Abstract
Herpesvirus saimiri is known to encode a homolog of human complement regulators named complement control protein homolog (CCPH). We have previously reported that this virally encoded inhibitor effectively inactivates complement by supporting factor I-mediated inactivation of complement proteins C3b and C4b (termed cofactor activity), as well as by accelerating the irreversible decay of the classical/lectin and alternative pathway C3 convertases (termed decay-accelerating activity). To fine map its functional sites, in the present study, we have generated a homology model of CCPH and performed substitution mutagenesis of its conserved residues. Functional analyses of 24 substitution mutants of CCPH indicated that (i) amino acids R118 and F144 play a critical role in imparting C3b and C4b cofactor activities, (ii) amino acids R35, K142, and K191 are required for efficient decay of the C3 convertases, (iii) positively charged amino acids of the linker regions, which are dubbed to be critical for functioning in other complement regulators, are not crucial for its function, and (iv) S100K and G110D mutations substantially enhance its decay-accelerating activities without affecting the cofactor activities. Overall, our data point out that ionic interactions form a major component of the binding interface between CCPH and its interacting partners.
INTRODUCTION
The complement system functions as the first line of immunological defense against various pathogens, including viruses, and thus creates a hostile environment for their survival (1–3). It is now clear that complement not only neutralizes viruses directly as a result of opsonization, aggregation, and lysis but also supports their control by augmenting virus-specific antibody as well as cell-mediated immune responses (4–6). It is, therefore, a prerequisite for viruses that they thrive in this humoral system before gaining entry into the host cells. Consistent with this argument, viruses are known to have developed multiple strategies to counteract and bypass the host complement system (7–10). One of the central stratagems utilized by the large DNA viruses like herpes- and poxviruses is the molecular mimicry of human complement regulators: they encode homologs of human regulator-of-complement-activation (RCA) proteins (11, 12).
Herpesvirus saimiri, a T-lymphotropic gammaherpesvirus type 2, is known to cause fulminant lymphomas, lymphosarcomas, and leukemias of T cell origin in New World monkeys such as tamarins, common marmosets, and owl monkey (13–15). It, however, does not cause any apparent disease symptoms in its natural host, the squirrel monkey (16). Interestingly, unlike any other known virus, it encodes two different functional homologs of complement regulatory proteins to subvert the complement system. One is a homolog of RCA proteins named complement control protein homolog (CCPH; encoded by ORF4) (17, 18) that targets the early steps of complement activation, and the other is a homolog of CD59 (encoded by ORF15) that targets the late step of complement activation (19, 20). Structurally, CCPH is composed of four short consensus repeats (SCRs), complement control protein (CCP) domains, or sushi domains connected by short linkers of four amino acids. Analysis of its transcripts revealed that the protein is expressed in soluble as well as membrane-anchored forms (sCCPH and mCCPH, respectively) as a result of differential splicing of the primary transcript (17).
Initial functional characterization of CCPH showed that it is an efficient inhibitor of the classical pathway (CP) as well as the alternative pathway (AP) of complement activation (18). Later, mechanistic studies revealed that it inhibits complement by targeting classical/lectin (C4b,2a) as well as alternative (C3b,Bb) pathway C3 convertases by two mechanisms: (i) by accelerating their decay into their subunits, which is designated decay-accelerating activity (DAA), and (ii) by assisting factor I in degrading C3 convertase subunits C3b and C4b into their inactive forms, which is designated cofactor activity (CFA) (21). A comparison of its DAAs with those of the human complement regulators have shown that sCCPH is about 18-fold and 2,000-fold less active in decaying the classical and alternative pathway C3 convertases than soluble CR1 (sCR1), which suggests that it possesses a very limited AP DAA. Its CFAs, however, are more similar to the human complement regulators, as it is only 2.5-fold less active in inactivating C3b than the human complement regulators factor H and sCR1 and 8-fold less active in inactivating C4b than sCR1 (21). The strategy of C3 convertase inhibition for immune evasion is clearly ingenious, as inhibition at this level would not only result in inhibition of the effector functions of complement, viz., opsonization, direct lysis, and recruitment of inflammatory cells, but would also result in inhibition of complement-mediated enhancement of antiviral adaptive immunity (4, 5, 22). sCCPH has also been mapped for identification of its functional domains using deletion mutagenesis. The results demonstrated that the SCR2 module is the smallest structural unit that can impart CFAs and DAAs, but all four of its modules are required for optimal function (23).
In the present study, we sought to identify the structural determinants in sCCPH that are important for its functional activities. Since the three-dimensional structure of sCCPH is not available, we built a homology model of sCCPH and employed it to identify the putative functional sites in sCCPH on the basis of the previous knowledge of active sites in human RCA proteins and their conservation and solvent accessibility in sCCPH. Overall, we designed 16 Ala substitution mutants and analyzed their functional activities to define the amino acids that play a critical role in its function. In addition, we also designed 8 gain-of-function mutants on the basis of the earlier knowledge of gain of function in other human and viral RCA proteins and educated guesses. The rationale was that if the molecular basis of sCCPH function is identical to that of human or other viral RCA proteins, then such mutations would result in enhancement of sCCPH activities.
(This work was done in partial fulfillment of a Ph.D. thesis of M. J. Reza to be submitted to the University of Pune, Pune, India.)
MATERIALS AND METHODS
Reagents and buffers.
Antibody-sensitized sheep erythrocytes (EAs) were prepared by incubating the erythrocytes with anti-sheep erythrocyte antibodies from ICN Biomedicals Inc. (Irvine, CA). Biotinylated C3b and C4b were generated by site-specific labeling of their free —SH group with EZ-Link polyethylene oxide-maleimide-activated biotin (Pierce, Rockford, IL) (24). Buffers used for various complement assays were as follows. Veronal-buffered saline (VBS) contained 5 mM barbital, 145 mM NaCl, pH 7.4; gelatin veronal-buffered saline (GVB) was VBS containing 0.1% gelatin; dextrose gelatin veronal-buffered saline (DGVB++) was half-ionic-strength GVB containing 2.5% dextrose, 0.5 mM MgCl2, and 0.15 mM CaCl2; and phosphate-buffered saline (PBS)–Tween (PBS-T) contained 10 mM sodium phosphate, 145 mM sodium chloride, and 0.05% Tween 20, pH 7.4.
Complement proteins and their proteolytically activated products.
Human complement protein C3 was purified from plasma as described by Hammer et al. (25) with minor modification (26). For performing alternative pathway DAA, native C3 was separated from C3 (H2O) by passing it onto a Mono S column (Amersham Pharmacia Biotech, Uppsala, Sweden) (27). C3 purified as described above was also used for generation of C3b by limited trypsinization, which was then purified using a Mono Q column (Amersham Pharmacia Biotech) (28). Factor B was purified from human plasma using a 3-step method described earlier (21). All the other complement proteins, viz., C1, C4, C2, C4b, factor D, and factor I, were purchased from Calbiochem (La Jolla, CA).
Homology modeling.
We built a homology model of sCCPH utilizing the experimentally solved structure of Crry (CCP1 to CCP4) as the template (Protein Data Bank accession number 2xrb) and the SWISS-MODEL server (http://swissmodel.expasy.org) (29–31). The rationale for using the Crry structure as the template was that (i) it exhibited better homology than the experimentally solved structures of other complement regulators, (ii) the modeled structure showed the least deviation with the template, (iii) the conserved cysteine residues of the modeled structure matched those of the template, and (iv) Crry, like sCCPH, possesses both DAA and CFA. The modeled structure generated was inspected for van der Waals clashes and disulfide bond correctness and used to determine the solvent accessibility of the residues of interest.
Construction of substitution mutants.
The soluble form of herpesvirus saimiri CCPH was PCR amplified from the full-length CCPH clone (21) using the specific primers 5′-GGAATTCCATATGAGCTGTCCTACACGTAACCAGTATG-3′ (the NdeI site is underlined) and 5′-CCGCTCGAGCATACATTCAGGAATAGCTGGAGAC-3′ (the XhoI site is underlined) and cloned into the pGEM-T Easy vector (Promega). This clone was then digested with NdeI and XhoI and subcloned into pET29 for expression.
The pGEM-T clone generated as described above was used as the template to generate substitution mutants using a QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA). Two sets of mutants were generated. In the first set, Ala substitution mutants were generated to identify the protein-protein interaction sites. The Ala substitution mutants generated were R35A, K40A, K61A, K62A, Y83A, N92A, S100A, L106A, V113A, R118A, K126A, K128A, K142A, F144A, N156A, and K191A. In the second set of mutants, the mutations were guided by the earlier knowledge of gain of function in other RCA proteins and educated guesses. The gain-of-function mutants generated were S96Y, S100E, S100K, T101Y, A104F, A104Y, G110D, and Y225K. Since all the above-described mutants were generated using the pGEM-T clone as the template, they were subcloned into pET29 and transformed into Escherichia coli BL21 cells for expression. The fidelity of the pGEM-T as well as the pET29 clones was verified by DNA sequencing using an automated ABI 3730 DNA analyzer.
Expression, purification, and refolding of sCCPH and its substitution mutants.
