Abstract
Influenza A viruses commonly cause pancreatitis in naturally and experimentally infected animals. In this study, we report the results of in vivo investigations carried out to establish whether influenza virus infection could cause metabolic disorders linked to pancreatic infection. In addition, in vitro tests in human pancreatic islets and in human pancreatic cell lines were performed to evaluate viral growth and cell damage. Infection of an avian model with two low-pathogenicity avian influenza isolates caused pancreatic damage resulting in hyperlipasemia in over 50% of subjects, which evolved into hyperglycemia and subsequently diabetes. Histopathology of the pancreas showed signs of an acute infection resulting in severe fibrosis and disruption of the structure of the organ. Influenza virus nucleoprotein was detected by immunohistochemistry (IHC) in the acinar tissue. Human seasonal H1N1 and H3N2 viruses and avian H7N1 and H7N3 influenza virus isolates were able to infect a selection of human pancreatic cell lines. Human viruses were also shown to be able to infect human pancreatic islets. In situ hybridization assays indicated that viral nucleoprotein could be detected in beta cells. The cytokine activation profile indicated a significant increase of MIG/CXCL9, IP-10/CXCL10, RANTES/CCL5, MIP1b/CCL4, Groa/CXCL1, interleukin 8 (IL-8)/CXCL8, tumor necrosis factor alpha (TNF-α), and IL-6. Our findings indicate that influenza virus infection may play a role as a causative agent of pancreatitis and diabetes in humans and other mammals.
INTRODUCTION
Influenza A viruses (IAVs) originate from the wild-bird reservoir and infect a variety of hosts, including domestic birds. These viruses are also able to infect a significant number of mammals, in which they may become established. Among these are pigs, equids, mustelids, sea mammals, canids, felids, and humans. IAVs cause systemic or nonsystemic infection, depending on the strain involved. The systemic disease occurs mostly in avian species and is known as highly pathogenic avian influenza (HPAI). It is characterized by extensive viral replication in vital organs and death within a few days after the onset of clinical signs in the majority of infected animals. The nonsystemic form, which is by far the most common, occurs in birds and mammals and is characterized by mild respiratory and enteric signs. To differentiate it from HPAI, in birds it is known as low-pathogenicity avian influenza (LPAI). The different clinical presentation results from the fact that nonsystemic influenza A viruses are able to replicate only in the presence of trypsin or trypsin-like enzymes, and thus, their replication is believed to be restricted to the respiratory and enteric tracts. Avian IAVs have a tropism for the pancreas (1–4). Necrotizing pancreatitis is a common finding in wild and domestic birds infected with HPAI virus (5–8), and the systemic nature of HPAI is in line with these findings. In contrast, it is difficult to explain pancreatic colonization by LPAI viruses, which is a common finding in infected chickens and turkeys (9–14). Previous studies have reported that certain IAVs can also cause pancreatitis in mammals following natural or experimental infection (15–18). Recently, there have been reports of pancreatic damage in human cases associated with H1N1pdm influenza A virus infection, including both acute pancreatitis and the onset of type 1 diabetes (T1D) (19–23).
To date, there has been no attempt to establish whether influenza viruses are able to grow in pancreatic cells in vitro, and no data are available on the consequences of influenza virus replication in the pancreas in vivo. In this study, we explored the implications of influenza virus infection for pancreatic endocrine function in an animal model, and we performed in vitro experiments aiming to establish the occurrence, extent, and implications of influenza A virus infection in human cells of pancreatic origin. For the in vivo studies, we selected the turkey as a model, due to the fact that turkeys are highly susceptible to influenza virus infection and pancreatic damage is often observed as a postmortem lesion. Experimental infections were performed with two LPAI viruses, A/turkey/Italy/3675/1999 (H7N1) and A/turkey/Italy/2962/2003 (H7N3), as both viruses had been associated with pancreatic lesions in naturally infected birds. For the in vitro studies, in addition to the previously mentioned avian strains, we selected A/New Caledonia/20/99 (H1N1) and A/Wisconsin/67/05 (H3N2), as these viruses have circulated for extensive periods in humans, and existing epidemiological data would be suitable for a retrospective study. The strains were used to infect two established human pancreatic cell lines (including human insulinoma and pancreatic duct cell lines) and primary cultures of human pancreatic islets.
MATERIALS AND METHODS
In vivo experiment.
The aim of this study was to establish whether two natural nonsystemic avian influenza viruses obtained from field outbreaks, without prior adaptation, could cause endocrine or exocrine pancreatic damage following experimental infection of young turkeys. The study was performed in strict accordance with the relevant national and local animal welfare bodies [Convention of the European Council no. 123 and National Guidelines (Legislative Decree 116/92)]. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Istituto Zooprofilattico Sperimentale delle Venezie (IZSVe) (permit number CE.IZSVE.022012).
Animals.
Sixty-eight female meat turkeys obtained at 1 day of age from a commercial farm were used in this study. The birds were housed in negative-pressure, HEPA-filtered isolation cabinets for the duration of the experimental trial. Before carrying out the infection, the animals were housed for 3 weeks to allow adaptation and growth, received feed and water ad libitum, and were identified by means of wing tags.
Viruses.
Two LPAI viruses isolated during epidemics in Italy were used for the experimental infection: A/turkey/Italy/3675/1999 (H7N1) and A/turkey/Italy/2962/2003 (H7N3). Stocks of AI viruses were produced by a single passage in 9-day-old embryonated specific-pathogen-free (SPF) chicken eggs via the allantoic cavity, according to EU Council Directive 2005/94/EC (24). The allantoic fluid was harvested 48 h postinoculation, aliquoted, and stored at −80°C until use. For viral titration, 100 μl of 10-fold-diluted viral suspension was inoculated in SPF embryonated chicken eggs, and the median embryo infectious dose (EID50) was calculated according to the Reed and Muench formula (54).
Experimental design.
Birds were divided into three experimental groups (A [H7N1], B [H7N3], and K [control]). Groups A and B, each consisting of 24 animals, were infected via the oral-nasal route with 0.1 ml of allantoic fluid containing 106.83 EID50 of the A/turkey/Italy/3675/1999 (H7N1) virus and 106.48 EID50 of the A/turkey/Italy/2962/2003 (H7N3) virus. Group K, consisting of 20 animals, received 0.1 ml of negative allantoic fluid via the oral-nasal route and served as a negative control. All birds were observed twice daily for clinical signs. On days 0, 3, 6, 9, 13, 15, 20, 23, 27, 31, 34, 41, and 45 postinfection (p.i.), blood was collected from the brachial veins of all animals using heparinized syringes in order to determine glucose and lipase levels in plasma. On days 2 and 3 p.i., tracheal swabs were collected to evaluate viral replication. On day 3 p.i., blood was also collected to determine the presence of viral RNA in the blood. On days 4 and 7 p.i., two birds from each infected group were humanely sacrificed, and the pancreas and lungs were processed for the detection of viral RNA and for histopathology and immunohistochemistry (IHC). Similarly, on days 8 and 17 p.i., one subject from each experimental group was euthanized, and the pancreas was collected for histological and immunohistochemical studies. For this purpose, we selected hyperglycemic subjects that had also shown an increase in lipase levels.
Biochemical analyses.
Blood samples were collected in Gas Lyte 23 G pediatric syringes containing lyophilized lithium heparin as an anticoagulant. At each sampling, 0.3 ml of blood was collected and refrigerated at 4°C until it was processed. To obtain plasma, samples were immediately centrifuged at 1,500 × g for 15 min at 4°C. To determine the levels of glucose and lipase in plasma, commercially available kits (Glucose HK and LIPC; Roche Diagnostics GmbH, Mannheim, Germany) were applied to the computerized system Cobas c501 (F. Hoffmann-La Roche Std., Basel, Switzerland). The Glucose HK test is based on a hexokinase enzymatic reaction. The linearity of the reaction is 0.11 to 41.6 mmol/liter (2 to 750 mg/dl), and its analytic sensitivity is 0.11 mmol/liter (2 mg/dl). The LIPC test is based on a colorimetric enzymatic reaction with a linearity of 3 to 300 U/liter and an analytic sensitivity of 3 U/liter.
Molecular tests.
Tracheal swabs, blood samples, and organs (pancreas and lungs) were tested for viral RNA by means of real-time reverse transcriptase PCR (RRT-PCR) for the identification of the influenza virus matrix (M) gene.
RNA extraction.
Viral RNA was extracted from 100 μl of blood using the commercial NucleoSpin RNA II kit (Macherey-Nagel) and from 50 μl of phosphate-buffered saline (PBS) containing a tracheal-swab suspension using the Ambion MagMax-96 AI-ND Viral RNA Isolation Kit for the automatic extractor. One hundred fifty milligrams of homogenized lung and pancreas tissues was centrifuged, and viral RNA was extracted from 100 μl of clarified suspension using the NucleoSpin RNA II kit (Macherey-Nagel).
One-step RRT-PCR.
