Abstract
Residues Arg283, Arg285, and Ile287 are highly conserved amino acids in bovine viral diarrhea virus RNA polymerase (BVDV RdRp) and RdRps from related positive-strand RNA viruses. This motif is an important part of the binding pocket for the nascent RNA base pair during initiation and elongation. We found that replacement of the arginines with alanines or more conserved lysines or replacement of isoleucine with alanine or valine alters the ability of the mutant RdRps to incorporate ribonucleotides efficiently. The reduced RdRp activity stems from both decreased ribonucleotide binding and decreased catalytic efficiency in both primer-dependent and de novo initiation, as shown by kinetic studies. In line with other studies on flaviviral RdRps, our data suggest that Arg283 and Ile287 may be implicated in ribonucleotide binding and positioning of the template base in the active site. Arg285 appears to be involved directly in the selection of cognate nucleotide. The findings for Arg285 and Ile287 mutants also agree with similar data from picornavirus RdRps.
INTRODUCTION
Pestiviruses are known to cause many significant diseases in livestock, such as bovine viral diarrhea in cattle, border disease in sheep, and classical swine fever in pigs. Two biotypes of pestiviruses have been identified: those that result in lysis of in vitro-infected cells, named cytopathogenic (CP) viruses, and those that do not cause cell lysis, termed noncytopathogenic (non-CP) viruses (1). In the United States alone, the economic impact due to bovine viral diarrhea virus (BVDV) infections in cattle ranges from $10 to $40 million per million calves (2, 3). All members of the Flaviviridae, which include important human pathogens such as hepatitis C virus (HCV), dengue fever virus (DFV), Japanese encephalitis virus, and West Nile virus (4), presumably share a similar overall genome organization, replication cycle, and virion structure (1, 5).
The RNA genome of BVDV is one of the largest (12.5 kb) in the Flaviviridae (6). The polyprotein of approximately 3,900 amino acids is processed co- and posttranslationally into at least 12 functional viral proteins (7). The last gene product, NS5B, is the viral RNA-dependent RNA polymerase (RdRp), responsible for transcription and replication of the viral genome. RdRps have been shown to function as part of a larger, membrane-associated replication complex (1, 8–14). The crystal structures of BVDV RdRp (15, 16) and the ternary complexes of various other viral RdRp–double-stranded RNA (dsRNA) cocrystal structures have given further insights into the molecular mechanisms of viral replication (8, 9, 15, 17–26). For reviews of the structure-function relationships in flaviviral and picornaviral RdRps, see the works of Choi and Rossmann and Ferrer-Orta et al. (27, 28).
The ribonucleotide triphosphate (rNTP) binding channel is characterized in part by a highly conserved basic sequence motif, termed “motif F” (5, 29). In this study, we applied steady-state and pre-steady-state kinetics to probe the functional roles of residues R283, R285, and I287 (Fig. 1), all of which are hallmarks of motif F. Through close examination of multiple structure-based alignments by use of relevant RdRp crystal structures (8, 15, 16, 19, 20) and other ternary RdRp complexes (19–21, 30–32, 49, 50), we wanted to investigate the effects that these residues at the fingertips of the RdRp domain exert on rNTP import, formation of the preinitiation complex, and catalytic turnover of BVDV NS5B. Other residues in motif F of BVDV RdRp (K263, R267, and K282) are not considered in this investigation, since they are either missing from the models, poorly defined in the crystal structure, or too far away from the location where the ultimate base pair is expected to bind. The molecular basis for nucleotide selection by RdRps has been a topic of considerable attention (31), and although new structural insights into the interactions of RdRps and conserved RNA and NTP in the active site have been defined for Norwalk virus (32), foot-and-mouth disease virus (FMDV) (19, 20), and poliovirus (PV) (21), no details have been reported for BVDV NS5B. Our data indicate that R283, R285, and I287 are involved in the formation of the preinitiation complex and in catalytic turnover via either primer-dependent or de novo initiation and that they also affect the fidelity of BVDV RdRp.
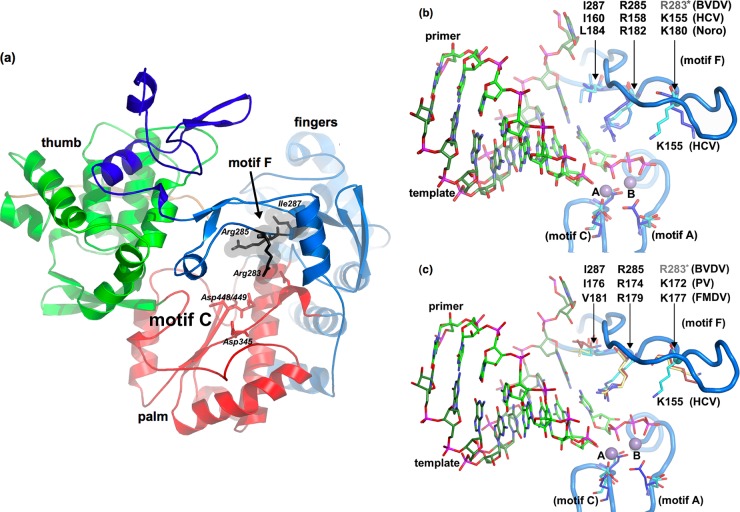
Fig 1.
(a) Ribbon diagram of BVDV NS5B, highlighting three conserved residues in motif F. The backbone of BVDV NS5B is colored according to subdomains, as follows: blue, fingers; red, palm; and green, thumb. Residues R283, R285, and I287 are highlighted in dark gray. The basic residues at positions 283 and 285 form the roof of the rNTP channel. Residue I287 is located further toward the interior of the active site lining the binding pocket for the nascent base pair. (b) Homology modeling of related RdRps from BVDV (PDB accession no. 2CJQ), HCV (PDB accession no. 1NB6), and norovirus (PDB accession no. 3BSO). Superposition of the norovirus structure onto BVDV and HCV NS5Bs reveals that norovirus K181 is bulged out in the RNA cocrystal structure of norovirus RdRp and does not match up with the BVDV K282 and HCV R154 positions. (c) Homology modeling of BVDV, PV (PDB accession no. 1RA7; dark red), and FMDV (PDB accession no. 2E9R; light yellow) RdRps. Comparison of the respective biochemical and structural data of these plus-strand RdRps indicates that the conserved and highlighted residues are involved in rNTP binding and catalysis. Based on these homologies, three highly conserved residues in BVDV RdRp motif F, namely, R283, R285, and I287, were selected for detailed kinetic studies.
MATERIALS AND METHODS
Materials.
