Abstract
Previous studies from our laboratory revealed that cellular poly(C) binding protein 2 (PCBP2) downregulates vesicular stomatitis virus (VSV) gene expression. We show here that VSV infection induces the formation of granular structures in the cytoplasm containing cellular RNA-binding proteins, including PCBP2, T-cell-restricted intracellular antigen 1 (TIA1), and TIA1-related protein (TIAR). Depletion of TIA1 via small interfering RNAs (siRNAs), but not depletion of TIAR, results in enhanced VSV growth and gene expression. The VSV-induced granules appear to be similar to the stress granules (SGs) generated in cells triggered by heat shock or oxidative stress but do not contain some of the bona fide SG markers, such as eukaryotic initiation factor 3 (eIF3) or eIF4A, or the processing body (PB) markers, such as mRNA-decapping enzyme 1A (DCP1a), and thus may not represent canonical SGs or PBs. Our results revealed that the VSV-induced granules, called SG-like structures here, contain the viral replicative proteins and RNAs. The formation and maintenance of the SG-like structures required viral replication and ongoing protein synthesis, but an intact cytoskeletal network was not necessary. These results suggest that cells respond to VSV infection by aggregating the antiviral proteins, such as PCBP2 and TIA1, to form SG-like structures. The functional significance of these SG-like structures in VSV-infected cells is currently under investigation.
INTRODUCTION
Mammalian cells respond to various environmental stresses, such as heat shock, amino acid deprivation, oxidative stress, and hypoxia, as well as to viral infections, by inducing the formation of nonmembranous cytoplasmic structures called stress granules (SGs) (1–3). The SGs regulate mRNA metabolism by blocking the translation of housekeeping genes and facilitating the synthesis of mostly stress response proteins (4). These responses help the cells survive under stress conditions (1, 2). Several families of viruses influence the formation of SGs in infected cells (5–20). These viruses either induce or suppress SG formation to promote their replication. For example, orthoreoviruses induce SG formation early in the infection cycle, which has been assumed to be beneficial for virus replication (18). Poliovirus induces SG formation early in infection, but the protein composition of these SGs changes at later stages of infection due to the cleavage of several SG marker proteins by the viral proteases (15, 19, 20). On the other hand, several other viruses have been shown to interfere with SG formation (5, 6, 8–10, 12, 16, 17, 19), and the results of these studies emphasize the important role of SGs in the cellular antiviral response. Individual protein components of the SGs or the so-called “antiviral granules” (AVGs), such as T-cell-restricted intracellular antigen 1 (TIA1) and RasGAP SH3 domain binding protein 1 (G3BP1), have been found to inhibit the replication of a number of viruses, indicating that not only the SGs but also the individual components of the SGs may serve as antagonistic factors in viral replication processes (17, 19). Thus, SGs and/or AVGs formed in response to viral infection may represent manifestations of interactions between host cells and viruses, a possibility that is also supported by the observations that (i) Sendai virus trailer RNA binds and sequesters TIA1-related protein (TIAR) in order to reduce the virus-induced apoptosis mediated by TIAR (21) and (ii) the interaction of TIA1/TIAR with the 3′-terminal stem-loop of West Nile virus (WNV) minus-strand RNA suppresses the formation of SGs and processing bodies (PBs) to favor virus replication (6).
Infection of TIA1-knockout murine embryonic fibroblasts (MEFs) with vesicular stomatitis virus (VSV), Sindbis virus, herpes simplex virus 1, or vaccinia virus results in increased virus growth, whereas WNV replication is suppressed in these cells (10), indicating that TIA1 has both antiviral and proviral properties. However, the mechanisms of TIA1 involvement in these functions remain poorly understood. TIA1 is known to interact and colocalize with poly(C) binding protein 2 (PCBP2) in cytoplasmic SGs (22). Since we have demonstrated previously that PCBP2 exerts antiviral activity against VSV (23), it is possible that the inhibitory activity of PCBP2 may be mediated through cooperation between these cellular proteins and/or through the formation of SGs or similar structures in infected cells.
VSV is an enveloped, nonsegmented, negative-stranded RNA virus in the family Rhabdoviridae that initiates its infection cycle by entering cells through receptor-mediated endocytosis, followed by the delivery of its nucleocapsid (NC) template into the cytoplasm. Viral genome transcription and replication occur in the cytoplasm. Unlike those for many positive-stranded and double-stranded RNA viruses, whose replication and transcription usually occur in cytoplasmic factories or in association with membranous organelles specifically induced in virus-infected cells (24–26), the cytoplasmic sites for VSV replication are poorly characterized. In VSV-infected cells, the viral replication proteins (N, P, and L) and nascently synthesized viral RNAs have been seen to be colocalized to “granular structures” or “inclusions” in the cytoplasm (27, 28). Further, these granular structures or inclusions have been suggested to be the sites of viral mRNA synthesis (27, 28). It has been proposed that these inclusions might reflect a cellular response to the viral infection (28). While these granules or inclusions contain viral proteins and RNA, their host cell protein composition, if any, has not been characterized yet. SG formation has been demonstrated previously in cells infected with Sendai virus (21) and, more recently, in respiratory syncytial virus-infected cells (7, 11). Whether the granules or inclusions seen in VSV-infected cells are similar to the SGs observed in cells infected with other viruses remains a matter of further investigation.
In this communication, we report the findings of our extended studies on the role of PCBP2 in suppressing VSV infection and on its relationship to TIA1 and TIAR. Our results show that TIA1 and TIAR, together with PCBP2, are redistributed to cytoplasmic VSV-induced SG-like structures that also contain the viral replication proteins and RNA. Like PCBP2, TIA1 displays an inhibitory effect on viral gene expression and/or replication in VSV-infected cells. The formation of SG-like structures containing TIA1/PCBP2 is independent of the cellular microtubule and microfilament network and of cellular mRNA synthesis but requires viral replication and ongoing protein synthesis. These SG-like structures appear simultaneously with the phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α) and the host cell translational shutoff, without a significant inhibitory effect on viral mRNA translation.
MATERIALS AND METHODS
Cell culture and reagents.
Monolayer cultures of HeLa (ATCC catalog no. CCL2), Huh7, MCF-7, and HEK293 cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 8% fetal bovine serum (FBS) and the antibiotics penicillin (100 U/ml), kanamycin (20 U/ml), and streptomycin (20 U/ml) (PKS). Baby hamster kidney (BHK-21) cells were maintained in MEM supplemented with 5% FBS and PKS. Immortalized murine embryonic fibroblasts (MEFs) from wild-type (wt) and TIA1-knockout mice (29) were obtained from P. Anderson (Harvard University) as well as M. Brinton (Georgia State University) and were maintained as described previously (30). Sodium arsenite (SA), nocodazole (Noc), cytochalasin D (CytoD), actinomycin D (ActD), 5-bromouridine 5′-triphosphate sodium salt (BrUTP), cycloheximide (CHX), and a protease inhibitor cocktail were obtained from Sigma-Aldrich and were used at concentrations of 20 mM, 10 μg/ml, 10 μg/ml, 15 μg/ml, 50 mM, 100 μg/ml, and 1×, respectively. A 35S-labeled Met-Cys mixture for protein labeling was obtained from Perkin-Elmer.
Viruses and VSV nucleocapsid (NC or RNP) preparation.
Stocks of wt VSV were prepared as described earlier (27, 31). VSVΔG virus and VSV NCs (RNPs) were prepared as described previously (23).
Virus titration and infection.
Virus titers were determined by plaque assays on BHK-21 or HeLa cells. In each experiment, virus infection was performed at a multiplicity of infection (MOI) of 1 or 0.01 PFU per cell, except where indicated otherwise.
Antibodies.
Anti-M (23H12) and anti-N (10G4) monoclonal antibodies were kindly provided by D. Lyles. The mouse anti-VSV (serotype Indiana) polyclonal antibody, the rabbit anti-P polyclonal antibody, and the rabbit polyclonal antibody against the NH2-terminal region of the VSV L protein have been described previously (32, 33). Antibodies against actin (mouse monoclonal; Sc-47778), TIA1 (goat polyclonal; Sc-1751), TIAR (goat polyclonal; Sc-1749), eIF3η (mouse monoclonal; Sc-137214), and eIF4AII (mouse monoclonal; sc-137148) were purchased from Santa Cruz Biotechnology, Inc. A rabbit polyclonal antibody against DCP1a (ab47811) was purchased from Abcam. A mouse monoclonal antibody against bromodeoxyuridine (BrdU) (clone BMC 9318) was obtained from Roche Diagnostics. Anti-tubulin (clone DM 1A), anti-hemagglutinin (anti-HA) monoclonal antibody HA-7 (H3663), horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (IgG) (A6154), horseradish peroxidase-conjugated goat anti-mouse IgG (A4416), and horseradish peroxidase-conjugated rabbit anti-goat IgG (A4174) were obtained from Sigma-Aldrich. Alexa Fluor 594-labeled donkey anti-goat IgG (A-11058), Alexa Fluor 488-labeled donkey anti-mouse IgG (A-21202), Alexa Fluor 488-labeled donkey anti-rabbit IgG (A-21206), Alexa Fluor 647-labeled donkey anti-rabbit IgG (A-31573), Alexa Fluor 594-labeled goat anti-mouse IgG (A-11005), and Alexa Fluor 488-labeled goat anti-rabbit IgG (A-11034) were obtained from Invitrogen.