The isolated RNA was amplified using the published primers and probes from Spackman et al. (25), targeting the conserved matrix (M) gene of type A influenza virus. Five microliters of RNA was added to the reaction mixture, composed of 300 nM forward and reverse primers (M25F and M124-R, respectively) and 100 nM fluorescent-label probe (M+64). The amplification reaction was performed in a final volume of 25 μl using the commercial QuantiTect Multiplex RT-PCR kit (Qiagen, Hilden, Germany). The PCR was performed using the following protocol: 20 min at 50°C and 15 min at 95°C, followed by 40 cycles at 94°C for 45 s and 60°C for 45 s. In vitro-transcribed target RNA was obtained using Mega Short Script 7 (high-yield transcription kit; Ambion) according to the manufacturer's instructions, quantified by NanoDrop 2000 (Thermo Scientific), and used to create a standard calibration curve for viral-RNA quantification. To check the integrity of the isolated RNA, the β-actin gene was also amplified using a set of primers designed in house (primer sequences are available upon request). The reaction mixture was composed of 300 nM forward and reverse primers and 1× EvaGreen (Explera, Jesi, Italy). The amplification reaction was performed in a final volume of 25 μl using the commercial Superscript III kit (Invitrogen, Carlsbad, CA). The PCR was performed using the following protocol: 30 min at 55°C and 2 min at 94°C, followed by 45 cycles at 94°C for 30 s and 60°C for 1 min.
Histology and immunohistochemistry.
Formalin-fixed, paraffin-embedded pancreas sections were cut (3 μm thick). Slides were stained with hematoxylin and eosin (H&E) (Histoserv, Inc., Germantown, MD). Representative photographs were taken with SPOT Advanced software (version 4.0.8; Diagnostic Instruments, Inc., Sterling Heights, MI). The reagents and methodology for influenza virus IHC were as follows: polyclonal antibody anti-type A influenza virus nucleoprotein (NP) and mouse anti-influenza virus A (NP subtype A, clone EVS 238; European Veterinary Laboratory), 1:100 in PBS-2.5% bovine serum albumin (BSA) for 1 h at room temperature; secondary antibody, goat anti-mouse IgG2a horseradish peroxidase (HRP) (Southern Biotech), 1/200 in PBS-2.5% BSA for 1 h at room temperature. Antigen retrieval was performed by incubating the slides for 10 min at 37°C in trypsin (Digest-all Kit; Invitrogen). Endogenous peroxidase was blocked with 3% H2O2 for 10 min at room temperature. Before incubation with primary antibody, a blocking step was performed with PBS-5% BSA for 20 min at room temperature. Diaminobenzidine (DAB) was applied as a chromogen (DakoCytomation; code K3468). IHC for insulin and glucagon was performed with polyclonal guinea pig anti-swine insulin, 1:50 (A0564; Dako, Carpinteria, CA), and polyclonal rabbit anti-glucagon, 1:200 (NCL-GLUC; Novocastra, Newcastle, United Kingdom), using as a detection system the En Vision Ap (K1396; Dako, Carpinteria, CA) and nuclear fast red (K139;6 Dako) for influenza virus A staining and En Vision+System HRP-labeled polymer anti-rabbit (K4002; Dako, Carpinteria, CA) and DAB (K3468; Dako, Carpinteria, CA) for insulin and glucagon staining.
In vitro experiment.
The aim of these experiments was to establish whether human and avian influenza viruses could grow on cell lines derived from the human pancreas and to investigate the effect of human influenza virus replication in human pancreatic islets.
Cell lines.
Madin-Darby canine kidney (MDCK) cells were maintained in Alpha's modified Eagle medium (AMEM) (Sigma) supplemented with 10% fetal bovine serum (FBS), 1% 200 mM l-glutamine, and a 1% penicillin-streptomycin-nystatin (pen-strep-nys) solution. The human insulinoma cell line hCM (26) and immortalized human ductal epithelial cell line HPDE6 (27) were maintained in RPMI (Gibco) supplemented with 1% l-glutamine, 1% antibiotics, and FBS (5% and 10%, respectively). MDCK and HPDE6 cells were passaged twice weekly at a subcultivation ratio of 1:10 and 1:4, while hCM cells were split three times per week at a ratio of 1:4. All cells were maintained in a humidified incubator at 37°C with 5% CO2.
Primary cells.
Pancreatic islets were isolated and purified at San Raffaele Scientific Institute from pancreases of multiorgan donors according to Ricordi's method. Briefly, after cannulation of the pancreatic duct, collagenase solution (2,000 U; Serva, Germany) at 4°C was injected through the duct (perfusion). Subsequently, the pancreas was cut into small pieces and loaded into a digestion chamber, named Ricordi's chamber, for an enzymatic and mechanical digestion at 37°C. Final purification of digested pancreas was performed using a continuous gradient (Ficoll; Biochrom, Berlin) in a computerized centrifuge system (COBE 2991 cell processor). Islet preparations with purities of >80% ± 8% (mean ± standard deviation [SD]; n = 6) not suitable for transplantation were used after approval by the local ethical committee. Cells were seeded in 24-well plates and 25-cm2 flasks at 150 islets/ml and maintained in final wash culture medium (Mediatech, Inc., Manassas, VA) at 37°C with 5% CO2.
Sialic acid receptor characterization on hCM and HPDE6 cells.
The presence of alpha-2,3- and alpha-2,6-linked sialic acid residues was determined by flow cytometry. Following trypsinization, 1 × 106 cells were washed twice with PBS-10 mM HEPES (PBS-HEPES) for 5 min at 1,200 rpm and then treated with an avidin/biotin-blocking kit (Vector Laboratories) according to the manufacturer's instructions, with cells incubated for 15 min with 100 μl of each solution. Alpha-2,3 and alpha-2,6 sialic acid linkages were detected by incubating cells for 30 min with 100 μl of biotinylated Maackia amurensis (MAA) lectin II (Vector Laboratories; 5 μg/ml), followed by 100 μl of phycoerythrin (PE)-streptavidin (BD Biosciences; 10 μg/ml) for 30 min at 4°C in the dark or with 100 μl of fluorescein-conjugated Sambucus nigra (SNA) lectin (Vector Laboratories; 5 μg/ml). Cells were washed twice with PBS-HEPES between all blocking and staining steps and resuspended in PBS with 1% formalin prior to analysis. To confirm the specificity of lectins, cells were pretreated with 1 U per ml of neuraminidase (NA) from Clostridium perfringens (Sigma) for 1 h prior to the avidin/biotin block. Samples were analyzed on a BD Facscalibur or BD LSR II (BD Biosciences), and a minimum of 5,000 events were recorded.
Viruses and viral titration.
Stocks of A/New Caledonia/20/99 (H1N1) and A/Wisconsin/67/05 (H3N2) viruses were kindly provided to San Raffaele Hospital (HSR) by Nadia Naffakh, Pasteur Institute, CNR Virus Influenza (Paris, France), and were serially expanded 2 times in MDCK cells prior to use.
To propagate IAV, monolayer-cultured MDCK cells were washed twice with PBS and infected with A/NewCaledonia/20/99 (H1N1) or A/Wisconsin/67/05 (H3N2) at a multiplicity of infection (MOI) of 0.001. After virus adsorption for 1 h at 35°C, the cells were washed twice and incubated at 35°C with Dulbecco's modified Eagle's medium (DMEM) without serum supplemented with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (1 μg/ml; Worthington Biomedical Corporation, Lakewood, NJ). Supernatants were recovered 48 h postinfection.
LPAI H7N1 A/turkey/Italy/3675/1999 and H7N3 A/turkey/Italy/2962/2003 viruses isolated during epidemics in Italy were grown in 9-day-old embryonated SPF chicken eggs as described above. The sequences for A/New Caledonia/20/99 (H1N1), A/Wisconsin/67/05 (H3N2), and A/turkey/Italy/3675/1999 (H7N1) isolates were previously published.
For viral titration, plaque assays were performed as previously described (28). Briefly, MDCK monolayer cells, plated in 24-well plates at 2.5 × 105 cells/well, were washed twice with DMEM without serum, and serial dilutions of virus were adsorbed onto cells for 1 h. The cells were covered with 2× minimal essential medium (MEM)-Avicel (FMC Biopolymer, Philadelphia, PA) mixture supplemented with TPCK-treated trypsin (1 μg/ml). Crystal violet staining was performed 48 h postinfection, and visible plaques were counted.
Virus replication kinetics in pancreatic-cell lines.
Semiconfluent monolayers of HPDE6 and hCM cells seeded on 24-well plates were washed twice with PBS and then infected with human viruses (H1N1 and H3N2) at an MOI of 0.001 and with avian viruses (H7N1 and H7N3) at an M.O.I of 0.01 using 200 μl of inoculum per well. The inoculum was removed after 1 h of absorption and replaced with 1 ml of serum-free medium containing 0.05 μg/ml TPCK-trypsin (Sigma). At 1, 24, 48, and 72 h postinfection, supernatants from three infected wells and one control well were harvested, and viral titers were determined by virus isolation using the 50% tissue culture infectious dose (TCID50) assay, as well as by real-time RT-PCR detection of the matrix gene. All replication kinetics experiments were repeated three times.
TCID50.
Confluent monolayers of MDCK cells seeded onto 96-well plates were washed twice in serum-free medium and inoculated with 50 μl of 10-fold serially diluted samples in serum-free MEM. After 1 h of adsorption, an additional 50 μl of serum-free medium containing 2 μg/ml TPCK-trypsin was added to each well. Cytopathic-effect (CPE) scores were determined after 3 days of incubation at 37°C by visual examination of infected wells using a light microscope. The TCID50 value was determined using the method of Reed and Muench.