The protocol used for expression and purification of the wild-type and mutant proteins was as previously described (32, 33), with minor modifications. In brief, 5 ml of the culture grown overnight was inoculated into 500 ml of LB-kanamycin broth and kept at 37°C for incubation until the optical density at 600 nm reached 0.6. Protein expression was then induced by adding 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the culture was left for an additional 4 h for incubation. Thereafter, the cells were harvested by centrifugation at 8,000 rpm at 4°C and lysed in buffer containing 100 mM Tris, pH 8.0, 5 mM EDTA, 100 mM NaCl, 0.5% Triton X-100, 0.1 mM phenylmethylsulfonyl fluoride, and 0.1% NaN3. The cell lysate thus obtained was then subjected to sonication for 5 min with brief pulses of 5 s each and mixed with 2 M MgSO4 to achieve a final concentration of 10 mM. This lysate was then left for 5 min, treated with 0.1 mg/ml of lysozyme for 20 min at 37°C, and subjected to centrifugation at 8,000 rpm for 15 min at 4°C to collect the inclusion bodies. The pellet obtained was resuspended in 100 mM Tris and 50 mM glycine, sonicated for 5 min, added dropwise to the dispersion medium (8 M urea, 100 mM Tris, 50 mM glycine, 5 mM l-glutathione reduced, and 0.5 mM l-glutathione oxidized), and left overnight at 4°C under stirring conditions. On the next day, protein solution was loaded onto an Ni-nitrilotriacetic acid (NTA) column (Qiagen, Hilden, Germany) preequilibrated with binding buffer (100 mM NaH2PO4, 10 mM Tris, pH 8.0, containing 8.0 M urea), rotated for 15 min at room temperature, and then washed with binding buffer containing 40 mM imidazole. The bound protein was eluted with 200 mM imidazole made in the same buffer, concentrated, and refolded by a rapid dilution method (34). Finally, purified proteins were subjected to SDS-PAGE and circular dichroism (CD) analysis (21, 35).
Factor I cofactor activity assay.
The factor I cofactor activity of sCCPH and its mutants for C3b and C4b was assessed by using the previously standardized fluid-phase assay (24). For the C3b cofactor assay, 500 ng of sCCPH or its mutants was mixed with 3 μg of C3b and 10 ng of factor I in a total volume of 15 μl PBS, pH 7.4. The mixture was then incubated at 37°C for various time intervals and subjected to SDS-PAGE under reducing conditions. The C3b cleavages that occurred were visualized by staining the gel with Coomassie blue. For quantitation of C3b cleavage, gels were scanned and the percentage of the α′ chain that remained was quantitated by using a VersaDoc XRS system (Bio-Rad, Segrate, Italy). Data were normalized by considering 100% α′ chain to be equal to the α′ chain intensity obtained in the absence of factor I (control). C4b cofactor assays were performed in an essentially similar manner, with the difference that 3 μg of C4b instead of C3b was used and the factor I concentration used was 50 ng. Activity differences of ≥3-fold were considered significant (33, 37).
Decay-accelerating activity assay.
The CP and AP decay-accelerating activities of sCCPH and its mutants were determined by forming the respective convertases on the erythrocytes and measuring their decay (26, 38). The classical pathway C3 convertase C4b,2a was formed as described below. One hundred microliters of EAs (4 × 109/ml) was pelleted by centrifugation and resuspended in 30 μl of DGVB++, and the mixture was incubated with 5 μl of C1 (0.2 mg/ml) at 30°C for 20 min (mixed every 3 min). The coated cells were then washed with 200 μl of ice-cold DGVB++, resuspended in 60 μl of freshly diluted C4 (50 μg/ml), and incubated for 20 min at 30°C to coat the cells with C4b. These cells were then further incubated with 2 μl of C2 (0.5 mg/ml) for 4 min at 30°C to form the CP C3 convertase. The convertase formation was stopped by addition of 10 mM EDTA. To measure the decay activities of sCCPH and the mutants, 10 μl of the CP C3 convertase-coated cells (1 × 109/ml) was mixed with the indicated concentration of sCCPH or its mutants in a total volume of 25 μl and the mixture was incubated for 5 min at 22°C. This reaction mixture was then mixed with 125 μl of DGVB++ and 100 μl of guinea pig serum diluted 1:100 in DGVB++ containing 10 mM EDTA (a source of C3 to C9) and incubated at 37°C for 30 min. The reaction mixtures were then centrifuged, and the percent lysis was determined by measuring the absorbance at 405 nm. Each inhibitor was tested at three different concentrations, and the data obtained were normalized by setting 100% C3 convertase activity equal to the lysis that occurred in the absence of an inhibitor. The average activity (mean of three independent experiments) was then plotted against the concentration to determine the inhibitor concentration required to inhibit 50% of enzyme activity (IC50s). Activity differences of ≥3-fold were considered significant (33, 36, 37).
To measure the alternative pathway C3 convertase decay, the erythrocytes were first coated with C3b and then these cells were utilized to form the AP C3 convertase and measure decay. Rabbit erythrocytes coated with C3b were generated as described below. One hundred fifty microliters of rabbit erythrocytes (1 × 109/ml) was centrifuged, and the packed cells were mixed and incubated for 30 min at 30°C with 50 μl C3 (3 mg/ml), 45 μl factor B (1 mg/ml), 8.4 μl factor D (0.1 mg/ml), and 8.4 μl NiCl2 (50 mM). These cells were then washed twice with 200 μl ice-cold GVB; pelleted down; mixed with 11.2 μl factor B (1 mg/ml), 2 μl factor D (0.1 mg/ml), 2 μl NiCl2 (50 mM), and 13.2 μl GVB; and incubated for 5 min at 30°C. Thereafter, the cells were mixed with 10 mM EDTA and 1 μl C3 (3 mg/ml) and incubated for 30 min at 30°C. The cells obtained as described above were washed twice with ice-cold GVB and resuspended in 100 μl GVB.
To generate C3b,Bb on rabbit erythrocytes, 100 μl of C3b-coated rabbit erythrocytes was mixed with 28 μg factor B and 0.51 μg factor D in a total volume of 145 μl GVB containing 5 mM NiCl2 and the mixture was incubated for 5 min at 30°C. The enzyme formation was then stopped by adding 5 μl of 300 mM EDTA. To measure the decay activities of sCCPH and the mutants, 7 μl of the enzyme-coated cells (1 × 109/ml) was mixed with the indicated concentration of sCCPH or its mutants in a total volume of 25 μl, and the mixture was incubated for 10 min at 37°C. The reaction mixture was then mixed with 25 μl of human serum diluted 1:5 in GVB containing 20 mM EDTA (GVBE), and the mixture was incubated at 37°C for 20 min. After incubation, the reaction mixtures were mixed with 100 μl GVBE and centrifuged, and the percent lysis was determined by measuring the absorbance at 405 nm. Each inhibitor was tested at three different concentrations, and the IC50s were determined as described above for the CP C3 convertase decay-accelerating activity. Activity differences of ≥3-fold were considered significant (33, 36, 37).
SPR measurements.
Binding of sCCPH and its mutants to C3b and C4b was analyzed on a surface plasmon resonance (SPR)-based Biacore 2000 biosensor (Biacore AB, Uppsala, Sweden) as described previously (24). Two separate chips were used for analysis of C3b and C4b binding. In brief, C4b (∼1,900 response units [RUs]) and C3b (∼4,000 RUs) biotinylated at their free —SH group were immobilized onto flow cell 2 of streptavidin chips (Sensor Chip SA; Biacore AB), and flow cell 1, which was immobilized with bovine serum albumin-biotin (Sigma), served as a control. Binding of all the proteins was performed at 25°C in PBS-T at a 50-μl/min flow rate. The association phase was measured by injecting the analyte for 120 s, and the dissociation phase was measured by replacing the analyte with the buffer for 180 s. Sensograms were normalized by subtracting the nonspecific signal measured using flow cell 1. The sensor chip was regenerated between the runs by injecting brief pulses of 0.2 M sodium carbonate, pH 9.5.
Complement-mediated hemolytic assays.
The inhibitory effect of graded concentrations of sCCPH and its mutants on activation of human, monkey (rhesus), and rat classical and alternative complement pathways was measured by using hemolytic assays as previously described (28). The data obtained were fitted using nonlinear regression analysis (GraFit; Erithacus Software, London, United Kingdom), and a four-parameter analysis was performed to identify the best-fit IC50s (inhibitor concentrations required to inhibit 50% hemolytic activity).
RESULTS
Design, expression, and characterization of sCCPH substitution mutants.
Identification of functional sites in sCCPH required knowledge of the three-dimensional structure of the molecule at high resolution. Since such a structure of sCCPH is not available, we generated a homology model utilizing the experimentally determined structure of Crry. The generated model showed the least deviation from the template (deviations were mainly found in the SCR3 module between residues S172 and S176, W177 and K184, and E158 and K160). We then utilized the modeled structure to identify the putative protein-protein interaction sites in sCCPH on the basis of previous knowledge of functionally important amino acids in human and other viral RCA proteins and their solvent accessibility in the sCCPH structure (Fig. 1). The mutations selected by us did not belong to the region that showed deviations.
Fig 1.
Model of sCCPH and gel analysis of purified sCCPH and its mutants. (A) Overlap of ribbon models of sCCPH (red) and Crry (green) structures showing side chains of mutated residues (blue). (B) Corey-Pauling-Koltun representation of sCCPH model showing positions of Ala substitution mutations in red and predicted gain-of-function mutations in yellow. Amino acid S100, labeled in orange, was mutated to Ala as well as Glu and Lys. (C) SDS-PAGE analysis of purified proteins. Wild-type and mutant proteins were subjected to electrophoresis on 11.5% SDS-polyacrylamide gels under reducing conditions and stained with Coomassie blue. The molecular weights (MW) of purified proteins (indicated on the left, in thousands) ranged from 26,700 to 27,400.