The homopolymeric poly(C) RNA and rNTPs were purchased from Sigma, and the short heteropolymeric RNA templates were synthesized by Dharmacon Inc. (Table 1). Radioactively labeled nucleotides (3,000 mCi/mmol) were obtained from Perkin-Elmer.
Table 1.
Sequences of oligonucleotides used in RdRp activity assays
| Template | Oligonucleotide sequence |
|---|---|
| Duplex A | 5′-AGCUCGCGGUACC |
| 3′-GCGCCAUGG | |
| Duplex B | 5′-CAGACCAUCAGCACCAUAUGCGGAUACCGCGAGCU |
| 3′-UACGCCUAUGGCGCUCGA | |
| Template C | 5′-CCUUUUCUAAUUCUCGUAUAC-ddC |
Mutagenesis, expression, and purification.
The BVDV NS5B protein (from strain NADL), lacking 24 hydrophobic, C-terminal amino acids, was cloned into the expression plasmid pET28a encoding a C-terminal hexahistidine tag. All single point mutations of NS5B were introduced using the QuikChange method (Stratagene). The strategy for mutagenesis was based upon three-dimensional alignments in COOT (http://www.biop.ox.ac.uk/coot) and PyMOL (http://www.pymol.org) of RdRps from BVDV, HCV, DFV, norovirus, PV, FMDV, reovirus (RV), and φ6 bacteriophage. The superpositions were initially carried out by aligning secondary structure elements in the RdRp palm domain (COOT) and were further optimized by aligning catalytic aspartate residues and the critical residues in motif F (PyMOL) (Fig. 1 and Table 2). Expression of wild-type BVDV NS5B and its mutants was carried out in Escherichia coli BL21(DE3) Star (Invitrogen). Cells were grown to exponential phase (optical density at 600 nm [OD600] of <0.8), and protein expression was induced at 37°C for 8 h by adding isopropyl-β-d-thiogalactoside to a final concentration of 0.4 mM. The cell pellets were stored at −20°C until further analysis. The cell pellets were thawed and resuspended in lysis buffer containing 50 mM Tris-HCl, pH 8.0, 600 mM NaCl, 0.2% Triton X-100, 20 mM MgCl2, 1 U/ml DNase I, and 2 tablets of EDTA-free Complete protease inhibitor (Roche). After sonication, the cell lysate was centrifuged and the cytosolic fraction applied to a Ni-nitrilotriacetic acid (Ni-NTA) column (Superflow; Qiagen) preequilibrated with buffer A (50 mM Tris-HCl, pH 8.0, 600 mM NaCl). The protein was eluted with a linear gradient of imidazole ranging from 20 to 350 mM, dialyzed overnight in 50 mM Tris-HCl, pH 7.5, 600 mM NaCl, and 5 mM dithiothreitol (DTT), and applied to a gel filtration column (Superdex High Load 26/60 S200 column; Amersham Bioscience). Wild-type and mutant NS5B proteins were purified to near homogeneity (<95%) as judged by SDS-PAGE and dynamic light scattering. The eluted material was dialyzed into 50 mM Tris-HCl, pH 7.5, 600 mM NaCl, 5 mM DTT, and 30% glycerol and stored in small aliquots at −80°C.
Table 2.
Alignment and root mean square deviations of BVDV, HCV, norovirus, PV, and FMDV RdRp cocrystal structuresa
| Target PDB accession no. (virus) | Alignment length (no. of residues) | %SSEb matched to query PDB structure | Overall RMSD (RMSD for motifs A, C, and F only)c (Å) |
|---|---|---|---|
| 1NB6 (HCV) | 412 | 65 | 2.90 (0.67) |
| 3BSO (norovirus) | 365 | 76 | 3.05 (0.77) |
| 2E9R (FMDV) | 354 | 62 | 2.90 (1.09) |
| 1RA7 (PV) | 336 | 71 | 2.99 (1.14) |
The query structure for the BVDV RdRp is found under PDB accession no. 2CJQ.
Percentage of secondary structure elements matched by COOT or PDBeFold.
Includes residues A341 to V353, R442 to 1453, and P284 to Y288 in query structure (PDB accession no. 2CJQ). RMSD, root mean square deviation.
Steady-state kinetic polymerase assays using poly(rC)-oligo(rG)12.
In order to compare the activities of wild-type NS5B and its mutants, a variety of steady-state assays were performed. Reaction conditions were optimized according to assay temperature, pH, ionic strength, and metal ion concentration, and reaction mixtures were set up in 20 mM Tris-HCl, pH 7.5, 5 mM MgCl2 (or 2 mM MnCl2), 5 mM DTT, 10% (vol/vol) glycerol, 50 μg/ml bovine serum albumin (BSA), and 0.25% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), with 0.4 μCi/μl [α-32P]rGTP, 0.250 μg of poly(C) template, 0.125 μg of oligo(G)12 primer, and 50 nM NS5B, in a total volume of 25 μl. Small aliquots were removed at specified time intervals and quenched by addition of 5 μl of 0.5 M EDTA. Ten-microliter samples of the quenched reaction mixture were then blotted onto DE81 filter paper discs and dried. The discs were washed three times for 10 min in 250 ml of 5% dibasic sodium phosphate and 2% sodium pyrophosphate, rinsed with 95% ethanol, and air dried. Bound radioactivity was quantified by using a scintillation counter (Beckman).
Kinetics of single nucleotide incorporation.
The rates of single nucleotide incorporation were determined under pre-steady-state conditions. The primer-template duplex (duplex A) (Table 1) used in this RdRp assay was prepared by annealing the oligonucleotides to form a duplex with a 4-nucleotide (nt) overhang. Prior to annealing, the shorter oligonucleotide was 5′-end labeled using [γ-32P]ATP and T4 polynucleotide kinase, with excess nucleotide removed by passing the sample over a Sephadex G-25 spin column (Amersham Biosciences). The RdRp assay reaction mixtures contained 2.5 μM enzyme, 0.1 μM 5′-end-labeled RNA, and up to 100 mM NaCl. Reaction mixtures were incubated for 5 to 60 min at 37°C in 20 mM Tris-HCl, pH 7.5, 5 mM MgCl2 (or 2 mM MnCl2), 0.25% (wt/vol) CHAPS, 5 mM DTT, 10% (vol/vol) glycerol, and 50 μg/ml BSA in a 20-μl volume. Reaction mixtures were set up manually by pipetting ATP at concentrations ranging from 0 to 5 mM and were incubated at 37°C. Samples (1.5 μl) were removed at various time points and quenched by adding 4 μl of gel loading buffer containing 90% (vol/vol) formamide, 30 mM EDTA, 0.2% (wt/vol) SDS, 0.025% (wt/vol) bromophenol blue, and 0.025% (wt/vol) xylene cyanol. The RNA products were separated in a 12% polyacrylamide-7.5 M urea gel and quantified by phosphorimaging analysis (Fuji BAS-2000).