Plasmid constructs.
Coding sequences of TIA1a (variant 2; NM_022173.2) and TIARb (variant 1; NM_003252.3) were amplified using total RNA from HeLa cells and a LongAmp kit (New England Biolabs) with the specific primers shown in Table 1. The PCR products were digested with KpnI and EcoRI and were cloned into pcDHA (a plasmid carrying an HA epitope tag) as described previously (23). The resulting constructs express the inserted genes with an HA epitope at the N-terminal ends of the proteins. The expression of the proteins can be driven by either a cytomegalovirus (CMV) promoter or a T7 promoter located upstream of the coding sequences.
Table 1.
Primers used in this study
| Primer name | Primer sequence (5′ to 3′)a | Use |
|---|---|---|
| TIA1-KpnI For | ATATGGTACCATGGAGGACGAGATGCCCAAG | Amplification and cloning of the TIA1a isoform |
| TIA1-EcoRI Rev | ATATGAATTCTCACTGGGTTTCATACCCTGCC | |
| TIAR-KpnI For | ATATGGTACCATGATGGAAGACGACGGGCAG | Amplification and cloning of the TIARb isoform |
| TIAR-EcoRI Rev | ATATGAATTCTCACTGTGTTTGGTAACTTGCCATAC |
Underlined sequences show restriction enzyme sites.
siRNA-mediated silencing.
For depletion of TIA1 and TIAR, small interfering RNAs (siRNAs) (pools of four different siRNAs) targeting these genes (catalog no. L-013042-00-0005 and L-011405-00-0005, respectively; Dharmacon) were transfected at a final concentration of 10 nM (except where indicated otherwise) by following the protocol of reverse transfection with Lipofectamine RNAiMax (Invitrogen) as recommended by the manufacturer. At 24 h posttransfection (hpt), the transfection mixture was replaced with DMEM containing 8% FBS and PKS, and the cells were incubated further for 40 to 48 h to allow knockdown of the gene. A nontargeting (NT) siRNA (catalog no. 1027281; Qiagen), which does not target any of the known mammalian genes, was used as a control siRNA.
Plasmid DNA and NC (RNP) transfection.
Plasmids were transfected by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. At 4 hpt, the transfection mixture was replaced with complete growth medium, and the culture was incubated for 40 to 44 h before being used for an assay. Viral RNPs were also transfected by the same procedure as that used for plasmid transfection and as described previously (23).
WB.
Western blotting (WB) and quantification of the protein bands were performed as described previously (23). The following antibodies were used: anti-M (dilution, 1:1,000 to 1:3,000), anti-actin (1:3,000 to 1:5,000), anti-TIA1 (1:500), anti-TIAR (1:500), and anti-HA (1:10,000). Horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (IgG), horseradish peroxidase-conjugated goat anti-mouse IgG, and horseradish peroxidase-conjugated rabbit anti-goat IgG, at dilutions of 1:500 to 10,000, were used as secondary antibodies.
Immunofluorescence (IF) microscopy.
HeLa cells grown on glass coverslips were infected with wt VSV at an MOI of 1 for the indicated time (see figure legends), followed by fixation with methanol-acetone (1:1) for 15 min at −30°C. After the cells were washed twice with phosphate-buffered saline (PBS), they were stained with specific primary antibodies against the proteins shown in each figure. The dilutions of the primary antibodies used were as follows: 1:4,000 for anti-VSV N and anti-VSV L, 1:300 for anti-VSV P, 1:100 for anti-TIA1, anti-TIAR, and anti-eIF3η, 1:3,000 for anti-HA, and 1:500 for anti-tubulin and anti-actin. The dilutions of secondary antibodies were 1:300 to 1:500 for Alexa Fluor 594-labeled donkey anti-goat IgG, 1:2,000 for Alexa Fluor 488-labeled donkey anti-mouse IgG, 1:1,000 for Alexa Fluor 488-labeled donkey anti-rabbit IgG, Alexa Fluor 647-labeled donkey anti-rabbit IgG, and Alexa Fluor 488-labeled goat anti-rabbit IgG, and 1:2,000 for Alexa Fluor 594-labeled goat anti-mouse IgG. Images were obtained with an Olympus FV500/IX81 inverted laser scanning confocal microscope.
qRT-PCR.
The primers and probes for quantification of VSV P mRNA and antigenome RNA by quantitative real-time PCR (qRT-PCR) were used as described previously (34).
Metabolic labeling of proteins.
Proteins in cells were labeled and detected as described previously (35).
Statistical analysis.
Statistical analyses were performed using the MIXED procedure (SAS Institute, Inc., Cary, NC). The statistical model included Treatment as a fixed effect. Data are presented as means ± standard errors of the means. A P value of <0.05 was considered statistically significant.
RESULTS
Cellular TIA1 downregulates VSV growth.
Previously, we reported that cellular PCBP2 inhibits VSV infection at the level of viral gene expression (23). Recent studies have demonstrated that PCBP2 is a component of SGs and that it interacts and colocalizes with TIA1 in SGs induced by oxidative stress with agents such as sodium arsenite (SA) (22). TIA1 and TIAR are well-known SG markers (1, 2). The observations that MEFs from TIA1−/− mice support increased VSV growth (10) and that the putative SG inducer thapsigargin inhibits VSV protein synthesis (36) led us to hypothesize that the TIA1 and PCBP2 proteins may have related functions in controlling VSV infection and may be involved in the induction of SGs during VSV infection. To test this hypothesis, we first examined the effects of depletion or overexpression of TIA1 on VSV growth in infected cells. Treatment of the cells with siRNA for TIA1 resulted in >95% depletion of TIA1 protein (Fig. 1A). In TIA1-depleted cells infected with VSV, virus growth was enhanced (>3-fold) over that in cells treated with the nontargeting (NT) siRNA (Fig. 1B). Viral gene expression, examined by monitoring the levels of M protein in infected cells, was also increased similarly upon TIA1 depletion (Fig. 1C).
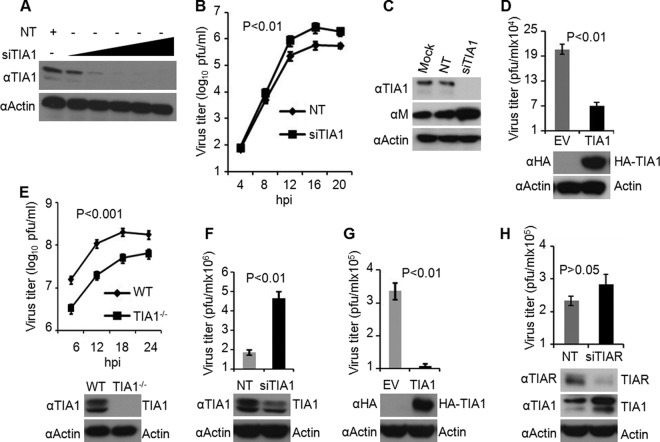
Fig 1.
TIA1 inhibits VSV growth. (A) Depletion of TIA1 by siRNA. HeLa cells were treated with siRNA targeting TIA1 at final concentrations of 0.1, 1, 5, 10, and 20 nM or with 20 nM nontargeting (NT) siRNA. TIA1 protein levels were determined by WB with an anti-TIA1 polyclonal antibody at 72 hpt. Actin served as a loading control. (B) VSV growth is enhanced in TIA1-depleted cells. HeLa cells treated with NT siRNA or TIA1 siRNA at 10 nM for 68 to 72 h were infected with VSV at an MOI of 0.01. Virus titers in the culture supernatants at the indicated hours postinfection were determined by a plaque assay. Statistical significance (P value) was determined for virus titers at 12 and 16 hpi. (C) VSV M protein levels in TIA1-depleted cells. siRNA treatment and virus infection were performed as described for panel B. VSV M protein and endogenous TIA1 in cell lysates prepared at 12 hpi were detected by WB with corresponding antibodies. (D) Ectopic expression of TIA1 resulted in reduced levels of VSV growth. HeLa cells transfected with 1.5 μg of an empty vector (EV) or an HA-TIA1-encoding plasmid for 48 h were infected with 0.01 MOI of VSV. At 12 hpi, culture supernatants were collected and analyzed by a plaque assay for infectious virus. Cell lysates were examined by WB for HA-TIA1 expression. (E) Growth of VSV in wt and TIA1-knockout MEFs. MEFs were infected with VSV at an MOI of 1, and at the indicated hours postinfection, culture supernatants were collected and titrated for infectious virus. Statistical significance was determined for virus titers at 12 and 18 hpi. TIA1 protein levels were determined by WB with an anti-TIA1 polyclonal antibody. (F) Depletion of TIA1 by siRNA in wt MEFs results in enhanced growth of VSV. Transfection of siRNA and virus infection were performed as described for panel B. Virus titers in the supernatants were determined at 12 hpi, and endogenous TIA1 levels in the cells were determined by WB. (G) Ectopic expression of TIA1 in TIA1-knockout MEFs results in reduced virus growth. Plasmid transfection, VSV infection, virus titration, and HA-TIA1 detection were carried out as described for panel D. (H) TIAR has no significant effect on VSV growth. The conditions of siRNA transfection, virus infection, virus growth (at 16 hpi), and WB to detect TIAR and TIA1 are as described above for panels B and C. Error bars represent the standard errors of the means from 3 independent experiments.