Overall, we identified 21 amino acids likely to be involved in sCCPH function. These residues belonged to SCR modules 1 to 4 as well as the linkers between SCRs 1 and 2 and SCRs 2 and 3. We mutated these residues either to Ala (for loss-of-function analysis) or to the residue of interest (for gain-of-function analysis) to generate 24 substitution mutants. All the generated mutants were then expressed in E. coli using the pET expression system, purified over an Ni-NTA matrix, and refolded using the rapid dilution method (34). As evident from SDS-PAGE analysis, all the purified mutants were >95% pure (Fig. 1). In addition, they also yielded a characteristic peak at 230 nm (21, 35) when analyzed by CD, suggesting that they maintained the proper conformation.
Amino acids R118 and F144 play a critical role in factor I cofactor activity of sCCPH.
In our earlier study on characterization of the cofactor activity of sCCPH employing various truncation mutants, we observed that domains 1 and 2 play a critical role in imparting the cofactor activities and domains 3 and 4 contribute to its optimal activity (23). We therefore selected residues for mutation that belonged to all four domains of sCCPH. In addition, we also selected residues that are located in the linker regions between SCRs 1 and 2 and SCRs 2 and 3, as charged residues in these linkers have been shown to be crucial in mediating the cofactor activities in other RCA proteins (32, 36, 39, 40). Altogether, eight residues (R35, K40, N92, S100, R118, K128, F144, and K191) that are located in the different domains of sCCPH and three residues (K61, K62, and K126) that are part of the linkers were chosen for mutation to Ala (Fig. 1). The mutants were then examined for C3b and C4b cofactor activities using a fluid-phase assay wherein C3b or C4b was incubated with sCCPH or the mutant of interest and factor I and inactivation of C3b/C4b was assessed by measuring the cleavage of their α′ chains.
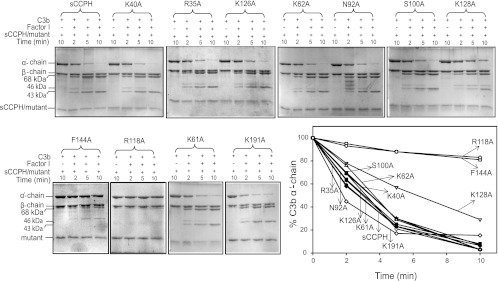
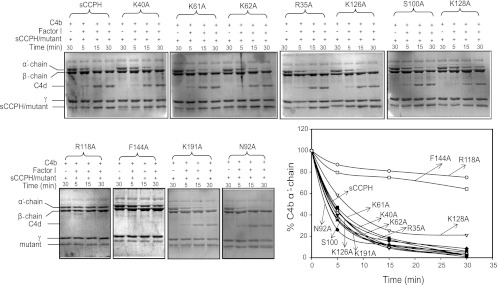
Of the 11 mutants described above, only R118A and F144A showed a significant decrease in the C3b cofactor activity compared to the wild-type protein (Fig. 2 and Table 1). It is noteworthy that removal of a positive charge at position 35 (implicated in factor H CFA [41]) and the linker between SCRs 1 and 2 and SCRs 2 and 3, which was expected to have a large effect on the cofactor activities, showed no effect. Next, the above-described mutants were also evaluated for their cofactor activity against C4b. The results were essentially similar (Fig. 3 and Table 1).
Fig 2.
Factor I cofactor activity of sCCPH and its Ala substitution mutants for complement protein C3b. For assessing C3b CFA, sCCPH or each of the mutants was incubated with C3b in the presence or absence of factor I in PBS, pH 7.4, at 37°C. The reactions were stopped at the indicated times by addition of sample buffer containing dithiothreitol, and the cleavage products were visualized by subjecting the reaction mixtures to 10% SDS-PAGE under reducing conditions, followed by staining with Coomassie blue. Cleavage of the α′ chain of C3b by factor I results in the generation of N-terminal 68-kDa and C-terminal 46-kDa fragments, indicating inactivation of C3b. The C-terminal fragment is subsequently cleaved into a 43-kDa fragment. The amount of the α′ chain that remained after various time intervals was quantitated by densitometric analysis and is represented graphically against time (lower right).
Table 1.
Summary of complement regulatory activities of sCCPH and its mutants
| sCCPH | Mutation location | C3b CFA |
C4b CFA |
AP DAA |
CP DAA |
||||
|---|---|---|---|---|---|---|---|---|---|
| Time (min) for 50% cleavage of C3b α′ chain | Relative % of C3b CFAa | Time (min) for 50% cleavage of C4b α′ chain | Relative % of C4b CFAa | DAA IC50 (μM) | Relative % of AP DAAa | DAA IC50 (μM) | Relative % of CP DAAa | ||
| Wild type | 2.8 | 100 | 4.5 | 100 | 0.55 | 100 | 0.13 | 100 | |
| R35Ab | SCR1 | 2.7 | 104 | 3.8 | 118 | 2.85 | 19 | 0.73 | 18 |
| K40A | SCR1 | 3.2 | 88 | 4.3 | 105 | 0.35 | 157 | 0.22 | 59 |
| K61A | SCR1-SCR2 linker | 3.1 | 90 | 4.8 | 94 | 0.29 | 190 | 0.1 | 130 |
| K62A | SCR1-SCR2 linker | 3.6 | 78 | 4.5 | 100 | 0.47 | 117 | 0.22 | 59 |
| Y83A | SCR2 | 2.9 | 97 | 4.6 | 97 | 0.49 | 112 | 0.29 | 45 |
| N92A | SCR2 | 3.0 | 93 | 3.4 | 132 | 0.49 | 112 | 0.25 | 52 |
| S100A | SCR2 | 3.5 | 80 | 4.0 | 113 | 0.36 | 153 | 0.3 | 43 |
| L106A | SCR2 | 1.7 | 165 | 3.4 | 132 | 0.84 | 66 | 0.17 | 77 |
| V113A | SCR2 | 2.7 | 104 | 3.7 | 122 | 1.3 | 42 | 0.24 | 54 |
| R118A | SCR2 | 49 | 6 | 270 | 2 | 1.45 | 38 | 0.29 | 45 |
| K126A | SCR2-SCR3 linker | 3.6 | 78 | 4.0 | 113 | 1.5 | 37 | 0.1 | 130 |
| K128A | SCR3 | 6.3 | 44 | 7.5 | 60 | 0.09 | 611 | 0.15 | 87 |
| K142A | SCR3 | 1.7 | 165 | 4.3 | 105 | 2.45 | 22 | 0.51 | 26 |
| F144A | SCR3 | 47 | 6 | 120 | 4 | 0.08 | 688 | 0.23 | 57 |
| N156A | SCR3 | 4.2 | 68 | 4.8 | 94 | 1.55 | 35.5 | 0.34 | 38 |
| K191A | SCR4 | 1.8 | 156 | 3.8 | 118 | 1.75 | 31 | 0.24 | 54 |
| S96Y | SCR2 | 4.0 | 70 | 11.5 | 39 | 1.51 | 36 | 0.17 | 77 |
| S100E | SCR2 | 16.5 | 17 | 58.2 | 8 | >5c | <11 | 0.82 | 16 |
| S100K | SCR2 | 3.6 | 78 | 4.1 | 110 | 0.09 | 611 | 0.005 | 2,600 |
| T101Y | SCR2 | 3.7 | 76 | 6.6 | 68 | 1.85 | 30 | 0.037 | 351 |
| A104F | SCR2 | 3.7 | 76 | 7.9 | 57 | 2.35 | 23 | 0.08 | 163 |
| A104Y | SCR2 | 92 | 3 | 60.3 | 8 | >30c | <2 | 0.19 | 68 |
| G110D | SCR2 | 3.9 | 72 | 4.6 | 98 | 0.85 | 65 | 0.006 | 2,167 |
| Y225K | SCR4 | 73 | 4 | 69 | 7 | >5c | <11 | 0.1 | 130 |
Relative activity compared to sCCPH: >33%, no effect; 17 to 33%, moderate reduction; 5 to 16%, considerable reduction; <5%, abolished activity; >300%, significant increase in activity.
Boldface indicates the mutants and data with a >3-fold difference in activity, which was considered significant.
The highest concentration of mutant tested in the assay.
Fig 3.
Factor I cofactor activity of sCCPH and its Ala substitution mutants for complement protein C4b. For assessing C4b CFA, sCCPH or each of the mutants was incubated with C4b in the presence or absence of factor I in PBS, pH 7.4, at 37°C. The reactions were stopped at the indicated times by addition of sample buffer containing dithiothreitol, and the cleavage products were visualized by subjecting the reaction mixtures to 10% SDS-PAGE under reducing conditions, followed by staining with Coomassie blue. Cleavage of the α′ chain of C4b by factor I results in the generation of N-terminal 27-kDa and C-terminal 16-kDa (not visible on the gel) and central C4d fragments, indicating inactivation of C4b. The amount of the α′ chain that remained after various time intervals was quantitated by densitometric analysis and is represented graphically against time (lower right).
The cofactor activity is a result of the interaction between the regulator, target protein (C3b or C4b), and factor I (42). It is therefore conceivable that the decrease in cofactor activity owing to mutation could be a result of a decrease in binding to the target protein or factor I. We therefore measured binding of the above-described mutants to C3b and C4b tethered onto the SPR sensor chips in their physiological orientation (Fig. 4). Mutants R118A and F144A, which showed considerable decreases in C3b cofactor activity, did not show any decrease in C3b binding. Similarly, there was only a moderate effect on the binding of these mutants to C4b. It is therefore likely that loss of the cofactor activity of these mutants is primarily due to a decrease in their interaction with factor I.
