.
Vero cells on 6-well plates were transfected with 6 μg of pCG-EdmHn3xFLAG or pCG-EdmH473-477An3xFLAG (37). At 36 h posttransfection, the cells were lysed in M2 lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, protease inhibitors [Roche], 1 mM phenylmethylsulfonyl fluoride [PMSF]), cleared by centrifugation for 30 min at 20,000 × g and 4°C, and incubated with MV H protein ectodomain Abs (CV1/CV4 at 1:1,000 or E128 at 1:2,000). Immunoprecipitation, gel fractionation, and detection of Flag-tagged H protein antigenic material was performed as described previously (34).
Pulse-labeling and immunoprecipitation of MV proteins.
At 36 h postinfection, Vero/hSLAM cells infected with recombinant MVs were cultured in methionine-cysteine-deficient medium for 1 h, pulse-labeled with [35S]methionine-cysteine using EasyTag EXPRE35S35S protein labeling mix (PerkinElmer), and then lysed in radioimmunoprecipitation assay (RIPA) buffer. Polypeptides in the cell lysate were immunoprecipitated with a rabbit polyclonal Ab raised against the Toyoshima MV strain and analyzed by SDS-PAGE as reported previously (38). For endoglycosidase H (Endo-H) digestion, immunoprecipitated samples were eluted in 50 mM Tris-HCl (pH 7.4) containing 0.5% SDS by boiling for 4 min. The supernatants were then mixed with 0.1 M sodium citrate buffer (pH 5.3) containing Endo-H and incubated overnight at 37°C.
Surface plasmon resonance assay.
Surface plasmon resonance assays were performed using a Biacore 3000 (GE Healthcare) as reported previously (19). Briefly, the biotinylated H protein head domain was immobilized using a biotin capture kit (GE Healthcare) at 400 response units for binding experiments. A solution including each MAb was applied to the chip at a saturated state. Next, soluble human SLAM ectodomain comprising the N-terminal Ig V set domain tandemly connected to the corresponding Ig C2 set domain of mouse SLAM, which largely improves the protein stability, was placed on the chip at a flow rate of 10 ml/min at 25°C. Biotinylated bovine serum albumin (BSA) was used as a negative control. It was difficult to determine the proper regeneration conditions, because the immobilized MV H protein head domain was not as stable under typical regeneration conditions, such as acidic pH, basic pH, or other organic solutions. Therefore, we immobilized the same level of the H protein head domain for each binding experiment.
RESULTS
Antigenic sites I, II, and vi are effective neutralizing epitopes, and all except antigenic site II are conserved among different genotypes.
In 1985, Sato et al. (25) reported various sets of MAbs against MV structural proteins. In the study, 11 MAbs recognizing the H protein were characterized in detail. Based on competitive ELISA data, three nonoverlapping or partially overlapping antigenic sites (I, II, and III) in the H protein were predicted (25). In addition to these 11 MAbs, we analyzed a further five MAbs (E81, E128, E185, E39, and E103) directed against the H protein by competitive binding ELISAs (Table 1). E81 and E128 completely inhibited the binding of MAbs recognizing antigenic sites I and II, respectively (Table 1). On the other hand, neither E185, E39, nor E103 showed complete inhibition against the binding of MAbs recognizing antigenic sites I, II, and III. E39 and E185 only partially inhibited the binding of MAbs recognizing antigenic sites I and III, respectively (Table 1). E103 showed no competitive inhibition against the binding of MAbs recognizing antigenic sites I, II, and III. Based on these data, we tentatively predicted antigenic sites iv, v, and vi, which were recognized by E185, E39, and E103, respectively (Table 1). The newly tested MAbs (E81, E128, E185, E39, and E103) and B5 reported by Sato et al. (25) were then used for MV neutralizing assays. The antigenic site recognized by each MAb is shown in parentheses: B5(I), E81(I), E128(II), E185(iv), E39(v), and E103(vi). Eight distinct recombinant MVs were used as neutralization targets (Table 2; see also Table S1 in the supplemental material). These recombinant MVs were based on the IC323 genomic background and encoded a Renilla luciferase reporter gene and an H gene derived from different MV genotypes (A, B3, D3, D4, D5, D8, D9, and H1) (see Table S1) (28, 39). They were named on the basis of the genotype of the H gene: genotype A, B3, D3, D4, D5, D8, D9, and H1 viruses. MAbs B5(I), E81(I), E128(II), and E103(vi) showed high neutralizing titers in SLAM+ B95a and/or nectin4+ II-18 cells (20, 21). While B5(I), E81(I), and E103(vi) neutralized all of the MV genotypes tested, E128(II) was only effective against genotype A, B3, D8, and H1 viruses (Table 2). The neutralizing titers of E185(iv) and E39(v) were significantly lower than those of B5(I), E81(I), E128(II), and E103(vi) (Table 2). These data suggest that antigenic sites I, II, and vi are effective neutralizing epitopes and that, with the exception of antigenic site II, all epitopes are conserved among the different MV genotypes.
Table 1.
Summary of competitive binding ELISAs of anti-H protein MAbs
| Unlabeled Aba | Antigenic site | Antigenic site, peroxidase-labeled Abb |
|||||
|---|---|---|---|---|---|---|---|
| I, B5a | I, B69 | I, B12 | II, A2 | II, A26 | III, C146 | ||
| B5 | I | ++ | ++ | ++ | − | + | − |
| E81 | I | ++ | ++ | ++ | − | − | − |
| E128 | II | − | − | − | ++ | − | − |
| E185 | iv | − | − | − | − | − | + |
| E39 | v | − | − | + | − | − | − |
| E103 | vi | − | − | − | − | − | − |
MAbs used for neutralizing assays in the present study.
++, complete inhibition; +, partial inhibition; −, no inhibition.
Table 2.