Growth assay in pancreatic islets.
Islets were infected with H1N1 and H3N2 influenza viruses by adding 4.8 × 102 or 4.8 × 103 PFU/well, respectively. Viral growth was performed with and without the addition of TPCK-trypsin (Sigma) (1 μg/ml). Uninfected islets were left as a negative control. Samples were collected every 48 h from the day of infection (time zero [t0]) until day 10 (t5, the fifth time point that occurred at 10 days postinfection). Each sample was centrifuged at 150 × g for 5 min. The supernatant was collected and stored at −80°C for quantitative real-time PCR, virus titration, and cytokine expression profiling. The pellet was washed twice with PBS, stored at −80°C, and subsequently processed for real-time PCR. All pellets and supernatants were tested by real-time PCR in triplicate.
Detection of viral RNA from pancreatic tissue.
The total RNAs from pancreatic islet pellets and supernatants were isolated using the commercial NucleoSpin RNA II kit (Macherey-Nagel) according to the manufacturer's instructions. The RNAs were eluted in 60 μl of elution buffer and tested using one-step RRT-PCR for the influenza virus matrix gene to evaluate viral growth.
A quadratic regression model (CT [threshold cycle] = β0 + β1TPCK-trypsin + β2time + β3time2 + β4time × TPCK-trypsin + β5time2 × TPCK-trypsin) for each virus and specimen was used to analyze the trend of CT values over time. The influence of TPCK-trypsin presence and the interaction between its presence and the time point were evaluated. The regression model took into account the influence of the intragroup correlation among repeated measurements for each observed time in the confidence interval (CI) calculation. A residual postestimation analysis was performed to verify the validity of the model.
One-step RRT-PCR.
Quantitative real-time PCR targeting the conserved M gene of type A influenza virus was applied according to the protocol described above. To check the integrity of the isolated RNA, the β-actin gene was also amplified using the primers and probe previously described (29). The reaction mixture consisted of 400 nM forward and reverse primers (Primer-beta act intronic and Primer-beta act reverse, respectively) and 200 nM fluorescent-label probe (5′ Cy5 or 3′ black hole quencher 1 [BHQ1]). The amplification reaction was performed in a final volume of 25 μl using the commercial QuantiTect Multiplex RT-PCR kit (Qiagen, Hilden, Germany). The PCR protocol was 20 min at 50°C and 15 min at 95°C, followed by 45 cycles at 94°C for 45 s and 55°C for 45 s.
Western blot analysis.
Cellular pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 8, 1.0% SDS, 350 mM NaCl, 0.25% Triton-X, proteases inhibitor cocktail) and then mixed and incubated on ice for 30 min. The suspension was sonicated three times for 5 min each time and then centrifuged at maximum speed for 10 min. A Bradford test was performed in order to calculate the total protein concentration for each sample. Based on this calculation, the same amount of protein per sample was loaded and electrophoresed in 12% polyacrylamide gels. Following SDS-PAGE, the proteins were transferred from the gel onto immunoblot polyvinylidene difluoride (PVD) membranes (Bio-Rad) by electroblotting. The membranes were saturated overnight at 4°C in 5% grade blocker nonfat dried milk (Bio-Rad) in PBS and then incubated for 1 h at room temperature under constant shaking in PBS containing 0.05% Tween 20 (Sigma), 5% dried milk, and mouse monoclonal influenza A virus nucleoprotein antibody (Abcam). β-Actin antibody (Abcam) was used as a loading control. After incubation with the primary antibody, the membranes were exposed for 1 h to HRP-rabbit polyclonal secondary antibody to mouse IgG (Abcam), followed by visualization of positive bands by enhanced chemiluminescence (ECL) using Hyperfilm ECL (Amersham Biosciences).
Visualization of viral growth in pancreatic cell lines.
HPDE6 and hCM cells were grown on slides to 80% confluence and infected with either H1N1or H3N2 virus at an MOI of 0.1 with 0.05 mg/ml of TPCK-trypsin. The cells were fixed and permeabilized at 0, 24, 48, and 72 h p.i. with chilled acetone (80%). After blocking with PBS containing 1% BSA, the cells were incubated for 1 h at 37°C in a humidified chamber with mouse monoclonal antibody to fluorescein isothiocyanate (FITC)-conjugated influenza A virus nucleoprotein (Abcam) in PBS containing 1% BSA and 0.2% Evan's Blue. The staining solution was decanted, and the cells were washed three times. Nuclei of negative-control cells were stained with DAPI (4′,6-diamidino-2-phenylindole) (Sigma) and then washed with PBS and observed under UV light.
In situ visualization of viral RNA in pancreatic islets.
To visualize viral RNA localized within cells, purified human pancreatic islets were harvested at 2, 5, and 7 days postinfection with human influenza viruses. The islets were then incubated for 24 h in methanol-free 10% formalin, deposited at the bottom of flat-bottom tubes, embedded in agar to immobilize them, dehydrated, and finally embedded in paraffin. Islet samples were sectioned at 4 μm. For colocalization experiments, islets were enzymatically digested into single cells with a trypsin-like enzyme (12605-01; TrypLE Express; Invitrogen, Carlsbad, CA) and cytocentrifuged onto glass slides. In situ hybridization was performed using the Quantigene ViewRNA technique, based on multiple oligonucleotide probes and branched-DNA signal amplification technology, according to the manufacturer's instructions (Affymetrix, Santa Clara, CA). The probe set used was designed to hybridize the H1N1/A/New Caledonia/20/99 virus (GenBank sequence DQ508858.1). Due to sequence homology in the region covered by the probes, the same set also recognized the H3N2 virus RNA, as confirmed in pilot experiments. To identify cell types within islets, the following Quantigene probes were used: insulin for beta cells (INS gene; NCBI reference sequence NM_000207), alpha-amylase 1 for exocrine cells (AMY1A gene; NCBI reference sequence NM_004038), and cytokeratin 19 for duct cells (KRT19 gene; NCBI reference sequence NM_002276). Quantification of cells positive for each probe was performed within 8 randomly chosen fields using the IN Cell Investigator software (GE Healthcare United Kingdom Ltd.).
Determination of insulin secretion in infected islets.
Aliquots of 100 islet equivalents (uninfected or infected with H1N1/A/New Caledonia/20/99 and H3N2/A/Wisconsin/67/05) per column were loaded onto Sephadex G-10 columns with medium at low glucose concentration (2 mM) and preincubated at 37°C for 1 h. After preincubation, the islets were exposed to sequential 1-h incubations at low (2 mM) and high (20 mM) glucose concentrations. Supernatants were collected with protease inhibitor cocktail (Roche Biochemicals, Indianapolis, IN) and stored at −80°C at the end of each incubation. The insulin content was determined with an insulin enzyme-linked immunoassay kit (Mercodia AB, Uppsala, Sweden) following manufacturer's instructions. Insulin secretion indices were calculated as the ratio between the insulin concentration at the end of high-glucose incubation and the insulin concentration at the end of low-glucose incubation.
Cytokine expression profile.
The capability of H1N1 and H3N2 viruses to induce cytokine expression in human pancreatic islets was measured using multiplex bead-based assays based on xMAP technology (Bio-Plex; Bio-Rad Laboratories, Hercules, CA). The parallel wells of pancreatic islets were infected with viruses or were mock infected. The culture medium supernatant was collected before infection and 2, 4, 6, 8, and 10 days postinfection and assayed for 48 cytokines. Selected cytokines, the limits of detection, and the coefficients of variability (intra-assay percent coefficient of variation [%CV] and interassay %CV) of the cytokine/chemokine are reported in Table S5 in the supplemental material.
Evaluation of cell death following infection (LIVE/DEAD assay).
The viability of islet cells after infection with human viruses was measured using the LIVE/DEAD cell assay kit (L-3224; Molecular Probes, Inc., Leiden, The Netherlands). The assay is based on the simultaneous determination of live and dead cells with two fluorescent probes. Live cells are stained green by calcein due to their esterase activity, and nuclei of dead cells are stained red by ethidium homodimer 1. Islets harvested after 5 days of culture were further enzymatically digested into single cells with trypsin-like enzyme (12605-01; TrypLE Express; Invitrogen, Carlsbad, CA). According to the manufacturer's instructions, single cells were incubated with the labeling solution for 30 min at room temperature in the dark, cytocentrifuged onto glass slides, and assessed with a Carl Zeiss Axiovert 135TV fluorescence microscope. Analysis of dead cells was performed on cytospin preparations using IN Cell Investigator software (GE Healthcare United Kingdom Ltd.). Positive cells in each category were quantified with 10 systematically random fields.
Statistical analysis.
Data were generally expressed as mean ± standard deviation or median (minimum-maximum). Differences between parameters were evaluated using Student's t test when parameters were normally distributed and a Mann-Whitney U test when parameters were not normally distributed. Kaplan-Meier and/or Cox regression analysis was used to analyze the incidence of events during the time. A P value of less than 0.05 was considered an indicator of statistical significance. Analysis of data was done using the SPSS statistical package for Windows (SPSS Inc., Chicago, IL).
Nucleotide sequence accession numbers.
The sequences of all eight influenza virus genome segments belonging to H7N3 A/turkey/Italy/2962/2003 were submitted to GenBank and assigned accession numbers JX515660, JX515661, JX515662, JX515663, JX515664, JX515665, JX515666, and DQ090062.
