Abstract
Two transcription factors, ZEBRA and Rta, switch Epstein-Barr virus (EBV) from the latent to the lytic state. While ZEBRA also plays an obligatory role as an activator of replication, it is not known whether Rta is directly required for replication. Rta is dispensable for amplification of an oriLyt-containing plasmid in a transient-replication assay. Here, we assessed the requirement for Rta in activation of viral DNA synthesis from the endogenous viral genome, a function that has not been established. Initially, we searched for a ZEBRA mutant that supports viral replication but not transcription. We found that Z(S186A), a mutant of ZEBRA unable to activate transcription of Rta or viral genes encoding replication proteins, is competent to bind to oriLyt and to function as an origin recognition protein. Ectopic expression of the six components of the EBV lytic replication machinery failed to rescue replication by Z(S186A). However, addition of Rta to Z(S186A) and the mixture of replication factors activated viral replication and late gene expression. Deletion mutagenesis of Rta indicated that the C-terminal 10 amino acids (aa) were essential for the function of Rta in replication. In vivo DNA binding studies revealed that Rta interacted with the enhancer region of oriLyt. In addition, expression of Rta and Z(S186A) together, but not individually, activated synthesis of the BHLF1 transcript, a lytic transcript required for the process of viral DNA replication. Our findings demonstrate that Rta plays an indispensable role in the process of lytic DNA replication.
INTRODUCTION
Lytic infection is intrinsic to Epstein-Barr virus (EBV) pathogenesis. Viral particles are synthesized and assembled exclusively during the lytic state. Activation of the lytic cycle gene expression program is the only route for virus propagation. Activation of the lytic program is mediated by two transcription factors, ZEBRA and Rta, encoded by the viral BZLF1 and BRLF1 genes (1–4). Both proteins are required to trigger sequential events that include expression of replication proteins (RPs), viral genome amplification, and synthesis of late structural proteins (1, 2, 5–14).
A complete linear viral genome contains two copies of the origin of lytic DNA replication (oriLyt), which regulate the process of genome amplification. These oriLyt sequences are ∼105 kbp apart and are located in the left and right duplicated sequences of the genome (DSL and DSR) (15). Naturally occurring EBV strains with deletions of one copy of either origin, such as the B95-8 and P3HR1 virus strains, still maintain the capacity to replicate the entire viral genome (16, 17). Each origin of replication contains the DNA regulatory elements sufficient to replicate a surrogate oriLyt plasmid in cis (15). Previous studies with oriLyt plasmids characterized three important cis elements present in the DSL origin, also known as BamHI-H oriLyt (18). These cis elements are the upstream and downstream elements, which are essential for genome amplification, and a dispensable enhancer element (15, 18). The upstream element overlaps the promoter controlling expression of the BHLF1 gene. The DNA sequences overlapping the BHLF1 promoter and the upstream region of oriLyt contain four ZEBRA response elements (ZRE-1 to -4) (9). ZRE-1 to -4 are necessary for transcription of BHLF1 and replication; deletion of these four ZREs or substitution with bovine papillomavirus E2 or Gal4 binding sites disrupts the function of the BHLF1 promoter in activation of transcription and the function of oriLyt in replication (12, 13). In addition to the ZREs, the upstream region contains a functional C/EBP binding site located between ZRE-2 and ZRE-3. Deletion of the C/EBP binding site in an oriLyt plasmid has a deleterious effect on the capacity of oriLyt to support replication (19).
The 40-bp downstream element of oriLyt is separated from the upstream region by approximately 530 nucleotides (18). The downstream element lacks ZEBRA and Rta binding sites but contains Sp1 and ZBP-89 binding motifs (20, 21). In vitro DNA binding studies revealed that Sp1 and ZBP-89 bind to the downstream component of oriLyt in a cooperative manner (20). Overexpression of Sp1 or ZBP-89 enhanced replication of an oriLyt plasmid in EBV-positive cells. Sp1 and ZBP-89 were shown to interact with the viral polymerase holoenzyme that consists of the catalytic subunit (BALF5) and the polymerase processivity factor (BMRF1). Thus, Sp1and ZBP-89 might contribute to the process of lytic DNA replication by tethering replication proteins to oriLyt (20).
Although the enhancer element in oriLyt is categorized as nonessential, its presence increases the replication efficiency of an oriLyt-containing plasmid. The enhancer element contains three ZEBRA response elements (ZRE-5, -6, and -7), two Rta response elements (RRE-1 and -2), and a ZBRK1 binding site (18, 22). Deletion of the Rta or ZBRK1 binding sites reduced but did not abolish replication of an oriLyt plasmid (18, 22).
In a recent reexamination of the minimal sequences of EBV DNA that are sufficient to mediate oriLyt plasmid amplification, Rennekamp and Lieberman found that either the BHLF1 or the BHRF1 open reading frames (ORFs) and transcripts from one of these regions were essential for oriLyt function (23). Synthesis of the GC-rich BHLF1 transcript seemed to be necessary for the formation of an RNA-DNA hybrid with oriLyt DNA during initiation of lytic replication. This hybrid was thought to promote initial strand separation and recruitment of the viral single-stranded DNA (ssDNA) binding protein, BALF2, to oriLyt. However, it is not understood how the BHLF1and BHRF1 transcripts compensate for each other's role in supporting DNA replication. These new findings implicate transcription factors that mediate activation of the BHLF1and BHRF1 promoters in the process of viral genome amplification.
Rta plays an essential role in the transition of EBV from latency to the lytic cycle in infected B lymphocytes and epithelial cells (6, 11, 14). The Rta protein has three functional domains, a DNA binding domain (positions 1 to 280), a dimerization domain (positions 1 to 232), and a transcription activation domain (positions 416 to 605) (24). The DNA binding activity of Rta is regulated by an intrinsic 55-amino-acid sequence (positions 555 to 605) referred to as the DNA binding-inhibitory sequence (DBIS). The DBIS blocks the capacity of wild-type (wt) Rta or the minimal DNA binding region of Rta (positions 1 to 350) to interact with DNA (25). Deletion of the last 10 amino acids in Rta or alanine substitution of phenylalanines 600 and 605 disrupted the action of the DBIS. While these mutations enhance the capacity of Rta to interact with DNA in vitro, the mechanism by which the inhibitory effect of the DBIS is suppressed to allow Rta to interact with DNA in vivo is not known.
While the role of Rta in activation of transcription is well established, evidence for its involvement in the process of viral DNA replication is conflicting. Rta is dispensable for replication of an oriLyt-containing plasmid. However, several lines of indirect evidence suggest that Rta plays an important role in viral DNA replication. Rta colocalizes with other replication proteins such as BMRF1 to viral replication compartments in the nuclei of lytically infected cells (26, 27). Rta interacts with the primase-associated factor BBLF2/3, one of the main components of the viral replication machinery (22). Rta partially rescues the replication function of Z(m12/13), a ZEBRA mutant that fails to support replication, presumably due to its inability to interact with BBLF2/3-BSLF1 primase subcomplex (28). In Kaposi's sarcoma-associated herpesvirus (KSHV), the ORF50 protein, a homolog of Rta, plays an essential role in the process of viral DNA replication (29).
In this report, we employed replication-defective ZEBRA mutants to investigate the role of Rta in replicating the endogenous viral genome. We found that providing the origin recognition function of ZEBRA plus all six replication proteins was insufficient to activate viral replication of the endogenous viral genome. However, ectopic expression of Rta was sufficient to rescue both viral DNA replication and late gene expression. Our findings established an essential role for the Rta protein in the process of lytic viral DNA replication. Plausible mechanisms for the role of Rta in replication included binding to the enhancer region of oriLyt and activation of transcription of BHLF1.
MATERIALS AND METHODS
Cell culture and transfection.
BZKO (Bam Z knockout) cells are 293 human embryonic kidney cells (30, 31) stably transfected with the EBV B95-8 bacmid containing a deletion in the BZLF1 gene, encoding the ZEBRA protein (6). BZKO cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 μg/ml hygromycin B (Calbiochem), and antibiotics (penicillin-streptomycin at 50 units/ml and amphotericin B at 1 μg/ml). DNA transfection was performed in 25-cm2 flasks using 36 μl DMRIE-C reagent (Invitrogen) and 18 μg of total DNA, which included 3 μg of ZEBRA or Rta expression vectors and 2 μg of each construct encoding a replication protein. Empty vectors were used to adjust the final DNA concentration to 18 μg. All transfected cells were harvested at 48 h after transfection. Cells were maintained at 37°C in a humidified 5% CO2 incubator.