To determine whether overexpression of TIA1 in HeLa cells would affect VSV growth, we generated a plasmid encoding HA-tagged TIA1 (variant a) under the control of the CMV promoter. TIA1 variant a is the major isoform of TIA1 expressed in HeLa cells (30). When HeLa cells were transfected with the plasmid encoding HA-TIA1, the protein was readily detectable in the cells, and VSV growth in these cells was inhibited >3-fold (Fig. 1D).
The availability of TIA1-knockout MEFs allowed us to further examine the role of TIA1 in VSV replication. A previous study (10) reported enhanced growth (up to 6-fold) of VSV in TIA1-knockout MEFs. In contrast to the results of that study and those from our siRNA-mediated depletion study, described above, we found that VSV growth in TIA1−/− MEFs was significantly reduced from that in wt MEFs. Viral growth kinetics analysis revealed that VSV growth was dampened at least 5-fold in TIA1-knockout MEFs, compared to that in wt MEFs, at all times examined (Fig. 1E). This result was unexpected given the previous report (10) as well as our studies with transient depletion of TIA1 by siRNA (Fig. 1B) but was consistently reproducible in at least four repeat experiments using TIA1-knockout MEFs obtained from both P. Anderson (clone α44) and M. Brinton (clone α43). Strikingly, depletion of TIA1 from wt MEFs by siRNA resulted in enhanced VSV growth (Fig. 1F), whereas ectopic expression of HA-TIA1 in TIA1−/− MEFs resulted in significant inhibition of VSV growth (Fig. 1G). Taking these findings together, although these studies point toward an inhibitory role of TIA1 in VSV growth, there appears to be a discrepancy between the effects of TIA1 when the protein is depleted transiently (siRNA mediated) versus permanently (knockout).
Since TIA1 was found to inhibit VSV growth, we determined whether TIAR, a TIA1-related protein, would also affect VSV growth similarly. The results showed that depletion of TIAR by siRNA did not enhance VSV growth significantly, although >80% of the protein was depleted from the cells (Fig. 1H). Since TIAR depletion resulted in increased TIA1 levels (Fig. 1H), consistent with previous observations (10, 30), it is possible that the nonsignificant increase in VSV growth in TIAR-depleted cells is due to higher levels of TIA1 in these cells.
TIA1 exerts its effect on VSV growth at the level of viral gene expression and/or genome replication.
We next examined the step(s) of VSV infection that is affected by TIA1. Since transfection of viral nucleocapsids (NCs) into cells bypasses the virus entry steps and results in the expression of viral genes (23, 34), we examined the effect of TIA1 depletion on viral gene expression in cells transfected with viral NCs. The level of viral M protein in NC-transfected cells treated with siRNA specific for TIA1 was approximately 3-fold higher than that in NC-transfected cells treated with NT siRNA (Fig. 2A, lanes 5 and 6). Furthermore, in cells infected with VSVΔG (a virus that lacks the G gene and cannot produce infectious virions, so that infection with this virus is limited to a single cycle), the relative levels of VSV M expression increased to a similar extent, approximately 3-fold, in TIA1 siRNA-treated cells compared to NT siRNA-treated cells (Fig. 2A, lanes 3 and 4). Overall, the results show that TIA1 depletion results in a 3-fold increase in viral gene expression irrespective of the use of a wt VSV, VSVΔG, or viral NC template, indicating that the inhibitory effect of TIA1 on VSV infection takes place not at the entry or assembly/budding step but likely at the viral gene expression and/or replication step(s).
Fig 2.
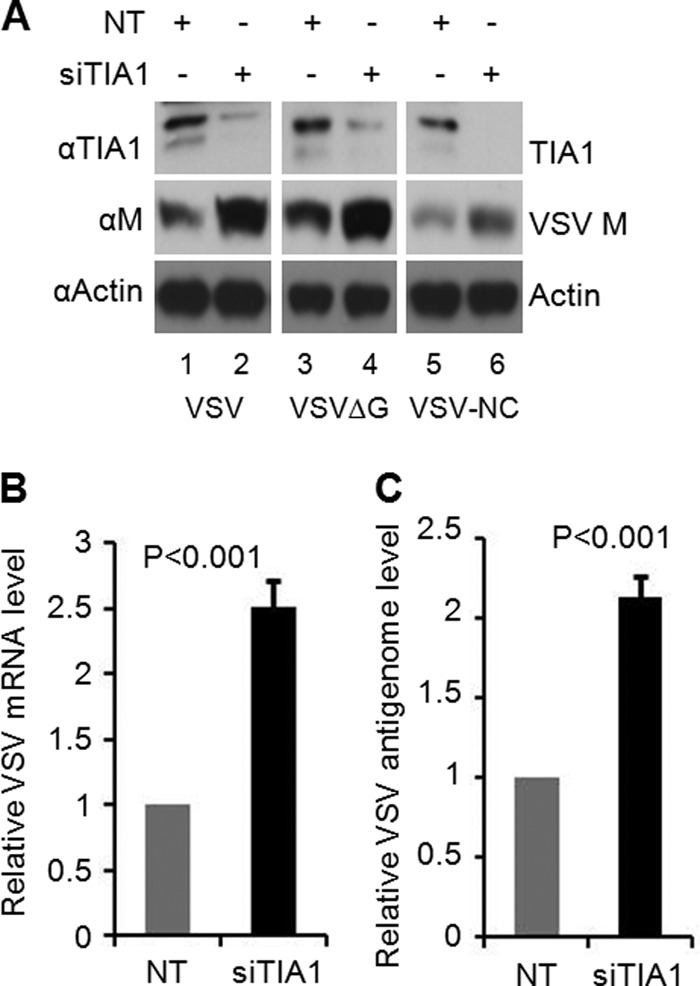
TIA1 negatively regulates VSV gene expression and/or replication. (A) HeLa cells were transfected with 10 nM NT or TIA1 siRNA for 72 h. Then the cells either were infected with VSV at an MOI of 0.01 for 12 h (lanes 1 and 2) or with VSVΔG at an MOI of 0.5 for 8 h (lanes 3 and 4) or were supertransfected for 6 h with viral NC prepared from VSV (lanes 5 and 6). Cell lysates corresponding to equal amounts of total proteins were analyzed by WB with anti-TIA1 and anti-M antibodies. Actin served as a loading control. (B and C) VSV mRNA (B) and antigenomic RNA (C) levels in TIA1-depleted cells were determined by qRT-PCR. The conditions of virus infection were the same as those described for panel A, lanes 1 and 2. Values are means ± standard errors of means for duplicate reactions from two independent experiments after normalization to the values for the NT control.
To more conclusively determine the effects of TIA1 on VSV gene expression or replication, we used real-time PCR to assess the levels of viral mRNA and antigenomic RNA in infected cells depleted of TIA1. The results show that silencing of TIA1 protein led to increased levels of both VSV mRNA (Fig. 2B) and antigenomic replicative RNA (Fig. 2C). The increase in mRNA and antigenomic RNA levels is similar to that seen with the viral M protein levels (Fig. 2A). Thus, the results further suggest that TIA1 restricts VSV infection at the level of viral gene expression and/or replication.
VSV infection induces the formation of SG-like structures that colocalize with viral replication proteins and RNA.
Since TIA1 is a common cytoplasmic SG marker (1), our observation that TIA1 inhibits VSV growth at gene expression steps prompted us to investigate whether cytoplasmic SGs are induced in VSV-infected cells. For this purpose, we infected cells with VSV and performed an IF assay using antibodies to detect endogenous TIA1 at different times postinfection. In mock-infected cells, the majority of TIA1 was detected inside the nucleus (Fig. 3A, top row). Following infection of cells with VSV, TIA1 was detected readily in the cytoplasm at 3 h postinfection (hpi), with a concomitant decrease in the levels of proteins in the nucleus (data not shown). By 7 hpi, the majority of TIA1 was seen in the cytoplasmic granular structures (Fig. 3A, bottom row). Interestingly, the VSV N protein was found to colocalize with these granular structures (Fig. 3A, bottom row). The formation of these granules was readily detected as early as 3 hpi, and the percentage of cells that contained these granules continued to increase with time up to 7 hpi, the last time point examined in these studies (see Fig. 7B). The viral N protein was seen associated with these structures at all times examined. Similar induction of TIA1-containing granules was also observed in other cell lines, such as Huh7, MCF7, and BHK-21, upon VSV infection (data not shown). Since these granular structures induced in VSV-infected cells did not contain some of the canonical SG marker proteins (see below), we termed these “SG-like structures.”
