Abstract
Although precluded from using splicing to produce multiple small Rep proteins, adeno-associated virus type 5 (AAV5) generates a Rep40-like protein by alternative translation initiation at an internal AUG. A defined region upstream of the internal AUG was both required and sufficient to program internal initiation within AAV5 and may act similarly in heterologous contexts. The internally initiated AAV5 Rep40-like protein was functional and had helicase activity similar to that of AAV2 Rep40. Surprisingly, both the AAV5 Rep40-like protein and Rep52 were able to be translated from the AAV5 upstream P7-generated RNAs; however, the relative level of small to large Rep proteins was reduced compared to that of the wild type. A P19 mutant AAV5 infectious clone generated near-wild-type levels of the double-stranded monomer replicative form (mRF) replicative intermediate but reduced levels of virus, consistent with the previously defined role of Rep40-like proteins in genome encapsidation. Levels of mutant virus were dramatically reduced upon amplification.
INTRODUCTION
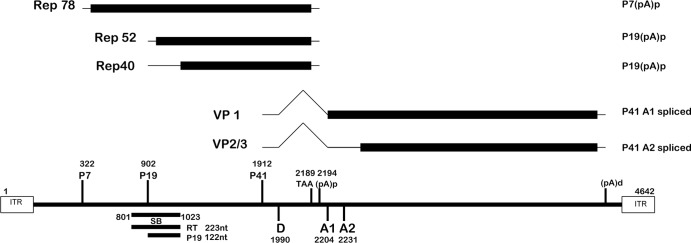
The transcription maps of adeno-associated virus type 5 (AAV5) and many of the animal AAVs are quite different from those of AAV2 and other human AAVs. While all AAV2 RNAs extend to a polyadenylation site near the right-hand end of the genome and are exported to the cytoplasm as both spliced and unspliced species, AAV5 RNAs generated by the viral P7 and P19 promoters are predominately polyadenylated at a site within the central intron, and thus, splicing is precluded (1, 2) (Fig. 1). AAV2 encodes two versions of its large (Rep78 and Rep68) and small (Rep52 and Rep40) Rep proteins from unspliced and spliced P5- and P19-generated RNAs, respectively (3–6). Because AAV5 P7 and P19 RNAs are not spliced, they were predicted to encode only Rep78 and Rep52 (Fig. 1); however, we have shown that while an AAV5 Rep68 protein was absent, as expected, AAV5 encodes an abundant Rep40-like protein from an in-frame internal initiation AUG 150 nucleotides (nt) downstream of the Rep52 initiator (7). That AAV5 uses another genetic mechanism to generate a Rep40-like protein is consistent with it playing an essential role during infection.
Fig 1.
Transcription profile of AAV5. The AAV5 genome (nt 1 to 4642) is shown, depicting the promoters (P7, P19, P41), the central intron (nt 1990 to 2204/2231), and the polyadenylation sites [the proximal polyadenylation site (pA)p, utilized by P7 and P19 transcripts, and the distal site (pA)d, utilized by P41 transcripts]. The open reading frames of the replication proteins (Rep78, Rep52, and Rep40-like protein) and capsid proteins (VP1, VP2/VP3) are indicated. The previously described (23) SB probe (nt 801 to 1023) utilized for RNase protection assays and the potential P7-derived read-through (223-nt) and smaller, P19-initiated read-through (122-nt) products are also depicted.
Work from a number of labs has demonstrated that the small replication proteins of AAV2 are essential for packaging the genome into the viral capsids (8–10). This is likely to be dependent upon their potent 3′ to 5′ helicase activity (10, 11). All AAV Rep proteins have a central helicase domain characterized by a 100-amino-acid stretch of residues containing Walker motifs A, B, B′, and C (12, 13). AAV Rep proteins are classified as SF3 helicases (11, 14), which also include a number of other viral proteins involved in DNA replication and packaging (14–17). The AAV2 Rep78 and Rep68 proteins have been shown to exist as hexamers in solution in the presence of double-stranded DNA, similar to other SF3 helicases (18–21). AAV2 Rep40 is a bimodular protein with a small helical bundle at the amino terminus and a large α/β domain at the C terminus (9, 11); there is little evidence to suggest that it exists other than as a monomer in solution (22). Interestingly, the AAV5 Rep40-like protein lacks the helical bundle at the amino terminus present in AAV2 Rep40 (7, 9).
Like AAV5, AAV2 contains both a similarly positioned internal polyadenylation site and an in-frame AUG downstream of its Rep52 initiating codon (23); however, neither the internal polyadenylation signal nor the internal AUG is used in the AAV2 context. In this article, we have demonstrated that the region between the two AAV5 small-Rep initiation sites was required and sufficient to both stimulate its usage within AAV5 and program internal initiation in heterologous systems. We have shown that the AAV5 Rep40-like protein, which has architecture different from that of its AAV2 counterpart, is functional and retains helicase activity. Surprisingly, we also observed that all three AAV5 Rep proteins can be encoded by AAV5 P7-generated mRNAs. An AAV5 P19 mutant infectious clone was found to replicate and generate virus even when the P19 promoter was dramatically debilitated; however, the ratio of small to large Rep proteins was considerably less than that produced from P19-replete wild-type virus. Virus production was concomitantly reduced and found to be reduced substantially further during amplification, consistent with a required role of the small Rep proteins in genome encapsidation.
MATERIALS AND METHODS
Cells and viruses/infections, titering, and transfections.