Expression vectors.
The expression vectors for ZEBRA (Z), Z(S186A), Z(S173A), BALF2, and BMRF1 were prepared as previously described (32–34). Expression vectors for the viral open reading frames encoding Rta (R), BALF5, BBLF2/3, BBLF4, and BSLF1 were a kind gift from Diane Hayward (2, 7). DNA fragments corresponding to Rta amino acid positions 1 to 350, 1 to 450, 1 to 550, and 1 to 595 were generated by PCR. The PCR fragments were cloned into the EcoRI and XbaI sites of the pRTS eukaryotic expression vector. Construction of the pRTS-VP16 plasmid and Rta(1-550)-VP16 has been described previously (35). PCR fragments encoding Rta(1-595) and Rta(F600A/F605A) were cloned into the pRTS-VP16 construct using XbaI and PstI cloning sites.
Antibodies.
Anti-FLAG is a mouse monoclonal antibody (Sigma). The monoclonal BMRF1 antibody (R3.1) was kindly provided by G. Pearson (36). Anti-ZEBRA (S1605-F), anti-BFRF3 (S1931-1), and anti-Rta (S2454) are polyclonal rabbit sera developed in our laboratory against full-length viral proteins expressed in Escherichia coli using the pET expression system. The proteins were purified from E. coli using nickel affinity chromatography.
SDS-PAGE and immunoblotting.
Expression of viral proteins was assessed using Western blotting. Cell lysates were prepared 48 h after transfection by resuspending cells in sodium dodecyl sulfate (SDS) sample buffer at 106 cells per 10 μl. Samples were sonicated for 20 s and boiled for 5 min. Proteins were separated in a 10% SDS-polyacrylamide gel by electrophoresis and transferred to nitrocellulose membranes (Bio-Rad). The Western blot was incubated with the indicated antibodies. For mouse monoclonal antibodies, a rabbit anti-mouse bridge (Invitrogen) was used. Protein bands were detected using 125I-labeled protein A (Perkin-Elmer) and autoradiography.
Quantitative RT-PCR (qRT-PCR).
RNA was purified from BZKO cells at 48 h after transfection using Qiagen products. The level of EBV transcripts encoding lytic viral replication proteins was determined using the iScript SYBR green RT-PCR kit (Bio-Rad). The amount of RNA present in each sample was normalized to 18S rRNA or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) RNA. Assays on individual samples were performed in triplicate. The efficiency of each primer set was determined by quantitative PCR using 10-fold serial dilutions of template DNA. The following DNA sequences were used as primers: to detect BALF2, forward 5′-CACCACATCTATGACGTTGC-3′ and reverse 5′-CCGGACACTAAGCTCTCAAG-3′; BALF5, forward 5′-CCGTCTACCAGAAGTTCGTC-3′ and reverse 5′-GGAGCAGCTTGTCGAAATAA-3′; BBLF2/3, forward 5′-GAGACCGCAGACTCTTGTCC-3′ and reverse 5′-AACTGTCCGGGAGACTCAGA-3′; BBLF4, forward 5′-CAGGGTCTGTCCCTAAACAA-3′ and reverse 5′-GATAGGGGATTCCTGTCCAT-3′; BSLF1, forward, 5′-TTTCTCCATGTCACGTTTGA-3′ and reverse 5′-ACACCCTCACATTCTTGGAG-3′, and BHLF1, forward, 5′-GCCCATTCGAACCCTACC-3′ and reverse, 5′-CTCCACTGCACCTGGAAT-3′.
EBV lytic replication assay.
BZKO cells were lysed and sonicated in 50 mM Tris-HCl (pH 8.1), 1% SDS, and 10 mM EDTA. Cell lysates were centrifuged, and supernatants were diluted 10-fold in 16.7 mM Tris-HCl (pH 8.1), 0.01% SDS, 1.1% Triton X-100, 167 mM NaCl, and 1.2 mM EDTA. Proteins were digested using proteinase K (Roche). The DNA concentration was measured by quantitative PCR.
Chromatin immunoprecipitation (ChIP).
Immunoprecipitation of viral DNA was performed as previously described (32). Rta-associated DNA was immunoprecipitated using two different antibodies, a mouse monoclonal anti-Rta antibody (Argene) and a rabbit polyclonal antibody (S2454). ZEBRA-associated DNA was immunoprecipitated using a rabbit polyclonal antibody (S1605-F) generated against the full-length protein.
RESULTS
Z(S186A), a mutant that fails to disrupt EBV latency, binds efficiently to the origin of lytic replication.
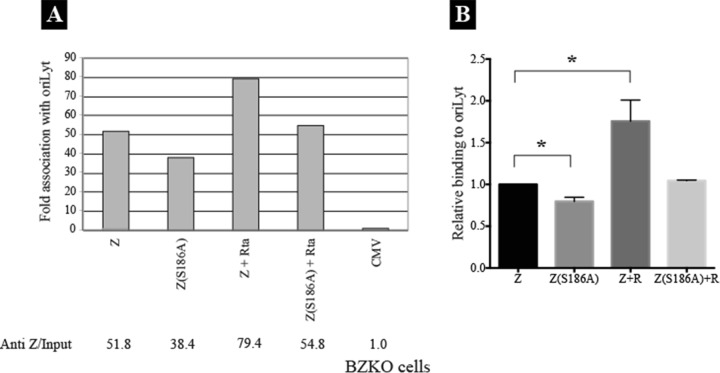
Z(S186A) is defective at activating expression of Rta (34, 37) because of a defect in binding to methylated CpGs that are embedded in two ZEBRA response elements (ZREs) of Rp (38, 39). Initially, using ChIP, we examined the capacity of Z(S186A) to bind to the upstream region of oriLyt in vivo, and we assessed the effect of overexpression of Rta on this interaction. BZKO cells, a 293 cell line harboring an EBV bacmid that lacks a functional gene for ZEBRA, were transfected with empty vector (cytomegalovirus [CMV]) or expression vectors encoding Z(S186A) or wt ZEBRA in the absence and presence of Rta. After 48 h, cells were cross-linked with formaldehyde and ZEBRA was immunoprecipitated using a specific antibody. Using real-time PCR to determine the amount of oriLyt coimmunoprecipitated with the ZEBRA protein, we found that Z(S186A) maintained 70% of the capacity of wt ZEBRA to interact with oriLyt (Fig. 1). Coexpression of Rta enhanced association of Z(S186A) with oriLyt to a level equivalent to that observed with the wt ZEBRA protein alone. The effect of Rta on the interaction of ZEBRA with oriLyt was similar for both Z(S186A) and wt ZEBRA; coexpression of Rta increased the amount of oriLyt precipitated with Z(S186A) or wt ZEBRA by 43% and 53%, respectively. These results showed that Z(S186A) has the capacity to interact with oriLyt with high efficiency and that overexpression of Rta modestly, but reproducibly, enhances this interaction. Since Z(S186A) can recognize oriLyt in the absence of any other EBV replication protein, in several subsequent experiments (see Fig. 3, 5, and 6), Z(S186A) was provided as an origin binding protein.
Fig 1.
Rta enhances association of wt Z and Z(S186A) to oriLyt. (A) Chromatin immunoprecipitation (ChIP) was performed on BZKO cells transfected with empty vector (CMV), wt ZEBRA (Z), or Z(S186A) with and without Rta. The cells were treated with phosphonoacetic acid to block viral replication. Cells were collected after 48 h and cross-linked with formaldehyde. The amount of ZEBRA bound to the upstream region of oriLyt was determined by real-time PCR using the standard curve method. The AntiZ/Input ratio represents the amount of oriLyt immunoprecipitated with a specific antibody to ZEBRA relative to that present in the corresponding input sample. Fold association of ZEBRA with EBV oriLyt DNA was normalized to the AntiZ/Input ratio present in cells transfected with empty vector. (B) Bar graph demonstrating the efficiency of Z(S186A), ZEBRA+Rta, and Z(S186A)+Rta in binding to oriLyt relative to wild-type ZEBRA. The data were compiled from three independent ChIP experiments. Asterisks denote statistically significant changes (P < 0.05). ChIP of Z(S186A) to oriLyt in the presence of Rta was repeated twice, and thus no statistical analysis is available for this sample.