Single nucleotide omission assay.
Primer extension assays were performed using the preannealed dsRNA as the substrate (duplex B) (Table 1). Nucleotide omission assays were carried out using the same protocol as the single nucleotide incorporation assays, but only three nucleotides at a time were used to initiate the reactions. Each protein (2.5 μM) was mixed with 0.1 μM RNA, and reaction mixtures were incubated for 1 h before quenching and PAGE analysis (as described above).
De novo polymerization assay.
A synthetic 22-mer RNA (template C) (Table 1) was used as the template for de novo initiation reactions. In order to avoid self-priming, the RNA template was dideoxy terminated using 3′-ddC. Reactions were carried out in the same reaction buffer as that mentioned above, but in the presence of 1 μM 22-mer RNA, 1 μM NS5B, 3 μCi [α-32P]UTP, 10 μM UTP, 250 μM GTP, and a 200 μM concentration of each remaining NTP at 37°C for 2 h. The RNA products were ethanol precipitated, and the amount of radiolabeled UTP incorporated into RNA was measured by liquid scintillation counting. RNA products were also analyzed in 20% polyacrylamide-7 M urea gels. Gels were dried under a vacuum and quantified using a phosphorimager (Fuji BAS-2000).
Data analysis.
Steady-state kinetic data were plotted by nonlinear regression using the program KaleidaGraph (Synergy Software, Reading, PA). Data from steady-state kinetic experiments were fitted to the Michaelis-Menten equation, i.e., v = {(Vmax × [rNTP])/(Km + [rNTP])}, where v is the velocity measured experimentally before more than approximately 10% of the substrate has been converted to product, Vmax is the maximum rate of NTP incorporation, and Km is the Michaelis-Menten constant. The kcat was calculated from the Vmax value by assuming that all enzyme molecules were active. The pre-steady-state single nucleotide incorporation kinetic data were fitted to the following single exponential equation: product = A[1 − exp(−kobst[r])], where t represents reaction time, A the reaction amplitude, and kobs the observed apparent single turnover rate. The maximum rate of incorporation (kpol) and the apparent binding constant (Kd) were determined by plotting the measured rates of single nucleotide incorporation (kobs) as a function of nucleotide concentration and by fitting the data to the hyperbolic equation kobs = (kpol[rNTP])/(Kd + [rNTP]), where kobs is the observed reaction rate, kpol is the maximum rate of phosphodiester bond formation, and Kd is the substrate concentration at which the reaction rate is half its maximum speed. The data reported in Tables 3 and 4 represent the averages for at least three independent determinations, with the exception of the steady-state kinetics, where two independent experiments were averaged. Gel images shown in Fig. 3, 5, and 6 are representative.
Table 3.
Steady-state kinetic parameters for wild-type BVDV NS5B and its mutants, using poly(C)-oligo(G)12 as the substrate
| Mutant | Km (μM) | Vmax (pmol/min/μM) | kcat (min−1) | kcat/Km (min−1 M−1) | Ratio of (kcat/Km)WT to (kcat/Km)mutant |
|---|---|---|---|---|---|
| R283A | 0.81 ± 0.015 | 0.32 ± 0.006 | 0.05 ± 0.001 | 64,596 | 3.3 |
| R283K | 0.08 ± 0.032 | 0.08 ± 0.003 | 0.01 ± 0.005 | 154,761 | 1.4 |
| R285A | 2.8 ± 0.928 | 0.12 ± 0.022 | 0.02 ± 0.003 | 6,749 | 31.3 |
| R285K | 0.14 ± 0.027 | 0.05 ± 0.016 | 0.007 ± 0.002 | 51,851 | 4.1 |
| I287A | 0.07 ± 0.026 | 0.01 ± 0.004 | 0.001 ± 0.0001 | 14,285 | 14.8 |
| I287V | 0.16 ± 0.016 | 0.10 ± 0.00 | 0.016 ± 0.004 | 101,265 | 2.1 |
| WT | 0.3 ± 0.058 | 30.39 ± 0.019 | 0.06 ± 0.003 | 211,401 |
Table 4.
Pre-steady-state kinetic parameters for single nucleotide incorporation using duplex A (14-nt–10-nt template-primer) for wild-type NS5B and its mutant derivatives
| Mutant | Km (μM)a | kpol (min−1)a | kpol/Km (min−1 M−1) |
|---|---|---|---|
| R283A | 604 ± 24 | 0.143 ± 0.002 | 236 |
| R283K | 909 ± 33 | 0.165 ± 0.027 | 181 |
| R285A | ND | ND | |
| R285K | 919 ± 27 | 0.087 ± 0.005 | 95 |
| I287A | ND | ND | |
| I287V | 614 ± 64 | 0.099 ± 0.005 | 161 |
| WT | 378 ± 82 | 0.122 ± 0.006 | 322 |
ND, near or at the detectable limit of NTP incorporation under the assay conditions tested.
Fig 3.
Single nucleotide incorporation. (a) Single nucleotide incorporation by wild-type BVDV NS5B and its mutant derivatives, using duplex A (Table 1) as the template primer pair. NS5B (2.5 μM) and the RNA duplex (0.1 μM) were mixed under standard reaction conditions with 500 μM ATP in the presence of 5 mM MgCl2 and incubated at 37°C for 30 min. Samples (1.5 μl) were withdrawn at various time points and quenched by adding denaturing buffer prior to PAGE analysis. (b) Single nucleotide incorporation by wild-type BVDV NS5B under the same reaction conditions as those described for panel A, but in the presence of MnCl2. The results shown are representative; each assay was reproduced a minimum of three times.
Fig 5.
De novo RNA synthesis. (a) De novo RNA synthesis of wild-type BVDV NS5B and its mutant derivatives. The enzymes (1 μM) were incubated with 1 μM 22-mer RNA. Reactions were initiated by adding 3 μCi [α-32P]UTP, 10 μM UTP, 250 μM GTP, and a 200 μM concentration of each NTP. Reactions were carried out for 2 h at 37°C, and the RNA product was quantified by scintillation counting. Reactions were performed in the presence of MgCl2 (white bars) or MnCl2 (gray bars) as the divalent metal ion. (b) Gel analysis of the reaction products of de novo RNA synthesis by BVDV NS5B mutants, carried out in the presence of MgCl2. (c) Gel analysis of the reaction products of de novo RNA synthesis by BVDV NS5B mutants, carried out in the presence of MnCl2.
