Abstract
Polyadenylate-binding protein cytoplasmic 1 (PABPC1) is a cytoplasmic-nuclear shuttling protein important for protein translation initiation and both RNA processing and stability. We report that PABPC1 forms a complex with the Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 protein, which allows ORF57 to interact with a 9-nucleotide (nt) core element of KSHV polyadenylated nuclear (PAN) RNA, a viral long noncoding RNA (lncRNA), and increase PAN stability. The N-terminal RNA recognition motifs (RRMs) of PABPC1 are necessary for the direct interaction with ORF57. During KSHV lytic infection, the expression of viral ORF57 leads to a substantial decrease in overall PABPC1 expression, along with a shift in the cellular distribution of the remaining PABPC1 to the nucleus. Interestingly, PABPC1 and ORF57 have opposing functions in modulating PAN steady-state accumulation. The suppressive effect of PABPC1 specific to PAN expression is alleviated by small interfering RNA knockdown of PABPC1 or by overexpression of ORF57. Conversely, ectopic PABPC1 reduces ORF57 steady-state protein levels and induces aberrant polyadenylation of PAN and thereby indirectly inhibits ORF57-mediated PAN accumulation. However, E1B-AP5 (heterogeneous nuclear ribonucleoprotein U-like 1), which interacts with a region outside the 9-nt core to stimulate PAN expression, does not interact or even colocalize with ORF57. Unlike PABPC1, the nuclear distribution of E1B-AP5 remains unchanged by viral lytic infection or overexpression of ORF57. Together, these data indicate that PABPC1 is an important cellular target of viral ORF57 to directly upregulate PAN accumulation during viral lytic infection, and the ability of host PABPC1 to disrupt ORF57 expression is a strategic host counterbalancing mechanism.
INTRODUCTION
In humans, polyadenylate-binding protein 1 (PABP1) is encoded by the poly(A)-binding protein cytoplasmic 1 (PABPC1) gene (1) (also known as PAB1, PABP, PABPC2, and PABPL1) and is a cytoplasmic protein involved in mRNA translation initiation and stability (2–5). In the cytoplasm, PABPC1 binds to the 3′ poly(A) tail of eukaryotic mRNAs through its RNA recognition motifs (RRMs) and interacts with the N terminus of eukaryotic initiation factor 4G (eIF4G), part of the eIF4F complex associated with the 5′ cap structure. The interactions of PABPC1 with RNA and elF4G cause the mRNA to form a closed loop (6–9), thereby stabilizing the RNA and promoting ribosome recruitment and translation initiation (8, 10). Through interaction with eukaryotic release factor 3 (eRF3) and an exon junction complex, PABPC1 is also involved in nonsense-mediated decay (NMD) (11–13).
The activities of PABPC1 are regulated by PABPC1-interacting proteins (7). PABPC1 interacts with GW182 (14), an essential component of the microRNA pathway in animal cells (15). The binding of GW182 to PABPC1 seems to repress translation by interfering with the formation of an mRNA closed loop (14). Although PABPC1 is mainly cytoplasmic, it shuttles between the nucleus and cytoplasm (16). When present in the nucleus, PABPC1 associates with intron-containing pre-mRNAs undergoing polyadenylation and interacts with poly(A) polymerase to engage in nuclear RNA processing (3, 16, 17). Nuclear export of PABPC1 protein is coupled to active mRNA export since blocking RNA export results in the nuclear accumulation of PABPC1 (18). Although PABPC1 has a general protective influence on the poly(A) tail, PABPC1 is also involved in RNA deadenylation by interacting with deadenylation complexes, Pan3 (19), Tob (20, 21), Caf1-Ccr4, and eRF1-eRF3 (22).
The normal host activity of PABPC1 can be altered by both DNA and RNA viruses during viral infections (4). Numerous studies have indicated that viral machinery induces host translational shutoff by altering the cellular distribution (4, 23–29) or specific cleavage (30–38) of the PABPC1 protein. The redistribution of PABPC1 to the nucleus is accompanied by a reduction in global protein synthesis but is not linked to apoptosis (18). Viruses are capable of maintaining the translatability of their own mRNAs with a reduced level of cytoplasmic PABPC1, although the mechanisms remain to be understood (4, 23).
Kaposi's sarcoma-associated herpesvirus (KSHV) infection induces nuclear localization of PABPC1 (28, 39, 40). This was initially observed with KSHV K10/K10.1 protein, a viral homologue of interferon regulatory factors, which interacts with PABPC1 via protein-protein interaction (40). KSHV ORF37 (SOX) (41), a viral alkaline exonuclease with intrinsic RNase activity (42), shuts off host gene expression by accelerating host mRNA turnover in the cytoplasm (43) and, when overexpressed, induces nuclear localization of PABPC1 (28). Whether SOX promotes the nuclear import of PABPC1 and shuts off host gene expression in the context of the KSHV genome remains to be determined. Subsequent studies suggest that SOX-induced nuclear accumulation of PABPC1 promotes nuclear retention and hyperadenylation of mRNAs (27, 28). However, a lack of a functional connection between viral SOX and PABPC1 in these studies suggests that SOX must stimulate hyperadenylation indirectly through unknown cellular cofactors.
During lytic viral infection, KSHV expresses nuclear ORF57 (Mta [mRNA transcript accumulation]) to promote the accumulation of viral intronless transcripts (44–48) and RNA splicing of intron-containing viral RNAs (49, 50). We and others recently described the ORF57-mediated nuclear accumulation and stability of a KSHV long noncoding RNA (lncRNA), polyadenylated nuclear (PAN) RNA, by ORF57 interacting with an Mta-responsive element (MRE) containing a 9-nucleotide (nt) core at the 5′ end of PAN RNA (51, 52). The MRE at the 5′ PAN also interacts with PABPC1 and a heterogeneous nuclear ribonucleoprotein U-like 1 (hnRNPUL1 or E1B-AP5 [E1B 55-kilodalton-associated protein 5]) (51). It has been shown that the binding of nuclear PABPC1 to the poly(A) tail of PAN is required for SOX-mediated accumulation of nuclear PAN (28, 53). It is now recognized that PAN is a regulatory lncRNA important for viral gene expression, replication, and immune modulation (53–55). The observation that KSHV ORF57, PABPC1, and E1B-AP5 interact with the PAN 5′-end motif (51) highlights the importance of these new discoveries regarding the nuclear accumulation and function of PAN RNA during KSHV lytic infection. In this study, we demonstrate that PABPC1 directly interacts with the 9-nt core in MRE stem-loop II (MRE-II) at the PAN 5′ end and this interaction is necessary for ORF57 association with the MRE core. Interestingly, we found PABPC1 to be suppressive toward PAN expression, while ORF57 was able to relieve this inhibition by PABPC1 to promote PAN RNA accumulation.
MATERIALS AND METHODS
Cells.
JSC-1 cells (KSHV positive [KSHV+]/Epstein-Barr virus positive) and BCBL1 cells (KSHV+) were grown in RPMI 1640 containing 10% fetal bovine serum (FBS; HyClone, Logan, UT) and were induced, respectively, with butyrate (Bu; 3 mM) and valproate (VA; 1 mM) for lytic infection. TREx BCBL1-vector (noninducible) and TREx BCBL1-Rta (doxycycline-inducible) cells (56) were cultivated in RPMI 1640 supplemented with 10% FBS and hygromycin B (50 μg/ml). To induce the expression of KSHV lytic genes, TREx BCBL1-Rta or TREx BCBL1-vector (a negative control) cells were seeded at 5 × 105 cells/ml and cultivated for 24 h in the presence of 1 μg/ml of doxycycline (Dox). Human HEK293 and HeLa cells were cultivated in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% FBS. HEK293 cell lines stably harboring a KSHV wild-type (wt) genome (Bac36-wt) or a KSHV genome with disrupted ORF57 (Bac36-Δ57) (57) were induced with 3 mM Bu or 1 mM VA for lytic infection.
Mammalian expression vectors, transient-transfection assays, and RNA interference.
Mammalian expression vectors pVM7 for FLAG-tagged ORF57, pVM68 for 3× FLAG-tagged ORF57, pJM1 for PAN RNA, pJM5 for K5, and pJM6 for viral G-protein-coupled receptor (vGPCR) were described in our previous reports (47, 51, 58). PABPC1-myc-expressing plasmid was purchased from Origene (Rockville, MD). All hemagglutinin (HA)-tagged wt and deletion mutant (mt) PABPC1 expression vectors were gifts from Britt Glaunsinger (27).
Cotransfection of expression vector PAN, vGPCR, or K5 with PABPC1 was performed in HEK293 or HeLa cells (5 ×105 cells/well) in a six-well plate with 1 μg of individual vector together with 0.2 μg (5:1 ratio) of pVM7 (ORF57-FLAG fusion) (58) or an empty pFLAG-CMV-5.1 vector using Lipofectamine 2000 (Invitrogen). In PABPC1 cotransfection assays, a lower dose of the PABPC1 expression vector was titrated.
Small interfering RNA (siRNA) targeting endogenous PABPC1 (siPABPC1), E1B-AP5 (siE1B-AP5), or a nonspecific target (siControl) was purchased from Dharmacon (Lafayette, CO) as an siGenome SMARTpool (a mixture of 4 individual siRNAs). Each siRNA was used at 40 nM with Lipofectamine 2000 for transfection of HEK293 or HeLa cells (2.5 × 105 cells per well in a 24-well plate). At 18 h after transfection, cells were passed to a six-well plate and transfected again with the same amount of siRNA and 6 to 8 h later were ready for transfection with 1 μg of PAN together with 0.2 μg of pVM7 (ORF57-FLAG fusion) or empty pFLAG-CMV-5.1. The same strategy of PABPC1 knockdown by siRNA was used in Bac36-wt or Bac36-Δ57 cells before virus lytic induction.
Western blot analyses.
Protein samples for Western blot analyses were prepared by direct lysis of the cells in 2× SDS sample buffer (Quality Biological, Inc., Gaithersburg, MD) plus 5% 2-mercaptoethanol. Samples were boiled and resolved in a 4% to 12% SDS-polyacrylamide gel by electrophoresis. The following antibodies were used in Western blot analyses: a rabbit polyclonal anti-ORF57 antibody (57); mouse monoclonal anti-ORF57 (unpublished data), anti-NXF1 (clone 53H8; ab50609; Abcam, Cambridge, MA), anti-KSHV K8α (ProMab, Albany, CA), anti-green fluorescent protein (anti-GFP; JL-8; BD Biosciences Living Colors, Mountain View, CA), and anti-β-tubulin (T5201; Sigma, St. Louis, MO) antibodies; and rabbit polyclonal anti-PABPC1 (ab21060; Abcam), anti-E1B-AP5 (ab68480; Abcam), anti-CBP80 (ab42389; Abcam), and anti-cyclophilin A (Upstate, Lake Placid, NY) antibodies, together with corresponding peroxidase-conjugated secondary antibodies (Sigma). The signal on the Western blot was detected with WestPico chemiluminescence substrate (Pierce, Rockford, IL).
RNA preparation and Northern blot analyses.