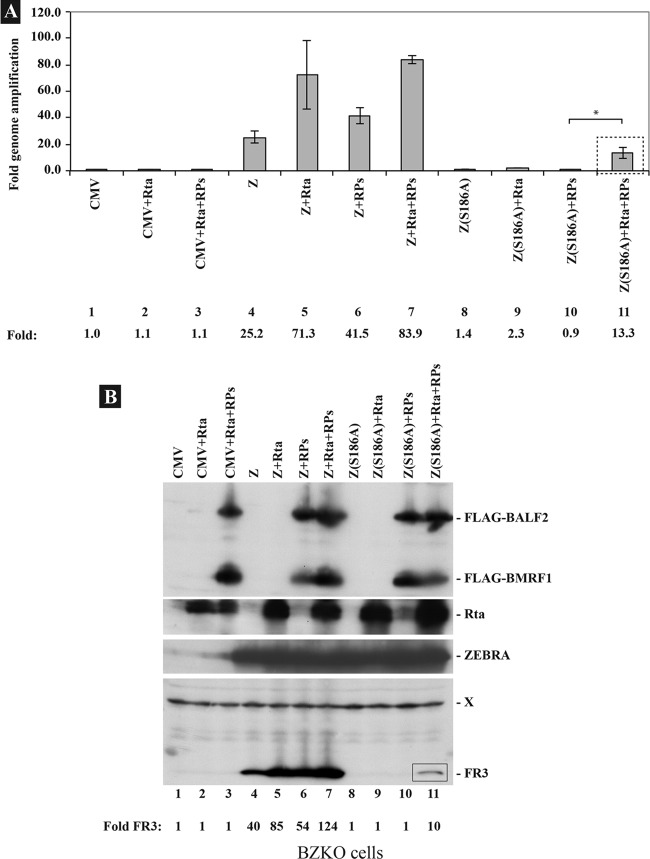
Fig 3.
Rta rescues activation of late gene expression and EBV lytic DNA replication by Z(S186A). (A) DNA purified from BZKO cells transfected with empty vector (CMV) or plasmids encoding wt ZEBRA (Z) or Z(S186A) with or without expression vectors for Rta, EBV replication proteins, or Rta plus EBV replication proteins was analyzed for its EBV genome content by quantitative PCR using primers specific for the upstream region of oriLyt. The extent of EBV genome amplification was normalized to the amount of DNA detected in cells transfected with empty vector. RPs, 2 μg of each of the six EBV replication proteins (BBLF4, BBLF2/3, BSLF1, BALF2, BMRF1, and BALF5). (B) Western blot analysis of the same samples. The membrane was blotted with antibodies against the late protein FR3, the ZEBRA protein, and the FLAG epitope. Both BALF2 and BMRF1 were expressed with a FLAG tag at their N termini. The asterisk denotes statistically significant changes (P < 0.05). The P value was calculated from four independent experiments.
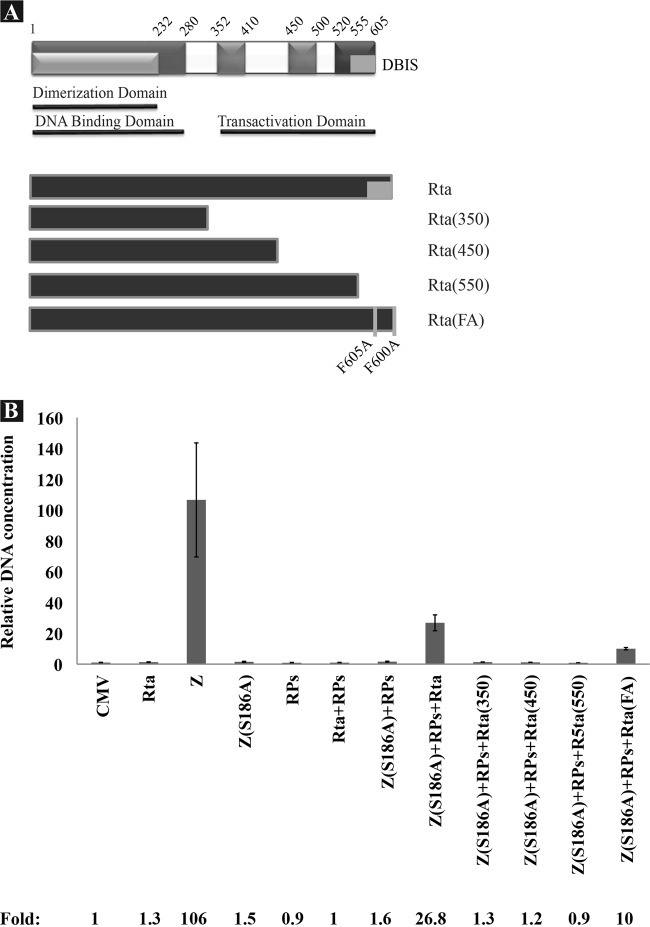
Fig 5.
The C-terminal 55 amino acids of Rta are necessary for its role in lytic DNA replication. (A) Schematic diagram of Rta and various mutants. (B) Quantitative PCR was used to measure the extent of EBV genome amplification in BZKO cells coexpressing Z(S186A) together with a mixture of the six EBV replication proteins and either wt Rta or mutant Rta. Viral genome synthesis was monitored using primers specific to the upstream region of oriLyt. Fold genome amplification was normalized to the amount of oriLyt DNA detected in BZKO cells transfected with empty vector (CMV). RPs represent all six EBV replication proteins (BBLF4, BBLF2/3, BSLF1, BALF2, BMRF1, and BALF5).
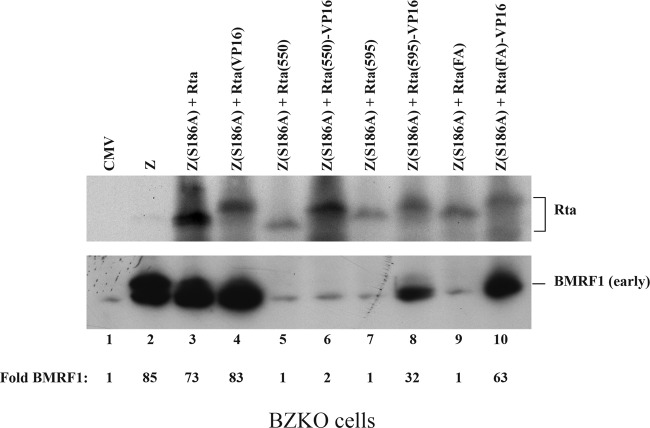
Fig 6.
Addition of the heterologous VP16 transactivation domain to Rta deletion mutants restores the capacity of Rta to activate expression of BMRF1 by synergy with Z(S186A). Western blot analysis of BZKO cells expressing Z(S186A) together with full-length or mutated Rta with or without VP16 fusions is shown. Cells were harvested after 48 h. The membrane was blotted with antibodies against Rta and BMRF1.
Role of Rta in activating expression of genes encoding the EBV replication proteins.