Fig 6.
Nucleotide omission assays. Misincorporation assays were performed in the presence of MgCl2 and MnCl2, using wild-type BVDV NS5B and 6 motif F mutants. The 5′-radiolabeled 18-mer primer annealed to a 35-mer RNA template was extended by 2.5 μM BVDV NS5B. The completed sequence of duplex B is shown in gray, with the expected fully extended primer underlined in black. The extension reactions were carried out for up to 30 min in the presence of a 500 μM concentration of each of the four nucleotides (+) or in the presence of only three complementary nucleotides, i.e., minus ATP (−A), minus CTP (−C), minus GTP (−G), or minus UTP (−U). The extension reactions were analyzed by 20% denaturing gel electrophoresis.
RESULTS
Structure and amino acid sequence comparison.
As a first step toward elucidating the structure-function relationships of BVDV NS5B (Fig. 1), we aligned the crystal structures of BVDV RdRp with those of relevant viral RdRp cocrystal structures by using COOT and PyMOL (Fig. 1 and Table 2). Previous studies have proposed structural homologies between these RdRps, despite the low sequence homology between the proteins (33, 34). The structural similarity is not confined to the overall fold of the protein but, predictably, extends to features in the active sites, where important amino acids such as those in motif F are functionally and spatially quite well conserved (Fig. 1). Using several models (Protein Data Bank [PDB] accession numbers 2CJQ [BVDV], 1NB6 [HCV], 3BSO [norovirus], 2E9R [FMDV], and 1RA7 [PV]) as a guide, we structurally aligned highly conserved residues in the active site of BVDV NS5B (Fig. 1 and Table 2) with equivalent residues in the RNA-dependent RNA polymerases of other viruses of the Flavi-, Noro-, and Picornaviridae families. We used site-directed mutagenesis and kinetic assays to examine more closely the functional role of motif F residues in BVDV NS5B with respect to the formation of the preinitiation complex and the remainder of the RdRp catalytic cycle. We also compared and contrasted these data with similar studies carried out with DFV and FMDV RdRps (35). R283, R285, and I287, which are located above the rNTP binding pocket in the BVDV NS5B fingertips (Fig. 1), were chosen because they form potentially critical interactions between the protein and the incoming nucleotides in the active site in the available three-dimensional structures. The side chains at positions 283 and 285 were changed to those of Ala and Lys, respectively, and I287 was replaced by Ala or Val.
Optimization of temperature, pH, and metal ions for RdRp activity.
In vitro, BVDV NS5B can utilize homopolymeric templates and primers as substrates to catalyze incorporation of nucleotides (14). Moreover, as BVDV NS5B exhibits a preference toward poly(C)-oligo(G)12 (36), this template-primer RNA was also used as a substrate for steady-state experiments to determine optimal reaction conditions. Since previous reports are inconsistent with respect to optimal NS5B reaction conditions (14, 36), we reevaluated the effects of temperature, pH, and divalent metal ion concentration on the RdRp activity of the wild-type BVDV NS5B protein.
In agreement with the data of Zhong and coworkers (14, 36), 37°C was the optimum temperature. BVDV RdRp activity was increased about 6-fold when Mn2+ was used as the metal cofactor. For completeness, Ca2+, Co2+, Fe2+, Cu2+, Zn2+, and Ni2+ were also tested for the ability to support polymerase activity of BVDV NS5B at concentrations ranging from 1 to 10 mM. Consistent with the data for HCV NS5B (25, 37), none of these divalent metals induced any detectable turnover in BVDV NS5B.
We also examined the RdRp activity of wild-type BVDV NS5B and its mutants to determine whether they exhibited any differences in the general ability to synthesize RNA by use of a homopolymeric template (Fig. 2). Compared to wild-type BVDV NS5B, all mutants showed lowered polymerase activity with the poly(C)-oligo(G)12 substrate, but to various degrees. The R283A, R285A, and I287A mutants were most seriously affected, with 50% or less of the wild-type NS5B activity. Predictably, the more conservative mutations, R283K, R285K, and I287V, had much smaller detrimental effects on the overall activity, thus providing some mechanistic insights into the functional roles of the residues at these positions. The biochemical basis for the reduced polymerase activity of each of the mutant proteins was investigated in greater detail by determining the steady-state and pre-steady-state kinetic parameters, which should provide important RdRp structural and functional insights about the catalytic events, such as formation of the preinitiation complex, rNTP ground-state binding affinity, and turnover rate.
Fig 2.
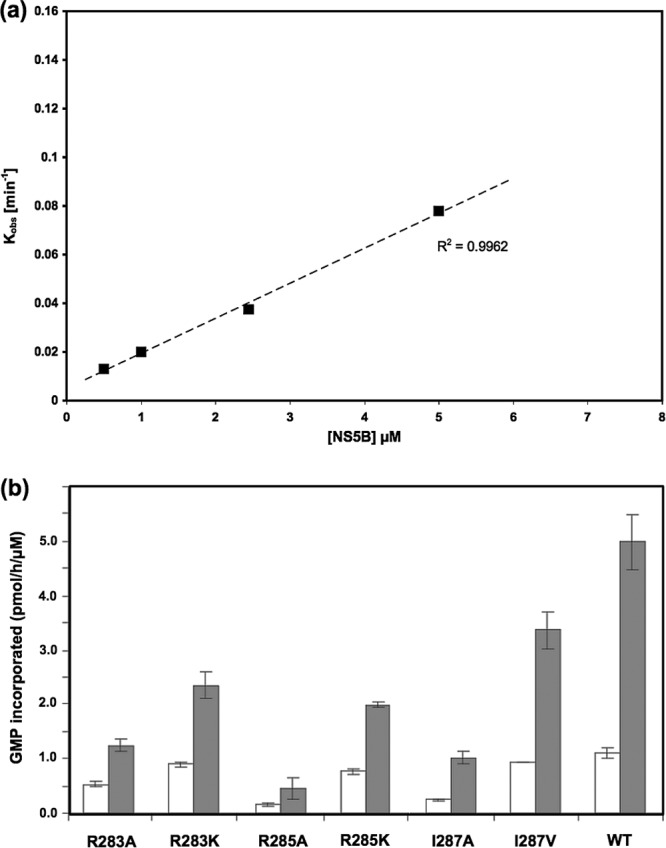
Primer-dependent RNA synthesis. (a) Concentration dependence of primer-dependent RNA synthesis by wild-type BVDV NS5B. A 3′-recessed RNA primer-template duplex (duplex A) was used in this RdRp assay. The RdRp assay reaction mixtures contained between 0.5 and 5 μM wild-type BVDV NS5B, 0.1 μM 5′-end-labeled RNA, and 50 mM NaCl. (b) Primer extension by wild-type (WT) BVDV NS5B and its mutant derivatives in the presence of MgCl2 (white bars) or MnCl2 (gray bars) as the divalent metal ion. Reactions were carried out at 37°C for 2 h, using 0.250 μg of poly(C) template, 0.125 μg oligo(G)12 primer, 0.4 μCi/μl [α-32P]rGTP, and 50 nM NS5B in a total volume of 25 μl. [α-32P]rGMP incorporation was quantified by a DEAE filter binding scintillation assay.