Total cell RNA samples were prepared 24 h after transfection (HEK293, HeLa) or after induction (B cells) by the addition of 1 ml of TRIzol reagent according to the TRIzol protocol (Invitrogen). Cytoplasmic and nuclear total RNAs were fractionated as described previously (47). RNA (∼5 μg) was separated in a 1% agarose gel and analyzed by Northern blotting (47). PAN, K5, and vGPCR RNAs expressed from individual plasmids were detected with 2 × 106 cpm of a γ-32P-labeled T7 probe, oZMZ243 (5′-CTATAGTGAGTCGTATTAAT-3′). PAN was also detected with a PAN-specific probe, oJM7 (5′-GTTACACAACGCTTTCACCTACA-3′). For other mRNAs, ORF57 was detected with oligonucleotide probe oVM11 (5′-CTCGTCTTCCAGTGTCGGTG-3′), PABPC1 was detected with oligonucleotide probe oJM85 (5′-TCATCCACAGCTTTCTGTGC-3′), GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was detected with oligonucleotide probe oZMZ270 (5′-TGAGTCCTTCCACGATACCAAA-3′), neomycin (Neo) was detected with oligonucleotide probe oJM22 (5′-GAAGGCGATAGAAGGCGATGC-3′), K8β was detected with oligonucleotide probe oST3 (5′-GTACTCACCCC/CACACAAAGTCTGGCATGGTTCTCCC-3′), GFP was detected with oligonucleotide probe oZMZ296 (5′-GCATGGCGGACTTGAAGAA-3′), and human U6 was detected with oligonucleotide probe oST197 (5′-AAAATATGGAACGCTTCACGA-3′). The hybridization signal was captured using a Molecular Dynamics PhosphorImager Storm 860 phosphorimager and analyzed with ImageQuant software (GE Healthcare, Piscataway, NJ).
RNA-protein pulldown assays.
Total cell extracts used in RNA pulldown assays were prepared from ∼5 × 106 TREx BCBL1-Rta or -vector cells as described previously (51). An RNA pulldown assay using biotinylated RNA oligomers was performed as described previously (59, 60). RNA oligomers oJM35 and oJM68 derived from PAN RNA (Fig. 1A) (51) and RNA oligomers oNP41 (biotin-GCUUCUGACGAAGACCUUAGGAU) and oNP42 (biotin-CUUAGGAUGGGACAUACAGGAAGAG) derived from viral interleukin-6 (vIL-6) RNA (60) were used in the pulldown assays. The proteins in the pulldown assays were analyzed by Western blotting.
Fig 1.
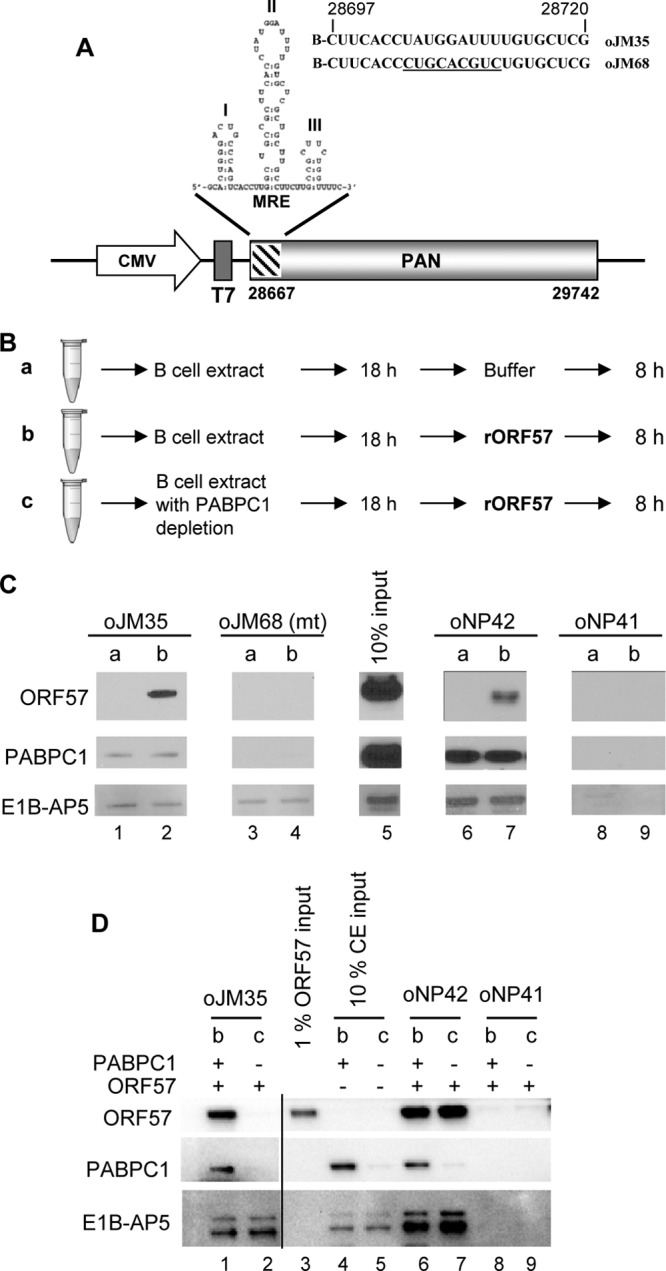
The interaction of ORF57 with the PAN MRE core in vitro depends on PABPC1 but not E1B-AP5. (A) PAN RNA structure and corresponding expression vector pJM1. The diagram shows the MRE (Mta-responsive element) at the PAN 5′ end consisting of three stem-loop structures and the MRE-II loop containing a 9-nt core responsible for ORF57 accumulation of PAN (51). Sequences of biotin (B)-labeled RNA oligonucleotides oJM35 and oJM68 covering the MRE-II loop are shown with the mutant 9-nt core sequences underlined in oJM68. CMV, cytomegalovirus immediate-early promoter. (B) A strategy to dissect the dependence of ORF57 binding to the PAN MRE core by using B cell extracts with or without PABPC1 depletion. RNA oligomers oJM35 (a wt MRE core), oJM68 (a mt MRE core), oNP42 (a vIL-6 MRE), and oNP41 (a negative control MRE), described in the text, were examined under three different pulldown conditions, conditions a, b, and c. Bead-bounded oligomers were incubated overnight with either TREx BCBL1-vector cell extract (conditions a and b) or the same extract depleted of PABPC1 (condition c) and then further incubated with either 100 μl of pulldown buffer (condition a) or rORF57 protein (250 ng/100 μl) (conditions b and c) for an additional 8 h. (C) Western blots were used to examine the presence of ORF57, PABPC1, and E1B-AP5 proteins in the pulldown assays. (D) PABPC1 association with the MRE core is a prerequisite for ORF57-RNA interaction. RNA oligomers oJM35, oNP42, and oNP41 were examined as described for panel A with pull-down condition b or c using cell extracts (CE) in the presence (+) or absence (−) of PABPC1. The depletion of PABPC1 from the BCBL1 cell extract was performed by three runs of IP depletion with a PABPC1-specific antibody and verified by Western blotting (compare input lane 5 versus lane 4 for depletion efficiency). The ORF57, PABPC1, and E1B-AP5 proteins in the pulldown assays were detected by Western blotting.
IP.
Immunoprecipitation (IP) was performed as described previously (47). Protein samples for IP were prepared from TREx BCBL1 cells after Dox induction, by incubating cells with IP lysis buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2.5 mM MgCl2, 0.1% NP-40 or 1× buffer RSB-150 plus 0.1% NP-40) (61) containing protease inhibitors (1× complete mini-EDTA-free protease inhibitor cocktail; Roche) and phosphatase inhibitors (1 mM sodium fluoride, 1 mM β-glycerol phosphate). The extracts were treated with an RNase A-T1 mixture (1:100; Ambion) for 10 min at 25°C before IP. Rabbit anti-PABPC1 antibody (ab21060; Abcam) and anti-E1B-AP5 antibody (ab68480; Abcam) were used for IP. The IP buffer used for pulldown and subsequent washes was composed of 50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, and 0.1% NP-40 with the addition of Roche's protease inhibitor cocktail. Proteins from IP were identified by Western blotting.
HEK293 cells were transfected with HA-tagged wt or mt PABPC1 expression vectors. Empty FLAG (negative control) and ORF57-FLAG expression vectors were separately transfected for another set of HEK293 cells to avoid a negative effect of PABPC1 on ORF57 expression. After 24 h of transfection, the transfected cells were harvested in RSB-200 IP buffer plus 0.1% NP-40 (61) and 1× EDTA-free complete protease inhibitors. Anti-FLAG-covered agarose beads resuspended in RSB-200 IP buffer were first incubated for 2 h at 4°C with the HEK293 cell lysates containing FLAG only or ORF57-FLAG protein. After binding, the beads were washed once with RSB-200 IP buffer and equally divided into six tubes. An equal amount of the cell lysates prepared from individual PABPC1 vector transfection was added to the beads with FLAG only or ORF57-FLAG protein, and the mixtures were incubated overnight at 4°C. After extensive washing (5 times each with 0.5 ml of the IP buffer), the immunoprecipitated complexes were eluted with 40 μl of the IP buffer containing 100 μg/ml of 3× FLAG peptide (Sigma) for 1 h at 4°C. Western blotting was carried out to detect the presence of ORF57-FLAG and HA-PABPC1 proteins with anti-FLAG and anti-HA antibody (Sigma), respectively.
For PABPC1 depletion, TREx-vector cells were harvested 24 h after seeding (without induction) and the cell pellet was washed with phosphate-buffered saline (PBS) and resuspended in 1× buffer RSB-150 plus 0.1% NP-40 (61) with the addition of protease inhibitor cocktail. The samples were incubated for 5 min on ice, briefly sonicated, and centrifuged for 10 min at 10,000 × g and 4°C. The extract from the supernatant was split, with half used for depletion and half used as a control. The extracts were first precleared with 100 μl of prewashed protein A/G agarose beads (Upstate, Waltham, MA) for 1 h at 4°C and split in half. PABPC1 depletion was carried out with anti-PABPC1 antibody-coated protein A/G beads overnight at 4°C. The second half was incubated with rabbit IgG-coated protein A/G beads and used as a negative control. Three rounds of depletions were performed before the extracts were used for the assays.
Confocal microscopy.
TREx BCBL1-Rta or -vector cells induced by Dox, JSC-1 cells induced by Bu, and BCBL1 cells induced by VA were washed twice with PBS, spotted onto poly-d-lysine-treated glass coverslips, and fixed with 4% paraformaldehyde in PBS before staining. HeLa cells grown on coverslips were transfected with 500 ng of FLAG-ORF57 or an empty vector control. Bac36-wt and Bac36-Δ57 cells grown on coverslips were induced with Bu. Immunofluorescence staining was performed as described previously (49, 60). Mouse monoclonal anti-ORF57 antibody and rabbit polyclonal anti-PABPC1 or rabbit polyclonal anti-E1B-AP5 antibody were used for double staining, together with Alexa Fluor-conjugated secondary antibodies (Invitrogen). Confocal fluorescence images were collected using a Zeiss LSM510 META laser-scanning microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY) equipped with ×20 Plan-Aprochromat (numerical aperture, 0.8) and ×63 Plan-Apochromat (numerical aperture, 1.4) objective lenses. For image fields, confocal optical slices with a thickness of 1.9 μm and x-y pixel sampling of 0.44 μm were collected using the ×20 objective lens, whereas higher-resolution confocal images with an optical slice thickness of 1.0 μm and x-y pixel sampling of 0.1 μm were acquired using the ×63 objective lens. The profile module of the LSM510 software was used to draw straight-line profiles across selected cells, with the pixel intensities for each fluorescence channel along the length of the line displayed as an intensity plot. The histogram module of the LSM510 software was used to generate scatterplots of the pixel intensities with the individual confocal images. Threshold intensity values for each fluorescence channel were applied to the scatterplot to gate the pixels with intensity above the background. The locations of the gated pixels were highlighted in the confocal image as a white mask. To measure the mean pixel intensities, regions of interest were drawn to include only the cytoplasm or nucleus, the background was subtracted, and the mean pixel intensity within the region was calculated. Individual confocal images were subsequently saved in TIFF format, and Adobe Photoshop (version 6.0) software (Adobe Systems, San Jose, CA) was used to process the images into composite figures. Cells were counted in five different fields, randomly chosen from cell populations in two different experiments.