Neutralizing titers against MV strains possessing H genes from different genotypes
| Cell line | Genotype, year isolated | Neutralizing titer against recombinant MV straina: |
|||||
|---|---|---|---|---|---|---|---|
| B5(I) | E81(I) | E128(II) | E185(iv) | E39(v) | E103(vi) | ||
| B95a | A, 1954 | 30,144 | 863 | 1,968 | 494 | <27 | 2,558 |
| B3, 2009 | 60,288 | 1,727 | 1,968 | 987 | <27 | 10,231 | |
| D3, 1984 | 7,536 | 1,727 | 15 | 31 | <27 | 10,231 | |
| D4, 2009 | 30,144 | 1,727 | 31 | 62 | <27 | 20,462 | |
| D5, 2001 | 30,144 | 863 | 15 | 494 | <27 | 10,231 | |
| D8, 2009 | 60,288 | 1,727 | 1,968 | 987 | <27 | 20,462 | |
| D9, 2010 | 60,288 | 1,727 | 15 | 987 | <27 | 20,462 | |
| H1, 2009 | 30,144 | 1,727 | 1,968 | 494 | <27 | 20,462 | |
| II-18 | A, 1954 | 30,144 | 27,631 | 62,977 | 124 | 1,750 | 10,231 |
| B3, 2009 | 30,144 | 27,631 | 62,977 | 1,974 | 1,750 | 10,231 | |
| D3, 1984 | 3,768 | 27,631 | 123 | 62 | <27 | 20,462 | |
| D4, 2009 | 30,144 | 27,631 | 31 | <8 | <27 | 10,231 | |
| D5, 2001 | 30,144 | 27,631 | <4 | 1,974 | <27 | 20,462 | |
| D8, 2009 | 60,288 | 27,631 | 62,977 | 987 | <27 | 10,231 | |
| D9, 2010 | 60,288 | 27,631 | 123 | 1,974 | <27 | 10,231 | |
| H1, 2009 | 15,072 | 13,815 | 62,977 | 494 | 1,750 | 10,231 | |
Neutralizing titers for 1 mg/ml of Ig.
Antigenic site I is located near the RBS, while antigenic site vi is located at a position distant from the RBS.
To identify the locations of antigenic sites on the H protein, EGFP-expressing MVs (IC323-EGFP) (30) that escaped from neutralization were isolated, and the amino acid changes in the H protein were identified by sequence analysis. Of the two escape mutants obtained, one escaped from neutralization by E81(I) through a Q391R mutation, and the other escaped E103(vi) through a Q311R mutation. These mutations were rebuilt in the luciferase gene-encoding recombinant IC323-Luci genome (24) by site-directed mutagenesis and reverse genetics (see Table S1 in the supplemental material). The resulting IC323-Luci-H(Q391R) was largely resistant to B5(I) but neutralized by E103(vi) with similar efficiency to that found for the parental virus (Table 3). The IC323-Luci-H(Q311R) recombinant was neutralized efficiently by B5(I) and E81(I) but escaped from neutralization by E103(vi) (Table 3). The amino acid residues at positions 391 and 311 are located within previously recognized epitopes on the H protein (10, 18, 19), which apparently correspond to antigenic sites I and vi (Fig. 1B and Table 4). The amino acid changes observed in the escape mutants indicate that antigenic site I is located near the SLAM-binding site, while antigenic site vi is located at a more distal position, although both act as effective neutralizing epitopes (Fig. 1B and Table 2). Mutagenesis of the H protein confirmed that the nectin4-binding site is distinct from the SLAM-binding site but probably shows a partial overlap (32, 40). This is consistent with the neutralization of MV by B5(I) and E81(I) on II-18 cells (Table 2). Epitope vi recognized by E103 corresponds to the previously predicted epitope recognized by BH38 and I-29 (Table 4) (10, 41, 42). Importantly, H mutants with Q391R or Q311R replicated somewhat less efficiently than the parental virus in cultured cells (Fig. 2), suggesting conformational effects of these changes on the H protein structure.
Table 3.
Neutralizing titers against recombinant MVs possessing the H protein of genotye D3 virus with amino acid substitutions
| Cell line | Mutation | Neutralizing titer against recombinant MVa: |
||
|---|---|---|---|---|
| B5(I) | E81(I) | E103(vi) | ||
| B95a | wt: (−) | 7,536 | 1,727 | 10,231 |
| Q391R | <471 | <14 | 20,462 | |
| Q311R | 1,884 | 1,727 | 80 | |
| II-18 | wt: (−) | 3,764 | 27,631 | 20,462 |
| Q391R | <4 | <3 | 20,462 | |
| Q311R | 1,884 | 27,631 | 20 | |
Neutralizing titers for 1 mg/ml of Ig.
Fig 1.