293 and 293T cells were propagated as previously described (24) in Dulbecco's modified Eagle medium (DMEM) with 5% fetal calf serum. Transfections were done using either Lipofection Plus (Invitrogen, CA) or LipoD293 (SignaGen, Rockville, MD). Cell-associated virus isolated from transfections of wild-type or P19 mutant constructs was assayed for genome content by Southern blot analysis, and equivalent genome copies were used for subsequent infections.
Plasmid construction.
(i) The chimera constructs MVM/5AUG2 and MVM/2AUG2 were generated from the previously described cytomegalovirus (CMV) AAV5 and AAV2 Rep52 expression plasmids (7) by overlapping PCR mutagenesis, which replaced the intervening sequence between the Rep52-initiating AUG and the internal in-frame AUG (which is 150 nt downstream) in both AAV5 and AAV2 with minute virus of mice (MVM) capsid sequences (nt 4201 to 4338). Furthermore, AAV5Cap/5AUG2 and AAV5Cap/2AUG2 chimera constructs were also generated by overlapping PCR mutagenesis, which replaced the intervening sequence between the AUGs with AAV5 capsid sequences (nt 4201 to 4338). These constructs retained the extended −6 to +3 sequences associated with both AUGs.
(ii) 5(1-150)TIA-1, 2(1-150)TIA-1, and 5(1-150)GFP chimera constructs were cloned into pcDNA3.1(+) (Invitrogen, CA) by placing the first 150 nt of AAV5 Rep52, and the analogous sequences from AAV2 Rep52, upstream of either T cell-interacting antigen 1(TIA-1) or green fluorescent protein (GFP).
(iii) The constructs used for the mapping experiments shown in Fig. 2 were generated by site-directed mutagenesis, which resulted in the replacement of the various sequences of AAV2 Rep52 with analogous sequences from AAV5 Rep52, as illustrated in Fig. 2A and B.
Fig 2.
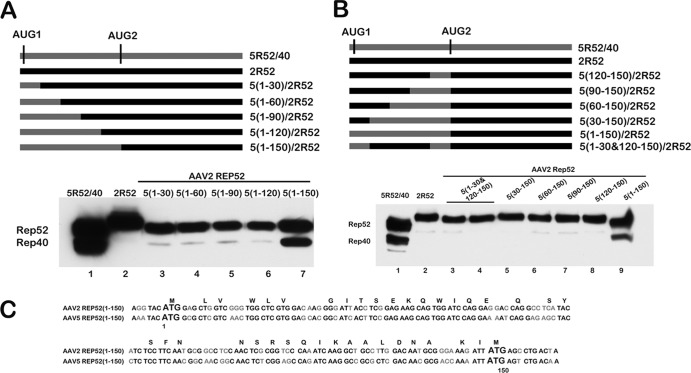
The full 5(1-150) intervening motif was required to enhance internal initiation within the AAV2 background. (A) A schematic of CMV 5R52/40 and CMV AAV5/AAV2 Rep52 chimera constructs [5(1-30)/2R52, 5(1-60)/2R52, 5(1-90)/2R52, 5(1-120)/2R52, and 5(1-150)/2R52)], containing replacements of AAV2 sequences with analogous sequences of the AAV5 5(1-150) intervening motif in the 5′ to 3′ direction, as described in the text, is shown. The positions of the AUGs are indicated as described in the text, while AAV2 sequences and AAV5 sequences are depicted using black and dark gray bars, respectively. Expression of Rep52 and Rep40-like proteins from these constructs following transfection of 293T cells is shown in a Western blot using antibodies against HA (Sigma, St. Louis, MO) below the diagram. (B) A schematic of CMV 5R52/40 and CMV AAV5/AAV2Rep52 chimera constructs [5(120-150)/2R52, 5(90-150)/2R52, 5(60-150)/2R52, 5(30-150)/2R52, and 5(1-30 &120-150)/2R52], containing replacements of AAV2 sequences with analogous sequence from the AAV5 5(1-150) intervening motif in the 3′ to 5′ direction, as described in the text, is shown. The positions of the AUGs are indicated as described in the text, while AAV2 sequences and AAV5 sequences are depicted using black and dark gray bars, respectively. Expression of Rep52 and Rep40-like proteins from these constructs following transfection of 293T cells is shown in a Western blot using antibodies against HA (Sigma, St. Louis, MO) below the diagram. (C) Alignment of AAV5 5(1-150) and AAV2 2(1-150) nucleotide sequences between the two AUGs of the small replication proteins, including an additional 6 nt upstream and 9 nt downstream, is shown. Codons that are translated into the same amino acid residues in both AAV5 and AAV2 are indicated on top of the sequences, while nucleotides that are conserved in both AAV5 and AAV2 are shown in black and dissimilar nucleotides are shown in gray.
(iv) The AAV5 P19 mutant was generated by site-directed mutagenesis that resulted in the mutation of the P19 TATA box from GAGTATAAATTGG to GAGTACAAGTTGG and the P19 initiation site and sequences surrounding it from GAGGAGCGCAAAC to GAAGAAAGAAAGC, with the changes shown in bold.
Immunoblots, antibodies, RNase protections, and Southern blots.
Immunoblots were performed as previously described (24) using antibody against either hemagglutinin (HA) (Sigma, St. Louis, MO) or the AAV5 Rep proteins (ARP, Belmont, MA). RNase protection assays were performed as previously described (25) using the probes indicated in the text. Southern blot analysis of transfected and infected cells was done as previously described (26) using equivalent amounts of total DNA as determined by NanoDrop quantification.
Protein purification/helicase assays.