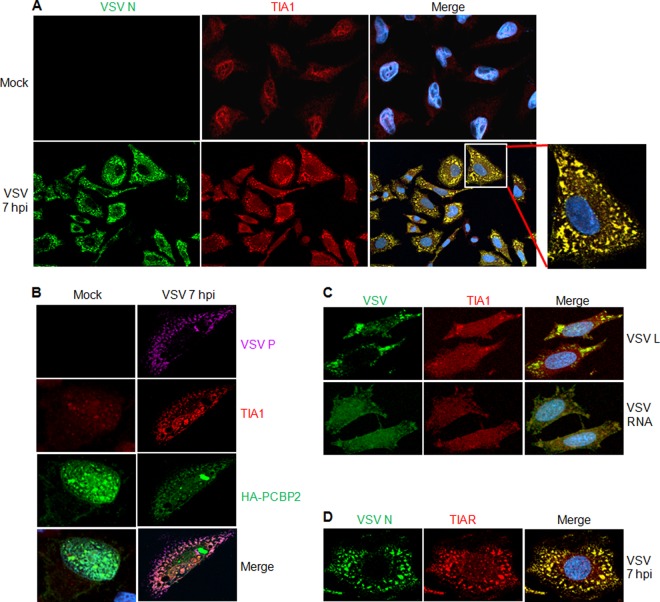
Fig 3.
VSV infection induces the formation of SG-like structures. (A) VSV N protein colocalizes with SG-like structures containing cellular TIA1 protein. HeLa cells grown on glass coverslips were either mock infected or infected with VSV at an MOI of 1. Coverslips were collected at 7 hpi and were immunostained for VSV N and cellular TIA1. The secondary antibodies used were Alexa Fluor 594-labeled donkey anti-goat IgG for TIA1 (red) and Alexa Fluor 488-labeled donkey anti-mouse IgG for VSV N (green). Nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). (B) PCBP2 colocalizes with VSV-induced SG-like structures. HeLa cells transfected with a plasmid carrying HA-PCBP2 for 48 h were infected with wt VSV at an MOI of 1. Coverslips containing VSV-infected or mock-infected cells were processed for IF analysis with anti-HA (for HA-PCBP2), anti-VSV P, and anti-TIA1 antibodies at 7 hpi. The secondary antibodies used were Alexa Fluor 594-labeled donkey anti-goat IgG for TIA1 (red), Alexa Fluor 488-labeled donkey anti-mouse IgG for HA-PCBP2 (green), and Alexa Fluor 647-labeled donkey anti-rabbit IgG for VSV P (purple). Nuclei were stained with 4′,6-diamidino-2-phenylindole. (C) VSV-induced SG-like structures contain viral L protein and RNA. HeLa cells were infected with VSV at an MOI of 1. At 2 hpi, cells were treated with ActD, and incubation was continued for a further 5 h. The cells were then fixed and processed for IF to detect TIA1 protein (red) and VSV L protein (green). To detect viral RNA, cells similarly infected with VSV were treated with ActD at 2 hpi for 1 h and were then transfected with BrUTP for 4 h in the presence of ActD. The cells were subjected to IF with antibodies against BrdU to detect newly synthesized RNA (green) and TIA1 (red). Secondary antibodies were Alexa Fluor 594-labeled donkey anti-goat IgG for TIA1 (red), Alexa Fluor 488-labeled donkey anti-mouse IgG for BrUTP (green), and Alexa Fluor 488-labeled donkey anti-rabbit IgG for VSV L protein (green). Nuclei were stained with 4′,6-diamidino-2-phenylindole. (D) VSV N protein colocalizes with SG-like structures containing cellular TIAR protein. Experimental conditions were the same as those described for panel A.
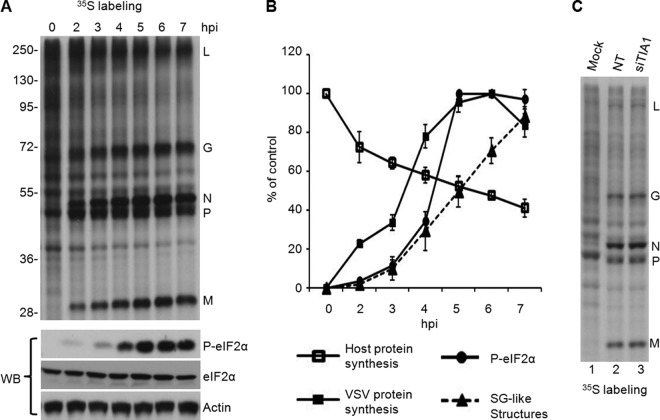
Fig 7.
Viral protein synthesis remains unaffected by the formation of SG-like structures despite eIF2α phosphorylation. (A) Protein synthesis in VSV-infected cells. Two sets of HeLa cells were infected with wt VSV at an MOI of 1. (Top) One set was pulse-labeled with [35S]Met-Cys for 10 min before lysis at 0, 2, 3, 4, 5, 6, and 7 hpi. The viral proteins and the sizes of molecular markers (in kilodaltons) are shown. (Bottom) The other set of cells was directly harvested at each time point mentioned above for WB analysis with antibodies to detect eIF2α and its phosphorylated form (P-eIF2α) as indicated. Actin served as a loading control. (B) Quantitative analysis of VSV-induced SG-like structures, levels of viral and cellular protein synthesis, and phosphorylation of eIF2α (P-eIF2α) at various times postinfection. The levels of cellular and viral protein synthesis and of P-eIF2α were determined from the results shown in panel A, based on the intensity of the bands in the gels, by using Quantity One software (Bio-Rad). The rate of cellular protein synthesis was determined by the radioactivity between the L and G bands, the G and N bands, and the P and M bands. The total cellular protein level at 0 hpi was set at 100%. The rate of viral protein synthesis was determined by quantification of the L, G, N, P, and M bands. The total viral protein level at 6 hpi was set at 100%. The rate of eIF2α phosphorylation was determined by setting the P-eIF2α level to 100% at 6 hpi. The percentage of infected cells (indicated by staining for the VSV N protein) containing SG-like structures was determined by random examination of 150 cells from each experiment. Error bars represent the standard errors of the means from three independent experiments. (C) Protein synthesis in VSV-infected cells depleted of TIA1. HeLa cells transfected with NT siRNA or TIA1 siRNA for 68 to 72 h were infected with wt VSV at an MOI of 1. At 6 hpi, cells were pulse-labeled with [35S]Met-Cys for 10 min before lysis. Equal amounts of proteins were analyzed on a sodium dodecyl sulfate-polyacrylamide gel by electrophoresis, and the labeled proteins were detected by fluorography. Viral proteins are identified on the right.
Previously, we reported that PCBP2 interacts with VSV P protein and inhibits VSV growth and gene expression (23). Thus, we next determined whether PCBP2 is present in these TIA1-containing SG-like structures. IF staining of VSV-infected cells expressing HA-tagged PCBP2 (HA-PCBP2) showed that HA-PCBP2 colocalizes with endogenous TIA1 as well as with the viral P protein in the SG-like structures (Fig. 3B). In mock-infected cells, both HA-PCBP2 and TIA1 were detected mostly in the nucleus, where they partially colocalized. These results indicate that PCBP2 associates with TIA1 and VSV P protein in these virus-induced SG-like structures.
In uninfected cells, TIA1 plays a key role in the induction of SG formation and translational shutoff by replacing eIF2-GTP-tRNAMet in the canonical preinitiation complex, leading to translational silencing (37). We found that in VSV-infected cells, TIA1 suppresses viral infection, colocalizes with the viral N and P proteins, and is localized to the virus-induced SG-like structures. Previous studies showed that VSV infection results in the formation of cytoplasmic granular structures (27, 28) that contain not only the viral N, P, and L replicative proteins but also viral RNA, and these structures were described as cytoplasmic granules or replication and transcription inclusions (27, 28). Therefore, we next determined whether the virus-induced SG-like structures contain the other VSV replication proteins, such as the L protein, and viral RNA. To address this question, we infected cells with VSV and, at 2 hpi, incubated the cells in actinomycin D (ActD) for 1 h prior to the use of BrUTP for 4 h to label the newly synthesized viral RNA as described previously (27). Under these conditions, cellular DNA-dependent RNA synthesis is inhibited without any effect on viral RNA-dependent RNA synthesis, and the newly synthesized viral RNAs that have incorporated BrUTP can be specifically detected with an antibody against bromodeoxyuridine (BrdU) (27). IF staining of TIA1 and the VSV L protein or the BrUTP-labeled viral RNA showed that TIA1 colocalized with the viral L protein and the newly synthesized viral RNA (Fig. 3C). The TIAR protein was also found to colocalize with VSV N, P, L, and RNA in the same pattern as that observed for TIA1 (Fig. 3D and data not shown). These results suggest that VSV infection induces SG-like structures containing the viral replicative proteins and viral RNA.