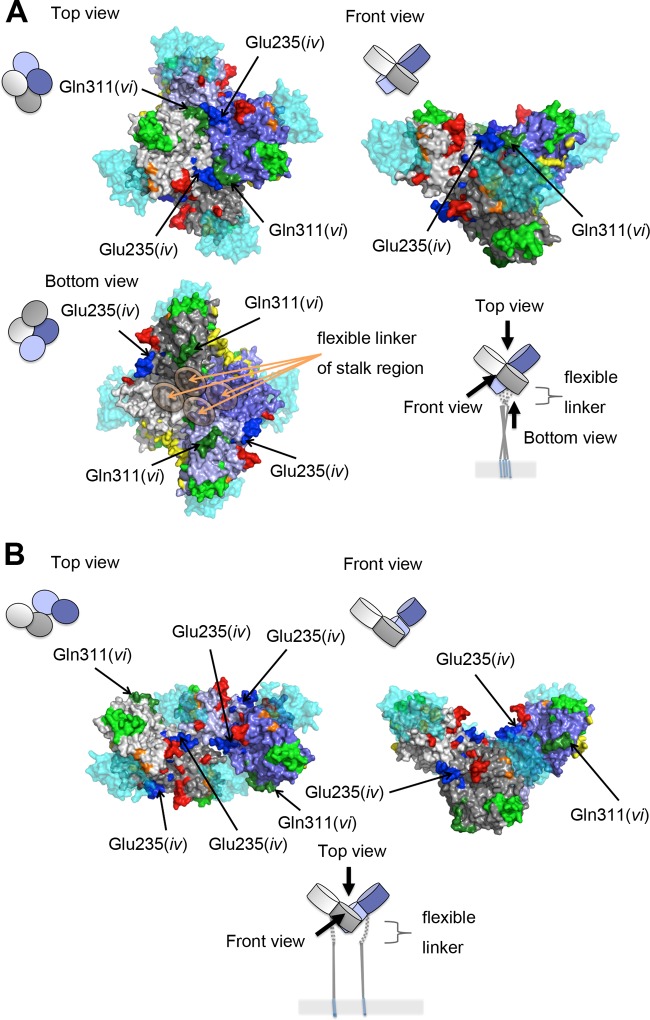
Locations of epitopes on the H protein dimeric structure. (A) Diagrams of a dimer of H protein dimers. The four H protein molecules are shown in gray, light gray, purple, and light purple. SLAM is shown in cyan. (B) Locations of epitopes I, iv, and vi. SLAM and predicted N-linked sugars are shown in translucent cyan and magenta, respectively. The amino acid residues demonstrated or suggested to constitute a portion of an epitope are shown in colors: residues on β-sheets 1, 2, 3, 4, 5, and 6 (18) are shown in blue, green, light green, yellow, orange, and red, respectively. (C) Location of epitope II. H protein dimers without (left) and with (right) the N-linked sugar at position 416 are shown. The figures were produced using PyMOL (Schrödinger; http://www.pymol.org).
Table 4.
Relation between epitopes and Absa
| Epitope name |
Region or amino acids which constitute an epitope | Ab which recognizes the epitope |
||
|---|---|---|---|---|
| Present study | Previous papers | Present study | Previous papers | |
| 126–135 | MAb48 | |||
| 185–195 | I-44 | |||
| iv | 233–240 | E185 | BH1 | |
| NE | 244–250 | BH47, BH59, BH129 | ||
| v | 302 | E39 | ||
| vi | 309–318 | E103 | I-29, BH38 | |
| I | HNE | 380–400 | E81, B5 | BH6, BH21, BH216 |
| 377 | BH171 | |||
| 378 | BH168 | |||
| II | 473–477 | E128 | ||
| 491 | 16-CD-11 | |||
| 505–506 | 80-II-B2 | |||
| 552 | I-41 | |||
| 587–596 | MAb18 | |||
Fig 2.
Replication kinetics of two recombinant MVs possessing Q311R or Q391R mutations. (A) Replication kinetics of two escape mutants in II-18 cells. II-18 cells were infected with the MVs at an MOI of 0.01. At various time intervals, cells were harvested in the culture medium and their CIU values were determined in Vero/hSLAM cells. (B) EGFP autofluorescence in MV-infected monolayers of II-18 cells. Panels show representative images obtained with a fluorescence microscope at 6 days postinfection.
Antigenic site II is located near the bottom surface of the H protein head domain and is shielded by N-glycans attached to residue 416 in recent outbreak strains of MV.
As described above, E128(II) neutralized several MV genotypes but failed to neutralize genotype D3, D4, D5, and D9 viruses (Table 2). Comparison of the H protein sequences of various MV genotypes indicated that the H proteins of MV genotypes D3, D4, D5, and D9, but not of genotypes A, B2, D8, and H1, harbor an additional potential site for N-linked glycosylation at residue 416 (see Fig. S1 in the supplemental material) (17, 43, 44). The electrophoretic mobility of the H proteins of genotype D3, D4, D5, and D9 was markedly reduced compared with that of the H proteins of genotypes A, B3, D8, and H1 (Fig. 3A), suggesting that an additional N-glycan moiety may be present on the H proteins of the former genotypes. Follow-up analyses with recombinant MV variants featuring a chimeric H gene combining fragments of the IC323 (genotype D3) and Edmonston (genotype A) strains (see Table S1) (31) showed that the amino acid difference at position 416 served as a determinant for the differential response to neutralization by E128(II) (Fig. 2B and C and Table 5). The addition of an N-glycan was confirmed by endoglycosidase H (Endo-H) treatment (Fig. 3D) as reported previously (17). In addition, the H protein of the Edmonston strain entirely lost its reactivity with E128(II) when carrying amino acid substitutions at positions 473 to 477 (Fig. 3E) (34). The epitope II likely constitutes a portion of RBS, since the amino acid region at positions 473 to 477 is involved in interaction with CD46, a receptor for MV vaccine strains (10, 45). Taken together, these data underscore that the N-glycan at position 416 masked epitope II, one of the major antigenic sites, which is located in the vicinity of a CD46-binding site (Fig. 1C). However, it was previously reported that serum samples derived from vaccine recipients neutralized all MV strains efficiently, regardless of the glycosylation status at residue 416 (17). Thus, this glycosylation site does not amount to a serious concern with regard to current MV vaccines. However, the additional carbohydrate moiety may provide some advantage for the endemic spread of MV, since the MV strains associated with recent large outbreaks in Europe and Southeast Asia (D4 and D9, respectively) possess this glycosylation site (see Fig. S1 and S2 in the supplemental material) (46). On the other hand, the genotype H1 and D8 strains, which are currently endemic in China and India, respectively, do not have this extra glycosylation site (see Fig. S1 and S2).
Fig 3.