Fig 4.
Binding of sCCPH and its Ala substitution mutants to human complement proteins C3b and C4b. Sensogram overlays for the interaction of sCCPH and its mutants with C3b (left) and C4b (right) are shown. C3b or C4b biotinylated through its free —SH group was oriented on a streptavidin sensor chip, and 50 nM sCCPH or its mutants (for C3b) or 500 nM sCCPH or its mutants (for C4b) was flown over it to measure binding.
Among the other mutants that did not show any significant change in CFAs, mutants R35A, N92A, K191A, and K126A demonstrated moderate to considerable decreases in C3b binding and mutants R35A, K40A, N92A, S100A, K191, K62A, and K126A showed moderate to considerable decreases in C4b binding (Fig. 4). This, however, was not surprising, as there is a large body of data which indicates that target binding does not always correlate with cofactor activity (26, 43, 44). A likely explanation for this is that factor I, which docks onto the C3b/C4b-sCCPH complex, stabilizes the complex by interacting with both partners (41, 42).
Positively charged residues R35, K142, and K191 are vital for DAA of sCCPH.
Previous mapping of domains required for DAAs in sCCPH using truncated mutants indicated that SCRs 1 to 3 are primarily responsible for driving CP DAA, whereas all four SCRs are necessary for driving the AP DAA (23). Here, for mapping residues critical for DAAs, we selected residues from SCRs 1 to 3 as well as linkers between SCRs 1 and 2 and SCRs 2 and 3, which were shown to be vital for DAAs in other RCA proteins (32, 36, 39, 40). The mutants generated were R35A, K40A, K61A, K62A, Y83A, L106A, V113A, K126A, K142A, and N156A (Fig. 1). These were then examined for their ability to decay the preformed classical/lectin (C4b,2a) as well as alternative (C3b,Bb) pathway C3 convertases made on sheep and rabbit erythrocytes, respectively.
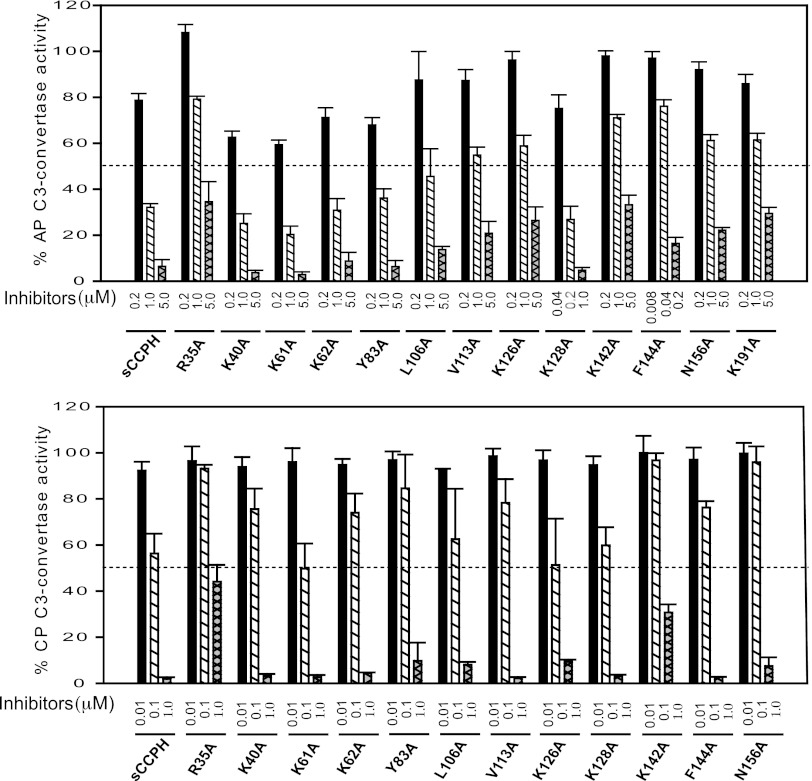
Among the 10 mutants described above that were expected to show effects on DAAs, mutants R35A and K142A displayed a loss of AP DAA as well as CP DAA (Fig. 5 and Table 1). In addition, mutant K191A, which was generated to determine the effect of this residue on CFAs, also showed a loss of AP but not CP DAA (Table 1). Interestingly, similar to the effect on CFAs, linker mutants that were expected to demonstrate a loss of DAAs did not show any effect on the AP or CP DAAs (Fig. 5 and Table 1).
Fig 5.
Decay-accelerating activity of sCCPH and its Ala substitution mutants for classical and alternative pathway C3 convertases. (Top) AP DAA of sCCPH and its indicated mutants was measured by assessing the decay of AP C3 convertase formed on rabbit erythrocytes; (bottom) CP DAA of sCCPH and its indicated mutants was measured by assessing the decay of CP C3 convertase formed on sheep erythrocytes. Data obtained were normalized by considering 100% C3 convertase activity to be equal to the average activity in the absence of inhibitor.
Like cofactor activity, DAA is also a result of trimolecular interaction. Herein, interaction of the regulator with both subunits of the C3 convertases (C3b and Bb or C4b and C2a) results in dissociation of the protease subunit (Bb or C2a) from the bimolecular enzyme (45, 46). To understand whether decreases in the DAAs of the R35A, K142A, and K191 mutants are owing to decreases in their binding to C3b or C4b, we looked at their direct binding to these molecules (Fig. 4). Mutant R35A showed a moderate decrease in C3b binding, whereas the other two mutants displayed substantial decreases in C3b binding. Further, mutants R35A and K142A also displayed substantial decreases in C4b binding. In sum, mutants that showed decreased DAAs also showed decreases in their abilities to bind to the target proteins, thereby suggesting that the loss of DAAs is likely due to decreases in target recognition.
There were, however, mutants which did not show any change in DAAs but showed decreased binding to C3b (e.g., K126A, N156A, and mutants generated for studying CFA) and C4b (K40A, K62A, L106A, K126A, N156A, and mutants generated for studying CFA) (Fig. 4). It is likely that these mutants possess a higher affinity for the convertases than their noncatalytic subunits (C3b or C4b), as has been seen in the case of decay-accelerating factor (DAF) (47). It is intriguing to note that two mutants (K128A and F144A) generated to investigate the role of these residues in cofactor activity showed substantial gains in AP DAA (Fig. 5 and Table 1). Admittedly, the precise reason for this is not clear.
Functional characterization of predicted gain-of-function mutants.
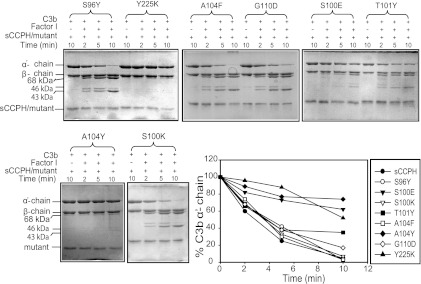
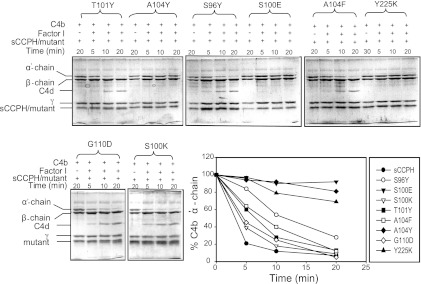
We designed eight gain-of-function mutants to determine whether mutations based on gain of function in other RCA proteins would result in enhancement in sCCPH function (Fig. 1). We mutated S96 to Y, as Y at the comparable position in SPICE (smallpox inhibitor of complement enzymes) is one of the four residues responsible for its 100-fold better cofactor activity compared to that of vaccinia virus complement control protein (VCP) (33) and mutation of Y to A at this position in membrane cofactor protein (MCP) results in loss of its C4b cofactor activity (48). This mutation, however, did not result in any enhancement in the cofactor activities of sCCPH, as expected (Fig. 6 and 7); no effect on CP and AP DAAs was observed either (Fig. 8). A similar T101-to-Y mutation based on SPICE was also performed, as Y present at the comparable position in SPICE is another residue responsible for its robust activity (33), but this change also did not affect the cofactor activities (Fig. 6 and 7); there was a moderate decrease in AP DAA and an increase in CP DAA (Fig. 8).
Fig 6.
Factor I cofactor activity of sCCPH and its predicted gain-of-function mutants for complement protein C3b. The activity was assessed by measuring the cleavage of the α′ chain of C3b. The amount of the α′ chain that remained after various time intervals was quantitated by densitometry and plotted against time (bottom right).
Fig 7.
Factor I cofactor activity of sCCPH and its predicted gain-of-function mutants for complement protein C4b. The activity was assessed by measuring the cleavage of the α′ chain of C4b. The amount of the α′ chain that remained after various time intervals was quantitated by densitometry and plotted against time (bottom right).
Fig 8.
Decay-accelerating activity of sCCPH and its predicted gain-of-function mutants for classical and alternative pathway C3 convertases. (Top) AP DAA of sCCPH and its indicated mutants; (bottom) CP DAA of sCCPH and its indicated mutants. Data obtained were normalized by considering 100% C3 convertase activity to be equal to the average activity in the absence of inhibitor.