GST pulldown.
Purified full-length PABPC1 protein with an N-terminal glutathione S-transferase (GST) tag (catalog number H00026986-P01) was obtained from Abnova (Taipei, Taiwan). Untagged GST was expressed in Escherichia coli cells transformed with the pGEX-6P-1 vector and purified on glutathione-Sepharose 4B beads (GE Healthcare). Recombinant full-length KSHV ORF57 protein with a C-terminal FLAG tag from baculovirus was described in our previous study (49). Briefly, 400 ng of untagged GST or GST-PABPC1 fusion protein was first immobilized onto glutathione-Sepharose 4B for 1 h at 4°C in IP buffer. After binding, 800 ng of ORF57-FLAG protein was added to each tube, and the mixture was incubated overnight at 4°C. Beads without any GST protein were used as an additional negative control. The beads were then washed 5 times each with 0.5 ml of IP buffer, resuspended in SDS protein sample buffer, and analyzed by Western blotting. GST fusion proteins were detected with anti-GST antibody (GE Healthcare).
CLIP.
UV cross-linking and immunoprecipitation (CLIP) assays were performed as described previously (60, 62). Briefly, HEK293 cells were cotransfected with ORF57-FLAG and HA-PABPC1 expression vectors. At 24 h of transfection, the cells were irradiated by UV light to cross-link RNA to proteins, followed by lysis in radioimmunoprecipitation assay (RIPA) buffer containing protease and RNase inhibitors. The prepared cell lysates were precleared with prewashed protein A agarose beads for 2 h at 4°C. After binding, the precleaned cell lysates were incubated with antibody-covered beads overnight at 4°C in RIPA buffer. After washing (5 times each with 1 ml of RIPA buffer), the beads bound with RNA-protein complexes were treated with proteinase K for 30 min at 37°C, followed by phenol-chloroform extraction. Purified RNA was precipitated and treated with Turbo-free DNase (Ambion). Input RNA was isolated from 100 μl of precleaned extracts by TRIpure reagent (Roche). ORF57 RNA was detected by reverse transcription (RT)-PCR using oligonucleotides oVM1 (5′-CACC/ATGGTACAAGCAATGATAGACATGG-3′) and oVM11 (5′-CTCGTCTTCCAGTGTCGGTG-3′).
RESULTS
PABPC1 is required for ORF57 interaction with the PAN MRE core.
We previously reported that PAN stability requires ORF57 and a functional MRE core at the 5′ PAN and that the MRE core is responsible for interacting with both PABPC1 and ORF57 (51). However, whether interaction of PABPC1 with the MRE core is required for ORF57-mediated accumulation of PAN RNA remains unknown. To further examine the molecular interplay between PABPC1 and ORF57 in association with the MRE core, we performed RNA pulldown assays using a PAN RNA oligomer, oJM35, that covers the MRE 9-nt core region in MRE stem-loop II (Fig. 1A). As a positive control for ORF57 binding, we used a vIL-6 MRE RNA oligomer, oNP42 (60), which has previously been shown to be bound strongly by ORF57. Negative binding controls, such as an MRE core mutant RNA oligomer (oJM68) and a known non-ORF57 binding RNA sequence (oNP41) (60), indicated that in the presence of cellular proteins the interaction of ORF57 with PAN was specific to the MRE core sequence of PAN (51). Because cellular proteins were proven previously to be necessary for the specific interaction of ORF57 with its targeted RNA (47, 63), we examined whether PABPC1 is required for the interaction of ORF57 with the MRE core of PAN. In our interaction assays, we used a TREx BCBL1-vector cell extract lacking ORF57 but supplemented with baculovirus recombinant ORF57 (rORF57) protein. The addition of cell extract plus ORF57 to the RNA oligomer samples was performed using three different conditions, as diagramed in Fig. 1B: condition a, no rORF57 protein added; condition b, rORF57 added 18 h after incubation of the RNA oligomers with cell extract; condition c, similar to condition b but with cell extract depleted of PABPC1. We did not perform a control interaction assay with ORF57 alone because it is known that rORF57, in the absence of cellular proteins, binds nonspecifically to all RNAs tested, as shown in our previous study (47). In the RNA pulldown experiments, Western blot analysis was used to examine the presence of ORF57, PABPC1, or E1B-AP5 binding to specific RNA oligomers under the different interaction assay conditions. When rORF57 was added to the interaction assay, as under condition b, we observed that rORF57 bound to the MRE 9-nt core RNA sequence (oJM35), as well as to the positive-control vIL-6 sequence (oNP42) (Fig. 1C, lanes 2 and 7), but not to the mutant MRE core sequence (oJM68) or the negative binding control sequence (oNP41). Similarly, PABPC1 also bound to oJM35, though with less affinity than ORF57, and to oNP42, but not to oJM68 or oNP41, under assay conditions when either ORF57 was present (condition b) or absent (condition a). E1B-AP5, which interacts with another region of MRE-II (51), was used as a binding control. We found that under both assay conditions (a and b) E1B-AP5 bound to oJM35, including its mutant (oJM68), and oNP42 oligomers. These data suggest that both rORF57 and PABPC1 interact with the 9-nt core but E1B-AP5 binds to the sequences outside the MRE core.
After demonstrating that both PABPC1 and ORF57 bind specifically to the MRE core, we next determined whether the interaction of PABPC1 with the MRE core is important for facilitating ORF57-MRE core binding. First, we prepared a PABPC1-depleted BCBL1 cell total extract using anti-PABPC1 antibody-immobilized beads. We then compared the binding of ORF57 to the MRE core-containing RNA oligomer oJM35 in the PABPC1-depleted extract with that in the non-PABPC1-depleted extract (Fig. 1B, conditions b and c). As shown in Fig. 1D, efficient depletion of PABPC1 from the cell extract (compare lane 5 to lane 4) influenced ORF57 binding, but not E1B-AP5 binding, to the MRE core (compare lane 2 to lane 1). This was in contrast to ORF57 binding to a vIL-6 MRE (oNP42), which is independent of PABPC1 (compare lane 7 to lane 6). Again, these interactions with the RNA target were specific because a negative-control RNA oligomer, oNP41, derived from a vIL-6 RNA did not show any protein binding under either assay condition (b or c). Together, these data clearly indicate that the interaction of ORF57 with the PAN MRE core is distinguishable from its interaction with the vIL-6 MRE and requires the cellular protein PABPC1.
PABPC1, but not E1B-AP5, interacts directly with ORF57.
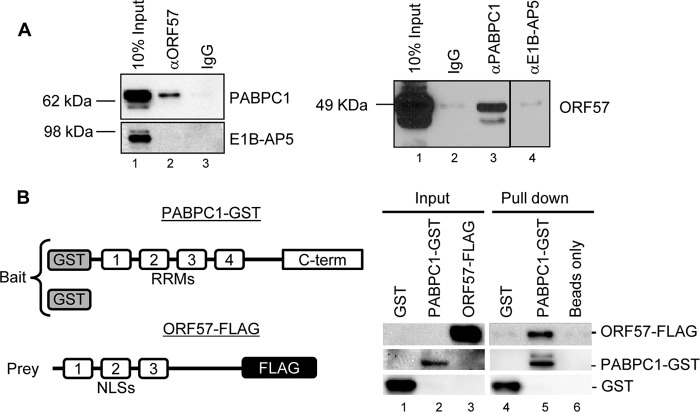
To understand how the PABPC1 association with the PAN MRE may facilitate ORF57 binding to the MRE core, we investigated whether PABPC1 could interact with ORF57 via direct protein-protein interactions. Using an RNase-treated cell extract prepared from Dox-induced TREx BCBL1-Rta cells, we found that PABPC1 and ORF57 could be coimmunoprecipitated by using either anti-ORF57 or anti-PABPC1 antibodies (Fig. 2A), and this interaction in B cells was found to be independent of RNA. In contrast, we were unable to observe such an interaction between ORF57 and E1B-AP5 by the same approach (Fig. 2A, lane 4). To exclude the possibility that other proteins might mediate the interaction of PABPC1 with ORF57, we conducted GST pulldown assays by using a recombinant full-length PABPC1 with a GST tag at the N terminus and a recombinant full-length ORF57 with a FLAG tag at the C terminus (47, 49) (Fig. 2B). We found that PABPC1-GST directly interacts with ORF57-FLAG and could efficiently pull down ORF57-FLAG in the GST pulldown assays (Fig. 2B, lane 5).
Fig 2.
KSHV ORF57 interacts with PABPC1. (A) ORF57 interacts with PABPC1, but not with E1B-AP5, as shown by coimmunoprecipitation assays. Total cell extracts from TREx BCBL1-Rta cells induced by Dox (1 μg/ml) and treated with a mixture of RNase A and T1 were immunoprecipitated with preimmune mouse IgG- or monoclonal anti-ORF57 antibody-coated beads and examined for PABPC1 and E1B-AP5 by Western blotting (left). Conversely, the same extracts treated with a mixture of RNase A and T1 were immunoprecipitated with nonspecific rabbit IgG- or polyclonal anti-PABPC1 or anti-E1B-AP5 antibody-coated beads and blotted for ORF57 by Western blotting (right). (B) PABPC1 directly interacts with ORF57, as shown by GST pulldown assays. Full-length PABPC1 protein with an N-terminal GST tag or untagged GST was immobilized on glutathione-Sepharose 4B beads and incubated at 4°C overnight in IP buffer complemented with KSHV ORF57 protein containing a C-terminal FLAG tag that was expressed from baculovirus. The glutathione-Sepharose 4B beads without any GST protein were used as an additional negative control. After binding, the beads were washed five times with IP buffer, resuspended in 2× SDS sample buffer, and analyzed by Western blotting with two antibodies, an anti-GST antibody recognizing PABPC1 and an anti-FLAG antibody that recognizes the epitope-tagged ORF57.