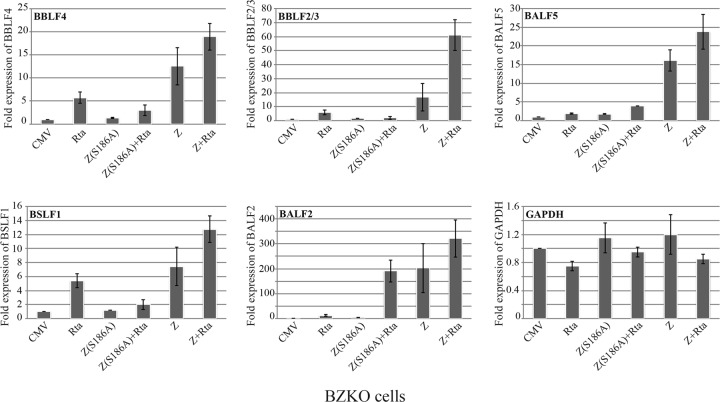
Previous reports demonstrated that Rta synergizes with ZEBRA to activate expression of BALF2, the ssDNA binding protein, and BMRF1, the DNA polymerase processivity factor, two essential components of the EBV replication machinery (37, 40, 41). However, the role of Rta in activating expression of genes encoding other viral replication proteins was unknown. To determine if Rta, either alone or synergistically with Z(S186A), activates transcription of genes encoding EBV replication proteins, we isolated RNA from BZKO cells and used quantitative RT-PCR (qRT-PCR) to assess the transcript levels of five genes, i.e., those for BBLF4, BBLF2/3, BSLF1, BALF2, and BALF5, that encode replication proteins. wt ZEBRA activated transcription of all five genes, while expression of Z(S186A) alone failed to activate transcription of any of the genes examined (Fig. 2). Expression of Rta activated four of the five genes, namely, BALF2, BBLF4, BBLF2/3, and BSLF1. However, the level of transcripts induced by Rta alone was always less than that obtained as a result of expressing wt ZEBRA, ZEBRA plus Rta, or, in the case of BALF2, Z(S186A) plus Rta. Since ZEBRA activates Rta in BZKO cells, the effects of ZEBRA are likely to result from the combined action of ZEBRA and Rta. Although previous reports indicated that Rta alone activates the BALF5 promoter by an indirect mechanism of interaction with the cellular proteins E2F and USF (42, 43), we found that Rta alone did not significantly increase expression of BALF5 in BZKO cells. Consistent with a previous report showing that Z and Rta synergize to activate the BALF2 promoter (41), coexpression of Rta and Z(S186A) increased the level of BALF2 mRNA by 16-fold relative to that with Rta alone. Based on these results, genes encoding the viral replication proteins could be divided into three groups: BALF5 was activated primarily by ZEBRA; BBLF2/3, BBLF4, and BSLF1 were activated by the additive action of ZEBRA and Rta; and BALF2 and BMRF1 were activated by synergy between Rta and Z(S186A) (Fig. 2 and 3) (37, 41). For the purpose of using Z(S186A) to demonstrate a role of Rta in lytic replication, it is essential to note that this ZEBRA mutant did not synergize with Rta to activate expression of four of the six viral replication genes (those for BALF2, BALF5, BBLF2/3, BBLF4, BMRF1, and BSLF1). Therefore, since Rta was unable to provide a function that leads to transcription of the full complement of replication proteins, we provided these RPs in trans in subsequent experiments.
Fig 2.
Effects of Rta, wt Z, and Z(S186A) on the expression levels of mRNAs transcribed from EBV genes encoding lytic replication proteins. cDNA was synthesized from RNA samples isolated from BZKO cells transfected with the indicated expression vectors. Cells were harvested at 48 h after transfection. The relative concentration of each transcript was quantitated by real-time PCR using the standard curve method. GAPDH cDNA was used as an internal control for the total amount of cDNA used in each reaction. BBLF4, helicase; BBLF2/3, primase-associated factor; BALF5, DNA polymerase; BSLF1, primase; BALF2, single-stranded DNA binding protein (SSB).
Z(S186A), functioning as an origin binding protein, reveals a necessary role for Rta in replication.
The previous experiments demonstrated that Z(S186A) could bind to oriLyt (Fig. 1), but neither Z(S186A) nor Rta alone could activate the full complement of RPs (Fig. 2). Therefore, we conducted an experiment to determine whether providing an exogenous source of RPs with or without Rta could allow Z(S186A) to support viral replication from the endogenous viral genome. BZKO cells were transfected with either Z(S186A) or wt ZEBRA in the absence and presence of RPs. Quantitative PCR was used to determine the extent of EBV replication under each condition (Fig. 3A). Coexpression of RPs stimulated the capacity of wt ZEBRA to induce viral DNA synthesis by 1.6-fold, as previously described (33); however, RPs alone failed to support viral DNA replication when coexpressed with Z(S186A). This result suggested that wt ZEBRA, but not Z(S186A), activated expression of an additional protein that was essential for activation of viral DNA replication from the endogenous viral genome. A likely candidate for this additional protein is Rta, since Z(S186A) is known to be defective in activating expression of Rta (34). To test whether Rta was essential for activation of viral replication from the endogenous origin of lytic replication, Rta was expressed in BZKO cells together with Z(S186A) or Z(S186A) plus RPs. EBV genome amplification was detected when Rta, Z(S186A), and a mixture of replication proteins were coexpressed together. The level of genome amplification achieved with this mixture was about 50% of that with wt ZEBRA (Fig. 3A). In this experiment, expression of Rta plus Z(S186A) or Z(S186A) plus RPs was insufficient to activate lytic DNA synthesis.
Since late gene expression is contingent upon viral DNA replication, expression of a late protein, FR3, the smallest capsid protein, was used as an additional assay for the occurrence of viral DNA replication. Overexpression of either Rta or the replication proteins enhanced expression of the late FR3 protein by wt ZEBRA to an extent that was similar to the effect of the same proteins on DNA replication. However, FR3 was detected only when both Rta and RPs were coexpressed with Z(S186A). These results support a model in which Z(S186A), though unable to activate transcription of Rta or replication genes, can function as an origin binding protein and enable lytic viral replication and late gene expression provided that both RPs and Rta are provided in trans. The experiments illustrated in Fig. 3 thus show that expression of Rta promotes activation of EBV lytic DNA replication and late gene expression.
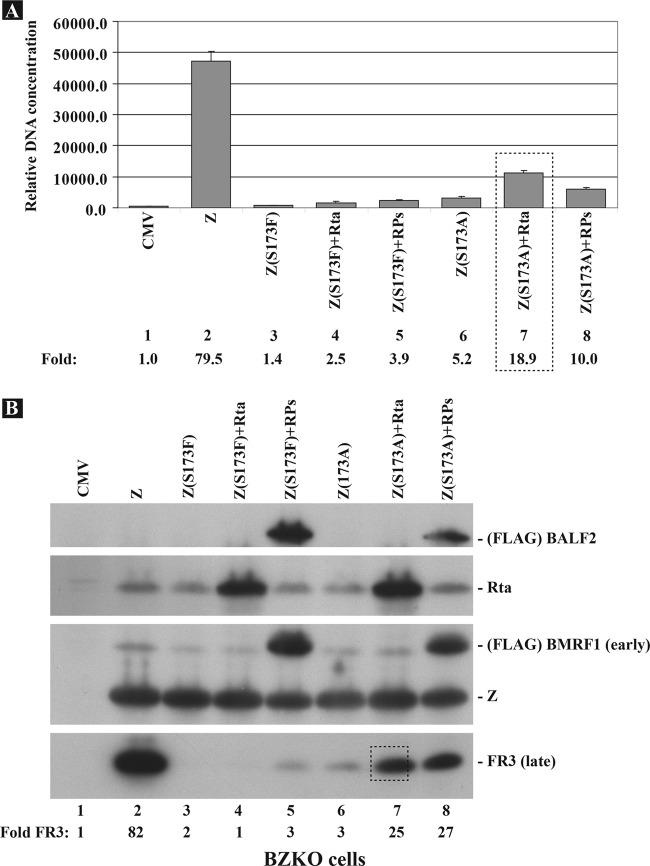
Rta restores the capacity of a ZEBRA replication-defective mutant, Z(S173A), to activate viral replication and late gene expression.