RESULTS
In vivo experiment. (i) Clinical disease.
Turkeys from both H7N1-challenged (A) and H7N3-challenged (B) groups showed clinical signs typical of LPAI infection, such as conjunctivitis, sinusitis, diarrhea, ruffled feathers, and depression on day 2 p.i. Mild symptoms regressed by day 20 p.i. Only two subjects from group A showed sinusitis until day 30 p.i. Mortality rates were low in both groups: one subject in group A died on day 8 p.i., and one subject in group B died on day 19 p.i.
(ii) Detection of viral RNA.
Viral RNA was detected from the tracheal swabs collected from 17/20 birds infected with H7N1 virus and 19/20 birds infected with H7N3 virus on day 2 p.i. and in all animals on day 3 p.i. Viral RNA was also detected from the blood of two birds of group A H7N1 and four birds of group B H7N3 on day 3 p.i. and from the pancreases and lungs collected on days 4 and 7 p.i. (see Table S4 in the supplemental material). No viral RNA was detected from the uninfected controls.
(iii) Biochemical analyses.
In blood samples collected intra vitam to reveal metabolic alterations, a significant increase in plasma lipase levels (10 to 100 times the values of the control birds) was evident in H7N1-challenged (12/20) and H7N3-challenged (10/20) turkeys between days 3 and 9 p.i. (Fig. 1A), while no uninfected controls showed modification of lipase levels (20/20; P < 0.001; Pearson chi-squared test). A clear trend between the presence of viral RNA in blood at day 3 and the increase in lipase was evident in infected animals (hazard ratio, 2.51; 95% confidence interval, 0.92 to 6.81; P = 0.07). Lipase levels within the normal range were rapidly reestablished in all cases, and it was decided to no longer evaluate this parameter on day 23 (see Tables S1 and S2 in the supplemental material). After day 9 p.i., 5 birds in group A and 5 birds in group B developed hyperglycemia (Fig. 1B). Of these, two birds maintained the hyperglycemic status throughout the experiment, while in all the other birds, the levels of blood glucose returned to levels similar to those of controls (see Table S3 in the supplemental material). A clear association between the increase in lipase between days 3 and 9 p.i. and the development of hyperglycemia after day 9 p.i. was evident. In fact, hyperglycemia was present only in the birds that developed high lipase values postinfection and never appeared in birds with normal lipase levels (10/22 and 0/18, respectively; P = 0.001), with a median time between development of hyperlipasemia and hyperglycemia of 4.5 days (minimum-maximum, 3 and 7).
Fig 1.
Biochemical analysis. Kaplan-Meier analysis for the appearance of hyperlipasemia (A) and hyperglycemia (B) (plasma glucose, >27.78 mmol/liter) among mock-, H7N1-, and H7N3-infected turkeys. Differences were tested using the log rank statistic. The bar graphs show the frequency of events in relation to hyperlipasemia, hyperglycemia, and presence of viremia.
(iv) Histopathology and immunohistochemistry.
None of the control turkeys showed significant histological changes or positive immunohistological reactions to avian influenza virus (AIV) (Fig. 2A). In all infected birds, histopathologic lesions were evident, although markedly different in samples collected at different times postinfection. At early stages (days 4 to 8 p.i.), acute pancreatitis with necrotic acinar cells and massive inflammatory infiltration composed of macrophages, heterophils, lymphocytes, and plasma cells dominated over areas of healthy/uninvolved/spared tissue (Fig. 2B). From day 8 p.i., these necrotic inflammatory lesions/necrotic inflammatory areas were gradually replaced by ductule hyperplasia and lymphocytic infiltration with a mild degree of fibroplasia. At later stages (day 17 p.i), extensive fibrosis with lymphoid nodules replaced pancreatic parenchyma, and disruption of the normal architecture of the organ was evident (Fig. 2C). Variable numbers of necrotic acinar cells were observed during the entire experimental period. Obstructive ductal lesions were not seen in any case or stage.
Fig 2.
Histopathology and immunohistochemistry. (A) Turkey pancreas. Normal tissue is shown. Acinar cells containing zymogen granules in their cytoplasm are evident, associated with two nests of normal islet cells and a ductal structure (H&E). (B) Turkey pancreas. Shown is diffuse and severe necrosis of acinar cells (arrows) with severe inflammatory infiltrate (asterisks) (H&E). (C) Turkey pancreas. Most of the pancreas has been replaced by foci of fibrous connective tissue and lymphoid nodules, with some ductular proliferation. (D) Turkey pancreas. Shown is immunohistochemistry for avian influenza virus NP, with an area of necrosis, and positive nuclei and cytoplasm in both acinar and ductal cells.
By immunohistochemistry, degenerating and necrotic acinar cells showed specific reaction to virus nucleoprotein antigen antibody during the experimental period (Fig. 2D). Some of the vascular endothelial cells also showed a positive reaction, as well as occasional ductal epithelial cells. In uninfected controls, the insulin-positive tissues of the pancreas were scattered singly or in small groups of islets of various shapes and sizes in the interstitium of the exocrine part (Fig. 3A). At day 8 p.i., the normal structure of islets was partially destroyed and the number of islet cells was reduced. The remaining islets were smaller and distorted, with irregular outlines or dilated intraislet capillaries; the number of cells staining for insulin was also reduced, and these cells presented enlarged cytoplasm and sometimes appeared to have granular degeneration and even necrosis. Fibrous bands appeared inside the islet, with islet fragmentation and dislocation of small and large clusters of endocrine cells (Fig. 3B). At day 17 p.i., separate large clusters of endocrine insulin-positive cells were evident embedded in or close to the extensive fibrosis that replaced the acinar component (Fig. 3C). Beyond day 17 p.i., groups of very large (>200 μm in diameter), usually irregular islet-like areas of mainly insulin immunoreactivity were clearly present scattered in extensive acinar fibrosis (Fig. 3D and E).
Fig 3.
Immunohistochemistry for insulin. Shown is turkey pancreas with representative islet structures before and after H7N3 infection at different time points.
In vitro experiment. (i) Susceptibility of human pancreatic cell lines to human influenza A viruses.
The susceptibility of endocrine (hCM insulinoma) and ductal (HPDE6) cell lines to H1N1/A/New Caledonia/20/99 and H3N2/A/Wisconsin/67/05 infections was investigated.
(ii) Receptor distribution.
Our investigation using lectin staining for receptor detection on both the hCM and HPDE6 cell lines revealed high levels of alpha-2,6 sialic acid-linked sialic acid molecules (required by human-tropic viruses), as well as alpha-2,3-linked residues (used by avian-tropic viruses). The mean peak intensities of hCM incubated with MAA lectin II (alpha-2,3 specific) and SNA lectin (alpha-2,6 specific) were nearly identical at approximately 2.6 × 104 for both receptors. HPDE6 also had high-level expression of both receptor types, with 3.7 × 104 for SNA and 1.6 × 104 for MAA. MDCK cells were also included as a positive-control line for both receptor types, as these cells are widely used for the isolation of viruses of human and avian origin. Fluorescence-activated cell sorter (FACS) analysis showed that MDCK cells expressed levels of alpha-2,3 receptors similar to those of HPDE6 cells, with the mean peak intensity near 1.8 × 104, while alpha-2,6 expression was equal to that of hCM cells, with mean fluorescence at 2.5 × 104. Therefore, both pancreatic cell lines can be said to express sialic acid receptors at levels comparable to those of MDCK cells, and in the case of hCM cells, expression of the human-virus receptors was even higher (see Fig. S1 in the supplemental material). Pretreatment of all cells with 1 U/ml of NA from Clostridium perfringens resulted in decreased fluorescence for both lectin types, confirming specificity (data not shown).
(iii) Virus replication kinetics in pancreatic cell lines.
hCM and HPDE6 cells were infected with human influenza H1N1 and H3N2 viruses at an MOI of 0.001. Visual examination of the infected cells by light microscopy revealed no cytopathic effect at any time point postinfection on hCM or HPDE6 cells. TCID50 results revealed a continued increase in viral titers in HPDE6 cells over the 72-h course, though the H1N1 viral titers were only slightly higher at 72 h than at 48 h postinfection. In contrast, viral titers reached in hCM cells remained quite similar from 48 to 72 h postinfection for both H1N1 and H3N2 isolates (Fig. 4A and B). An examination of viral-RNA replication by qRRT-PCR showed a continued increase in viral replication up to 72 h postinfection in both cell lines and for both human viruses tested (Fig. 4C and D).
Fig 4.
Human influenza virus replication kinetics in pancreatic cell lines. Shown is the replication kinetics of A/New Caledonia/20/99 (H1N1) and A/Wisconsin/67/2005 (H3N2) in hCM and HPDE6 cells. hCM and HPDE6 cells were infected with each virus at an MOI of 0.001, and at 24, 48, and 72 h postinfection, supernatants from three infected and one mock-infected control well were harvested for virus isolation and qRRT-PCR. (A) Virus isolation results of H1N1 in hCM and HPDE6 cells. (B) Virus isolation results for H3N2 in hCM and HPDE6 cells. (C) qRRT-PCR results for H1N1 in hCM and HPDE6 cells. (D) qRRT-PCR results for H3N2 in hCM and HPDE6 cells. All results represent means plus standard deviations for three independent experiments.