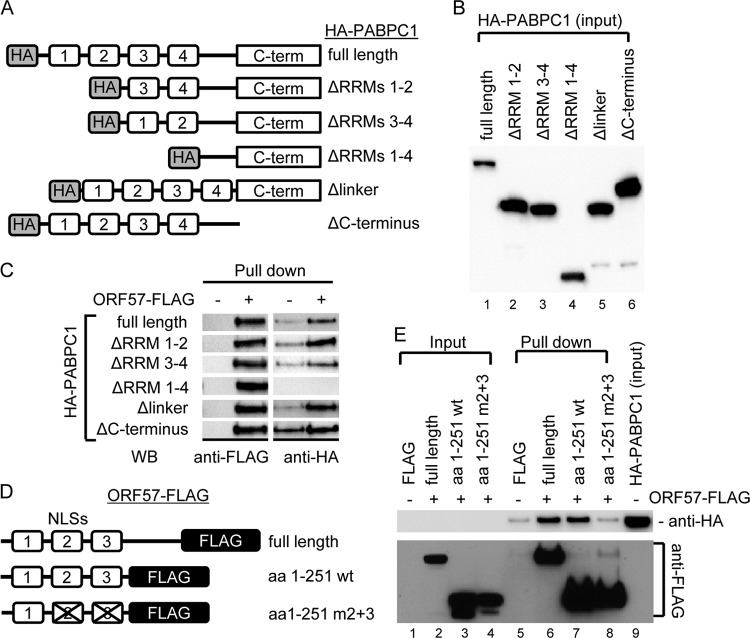
To further define the interaction domains of PABPC1 and ORF57, we expressed wt PABPC1 and deletion mutants with an HA tag at the N terminus (Fig. 3A and B) in HEK293 cells separately from wt ORF57 and its corresponding mutants with a FLAG tag at the C terminus (Fig. 3D). The cell extracts containing HA-PABPC1 or ORF57-FLAG from separate transfections were then used for coimmunoprecipitation with anti-FLAG beads. As shown in Fig. 3C and E, the RRMs at the N terminus of PABPC1 are needed to interact with the N-terminal region of ORF57 containing nuclear localization signal 2 (NLS2) and NLS3 (47).
Fig 3.
Mapping the interacting regions of PABPC1 and ORF57. (A) Structures of HA-tagged wt PABPC1 and its deletion mutants. (B and C) RRMs of PABPC1 interact with ORF57. HEK293 cell lysates derived from HA-tagged wt or mt PABPC1-transfected cells (B) were incubated with FLAG only-coated (ORF57 −) or ORF57-FLAG-coated (ORF57 +) agarose beads at 4°C overnight. The IP complexes present in the pulldown assays were examined by Western blotting (WB) using anti-FLAG to recognize ORF57 and anti-HA for PABPC1 (C). (D) Structures of FLAG-tagged wt and mutant ORF57. aa, amino acids; m2+m3, mutations in both NLS2 and NLS3. (E) NLS2 and NLS3 of ORF57 interact with PABPC1. The agarose beads immobilized with wt or mt ORF57 or FLAG alone were used in the IP pulldown assays of wt PABPC1 described above, and the protein complexes present in the pulldown assays were detected with anti-HA for PABPC1 and anti-FLAG for ORF57.
Reduction of cytoplasmic PABPC1 and nuclear redistribution of PABPC1 during KSHV lytic infection are attributable to the expression of viral ORF57.
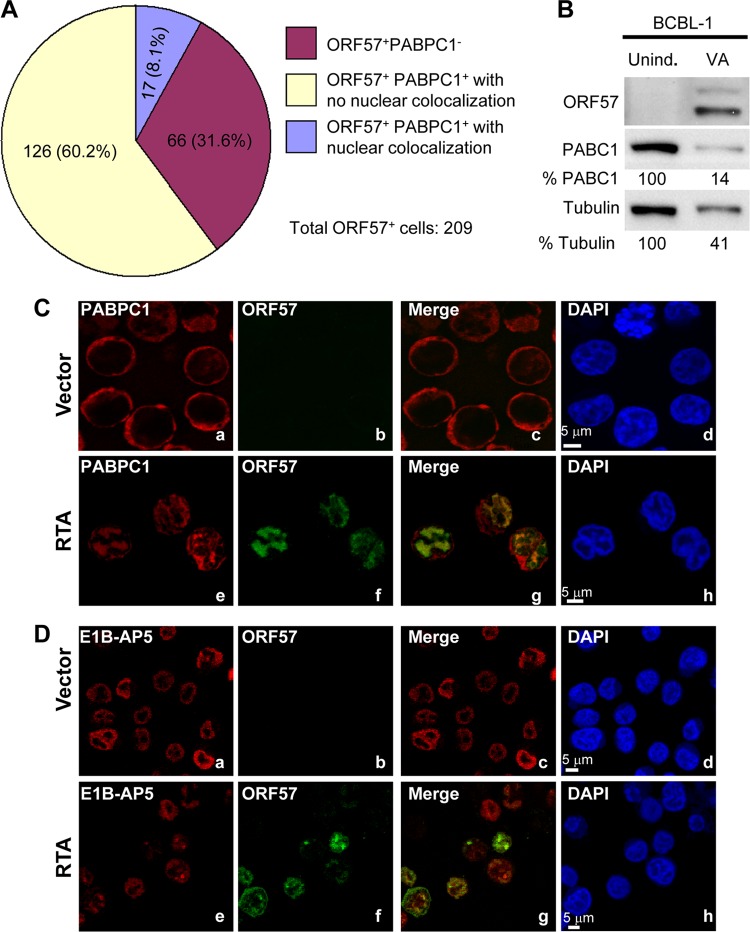
PABPC1 is a cytoplasmic protein capable of shuttling between the nucleus and cytoplasm (16). We examined the distribution of ORF57, PABPC1, and E1B-AP5 in KSHV-infected cells by confocal microscopy. We observed that 24 h after Dox induction, 8.1% of TREx BCBL1-Rta cells displayed PABPC1 present in the nucleus and colocalized with ORF57 (Fig. 4A and C; compare Fig. 4Ca to d with e to h). In addition, we found that B cells with KSHV lytic induction exhibited a much reduced level of PABPC1, as shown in BCBL1 cells by Western blotting (Fig. 4B). We also observed reduced cytoplasmic PABPC1 in three different cell lines (JSC-1, BCBL1, and TREx BCBL1-Rta) when examined by confocal microscopy (see Fig. S1 in the supplemental material), which is consistent with previous reports (39). The overall reduction in cellular PABPC1 and the accompanying shift in the distribution of PABPC1 to the nucleus, along with KSHV lytic infection and ORF57 expression, were confirmed in Bac36-wt cells containing a wt KSHV genome (see Fig. S2 in the supplemental material). Bac36-Δ57 cells, containing a KSHV genome with disrupted ORF57, did not exhibit the same changes in PABPC1. Furthermore, Bac36-wt cells expressing ORF50/RTA displayed a nuclear distribution of PABPC1 (see the line profile of nucleus 2 in Fig. S3A in the supplemental material), whereas the Bac36-Δ57 cells expressing ORF50/RTA did not (see the line profile of nucleus 2 in Fig. S3B in the supplemental material), suggesting that the nuclear distribution of PABPC1 is induced by other viral proteins downstream of ORF50/RTA. The altered PABPC1 nucleocytoplasmic distribution observed in B cells with KSHV lytic infection was not observed for E1B-AP5, which has no direct interaction with ORF57, as shown in our co-IP experiments. Although E1B-AP5 is present along with ORF57 in the nucleus, KSHV lytic infection did not influence the distribution of E1B-AP5 (Fig. 4De to h).
Fig 4.
KSHV lytic infection induces host gene shutoff and nuclear distribution of cytoplasmic PABPC1. (A) Pie chart summary of PABPC1 status in ORF57-expressing BCBL1 cells after KSHV lytic induction. TREx BCBL1-Rta cells induced with Dox for 48 h were fixed and double stained with anti-PABPC1 and anti-ORF57 antibodies and imaged by confocal microscopy. A total of 282 cells, with or without PABPC1 and ORF57, were randomly counted in multiple microscope image fields, and only 209 cells with ORF57 staining were calculated for PABPC1 status. (B) Reduction of PABPC1 and β-tubulin in BCBL1 cells with lytic KSHV induction. Total protein was prepared from BCBL1 cells 48 h after lytic KSHV infection induced by 1 mM VA and host gene expression of PABPC1 and β-tubulin, and the presence of ORF57 was examined by Western blotting. BCBL1 cells without lytic induction (uninduced [unind.]) served as a control. (C and D) ORF57-expressing B cells undergoing lytic KSHV infection exhibit redistribution of PABPC1 but not E1B-AP5. TREx BCBL1-vector (a to d) or TREx BCBL1-Rta (e to h) cells induced by Dox were stained with either rabbit anti-PABPC1 (C) or E1B-AP5 (D) antibodies along with a mouse monoclonal anti-ORF57 antibody. Confocal imaging was performed as described in Materials and Methods. The nuclear distributions of cytoplasmic PABPC1 (red) (C) and nuclear E1B-AP5 (red) (D) were seen in ORF57-positive (green) TREx BCBL1-Rta cells. DAPI, 4′,6′-diamino-2-phenylindole nuclear staining.
Reduction of cytoplasmic PABPC1 expression in B cells appeared to be associated with the viral lytic infection. By counting JSC-1 cells (n = 311) at 24 h after lytic induction by butyrate, we found that 35.7% cells expressed ORF57, indicating viral lytic induction, and 12.6% of them had minimal cytoplasmic PABPC1. By counting TREx BCBL1-Rta cells (n = 282) at 48 h after lytic induction by Dox, we found that 74.1% cells expressed ORF57, of which 31.6% showed no detectable cytoplasmic PABPC1 (Fig. 4A).
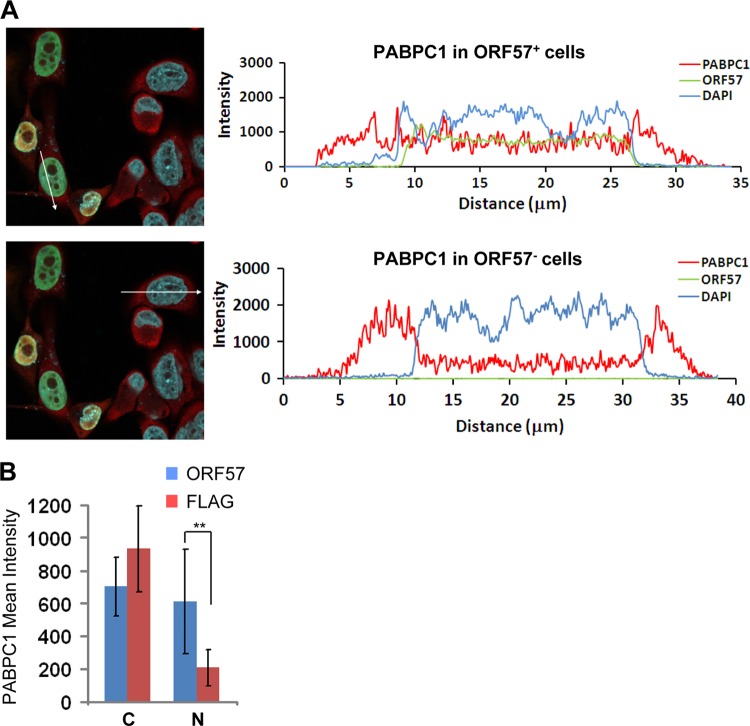
To differentiate the global effect of virus host shutoff in B cells with KSHV lytic infection from a direct role of ORF57 itself in mediating the cytoplasmic reduction and altered nuclear distribution of PABPC1, HeLa cells with transient expression of ORF57 were used to quantify PABPC1 intensity and cellular distribution. We confirmed that a large proportion of ORF57-expressing HeLa cells exhibited a shift in the accumulation of PABPC1 in the nucleus compared with the cells not expressing ORF57 (Fig. 5). In contrast, ORF57 did not affect the nucleocytoplasmic distribution of E1B-AP5 in this assay (see Fig. S4 in the supplemental material). The finding of the altered PABPC1 distribution by ORF57 alone in HeLa cells suggests that ORF57 modulation of the nuclear redistribution of PABPC1 is independent of other viral proteins, including K10/K10.1 (40), and a general host-shutoff function of viral SOX protein (41).