VSV replication and ongoing protein synthesis are required for the generation and maintenance of SG-like structures.
The observation that TIA1-containing SG-like structures increase in number during the course of VSV infection led us to ask whether the formation of these SG-like structures required ongoing viral protein synthesis as well as viral replication. To address this question, we infected cells with UV-inactivated VSV and performed IF assays for the VSV N protein and TIA1 at 7 hpi. In cells infected with UV-inactivated VSV, the majority of TIA1 was still present in the nucleus, and cytoplasmic aggregates of TIA1 similar to SG-like structures, typically seen in VSV-infected cells (Fig. 4A, middle row), were not detected (Fig. 4A, bottom row). The absence of detectable levels of N protein even at 7 hpi suggested that UV treatment was sufficient to inactivate the virus and the induction of SG-like structures. These results indicate that VSV replication is critical for the formation of SG-like structures.
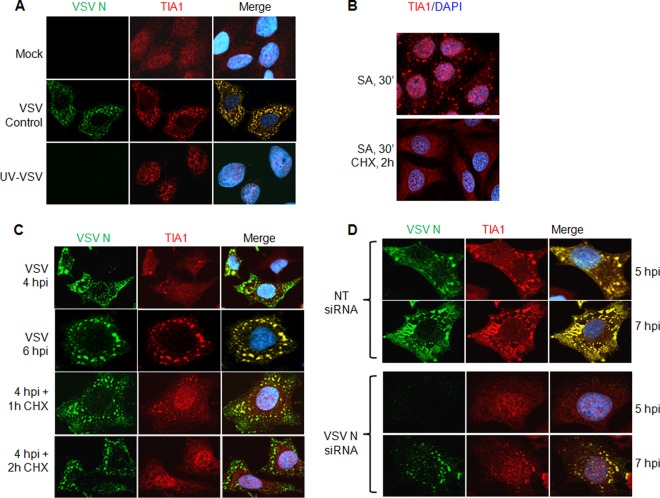
Fig 4.
VSV replication and ongoing protein synthesis are required for the generation and maintenance of SG-like structures. (A) UV-inactivated VSV does not induce SG-like structures. HeLa cells were infected with untreated (VSV Control) or UV-treated (UV-VSV) VSV at an MOI of 1 for 7 h. Cells were processed for IF to detect the VSV N (green) and TIA1 (red) proteins. The secondary antibodies were the same as those described for Fig. 3A. Nuclei were stained with 4′,6-diamidino-2-phenylindole. (B) SGs induced by SA are disrupted by the protein synthesis inhibitor CHX. HeLa cells were treated with SA for 30 min and subsequently were either left untreated (top) or treated with CHX (bottom) for another 2 h before processing for IF. TIA1 was immunostained in red, and nuclei were stained with 4′,6-diamidino-2-phenylindole. (C) VSV-induced SG-like structure formation and maintenance require ongoing protein synthesis. HeLa cells were infected with VSV as described in panel A above for 4 or 6 h before IF (top two rows) or were treated with CHX at 4 hpi for 1 or 2 h and were then processed for IF (bottom two rows) to detect the VSV N and cellular TIA1 proteins. Staining with secondary antibodies and 4′,6-diamidino-2-phenylindole was carried out as in the experiment for which results are shown in panel A. (D) Either SG-like structures are not formed, or their numbers are reduced and their formation delayed, when VSV replication and viral protein synthesis are inhibited by siRNA targeting VSV N mRNA. HeLa cells were transfected either with NT siRNA (top two rows) or with siRNA for VSV N mRNA (bottom two rows) for 72 h and were then infected with VSV at an MOI of 1. IF was performed at 5 and 7 hpi for VSV N and TIA1 proteins. The secondary antibodies used in these experiments were the same as those described for Fig. 3A. Nuclei were stained with 4′,6-diamidino-2-phenylindole.
Since viral protein synthesis is necessary for VSV replication, we next explored whether viral protein synthesis is required for the maintenance of the SG-like structures in infected cells. Cycloheximide (CHX), a translational inhibitor of protein synthesis, was used in VSV-infected cells to examine the formation of SG-like structures. CHX is also known to be an inhibitor of SG formation, since it stalls the elongation of translation without allowing the disassembly of the polysomes and thus prevents the formation and/or maintenance of SGs (2). In agreement with previous observations (2, 17), the SGs induced by SA were disrupted when cells were incubated with CHX for 2 h (Fig. 4B). Although SG-like structures were readily seen in VSV-infected cells at 4 hpi and had developed throughout the cells by 6 hpi, the number and size of these structures were markedly reduced by incubation of the infected cells with CHX at 4 hpi for an additional 1 or 2 h (Fig. 4C). The reductions in the size and number of SG-like structures in the prolonged presence of CHX are concomitant with increased translocation of TIA1 protein back to the nucleus (Fig. 4C), suggesting that the induction and maintenance of VSV-induced SG-like structures require active viral and/or cellular translation. This hypothesis was further confirmed when we used siRNA targeting VSV N mRNA to specifically inhibit translation, deplete the viral N protein pool in infected cells, and block viral replication. Transfection of N mRNA-specific siRNAs into cells inhibits VSV infection by >99% (34) and results in undetectable levels of viral proteins and virus replication. Under these conditions, no formation of TIA1-containing SG-like structures was detected at 5 hpi, although at 7 hpi, a very small number of cells expressed low levels of the viral N protein and exhibited a few small SG-like structures (Fig. 4D, bottom two rows). These structures, however, could be readily detected in infected cells transfected with a nontargeting siRNA at 5 or 7 hpi (Fig. 4D, top two rows). These results suggest that viral protein synthesis and viral replication are required for the induction and development of TIA1-containing SG-like structures in VSV-infected cells.
VSV induced SG-like structure formation is not dependent on a cellular microtubule or microfilament network.
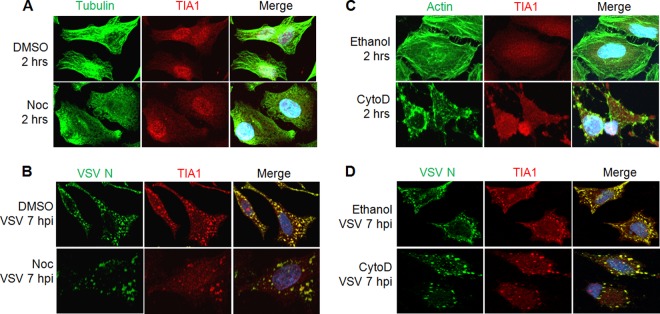
Cytoskeletal networks, particularly the microtubules and microfilaments, have been shown to play different roles for the formation of SGs. Microtubules are required for the induction, but not for the maintenance, of already formed SGs. In contrast, microfilaments are dispensable for SG formation (38–41). Since microtubules and microfilaments are important for VSV entry and the trafficking of viral components in infected cells (27, 28, 42), we explored whether microtubules and microfilaments are involved in the formation of VSV-induced SG-like structures. In uninfected cells treated with nocodazole (Noc), a drug known to inhibit microtubule polymerization, complete depolymerization of microtubules was observed after 2 h of treatment (Fig. 5A), and under these conditions, SA-induced SG formation was inhibited, as reported previously (39, 40). In cells treated with Noc for 2 h and subsequently infected with VSV for 7 h, SG-like structures containing TIA1 could be detected readily, although they were somewhat less numerous than those in control cells treated with dimethyl sulfoxide (DMSO) (Fig. 5B). The reduced number of SG-like structures could be a result of reduced and/or delayed growth of VSV in the presence of Noc (27, 28). These results indicate that VSV-induced SG-like structures containing TIA1 are not dependent on a microtubule network.
Fig 5.
The formation of VSV-induced SG-like structures is not dependent on a cellular microtubule or microfilament network. (A and B) A microtubule network is dispensable for the formation of VSV-induced SG-like structures. Cells were treated with DMSO (vehicle control) or Noc for 2 h before mock infection (A) or infection with VSV at an MOI of 1 (B). Mock-infected cells treated with Noc were used for IF to detect tubulin (green) and TIA1 (red). VSV-infected cells were harvested at 7 hpi (9 h after Noc treatment), and specific antibodies were used to detect VSV N (green) and TIA1 (red). (C and D) The formation of VSV-induced SG-like structures does not require a microfilament network. Cells were treated with ethanol (vehicle control) or CytoD for 2 h before mock infection (C) or infection with VSV at an MOI of 1 (D). Mock-infected cells treated with CytoD were used for IF to detect actin (green) and TIA1 (red). VSV-infected cells were harvested at 7 hpi (9 h after CytoD treatment), and specific antibodies were used to detect VSV N (green) and TIA1 (red). The secondary antibodies used in these experiments were Alexa Fluor 594-labeled donkey anti-goat IgG for TIA1 (red) and Alexa Fluor 488-labeled donkey anti-mouse IgG for VSV N, tubulin, or actin (green).