The AAV5 Rep40-like protein and AAV2 Rep40 were cloned into the pET-28a vector using NdeI and XhoI restriction sites at the 5′ and 3′ ends, respectively. The proteins were expressed in Escherichia coli BL21(DE3)-pLysS cells. Cultures were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 1.5 h at 37°C. The cell pellets were lysed in buffer containing 25 mM Tris-Cl (pH 7.5), 1 mM dithiothreitol (DTT), 0.01% NP-40, 5% glycerol, 250 mM NaCl, and protease inhibitors. The lysates were purified with 1-ml nickel affinity columns and washed with the lysis buffer containing increasing concentrations of imidazole (25 mM, 50 mM, and 100 mM). The proteins were eluted in lysis buffer containing 300 mM imidazole and dialyzed overnight into storage buffer containing 25 mM Tris-Cl (pH.7.5), 50 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.1% NP-40, and 20% glycerol. The proteins were further purified by size exclusion chromatography for optimal purity and stored at −80°C.
Helicase assays were performed with a double-stranded 25-mer DNA oligonucleotide that had a 20-nt 3′ overhang and a Cy3-labeled 5′ end as the substrate. A 1 μM substrate was incubated with 0.75 μM purified AAV5 Rep40-like protein in helicase reaction buffer for 45 min as previously described (9), and the reaction mixture was resolved in a native gel. The result was analyzed on a FLA9000 imager (GE).
RESULTS
The region between the two AAV5 small-Rep AUGs stimulates downstream AUG usage.
As mentioned, AAV2 contains a silent internal AUG at essentially the same position as that which is used in AAV5 to initiate its Rep40-like protein (Fig. 2C). We have previously shown that when the 150-nt region between the two AAV5 small-Rep AUGs (here termed AUG1 and AUG2, respectively) was replaced with the analogous region from AAV2, the downstream AAV5 AUG (AUG2) was not used at detectable levels (7). As can be seen in Fig. 3, when the AAV5 intervening sequence containing both AUGs was used to replace the analogous region of AAV2, we observed that the internal AAV2 AUG was now efficiently used [5(1-150)/2AUG2] (Fig. 3A, lane 5). For these experiments, we utilized a system in which the small rep genes from AAV5 and AAV2 were expressed from a CMV expression vector and were tagged at their carboxyl termini with HA, as previously described (7). Genes within these constructs were expressed similarly to their expression from within full-length viral constructs (7) or from the isolated P19 transcription unit (data not shown). The −6 to +3 extended AUG1 and AUG2 motifs of AAV2 are somewhat different from those of AAV5 (Fig. 2C); however, changing the sequences of the two extended AAV2 AUG motifs to those of AAV5 had no effect on relative levels of initiation (data not shown).
Fig 3.
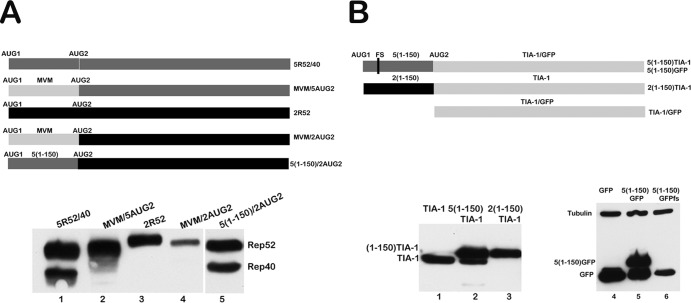
The region between the two AAV5 small-Rep AUGs stimulates downstream AUG usage. (A) A schematic of constructs 5R52/40, MVM/5AUG2, 2R52, MVM/2AUG2, and 5(1-150)/2AUG2, as described in the text, is shown. The positions of the AUGs are indicated, and all constructs were tagged at their C terminus with HA. AAV2 sequences, AAV5 sequences, and MVM heterologous sequences are depicted with black, dark gray, and light gray bars, respectively. A Western blot showing expression of the Rep52 and Rep40-like proteins from these constructs following transfection of 293T cells using antibodies against HA (Sigma, MO) is shown below the diagram. (B) A schematic of the constructs 5(1-150)TIA-1 and 5(1-150)GFP, the position of the frameshift insertion (fs) in 5(1-150)GFP and 2(1-150)TIA-1, and TIA-1 and GFP, as described in the text, is shown. All constructs were tagged at their C terminus with HA. AAV2 sequences, AAV5 sequences, and GFP and TIA-1 heterologous sequences are depicted with black, dark gray, and light gray bars, respectively. Expression of Rep52 and Rep40-like proteins from these constructs following transfection of 293T cells is shown in a Western blot using antibodies against HA (Sigma, St. Louis, MO) below the diagram.
To determine whether the intervening region between the two AAV5 initiating AUGs was itself stimulatory or, alternatively, whether the analogous AAV2 intervening region was inhibitory for internal initiation, we replaced the regions between the two AUGs within the two CMV expression parental constructs (AAV5, 5R52/40; AAV2, 2R52) (Fig. 3A) with heterologous sequences that have no known translational initiating activity. These were taken from either the MVM capsid gene region (Fig. 3A) or capsid gene sequences from AAV5 (data not shown). In these constructs, the extended parental AUG sequences from −6 to +3 were kept intact. Within the normally active AAV5 background, this heterologous substitution prevented detectable internal initiation (Fig. 3A shows the MVM insertion constructs MVM/5AUG2 and parent 5R52/40, diagram at top, compare lane 2 to lane 1), while within the normally silent AAV2 background, heterologous substitution did not allow internal initiation (Fig. 3A shows the MVM insertion constructs MVM/2AUG2 and parent 2R52, diagram at top, compare lane 4 to lanes 3 and 1). These results are consistent with a model in which the AAV5 intervening region between AUG1 and AUG2 provided a stimulatory effect on internal initiation, rather than the AAV2 intervening region being inhibitory.