Steady-state kinetics of RNA polymerization.
To establish whether the decrease in RdRp activity of the mutants was due to a change in catalytic efficiency (kcat/Km), i.e., the concentration of nucleotide at which the turnover is half maximum, the wild type and the mutants were assessed for the ability to extend an oligo(G)12 primer opposite a poly(C) template by use of a continuous steady-state assay. Wild-type and mutant NS5Bs all obeyed simple Michaelis-Menten kinetics. The BVDV NS5B mutants showed differences in catalytic efficiency of up to 4-fold from that obtained for the parent protein (Table 3), with the exception of the R285A mutant, which showed a decrease in catalytic efficiency of >30-fold over that of wild-type NS5B. In terms of the polymerization rate, the I287A mutant was the most impaired, followed by the R285K mutant, with an approximately 10-fold decrease. This indicates that in this assay, with a 3′-recessed dsRNA, RdRp residues closer to the incoming base and RNA template binding site are more critical for catalytic turnover with short dsRNA templates (Table 1).
Kinetics of single nucleotide incorporation.
Pre-steady-state kinetic reactions were carried out as a useful means of measuring the effects of mutations on nucleotide incorporation. A short RNA duplex with a 4-nt overhang (duplex A) (Table 1) was used to test primer extension by a single rNTP, rAMP. Reactions were performed using a 25-fold excess of each protein over dsRNA to ensure that the maximum number of substrate molecules was bound by NS5B and that the nucleotide incorporation reactions were synchronized. During optimization of substrate binding and assay conditions, it was found that further increases in protein concentration did not result in any significant increase of the polymerization rate (Fig. 2a), suggesting that all dsRNA was bound by the enzyme and that polymerase cooperativity did not play a role under the chosen reaction conditions. It is important, however, that the in vitro activity of many flaviviral RdRps in the presence of magnesium is distributive or shows a tendency for template switching (29). Since the R285A and I287A mutants proved remarkably inefficient at incorporating rNTPs, even in the presence of Mn2+ and after long extension times (Fig. 2b), only the remaining five NS5B mutants and wild-type NS5B were considered for further pre-steady-state kinetic analyses (Fig. 3 and 4).
Fig 4.
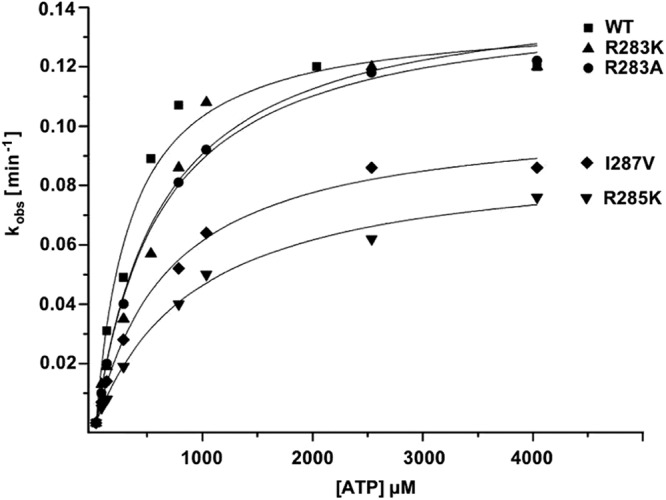
Pre-steady-state kinetics of nucleotide incorporation. The dependence of the reaction rate (kobs) on ATP concentration was fit to a hyperbola according to the Michaelis-Menten equation, i.e., kobs = (kpol[rNTP])/(Km + [rNTP]), where kobs is the observed reaction rate, kpol is the maximum rate of phosphodiester bond formation, and Km is the equilibrium dissociation constant. The fitted curves are overlaid for comparison of observed reaction rates of wild-type and mutant BVDV NS5Bs.
The ground-state binding affinity of rNTPs (Km) for the binary enzyme-RNA complex was determined by following the rNTP concentration dependence of the single turnover rate (kobs). Under these conditions, the maximal reaction rate reflects kpol and the rNTP concentration dependence of the reaction rate gives Km(rNTP). The kpol and Km values were derived by analysis of the data (Fig. 3 and 4), and their kinetic parameters are summarized in Table 4. All mutant enzymes showed Km values for rATP of up to 3-fold over that for wild-type NS5B, except for the R285A and I287A mutants, for which rAMP incorporation was not detectable even in the presence of Mn2+. The mutants with more conservative amino acid substitutions did not show any significant changes in polymerization rates (kpol) (Table 4).
De novo RNA synthesis.
Previous reports have demonstrated that flavivirus NS5Bs are capable of initiating RNA synthesis via a de novo mechanism (29, 36, 38, 39). To probe the ability of the mutant NS5B proteins to initiate RNA synthesis on a single-stranded RNA molecule in the absence of a primer, we used an assay format where the synthetic RNA template carried a dideoxy group on the 3′ cytidylate (template C) (Table 1). The lack of 2′- and 3′-hydroxyls prevents the RNA synthesis from occurring via self-priming. The RNA was designed in order to correspond to the 22 nucleotides at the 3′ end of the negative strand of the BVDV genome. The cytidylate at the 3′ end is essential for de novo initiation (36).
With the exception of the R283A and R283K mutants, all BVDV NS5B mutants displayed 3- to 10-fold reductions in the ability to mediate RNA synthesis de novo (Fig. 5a). As already seen in the primer-dependent RdRp assays, a burst of de novo initiation activity was observed when Mn2+ was used as the metal ion. The R285A and I287A mutants remained devoid of de novo synthesis even in the presence of MnCl2 (Fig. 5b).
Single nucleotide omission assay.
We further analyzed the functional roles of R283, R285, and I287 by testing the ability of the relevant mutants to discriminate against the incorrect nucleotide in a biased pool of nucleotides. Using duplex B (Table 1), primer extension activity was assayed both in the presence of Mg2+ and in the presence of Mn2+, but with only three of the four rNTPs complementary to the RNA template present in the reaction mix. Under these conditions, elongation of the primer opposite a template base required the insertion of an incorrect nucleotide followed by extension beyond the mismatched base pair, revealing mutants with a fidelity lower than that of wild-type NS5B.