Despite the higher MOI used to perform the infections (MOI = 0.01), both H7N1 and H7N3 avian influenza viruses showed lower levels of replication in both pancreatic cell lines than the human viruses (see Fig. S2 in the supplemental material), with a trend characterized by steady levels of virus RNA up to 48 h p.i. and a decrease for both cell lines at 72 h p.i. Based on the RRT-PCR results, hCM cells appeared to be more sensitive to avian viruses, since the total amount of “M gene” RNA on average was 2 log units higher than in HPDE6 cells (see Fig. S2A and B in the supplemental material). This was also confirmed by TCID50 results (see Fig. S2C and D in the supplemental material), which for both viruses reached higher titers in hCM. In the latter, however, the H7N1 strain exhibited a higher replication efficacy in hCM than in H7N3. This result is not reflected in the RRT-PCR results, in which comparable amounts of viral RNA were detected for both viruses.
No significant differences in viral replication between the two avian viruses were observed in HPDE6 cells.
(iv) Western blot analysis for detection of virus nucleoprotein.
Results of H1N1 and H3N2 influenza virus nucleoprotein expression in the hCM and HPDE6 cell lines are reported in Fig. 5a, b, e, and f. No differences relating to the viral strain or the cell type were shown in the trend of NP expression. As expected, influenza virus nucleoprotein was not observed at t0 (before infection), while it was detected at 24 (t24), 48 (t48), and 72 (t72) hours p.i. for both viruses in hCM and HPDE6 cells. Comparing the bands obtained from samples at t24 to those obtained at t48 and t72, an increase in the NP expression was observable. On the other hand, the amount of β-actin, used as loading control, was at the same level in all the samples tested (Fig. 5c, d, g, and h).
Fig 5.

Western blot analysis for the detection of viral nucleoprotein in pancreatic cell lines. Western blot analysis of H1N1 (a and b) and H3N2 (e and f) influenza virus NP (56 kDa) in hCM and HPDE6 cells. (c, d, g, and h) Samples were collected before infection (t0) and 24 (t24), 48 (t48), and 72 (t72) hours postinfection. β-Actin (42 kDa) was used as a loading control in order to ensure that the same amount of protein was tested for each sample.
(v) Immunofluorescence targeting the NP protein.
Human influenza virus replication was also detected by a fluorescent signal derived from FITC conjugate in hCM cells at 24 h p.i. (see Fig. S3A and B in the supplemental material) for both viruses tested, which increased at 48 and 72 h p.i. No differences were observed between the viral strains tested. The fluorescent signal for both viruses was also observed at 24 h postinfection in HPDE6 cells (see Fig. S3C and D in the supplemental material). In this case also, the number of cells marked continued to increase at 48 and 72 h p.i., demonstrating the enhancement of the nucleoprotein expression over time (data not shown).
(vi) Susceptibility of human pancreatic islets to human influenza A viruses.
The regression model indicated that the CT values for both viruses, in the presence or absence of TPCK-trypsin, tested both in pellets and in supernatant specimens, decreased significantly over time (P < 0.05) (Fig. 6). The statistical analysis showed that the virus titer increased over time independently of the virus subtype and type of sample (pellet or supernatant). Interestingly, CT values for the viruses grown with TPCK-trypsin decreased significantly more than those obtained without the exogenous proteases (P < 0.01) only for H1N1 pellets and supernatant samples (Fig. 6A and C). TPCK-trypsin seemed to enhance H3N2 virus growth, but the difference did not reach statistical significance (P > 0.10) (Fig. 6B and D). The residual postestimation analysis indicated that the model used was appropriate (data not shown). Analysis of data was done using STATA 12.0 statistical software.
Fig 6.
Viral-RNA detection by RRT-PCR of the M gene in human pancreatic islets. Shown are two-way quadratic prediction plots with CIs for RRT-real-time CT values (ct) obtained from H1N1 (A and C) and H3N2 (B and D) (4.8 × 103 PFU/well) pancreatic islet infection. For each virus, the CT trends in pancreatic islet pellets and supernatants from the day of infection (t0) until day 10 (t5) in the presence (left) or absence (middle) of TPCK-trypsin and as an average of the previous two conditions (right) are represented.
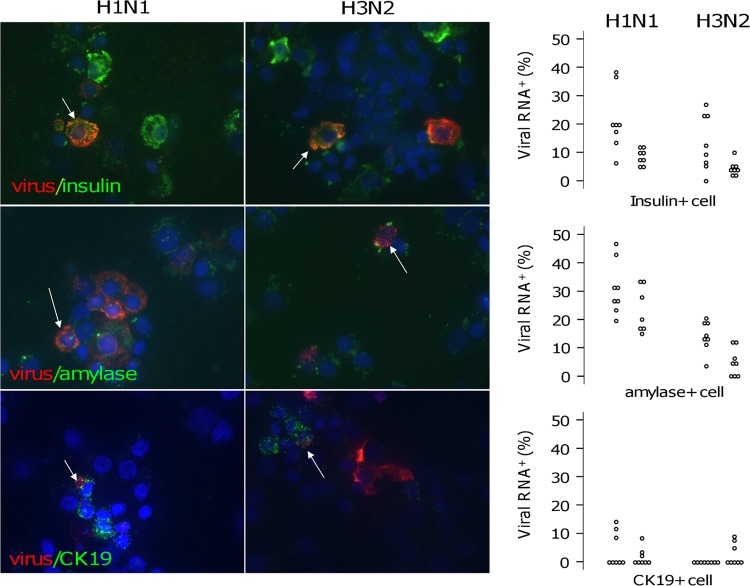
In situ hybridization was performed to visualize viral RNA localized within islet cells. The results clearly demonstrate the presence of viral RNA after both H1N1 and H3N2 virus infection (Fig. 7A). Since human islet primary cultures contain both endocrine and exocrine cells, a fluorescence-based multiplex in situ hybridization strategy was applied to determine which and how many cells were infected in the islets. For this purpose, distinctly labeled probes were combined to analyze viral RNA and insulin, amylase, or cytokeratin in 19 transcripts simultaneously, and after hybridization, human islets were disaggregated and cell positivity was quantified. Five days after infection, 0%, 10.8%, and 4.3% of total cells were positive for viral RNA in mock-, H1N1-, and H3N2-infected islets, respectively (P < 0.001) (Fig. 7B). Of the H1N1-positive cells, 49% ± 27% stained positive for insulin, 26% ± 16% for amylase, and 1.6 ± 2.4% for CK19, and 25% ± 21% were negative for the tested transcripts. Of the H3N2-positive cells, 40% ± 23% stained positive for insulin, 20% ± 20% for amylase, and 2.3 + 5% for CK19, and 41% ± 45% were negative for the tested transcripts (Fig. 7C). On the other hand, of the insulin-positive cells, 14% ± 10% and 8% ± 8% were positive for viral RNA 5 days after H1N1 and H3N2 infection, respectively (P = 0.023). Of the amylase-positive cells, 27% ± 9% and 9% ± 6% were positive for viral RNA after H1N1 and H3N2 infection, respectively (P < 0.001). Of the CK19-positive cells, 3% ± 4% and 1.3% ± 3% were positive for viral RNA after H1N1 and H3N2 infection, respectively (P = 0.36) (Fig. 8).
Fig 7.
Viral-RNA detection by in situ hybridization in human pancreatic islets. Islets were infected with H1N1 and H3N2 by adding 100 μl of viral suspension containing a viral dilution of 4.8 × 103 PFU/well. Mock-infected islets were left as a negative control. (A) Two days after infection, the presence of the viral-RNA molecules was detected on cytoembedded pancreatic islets upon addition of the Fast Red alkaline phosphatase substrate due to the formation of a colored precipitate. Bound viral mRNA was then visualized using either a standard bright-field or a fluorescence microscope (magnification, ×40). Arrows, viral-mRNA-positive cells. (B and C) Five days after infection, multiplex fluorescence-based in situ hybridization was performed, and after disaggregation, islet cells were cytocentrifuged onto glass slides. Viral-RNA-, insulin-, amylase-, and CK19-positive cells were assessed with a Carl Zeiss Axiovert 135TV fluorescence microscope. Quantification was performed using IN Cell Investigator software. Each dot represents the percentage of positive cells quantified on one systematically random field. Results from two experiments are shown. A Mann-Whitney U test was used for statistical analysis.
Fig 8.
Virus RNA and insulin/amylase/CK19 localization. Islets were infected with H1N1 and H3N2 by adding 100 μl of viral suspension containing a viral dilution of 4.8 × 103 PFU/well. Five days after infection, multiplex fluorescence-based in situ hybridization was performed using the Quantigene ViewRNA technique, based on multiple oligonucleotide probes and branched-DNA signal amplification technology, according to the manufacturer's instructions. (Left) The red signal corresponds to the presence of influenza virus RNA, and the green signal corresponds to the presence of insulin, amylase, or CK18 transcripts (magnification, ×63). White arrows, double-positive cells. (Right) Viral-RNA-, insulin-, amylase-, and CK19-positive cells were assessed with a Carl Zeiss Axiovert 135TV fluorescence microscope. Quantification was performed using IN Cell Investigator software. Each dot represents the percentage of positive cells quantified on one systematically random field. Results from two experiments are shown.
(vii) Modulation of survival, insulin secretion, and innate immunity in human pancreatic islets infected with human influenza A viruses in vitro.