Masking of epitope II by the N416-linked sugar. (A) SDS-PAGE analyses for the mobility of H proteins of different genotypes. MV-infected Vero/hSLAM cells at 36 h postinfection were labeled with [35S]methionine-cysteine and lysed in RIPA buffer. Polypeptides were then immunoprecipitated with a polyclonal Ab against MV and resolved by SDS-PAGE. (B) SDS-PAGE analyses for the mobility of H proteins of previously reported recombinant MVs possessing a chimeric H gene between the IC323 (genotype D3) and Edmonston (genotype A) strains or various point mutations. (C) Neutralizing assays of E128 against EGFP-expressing MV strains possessing a chimeric H gene between the IC323 (genotype D3) and Edmonston (genotype A) strains or various point mutations. The CIU of each virus was determined in II-18 cells in the presence or absence of E128. The CIU determined in the presence of E128 was compared with that in the absence of E128. The CIU in the absence of E128 was set to 100%. (D) SDS-PAGE analyses for the mobility of Endo-H-treated H proteins of different genotypes. (E) A five-residue alanine substitution in the H protein at residues 473 to 477 prevents binding of MAb E128. Immunoprecipitation of MV HFlag and CD46-binding defective MV HFlag-(473-477A) with MAbs CV1/CV4 and E128. The immunoprecipitated material was gel fractionated, followed by immunoblotting and detection of HFlag with an anti-Flag M2 antibody. IgG ctrl; control IgG.
Table 5.
Amino acid differences in recombinant MVs
| Amino acid position | Amino acid difference in recombinant MVa: |
||||||
|---|---|---|---|---|---|---|---|
| A | D3 | β12(N481Y) | H8 | H9 | H10 | H11 | |
| 174 | T | A | T | A | A | A | A |
| 176 | T | A | T | A | A | A | A |
| 211 | G | S | G | S | S | S | S |
| 235 | E | G | E | G | G | G | G |
| 243 | K | G | K | G | G | G | G |
| 252 | Y | H | Y | H | H | H | H |
| 276 | L | F | L | F | F | F | F |
| 284 | L | F | L | F | F | F | F |
| 296 | L | F | L | F | F | F | F |
| 302 | G | R | G | R | R | R | R |
| 334 | Q | R | Q | R | R | R | R |
| 390 | I | N | N | I | I | N | I |
| 416 | D | N | N | D | N | D | D |
| 446 | S | T | T | T | S | S | S |
| 481 | Y | N | Y | Y | Y | Y | Y |
| 492 | G | E | E | G | G | G | E |
| 575 | Q | K | K | K | K | K | K |
These recombinant MVs were reported previously (31).
Antigenic sites iv and v, located at the lateral surface of the H protein head dimer, are less important for neutralization.
In contrast to genotype D3, MV strains of genotype A were neutralized by E185(iv) and E39(v) in SLAM+ B95a cells and nectin4+ II-18 cells, respectively (Table 2). We further tested two recombinant MVs possessing chimeric H genes derived from genotypes A and D3 or featuring point mutations [MV323-EGFP-H-β12(N481Y) and MV323-EGFP-H8] (31) for neutralization by E185(iv) and E39(v) (Fig. 4 and Table 5; see also Table S1 in the supplemental material). The neutralization data demonstrated that amino acids within residues 174 to 334 determined the difference in neutralization by E185(iv) and E39(v) between MVs with genotypes A and D3. Amino acid substitutions present in this region and predicted to be exposed on the H protein surface (19) were individually introduced into a genotype A virus, resulting in the generation of six additional MVs (Table 6; see also Table S1 in the supplemental material). These recombinant MVs were subjected to neutralization analyses. H protein mutations E235G and G302R rendered the recombinant MVs resistant to neutralization by E185(iv) and E39(v), respectively (Table 6). These data suggested that residues 235 and 302 are part of epitopes iv and v, respectively. Epitope iv is therefore likely to correspond to the previously reported epitope recognized by BH1 (amino acids 233 to 240 are critical for BH1 binding) (Fig. 5 and Table 4) (47). Both residues are located at the lateral surface of the H protein head dimer, distal from the RBS (Fig. 1B). Furthermore, these epitopes are quite close to the N-linked sugars, which may exert some steric hindrance to reduce the binding activities of the Abs (Fig. 1B). These observations are consistent with the weak neutralizing phenotypes of E185(iv) and E39(v) (Table 2).
Fig 4.
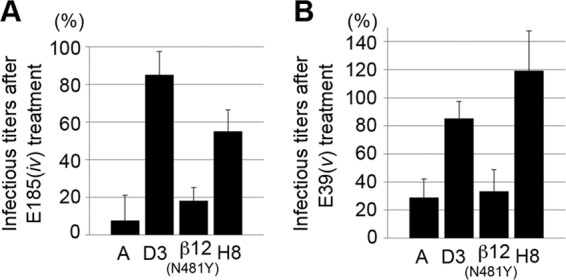
Neutralizing assays of E185 and E39 against EGFP-expressing MV strains with different H protein sequences. (A) The CIU of each virus was determined in B95a cells in the presence or absence of E185. (B) The CIU of each virus was determined in II-18 cells in the presence or absence of E39. The CIU determined in the presence of the Abs was compared with that in the absence of the Abs. The CIU in the absence of the Abs was set to 100%.
Table 6.
Neutralizing titers against recombinant MV possessing the Edmonston H protein with various amino acid substitutionsa
| Mutation | Neutralizing titer against recombinant MV: |
|
|---|---|---|
| E185(iv) in B95a | E39(v) in II-18 | |
| (−) | 494 | 1,750 |
| G211S | 494 | 875 |
| E235G | 62 | 1,750 |
| Y252H | 494 | 1,750 |
| L284F | 494 | 1,750 |
| L296F | 494 | 1,750 |
| G302R | 494 | <27 |
Neutralizing titers for 1 mg/ml of Ig. (−), parental virus.
Fig 5.

Amino acid sequence of epitope iv. The amino acid residues at positions 231 to 252 are shown. The residues known to constitute part of the epitope are shown in blue characters (top). The amino acid residues represented in the crystal structures are shown. Missing residues are indicated by dashes.
Neutralizing Abs that recognize antigenic sites I and II inhibit receptor binding, while neutralizing Abs specific for antigenic site vi interfere with the H-F protein interaction.