In most reactions, in the presence of both wild-type NS5B and its mutants, a substantial accumulation of RNA products at the site corresponding to the missing nucleotide (Fig. 6, site +0 in lanes −A, +2 in lanes −C, +4 in lanes −G, and +1 in lanes −U) was observed, and addition of nucleotides beyond a mismatched pair occurred in only a few cases. As shown in Fig. 6, the primer extension efficiency of the wild-type enzyme in the presence of all nucleotides and MgCl2 was very low. Only a few nucleotides were incorporated before the enzyme presumably stalled or dissociated from the duplex RNA, suggesting that BVDV NS5B is a distributive enzyme in vitro. A higher degree of primer extension activity was observed when MnCl2 was used as the catalytic metal ion, which is in line with other RdRp kinetic studies (25, 29, 38). Full-length products were observed in the presence of Mn2+ only. Furthermore, wild-type NS5B showed little misincorporation in the presence of either of the metal ions. A similar pattern was observed for the R283A and R283K mutants. No misincorporation was observed in the presence of Mg2+, but these mutants showed a small but noticeable decrease in fidelity in the presence of Mn2+ compared to that of wild-type NS5B. The lowered fidelity was observed consistently, as all experiments were repeated at least in triplicate.
The R285A mutant was more prone to misincorporation than the other mutants (Fig. 6). Misincorporation followed by extension of the mismatched termini was observed in the presence of Mg2+ and Mn2+. In contrast to the R285A mutant, the R285K mutant was not misincorporated in the presence of Mg2+; however, high misincorporation levels were observed in the presence of Mn2+. Most of the product synthesized by the R285K mutant in the presence of Mg2+ accumulated at the position complementary to the missing nucleotide, indicating that this mutant was unable to extend beyond the mismatch. The I287A mutant had properties similar to those of the R285K mutant; less significant effects were observed with the I287V mutant.
DISCUSSION
In this study, we characterized a series of BVDV NS5B mutants in which the highly conserved active site residues of motif F have been mutated. Both more and less conservative changes (Arg to Ala or Lys, and Ile to Ala or Val) in the side chains at positions 283, 285, and 287 were used to evaluate the effects of these substitutions on BVDV NS5B function. Other residues in motif F of BVDV RdRp (K263, R267, and K282) were not investigated here, since either they are less well conserved in RdRps or their structural and functional roles are less well defined in the active sites or, in fact, disordered in the BVDV NS5B crystal structure. Our data show that the conserved residues in the nucleotide binding pocket play an essential role for cognate rNTP binding and efficient incorporation by BVDV NS5B. The most pronounced effects of motif F mutants on RdRp activity were detected in steady-state kinetic assays using homopolymeric primer-template pairs. In this type of assay, the incorporation rate and the overall activity are far greater than in the case of heteropolymeric substrates, presumably due to either multiple priming events or more efficient elongation of C·G duplex RNA. Our results also provide a detailed kinetic analysis of the effects of these substitutions in motif F, while previous publications reported that Ala substitutions at positions 285 and 287 largely abrogated the ability of the mutants to incorporate nucleotides (36). Similar results have been observed with Ala substitutions at the equivalent positions of DFV NS5. For example, the motif F mutants M10 (K456/R457 → A) and M11 (K456/R457/K459/K460 → A) displayed <10% of wild-type DFV NS5 activity on the heteropolymeric 5′ DFV template RNA (35). As a hydrophobic residue, L461 (BVDV I287) (Fig. 1) was not considered in this otherwise wide-ranging and complete study of motif F residues in DFV NS5 (35).
Replacements of R283 with Ala and Lys in BVDV NS5B had little effect on the primer-dependent or de novo RNA polymerase activity, as both mutants were able to synthesize RNA by both primer-dependent and de novo initiation modes. Both the Lys and Ala mutants, however, showed up to 3-fold decreases in catalytic efficiency compared to wild-type BVDV NS5B. These differences between R283 mutants and wild-type NS5B could be sufficient to indicate a role for this residue in the interaction with incoming nucleotides. In addition, since the fidelity of R283 mutants was comparable to that of the wild-type enzyme when Mg2+ was used as the catalytic metal, it is plausible that R283 may interact with the phosphate groups rather than in the selection of the base. Homology modeling of BVDV NS5B with norovirus, FMDV, PV, HCV, and φ6 RdRp substrate cocrystal structures (8, 19–21, 24, 25) suggests that in BVDV NS5B, R283 (equivalent to K155 in HCV NS5B) could also interact with the γ-phosphate of the incoming nucleotide (Fig. 1) (40). This interaction could contribute to a stabilization of the preinitiation complex prior to the nucleotide moving into the priming site. The K155A and K155R mutants were less active than wild-type NS5B (41). Incidentally, the K155A mutant has also been shown to be impaired in HCV replicon assays (42).
The importance of residue R285 in determining the turnover rate and in rNTP binding is demonstrated by the overall low catalytic efficiencies of both the R285K and R285A mutants in primer-dependent and de novo initiation (Fig. 2, 3, and 5a). The 30-fold change in the kcat/Km value for rNTP measured for the R285A mutant in primer extension reactions suggests the importance of the interactions between this residue and the incoming nucleotides. This idea was also confirmed by the altered nucleotide incorporation efficiency of the R285K mutant under pre-steady-state conditions (Table 4). Position 285 in BVDV NS5B structurally overlaps with R158 in HCV NS5B, R182 in norovirus RdRp, R174 in PV 3Dpol, and R72 in HIV-1 RT, as shown by superimposition of conserved polymerase motifs (A and C) in these RdRp structures (Fig. 1). In the complete ternary complex of norovirus RdRp, R182 stacks perfectly against the base of the incoming rCTP and comes within 3.2 Å of the α- and β-phosphates. This type of interaction is also preserved in the evolutionarily more distantly related HIV-1 RT ternary complex. In this case, the base of the incoming nucleotide stacks against the guanidinium group of R72, which is also involved in charged interactions with the α-phosphate (30). The stacking interaction of R72 with the base of the incoming nucleotide triphosphate positions the base correctly for cognate hydrogen bonding to the template strand (43).