Serine 173 is a major phosphorylation site located upstream of the DNA binding domain of ZEBRA (44). Failure to phosphorylate ZEBRA at this residue as a result of the Z(S173A) mutation abolishes lytic viral DNA replication; however, Z(S173A) maintains the ability to activate transcription of early genes encoding replication proteins at wt levels (32). Previously we demonstrated that overexpression of a mixture of the EBV replication proteins enhanced the interaction of Z(S173A) with oriLyt and enhanced the capacity of Z(S173A) to activate viral DNA replication and late gene expression (33). We next assessed the capacity of Rta to rescue the phenotype of two ZEBRA mutants with mutations at the S173 phospho-acceptor site that fail to activate viral replication (Fig. 4). Activation of viral replication was assessed by qPCR using primers specific to oriLyt (Fig. 4A), while expression of late genes was detected by Western blot analysis using a specific antibody against the late FR3 protein (Fig. 4B). Transfection of wt ZEBRA in BZKO cells activated viral replication and expression of FR3 (80-fold) compared to that in cells transfected with empty vector (CMV) (Fig. 4, compare lanes 1 and 2). The Z(S173A) mutation reduced viral replication by 15-fold and late gene expression by 27-fold (Fig. 4, lanes 3 and 6). Coexpression of Z(S173A) with Rta enhanced the capacity of Z(S173A) to activate viral replication by 3.6-fold and synthesis of FR3 by 8-fold (Fig. 4, lanes 7 and 8). The mixture of RPs enhanced replication by Z(S173A) by 2-fold and late gene expression by 9-fold. The mutant in which S173 was replaced with phenylalanine activated expression of Rta and EA-D but not viral replication and late gene expression (Fig. 4, lane 3, and data not shown). Expression of Rta or RPs together with Z(S173F) did not restore genome amplification or late gene expression (Fig. 4, lanes 4 and 5). These results show that Rta and replication proteins each can suppress the replication defect in the mutant Z(S173A) but not Z(S173F).
Fig 4.
Rta enhances the capacity of Z(S173A), a replication-defective ZEBRA regulatory domain mutant, to activate viral replication and late gene expression. (A) Total DNA prepared from BZKO cells transfected with wt ZEBRA, Z(S173F), and Z(S173A) in the absence and presence of Rta or a mixture of all the EBV replication proteins was analyzed by quantitative PCR for the level of the upstream oriLyt region as a marker for viral replication. The relative DNA concentration was calculated using the standard curve method. (B) Immunoblots of cell lysates prepared from the same cell lysates were probed with antibodies against FLAG, Rta, Z, and FR3. RPs, 2 μg of each of the six EBV replication proteins (BBLF4, BBLF2/3, BSLF1, BALF2, BMRF1, and BALF5).
Deletion of the last 55 amino acids of Rta abolishes its role in supporting viral DNA replication.
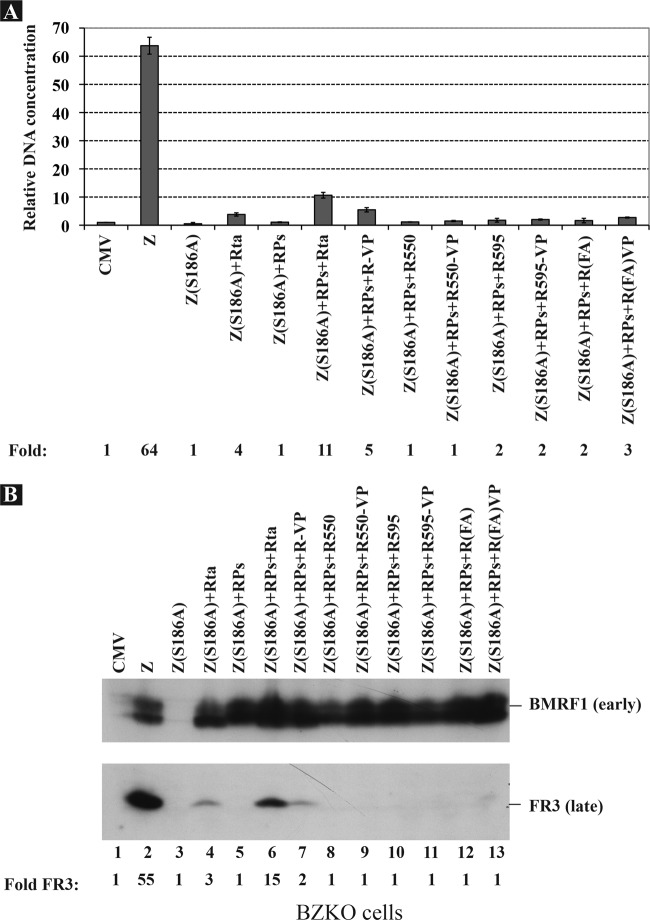
The next experiments were directed at determining whether the functions of Rta involved in replication overlap those involved in transcription. The transactivation domain of Rta, located at the C terminus, extends from amino acid 352 to the last residue in the protein, aa 605. Previous functional analysis of this domain revealed two important regions, a proline-rich region (aa 352 to 515) and an acidic region (aa 515 to 605) (24, 45). The proline rich region can be divided into two subregions, aa 352 to 410 and aa 450 to 500. In addition, a DNA binding-inhibitory sequence (DBIS) spans the region between aa 555 and 605 (25). Deletion of this region enhanced the capacity of Rta to bind to its corresponding response elements. To characterize the regions of Rta necessary for viral DNA replication, we assessed the capacity of progressive C-terminal deletions in Rta to support viral replication in BZKO cells when Z(S186A) was used as an origin binding protein and all six components of the replication machinery were supplied in trans to compensate for the loss of the capacity of the Rta deletion mutants to activate transcription of viral genes encoding replication proteins. Expression of wt and mutant Rta was confirmed by Western blot analysis (data not shown). While wt Rta plus Z(S186A) and RPs activated EBV lytic DNA replication to a level equal to 25.2% of that detected in cells transfected with wt ZEBRA, all three Rta deletion mutants, Rta(1-350), Rta(1-450), and Rta(1-550), failed to support viral replication under the same conditions.
Two phenylalanines, F600 and F605, were previously shown to contribute to the activity of the DBIS (25). Alanine substitutions at these positions, F600A and F605A, increased the DNA binding activity of full-length Rta compared to wt Rta. To assess the importance of these two phenylalanines in activation of viral DNA replication, we compared the amount of viral DNA detected in BZKO cells transfected with wt Rta or Rta(F600A/F605A) together with Z(S186A) and RPs using qPCR (Fig. 5). We found that these two phenylalanine-to-alanine substitutions reduced viral replication by 62.6% relative to that with wt Rta. In a separate experiment, we found that the R(F600A/F605A) mutant, when expressed together with Z(S186A) and RPs, was defective in activating late gene expression (see Fig. 7, lane 12). In summary, the deletion mutants showed that the last 55 amino acids of Rta were essential for its role in lytic viral DNA replication. The R(F600A/F605A) point mutant indicated that these two residues within the carboxy terminus of the protein facilitated replication.
Fig 7.
Addition of the heterologous VP16 transactivation domain to Rta deletion mutants fails to restore the role of Rta in replication. (A) The extent of viral DNA replication was measured using real-time PCR in BZKO cells transfected with the indicated combinations of expression vectors encoding Z(S186A), a mixture of EBV replication proteins (RPs), and full-length Rta or C-terminal deletion mutants of Rta without or with VP16 fusions. (B) Protein extracts were prepared from the same group of cells. The membrane was blotted using antibodies specific to the BMRF1 early protein and the late FR3 protein.
VP16 restores the capacity of Rta(1-595) and Rta(F600A/F605A) to activate expression of the early protein BMRF1.
The mutations of Rta that impair replication also impinge on functions of the protein that are required for transcription. In the next experiments, representing an attempt to distinguish effects of mutations on transcription from effects on replication, expression of the BMRF1 gene was used as a marker for the ability of Rta mutants to activate early gene expression. The BMRF1 open reading frame encodes the DNA polymerase processivity factor, also known as early antigen diffuse (EA-D). Activation of expression of the BMRF1 gene is mediated by synergy between ZEBRA and Rta; synergy in activation of BMRF1 expression is defined by the combined action of the mutant Z(S186A) and Rta, neither of which activates BMRF1 expression when present individually (34, 40). We found that Rta deletion mutants that lack the C-terminal 55 or 10 amino acids were defective in synergy with Z(S186A) to activate expression of the BMRF1 protein (Fig. 6, lanes 5 and 7). Similarly, the R(F600A/F605A) mutant also failed to activate expression of BMRF1 in the presence of Z(S186A). These mutants retain the DNA binding domain, the dimerization domain, the two accessory transactivation domains of Rta, and portions of the acidic activation domain. Fusion of the VP16 transactivation domain (positions 413 to 490) to Rta(1-595) and to Rta(F600A/F605A) restored the transcriptional function of these mutants, which thus regained the capacity to activate the BMRF1 protein when coexpressed with Z(S186A) (Fig. 6, lanes 8 and 10).