In further confirmation of this model, the AAV5, but not the AAV2, intervening region was able to program the internal initiation of a heterologous gene (the gene for the RNA processing factor TIA-1) [Fig. 3B, lanes 2 and 3, 5(1-150)TIA-1 and 2(1-150)TIA-1, respectively; see construct diagrams at the top of the figure]. Similarly, the AAV5 motif was able to program the internal initiation of the GFP gene [Fig. 3B, lane 5, 5(1-150)GFP]. Introduction of a frameshift (fs) mutation within the intervening region in the GFP construct prevented production of the fusion protein but did not affect production of the distally initiated smaller protein, confirming that the internal initiation signal was used and that the smaller protein was not a cleavage product of the larger one [Fig. 3B, lane 6, 5(1-150)GFPfs].
The full 5(1-150) intervening motif was required to enhance internal initiation within the AAV2 background.
The differences in function between the AAV5 and AAV2 intervening regions just described suggested a starting point to elucidate the cis-acting signals that governed the usage of the internal initiating AUG. As shown above (Fig. 3A, lane 5), the full AAV5 intervening sequence 5(1-150) was capable of conferring initiation of the internal AUG in both AAV2 and heterologous backgrounds. The level of internal AAV2 AUG usage in the chimeric constructs was similar to that seen for the AAV5 rep gene itself. In additional experiments not shown, the use of an intervening region that had been extended by as many as 60 additional nucleotides downstream [5(1-210)] had no additional effect. To delineate the minimum sequences within the 5(1-150) region that are both necessary and sufficient to confer initiation at the AAV2 internal AUG, we generated a series of chimeras in which increasing amounts of AAV5 intervening sequence were substituted within the AAV2 intervening region in the parent 2R52, increasing by 30 nt in both a 5′ to 3′ (Fig. 2A) and a 3′ to 5′ (Fig. 2B) manner. As can be seen in Fig. 2A, while substitution of the full first 150 nt of AAV5 supported initiation of the AAV2 internal AUG similarly to the full-length AAV5 construct [Fig. 2A, compare lane 7 to lane 1, 5(1-150)/2R52 and 5R52/40, respectively], substitutions in the 5′ to 3′ direction of up to nt 1 to 120 had no detectable effect (Fig. 2A, lanes 3 to 6). This indicated that the 5′-most 30 nt of the motif were required. Similarly, substitutions in the 3′ to 5′ direction up to nt 30 to 150 had no effect (Fig. 2B, lanes 5 to 8), suggesting that the first 30 nt were also required. However, a mutant in which both nt 1 to 30 and nt 120 to 150 of AAV2 were replaced by their AAV5 counterparts remained unable to generate an internally initiated product [Fig. 2B, lanes 3 to 4, 5(1-30 + 120-150)/2R52 (here in duplicate)]. This result demonstrated that although required, together these elements were not sufficient. We conclude that the full 5(1-150) region of AAV5 was both necessary and sufficient to program initiation at internal AUGs.
The AAV5 Rep40-like protein has efficient helicase activity.
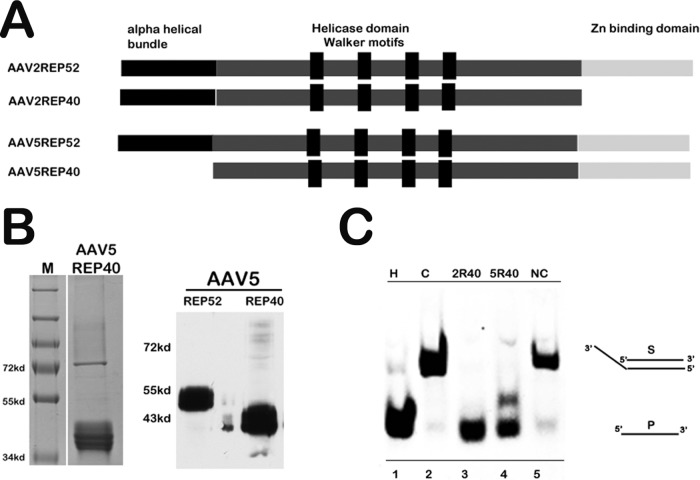
Because its coding sequence is contained totally within Rep52 and Rep78, it is difficult to determine by genetic means whether the AAV5 Rep40-like protein is specifically essential for AAV5 replication: mutations which affect either the Rep40-like protein itself or its expression are also present within the larger proteins. We therefore attempted to determine if the purified AAV5 Rep40-like protein was functional and had helicase activity similar to that of the AAV2 Rep40 protein. Although the AAV5 Rep40-like protein has a configuration different from that of the AAV2 Rep40 protein, lacking the amino terminal α-helical bundle, it retains the Walker box helicase motifs (Fig. 4A). The AAV5 Rep40-like open reading frame (ORF) was cloned into the bacterial expression vector pET28a, expressed in E. coli BL21lysS, and purified by fast protein liquid chromatography (FPLC) (Fig. 4B). AAV2 Rep40 and the xenotropic murine leukemia virus-related virus (XMRL) reverse transcriptase (RT) proteins were similarly cloned and purified for use as positive and negative controls, respectively. As can be seen in Fig. 4C, at the conditions described, the AAV5 Rep40-like protein had 3′ to 5′ helicase activity in in vitro assays that was similar to that of AAV2 Rep40.
Fig 4.