Visual examination of the infected islets by light microscopy and LIVE/DEAD assay revealed no significant cytopathic effect at any time point postinfection (days 0 to 7). Five days after infection, uninfected cells showed an overall mortality of 3.26%, H3N2-infected cells 5.21%, and H1N1-infected cells 7.38% (P, nonsignificant versus mock-infected cells) (see Fig. S4A in the supplemental material). Moreover, exposure of islets to either H1N1 or H3N2 virus did not affect their ability to respond to high glucose, as tested in a static perfusion system (see Fig. S4B in the supplemental material).
The capability of H1N1 and H3N2 viruses to induce cytokine/chemokine expression in human pancreatic islets was shown by detectable expression of all but three (interleukin 1b [IL-1b], IL-5, and IL-7) of the cytokines tested. In mock-infected cells, the highest concentrations were detected for CCL2/MCP1 (maximum, 25,558 pg/ml, day 4), ICAM-1 (maximum, 14,063, day 10), CXCL8/IL-8 (maximum, 11,635 pg/ml, day 10); IL-6 (8,452 pg/ml, day 4), CXCL1/GRO-α (maximum, 8,581 pg/ml, day 4), VCAM-1 (maximum, 5,566 pg/ml, day 6), VEGF (maximum, 3,225 pg/ml, day 10), SCGF-b (maximum, 1,427 pg/ml, day 6), hepatocyte growth factor (HGF) (maximum, 1,195 pg/ml, day 6). MIF (maximum, 806 pg/ml, day 6), granulocyte colony-stimulating factor (G-CSF) (maximum, 794 pg/ml day 6), CXCL9/MIG (maximum, 448 pg/ml, day 6), granulocyte-macrophage colony-stimulating factor (GM-CSF) (maximum, 280 pg/ml, day 4), IL-2Ra (maximum, 230 pg/ml, day 6), IL-12p40 (maximum, 215 pg/ml, day 6), macrophage colony-stimulating factor (M-CSF) (maximum, 212 pg/ml, day 10), LIF (maximum, 185 pg/ml, day 6), and CXCL4/SDF1 (maximum, 121 pg/ml, day 6) showed lower but consistent expression. CXCL10/IP-10, PDGF-BB, IL-1Ra, IL-12p70, CCL11/eotaxin, FGFb, CCL5/RANTES, CCL4/MIP-1β, CCL7/MCP-3, IL-3, IL-16, SCF, TRAIL, alpha 2 interferon (IFN-α2), IFN-γ, and CCL27/CTAK showed low but consistent expression (maximum, between 10 and 100 pg/ml). Very low (maximum, <10 pg/ml) but detectable expression was present for IL-2, IL-4, IL-9, IL-10, IL-13, IL-15, CCL3/MIP-1α, tumor necrosis factor alpha (TNF-α), IL-17, IL-18, IL-1a, β-NGF, and TNF-β. Two inflammatory cytokines (IL-6 and TNF-α) and six inflammatory chemokines (CXCL8/IL-8, CXCL1/GRO-α, CXCL9/MIG, CXCL10/IP-10, CCL5/RANTES, and CCL4/MIP-1β) showed over 5-fold increase in influenza virus-infected cell supernatants compared to mock-infected controls (Fig. 9A). Among these, the IFN-γ-inducible chemokines CXCL9/MIG and CXCL10/IP-10 showed the strongest response to H1N1 or H3N2 infection (over 100-fold increase). Both peaked 6 to 8 days p.i. and showed a stronger response to higher doses of the viruses (Fig. 9B).
Fig 9.
Cytokine/chemokine expression profile modification induced by human influenza A virus infection. Islets were infected with H1N1 and H3N2 by adding 100 μl of viral suspension containing two viral dilutions of 4.8 × 103 or 4.8 × 102 PFU/well. Mock-infected islets were left as a negative control. Samples were collected every 48 h from the day of infection (t0) until day 10 (t10). The supernatant was collected and assayed for 50 cytokines. (A) Virus-induced modification in the islet cytokine/chemokine profile. The data are expressed as the maximum fold increase for each factor detected during culture in relation to mock-infected islets (n = 2). Dashed line, 5-fold-increase threshold. The error bars indicate standard deviations. Red items are factors with a >5-fold increased threshold. (B) IFN-γ-inducible chemokine CXCL9/MIG and CXCL10/IP-10 concentrations during 10 days of culture in the presence (+) or absence (−) of H1N1 and H3N2.
DISCUSSION
The objective of our work was to assess IAV replication in pancreatic cells and to evaluate its consequences both at the cellular level in vitro and at the tissue level in vivo. In fact, despite previous reports on a potential role of IAV in pancreatic damage in both animals and humans (2–4, 19–23), to date, there has been no attempt to establish whether IAVs are able to grow in pancreatic cells, and no data are available on the consequences of IAV replication in the pancreas. Our studies have generated novel in vitro data indicating that human influenza A viruses are able to grow in human pancreatic primary cells and cell lines. The addition of exogenous trypsin appears to enhance but not to be essential for viral replication in human pancreatic islets. In vitro studies are corroborated and become of greater significance if combined with animal studies, in which two nonsystemic strains of IAVs were able to colonize the pancreases of experimentally infected young turkeys and to cause metabolic consequences reflecting endocrine and exocrine damage.
Colonization of the pancreas by IAV has been reported following a number of natural and experimental infections of animals, primarily in birds undergoing both systemic and nonsystemic infection. To date, there is no direct evidence of influenza virus infection of the pancreas in humans; however, several reports yield indirect evidence of its occurrence. Here, we have for the first time demonstrated that two nonsystemic avian influenza viruses cause severe pancreatitis resulting in a dysmetabolic condition comparable with diabetes as it occurs in birds. Literature is available on the clinical implications of endocrine and exocrine dysfunctions of the pancreas in birds, including poultry. With reference to endocrine function, several studies indicate that with a total pancreatectomy, birds suffer severe hypoglycemic crisis leading to death (30). If a residual portion of the pancreas as small as 1% of the pancreatic mass is left in situ, a transient (or reversible) hyperglycemic condition is observed in granivorous birds, in which normal glycemia is reestablished within a couple of weeks (31, 32). This indicates that the pancreatic tissue of birds has significant compensatory potential and is also influenced by the fact that there is evidence of the presence of some endocrine tissue able to secrete insulin outside the pancreas (33). Insulin is the dominant hormone in well-fed birds, while glucagon is the dominant hormone in fasting birds. In our experiment, which was carried out with food ad libitum, damage to the endocrine component of the pancreas would most likely manifest itself as hyperglycemia.
With reference to the exocrine function, pancreatitis in birds is characterized by malaise, reluctance to feed, enteritis, and depression. Intra vitam investigations have been based on increased hematic lipase concentration (32). In our study, pancreatitis was evaluated both in vivo, by measuring the lipase concentration in the blood, and postmortem, by histopathologic examination of pancreases collected at different time points. As it occurs in mammals, pancreatic damage results in a rapid increase of the hematic lipase levels that is transient, and values returned to normal by day 15 p.i. Interestingly, the birds that had shown increased lipase levels in the blood and thus supposedly the most severe pancreatic damage exhibited high blood glucose levels in subsequent days, which in only a few cases persisted until the termination of the experiment. This is in keeping with the clinical and metabolic presentation of diabetes in birds. The histological investigations clearly demonstrated viral replication in the exocrine portion of the pancreas, resulting in fibrosis and disruption of the architecture of the organ. While it is clear that both isolates under study replicated extensively in the acinar component of the pancreas, we were unable to determine whether viral replication also occurred in the islets. In any case, it is reasonable to believe that viral infection resulted in severe acute pancreatitis, which impaired both the endocrine and exocrine functions.
Current knowledge of influenza virus replication indicates that influenza viruses that do not exhibit a multibasic cleavage site on the hemagglutinin (HA) protein do not become systemic. However, in our in vivo experiments, the virus reached the pancreas, and we detected viral RNA on day 3 p.i. in the blood of 2/20 (group A, H7N1) and 4/20 (group B, H7N3) infected turkeys. It is therefore possible that following replication in target organs, such as the lung and the gut, in some individuals, a small amount of virus reaches the bloodstream and thus the pancreas. Although the detected CT values indicate low levels of viral RNA, this often resulted in the development of pancreatitis (detected in vivo by hyperlipasemia). This, in turn, in our experimental model, has resulted in a hyperglycemic condition, in keeping with the presentation of diabetes in granivorous birds.
The results of our in vitro experiments show that all IAVs tested, both of avian (H7N1A/turkey/Italy/3675/1999 and H7N3 A/turkey/Italy/2962/2003) and of human (H1N1 Caledonia/20/99 and H3N2 A/Wisconsin/67/2005) origin, are able to grow in established pancreatic cell lines and that human viruses also grow in pancreatic islets. Viral replication occurs in cells of both endocrine and exocrine origin. Our investigations also show that both alpha-2,3 and alpha-2,6 receptors are present in pancreatic cells, indicating that both human and avian influenza viruses could find suitable receptors in the organ. The human viruses used in this study did not induce significant mortality of islet cells, and insulin secretion did not appear to be affected by infection in this system. Whether other strains, such as H1N1pdm or viruses of high pathogenicity, may cause more significant damage remains to be established. On the other hand, it was clear from the cytokine expression profile that IAV infection is able to induce a strong proinflammatory response in human pancreatic islets. The IFN-γ-inducible chemokines MIG/CXCL9 and IP-10/CXCL10 showed the highest increase after infection. Large amounts of RANTES/CCL5, MIP1b/CCL4, Groα/CXCL1, IL-8/CXCL8, TNF-α, and IL-6 were also released.