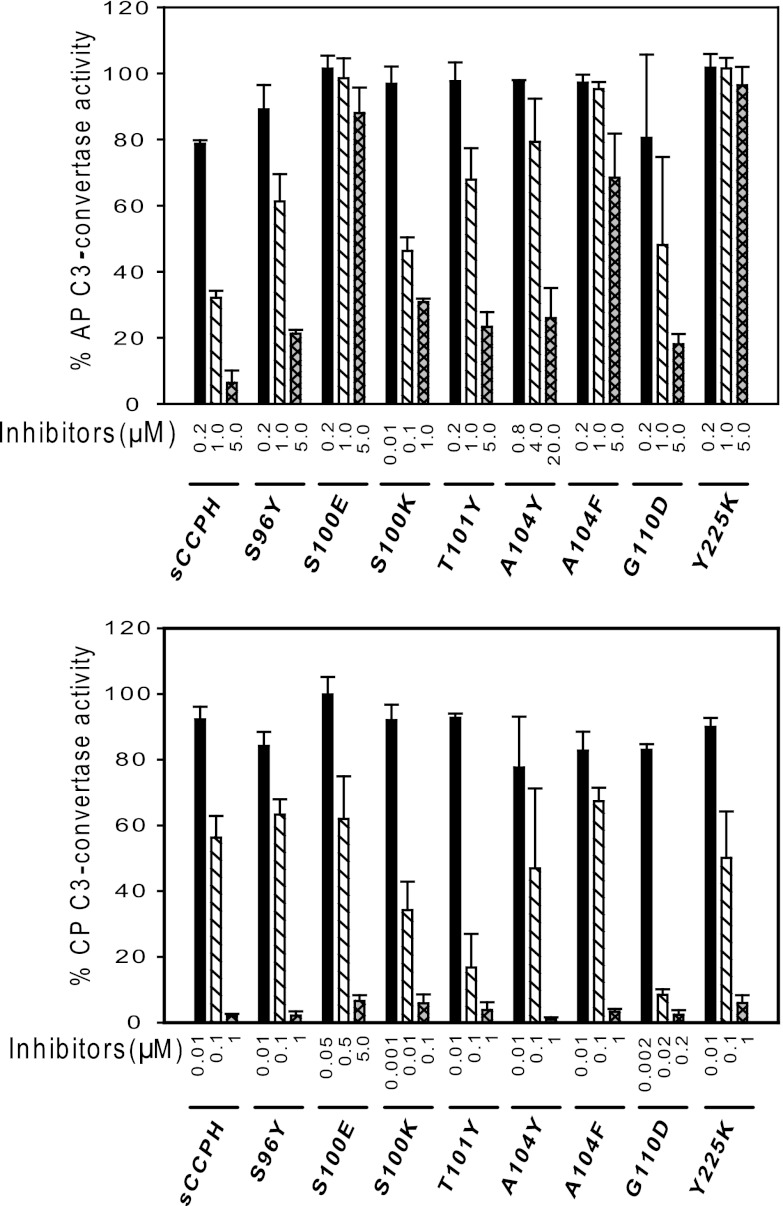
Mutation of S100 to E was based on MCP, as E at the equivalent position has been shown to be important for its cofactor activities (48). This mutation, however, resulted in moderate to substantial losses in the cofactor activities (Fig. 6 and 7 and Table 1) as well as AP and CP DAAs (Fig. 8). We thus next changed S100 to K, with the rationale that if abrogation of the activity is due to unfavorable electrostatic interactions, then substitution of a positively charged residue at this position might lead to a gain in activity. Moreover, a positively charged residue is present at this position in kaposica (Kaposi's sarcoma-associated herpesvirus inhibitor of complement activation). Though the S-to-K change did not affect the cofactor activities, it resulted in robust 6- and 26-fold increases in AP and CP DAAs, respectively (Fig. 8).
The basis of mutation of A104 to Y and F is as follows. Sequence alignment of RCA proteins showed that Y is highly conserved at the equivalent position in various human and viral complement regulators. Moreover, mutation of F to A at this position in DAF resulted in a significant decrease in AP and CP DAAs (36). These mutations in sCCPH, however, did not result in any increase in DAAs, as expected, and, if any change, there was a loss of CFAs and AP DAA (Fig. 6 to 8 and Table 1). Amino acid G110 in sCCPH corresponds to D109 in CR1, which seems to be important for its activity (49). Interestingly, D is also conserved at this position in factor H and the ateline herpesvirus RCA. We therefore mutated G110 to D. Though this mutation did not show any change in the cofactor activities and AP DAA, there was a 22-fold increase in CP DAA (Fig. 8).
Lastly, we mutated Tyr at position 225 to K, as a positively charged residue is conserved at this position in human and viral RCAs (e.g., CR1, C4bp, and kaposica). This mutation resulted in a complete loss of cofactor activities and AP DAA, with no effect on CP DAA (Fig. 6 to 8 and Table 1). In short, mutations predicted to result in a gain of function did not always demonstrate an increase in activity.
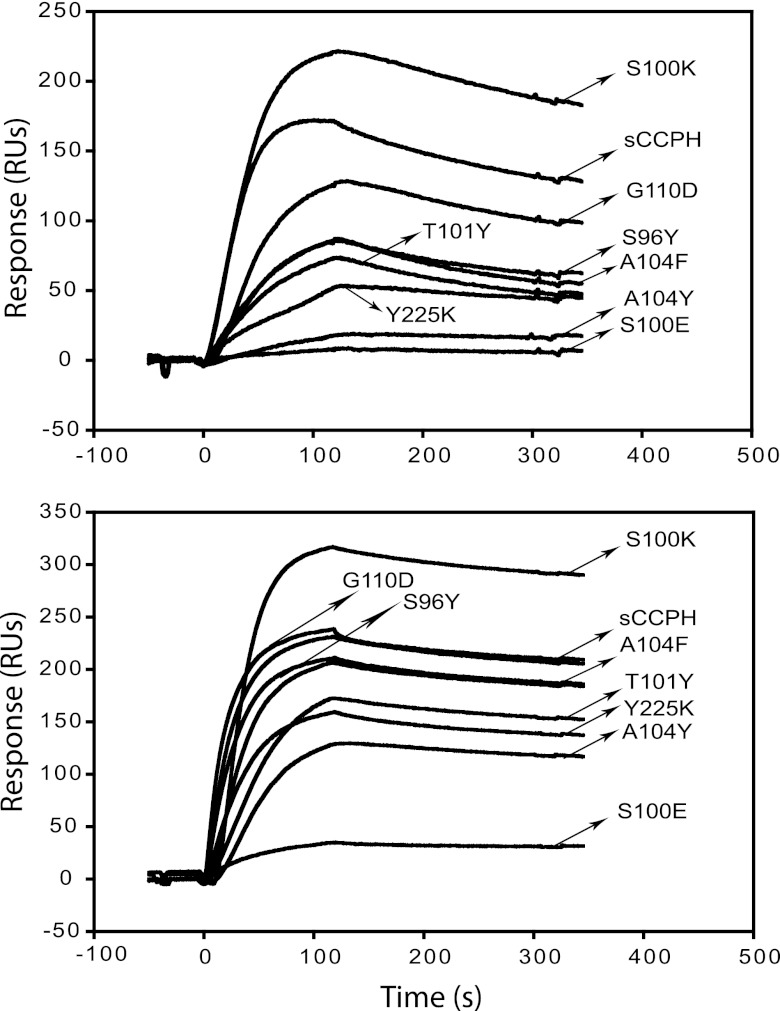
When we tried to correlate activities of the above-described mutants with their binding to C3b and C4b (Fig. 9), we observed that such a correlation was apparent only in the case of the S100K and S100E mutants: the S100K mutant, which exhibited 6- and 26-fold increases in AP and CP DAAs, respectively, showed an increase in binding to C3b and C4b, while the S100E mutant, which showed an overall loss of CFAs and DAAs, showed attenuated binding to the target proteins.
Fig 9.
Binding of sCCPH and its predicted gain-of-function mutants to human complement proteins C3b and C4b. Sensogram overlays for the interaction of sCCPH and its predicted gain-of-function mutants with C3b (top) and C4b (bottom) are shown.
Inhibition of human, monkey, and rat complement activity by sCCPH and its mutants.
In all the assays described above, we utilized human complement proteins to assess the activities. Because quite a few of the mutations gave unexpected results, we next asked if these results could be explained by assessing their effect on monkey complement, as herpesvirus saimiri is a monkey pathogen, or rat complement, as the sCCPH homology model that we have generated is based on rat Crry. Thus, we next compared the inhibitory activity of sCCPH and its selected mutants (which showed a loss or a gain in activity) against alternative and classical pathways of human, rhesus monkey, and rat complement using hemolytic assays. Though rhesus monkey is not a natural host of the virus, its complement components C3 and C4 (target proteins of sCCPH) are ≥87% identical to C3 and C4 of squirrel monkey, suggesting that the effect of sCCPH and the mutants on rhesus monkey complement that we observed here would be comparable to their effect on squirrel monkey complement.