The AAV5 Rep40-like protein has efficient helicase activity. (A) Schematic of the functional motifs of AAV2 and AAV5 small replication proteins. The N-terminal alpha helical bundle, the central helicase domain consisting of Walker motifs A, B, C, and C′, and the C-terminal Zn binding domain are indicated. The AAV5 Rep40-like protein lacks the alpha helical bundle that is present in the N terminus of AAV2 Rep40. The Zn binding domain present in the C terminus of AAV5 small replication proteins (Rep52 and Rep40-like proteins) and AAV2 Rep52 is absent in AAV2 Rep40. (B) Coomassie staining of purified His-tagged AAV5 Rep40-like protein is shown on the left and a Western blot of purified His-tagged AAV5 Rep40-like protein and AAV5 Rep52 using AAV5 Rep antibody (ARP, Belmont, MA) is shown on the right. (C) Helicase assay of the AAV5 Rep40-like protein. The assay was carried out with a Cy3-labeled AAV5 25-mer oligonucleotide containing a 20-nt 3′ overhang, as described in the text. A 1 μM substrate was incubated with 0.75 μM purified AAV5 Rep40-like protein in helicase reaction buffer as previously described (9). AAV2 Rep40 was used as a positive control (2R40; lane 3), and His-tagged XMRV reverse transcriptase was used as a negative control (NC; lane 5). Untreated substrate (C; lane 2), boiled, dissociated substrate (H; lane 1), and substrate incubated with purified AAV5 Rep40-like protein (5R40; lane 4) are shown. The duplex and single-stranded DNA species are indicated.
All AAV5 Rep proteins can be generated from P7-generated RNAs.
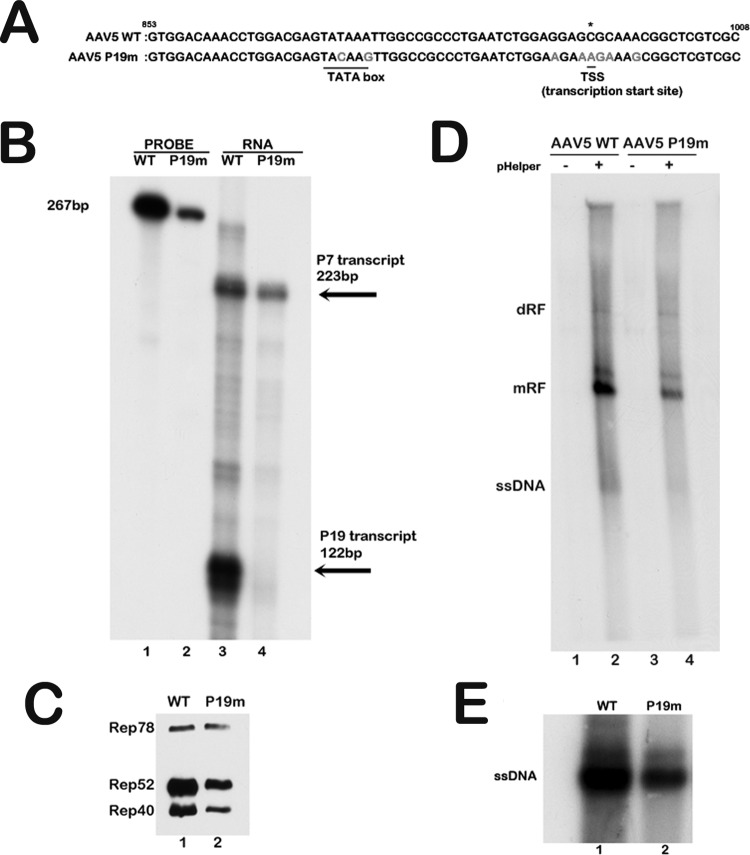
Since the Rep40-like protein was generated by internal initiation, we wondered whether it can also be expressed from the upstream P7-generated RNA. Initially, we debilitated the P19 TATA box and initiator (Inr) region within the AAV5 infectious clone using third-nucleotide mutations which did not alter the Rep78 amino acid sequence (Fig. 5A). Following transfection of 293 cells together with pHelper, this construct generated no detectable P19 transcripts, as assayed by an RNase protection assay (Fig. 5B, compare lanes 3 and 4), yet produced Rep78, Rep52, and the Rep40-like protein. The relative levels of Rep52 and the Rep40-like protein, compared to those for Rep78, were lower for the P19 mutant than those seen following transfection of a wild-type construct (Fig. 5C, compare lanes 1 and 2). This suggested that not only the Rep40-like protein but also Rep52 can be generated by the P7-generated RNA and that production of the small Rep proteins from this mutant cannot reach the levels generated by virus with a functional P19 promoter. The sequences that allow internal initiation of Rep52 from the P7-generated mRNA are currently being identified. The infectious clone bearing the P19 mutation was then assayed for replication. As can be seen in Fig. 5D, following transfection of 293 cells together with pHelper, this clone generated mRF forms efficiently. In our hands, detection of AAV5 single-stranded DNA (ssDNA) in Southern blots of transfected or infected cell extracts has always been variable; thus, the level of virus production from these transfections was directly assayed by Southern blotting. Quantification by genome copy number indicated that approximately 5-fold-less mutant virus was generated compared to the wild type 48 h posttransfection (Fig. 5E, compare lanes 1 and 2). Mutant virus was sequenced to ensure that the mutation that was introduced remained (data not shown).
Fig 5.