Addition of the heterologous VP16 transactivation domain to Rta deletion mutants fails to rescue their capacity to support viral DNA replication.
To investigate further whether the capacity of Rta to activate transcription was sufficient to induce lytic DNA replication from the endogenous viral genome, the three Rta mutants Rta(1-550), Rta(1-595), and R(F600A/F605A) without or with fusion to the transactivation domain of VP16 were compared to wt Rta for their capacity to activate viral replication. (Fig. 7). The assay was conducted in BZKO cells cotransfected with vectors encoding Z(S186A) and a mixture of the 6 known viral replication proteins. In agreement with data shown in Fig. 3, coexpression of Z(S186A) and replication proteins was insufficient to activate late gene expression and viral replication, but addition of Rta to this mixture activated both processes (Fig. 6). In this experiment, coexpression of Z(S186A) and Rta without replication proteins activated late gene expression and viral DNA amplification to low levels. Addition of VP16 to full-length Rta suppressed the ability of Rta to activate late gene expression and viral DNA replication (see Discussion). Fusion of VP16 to Rta(1-550), Rta(1-595) or Rta(F600A/F605A) failed to rescue the ability of these mutants to activate replication and late gene expression, although the chimeric mutants Rta595+VP16 and RtaFA+VP16 strongly activated expression of BMRF1 (Fig. 6). These results suggest that the ability of Rta to support viral replication was not solely linked to its capacity to activate transcription.
Rta interacts with oriLyt in vivo.
Our previous experiments provided genetic evidence for an independent function of Rta in supporting lytic viral DNA replication in the presence of ZEBRA mutants that are defective in this function. To begin to investigate a possible biochemical mechanism underlying the role of Rta in replication, we used chromatin immunoprecipitation to assess the capacity of Rta to interact physically with oriLyt in vivo and to determine whether ZEBRA influences such an interaction. BZKO cells were transfected with empty vector (CMV), Rta, or a combination of Rta and ZEBRA. Phosphonoacetic acid (PAA), an inhibitor of the viral DNA polymerase, was added to arrest DNA replication so that the amounts of input DNA would be comparable in all the samples. Cells harvested at 48 h after transfection were cross-linked with formaldehyde. The chromatin immunoprecipitation experiment was performed twice using mouse monoclonal and rabbit polyclonal antibodies to Rta. Quantitative PCR was used to analyze Rta-bound DNA. Two different regions of oriLyt were examined: the upstream region, which contains ZEBRA binding sites but no canonical Rta sites, and the enhancer region, which contains both ZEBRA and Rta binding sites. The two antibodies to Rta immunoprecipitated 3.7- and 2.4-fold more enhancer region in cells expressing Rta alone than in cells transfected with empty vector, (Fig. 8). Coexpression of ZEBRA and Rta markedly enhanced association of Rta with the enhancer region, by 24.6- and 13.8-fold. Using either of the two Rta-specific antibodies, we could not demonstrate an association between Rta and the upstream region of oriLyt when Rta alone or Rta plus ZEBRA was expressed (Fig. 8). In addition, no Rta-oriLyt complexes were immunoprecipitated using nonspecific antibodies, e.g., FLAG antibody (data not shown). These results provide strong evidence that Rta associates with oriLyt, presumably through the two Rta binding sites known to be present in the enhancer region. ZEBRA markedly enhances this interaction.
Fig 8.
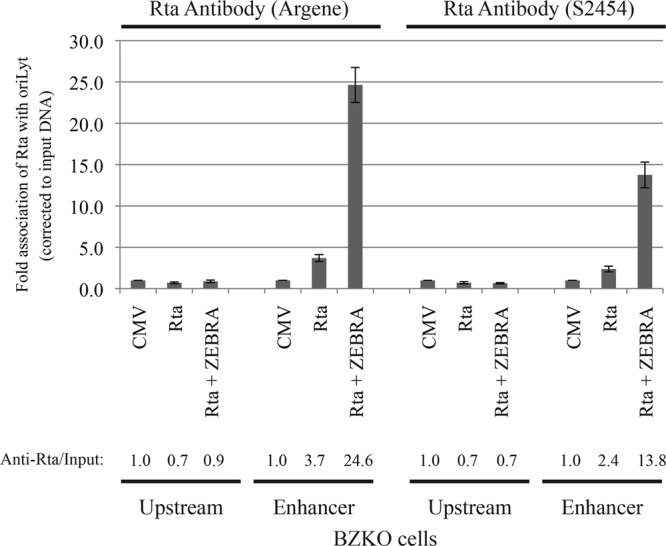
Chromatin immunoprecipitation demonstrates the association of Rta with the enhancer region of oriLyt. BZKO cells were transfected with empty vector (CMV) or expression vectors encoding Rta and ZEBRA proteins. Two different Rta antibodies, Argene and S2454, were individually used to immunoprecipitate Rta. Association of Rta with two regions of oriLyt was assessed using quantitative PCR. The two regions of oriLyt are the upstream region and the enhancer region. The extent of Rta association was calculated as the amount of oriLyt DNA specifically pulled down by Rta relative to the amount of oriLyt present in the corresponding input sample (Anti-Rta/Input). The fold association of Rta with EBV oriLyt DNA was normalized to the Anti-Rta/Input ratio present in cells transfected with empty vector (CMV).
ZEBRA and Z(S186A) promote the binding of Rta to the enhancer region of oriLyt.
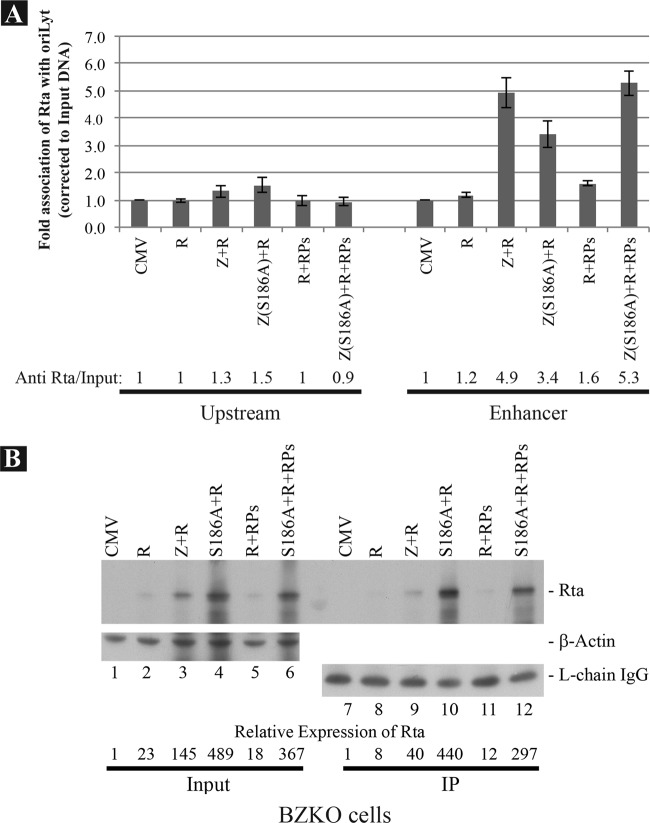
Since addition of Z(S186A) and a mixture of RPs to Rta promoted lytic viral DNA replication and late gene expression (Fig. 3), the next experiment addressed the question whether the interaction of Rta with oriLyt was augmented when Z(S186A) or RPs were coexpressed with Rta. In the ChIP experiment illustrated in Fig. 9A, Rta by itself only weakly interacted with the enhancer region of oriLyt; however, its interaction with oriLyt increased about 4.2-fold when ZEBRA was coexpressed. Coexpression of Z(S186A) also enhanced the interaction of Rta with oriLyt 2.9-fold. The interaction of Rta with oriLyt was minimally enhanced by coexpression of RPs (1.4-fold), but the combination of Z(S186A) and RPs promoted Rta binding by 4.5-fold, an effect similar to that observed when wild-type ZEBRA and Rta were coexpressed.