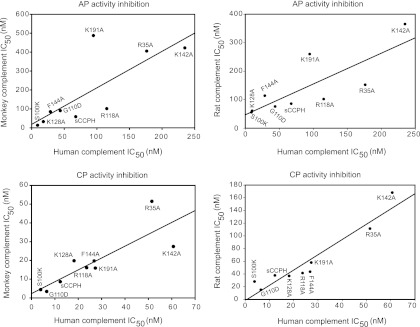
As depicted in Fig. 10, sCCPH showed comparable inhibitory activity against classical and alternative pathways of human and monkey complement, and there was a significant positive correlation between inhibition of human and monkey complement activity by sCCPH and its mutants (for AP, r = 0.78 and P = 0.013; for CP, r = 0.84 and P = 0.005). Interestingly, mutants that showed gains in DAAs showed increased inhibition of AP- and CP-mediated hemolytic activity and vice versa, but mutants that displayed decreased CFAs did not show appreciable decreases in inhibition of the hemolytic activity. Further, a similar trend of inhibitory activity against rat complement was also observed, and the positive correlation between inhibition of human and rat complement activity was apparent (for AP, r = 0.80 and P = 0.009; for CP, r = 0.95 and P < 0.0001). Together, these results suggest that the observed lack of expected phenotypes in certain CCPH mutants is likely owing to differences in the mechanism of complement inactivation between sCCPH and other RCA proteins and not due to examination of functional activities against human complement or the low fidelity of the structural model.
Fig 10.
Inhibition of human, monkey, and rat complement by sCCPH and its mutants. Inhibitory activities of sCCPH and its mutants against alternative and classical pathways of human, rhesus monkey, and rat complement were measured by employing hemolytic assays using rabbit erythrocytes and antibody-coated sheep erythrocytes as complement activators, respectively. (Upper left) Correlation between IC50s of sCCPH and its mutants for human and monkey alternative complement pathway (AP) (r = 0.78; P = 0.013); (upper right) correlation between IC50s of sCCPH and its mutants for human and rat alternative complement pathway (r = 0.80; P = 0.009); (lower left) correlation between IC50s of sCCPH and its mutants for human and monkey classical complement pathway (CP) (r = 0.84; P = 0.005); (lower right) correlation between IC50s of sCCPH and its mutants for human and rat classical complement pathway (r = 0.95; P < 0.0001). The correlation coefficient (r) was calculated using the Pearson product moment correlation.
DISCUSSION
It is now evident that viruses encode RCA homologs (8, 11) as well as non-RCA proteins (e.g., herpes simplex virus gC and herpesvirus saimiri CD59) (50, 51) to subvert the host complement attack. Among these, the viral RCA homologs, which are structurally as well as functionally similar to human RCA proteins, regulate complement by employing common mechanisms: irreversible decay of C3 convertases and proteolytic inactivation of C3b and C4b (21, 40, 52–57). This raises the following question: are functional sites similar and spatially conserved in viral and human RCA proteins?
Mutational characterization of human complement regulators (MCP, DAF, CR1, factor H, and C4BP) has led to the identification of many residues critical for imparting function in these proteins (36, 48, 49, 58). Likewise, mutational analyses in poxviral complement regulators SPICE (33, 59) and VCP (37) and Kaposi's sarcoma-associated herpesvirus complement regulator kaposica (32, 40) have also led to the identification of a few residues crucial for their function. Here, we have generated 24 sCCPH mutants on the basis of these human and viral RCA functional site residues to determine whether conserved functional site residues in sCCPH are also critical for its function and whether incorporation of functional site residues in sCCPH leads to gain of function. The study, however, did not look into the role of glycosylation in sCCPH, which may potentially affect its activity, as seen in the case of human complement inhibitor MCP (60).
Residues critical for CFA of sCCPH.
Our mutational dissection of functionally important residues in sCCPH in the present study resulted in identification of R118 and F144 as residues critical for imparting cofactor activities (Fig. 2 and 3 and Table 1); R118 was also identified to be critical in a previous study (21). Interestingly, we observed that binding of R118A and F144A mutants to C3b and C4b does not correlate with the CFAs of these mutants. Because CFA is a result of interaction of the regulator with C3b/C4b and factor I, we suggest that the decrease in CFAs of these mutants is due to decreases in their binding to factor I. This contention is also consistent with the cocrystal structure of C3b-factor H, which shows that residues of factor H equivalent to R118 and F144 are completely exposed to the solvent and are present on the putative factor I interaction face of factor H (41). Earlier, during mapping of functional domains in various viral complement regulators, we have identified SCRs 2 and 3 to be factor I-interacting domains (23, 26, 61). Notably, residues R118 and F144 are located in SCRs 2 and 3, respectively, and residues similar to these are also conserved in position in other viral complement regulators (see below). We therefore propose that these residues are two of the major spatially conserved determinants in viral RCAs crucial for their CFAs.
Unlike the present study, in our earlier study on kaposica (32), we observed a significant correlation between the binding response to C3b/C4b and CFAs. The difference between the sCCPH and kaposica mutants is that the mutations in the sCCPH mutants designed here were based on substitution of conserved functional site residues, whereas the kaposica mutants were designed with mutations to alter their electrostatic potential. Because electrostatic potential influenced binding of kaposica to C3b/C4b (32), we suggested that long-range electrostatic interactions drive the docking of the complement regulator onto C3b/C4b and this dictates its functional activities. It is, however, clear from the sCCPH data presented here that binding of a complement regulator to C3b/C4b does not always dictate its CFAs. Thus, the better correlation between C3b/C4b binding and CFAs in our earlier study could have been due to the influence of the electrostatic potential of kaposica on binding to C3b/C4b as well as factor I. However, whether long-range electrostatic interactions drive the docking of factor I onto the C3b/C4b-complement regulator complex is not clear and requires further investigation.
Sequence alignment of human and viral RCA proteins shows that charged amino acids are conserved at a position equivalent to R118 in human RCA proteins (CR1 site 2, K566; CR1 site 3, K1016; MCP, K119) as well as other viral RCA proteins (SPICE, K120; RCP-1, K124; ateline herpesvirus RCA, R117). It is pertinent to point out here that in CR1, mutation of K to E at the equivalent position in site 1 resulted in a decrease in CFA against human C3b and C4b, while mutation of E to K at the corresponding position in site 3 led to an increase in CFA (62). Intriguingly, charge reversal at this position is also observed in poxviral complement regulators SPICE and VCP; the former contains K, while the later contains E. Since SPICE and VCP function in a species-specific manner, we very recently looked into the functional relevance of this change. Our data revealed that charge reversal dictates the species-specific cofactor activity in SPICE and VCP: the presence of K in SPICE enhances its interaction with human factor I, while the presence of E in VCP enhances its interaction with bovine factor I (37). It therefore appears that the presence of a charged amino acid at a position colinear to R118 is critical for factor I interaction in human as well as viral complement regulators and the nature of the charge dictates their species specificity.
When we looked for conservation of F/Y at the position equivalent to F144 of sCCPH in other RCA proteins, we found that it is conserved in only one human regulator (C4BP). Among the viral complement regulators, however, it is highly conserved in various herpesviral as well as poxviral complement regulators (SPICE, F146; VCP, F146; MOPICE, F144; RCP-1, F150; RCP-H, F150; kaposica, Y150; ateline herpesvirus RCA, Y142). Among these examples, mutational analysis has been performed only in the case of C4BP, wherein substitution of F to S resulted in a substantial decrease in C3b and C4b CFAs (39). These data, along with our results showing a loss of CFAs in sCCPH as a result of mutation of F144 (Fig. 2 and 3, and Table 1), clearly suggest that F/Y at this position plays a paramount role in CFAs of herpesviral as well as poxviral complement regulators.
Although from the discussion presented above it seems that spatially conserved functional sites govern the activities in viral RCAs, this imperative is not always true. For example, earlier we have shown that Y98 and Y103 are among the four residues responsible for the 100-fold more potent C3b CFA of SPICE than that of VCP (33). Moreover, Y/F at the corresponding positions is highly conserved in various strains of variola virus and kaposica. When we substituted Y at the equivalent positions in sCCPH, however, we did not see any gain in activities (Fig. 6 and 7 and Table 1). Similarly, mutation of positively charged linker residues that are known to be important for CFAs in other viral RCAs (32, 36, 39, 40) does not seem to be important in sCCPH for its CFAs (Fig. 2, 3, and 5 and Table 1).
Residues critical for DAA of sCCPH.
In the present study, examination of DAA of Ala substitution mutants identified residues R35 and K142 to be critical for both AP and CP DAAs and K191 to be critical for AP DAA (Fig. 5 and Table 1). We believe that these are among the conserved major determinants in viral RCAs crucial for their DAAs.
The conservation of a positively charged residue at the position equivalent to R35 of sCCPH is observed in human complement regulators (CR1 site 1, R33; CR1 site 2, R483; CR1 site 3, R933; MCP, K31; DAF, R96; factor H, R35) and herpesviral complement regulators (kaposica, R35; RCP-1, R32; RCP-H, R32; gammaherpesvirus 68 RCA, R30; ateline herpesvirus RCA, R33) but not in poxviral complement regulators. The importance of the charge at this position has been looked into in the case of DAF, and its removal has been shown to considerably decrease the CP and AP DAAs (36). In the case of factor H, Arg35 is linked with an atypical hemolytic-uremic syndrome-related substitution (R35H) (63). Since this residue is solvent exposed in the C3b-factor H cocrystal structure and is present on the putative factor I interaction face of factor H, it was proposed that it might be involved in binding to factor I (41). Our data, along with the DAF data, show a decrease in DAAs as a result of the Ala substitution at this position. We therefore suggest that the presence of a positively charged residue at this position is vital for dissociation of the C3 convertase in human as well as herpesviral complement regulators.