The AAV5 Rep40-like and Rep52 proteins can be generated with Rep78 from the P7-generated RNA. (A) Alignment of AAV5 wild-type (WT) and AAV5 P19 knockout mutant (nt 853 to 1008) sequences showing the TATA box and transcription initiation site mutations in light gray. All mutations are silent third-nucleotide changes in the Rep open reading frame. The P19 transcription start site is shown with an asterisk. (B) RNase protection assay of 10 μg of total RNA extracted from 293T cells transfected with the AAV5 WT infectious clone and the AAV5 P19 mutant infectious clone together with additional adenoviral helper functions supplied by pHelper. The SB probe (nt 801 to 1023) shown in the schematic in Fig. 1 was utilized for this assay. The read-through protected product (P7 transcript) is 223 bp, and the P19-generated protected product is 122 bp. (C) Western blot of protein lysates from the 293T cell transfection described in panel B above. The AAV5 Rep antibody (ARP, Belmont, MA) was used to detect the Rep proteins. An equal concentration of total protein was loaded in each lane. (D) 293T cells were cotransfected with either the AAV5 WT or AAV5 P19 mutant infectious clone in either the absence or presence of the pHelper plasmid, which supplied the additional adenoviral helper functions as indicated. Total cellular DNA lysates were analyzed by Southern blotting for AAV5 replication forms 48 h after transfection. An equal amount of total DNA was loaded, as determined by a NanoDrop spectrophotometer. The positions of the dimer (dRF), monomer (mRF), and single-stranded DNA (ssDNA) replicative forms are indicated. (E) Shown is a Southern blot of cell-associated virus isolated from 293T cells following cotransfection of either the AAV5 WT or AAV5 P19 mutant infectious clone with pHelper. The single-stranded DNA genomes packaged by the AAV5 WT and AAV5 P19 mutant viral progeny produced 48 h posttransfection are indicated. The complete experiment was repeated three times; a representative experiment is shown.
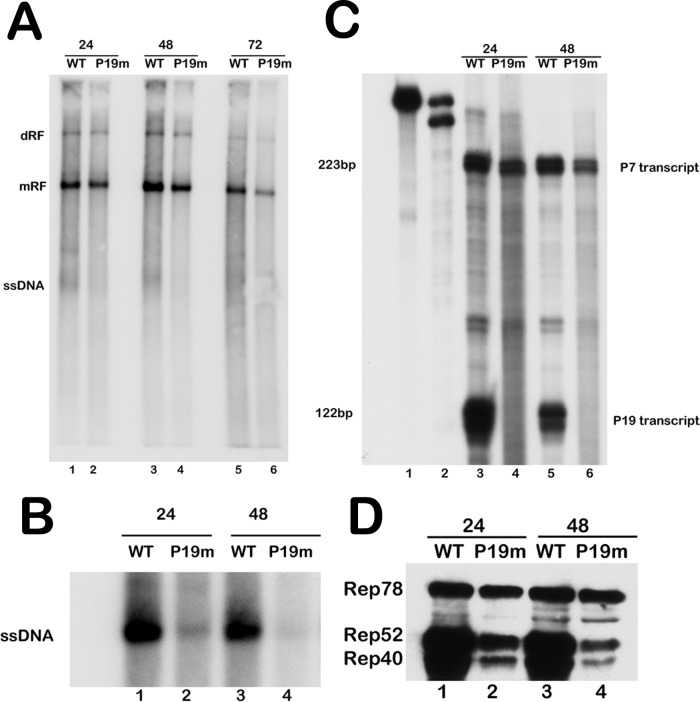
Equal genome equivalents of mutant and wild-type virus generated from the transfections shown in Fig. 5 were then used to infect 293 cells in the presence of adenovirus type 5. By 24 h postinfection, mutant and wild-type virus generated similar amounts of mRF; however, less mutant ssDNA was detected in the Southern blot assays (Fig. 6A, compare lanes 1 and 2). This was confirmed by analysis of cell-associated virus production (Fig. 6B, compare lanes 1 and 2). By 48 h postinfection, less mutant mRF had accumulated than the wild type, likely due to a decrease in mutant virus reinfection (Fig. 6A, compare lanes 3 and 4), and the amount of mutant virus produced at this point was substantially reduced (Fig. 6B, compare lanes 3 and 4). At 72 h postinfection, relative levels of mutant mRF DNA had diminished further (Fig. 6A, compare lanes 5 and 6). The P19 mutant virus used for these infections generated no detectable P19-initiated product (Fig. 6C, compare lanes 3 and 4 and lanes 5 and 6). As seen following transfection, at 24 h postinfection, when mRF levels were similar to those for the wild type, the mutant virus generated generally similar amounts of Rep78 while the small Rep proteins were produced at the relatively reduced levels of small to large that were previously noted, reflecting the lack of P19 promoter activity (Fig. 6D, compare lanes 1 and 2). At 48 h postinfection, the relative ratios of mutant small-to-large Rep proteins remained low and the total amount of protein was additionally reduced, reflecting the reduction in mutant virus production (Fig. 6D, compare lanes 3 and 4). These results suggested that AAV5 can generate Rep78, Rep52, and the Rep40-like protein from the P7-generated mRNA, and while this production was sufficient to allow initial replication and virus production, it was not enough to sustain efficient propagation upon amplification.
Fig 6.