Fig 9.
wt ZEBRA and Z(S186A) promote association of Rta with the enhancer region of oriLyt. BZKO cells were transfected with expression vectors for Rta, ZEBRA, Z(S186A), or a mixture of six replication proteins (RPs) as indicated. The cells were treated with PAA at time zero and harvested at 48 h after transfection. (A) Results of a ChIP experiment analyzing the upstream and enhancer regions of oriLyt, showing immunoprecipitation with monoclonal antibody to Rta. (B) Western blot demonstrating the level of Rta protein present in the input sample and in the immunoprecipitate used for ChIP. Rabbit polyclonal anti-Rta was used to detect Rta. The ratio of cell lysate analyzed in the input versus immunoprecipitate samples was 1:7.5. β-Actin and the light chain of the anti-Rta antibody were used as loading controls for input and immunoprecipitate samples, respectively.
The same cell lysates were analyzed for the amount of Rta protein in the input and in the immunoprecipitate (Fig. 9B). Coexpression of ZEBRA enhanced the level of Rta in the immunoprecipitate by 5-fold. Coexpression of the Z(S186A) mutant enhanced Rta expression 55-fold compared to Rta alone (Fig. 9B, compare lanes 8 and 10). RPs by themselves did not enhance Rta expression. The addition of RPs to the mixture of Rta and Z(S186A) also enhanced the level of Rta by 37-fold. Since both wt ZEBRA and Z(S186A) enhanced expression of Rta, the enhancing effect of ZEBRA and the Z(S186A) mutant could be attributed to a mixture of enhanced expression of Rta and independent enhancement of binding of Rta to oriLyt (see Discussion). Nonetheless, the results in Fig. 9 clearly demonstrated that Rta interacts preferentially with the enhancer region of oriLyt. Thus, one possible mechanism by which Rta enhances lytic replication is through interaction with the “enhancer” region of oriLyt.
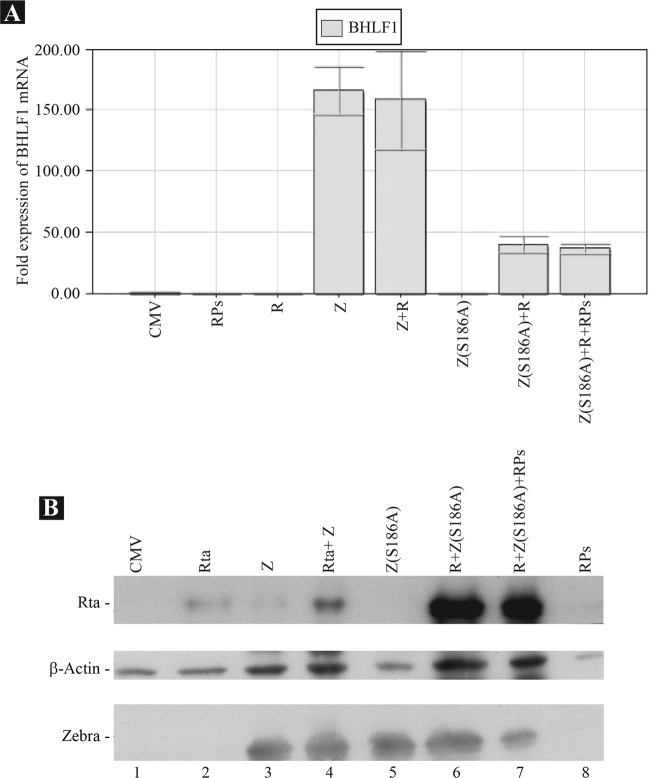
Z(S186A) and Rta synergize to activate transcription of the BHLF1 transcript.
The upstream component of oriLyt coincides with the BHLF1 promoter. Mutations in this region disrupt both oriLyt-mediated replication and activation of the BHLF1 promoter (13, 18). A recent report linked activation of oriLyt with transcription of the BHLF1 gene. The BHLF1 transcript played a role in recruiting the viral ssDNA binding protein, BALF2, to an oriLyt plasmid (23). In our experiments (Fig. 3, 4, 5, and 7), we found that Z(S186A), Rta, and a mixture of replication proteins activated viral DNA replication from the endogenous viral genome. This result suggested that Z(S186A), Rta, or a combination of both proteins might activate expression of the BHLF1 transcript. To examine this possibility, we transfected BZKO cells with either empty vector or expression vectors expressing the following proteins: Rta, ZEBRA, ZEBRA plus Rta, Z(S186A), Z(S186A) plus Rta, and Z(S186A) plus Rta and a mixture of the six replication proteins. The cells were harvested at 48 h after transfection. RNA was purified from transfected cells, and the concentrations were adjusted to 100 ng/μl. qRT-PCR was performed to assess the level of the BHLF1 transcript. The relative expression of BHLF1 was adjusted to the concentration of 18S RNA present in each sample. We found that expression of ZEBRA and ZEBRA plus Rta increased the level of BHLF1 mRNA by 166- and 158-fold, respectively, compared to that in CMV-transfected cells (Fig. 10A). Expression of Rta or Z(S186A) individually or a mixture of the six replication proteins did not induce synthesis of the BHLF1 transcript. However, coexpression of Z(S186A) plus Rta in the absence and presence of replication proteins increased the level of BHLF1 mRNA by 40- and 36-fold, respectively, relative to that with empty vector. Thus, Rta and Z(S186A) synergized to activate BHLF1, and the RPs did not affect the efficiency of this process.
Fig 10.
Rta and Z(S186A) synergize to activate expression of the BHLF1 gene. (A) BHLF1 mRNA levels measured by qRT-PCR in samples purified from transfected BZKO cells. (B) Expression of the transfected Rta and ZEBRA proteins in BZKO cells.
To assess expression of transfected plasmids, lysates were prepared from a second fraction of the same cells (Fig. 10B). The proteins were separated by 10% SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blotted with antibodies specific to ZEBRA, Rta, and β-actin, which served as a loading control. Similar to our findings in Fig. 3 and 9, expression of Rta was significantly enhanced when ZEBRA or Z(S186A) was coexpressed in BZKO cells (Fig. 10B, compare lane 2 with lanes 4, 6, and 7). While our results suggest that expression of BHLF1 mRNA requires Rta, the level of BHLF1 transcript was independent of the amount of Rta expressed in BZKO cells. For example, despite the higher level of Rta in cells transfected with expression vectors for ZEBRA and Rta relative to that in cells transfected with ZEBRA alone (Fig. 10B, lanes 3 and 4), both conditions resulted in equal levels of the BHLF1 transcript (Fig. 10A).
Although the BHLF1 promoter was previously reported to be activated solely by the ZEBRA protein using reporter assays (12), our result demonstrated that Rta was unequivocally necessary for activation of the BHLF1 transcript from the endogenous viral genome. The capacity of Rta to activate expression of BHLF1 mRNA might contribute to the function of the Rta protein in the process of viral DNA replication.
DISCUSSION
This report expands our understanding of the mechanisms that regulate EBV genome amplification. We provide several types of evidence that indicate an essential function of Rta in lytic EBV DNA replication. Although the ZEBRA mutant Z(S186A) is unable to activate expression of Rta and the six EBV-encoded lytic replication proteins, we found that it is capable of interacting with oriLyt and functioning as an origin binding protein. This unique phenotype of Z(S186A) allowed us to demonstrate that the known replication proteins together with a form of ZEBRA competent to bind oriLyt are incapable of activating replication of the endogenous EBV genome unless Rta was provided. These observations lead us to conclude that Rta plays an essential role in the process of viral DNA replication that can be separated from its capacity to activate transcription of viral replication genes. The role of Rta in replication requires the 10 C-terminal amino acids of the protein. Rta associates with the enhancer region of oriLyt. Coexpression of ZEBRA, Z(S186A), or the mixture of Z(S186A) plus replication proteins promotes association of Rta with the enhancer region. Rta associates only weakly or not at all with the upstream region, an essential element of oriLyt; however, Rta and Z(S186A) functionally interact at this site to activate expression of the BHLF1 transcript, presumably through activation of the BHLF1 promoter, which overlaps with the upstream element. In summary, our data strongly support a distinct and necessary function for Rta in the process of viral DNA replication.
Z(S186A) functions as an origin binding protein.