Fig 5.
ORF57-induced nuclear distribution of cytoplasmic PABPC1 in HeLa cells. (A) HeLa cells transfected with ORF57 were immunostained with antibodies recognizing ORF57 (green) and PABPC1 (red). The PABPC1 intensity and cellular distribution were profiled in a randomly selected confocal image showing different patterns of PABPC1 distribution with (upper left) or without (lower left) ORF57 expression. A straight-line profile was drawn across the cells in each image, and the intensity of each pixel along the length of the line was plotted, as shown on the right of each corresponding image. (B) PABPC1 mean intensity in the cytoplasm (C) or nucleus (N) in the absence (FLAG) or presence of ORF57. The mean intensity values were measured from regions of interest drawn to include only the nucleus or cytoplasm, and background values were subtracted. Bars represent means ± SDs (n = 11 for ORF57 group; n = 10 for FLAG group). **, P < 0.01 by the Tukey test.
Endogenous PABPC1 is an inhibitory protein for PAN expression.
To dissect the relationship between the ORF57-mediated redistribution of PABPC1 and PAN accumulation, we knocked down PABPC1 expression in HeLa cells and evaluated PAN expression in the presence or absence of ORF57. HeLa cells, despite not expressing high levels of ORF57 and PAN during transient transfection, were chosen for the knockdown because of their empirically determined high knockdown efficiency. As shown in Fig. 6A, we found that the cells with PABPC1 knockdown expressed a 70% higher level of PAN RNA, but not neomycin RNA from the same expression vector, than the cells receiving a nonspecific siRNA control (compare lane 1 to lane 2), suggesting an inhibitory effect of endogenous PABPC1 on PAN expression. ORF57 in the siRNA control cells did not affect the expression of neomycin RNA but increased PAN RNA expression by 2-fold compared to its counterpart control without ORF57 (compare lane 3 to lane 1). However, knocking down PABPC1 in HeLa cells in the presence of ORF57 promoted a further (∼32%) increase of PAN RNA expression over that for the siRNA control (compare lane 3 to lane 4). These data indicate that PABPC1 has a suppressive role toward PAN RNA expression and ORF57 can relieve the suppressive effect of PABPC1. On the other hand, knocking down the expression of E1B-AP5 in HeLa cells led to a reduced amount of PAN by ∼60% in the presence of ORF57 and by ∼40% in the absence of ORF57 compared to the amount for each corresponding siRNA control (Fig. 6B). Our results indicate that binding of E1B-AP5 to the other region of PAN MRE-II is synergistic with the ORF57 function to stabilize PAN.
Fig 6.
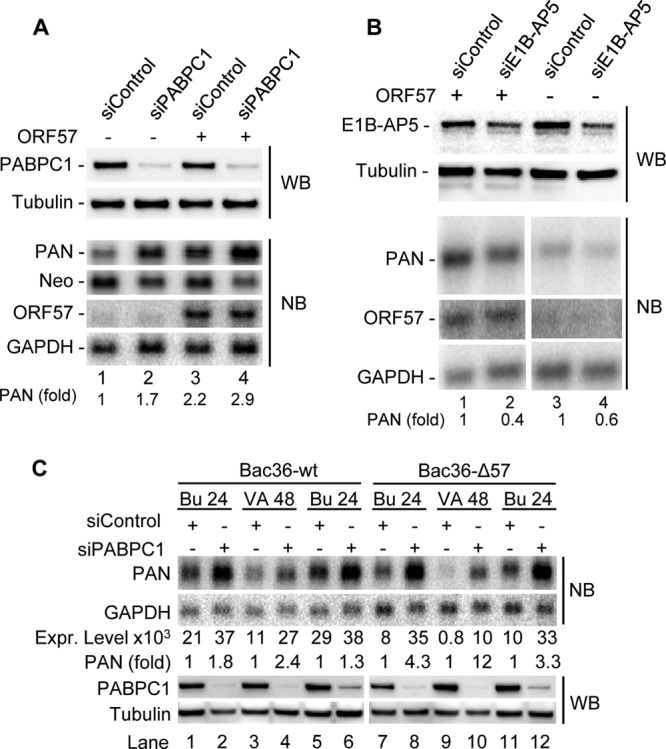
Roles of PABPC1 and E1B-AP5 in PAN expression. (A) Endogenous PABPC1 is inhibitory for PAN expression. PABPC1 in HeLa cells was knocked down using a PABPC1-specific siRNA (siPABPC1) in the presence or absence of ORF57. A nonspecific siRNA (siControl) was employed as a control. The PABPC1-knockdown efficiency was analyzed by Western blotting (WB). Total RNA of HeLa cells transfected with siPABPC1 or siControl, together with PAN-expressing vector pJM1 in the presence (+) or absence (−) of ORF57, was examined by Northern blotting (NB) with a 32P-labeled probe specific for PAN, neomycin, and ORF57 RNA. (B) E1B-AP5 enhances PAN accumulation. E1B-AP5 in HeLa cells was knocked down using E1B-AP5-specific siRNA (siE1B-AP5) in the presence or absence of ORF57. A nonspecific siRNA (siControl) was employed as a control. The E1B-AP5 knockdown efficiency was analyzed by Western blotting, and the expression of PAN and ORF57 was examined as described for panel A by Northern blotting. (C) Endogenous PABPC1 is a negative factor on PAN expression in the context of the KSHV genome. KSHV latently infected Bac36-wt cells containing a wild-type KSHV genome and Bac36-Δ57 cells containing an ORF57-null KSHV genome (57) were transfected twice, at intervals of 24 h, with 40 nM PABPC1-specific siRNA or a nonspecific control siRNA before viral lytic induction by either 3 mM butyrate for 24 h (Bu 24) or 1 mM valproate for 48 h (VA 48). The cells were harvested after virus induction for total protein and RNA preparation. The efficiency of PABPC1 knockdown was examined by Western blotting of total protein, and β-tubulin served as a loading control. PAN RNA was determined by Northern blotting of 3 μg of total RNA for Bac36-wt cells and 5 μg of total RNA for Bac36-Δ57 cells using the 32P-labeled, PAN-specific oligonucleotide probe oJM7. GAPDH RNA in panels A to C served as a sample loading control. The relative levels (signal intensity) of PAN RNA in each sample in panel C were calculated after normalization to the level of GAPDH RNA. A relative ratio (fold) of PAN in siPABPC1- to siControl-treated cells in panels A to C was determined according to the normalized levels in each sample pair.
To examine the negative effect of PABPC1 on PAN abundance in the context of the KSHV genome, we knocked down the expression of PABPC1 both in Bac36-wt cells containing a wt KSHV genome and in Bac36-Δ57 cells containing an ORF57-null KSHV genome and analyzed the expression level of PAN RNA by Northern blotting after virus lytic induction. The Northern blot analysis was performed by using 3 μg of total RNA from Bac36-wt cells and 5 μg of total RNA from Bac36-Δ57 cells with 24 h of butyrate induction or 48 h of valproate induction because Bac36-Δ57 cells express a smaller amount of PAN RNA (57). As shown in Fig. 6C, PAN expression was much reduced, as expected, in Bac36-Δ57 cells compared with that in Bac36-wt cells receiving control siRNA (compare lanes 7, 9, and 11, respectively, with lanes 1, 3, and 5) but was greatly enhanced in both types of cells upon PABPC1 being knocked down (compare lanes 2, 4, 6, 8, 10, and 12, respectively, with lanes 1, 3, 5, 7, 9, and 11), indicating a negative role of endogenous PABPC1 on PAN expression in the context of the KSHV genome. This was further proved by the percent (fold) increase of PAN in PABPC1-knockdown cells over that in the siRNA control cells. We found that PAN expression was increased even more in the Bac36-Δ57 cells than in the Bac36-wt cells upon knocking down PABPC1 (compare lanes 8, 10, and 12 with lanes 2, 4, and 6). Our data indicate that a function of ORF57, in the context of the KSHV genome, is to prevent PABPC1 from negatively regulating PAN abundance, whereas in the absence of ORF57, PABPC1 is more active in suppressing PAN expression.
Ectopic PABPC1 blocks ORF57-mediated accumulation of PAN RNA by reduction of steady-state ORF57 protein expression.
To further confirm the suppressive action of PABPC1 on PAN expression, we transiently expressed PABPC1 in HEK293 cells in the presence or absence of ORF57. We noticed that although ectopic PABPC1 at a low dose increased the RNA levels of both ORF57 (Fig. 7A; compare lanes 2 to 4 to lane 1) and PAN (Fig. 7A; compare lanes 8 to 10 to lane 7), PABPC1 exerted a strong suppression on ORF57-mediated accumulation of PAN in a dose-dependent manner (Fig. 7A; compare lanes 2 to 6 to lane 1). Ectopic PABPC1 at 600 ng slightly increased the levels of vGPCR (Fig. 7B, lane 8) and K5 RNA (Fig. 7B, lane 12), two ORF57 nonresponders (46, 51). It is worth noting that PAN in the presence of ectopic PABPC1 migrated as a doublet in the gel (Fig. 7A). This is due to its selection of two alternative poly(A) sites [a PAN poly(A) site and a growth hormone poly(A) site] separated by 181 nt in the expression vector (verified by 3′ rapid amplification of cDNA ends). Interestingly, a much slower migration of PAN RNA was also found to be increased in the cells transfected with 600 or 1,000 ng of PABPC1 (Fig. 7A, lanes 5 and 6 and lanes 11 and 12). This larger PAN RNA was produced by using a simian virus 40 (SV40) poly(A) site further downstream of the neomycin gene from the expression vector and was designated a chimeric PAN-Neo RNA (see Fig. S5 in the supplemental material). Further studies showed that the N-terminal RRMs of PABPC1 were most likely responsible for the induction of the aberrant polyadenylation of PAN RNA (Fig. 7C). Collectively, these data indicate that ectopic PABPC1 suppresses ORF57-mediated accumulation of PAN and stimulates the aberrant usage of alternative poly(A) sites for PAN polyadenylation from the expression vector. This is consistent with the studies showing that ectopic PABPC1 promotes aberrant polyadenylation of GFP RNA (27).
Fig 7.