The AAV5 P19 mutant virus expressed all replication proteins from the P7-generated RNA and replicated with reduced genome amplification as infection progressed. (A) 293T cells were coinfected with adenovirus type 5 (Ad5) (multiplicity of infection [MOI] of 3) and equilibrated genome copies of AAV5 WT or AAV5 P19 mutant virus. Total cellular DNA was extracted 24, 48, and 72 h postinfection and analyzed by Southern blotting for AAV5 replication. The locations of the replicative forms (dimer replicative form [dRF], monomer replicative form [mRF], and single-stranded DNA [ssDNA]) are indicated. Equivalent loading of total DNA was confirmed using a NanoDrop spectrophotometer. (B) AAV5 WT and AAV5 P19 mutant (P19m) viral genomes isolated from cell-associated virus collected from the 293T cell infections shown in panel A and analyzed by direct Southern blotting are shown. The single-stranded DNA genomes packaged by the AAV5 WT and AAV5 P19 mutant virus are indicated. (C) RNase protection assay of 10 μg total RNA isolated from the 293T cell infections described in panel A. RNA was isolated 24 and 48 h postinfection, and protections were done with the antisense SB probe (nt 801 to 1023) shown in Fig. 1. The wild-type and homologous P19 mutant probes are shown in lanes 1 and 2, respectively, while the bands protected by the P7 and P19 transcripts are shown on the right side of the panel. (D) Expression of AAV5 Rep proteins analyzed by Western blotting of cell lysate extracted from the 293T cell infection described in panel A, using anti-AAV Rep antibody (ARP, Belmont, MA). An equal concentration of total protein was loaded in each lane. The complete experiment was repeated three times; a representative experiment is shown.
DISCUSSION
While AAV2 generates Rep protein diversity by alternative splicing of its internal intron, the AAV5 Rep-encoding P7- and P19-generated RNAs are not spliced because they are polyadenylated at an internal site within the small intron (2, 5, 27). Although precluded from a splicing-dependent variability, AAV5, as well as all the animal AAVs so far examined (1, 2, 28), encodes a Rep40-like protein by an internal initiation event from an in-frame AUG 150 nt downstream of the initiating AUG of Rep52 (7). Interestingly, AAV2 retains in an inactive form both the internal polyadenylation signal and the internal AUG codon utilized by AAV5. In this article, we show that, in contrast to AAV2, the region between the two small-Rep AUGs in AAV5 contains a signal that is required and sufficient for AAV5 internal initiation and which can also program both the internal initiation of the AAV2 internal AUG and the AUGs of other heterologous genes. Our analysis was unable to delimit the required motif further.
It is difficult to make mutations in the AAV5 Rep40-like protein exclusively; mutations either within the coding region or in its expression signals also affect both Rep52 and Rep78. This makes it difficult to genetically confirm a role for the Rep40-like protein in infection. As an alternative approach, we have shown that the AAV5 Rep40-like protein was functional and has helicase activity similar to that of AAV2 Rep40. That the protein is functional and that AAV5 has adapted a second way to generate it is consistent with it having a critical role in replication.
Although the AAV5 Rep40-like protein contains the three Walker motifs characteristic of the AAV2 Rep proteins and of other SF3 helicases (12, 15), it is otherwise quite different from AAV2 Rep40, varying at both the amino and carboxyl termini. The AAV2 Rep40 protein is bimodular, with a small helical bundle at the amino terminus and a large α/β domain at the C terminus (9), while the AAV5 Rep40-like protein lacks this helical bundle at the amino terminus (7). The AAV5 Rep40-like protein retains significant helicase activity, and it will be interesting in the future to compare the biochemical properties and functions of AAV2 and AAV5 small Rep proteins.
The fact that the AAV5 Rep40-like protein (as well as the Rep40-like protein of all other animal AAVs examined) was generated by internal initiation led us to ask whether it may be generated from the upstream P7-generated RNA as well. We found that both the Rep40-like protein and, perhaps surprisingly, Rep52 can be generated from the P7-generated transcript. To examine the importance of P19-dependent expression of the small Rep proteins, we generated a specific mutant of the P19 promoter in an AAV5 infectious clone to determine its necessity in a viral context. AAV5 constructs that lack the P19 transcription unit would be expected to make normal levels of Rep78, and so, as expected, the mutant clone was found to efficiently generate the double-stranded mRF replication intermediates. However, the P19 mutant construct generated a reduced level of small Rep proteins relative to large compared to the level in the P19-replete infectious clone. Consistent with a role for the small Rep proteins in genome encapsidation, the mutant clone generated less virus as well. Subsequent infection with equilibrated levels of mutant virus showed similar results; however, the production of virus was dramatically reduced upon amplification. The mutant infectious clone generated more virus following transient transfection than did mutant virus infection, perhaps because of the higher level of protein expression under these conditions. This study also represents the first characterization of an AAV mutant that specifically lacks P19-generated RNAs, leaving the large Rep proteins unaltered. We confirm a role for the small Rep proteins in genome encapsidation, as pioneered by others (10).
The P19 transcription unit is, of course, the main generator of the small Rep proteins required for the high levels of virus production seen during infection. What is the role of the redundancy of their expression? Whether the amounts generated from the P7-generated mRNAs play a role at some point during AAV5 infection will be interesting to examine. AAV2 and all other primate AAVs examined utilize neither internal polyadenylation nor internal initiation for its small Rep proteins, although both the polyadenylation motif and in-frame internal AUG are present. This difference may shed light on the evolutionary relationships between the two virus groups.
ACKNOWLEDGMENTS
We thank Lisa Burger for excellent technical assistance and members of the lab for valuable discussion. We thank Stefan Sarafianos' lab for reagents and help with protein purification and helicase assays. We thank Jianming Qiu for a plasmid containing the full-length AAV5 terminal repeats.
This work was supported by PHS award RO1 046458 to D.J.P.