To mechanistically explore the basis for neutralization by the different MAbs, the effects of the MAbs on the interaction between the H protein (genotype A) and SLAM were analyzed by binding assays using surface plasmon resonance (Biacore assays). MAbs B5(I), E81(I), and E128(II) blocked the binding of the H protein to SLAM, whereas E103(vi) did not (Table 7), although all of the MAbs had high neutralization activities (Table 2). These data are consistent with the observations that epitopes I and II are located near the RBS (10, 45), while epitope vi has a more distal position (Fig. 1B). It remains uncertain how epitope vi can be a major neutralizing target site. It can be speculated that the epitope may be involved in one or some of the fusion steps other than the receptor binding. To examine whether the H-F protein interaction affects MAb binding, coimmunoprecipitation of MV envelope glycoprotein complexes was performed (36). Compared with a reference Ab cocktail, the amount of F protein coimmunoprecipitated with H protein was reduced by ∼50% when E103(vi) was used (Fig. 6). These data suggested that epitope vi is influenced by the H-F protein interaction. Interestingly, the higher order (tetrameric) structures of the H protein-SLAM complex proposed on the basis of the crystal structures suggest that epitope vi in two of the four H molecules (gray and light purple H molecules) is located at the bottom surface proximal to the stalk region, while that in the other two molecules (light gray and purple H molecules) forms part of the interface of the H protein dimers in form I, one of the tetrameric structures (Fig. 7A) (18), but not form II, the other tetrameric structure (Fig. 7B) (18). Furthermore, epitope vi appears to contact epitope iv at the interface (Fig. 7A). Epitope iv corresponds to the BH1-binding epitope (Table 4) (47), and most likely forms a single epitope together with a previously reported linear neutralizing epitope (NE) (amino acid positions 244 to 250) (Table 4) (47), although a definitive conclusion could not be made, because eight amino acids (positions 240 to 247) were so flexible that little electron density was observed in the H crystal structures (Fig. 5) (18).
Table 7.
Competitive binding of anti-H protein MAbs against SLAM
| Ab | Antigenic site | Competitive activitya |
|---|---|---|
| B5 | I | ++ |
| E81 | I | + |
| E128 | II | ++ |
| E103 | vi | − |
++, Complete inhibition; +, weak inhibition; −, no inhibition.
Fig 6.
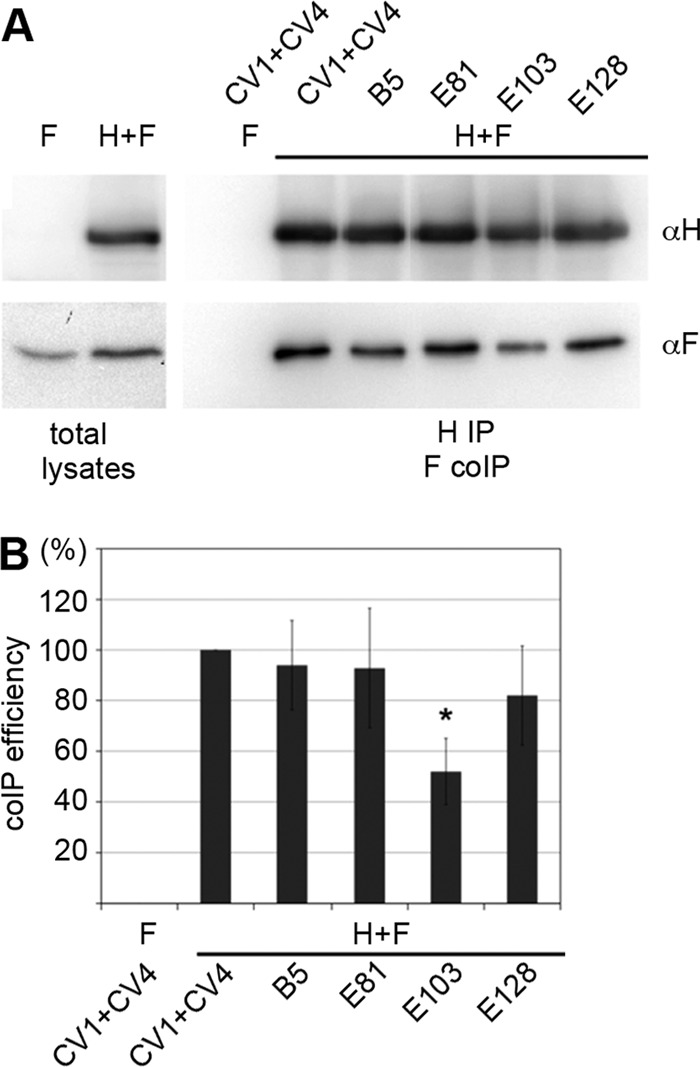
MV H and F protein coimmunoprecipitation. MAb E103 shows reduced immunoprecipitation efficiency for H/F hetero-oligomers. (A) Vero cells transiently transfected with expression plasmids encoding MV HFlag and FHA, or FHA only as a control, were immunoprecipitated with specific MAbs directed against the MV H protein ectodomain (CV1/CV4, B5, E81, E103, or E128 as indicated). Precipitated HFlag material was labeled with an anti-Flag M2 antibody, and the coprecipitated FHA was detected with an anti-HA 16b12 antibody. (B) For densitometric quantification, the ratio of FHA to HFlag signals was determined for each MAb, followed by normalization to the coimmunoprecipitation ratio obtained with the CV1/CV4 reference MAb mixture. Data represent the means ± standard errors of the means (SEM) of results from three experiments. The asterisk indicates that, in F densitometry, a t test returned P values of <0.05 for the difference in coimmunoprecipitation (CoIP) efficiency, with E103 relative to the CV1+CV4 reference.
Fig 7.
Locations of epitopes iv and vi on the H protein tetrameric structure. The four H protein molecules are shown in gray, light gray, purple, and light purple. SLAM is shown in translucent cyan. The amino acid residues demonstrated or suggested to constitute a portion of an epitope are shown in colors: residues on β-sheets 1, 2, 3, 4, 5, and 6 (18) are shown in blue, green, light green, yellow, orange, and red, respectively. (A) A tetrameric structure in form I (18). (B) A tetrameric structure in form II (18).