The lowered fidelity of the R285A mutant in BVDV is also consistent with the hypothesis of active site tightness as a means of regulating polymerase fidelity (44). According to this idea, the active site is shaped in such a way as to accommodate the correct Watson-Crick base pair. Replacement of amino acids that form the nucleotide binding pocket has been shown to alter nucleotide discrimination of many polymerases, such as DNA polymerase β (45, 46), Klenow fragment (47, 48), and HIV-1 RT (43). Despite the major effects caused by the Ala mutation, the properties of R285 were partially retained in the more conservative lysine substitution mutant. The R285K mutant showed smaller variations in steady-state and pre-steady-state kinetics compared to wild-type NS5B, and it maintained the ability to facilitate de novo synthesis. The similar fidelity of the R285K mutant to that of the wild type in the presence of Mg2+ suggests that the side chain of Lys in a fully extended conformation is still able to compensate for the lack of the guanidinium group. Similar molecular interactions between the structurally equivalent arginine residues and the incoming nucleotides are observed in the φ6, RV, norovirus, and PV polymerase initiation complexes (8, 20, 26, 38). Interestingly, the base-stacking interaction is not observed in the FMDV 3Dpol-ribavirin complex, perhaps indicating how this compound is able to insert and elongate the RNA primer (20, 40).
Our biochemical data indeed show that I287 is an essential residue for the enzyme's catalytic efficiency. The substantial decrease in kcat/Km under steady-state conditions (the alanine mutant was 15-fold less efficient than the wild type in a primer-dependent assay) and the inefficiency of de novo synthesis definitely prove a role for I287 in catalysis (Table 3; Fig. 5a). Less drastic effects on either primer extension or de novo initiation were observed in the case of the more conservative I287V mutant. Valine and leucine residues at this position in motif F of PV or FMDV RdRp have also been observed to interact prominently with the RNA template (20, 21). The I287V mutant of BVDV NS5B showed only a small decrease in catalytic efficiency compared with the wild type, with a Km for ATP similar to that of the wild type (Fig. 4). According to the information obtained by superimposing different RdRp structures, residues equivalent to I287 appear not to be in direct contact with the incoming nucleotides but are located in the 90° kink of the template, in a position apparently suited for stacking against the template base that is opposite the incoming nucleotide. Based on the crystallographic information available and on three-dimensional modeling (Fig. 1 and Table 2), I287 could be close enough to this template base to restrain its movement through steric hindrance. Such a constraint should favor geometrically compact Watson-Crick base pairing relative to incorrect base pairing and thus may contribute one mechanism for discrimination. The results obtained in this study suggest that the bulk of the I287 side chain confers a significant advantage in both the catalytic efficiency and the fidelity of this enzyme.
Conclusions.
Taking together the available crystallographic information from the Research Collaboratory for Structural Bioinformatics (RCSB) and the molecular modeling and kinetic data presented here, the functional roles of two potential NTP binding residues and a template-positioning residue have been characterized for BVDV NS5B motif F. Our data broadly confirm studies of motif F residues of FMDV and DFV RdRps, although only Ala substitutions rather than more conservative changes were characterized in the case of dengue fever virus (35). Our pre-steady-state and Michaelis-Menten kinetic data are compatible with the notion that R285 and I287 play important roles in both primer-dependent and de novo initiation. This implies that the side chains of R285 and I287 could be involved in the formation of the preinitiation complex. While R283 seems to contribute mainly to rNTP binding, R285 may more crucially affect the catalytic efficiency of the enzyme. According to our modeling studies and comparisons with noro- and picornavirus RdRp-RNA cocrystal structures, it is possible that the interaction of the R285 guanidinium group with the incoming (second) nucleotide also helps to align the α-phosphate and the 3′-hydroxyl group of the (first) bound nucleotide during de novo initiation (Fig. 1). R285 could also help to position the α-phosphate relative to the growing primer strand in a primer-dependent situation with the RdRp in elongation mode. In addition, mutation of R285 resulted in a significant effect on kpol, which can possibly be explained by contributions of this residue to phosphodiester bond formation. In the absence of a high-resolution crystallographic structure of a BVDV NS5B ternary complex, our findings begin to shed some light on the functional roles of highly conserved residues in motif F of BVDV RNA polymerase.
ACKNOWLEDGMENTS
Z. Hong is gratefully acknowledged for the kind gift of the plasmid harboring BVDV NS5B (strain NADL). We are also indebted to C. M. Joyce and her lab members at Yale for help with pre-steady-state kinetic assays. We thank D. J. Rowlands and J. D. Pata for critically reading the manuscript.
This work was supported by a Wellcome Trust studentship award to E.C. and by a Leeds University Research Fellowship (URF) and Health Research Inc./Wadsworth Center new investigator funds to J.J.
Footnotes
Published ahead of print 17 October 2012
REFERENCES
- 1. Lindenbach BD, Rice CM. 2003. Molecular biology of flaviviruses. Adv. Virus Res. 59:23–61 [DOI] [PubMed] [Google Scholar]
- 2. Lindberg A, Houe H. 2003. Characteristics in the epidemiology of bovine viral diarrhea virus (BVDV) of relevance to control. Prevent. Vet. Med. 72:55–73 [DOI] [PubMed] [Google Scholar]
- 3. Van Campen H. 2010. Epidemiology and control of BVDV in the U.S. Vet. Microbiol. 142:94–98 [DOI] [PubMed] [Google Scholar]
- 4. Westaway EG, Brinton MA, Gaidamovich S, Horzinek MC, Igarashi A, Kaariainen L, Lvov DK, Porterfield JS, Russell PK, Trent DW. 1985. Flaviviridae. Intervirology 24:183–192 [DOI] [PubMed] [Google Scholar]
- 5. Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Collett MS, Larson R, Gold C, Strick D, Anderson DK, Purchio AF. 1988. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology 165:191–199 [DOI] [PubMed] [Google Scholar]
- 7. Collett MS, Wiskerchen M, Welniak E, Belzer SK. 1991. Bovine viral diarrhea virus genomic organization. Arch. Virol. 3(Suppl):19–27 [DOI] [PubMed] [Google Scholar]
- 8. Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235–240 [DOI] [PubMed] [Google Scholar]