Footnotes
Published ahead of print 17 October 2012
REFERENCES
- 1. Qiu J, Cheng F, Pintel D. 2006. Molecular characterization of caprine adeno-associated virus (AAV-Go. 1) reveals striking similarity to human AAV5. Virology 356:208–216 [DOI] [PubMed] [Google Scholar]
- 2. Qiu J, Pintel DJ. 2004. Alternative polyadenylation of adeno-associated virus type 5 RNA within an internal intron is governed by the distance between the promoter and the intron and is inhibited by U1 small nuclear RNP binding to the intervening donor. J. Biol. Chem. 279:14889–14898 [DOI] [PubMed] [Google Scholar]
- 3. Green MR, Roeder RG. 1980. Transcripts of the adeno-associated virus genome: mapping of the major RNAs. J. Virol. 36:79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lusby EW, Berns KI. 1982. Mapping of the 5′ termini of two adeno-associated virus 2 RNAs in the left half of the genome. J. Virol. 41:518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mouw MB, Pintel DJ. 2000. Adeno-associated virus RNAs appear in a temporal order and their splicing is stimulated during coinfection with adenovirus. J. Virol. 74:9878–9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Srivastava A, Lusby EW, Berns KI. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farris KD, Pintel DJ. 2010. Adeno-associated virus type 5 utilizes alternative translation initiation to encode a small Rep40-like protein. J. Virol. 84:1193–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chejanovsky N, Carter BJ. 1989. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology 173:120–128 [DOI] [PubMed] [Google Scholar]
- 9. James JA, Escalante CR, Yoon-Robarts M, Edwards TA, Linden RM, Aggarwal AK. 2003. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure 11:1025–1035 [DOI] [PubMed] [Google Scholar]
- 10. King JA, Dubielzig R, Grimm D, Kleinschmidt JA. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. James JA, Aggarwal AK, Linden RM, Escalante CR. 2004. Structure of adeno-associated virus type 2 Rep40-ADP complex: insight into nucleotide recognition and catalysis by superfamily 3 helicases. Proc. Natl. Acad. Sci. U. S. A. 101:12455–12460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walker SL, Wonderling RS, Owens RA. 1997. Mutational analysis of the adeno-associated virus type 2 Rep68 protein helicase motifs. J. Virol. 71:6996–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoon-Robarts M, Blouin AG, Bleker S, Kleinschmidt JA, Aggarwal AK, Escalante CR, Linden RM. 2004. Residues within the B′ motif are critical for DNA binding by the superfamily 3 helicase Rep40 of adeno-associated virus type 2. J. Biol. Chem. 279:50472–50481 [DOI] [PubMed] [Google Scholar]
- 14. Hickman AB, Ronning DR, Kotin RM, Dyda F. 2002. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol. Cell 10:327–337 [DOI] [PubMed] [Google Scholar]
- 15. Hickman AB, Dyda F. 2005. Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 15:77–85 [DOI] [PubMed] [Google Scholar]
- 16. Patel SS, Picha KM. 2000. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69:651–697 [DOI] [PubMed] [Google Scholar]
- 17. Stenlund A. 2003. Initiation of DNA replication: lessons from viral initiator proteins. Nat. Rev. Mol. Cell Biol. 4:777–785 [DOI] [PubMed] [Google Scholar]
- 18. Dignam SS, Correia JJ, Nada SE, Trempe JP, Dignam JD. 2007. Activation of the ATPase activity of adeno-associated virus Rep68 and Rep78. Biochemistry 46:6364–6374 [DOI] [PubMed] [Google Scholar]
- 19. Li Z, Brister JR, Im DS, Muzyczka N. 2003. Characterization of the adeno-associated virus Rep protein complex formed on the viral origin of DNA replication. Virology 313:364–376 [DOI] [PubMed] [Google Scholar]
- 20. Mansilla-Soto J, Yoon-Robarts M, Rice WJ, Arya S, Escalante CR, Linden RM. 2009. DNA structure modulates the oligomerization properties of the AAV initiator protein Rep68. PLoS Pathog. 5:e1000513 doi:10.1371/journal.ppat.1000513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smith RH, Spano AJ, Kotin RM. 1997. The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J. Virol. 71:4461–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zarate-Perez F, Bardelli M, Burgner JW, II, Villamil-Jarauta M, Das K, Kekilli D, Mansilla-Soto J, Linden RM, Escalante CR. 2012. The interdomain linker of AAV-2 Rep68 is an integral part of its oligomerization domain: role of a conserved SF3 helicase residue in oligomerization. PLoS Pathog. 8:e1002764 doi:10.1371/journal.ppat.1002764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qiu J, Nayak R, Tullis GE, Pintel DJ. 2002. Characterization of the transcription profile of adeno-associated virus type 5 reveals a number of unique features compared to previously characterized adeno-associated viruses. J. Virol. 76:12435–12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nayak R, Pintel DJ. 2007. Adeno-associated viruses can induce phosphorylation of eIF2α via PKR activation, which can be overcome by helper adenovirus type 5 virus-associated RNA. J. Virol. 81:11908–11916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Venkatesh LK, Fasina O, Pintel DJ. 2012. RNAse mapping and quantitation of RNA isoforms. Methods Mol. Biol. 883:121–129 [DOI] [PubMed] [Google Scholar]
- 26. Miller CL, Pintel DJ. 2002. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J. Virol. 76:3257–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qiu J, Pintel DJ. 2002. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol. Cell. Biol. 22:3639–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qiu J, Cheng F, Pintel DJ. 2006. Expression profiles of bovine adeno-associated virus and avian adeno-associated virus display significant similarity to that of adeno-associated virus type 5. J. Virol. 80:5482–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]