Of interest, many of these factors are described as key mediators in the pathogenesis of type 1 diabetes (34). Recently IP10/CXCL10 was identified as the dominant chemokine expressed in vivo in the islet environment of prediabetic animals and type 1 diabetic patients, whereas RANTES/CCL5 and MIG/CXCL9 proteins were present at lower levels in the islets of both species (35). The chemokine IP-10/CXCL10 attracts monocytes, T lymphocytes, and natural killer (NK) cells, and islet-specific expression of CXCL10 in a mouse model of autoimmune diabetes caused by viruses (rat insulin promoter [RIP]-lymphocytic choriomeningitis virus [LCMV]) accelerates autoimmunity by enhancing the migration of antigen-specific lymphocytes (36). This is in line with other findings, in which neutralization of IP-10/CXCL10 (37) or its receptor (CXCR3) (38) prevents autoimmune disease in the same mouse model (RIP-LCMV). Studies in nonobese diabetic (NOD) mice have demonstrated elevated expression of IP-10/CXCL10 mRNA and/or protein in pancreatic islets during the prediabetic stage (39). Increased levels of MIP1b/CCL4 and IP-10/CXCL10 are present in the sera of patients who have recently been diagnosed as having type 1 diabetes mellitus (T1D) (40–42).
We speculate that if influenza virus finds its way to the pancreas, either through viremia, as detected in human patients (43–45), or through reflux from the gut through the pancreatic duct, the virus would find a permissive environment. Here, the virus would find appropriate cell receptors and susceptible cells belonging to both the endocrine and exocrine components of the organ. Viral replication would result in cell damage due to the activation of a cytokine storm similar to the one associated with various conditions linked to diabetes. Thus, we believe that influenza virus infections should be investigated as potential agents of pancreatic damage as reported previously (19, 20), resulting in acute pancreatitis and the onset of type 1 diabetes (21–23). The rapid worldwide increase in the incidence of T1D suggests a major role for environmental factors in the etiology. According to cross-sectional and prospective studies on T1D patients and/or prediabetic individuals, virus infections may be one of these. Possible viral involvement in the etiology of T1D has been suggested by several authors. Viruses associated with human T1D include enteroviruses, such as coxsackievirus B (CVB) (46–48), but also measles virus, congenital rubella virus, mumps virus, cytomegalovirus (49–51), and influenza B virus (52). Rotavirus and reovirus have additionally been shown to be diabetogenic in mice (49, 53).
In conclusion, our findings suggest that the pancreas could be a potential target of IAV infection, and thus, we propose that IAV could play a role in the etiopathogenesis of pancreatitis and diabetes in humans and perhaps in other mammals. Clearly, a thorough analysis of existing clinical data and prospective collection of human samples will be needed to fully address this issue. In the meantime, a mammalian preclinical model should be tested to evaluate the role of IAV infection directly or in the context of other immune-mediated conditions in etiopathogenesis of islet damage and beta cell autoimmunity leading to T1D. Similarly, IAV infection should be ruled out when clinical cases with symptoms of endocrine or exocrine pancreatic damage of unknown etiology occur.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge H. L. Shivaprasad (University of California, Davis) for his support with histopathology and immunohistochemistry and Marzia Mancin (IZSVe, Legnaro, Padova, Italy) for her support with statistical analysis of data.
Preliminary studies were funded through the EU FP6 project “Training and Technology Transfer of Avian Influenza Diagnostics and Disease Management Skills” (FLUTRAIN) (project no. 044212).
Footnotes
Published ahead of print 24 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00714-12.
REFERENCES
- 1. Capua I, Alexander DJ. 2009. Avian influenza and Newcastle Disease, a field and laboratory manual. Springer, Berlin, Germany [Google Scholar]
- 2. Harder TC, Vahlenkamp TW. 2010. Influenza virus infections in dogs and cats. Vet. Immunol. Immunopathol. 134:54–60 [DOI] [PubMed] [Google Scholar]
- 3. Kuiken T, Rimmelzwaan G, van Riel D, van Amerongen G, Baars M, Fouchier R, Osterhaus A. 2004. Avian H5N1 influenza in cats. Science 306:241. [DOI] [PubMed] [Google Scholar]
- 4. Rimmelzwaan GF, van Riel D, Baars M, Bestebroer TM, van Amerongen G, Fouchier RA, Osterhaus AD, Kuiken T. 2006. Influenza A virus (H5N1) infection in cats causes systemic disease with potential novel routes of virus spread within and between hosts. Am. J. Pathol. 168:176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abolnik C, Londt BZ, Manvell RJ, Shell W, Banks J, Gerdes GH, Akol G, Brown IH. 2009. Characterisation of a highly pathogenic influenza A virus of subtype H5N2 isolated from ostriches in South Africa in 2004. Influenza Other Respi. Viruses 3:63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bertran K, Perez-Ramirez E, Busquets N, Dolz R, Ramis A, Darji A, Abad FX, Valle R, Chaves A, Vergara-Alert J, Barral M, Hofle U, Majo N. 2011. Pathogenesis and transmissibility of highly (H7N1) and low (H7N9) pathogenic avian influenza virus infection in red-legged partridge (Alectoris rufa). Vet. Res. 42:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tanimura N, Tsukamoto K, Okamatsu M, Mase M, Imada T, Nakamura K, Kubo M, Yamaguchi S, Irishio W, Hayashi M, Nakai T, Yamauchi A, Nishimura M, Imai K. 2006. Pathology of fatal highly pathogenic H5N1 avian influenza virus infection in large-billed crows (Corvus macrorhynchos) during the 2004 outbreak in Japan. Vet. Pathol. 43:500–509 [DOI] [PubMed] [Google Scholar]
- 8. Teifke JP, Klopfleisch R, Globig A, Starick E, Hoffmann B, Wolf PU, Beer M, Mettenleiter TC, Harder TC. 2007. Pathology of natural infections by H5N1 highly pathogenic avian influenza virus in mute (Cygnus olor) and whooper (Cygnus cygnus) swans. Vet. Pathol. 44:137–143 [DOI] [PubMed] [Google Scholar]
- 9. Condobery PK, Slemons RD. 1992. Biological properties of waterfowl-origin type A influenza viruses in chickens. Avian Dis. 36:17–23 [PubMed] [Google Scholar]
- 10. Morales AC, Jr, Hilt DA, Williams SM, Pantin-Jackwood MJ, Suarez DL, Spackman E, Stallknecht DE, Jackwood MW. 2009. Biologic characterization of H4, H6, and H9 type low pathogenicity avian influenza viruses from wild birds in chickens and turkeys. Avian Dis. 53:552–562 [DOI] [PubMed] [Google Scholar]
- 11. Mutinelli F, Capua I, Terregino C, Cattoli G. 2003. Clinical, gross, and microscopic findings in different avian species naturally infected during the H7N1 low- and high-pathogenicity avian influenza epidemics in Italy during 1999 and 2000. Avian Dis. 47:844–848 [DOI] [PubMed] [Google Scholar]
- 12. Okamatsu M, Saito T, Yamamoto Y, Mase M, Tsuduku S, Nakamura K, Tsukamoto K, Yamaguchi S. 2007. Low pathogenicity H5N2 avian influenza outbreak in Japan during the 2005–2006. Vet. Microbiol. 124:35–46 [DOI] [PubMed] [Google Scholar]
- 13. Shinya K, Awakura T, Shimada A, Silvano FD, Umemura T, Otsuki K. 1995. Pathogenesis of pancreatic atrophy by avian influenza a virus infection. Avian Pathol. 24:623–632 [DOI] [PubMed] [Google Scholar]
- 14. Zanella A. 2003. Avian influenza attributable to serovar H7N1 in light layers in Italy. Avian Dis. 47:1177–1180 [DOI] [PubMed] [Google Scholar]
- 15. Klopfleisch R, Wolf PU, Wolf C, Harder T, Starick E, Niebuhr M, Mettenleiter TC, Teifke JP. 2007. Encephalitis in a stone marten (Martes foina) after natural infection with highly pathogenic avian influenza virus subtype H5N1. J. Comp. Pathol. 137:155–159 [DOI] [PubMed] [Google Scholar]
- 16. Lipatov AS, Kwon YK, Pantin-Jackwood MJ, Swayne DE. 2009. Pathogenesis of H5N1 influenza virus infections in mice and ferret models differs according to respiratory tract or digestive system exposure. J. Infect. Dis. 199:717–725 [DOI] [PubMed] [Google Scholar]