DISCUSSION
In the present study, we focused on neutralizing Abs directed against the H protein, because the protective immunity is predominantly humoral (48) and H protein-specific Abs mainly account for the neutralization activity in serum from vaccinated individuals (9–11). A key finding in the present study is that the antigenic site vi, which is unrelated to receptor binding but probably involved in the formation of a higher-order H-F protein oligomeric structure (34, 36, 49), is a major neutralizing epitope that is conserved among different genotype strains. This type of epitopes has been predicted by previous studies, although receptor binding has been tested for CD46, but not the wild-type receptors, SLAM, or nectin4 (10, 47, 50). One of them, known as NE, is recognized by BH47, BH59, and BH129 MAbs (Table 4) (10, 47). The other is recognized by I-29 (10, 50) and was determined as the epitope vi in the present study (Table 4). Therefore, our data are consistent with the previous studies that MAbs that recognize the epitope vi neutralize MV infection by inhibiting virus fusion without affecting the receptor binding (10, 50). Membrane fusion of MV is mediated by concerted actions of the H and F proteins. Binding of the H protein to a receptor triggers F protein-mediated membrane fusion. Although the triggering mechanism remains largely unknown, accumulated data indicate that rearrangement of an H-F protein oligomeric structure is a key for triggering fusion (18, 34, 36, 49). This epitope is fully conserved among different MV genotypes. A requirement for the formation of a functional fusion complex is likely to generate structural constraints that prevent substitutions in this epitope.
We also found that a large area containing epitopes I and II serves as a target for the humoral anti-MV response. Our data showed that MAbs B5 and E81 bind to an epitope (epitope I) corresponding to a previously reported hemagglutination and neutralization epitope (HNE) (amino acid positions 380 to 400), which is recognized by MAbs BH6, BH21, and BH216 (Table 4 and Fig. 8) (10, 41, 51). Six residues (positions 386, 387, 388, 391, 394, and 395) were shown to be critical for Ab binding (52). Accordingly, a Q391R substitution was sufficient for MV escape from neutralization by B5 and E81. On the other hand, epitope II maps to residues 473 to 477, which are located at the bottom surface of the H protein head domain and involved in interaction with CD46 (10, 45). In some of the current wild-type MV strains, this epitope is masked by an additional carbohydrate moiety (Asn416). Previous studies revealed that MV escape from MAb 16-CD-11 can be achieved through an amino acid substitution at position 491 in the H protein (41) and that escape mutants from MAbs BH171 and BH168 possess substitutions at positions 377 and 378, respectively (Table 4) (10, 53). The residues at positions 491, 377, and 378 in the H protein are located between the HNE (epitope I) and epitope II (Table 4 and Fig. 8). It was further shown that H protein binding of MAbs BH30 and BH99 competes with 16-CD-11 (10, 41). Taken together, these data support that a large area of the H protein head domain spanning from epitope I to epitope II can serve as a target for neutralizing Abs (Fig. 8). Our data suggest that the region around epitope I has structural constraints for change, since it is conserved among different genotype strains, thereby contributing to the single serotype nature of MV.
Fig 8.
A major epitope region covering a large area containing epitopes I and II. A side view of a dimer is shown. SLAM and predicted sugars are shown in translucent cyan and magenta, respectively. The amino acid residues demonstrated or suggested to constitute a portion of an epitope are shown in colors: residues on β-sheet 1, 2, 3, 4, 5, and 6 (18) are shown in blue, green, light green, yellow, orange, and red, respectively.
In summary, the H protein of MV possesses at least two conserved effective neutralizing epitopes. One, which is a previously recognized HNE epitope, is located near the RBS, and thus MAbs that recognize this epitope blocked the receptor binding of the H protein. On the other hand, the other epitope is located at the position distant from the RBS. Thus, MAbs that recognizes this epitope did not inhibit the receptor binding of the H protein but rather interfered with the H-F interaction. Based on the structural data of MV H protein, it was predicted that, when the H protein forms a tetramer, this epitope locates at two positions: the contact face of two H protein dimers and the bottom face of the head of the H protein tetramer. This epitope possibly plays a key role in the formation of a higher-order H-F protein oligomeric structure. Our data also demonstrated that one effective neutralizing epitope is not conserved, since the epitope has been masked by an N-linked sugar modification in some genotype MV strains. The data in the present study contribute to our understanding of the antigenicity of MV and support the global elimination program of measles.
Supplementary Material
ACKNOWLEDGMENTS
We thank T.A. Sato and T. Seya for providing MAbs, N. Ito and M. Sugiyama for providing the BHK/T7-9 cells, and K. Taira for providing an MV isolate. We also thank Y. Yanagi for invaluable suggestions and providing useful cell lines and all the members of the Department of Virology 3, NIID, Japan, for technical support.
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labor and Welfare of Japan and by a grant from The Takeda Science Foundation (to M.T.). The work of M.A.B. and R.K.P. was supported in part by Public Health Service grant AI083402 from the NIH/NIAID (to R.K.P.).
Footnotes
Published ahead of print 31 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02033-12.