To examine the involvement of microfilaments in the formation of SG-like structures, cells were treated for 2 h with cytochalasin D (CytoD) to inhibit the polymerization of actin before VSV infection. Treatment of cells with CytoD for 2 h resulted in complete disassembly of actin filaments (Fig. 5C). When cells treated with CytoD were infected with VSV, the SG-like structures were still formed, albeit at slightly reduced levels (Fig. 5D). The reduced levels of SG-like structures can be ascribed to the documented negative effects of actin depolymerization on VSV infection (42).
Overall, the results suggest that the formation of TIA1-containing SG-like structures in VSV-infected cells does not require intact cellular microtubules or microfilaments.
Canonical SGs are distinct from the SG-like structures induced by VSV and are not inhibited in VSV-infected cells.
The observations that (i) VSV infection induces the formation of SG-like structures containing the reliable SG marker TIA1 and (ii) the SG-like structures are not stable in infected cells treated with CHX (Fig. 4C) suggest that the VSV-induced SG-like structures are a form of SGs. However, the fact that the formation of these structures is not dependent on cellular transcription (Fig. 3D) or on a microtubule or microfilament network (Fig. 5) raises an important question: whether the SG-like structures and the authentic SGs are distinct. To address this question, we used IF microscopy to look for the presence of the cellular eIF3 and eIF4A proteins, two authentic markers of SGs (1), in the VSV-induced SG-like structures. Our results show that although a majority of the cellular TIA1 protein colocalized with the eIF3η subunit in SA-induced SGs in the cytoplasm (Fig. 6A, middle row) and TIA1 colocalized with VSV P protein in the SG-like structures (Fig. 6A, bottom row), a significant proportion of the eIF3η subunit did not colocalize with TIA1 or VSV P protein in these virus-induced structures in the cytoplasm (Fig. 6A, bottom row). Similar results were also obtained with eIF4A in VSV-infected cells (data not shown), suggesting that the VSV-induced SG-like structures are likely not the classical SGs generated in cells under stress conditions.
Fig 6.
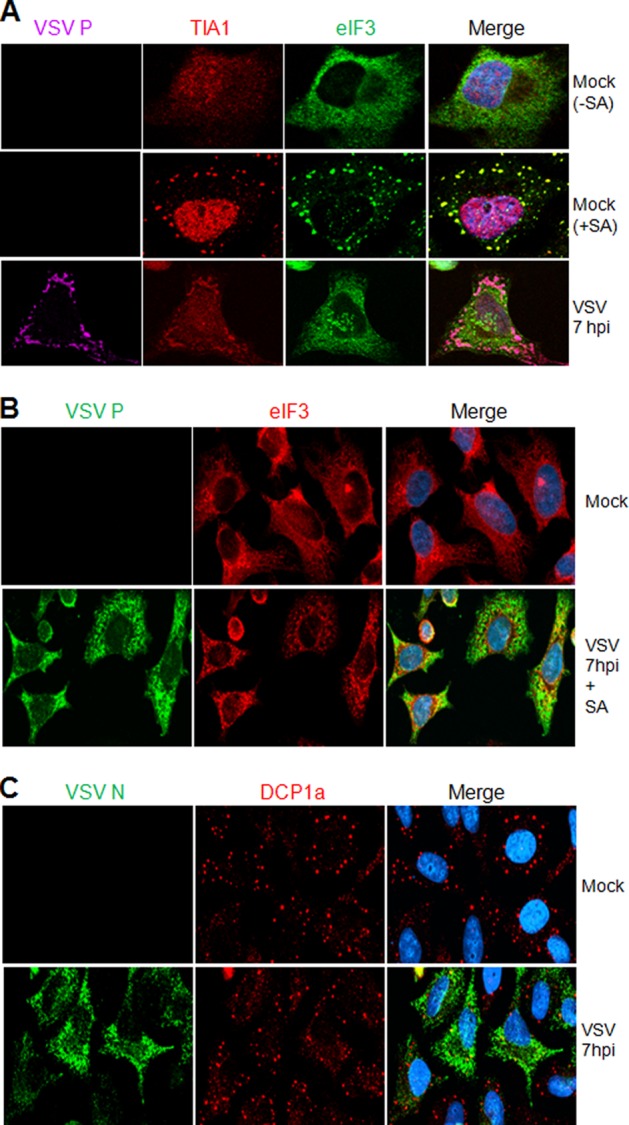
Canonical SGs are distinct from the SG-like structures induced by VSV. (A) VSV-induced SG-like structures do not contain eIF3η. HeLa cells were either mock infected (top two rows) or infected with wt VSV at an MOI of 1 (bottom row). Mock-infected cells were either left untreated (−) or treated with SA (+) for 30 min, after which the cells were processed for IF to detect TIA1 and eIF3. The bottom row shows VSV-infected cells at 7 hpi, immunostained for P, TIA1, and eIF3. The secondary antibodies used were Alexa Fluor 594-labeled donkey anti-goat IgG for TIA1 (red), Alexa Fluor 488-labeled donkey anti-mouse IgG for eIF3η (green), and Alexa Fluor 647-labeled donkey anti-rabbit IgG for VSV P (purple). (B) Canonical SGs are not inhibited in VSV-infected cells. HeLa cells were either mock infected (top row) or infected with wt VSV at an MOI of 1, and at 7 hpi, cells were further treated with SA for 30 min to induce SGs. Cells were immunostained for VSV P and cellular eIF3η. The secondary antibodies used here were Alexa Fluor 594-labeled goat anti-mouse IgG for eIF3η (red) and Alexa Fluor 488-labeled goat anti-rabbit IgG for VSV P (green). (C) VSV infection does not interfere with the formation of cellular PBs. HeLa cells were either mock infected or infected with VSV at an MOI of 1, and at 7 hpi, cells were stained for VSV N and the cellular PB marker DCP1a. The secondary antibodies used were Alexa Fluor 488-labeled goat anti-mouse IgG for VSV N (green) and Alexa Fluor 594-labeled goat anti-rabbit IgG for DCP1a (red).
Many viruses, including influenza A virus, Sendai virus, Junin virus, West Nile virus, dengue virus, orthoreovirus, picornaviruses, and vaccinia viruses, inhibit SG formation in infected cells (5, 6, 8, 12, 16, 17, 19, 21). In VSV-infected cells, we have observed that the virus-induced SG-like structures containing TIA1/PCBP2 are not canonical SGs. Thus, it was of interest to determine whether cells can form SGs induced by SA during VSV infection. To examine this, we infected cells with VSV and then induced SG formation by SA. We could readily observe that canonical SGs (indicated by the authentic marker eIF3η) formed at the same level in VSV-infected cells as in mock-infected cells at various time points postinfection. Figure 6B shows the formation of SGs induced by SA treatment of VSV-infected cells at 7 hpi. SGs induced by heat shock, as visualized by the eIF3η marker, also were not affected by VSV infection (data not shown). Further, we examined the formation of cellular processing bodies (PBs), another type of granule involved in RNA metabolism and known to function together with SGs in translational shutoff via mRNA sequestration or mRNA degradation (1, 2). As shown by IF staining for mRNA-decapping enzyme 1A (DCP1a) in the cytoplasm, PBs were not affected by VSV infection (Fig. 6C). These results indicate that VSV infection neither interferes with the physiological level of PBs nor hampers SG formation in response to oxidative stress or heat shock.
Viral protein synthesis is not significantly affected in the presence of SG-like structures despite eIF2α phosphorylation.
eIF2α phosphorylation and translational shutoff are the putative events concurrent with SG formation under stress conditions, including virus infections (1, 37). Cellular protein synthesis has been shown to be inhibited during VSV infection, while translation of viral proteins continued in parallel with the increasing level of eIF2α phosphorylation (36, 43), although induction of eIF2α phosphorylation before infection resulted in the inhibition of both cellular and VSV protein synthesis (36). Therefore, we examined the relationship between the phosphorylation of eIF2α, the formation of VSV-induced SG-like structures, and the synthesis of viral and cellular proteins. Our results show that the levels of phosphorylated eIF2α in infected cells increased steadily until 5 hpi and remained at the same level thereafter (Fig. 7A and B). The percentage of cells exhibiting SG-like structures increased steadily with time after virus infection (Fig. 7B). Cellular protein synthesis was inhibited with time after VSV infection, while viral protein synthesis levels increased (Fig. 7A and B) in spite of increased levels of phosphorylated eIF2α at different times postinfection. These results suggest that the formation of VSV-induced SG-like structures increased simultaneously with the phosphorylation of eIF2α and the shutoff of cellular protein synthesis.