- 9. Chinnaswamy S, Murali A, Li P, Kao CC. 2010. Regulation of de novo initiated RNA synthesis in the hepatitis C virus RNA-dependent RNA polymerase by intermolecular interactions. J. Virol. 84:5923–5935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao L, Aizaki H, He JW, Lai MMC. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo G, Hamatake RK, Mathis DM, Racela J, Rigat KL, Lemm J, Colonno RJ. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BE, Rice CM. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shirota Y, Luo H, Qin W, Kaneko S, Yamashita T, Kobayashi K, Murakami S. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277:11149–11155 [DOI] [PubMed] [Google Scholar]
- 14. Zhong W, Gutshall LL, Del Vecchio AM. 1998. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J. Virol. 72:9365–9369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi KH, Groarke JM, Young DC, Kuhn RJ, Smith JL, Pevear DC, Rossmann MG. 2004. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl. Acad. Sci. U. S. A. 101:4425–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choi KH, Gallei A, Becher P, Rossmann MG. 2006. The structure of bovine viral diarrhea virus RNA-dependent RNA polymerase and its amino-terminal domain. Structure 14:1107–1113 [DOI] [PubMed] [Google Scholar]
- 17. Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7:1417–1426 [DOI] [PubMed] [Google Scholar]
- 18. Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale RL, Mathieu M, De Francesco R, Rey FA. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 96:13034–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. 2004. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 279:47212–47221 [DOI] [PubMed] [Google Scholar]
- 20. Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. 2007. Sequential structures provide insights into the fidelity of RNA replication. Proc. Natl. Acad. Sci. U. S. A. 104:9463–9468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gong P, Peersen OB. 2010. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 107:22505–22510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lesburg CA, Cable MB, Ferrari E, Hong Z, Mannarino AF, Weber PC. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937–943 [DOI] [PubMed] [Google Scholar]
- 23. Ng KKS, Cherney MM, Vazquez AL, Machin A, Alonso JMM, Parra F, James MNG. 2002. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem. 277:1381–1387 [DOI] [PubMed] [Google Scholar]
- 24. Ng KKS, Pendas-Franco N, Rojo J, Boga JA, Machin A, Alonso JMM, Parra F. 2004. Crystal structure of Norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J. Biol. Chem. 279:16638–16645 [DOI] [PubMed] [Google Scholar]
- 25. O'Farrell D, Trowbridge R, Rowlands D, Jager J. 2003. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J. Mol. Biol. 326:1025–1035 [DOI] [PubMed] [Google Scholar]
- 26. Tao Y, Farsetta DL, Nibert ML, Harrison SC. 2002. RNA synthesis in a cage—structural studies of reovirus polymerase lambda3. Cell 111:733–745 [DOI] [PubMed] [Google Scholar]
- 27. Choi KH, Rossmann MG. 2009. RNA-dependent RNA polymerases from Flaviviridae. Curr. Opin. Struct. Biol. 19:746–751 [DOI] [PubMed] [Google Scholar]
- 28. Ferrer-Orta C, Arias A, Escarmís C, Verdaguer N. 2006. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 16:27–34 [DOI] [PubMed] [Google Scholar]
- 29. Ferrari E, Wright-Minogue J, Fang JW, Baroudy BM, Lau JY, Hong Z. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang H, Chopra R, Verdine GL, Harrison SC. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669–1675 [DOI] [PubMed] [Google Scholar]
- 31. Ng KKS, Arnold JJ, Cameron CA. 2008. Structure-function relationships among RNA-dependent RNA polymerases. Curr. Top. Microbiol. Immunol. 320:137–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zamyatkin DF, Parra F, Alonso JM, Harki DA, Peterson BR, Grochulski P, Ng KK. 2008. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J. Biol. Chem. 283:7705–7712 [DOI] [PubMed] [Google Scholar]
- 33. Delarue M, Poch O, Tordo N, Moras D, Argos P. 1990. An attempt to unify the structure of polymerases. Protein Eng. 3:461–467 [DOI] [PubMed] [Google Scholar]
- 34. Poch O, Sauvaget I, Delarue M, Tordo N. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iglesias NG, Filomatori CV, Gamarnik AV. 2011. The F1 motif of dengue virus polymerase NS5 is involved in promoter-dependent RNA synthesis. J. Virol. 85:5745–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lai VC, Kao CC, Ferrari E, Park J, Uss AS, Wright-Minogue J, Hong Z, Lau JY. 1999. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J. Virol. 73:10129–10136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lohmann V, Körner F, Herian U, Bartenschlager R. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kao CC, Del Vecchio AM, Zhong W. 1999. De novo initiation of RNA synthesis by a recombinant Flaviviridae RNA-dependent RNA polymerase. Virology 253:1–7 [DOI] [PubMed] [Google Scholar]
- 39. Kao CC, Singh P, Ecker DJ. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251–260 [DOI] [PubMed] [Google Scholar]
- 40. Ferrer-Orta C, Sierra M, Agudo R, de la Higuera I, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. 2010. Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin. J. Virol. 84:6188–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Curti E. 2004. Ph.D. thesis University of Leeds, Leeds, United Kingdom [Google Scholar]
- 42. Cheney IW, Naim S, Lai VCH, Dempsey S, Bellows D, Walker MP, Shim JH, Horscroft N, Hong Z, Zhong W. 2002. Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture. Virology 297:298–306 [DOI] [PubMed] [Google Scholar]
- 43. Lewis DA, Bebenek K, Beard WA, Wilson SH, Kunkel TA. 1999. Uniquely altered DNA replication fidelity conferred by an amino acid change in the nucleotide binding pocket of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 274:32924–32930 [DOI] [PubMed] [Google Scholar]
- 44. Kool ET. 2002. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 71:191–219 [DOI] [PubMed] [Google Scholar]
- 45. Ahn J, Werneburg BG, Tsai MD. 1997. DNA polymerase beta: structure-fidelity relationship from pre-steady-state kinetic analyses of all possible correct and incorrect base pairs for wild type and R283A mutant. Biochemistry 36:1100–1107 [DOI] [PubMed] [Google Scholar]
- 46. Beard WA, Osheroff WP, Prasad R, Sawaya MR, Jaju M, Wood TG, Kraut J, Kunkel TA, Wilson SH. 1996. Enzyme-DNA interactions required for efficient nucleotide incorporation and discrimination in human DNA polymerase beta. J. Biol. Chem. 271:12141–12144 [DOI] [PubMed] [Google Scholar]
- 47. Bell JB, Eckert KA, Joyce CM, Kunkel TA. 1997. Base miscoding and strand misalignment errors by mutator Klenow polymerases with amino acid substitutions at tyrosine 766 in the O helix of the fingers subdomain. J. Biol. Chem. 272:7345–7351 [DOI] [PubMed] [Google Scholar]
- 48. Minnick DT, Bebenek K, Osheroff WP, Turner RM, Jr, Astatke M, Liu L, Kunkel TA, Joyce CM. 1999. Side chains that influence fidelity at the polymerase active site of Escherichia coli DNA polymerase I. J. Biol. Chem. 274:3067–3075 [DOI] [PubMed] [Google Scholar]
- 49. Bressanelli S, Tomei L, Rey FA, De Francesco R. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Makeyev EV, Grimes JM. 2004. RNA-dependent RNA polymerases of dsRNA bacteriophages. Virus Res. 101:45–55 [DOI] [PubMed] [Google Scholar]