Analogous to position R118, reversal of the charge at the position corresponding to K142 of sCCPH seems to be associated with species specificity in viral complement regulators. SPICE, which is more potent against human complement, contains N (a neutral residue) at the equivalent position, while VCP, which is more bovine complement specific, contains E (a negatively charged residue). Mutational studies have shown that substitution of N in VCP (E144N) enhances its activity toward human complement (33), whereas substitution of E in SPICE (N144E) enhances its activity toward bovine complement (37). In fact, E144 in VCP is one of the three glutamates responsible for its specificity toward bovine complement (37). In the present study, substitution of Ala at K142 reduced the activity of sCCPH against human complement (Fig. 5 and Table 1). It could therefore be inferred that the presence of a positively charged (or neutral) residue at this position enhances the specificity of viral regulators toward human complement, whereas the presence of a negatively charged residue enhances their specificity toward bovine complement. This argument is also supported by substitution mutagenesis in DAF (R206A), which showed that removal of the charge at the equivalent position resulted in reduction in its activity against human complement (36). The other regulators wherein charge is conserved at this position are CR1 (site 1, R141; site 2, R591; site 3, R1041), kaposica (K148), ateline herpesvirus RCA (K141), and gammaherpesvirus 68 RCA (K145).
The analysis of charge conservation at a position equivalent to K191 of sCCPH in various human and viral regulators indicates that it is conserved in human complement regulators (MCP, R195; factor H, K193; C4BP, R192) and herpesviral complement regulators (kaposica, K194; ateline herpesvirus RCA, K190) but not in poxviral complement regulators. Mutational analysis at this position has been performed only in the case of MCP (R195A), which showed a reduction in CFAs as well as binding to C3b and C4b (48). Although substitution of K191 to Ala in sCCPH resulted in a decrease in its binding to C3b and C4b, the effect was principally on AP DAA and not CFAs (Fig. 4 and Table 1). Based on our data on sCCPH, we speculate that the presence of a positively charged residue at this location in herpesviral complement regulators is likely to be involved in the DAA. Having said this, further studies are clearly needed to validate our premise.
Our efforts to enhance the activity of sCCPH led to identification of at least two mutations (S100K and G110D) which lead to robust increases in DAAs (Fig. 8). Agreeably, these mutations were based on educated guesses and not prior knowledge of gain of function. Nevertheless, we clearly show that it is possible to enhance the functional activity of sCCPH. Since very limited knowledge of gain-of-function mutagenesis in RCA proteins exists, such leads would be useful in designing therapeutically important complement inhibitors.
In summary, our study has identified functional sites in CCPH critical for its CFAs and DAAs. The study, however, employed human complement components which are homologous to monkey complement components to examine these activities (37, 57, 64). Nevertheless, to put the various mutations in better perspective, we also examined and compared the activities of various mutants with human and monkey complement using hemolytic assays. Analysis of these data depicted significant positive correlations between the inhibitory potential of mutants against human and monkey complement (Fig. 10). Thus, the functional sites identified here using human complement components are relevant to the species target of CCPH.
ACKNOWLEDGMENTS
This work was supported by the Department of Biotechnology, India (grants BT/PR7683/MED/14/1062/2006 and BT/PR15155/MED/29/248/2011 to A.S.). We also acknowledge the financial assistance to M.J.R. and A.K. by the University Grants Commission, New Delhi, and the Indian Council of Medical Research, New Delhi, respectively.
We thank Jayati Mullick (National Institute of Virology, Pune, India) for her comments and critical reading of the manuscript and express our appreciation to Yogesh Panse, Sandeep Walujkar, and Girish Kulkarni for excellent technical assistance.
Footnotes
Published ahead of print 17 October 2012
REFERENCES
- 1. Judson KA, Lubinski JM, Jiang M, Chang Y, Eisenberg RJ, Cohen GH, Friedman HM. 2003. Blocking immune evasion as a novel approach for prevention and treatment of herpes simplex virus infection. J. Virol. 77:12639–12645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lachmann PJ. 2002. Microbial subversion of the immune response. Proc. Natl. Acad. Sci. U. S. A. 99:8461–8462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mullick J, Kadam A, Sahu A. 2003. Herpes and pox viral complement control proteins: ‘the mask of self.’ Trends Immunol. 24:500–507 [DOI] [PubMed] [Google Scholar]
- 4. Carroll MC. 2004. The complement system in regulation of adaptive immunity. Nat. Immunol. 5:981–986 [DOI] [PubMed] [Google Scholar]
- 5. Dunkelberger JR, Song WC. 2010. Complement and its role in innate and adaptive immune responses. Cell Res. 20:34–50 [DOI] [PubMed] [Google Scholar]
- 6. Pyaram K, Yadav VN, Reza MJ, Sahu A. 2010. Virus-complement interactions: an assiduous struggle for dominance. Future Virol. 5:709–730 [Google Scholar]
- 7. Finlay BB, McFadden G. 2006. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124:767–782 [DOI] [PubMed] [Google Scholar]
- 8. Lambris JD, Ricklin D, Geisbrecht BV. 2008. Complement evasion by human pathogens. Nat. Rev. Microbiol. 6:132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liang CY, Lee JS, Jung JU. 2008. Immune evasion in Kaposi's sarcoma-associated herpes virus associated oncogenesis. Semin. Cancer Biol. 18:423–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stoiber H, Pruenster M, Ammann CG, Dierich MP. 2005. Complement-opsonized HIV: the free rider on its way to infection. Mol. Immunol. 42:153–160 [DOI] [PubMed] [Google Scholar]
- 11. Ahmad M, Pyaram K, Mullick J, Sahu A. 2007. Viral complement regulators: the expert mimicking swindlers. Indian J. Biochem. Biophys. 44:331–343 [PubMed] [Google Scholar]
- 12. Cooper NR. 1998. Complement and viruses, p 393–407 In Volanakis JE, Frank MM. (ed), The human complement system in health and disease. Marcel Dekker, Inc, New York, NY [Google Scholar]
- 13. Fleckenstein B, Desrosiers RC. 1982. Herpesvirus saimiri and herpesvirus ateles, p 253–332 In Roizman B. (ed), The herpesviruses. Plenum Publishing Corporation, New York, NY [Google Scholar]
- 14. Jung JU, Choi JK, Ensser A, Biesinger B. 1999. Herpesvirus saimiri as a model for gammaherpesvirus oncogenesis. Semin. Cancer Biol. 9:231–239 [DOI] [PubMed] [Google Scholar]
- 15. Jung JU, Desrosiers RC. 1994. Herpesvirus saimiri and ateles, p 614–622 In Webster R, Granoff A. (ed), Encyclopedia of virology. Saunders Scientific Publications, Inc, Philadelphia, PA [Google Scholar]
- 16. Falk LA, Wolfe LG, Deinhardt F. 1972. Isolation of herpesvirus saimiri from blood of squirrel monkeys (Saimiri sciureus). J. Natl. Cancer Inst. 48:1499–1505 [PubMed] [Google Scholar]
- 17. Albrecht JC, Fleckenstein B. 1992. New member of the multigene family of complement control proteins in herpesvirus saimiri. J. Virol. 66:3937–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fodor WL, Rollins SA, Biancocaron S, Rother RP, Guilmette ER, Burton WV, Albrecht JC, Fleckenstein B, Squinto SP. 1995. The complement control protein homolog of herpesvirus saimiri regulates serum complement by inhibiting C3 convertase activity. J. Virol. 69:3889–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bramley JC, Davies A, Lachmann PJ. 1997. Herpesvirus saimiri CD59—baculovirus expression and characterisation of complement inhibitory activity. Biochem. Soc. Trans. 25:354S. [DOI] [PubMed] [Google Scholar]
- 20. Rother RP, Rollins SA, Fodor WL, Albrecht JC, Setter E, Fleckenstein B, Squinto SP. 1994. Inhibition of complement-mediated cytolysis by the terminal complement inhibitor of herpesvirus saimiri. J. Virol. 68:730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh AK, Mullick J, Bernet J, Sahu A. 2006. Functional characterization of the complement control protein homolog of herpesvirus saimiri: R118 is critical for factor I cofactor activities. J. Biol. Chem. 281:23119–23128 [DOI] [PubMed] [Google Scholar]
- 22. Kohl J. 2006. The role of complement in danger sensing and transmission. Immunol. Res. 34:157–176 [DOI] [PubMed] [Google Scholar]
- 23. Singh AK, Yadav VN, Pyaram K, Mullick J, Sahu A. 2009. Mapping of functional domains in herpesvirus saimiri complement control protein homolog: the complement control protein domain 2 is the smallest structural unit displaying cofactor and decay-accelerating activities. J. Virol. 83:10299–10304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bernet J, Mullick J, Panse Y, Parab PB, Sahu A. 2004. Kinetic analysis of the interactions between vaccinia virus complement control protein and human complement proteins C3b and C4b. J. Virol. 78:9446–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hammer CH, Wirtz GH, Renfer L, Gresham HD, Tack BF. 1981. Large scale isolation of functionally active components of the human complement system. J. Biol. Chem. 256:3995–4006 [PubMed] [Google Scholar]