Since TIA1 is a translational silencer (44–46), we determined whether TIA1 would affect the rate of viral protein synthesis in infected cells. To this end, cells treated with NT siRNA or siRNA specific for TIA1 were infected with VSV for 7 h, and viral protein synthesis was monitored by pulse-labeling with [35S]Met-Cys for 10 min. The results showed that the rate of viral protein synthesis was not significantly affected by TIA1 depletion in these cells (Fig. 7C). Although short pulse-labeling (10-min) studies failed to reveal significant differences in the rates of viral protein synthesis between cells with or without TIA1, it is possible that viral transcription, mRNA stability, protein stability, and/or viral replication could contribute to the increased levels of viral proteins detected in TIA1-depleted cells at 12 hpi as shown in Fig. 1C and 2A.
DISCUSSION
Previously, we showed that the cellular protein PCBP2 interacts with the VSV P protein and inhibits viral replication at the level of gene expression (23). In the present work, we conducted studies to understand how PCBP2 inhibits VSV replication. The rationale for these studies was derived from the observations that (i) PCBP2 is a component of cellular SGs and PBs (2, 22); (ii) PCBP2 interacts and colocalizes with TIA1 in SGs (22); and (iii) several viruses have been shown to induce and/or interfere with the formation of TIA1/TIAR-containing SGs (5, 6, 8, 9, 12, 16, 17, 19). We show here that TIA1, but not TIAR, inhibits VSV growth at the level of viral gene expression and/or replication. Following VSV infection, TIA1 translocates from the nucleus to the cytoplasm and aggregates into cytoplasmic granules similar to the SGs seen in the cytoplasm of cells under stress conditions. These granules also contain the viral replication proteins and RNA as well as PCBP2. The formation of these granules required viral replication and protein synthesis but was independent of cellular microtubule and microfilament networks or cellular transcription. The VSV-induced granules, termed “SG-like structures,” appear to be similar to those described previously as cytoplasmic granules or transcription and replication inclusions (27, 28). However, these studies could not unequivocally rule out the possibility that viral RNA synthesis occurred in the cytoplasm and some of the newly synthesized RNAs quickly accumulated at these sites. Therefore, the question of whether the cytoplasmic granules or inclusions represent the sites of viral RNA synthesis remains unresolved. The formation of the inclusions may reflect the host cell's response to VSV infection (28). This cellular response may depend on the synthesis of critical levels of viral proteins and/or RNA and their interactions with host cell components. Although the host cell proteins in these granules have not been identified, it is tempting to speculate that the granules are similar to the SG-like structures described here and may represent aggregation of translational silencing as well as RNA-binding proteins, such as TIA1 and PCBP2, that are involved in inhibiting viral gene expression. Since both TIA1 and TIAR are apoptotic proteins (47, 48), it is likely that sequestration of these molecules in SG-like structures in infected cells positively impacts cell survival and virus replication.
Although the induction of SG-like structures in infected cells required viral replication and protein synthesis, our inability to detect such structures (data not shown) in cells infected with VSV defective interfering particle-infected cells constitutively expressing the viral replication proteins (49) suggests that viral transcription and/or threshold concentrations of the viral proteins and RNAs are required for their formation. This interpretation is consistent with the delayed appearance (3 hpi) of the structures in infected cells, even though virus transcription/replication and protein synthesis can be detected within the first hour of infection (unpublished data).
Numerous viruses have been shown to either induce (11, 13, 15, 17, 18, 50) or suppress (6, 8, 12, 14, 51) SG formation in infected cells. It appears that the induction or suppression of SGs and of each of their component proteins plays divergent roles in either facilitating or inhibiting virus infections. Infection with poliovirus results in early induction of SGs (15, 19, 20) that are also maintained at later times of infection with compositionally different cellular SG proteins (15, 20). Interestingly, the compositionally different SGs that appear late in poliovirus infection, termed “pseudo-SGs,” contain mostly self-aggregated TIA1 and lack many of the canonical SG components (20). A mutant vaccinia virus lacking the double-stranded RNA-binding E3L protein induces cytoplasmic antiviral granules (AVGs) that inhibit virus replication (17). These AVGs, which are not generated in cells infected with wt vaccinia virus, contain TIA1, G3BP, and other RNA-binding proteins and restrict the replication of the ΔE3L mutant virus (17). On the other hand, hepatitis C virus (HCV)-induced SGs contain not only factors required for HCV replication (Rck1) but also antiviral factors (PCBP2, G3BP1, TIAR, Xrn1) (52). Thus, whether the SGs, AVGs, or pseudo-SGs are generated by the host cells in response to virus infection or are generated by the viral proteins and RNAs to facilitate or limit virus infection remains an open question at this time. Nevertheless, it appears that specific interactions between viral and cellular components over multiple pathways through the recruitment of common and unique SG markers may be crucial for the formation of these granular structures.
In the case of VSV, TIA1/TIAR and PCBP2, along with the viral proteins and RNAs, were found to be localized to the SG-like structures, whose induction was independent of the cellular microtubule or microfilament network. Unlike canonical SGs, whose formation is dependent not only on the cytoskeletal network but also on cellular transcription, the SG-like structures in VSV-infected cells formed in the absence of cellular transcription and were independent of the cytoskeleton. Additional studies will be necessary to determine the mechanism of formation of the SG-like structures in VSV-infected cells. VSV infection also did not interfere with the formation of canonical SGs containing eIF3 or eIF4A induced by SA treatment, suggesting that the SG-like structures induced in VSV-infected cells may have a unique cellular protein composition. Further work to identify the cellular proteins that accumulate in these SG-like structures will be important to determine the role played by these structures in the VSV infection cycle. The observations that (i) the SG-like structures are distinct from canonical SGs, (ii) the TIA1 protein, which blocks the translation of cellular mRNA in canonical SGs, did not inhibit VSV mRNA translation in infected cells (Fig. 7C), and (iii) VSV RNA synthesis is enhanced in TIA1-depleted cells suggest that TIA1 downregulates VSV infection through a mechanism other than translational suppression, its traditional role.
The involvement of eIF2α phosphorylation and the double-stranded RNA-dependent protein kinase (protein kinase R [PKR]) in the formation of VSV-induced SG-like structures has not been examined directly, although the level of SG-like structures increased concomitantly with the level of eIF2α phosphorylation. It will be of interest to explore whether PKR, which is activated in VSV infection and restricts virus replication (53), or other eIF2α-specific kinases play any role in the induction of the SG-like structures.
The observations that VSV replication is enhanced in cells when TIA1 is depleted transiently (by siRNA treatment) (Fig. 1B) but is inhibited in cells permanently depleted of TIA1 (knockout MEFs) (Fig. 1E) appear paradoxical. However, since TIA1 is involved in cellular gene expression, including translational suppression (29, 44–46) and mRNA processing (54–57), it is possible that the expression of a certain cellular gene(s) required for VSV growth may have been permanently compromised in MEFs derived from the knockout mouse. Alternatively, TIA1 depletion in MEFs may have resulted in the activation of an antiviral gene(s) to block VSV growth. In contrast to the knockout of TIA1 in MEFs, transient depletion of TIA1 in HeLa cells may not have permanently affected these cellular factors. Therefore, the true effects of TIA1 on VSV growth could be discerned in transient depletion experiments, whereas in cells stably depleted of the protein, the combined effects of TIA1 depletion and its subsequent effects on cellular proteins are observed. Such a contention is supported by the fact that siRNA-mediated depletion of TIA1 from wt MEFs resulted in increased VSV growth whereas ectopic expression of the protein in TIA1-knockout MEFs led to inhibition of VSV growth. Proteomic and transcriptomic analysis of wt and TIA1-knockout MEFs may reveal important clues for a better understanding of the role of TIA1 in VSV infection.
In summary, our studies presented here reveal that TIA1 inhibits VSV replication and colocalizes with PCBP2, another cellular protein shown previously (23) to inhibit VSV replication, to form SG-like structures in infected cells. Studies to determine the cellular protein composition of these SG-like structures and their roles in VSV-infected cells are ongoing.
ACKNOWLEDGMENTS
We thank Y. Zhou and Terry Fangman (University of Nebraska—Lincoln microscopy core facility) for help in fluorescence microscopy. We also thank P. Anderson (Harvard University) and M. Brinton (Georgia State University) for the kind gift of the immortalized MEFS from wt and TIA1 knockout mice. We appreciate the excellent assistance of Z. H. Gill in the laboratory.
This work was supported in part by National Institutes of Health grant R01 AI34956.