REFERENCES
- 1. Bryce J, Boschi-Pinto C, Shibuya K, Black RE. 2005. WHO estimates of the causes of death in children. Lancet 365:1147–1152 [DOI] [PubMed] [Google Scholar]
- 2. Strebel PM, Cochi SL, Hoekstra E, Rota PA, Featherstone D, Bellini WJ, Katz SL. 2011. A world without measles. J. Infect. Dis. 204(Suppl 1):S1–S3 [DOI] [PubMed] [Google Scholar]
- 3. Sanders R, Dabbagh A, Featherstone D. 2011. Risk analysis for measles reintroduction after global certification of eradication. J. Infect. Dis. 204(Suppl 1):S71–S77 [DOI] [PubMed] [Google Scholar]
- 4. Bellini WJ, a Rota PA. 2011. Biological feasibility of measles eradication. Virus Res. 162:72–79 [DOI] [PubMed] [Google Scholar]
- 5. Tatsuo H, Ono N, Tanaka K, Yanagi Y. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406:893–897 [DOI] [PubMed] [Google Scholar]
- 6. Noyce RS, Bondre DG, Ha MN, Lin LT, Sisson G, Tsao MS, Richardson CD. 2011. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 7:e1002240 doi:10.1371/journal.ppat.1002240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muhlebach MD, Mateo M, Sinn PL, Prufer S, Uhlig KM, Leonard VH, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, Ramachandran S, McCray PB, Jr, Cichutek K, von Messling V, Lopez M, Cattaneo R. 2011. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 480:530–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takeda M, Tahara M, Nagata N, Seki F. 2011. Wild-type measles virus is intrinsically dual-tropic. Front. Microbiol. 2:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Swart RL, Yuksel S, Osterhaus AD. 2005. Relative contributions of measles virus hemagglutinin- and fusion protein-specific serum antibodies to virus neutralization. J. Virol. 79:11547–11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bouche FB, Ertl OT, Muller CP. 2002. Neutralizing B cell response in measles. Viral Immunol. 15:451–471 [DOI] [PubMed] [Google Scholar]
- 11. de Swart RL, Yuksel S, Langerijs CN, Muller CP, Osterhaus AD. 2009. Depletion of measles virus glycoprotein-specific antibodies from human sera reveals genotype-specific neutralizing antibodies. J. Gen. Virol. 90:2982–2989 [DOI] [PubMed] [Google Scholar]
- 12. WHO 2012. Measles virus nomenclature update: 2012. Wkly. Epidemiol. Rec. 87:73–81 [PubMed] [Google Scholar]
- 13. Finsterbusch T, Wolbert A, Deitemeier I, Meyer K, Mosquera MM, Mankertz A, Santibanez S. 2009. Measles viruses of genotype H1 evade recognition by vaccine-induced neutralizing antibodies targeting the linear haemagglutinin noose epitope. J. Gen. Virol. 90:2739–2745 [DOI] [PubMed] [Google Scholar]
- 14. Shi J, Zheng J, Huang H, Hu Y, Bian J, Xu D, Li F. 2011. Measles incidence rate and a phylogenetic study of contemporary genotype H1 measles strains in China: is an improved measles vaccine needed? Virus Genes 43:319–326 [DOI] [PubMed] [Google Scholar]
- 15. Tamin A, Rota PA, Wang ZD, Heath JL, Anderson LJ, Bellini WJ. 1994. Antigenic analysis of current wild type and vaccine strains of measles virus. J. Infect. Dis. 170:795–801 [DOI] [PubMed] [Google Scholar]
- 16. Kuhne M, Brown DW, Jin L. 2006. Genetic variability of measles virus in acute and persistent infections. Infect. Genet. Evol. 6:269–276 [DOI] [PubMed] [Google Scholar]
- 17. Santibanez S, Niewiesk S, Heider A, Schneider-Schaulies J, Berbers GA, Zimmermann A, Halenius A, Wolbert A, Deitemeier I, Tischer A, Hebgek H. 2005. Probing neutralizing-antibody responses against emerging measles viruses (MVs): immune selection of MV by H protein-specific antibodies? J. Gen. Virol. 86:365–374 [DOI] [PubMed] [Google Scholar]
- 18. Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y. 2011. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat. Struct. Mol. Biol. 18:135–141 [DOI] [PubMed] [Google Scholar]
- 19. Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. 2007. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. U. S. A. 104:19535–19540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shirogane Y, Takeda M, Tahara M, Ikegame S, Nakamura T, Yanagi Y. 2010. Epithelial-mesenchymal transition abolishes the susceptibility of polarized epithelial cell lines to measles virus. J. Biol. Chem. 285:20882–20890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kobune F, Sakata H, Sugiura A. 1990. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J. Virol. 64:700–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ito N, Takayama-Ito M, Yamada K, Hosokawa J, Sugiyama M, Minamoto N. 2003. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol. Immunol. 47:613–617 [DOI] [PubMed] [Google Scholar]
- 23. Ono N, Tatsuo H, Hidaka Y, Aoki T, Minagawa H, Yanagi Y. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 75:4399–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takeda M, Tahara M, Hashiguchi T, Sato TA, Jinnouchi F, Ueki S, Ohno S, Yanagi Y. 2007. A human lung carcinoma cell line supports efficient measles virus growth and syncytium formation via a SLAM- and CD46-independent mechanism. J. Virol. 81:12091–12096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sato TA, Fukuda A, Sugiura A. 1985. Characterization of major structural proteins of measles virus with monoclonal antibodies. J. Gen. Virol. 66(Pt 7):1397–1409 [DOI] [PubMed] [Google Scholar]