- 26. Mullick J, Bernet J, Panse Y, Hallihosur S, Singh AK, Sahu A. 2005. Identification of complement regulatory domains in vaccinia virus complement control protein. J. Virol. 79:12382–12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pangburn MK. 1987. A fluorimetric assay for native C3. The hemolytically active form of the third component of human complement. J. Immunol. Methods 102:7–14 [DOI] [PubMed] [Google Scholar]
- 28. Sahu A, Isaacs SN, Soulika AM, Lambris JD. 1998. Interaction of vaccinia virus complement control protein with human complement proteins: factor I-mediated degradation of C3b to iC3b1 inactivates the alternative complement pathway. J. Immunol. 160:5596–5604 [PubMed] [Google Scholar]
- 29. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 30. Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37:D387–D392 doi:10.1093/nar/gkn750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peitsch MC. 1995. Protein modeling by e-mail. Biotechnology (NY) 13:658–660 [Google Scholar]
- 32. Pyaram K, Kieslich CA, Yadav VN, Morikis D, Sahu A. 2010. Influence of electrostatics on the complement regulatory functions of kaposica, the complement inhibitor of Kaposi's sarcoma-associated herpesvirus. J. Immunol. 184:1956–1967 [DOI] [PubMed] [Google Scholar]
- 33. Yadav VN, Pyaram K, Mullick J, Sahu A. 2008. Identification of hot spots in the variola virus complement inhibitor (SPICE) for human complement regulation. J. Virol. 82:3283–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. White J, Lukacik P, Esser D, Steward M, Giddings N, Bright JR, Fritchley SJ, Morgan BP, Lea SM, Smith GP, Smith RA. 2004. Biological activity, membrane-targeting modification, and crystallization of soluble human decay accelerating factor expressed in E. coli. Protein Sci. 13:2406–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kirkitadze MD, Krych M, Uhrin D, Dryden DT, Smith BO, Cooper A, Wang X, Hauhart R, Atkinson JP, Barlow PN. 1999. Independently melting modules and highly structured intermodular junctions within complement receptor type 1. Biochemistry 38:7019–7031 [DOI] [PubMed] [Google Scholar]
- 36. Kuttner-Kondo L, Hourcade DE, Anderson VE, Muqim N, Mitchell L, Soares DC, Barlow PN, Medof ME. 2007. Structure-based mapping of DAF active site residues that accelerate the decay of C3 convertases. J. Biol. Chem. 282:18552–18562 [DOI] [PubMed] [Google Scholar]
- 37. Yadav VN, Pyaram K, Ahmad M, Sahu A. 2012. Species selectivity in poxviral complement regulators is dictated by the charge reversal in the central complement control protein modules. J. Immunol. 189:1431–1439 [DOI] [PubMed] [Google Scholar]
- 38. Pan Q, Ebanks RO, Isenman DE. 2000. Two clusters of acidic amino acids near the NH2 terminus of complement component C4 alpha′-chain are important for C2 binding. J. Immunol. 165:2518–2527 [DOI] [PubMed] [Google Scholar]
- 39. Blom AM, Villoutreix BO, Dahlback B. 2003. Mutations in alpha-chain of C4BP that selectively affect its factor I cofactor function. J. Biol. Chem. 278:43437–43442 [DOI] [PubMed] [Google Scholar]
- 40. Mark L, Lee WH, Spiller OB, Proctor D, Blackbourn DJ, Villoutreix BO, Blom AM. 2004. The Kaposi's sarcoma-associated herpesvirus complement control protein mimics human molecular mechanisms for inhibition of the complement system. J. Biol. Chem. 279:45093–45101 [DOI] [PubMed] [Google Scholar]
- 41. Wu J, Wu YQ, Ricklin D, Janssen BJ, Lambris JD, Gros P. 2009. Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat. Immunol. 10:728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Soames CJ, Sim RB. 1997. Interactions between human complement components factor H, factor I and C3b. Biochem. J. 326:553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hourcade D, Liszewski MK, Krych-Goldberg M, Atkinson JP. 2000. Functional domains, structural variations and pathogen interactions of MCP, DAF and CR1. Immunopharmacology 49:103–116 [DOI] [PubMed] [Google Scholar]
- 44. Pangburn MK, Pangburn KL, Koistinen V, Meri S, Sharma AK. 2000. Molecular mechanisms of target recognition in an innate immune system: interactions among factor H, C3b, and target in the alternative pathway of human complement. J. Immunol. 164:4742–4751 [DOI] [PubMed] [Google Scholar]
- 45. Kuttner-Kondo LA, Dybvig MP, Mitchell LM, Muqim N, Atkinson JP, Medof ME, Hourcade DE. 2003. A corresponding tyrosine residue in the C2/factor B type A domain is a hot spot in the decay acceleration of the complement C3 convertases. J. Biol. Chem. 278:52386–52391 [DOI] [PubMed] [Google Scholar]
- 46. Lukacik P, Roversi P, White J, Esser D, Smith GP, Billington J, Williams PA, Rudd PM, Wormald MR, Harvey DJ, Crispin MD, Radcliffe CM, Dwek RA, Evans DJ, Morgan BP, Smith RA, Lea SM. 2004. Complement regulation at the molecular level: the structure of decay-accelerating factor. Proc. Natl. Acad. Sci. U. S. A. 101:1279–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pangburn MK. 1986. Differences between the binding sites of the complement regulatory proteins DAF, CR1, and factor H on C3 convertases. J. Immunol. 136:2216–2221 [PubMed] [Google Scholar]
- 48. Liszewski MK, Leung M, Cui W, Subramanian VB, Parkinson J, Barlow PN, Manchester M, Atkinson JP. 2000. Dissecting sites important for complement regulatory activity in membrane cofactor protein (MCP; CD46). J. Biol. Chem. 275:37692–37701 [DOI] [PubMed] [Google Scholar]
- 49. Krych-Goldberg M, Hauhart RE, Subramanian VB, Yurcisin BM, Crimmins DL, Hourcade DE, Atkinson JP. 1999. Decay accelerating activity of complement receptor type 1 (CD35). Two active sites are required for dissociating C5 convertases. J. Biol. Chem. 274:31160–31168 [DOI] [PubMed] [Google Scholar]
- 50. Kostavasili I, Sahu A, Friedman HM, Eisenberg RJ, Cohen GH, Lambris JD. 1997. Mechanism of complement inactivation by glycoprotein C of herpes simplex virus. J. Immunol. 158:1763–1771 [PubMed] [Google Scholar]
- 51. Lubinski JM, Wang L, Soulika AM, Burger R, Wetsel RA, Colten H, Cohen GH, Eisenberg RJ, Lambris JD, Friedman HM. 1998. Herpes simplex virus type 1 glycoprotein gC mediates immune evasion in vivo. J. Virol. 72:8257–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kapadia SB, Molina H, Bvan Speck VSH, Virgin HW. 1999. Murine gammaherpesvirus 68 encodes a functional regulator of complement activation. J. Virol. 73:7658–7670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liszewski MK, Leung MK, Hauhart R, Buller RM, Bertram P, Wang X, Rosengard AM, Kotwal GJ, Atkinson JP. 2006. Structure and regulatory profile of the monkeypox inhibitor of complement: comparison to homologs in vaccinia and variola and evidence for dimer formation. J. Immunol. 176:3725–3734 [DOI] [PubMed] [Google Scholar]
- 54. McKenzie R, Kotwal GJ, Moss B, Hammer CH, Frank MM. 1992. Regulation of complement activity by vaccinia virus complement-control protein. J. Infect. Dis. 166:1245–1250 [DOI] [PubMed] [Google Scholar]
- 55. Mullick J, Bernet J, Singh AK, Lambris JD, Sahu A. 2003. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) open reading frame 4 protein (kaposica) is a functional homolog of complement control proteins. J. Virol. 77:3878–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Okroj M, Mark L, Stokowska A, Wong SW, Rose N, Blackbourn DJ, Villoutreix BO, Spiller OB, Blom AM. 2009. Characterization of the complement inhibitory function of rhesus rhadinovirus complement control protein (RCP). J. Biol. Chem. 284:505–514 [DOI] [PubMed] [Google Scholar]
- 57. Rosengard AM, Liu Y, Nie Z, Jimenez R. 2002. Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc. Natl. Acad. Sci. U. S. A. 99:8808–8813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Manuelian T, Hellwage J, Meri S, Caprioli J, Noris M, Heinen S, Jozsi M, Neumann HP, Remuzzi G, Zipfel PF. 2003. Mutations in factor H reduce binding affinity to C3b and heparin and surface attachment to endothelial cells in hemolytic uremic syndrome. J. Clin. Invest. 111:1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liszewski MK, Leung MK, Hauhart R, Fang CJ, Bertram P, Atkinson JP. 2009. Smallpox inhibitor of complement enzymes (SPICE): dissecting functional sites and abrogating activity. J. Immunol. 183:3150–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liszewski MK, Leung MK, Atkinson JP. 1998. Membrane cofactor protein: importance of N- and O-glycosylation for complement regulatory function. J. Immunol. 161:3711–3718 [PubMed] [Google Scholar]
- 61. Mullick J, Singh AK, Panse Y, Yadav V, Bernet J, Sahu A. 2005. Identification of functional domains in kaposica, the complement control protein homolog of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J. Virol. 79:5850–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Krych M, Clemenza L, Howdeshell D, Hauhart R, Hourcade D, Atkinson JP. 1994. Analysis of the functional domains of complement receptor type 1 (C3b/C4b receptor, CD35) by substitution mutagenesis. J. Biol. Chem. 269:13273–13278 [PubMed] [Google Scholar]
- 63. Saunders RE, Goodship TH, Zipfel PF, Perkins SJ. 2006. An interactive web database of factor H-associated hemolytic uremic syndrome mutations: insights into the structural consequences of disease-associated mutations. Hum. Mutat. 27:21–30 [DOI] [PubMed] [Google Scholar]
- 64. Xu H, Kitano E, Sato Y, Kobayashi C, Firdawes S, Kitamura H, Fukuzawa M, Miyagawa S. 2008. Studies of monkey complement: measurement of cynomolgus monkey CH50, ACH50, C4, C2 and C3. Xenotransplantation 15:14–19 [DOI] [PubMed] [Google Scholar]