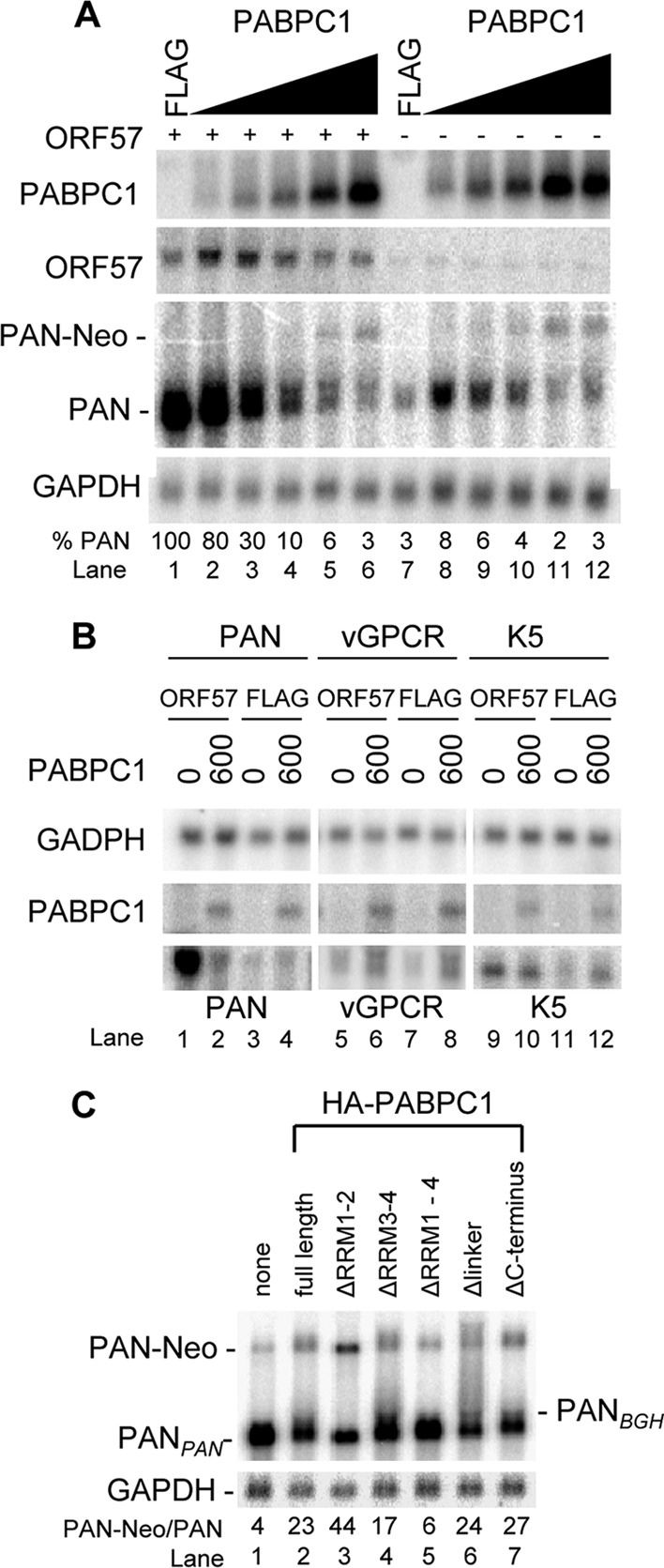
PABPC1 inhibits ORF57-mediated accumulation of PAN. (A) PABPC1 inhibits the expression of PAN in the presence of ORF57 in a dose-dependent manner. HEK293 cells (5 ×105) were transfected with a PAN construct (pJM1, 1 μg) together with 200 ng of empty FLAG vector (ORF57 −) or ORF57 expression vector pVM7 (ORF57 +) and an increased dose of 0 (600 ng FLAG), 50, 150, 300, 600, or 1,000 ng of the PABPC1 expression plasmid. Approximately 5 μg of total RNA from the cells transfected with ORF57 (ORF57 +) and 10 μg of total RNA from the cells transfected with the empty FLAG vector control (ORF57 −) were examined by Northern blotting (NB) with T7 probe oZMZ243 for PAN expression. The same membrane in each panel was stripped and reprobed separately with a 32P-labeled DNA oligonucleotide probe specific to PABPC1 (oJM85), ORF57 (oVM11), or GAPDH (oZMZ270). The relative level of PAN in each sample is shown at the bottom of the blots after normalization to the level of the corresponding GAPDH for sample loading. PAN-Neo is a PAN-Neo chimeric RNA using an alternative SV40 poly(A) site downstream of a neomycin-coding region for RNA polyadenylation (see Fig. S5 in the supplemental material). (B) PABPC1 slightly increases the expression of vGPCR and K5, which are not responsive to ORF57. HEK293 cells were transfected with a PAN, vGPCR, or K5 construct in the presence of ORF57 or FLAG control vector plus 600 ng of FLAG control vector (0 PABPC1) or PABPC1. Total RNA (5 μg) from each transfected cell was examined by Northern blotting for PABPC1 and GAPDH as described above and for T7-PAN, T7-vGPCR, and T7-K5 with T7 oligonucleotide probe oZMZ243. (C) The N-terminal RRMs of PABPC1 are responsible for the induction of aberrant usage of alternative pA sites for PAN expression. HEK293 cells were cotransfected with 200 ng of ORF57 (pVM68), 1 μg of PAN (pJM1), and 600 ng of wt or mt PABPC1 vector, and total cell RNA was examined by Northern blotting as described for panel A.
As PABPC1 at a low dose is able to increase ORF57 RNA levels (Fig. 7A, lanes 2 and 3), we reasoned that the suppression of ORF57-mediated accumulation of PAN RNA by PABPC1 might happen at either the ORF57 RNA export or the protein translational level. To address these possibilities, we first examined the PABPC1 association with ORF57 RNA in cells and found that ORF57 RNA could be pulled down by an HA antibody against HA-tagged PABPC1 or its deletion mutant ΔRRM1+2 by RNA-CLIP and with detection by RT-PCR assays (Fig. 8A, lanes 6 and 7), suggesting efficient binding of PABPC1 to ORF57 RNA. Subsequently, we examined whether PABPC1 associating with ORF57 RNA would prevent the export of ORF57 RNA. As shown in Fig. 8B and Fig. S5 in the supplemental material, from Northern blot analysis of total and fractionated nuclear and cytoplasmic RNA from transfected HEK293 cells, we observed that (i) ectopic PABPC1 increased the ORF57 RNA level but suppressed ORF57-mediated accumulation of PAN (Fig. 8B, compare lane 2 to lane 1), (ii) ectopic PABPC1 increased aberrant polyadenylation of PAN RNA at the SV40 poly(A) site to generate a PAN-Neo chimeric RNA (Fig. 8B; compare lanes 2, 5, and 6 to lanes 1, 3, and 4), (iii) ectopic PABPC1 did not inhibit ORF57 RNA export from the nucleus to the cytoplasm (Fig. 8B; compare lanes 5 and 6 to lanes 3 and 4), and (iv) the suppressive effect of ectopic PABPC1 was specific for PAN because ectopic PABPC1 enhanced the expression of neomycin RNA from the same expression vector (compare lanes 8, 11, and 12 to lanes 2, 5, and 6 in Fig. S5 in the supplemental material). Regardless of the circumstance, PAN was exportable in the presence of ORF57, as we have described previously (51). Together, we conclude that PABPC1-mediated reduction of PAN expression in the presence of ORF57 was not due to a defect in export of ORF57 RNA.
Fig 8.
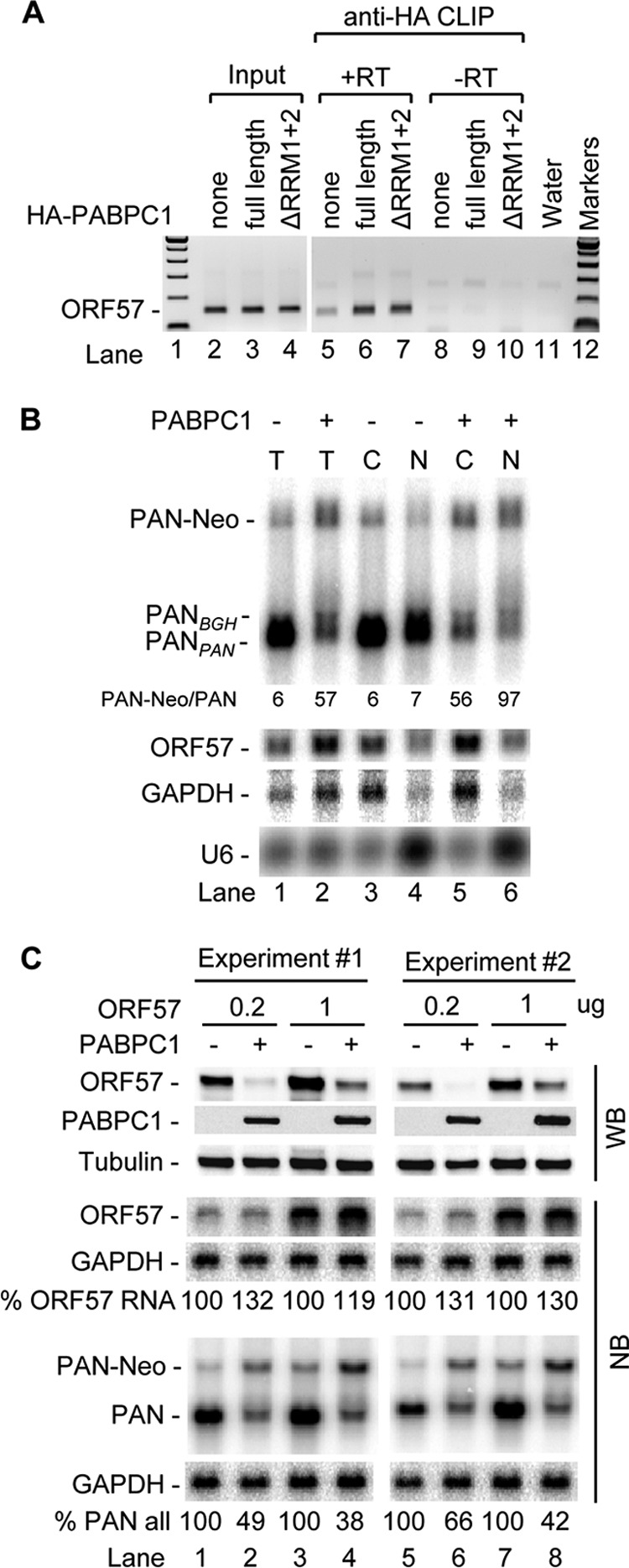
PABPC1 binds ORF57 RNA in vivo and inhibits translation of ORF57 protein but not transcription and export of ORF57 RNA. (A) PABPC1 binds ORF57 RNA in vivo. HEK293 cells cotransfected with ORF57-FLAG and HA-PABPC1 expression vectors at 24 h posttransfection were UV cross-linked for CLIP assays with an anti-HA antibody. The RNA-PABPC1 complexes present in the pulldown assay mixtures treated with proteinase K were extracted and digested with DNase. Input RNA was extracted from 100 μl of the cell extracts before anti-HA immunoprecipitation. ORF57 RNA was detected by RT-PCR using a primer pair of oVM1 and oVM11. +RT, reverse transcription before PCR; −RT, no reverse transcription before PCR. (B) PABPC1 inhibits PAN expression without affecting export of ORF57 RNA. HEK293 cells (5 × 105 cell/well in a 6-well plate, 2 wells per group) were cotransfected with 1 μg of ORF57-FLAG (pVM7) and 1 μg of PAN (pJM1) in the absence (FLAG) or presence of 600 ng PABPC1-myc expression vector. At 24 h after transfection, total (T) RNA (4 μg/lane) extracted from the cells in one well and fractionated nuclear (N) and cytoplasmic (C) RNA (4 μg/lane) extracted from the cells in another well were analyzed by Northern blotting for PAN, ORF57, and GAPDH RNA, as described in the legend to Fig. 7. U6 RNA served as a fractionation efficiency control. PAN-Neo chimeric RNA was derived from the usage of an alternative SV40 poly(A) site downstream of the neomycin-coding region (see Fig. S5 in the supplemental material). (C) PABPC1 inhibits steady-state expression or translation of ORF57 protein but not transcription of ORF57 RNA. HEK293 cells were cotransfected with 1 μg of PAN (pJM1) and 0.2 or 1 μg of ORF57-FLAG (pVM7) in the presence of 600 ng of PABPC1-myc or FLAG control vector (PABPC1 −). Total protein and RNA from transfected cells (24 h after transfection) were detected by Western blotting (WB) for ORF57, PABPC1, and β-tubulin protein by using anti-ORF57, anti-myc (for PABPC1), or anti-β-tubulin antibody or by Northern blotting (NB) for ORF57, PAN, and GAPDH RNA, as described in the legend to Fig. 7. The relative level of ORF57 or PAN (including PAN-Neo) RNA, normalized to the level of the corresponding GAPDH for sample loading, in each sample is shown at the bottom of the Northern blots.