Footnotes
Published ahead of print 17 October 2012
REFERENCES
- 1. Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150 [DOI] [PubMed] [Google Scholar]
- 2. Buchan JR, Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36:932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thomas MG, Loschi M, Desbats MA, Boccaccio GL. 2011. RNA granules: the good, the bad and the ugly. Cell. Signal. 23:324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nover L, Scharf KD, Neumann D. 1989. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell. Biol. 9:1298–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borghese F, Michiels T. 2011. The leader protein of cardioviruses inhibits stress granule assembly. J. Virol. 85:9614–9622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Emara MM, Brinton MA. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U. S. A. 104:9041–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hanley LL, McGivern DR, Teng MN, Djang R, Collins PL, Fearns R. 2010. Roles of the respiratory syncytial virus trailer region: effects of mutations on genome production and stress granule formation. Virology 406:241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khaperskyy DA, Hatchette TF, McCormick C. 2012. Influenza A virus inhibits cytoplasmic stress granule formation. FASEB J. 26:1629–1639 [DOI] [PubMed] [Google Scholar]
- 9. Khong A, Jan E. 2011. Modulation of stress granules and P bodies during dicistrovirus infection. J. Virol. 85:1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li W, Li Y, Kedersha N, Anderson P, Emara M, Swiderek KM, Moreno GT, Brinton MA. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76:11989–12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE., Jr 2010. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J. Virol. 84:12274–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Linero FN, Thomas MG, Boccaccio GL, Scolaro LA. 2011. Junin virus infection impairs stress granule formation in Vero cells treated with arsenite via inhibition of eIF2α phosphorylation. J. Gen. Virol. 92:2889–2899 [DOI] [PubMed] [Google Scholar]
- 13. McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljestrom P. 2005. Importance of eIF2α phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 16:3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Montero H, Rojas M, Arias CF, Lopez S. 2008. Rotavirus infection induces the phosphorylation of eIF2α but prevents the formation of stress granules. J. Virol. 82:1496–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Piotrowska J, Hansen SJ, Park N, Jamka K, Sarnow P, Gustin KE. 2010. Stable formation of compositionally unique stress granules in virus-infected cells. J. Virol. 84:3654–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qin Q, Carroll K, Hastings C, Miller CL. 2011. Mammalian orthoreovirus escape from host translational shutoff correlates with stress granule disruption and is independent of eIF2α phosphorylation and PKR. J. Virol. 85:8798–8810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simpson-Holley M, Kedersha N, Dower K, Rubins KH, Anderson P, Hensley LE, Connor JH. 2011. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J. Virol. 85:1581–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith JA, Schmechel SC, Raghavan A, Abelson M, Reilly C, Katze MG, Kaufman RJ, Bohjanen PR, Schiff LA. 2006. Reovirus induces and benefits from an integrated cellular stress response. J. Virol. 80:2019–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305 [DOI] [PubMed] [Google Scholar]
- 20. White JP, Lloyd RE. 2011. Poliovirus unlinks TIA1 aggregation and mRNA stress granule formation. J. Virol. 85:12442–12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iseni F, Garcin D, Nishio M, Kedersha N, Anderson P, Kolakofsky D. 2002. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 21:5141–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujimura K, Kano F, Murata M. 2008. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA 14:425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dinh PX, Beura LK, Panda D, Das A, Pattnaik AK. 2011. Antagonistic effects of cellular poly(C) binding proteins on vesicular stomatitis virus gene expression. J. Virol. 85:9459–9471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64:241–256 [DOI] [PubMed] [Google Scholar]
- 25. Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salonen A, Ahola T, Kaariainen L. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Das SC, Nayak D, Zhou Y, Pattnaik AK. 2006. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J. Virol. 80:6368–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heinrich BS, Cureton DK, Rahmeh AA, Whelan SP. 2010. Protein expression redirects vesicular stomatitis virus RNA synthesis to cytoplasmic inclusions. PLoS Pathog. 6:e1000958 doi:10.1371/journal.ppat.1000958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 19:4154–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Izquierdo JM, Valcarcel J. 2007. Two isoforms of the T-cell intracellular antigen 1 (TIA-1) splicing factor display distinct splicing regulation activities. Control of TIA-1 isoform ratio by TIA-1-related protein. J. Biol. Chem. 282:19410–19417 [DOI] [PubMed] [Google Scholar]
- 31. Das SC, Pattnaik AK. 2004. Phosphorylation of vesicular stomatitis virus phosphoprotein P is indispensable for virus growth. J. Virol. 78:6420–6430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Das SC, Pattnaik AK. 2005. Role of the hypervariable hinge region of phosphoprotein P of vesicular stomatitis virus in viral RNA synthesis and assembly of infectious virus particles. J. Virol. 79:8101–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schubert M, Harmison GG, Richardson CD, Meier E. 1985. Expression of a cDNA encoding a functional 241-kilodalton vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 82:7984–7988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Panda D, Das A, Dinh PX, Subramaniam S, Nayak D, Barrows NJ, Pearson JL, Thompson J, Kelly DL, Ladunga I, Pattnaik AK. 2011. RNAi screening reveals requirement for host cell secretory pathway in infection by diverse families of negative-strand RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 108:19036–19041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Das PB, Dinh PX, Ansari IH, de Lima M, Osorio FA, Pattnaik AK. 2010. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J. Virol. 84:1731–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Connor JH, Lyles DS. 2005. Inhibition of host and viral translation during vesicular stomatitis virus infection. eIF2 is responsible for the inhibition of viral but not host translation. J. Biol. Chem. 280:13512–13519 [DOI] [PubMed] [Google Scholar]
- 37. Anderson P, Kedersha N. 2002. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fujimura K, Katahira J, Kano F, Yoneda Y, Murata M. 2009. Microscopic dissection of the process of stress granule assembly. Biochim. Biophys. Acta 1793:1728–1737 [DOI] [PubMed] [Google Scholar]
- 39. Ivanov PA, Chudinova EM, Nadezhdina ES. 2003. Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp. Cell Res. 290:227–233 [DOI] [PubMed] [Google Scholar]
- 40. Ivanov PA, Chudinova EM, Nadezhdina ES. 2003. RNP stress-granule formation is inhibited by microtubule disruption. Cell Biol. Int. 27:207–208 [DOI] [PubMed] [Google Scholar]
- 41. Kwon S, Zhang Y, Matthias P. 2007. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 21:3381–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cureton DK, Massol RH, Whelan SP, Kirchhausen T. 2010. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog. 6:e1001127 doi:10.1371/journal.ppat.1001127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krishnamoorthy J, Mounir Z, Raven JF, Koromilas AE. 2008. The eIF2α kinases inhibit vesicular stomatitis virus replication independently of eIF2α phosphorylation. Cell Cycle 7:2346–2351 [DOI] [PubMed] [Google Scholar]
- 44. Dixon DA, Balch GC, Kedersha N, Anderson P, Zimmerman GA, Beauchamp RD, Prescott SM. 2003. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J. Exp. Med. 198:475–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. 2006. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell. Biol. 26:3295–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu Q, Cok SJ, Zeng C, Morrison AR. 2003. Translational repression of human matrix metalloproteinases-13 by an alternatively spliced form of T-cell-restricted intracellular antigen-related protein (TIAR). J. Biol. Chem. 278:1579–1584 [DOI] [PubMed] [Google Scholar]
- 47. Forch P, Valcarcel J. 2001. Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis 6:463–468 [DOI] [PubMed] [Google Scholar]
- 48. Kawakami A, Tian Q, Duan X, Streuli M, Schlossman SF, Anderson P. 1992. Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl. Acad. Sci. U. S. A. 89:8681–8685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Panda D, Dinh PX, Beura LK, Pattnaik AK. 2010. Induction of interferon and interferon signaling pathways by replication of defective interfering particle RNA in cells constitutively expressing vesicular stomatitis virus replication proteins. J. Virol. 84:4826–4831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA. 2007. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell. Microbiol. 9:2218–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Isler JA, Maguire TG, Alwine JC. 2005. Production of infectious human cytomegalovirus virions is inhibited by drugs that disrupt calcium homeostasis in the endoplasmic reticulum. J. Virol. 79:15388–15397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ruggieri A, Dazert E, Metz P, Hofmann S, Bergeest JP, Mazur J, Bankhead P, Hiet MS, Kallis S, Alvisi G, Samuel CE, Lohmann V, Kaderali L, Rohr K, Frese M, Stoecklin G, Bartenschlager R. 2012. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe 12:71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, Barber GN. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129–141 [DOI] [PubMed] [Google Scholar]
- 54. Del Gatto-Konczak F, Bourgeois CF, Le Guiner C, Kister L, Gesnel MC, Stevenin J, Breathnach R. 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20:6287–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J. 2002. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 21:6882–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Le Guiner C, Lejeune F, Galiana D, Kister L, Breathnach R, Stevenin J, Del Gatto-Konczak F. 2001. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 276:40638–40646 [DOI] [PubMed] [Google Scholar]
- 57. Zuccato E, Buratti E, Stuani C, Baralle FE, Pagani F. 2004. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J. Biol. Chem. 279:16980–16988 [DOI] [PubMed] [Google Scholar]