- 26. Sato TA, Enami M, Kohama T. 1995. Isolation of the measles virus hemagglutinin protein in a soluble form by protease digestion. J. Virol. 69:513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sakata H, Hishiyama M, Sugiura A. 1984. Enzyme-linked immunosorbent assay compared with neutralization tests for evaluation of live mumps vaccines. J. Clin. Microbiol. 19:21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Takeda M, Takeuchi K, Miyajima N, Kobune F, Ami Y, Nagata N, Suzaki Y, Nagai Y, Tashiro M. 2000. Recovery of pathogenic measles virus from cloned cDNA. J. Virol. 74:6643–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter MA. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hashimoto K, Ono N, Tatsuo H, Minagawa H, Takeda M, Takeuchi K, Yanagi Y. 2002. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J. Virol. 76:6743–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tahara M, Takeda M, Seki F, Hashiguchi T, Yanagi Y. 2007. Multiple amino acid substitutions in hemagglutinin are necessary for wild-type measles virus to acquire the ability to use receptor CD46 efficiently. J. Virol. 81:2564–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tahara M, Takeda M, Shirogane Y, Hashiguchi T, Ohno S, Yanagi Y. 2008. Measles virus infects both polarized epithelial and immune cells by using distinctive receptor-binding sites on its hemagglutinin. J. Virol. 82:4630–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seki F, Yamada K, Nakatsu Y, Okamura K, Yanagi Y, Nakayama T, Komase K, Takeda M. 2011. The SI strain of measles virus derived from a patient with subacute sclerosing panencephalitis possesses typical genome alterations and unique amino acid changes that modulate receptor specificity and reduce membrane fusion activity. J. Virol. 85:11871–11882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brindley MA, Plemper RK. 2010. Blue native PAGE and biomolecular complementation reveal a tetrameric or higher-order oligomer organization of the physiological measles virus attachment protein H. J. Virol. 84:12174–12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Plemper RK, Hammond AL, Gerlier D, Fielding AK, Cattaneo R. 2002. Strength of envelope protein interaction modulates cytopathicity of measles virus. J. Virol. 76:5051–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paal T, Brindley MA, St Clair C, Prussia A, Gaus D, Krumm SA, Snyder JP, Plemper RK. 2009. Probing the spatial organization of measles virus fusion complexes. J. Virol. 83:10480–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Corey EA, Iorio RM. 2009. Measles virus attachment proteins with impaired ability to bind CD46 interact more efficiently with the homologous fusion protein. Virology 383:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tahara M, Takeda M, Yanagi Y. 2005. Contributions of matrix and large protein genes of the measles virus Edmonston strain to growth in cultured cells as revealed by recombinant viruses. J. Virol. 79:15218–15225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takeda M, Ohno S, Tahara M, Takeuchi H, Shirogane Y, Ohmura H, Nakamura T, Yanagi Y. 2008. Measles viruses possessing the polymerase protein genes of the Edmonston vaccine strain exhibit attenuated gene expression and growth in cultured cells and SLAM knock-in mice. J. Virol. 82:11979–11984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leonard VH, Sinn PL, Hodge G, Miest T, Devaux P, Oezguen N, Braun W, McCray PB, McChesney MB, Cattaneo R. 2008. Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J. Clin. Invest. 118:2448–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ertl OT, Wenz DC, Bouche FB, Berbers GA, Muller CP. 2003. Immunodominant domains of the Measles virus hemagglutinin protein eliciting a neutralizing human B cell response. Arch. Virol. 148:2195–2206 [DOI] [PubMed] [Google Scholar]
- 42. Hu A, Sheshberadaran H, Norrby E, Kovamees J. 1993. Molecular characterization of epitopes on the measles virus hemagglutinin protein. Virology 192:351–354 [DOI] [PubMed] [Google Scholar]
- 43. Rota JS, Hummel KB, Rota PA, Bellini WJ. 1992. Genetic variability of the glycoprotein genes of current wild-type measles isolates. Virology 188:135–142 [DOI] [PubMed] [Google Scholar]
- 44. Sakata H, Kobune F, Sato TA, Tanabayashi K, Yamada A, Sugiura A. 1993. Variation in field isolates of measles virus during an 8-year period in Japan. Microbiol. Immunol. 37:233–237 [DOI] [PubMed] [Google Scholar]
- 45. Patterson JB, Scheiflinger F, Manchester M, Yilma T, Oldstone MB. 1999. Structural and functional studies of the measles virus hemagglutinin: identification of a novel site required for CD46 interaction. Virology 256:142–151 [DOI] [PubMed] [Google Scholar]
- 46. Rota PA, Brown K, Mankertz A, Santibanez S, Shulga S, Muller CP, Hubschen JM, Siqueira M, Beirnes J, Ahmed H, Triki H, Al-Busaidy S, Dosseh A, Byabamazima C, Smit S, Akoua-Koffi C, Bwogi J, Bukenya H, Wairagkar N, Ramamurty N, Incomserb P, Pattamadilok S, Jee Y, Lim W, Xu W, Komase K, Takeda M, Tran T, Castillo-Solorzano C, Chenoweth P, Brown D, Mulders MN, Bellini WJ, Featherstone D. 2011. Global distribution of measles genotypes and measles molecular epidemiology. J. Infect. Dis. 204(Suppl 1):S514–S523 [DOI] [PubMed] [Google Scholar]
- 47. Fournier P, Brons NH, Berbers GA, Wiesmuller KH, Fleckenstein BT, Schneider F, Jung G, Muller CP. 1997. Antibodies to a new linear site at the topographical or functional interface between the haemagglutinin and fusion proteins protect against measles encephalitis. J. Gen. Virol. 78(Pt 6):1295–1302 [DOI] [PubMed] [Google Scholar]
- 48. Duke T, Mgone CS. 2003. Measles: not just another viral exanthem. Lancet 361:763–773 [DOI] [PubMed] [Google Scholar]
- 49. Ader N, Brindley MA, Avila M, Origgi FC, Langedijk JP, Orvell C, Vandevelde M, Zurbriggen A, Plemper RK, Plattet P. 2012. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J. Biol. Chem. 287:16324–16334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Makela MJ, Salmi AA, Norrby E, Wild TF. 1989. Monoclonal antibodies against measles virus haemagglutinin react with synthetic peptides. Scand. J. Immunol. 30:225–231 [DOI] [PubMed] [Google Scholar]
- 51. Ziegler D, Fournier P, Berbers GA, Steuer H, Wiesmuller KH, Fleckenstein B, Schneider F, Jung G, King CC, Muller CP. 1996. Protection against measles virus encephalitis by monoclonal antibodies binding to a cystine loop domain of the H protein mimicked by peptides which are not recognized by maternal antibodies. J. Gen. Virol. 77(Pt 10):2479–2489 [DOI] [PubMed] [Google Scholar]
- 52. Putz MM, Hoebeke J, Ammerlaan W, Schneider S, Muller CP. 2003. Functional fine-mapping and molecular modeling of a conserved loop epitope of the measles virus hemagglutinin protein. Eur. J. Biochem. 270:1515–1527 [DOI] [PubMed] [Google Scholar]
- 53. Liebert UG, Flanagan SG, Loffler S, Baczko K, ter Meulen V, Rima BK. 1994. Antigenic determinants of measles virus hemagglutinin associated with neurovirulence. J. Virol. 68:1486–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.