We then examined the ORF57 protein level in HEK293 cells cotransfected with 600 ng of PABPC1 and PAN. As shown in Fig. 8C, ectopic PABPC1 had no effect on ORF57 RNA but reduced the steady-state protein expression of ORF57 (compare lanes 2, 4, 6, and 8 to lanes 1, 3, 5, and 7 for ORF57 RNA and protein), suggesting that PABPC1 acts at the level of protein translation. The reduction of ORF57 protein by ectopic PABPC1 was also accompanied by a lower level of PAN RNA (compare lanes 2, 4, 6, and 8 to lanes 1, 3, 5, and 7) and an increased level of aberrant PAN-Neo RNA in two separate experiments. This observation was further extended to the expression of the KSHV K8β protein and GFP. We found that ectopic PABPC1 has the same negative effect on the steady-state expression of KSHV K8β protein and GFP, while it increased their RNA levels in the cells (see Fig. S6 in the supplemental material).
DISCUSSION
We previously identified a 9-nt core sequence (UAUGGAUUU, KSHV nt 28704 to 28712) in the loop structure of the PAN MRE-II that ORF57 associates with to stabilize PAN RNA (51). The 9-nt core was found to be a binding site for ORF57 and PABPC1 but not for E1B-AP5, which binds to another region of the PAN MRE-II (51). In this study, we demonstrated that PABPC1 is required for ORF57 interaction with the PAN MRE core and functions as a suppressor/deregulator of PAN expression. Viral ORF57 promotes the nuclear distribution of PABPC1, binds to the PAN MRE core in the complex with PABPC1, and prevents the negative effect of PABPC1 on PAN RNA. However, ectopic PABPC1 is also suppressive to ORF57-mediated accumulation of PAN by blocking ORF57 protein production. Together, our data clearly indicate that although ORF57 interacts with PABPC1 for binding PAN RNA, the functions of two proteins on PAN RNA are mutually exclusive. In contrast, E1B-AP5 (64–68), which interacts with another region of the PAN MRE-II and has functions in transcription and RNA export (64, 67, 68), is a positive regulator for the expression of PAN RNA independent of ORF57 and does not directly bind to ORF57.
The finding that a functional connection exists between PABPC1 and ORF57 through protein-protein interaction and nuclear colocalization is consistent with the observations made of herpes simplex virus 1 ICP27 (25), an ORF57 homolog. The nuclear distribution of PABPC1 induced by virus infection is common for both RNA viruses and DNA viruses (4). In Bunyamwera orthobunyavirus, viral nucleocapsid protein binds to and colocalizes with PABP in the cytoplasm early during infection, followed by nuclear retention of PABP (23). In KSHV-infected cells, previous studies showed that KSHV K10/10.1 and SOX (ORF37) could be responsible for PABPC1 nuclear localization (28, 40). KSHV K10/10.1 interacts with PABPC1 and colocalizes with PABPC1 in the nucleus of PEL cells, but overexpression of K10/10.1 alone failed to redistribute PABPC1 to the nucleus (40). KSHV SOX in PEL cells or transiently expressed in HEK293 cells also colocalizes with PABPC1 in the nucleus (27, 28), but viral SOX does not directly interact with PABPC1. SOX-induced nuclear PABPC1 promotes aberrant hyperadenylation of GFP RNA (28). We previously demonstrated that one of the ORF57 functions is to stabilize ORF59 RNA by preventing aberrant ORF59 polyadenylation (62). In this report, PABPC1 is shown to be a negative regulator of the expression of PAN, an important regulator of KSHV gene expression, replication, and immune modulation (53–55). We believe that KSHV infection reduces PABPC1 expression to a minimal level for ORF57 to interact with an MRE of PAN and to promote PAN expression. Taken together, it is conceivable that KSHV, in addition to reducing PABPC1 expression during virus lytic infection, utilizes viral ORF57 to counteract PABPC1's adverse effect on viral RNA by direct interaction with PABPC1.
Strikingly, we found that ectopic PABPC1 has a negative effect on ORF57 stabilization of PAN RNA indirectly by reduction of ORF57 steady-state protein expression or translation. PABPC1 in the cytoplasm stimulates mRNA translation initiation and stability (3, 4, 11). When shuttled to the nucleus, PABPC1 engages in nuclear RNA processing (3, 16, 17). We found that knocking down PABPC1 leads to increased expression of PAN RNA (Fig. 6A and C). However, when expressed by transient transfection, the ectopic PABPC1 reduces the steady-state levels of ORF57 protein, presumably by inhibition of ORF57 translation, and induces aberrant polyadenylation of PAN RNA (Fig. 8B and C), in spite of its induction of a minimal increase of the ORF57 or PAN RNA level at a low dose (Fig. 7A). It is worth noting that the ectopic PABPC1 does not appear to affect the steady-state levels of endogenous cyclophilin A and β-tubulin proteins (Fig. 8C; see Fig. S6A in the supplemental material). PABPC1 is required to loop the mRNA 3′ end to the mRNA 5′ end for the establishment of steady-state translation in the cytoplasm via its interaction with eIF4G associated with the 5′ cap. Other studies indicate that GW182 binding to PABPC1 seems to repress translation by interfering with the formation of an mRNA closed loop (14). PABPC1 also interacts with eRF3 to facilitate the release of the nascent peptide chain from the terminating ribosome (69, 70). Whether ectopic PABPC1's reduction of steady-state ORF57 protein expression or translation is a result from interacting excessively with GW182 and/or eRF3 or from stalling the movement of the 40S ribosomal subunit (71) along the ORF57 5′ untranslated region is under active investigation.
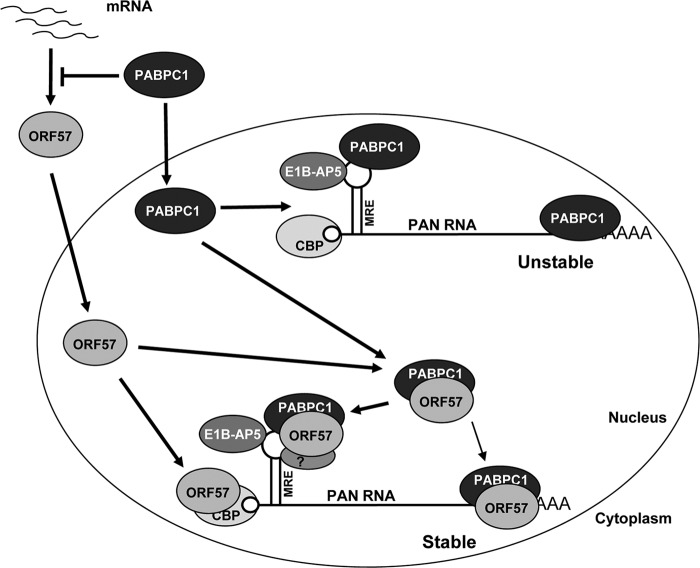
Collectively, our data support a model (Fig. 9) in which overwhelming ORF57 expression during virus lytic infection promotes PAN expression at least in part by modulating PABPC1 distribution and by counteracting the inhibitory activity of PABPC1 on PAN. Through interaction with the PAN 5′ cap (72) (see Fig. S7 in the supplemental material) and in complex with PABPC1 on the MRE-II core on the PAN 5′ end and presumably also on the PAN 3′ end (53), ORF57 along with E1B-AP5 binding to the PAN MRE-II exerts its maximal function in stabilization of PAN. The proposed model in Fig. 9 is similar to the observation that KSHV infection triggers PABPC1 relocation to the nucleus by a PABPC1-noninteracting viral protein, SOX (27, 28, 53). As nuclear accumulation of PABPC1 promotes hyperadenylation and nuclear retention of mRNA (27) and one of the functions of ORF57 is to prevent viral RNA from hyperadenylation (62), our data support a mechanistic link between PABPC1 and ORF57 in the accumulation of a viral lncRNA, PAN.
Fig 9.
Proposed model of PAN MRE-II in ORF57-, PABPC1-, and E1B-AP5-mediated PAN stability. Whereas it inhibits steady-state ORF57 protein expression or translation from its mRNA in the cytoplasm, PABPC1 protein, when shuttling to the nucleus, binds to the PAN MRE-II core on the 5′ end and the 3′ end of PAN and induces PAN instability. During virus lytic infection, ORF57 protein induces nuclear accumulation of PABPC1 and forms a complex with PABPC1 to prevent PABPC1's negative effect on PAN expression. The PABPC1-ORF57 complex interacts with the PAN MRE-II core on the 5′ end of PAN and a less functional expression and nuclear retention element (ENE)-containing region on the 3′ end of PAN (53) to promote PAN stability. E1B-AP5 binding to a region outside the PAN MRE core in the MRE-II stem-loop (51) provides additional strength to stabilize PAN. Recent studies indicate that ORF57 interacts with CBP80, a component of the CBP complex on the 5′ cap of PAN (see Fig. S7 in the supplemental material and reference 72). This model proposes that the PAN MRE-II-mediated interactions and cross-talks between 5′ capping and 3′ polyadenylation machineries via ORF57, PABPC1, and E1B-AP5 maximize PAN stability.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the NCI, Center for Cancer Research, National Institutes of Health.
We thank Jae Jung for providing Dox-inducible TREx BCBL1-Rta and -vector cell lines, Britt Glaunsinger for HA-tagged wt and mt PABPC1 expression vectors, Masahiko Ajiro for purified GST protein, Jun Zhu for his critical reading of our manuscript, and other members of the Z.-M. Zheng laboratory for their assistance and critical comments in the course of this study.
Footnotes
Published ahead of print 17 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01693-12.