- 17. Thiry E, Zicola A, Addie D, Egberink H, Hartmann K, Lutz H, Poulet H, Horzinek MC. 2007. Highly pathogenic avian influenza H5N1 virus in cats and other carnivores. Vet. Microbiol. 122:25–31 [DOI] [PubMed] [Google Scholar]
- 18. Yingst SL, Saad MD, Felt SA. 2006. Qinghai-like H5N1 from domestic cats, northern Iraq. Emerg. Infect. Dis. 12:1295–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blum A, Podvitzky O, Shalabi R, Simsolo C. 2010. Acute pancreatitis may be caused by H1N1 influenza A virus infection. Isr. Med. Assoc. J. 12:640–641 [PubMed] [Google Scholar]
- 20. Calore EE, Uip DE, Perez NM. 2011. Pathology of the swine-origin influenza A (H1N1) flu. Pathol. Res. Pract. 207:86–90 [DOI] [PubMed] [Google Scholar]
- 21. Cano M, Iglesias P, Perez G, Diez JJ. 2010. Influenza A virus (H1N1) infection as a cause of severe diabetic ketoacidosis in type 1 diabetes. Endocrinol. Nutr. 57:37–38 [DOI] [PubMed] [Google Scholar]
- 22. Nenna R, Papoff P, Moretti C, Pierangeli A, Sabatino G, Costantino F, Soscia F, Cangiano G, Ferro V, Mennini M, Salvadei S, Scagnolari C, Antonelli G, Midulla F. 2011. Detection of respiratory viruses in the 2009 winter season in Rome: 2009 influenza A (H1N1) complications in children and concomitant type 1 diabetes onset. Int. J. Immunopathol. Pharmacol. 24:651–659 [DOI] [PubMed] [Google Scholar]
- 23. Watanabe N. 2011. Conversion to type 1 diabetes after H1N1 influenza infection: a case report. J. Diabetes 3:103. [DOI] [PubMed] [Google Scholar]
- 24. European Union 2006. Community measures for the control of avian influenza and repealing directive 92/40/EEC. Council directive 2005/94/EC of 20 December 2005. Official J. Eur. Union Legis. L 49:16–65 [Google Scholar]
- 25. Spackman E, Senne DA, Myers TJ, Bulaga LL, Garber LP, Perdue ML, Lohman K, Daum LT, Suarez DL. 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 40:3256–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baroni MG, Cavallo MG, Mark M, Monetini L, Stoehrer B, Pozzilli P. 1999. Beta-cell gene expression and functional characterisation of the human insulinoma cell line CM. J. Endocrinol. 161:59–68 [DOI] [PubMed] [Google Scholar]
- 27. Ouyang H, Mou L, Luk C, Liu N, Karaskova J, Squire J, Tsao MS. 2000. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am. J. Pathol. 157:1623–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matrosovich M, Matrosovich T, Garten W, Klenk HD. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wakeley PR, Johnson N, McElhinney LM, Marston D, Sawyer J, Fooks AR. 2006. Development of a real-time, differential RT-PCR TaqMan assay for lyssavirus genotypes 1, 5 and 6. Dev. Biol. (Basel) 126:227–236, 326–327 [PubMed] [Google Scholar]
- 30. Hazelwood RL. 2000. Pancreas, p 539–555 In Whittow GC. (ed), Sturkie's avian physiology, 5th ed Academic Press, San Diego, CA [Google Scholar]
- 31. Laurent F, Mialhe P. 1978. Effect of free fatty acids and amino acids on glucagon and insulin secretions in normal and diabetic ducks. Diabetologia 15:313–321 [DOI] [PubMed] [Google Scholar]
- 32. Hoffmann WE, Solter PF. 2008. Diagnostic enzymology of domestic animals, p 365–366 In Kaneko J, Harvey JW, Bruss ML. (ed), Clinical biochemistry of domestic animals, 6th ed Elsevier Inc., New York, NY [Google Scholar]
- 33. Colca JR, Hazelwood RL. 1982. Persistence of immunoreactive insulin, glucagon and pancreatic polypeptide in the plasma of depancreatized chickens. J. Endocrinol. 92:317–326 [DOI] [PubMed] [Google Scholar]
- 34. Eizirik DL, Colli ML, Ortis F. 2009. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat. Rev. Endocrinol. 5:219–226 [DOI] [PubMed] [Google Scholar]
- 35. Sarkar SA, Lee CE, Victorino F, Nguyen TT, Walters JA, Burrack A, Eberlein J, Hildemann SK, Homann D. 2012. Expression and regulation of chemokines in murine and human type 1 diabetes. Diabetes 61:436–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rhode A, Pauza ME, Barral AM, Rodrigo E, Oldstone MB, von Herrath MG, Christen U. 2005. Islet-specific expression of CXCL10 causes spontaneous islet infiltration and accelerates diabetes development. J. Immunol. 175:3516–3524 [DOI] [PubMed] [Google Scholar]
- 37. Christen U, McGavern DB, Luster AD, von Herrath MG, Oldstone MB. 2003. Among CXCR3 chemokines, IFN-gamma-inducible protein of 10 kDa (CXC chemokine ligand (CXCL) 10) but not monokine induced by IFN-gamma (CXCL9) imprints a pattern for the subsequent development of autoimmune disease. J. Immunol. 171:6838–6845 [DOI] [PubMed] [Google Scholar]
- 38. Frigerio S, Junt T, Lu B, Gerard C, Zumsteg U, Hollander GA, Piali L. 2002. Beta cells are responsible for CXCR3-mediated T-cell infiltration in insulitis. Nat. Med. 8:1414–1420 [DOI] [PubMed] [Google Scholar]
- 39. Cardozo AK, Proost P, Gysemans C, Chen MC, Mathieu C, Eizirik DL. 2003. IL-1beta and IFN-gamma induce the expression of diverse chemokines and IL-15 in human and rat pancreatic islet cells, and in islets from pre-diabetic NOD mice. Diabetologia 46:255–266 [DOI] [PubMed] [Google Scholar]
- 40. Hanifi-Moghaddam P, Kappler S, Seissler J, Muller-Scholze S, Martin S, Roep BO, Strassburger K, Kolb H, Schloot NC. 2006. Altered chemokine levels in individuals at risk of type 1 diabetes mellitus. Diabet. Med. 23:156–163 [DOI] [PubMed] [Google Scholar]
- 41. Nicoletti F, Conget I, Di Mauro M, Di Marco R, Mazzarino MC, Bendtzen K, Messina A, Gomis R. 2002. Serum concentrations of the interferon-gamma-inducible chemokine IP-10/CXCL10 are augmented in both newly diagnosed type I diabetes mellitus patients and subjects at risk of developing the disease. Diabetologia 45:1107–1110 [DOI] [PubMed] [Google Scholar]
- 42. Shimada A, Morimoto J, Kodama K, Suzuki R, Oikawa Y, Funae O, Kasuga A, Saruta T, Narumi S. 2001. Elevated serum IP-10 levels observed in type 1 diabetes. Diabetes Care 24:510–515 [DOI] [PubMed] [Google Scholar]
- 43. Likos AM, Kelvin DJ, Cameron CM, Rowe T, Kuehnert MJ, Norris PJ, National Heart Lung Blood Institute Retrovirus Epidemiology Donor Study-II (REDS-II) 2007. Influenza viremia and the potential for blood-borne transmission. Transfusion 47:1080–1088 [DOI] [PubMed] [Google Scholar]
- 44. Oughton M, Dascal A, Laporta D, Charest H, Afilalo M, Miller M. 2011. Evidence of viremia in 2 cases of severe pandemic influenza A H1N1/09. Diagn. Microbiol. Infect. Dis. 70:213–217 [DOI] [PubMed] [Google Scholar]
- 45. Tse H, To KK, Wen X, Chen H, Chan KH, Tsoi HW, Li IW, Yuen KY. 2011. Clinical and virological factors associated with viremia in pandemic influenza A/H1N1/2009 virus infection. PLoS One 6:e22534 doi:10.1371/journal.pone.0022534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goldberg E, Krause I. 2009. Infection and type 1 diabetes mellitus—a two edged sword? Autoimmun. Rev. 8:682–686 [DOI] [PubMed] [Google Scholar]
- 47. Pino SC, Kruger AJ, Bortell R. 2010. The role of innate immune pathways in type 1 diabetes pathogenesis. Curr. Opin. Endocrinol. Diabetes Obes. 17:126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Richer MJ, Horwitz MS. 2009. Coxsackievirus infection as an environmental factor in the etiology of type 1 diabetes. Autoimmun. Rev. 8:611–615 [DOI] [PubMed] [Google Scholar]
- 49. Jun HS, Yoon JW. 2003. A new look at viruses in type 1 diabetes. Diabetes Metab. Res. Rev. 19:8–31 [DOI] [PubMed] [Google Scholar]
- 50. Richer MJ, Horwitz MS. 2008. Viral infections in the pathogenesis of autoimmune diseases: focus on type 1 diabetes. Front. Biosci. 13:4241–4257 [DOI] [PubMed] [Google Scholar]
- 51. von Herrath MG, Holz A, Homann D, Oldstone MB. 1998. Role of viruses in type I diabetes. Semin. Immunol. 10:87–100 [DOI] [PubMed] [Google Scholar]
- 52. Sano H, Terasaki J, Tsutsumi C, Imagawa A, Hanafusa T. 2008. A case of fulminant type 1 diabetes mellitus after influenza B infection. Diabetes Res. Clin. Pract. 79:e8–e9 doi:10.1016/j.diabres.2007.10.030 [DOI] [PubMed] [Google Scholar]
- 53. Graham KL, Sanders N, Tan Y, Allison J, Kay TW, Coulson BS. 2008. Rotavirus infection accelerates type 1 diabetes in mice with established insulitis. J. Virol. 82:6139–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.