REFERENCES
- 1. Grange T, de Sa CM, Oddos J, Pictet R. 1987. Human mRNA polyadenylate binding protein: evolutionary conservation of a nucleic acid binding motif. Nucleic Acids Res. 15:4771–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E. 2010. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 29:4146–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lemay JF, Lemieux C, St. Andre O, Bachand F. 2010. Crossing the borders: poly(A)-binding proteins working on both sides of the fence. RNA Biol. 7:291–295 [DOI] [PubMed] [Google Scholar]
- 4. Smith RW, Gray NK. 2010. Poly(A)-binding protein (PABP): a common viral target. Biochem. J. 426:1–12 [DOI] [PubMed] [Google Scholar]
- 5. Walters RW, Bradrick SS, Gromeier M. 2010. Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA 16:239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bag J, Bhattacharjee RB. 2010. Multiple levels of post-transcriptional control of expression of the poly (A)-binding protein. RNA Biol. 7:5–12 [DOI] [PubMed] [Google Scholar]
- 7. Derry MC, Yanagiya A, Martineau Y, Sonenberg N. 2006. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harbor Symp. Quant. Biol. 71:537–543 [DOI] [PubMed] [Google Scholar]
- 8. Hinnebusch AG. 2011. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75:434–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kahvejian A, Roy G, Sonenberg N. 2001. The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harbor Symp. Quant. Biol. 66:293–300 [DOI] [PubMed] [Google Scholar]
- 10. Kuhn U, Wahle E. 2004. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 1678:67–84 [DOI] [PubMed] [Google Scholar]
- 11. Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. 2007. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 26:1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 27:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singh G, Rebbapragada I, Lykke-Andersen J. 2008. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 6:e111 doi:10.1371/journal.pbio.0060111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. 2009. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 29:6220–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ding L, Han M. 2007. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 17:411–416 [DOI] [PubMed] [Google Scholar]
- 16. Afonina E, Stauber R, Pavlakis GN. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273:13015–13021 [DOI] [PubMed] [Google Scholar]
- 17. Hosoda N, Lejeune F, Maquat LE. 2006. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol. Cell. Biol. 26:3085–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burgess HM, Richardson WA, Anderson RC, Salaun C, Graham SV, Gray NK. 2011. Nuclear relocalisation of cytoplasmic poly(A)-binding proteins PABP1 and PABP4 in response to UV irradiation reveals mRNA-dependent export of metazoan PABPs. J. Cell Sci. 124:3344–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uchida N, Hoshino S, Katada T. 2004. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J. Biol. Chem. 279:1383–1391 [DOI] [PubMed] [Google Scholar]
- 20. Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CY, Zhong Z, Yamashita Y, Zheng D, Shyu AB. 2007. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol. Cell. Biol. 27:7791–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Okochi K, Suzuki T, Inoue J, Matsuda S, Yamamoto T. 2005. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes Cells 10:151–163 [DOI] [PubMed] [Google Scholar]
- 22. Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S. 2007. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 21:3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blakqori G, Kvan I, Elliott RM. 2009. Bunyamwera orthobunyavirus S-segment untranslated regions mediate poly(A) tail-independent translation. J. Virol. 83:3637–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Covarrubias S, Richner JM, Clyde K, Lee YJ, Glaunsinger BA. 2009. Host shutoff is a conserved phenotype of gammaherpesvirus infection and is orchestrated exclusively from the cytoplasm. J. Virol. 83:9554–9566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dobrikova E, Shveygert M, Walters R, Gromeier M. 2010. Herpes simplex virus proteins ICP27 and UL47 associate with polyadenylate-binding protein and control its subcellular distribution. J. Virol. 84:270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harb M, Becker MM, Vitour D, Baron CH, Vende P, Brown SC, Bolte S, Arold ST, Poncet D. 2008. Nuclear localization of cytoplasmic poly(A)-binding protein upon rotavirus infection involves the interaction of NSP3 with eIF4G and RoXaN. J. Virol. 82:11283–11293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumar GR, Glaunsinger BA. 2010. Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol. Cell. Biol. 30:4996–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee YJ, Glaunsinger BA. 2009. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol. 7:e1000107 doi:10.1371/journal.pbio.1000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Salaun C, MacDonald AI, Larralde O, Howard L, Lochtie K, Burgess HM, Brook M, Malik P, Gray NK, Graham SV. 2010. Poly(A)-binding protein 1 partially relocalizes to the nucleus during herpes simplex virus type 1 infection in an ICP27-independent manner and does not inhibit virus replication. J. Virol. 84:8539–8548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alvarez E, Castello A, Menendez-Arias L, Carrasco L. 2006. HIV protease cleaves poly(A)-binding protein. Biochem. J. 396:219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bushell M, Sarnow P. 2002. Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 158:395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gorgoni B, Gray NK. 2004. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief. Funct. Genomic. Proteomic. 3:125–141 [DOI] [PubMed] [Google Scholar]
- 33. Joachims M, Van Breugel PC, Lloyd RE. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kerekatte V, Keiper BD, Badorff C, Cai A, Knowlton KU, Rhoads RE. 1999. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J. Virol. 73:709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuyumcu-Martinez M, Belliot G, Sosnovtsev SV, Chang KO, Green KY, Lloyd RE. 2004. Calicivirus 3C-like proteinase inhibits cellular translation by cleavage of poly(A)-binding protein. J. Virol. 78:8172–8182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuyumcu-Martinez NM, Joachims M, Lloyd RE. 2002. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J. Virol. 76:2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol. 24:1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rivera CI, Lloyd RE. 2008. Modulation of enteroviral proteinase cleavage of poly(A)-binding protein (PABP) by conformation and PABP-associated factors. Virology 375:59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arias C, Walsh D, Harbell J, Wilson AC, Mohr I. 2009. Activation of host translational control pathways by a viral developmental switch. PLoS Pathog. 5:e1000334 doi:10.1371/journal.ppat.1000334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kanno T, Sato Y, Sata T, Katano H. 2006. Expression of Kaposi's sarcoma-associated herpesvirus-encoded K10/10.1 protein in tissues and its interaction with poly(A)-binding protein. Virology 352:100–109 [DOI] [PubMed] [Google Scholar]
- 41. Glaunsinger B, Ganem D. 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell 13:713–723 [DOI] [PubMed] [Google Scholar]
- 42. Bagneris C, Briggs LC, Savva R, Ebrahimi B, Barrett TE. 2011. Crystal structure of a KSHV-SOX-DNA complex: insights into the molecular mechanisms underlying DNase activity and host shutoff. Nucleic Acids Res. 39:5744–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Glaunsinger B, Chavez L, Ganem D. 2005. The exonuclease and host shutoff functions of the SOX protein of Kaposi's sarcoma-associated herpesvirus are genetically separable. J. Virol. 79:7396–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bello LJ, Davison AJ, Glenn MA, Whitehouse A, Rethmeier N, Schulz TF, Barklie CJ. 1999. The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol. 80:3207–3215 [DOI] [PubMed] [Google Scholar]
- 45. Gupta AK, Ruvolo V, Patterson C, Swaminathan S. 2000. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J. Virol. 74:1038–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kirshner JR, Lukac DM, Chang J, Ganem D. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Majerciak V, Yamanegi K, Nie SH, Zheng ZM. 2006. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 281:28365–28378 [DOI] [PubMed] [Google Scholar]
- 48. Nekorchuk M, Han Z, Hsieh TT, Swaminathan S. 2007. Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J. Virol. 81:9990–9998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Majerciak V, Kruhlak M, Dagur PK, McCoy JP, Jr, Zheng ZM. 2010. Caspase-7 cleavage of Kaposi sarcoma-associated herpesvirus ORF57 confers a cellular function against viral lytic gene expression. J. Biol. Chem. 285:11297–11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Majerciak V, Yamanegi K, Allemand E, Kruhlak M, Krainer AR, Zheng ZM. 2008. Kaposi sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes the expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J. Virol. 82:2792–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Massimelli MJ, Kang JG, Majerciak V, Le SY, Liewehr DJ, Steinberg SM, Zheng ZM. 2011. Stability of a long noncoding viral RNA depends on a 9-nt core element at the RNA 5′ end to interact with viral ORF57 and cellular PABPC1. Int. J. Biol. Sci. 7:1145–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sei E, Conrad NK. 2011. Delineation of a core RNA element required for Kaposi's sarcoma-associated herpesvirus ORF57 binding and activity. Virology 419:107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Borah S, Darricarrere N, Darnell A, Myoung J, Steitz JA. 2011. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog. 7:e1002300 doi:10.1371/journal.ppat.1002300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rossetto CC, Pari G. 2012. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog. 8:e1002680 doi:10.1371/journal.ppat.1002680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rossetto CC, Pari GS. 2011. KSHV noncoding PAN RNA interacts with virus and cellular-encoded proteins and suppresses expression of genes involved in immune modulation. J. Virol. 85:13290–13297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Majerciak V, Pripuzova N, McCoy JP, Gao SJ, Zheng ZM. 2007. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J. Virol. 81:1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Majerciak V, Yamanegi K, Zheng ZM. 2006. Gene structure and expression of Kaposi's sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J. Virol. 80:11968–11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jia R, Liu X, Tao M, Kruhlak M, Guo M, Meyers C, Baker CC, Zheng ZM. 2009. Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J. Virol. 83:167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kang JG, Pripuzova N, Majerciak V, Kruhlak M, Le SY, Zheng ZM. 2011. Kaposi's sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from microRNA-mediated suppression. J. Virol. 85:2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yong J, Golembe TJ, Battle DJ, Pellizzoni L, Dreyfuss G. 2004. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol. Cell. Biol. 24:2747–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Majerciak V, Uranishi H, Kruhlak M, Pilkington GR, Massimelli MJ, Bear J, Pavlakis GN, Felber BK, Zheng ZM. 2011. Kaposi's sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59. J. Virol. 85:1528–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Majerciak V, Zheng ZM. 2009. Kaposi's sarcoma-associated herpesvirus ORF57 in viral RNA processing. Front. Biosci. 14:1516–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bachi A, Braun IC, Rodrigues JP, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, Izaurralde E. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6:136–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barral PM, Rusch A, Turnell AS, Gallimore PH, Byrd PJ, Dobner T, Grand RJ. 2005. The interaction of the hnRNP family member E1B-AP5 with p53. FEBS Lett. 579:2752–2758 [DOI] [PubMed] [Google Scholar]
- 66. Blackford AN, Bruton RK, Dirlik O, Stewart GS, Taylor AM, Dobner T, Grand RJ, Turnell AS. 2008. A role for E1B-AP5 in ATR signaling pathways during adenovirus infection. J. Virol. 82:7640–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gabler S, Schutt H, Groitl P, Wolf H, Shenk T, Dobner T. 1998. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J. Virol. 72:7960–7971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kzhyshkowska J, Rusch A, Wolf H, Dobner T. 2003. Regulation of transcription by the heterogeneous nuclear ribonucleoprotein E1B-AP5 is mediated by complex formation with the novel bromodomain-containing protein BRD7. Biochem. J. 371:385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kozlov G, Gehring K. 2010. Molecular basis of eRF3 recognition by the MLLE domain of poly(A)-binding protein. PLoS One 5:e10169 doi:10.1371/journal.pone.0010169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhouravleva G, Frolova L, Le GX, Le GR, Inge-Vechtomov S, Kisselev L, Philippe M. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14:4065–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bag J. 2001. Feedback inhibition of poly(A)-binding protein mRNA translation. A possible mechanism of translation arrest by stalled 40 S ribosomal subunits. J. Biol. Chem. 276:47352–47360 [DOI] [PubMed] [Google Scholar]
- 72. Boyne JR, Colgan KJ, Whitehouse A. 2008. Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 4:e1000194 doi:10.1371/journal.ppat.1000194 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.