ZEBRA is an origin binding protein and a transcription activator that leads to expression of all lytic cycle proteins. Previous reports described several ZEBRA mutants that can segregate these two main functions of the protein. However, these mutants fall in one functional category; they are competent to activate transcription but defective in supporting viral DNA replication (32, 46, 47). Here we demonstrate that Z(S186A) has a unique phenotype; it supports viral DNA replication despite a specific defect in activation of transcription of Rta and other viral replication genes (Fig. 2 and 3) (34, 37, 40). Characterization of this transactivation-defective but replication-competent ZEBRA mutant allowed us to answer several unresolved questions about the process of EBV lytic replication. These questions include whether ZEBRA can bind to oriLyt in the absence of any other lytic cycle products, whether Rta can bind to oriLyt in vivo and the effect of ZEBRA on this interaction, and whether the viral replication proteins are sufficient to amplify the endogenous viral genome in the absence of Rta.
The DNA binding activity of ZEBRA is integral to its capacity to activate transcription and replication. The failure of Z(S186A) to disrupt latency has been attributed to changes in ZEBRA's conformation and to a change in DNA binding specificity, including a failure to bind methylated ZEBRA response elements present in viral promoters (34, 38, 48). Since none of the ZEBRA response elements present in oriLyt contain CpG motifs, we hypothesized that DNA methylation would not impede the capacity of Z(S186A) to recognize oriLyt. Indeed, in our experiments we found that Z(S186A) maintains the capacity of ZEBRA to interact with oriLyt (Fig. 1). Coexpression of Rta reproducibly increased association of Z(S186A) with oriLyt to levels equal or higher than that of wt ZEBRA. Since Z(S186A) failed to activate expression of any lytic cycle genes encoding replication proteins (Fig. 2), the initial interaction between ZEBRA and oriLyt does not require the six known virally encoded replication proteins. However, expression of replication proteins enhances the interaction of ZEBRA with the upstream region of oriLyt (33).
Rta interacts with oriLyt.
We demonstrate that Rta by itself weakly interacts with the enhancer region but not the upstream region of oriLyt (Fig. 8). Coexpression of ZEBRA markedly augmented the association of Rta with the enhancer region of oriLyt. The mixture of Z(S186A) and RPs that support viral replication in the presence of Rta also allows Rta to bind to the enhancer region of oriLyt. The efficiency of binding of Rta to oriLyt in the presence of ZEBRA or in the presence of Z(S186A) plus RPs is similar (Fig. 9A). While our experiments to study the capacity of Rta to associate with the distinct regions of oriLyt were in progress, Heilmann et al., using a ChIP-seq approach, reported that the bidirectional BHLF1/BHRF1 promoter that overlaps oriLyt was one of the high-confidence Rta binding sites (49).
Our inability to detect an interaction between Rta and the upstream region of oriLyt in the absence or presence of ZEBRA could be attributed either to the lack of such an interaction or to a defect in the ChIP assay used to detect such an interaction. For example, Rta might be involved in multiple protein-protein interactions at the upstream region of oriLyt that mask the epitope recognized by the Rta antibody. As a result of this steric hindrance, attempts to immunoprecipitate Rta complexed with the upstream region of oriLyt would fail. Assays other than ChIP, such as in vivo biotinylated DNA affinity assay (iBDAA), might help reveal an interaction between Rta and the upstream region of oriLyt.
How does ZEBRA or Z(S186A) plus RPs promote association of Rta with the enhancer region of oriLyt? The experimental observation (Fig. 9B) that both ZEBRA and Z(S186A) increased the level of Rta protein expressed from the transfected plasmid may partially explain the effect of ZEBRA on the binding of Rta to oriLyt. However, two additional observations make it unlikely that the expression level of Rta alone accounts for its increased binding to DNA in the presence of ZEBRA. First, in the experiment illustrated in Fig. 9, the increase in the expression level of Rta was 11-fold greater in the presence of S186A than in the presence of wt ZEBRA, but the Z(S186A) mutant was less efficient than wild-type ZEBRA at promoting the interaction of Rta with oriLyt. Second, RPs enhanced the binding of Rta to oriLyt in the presence of Z(S186A) but did not increase the Rta protein level.
Many other scenarios could account for the capacity of Z or Z(S186A) and RPs to facilitate binding of Rta to oriLyt. A direct interaction of ZEBRA with Rta might elicit a conformation of Rta that is more favorable for DNA binding (25). Signaling events induced by ZEBRA might cause posttranslational modifications that alter the DNA binding activity of Rta (50). ZEBRA might alter the chromatin structure, allowing Rta to interact with DNA (51, 52). ZEBRA might activate expression of cellular or viral proteins that regulate the DNA binding activity of Rta. ZEBRA might recruit replication proteins that interact with Rta in such a way as to promote binding of Rta to DNA (22).
Possible direct roles of Rta in replication.
Involvement of cellular or viral proteins in the process of EBV replication was previously assessed using a cotransfection replication assay employing a plasmid containing oriLyt and expression vectors for replication proteins (7, 12, 13, 19, 20, 23). However, to our knowledge, experiments that probe the role of Rta in replication of the endogenous EBV genome have not been reported. In order to gain insight into the role of Rta in viral DNA replication, one might first consider the differences between replicating an oriLyt-containing plasmid and the endogenous viral genome. An obvious distinction between these two replication systems would be differences in epigenetic regulation of the two types of templates. Unlike plasmids, the endogenous origin of lytic replication exists in a closed chromatin conformation during latency. Hence, one possible function for Rta during lytic genome amplification is to alter the chromatin structure at oriLyt, thereby providing access to other components of the replication machinery. Rta interacts with CREB binding protein (CBP), a transcription coactivator with intrinsic histone acetyltransferase activity (53). Acetylation of histones by CBP could result in an open chromatin structure at oriLyt, a condition that would favor recruitment of replication proteins to the origin. Both the amino- and carboxy-terminal regions of CBP independently interact with Rta (53). Conversely, multiple domains in Rta are necessary for its interaction with CBP. One of these domains is the C-terminal transactivation domain. In our study we found that mutations in the transcriptional activation domain of Rta, including deletion of the last 10 amino acids, abolished the capacity of Rta to activate transcription or to support viral DNA replication (Fig. 5, 6, and 7) (25). In addition to CBP, other chromatin-remodeling proteins might play a role in the effect of Rta on replication. While interactions between Rta and the SWI/SNF complex have yet to be described, the carboxyl region of ORF50, the KSHV homolog of Rta, interacts with the SWI/SNF complex and recruits it to lytic viral promoters (54).
The BHRF1 promoter overlaps with the enhancer region of oriLyt and is activated by both ZEBRA and Rta (2, 40, 55). Two Rta binding sites were mapped in this region using a gel retardation assay (56). In our results, we found that the weak association of Rta with this specific region of oriLyt was markedly enhanced in the presence of ZEBRA. Thus, an additional protein-protein interaction is likely to be required for Rta to interact with oriLyt (Fig. 8). Alternatively, the RREs in the BHRF1 promoter might function as a distant enhancer of BHLF1 transcription.
A third possible contribution of Rta in the process of viral genome amplification might involve stabilizing the formation of a replication complex or tethering replication proteins to oriLyt. For KSHV, Wang et al. proposed a model for recruitment of KSHV replication proteins to oriLyt in which both K-bZIP, the homolog of ZEBRA, and ORF50 interact with the core components of the replication machinery to form one complex. The whole complex then binds to the respective K8 and ORF50 sites on oriLyt in what was referred to as a two-point contact interaction between the replication complex and oriLyt (29). In our experiments we found that Rta enhanced the interaction of Z(S186A) with the upstream region of oriLyt by 2-fold in the absence of replication proteins (Fig. 1). Similarly, ZEBRA enhanced Rta interaction with the enhancer region of oriLyt by 4- to 6-fold (Fig. 8). Z(S186A) also enhanced association of Rta with the enhancer region by 2.8-fold (Fig. 9). The effect of ZEBRA on association of Rta with oriLyt could be a direct effect of ZEBRA itself or an effect of any other viral or cellular protein whose expression is stimulated by ZEBRA during the lytic cycle. For example, Rta has been shown to interact with the primase-associated factor BBLF2/3, a component of the heterotrimeric helicase-primase complex whose expression was strongly stimulated by the combination of ZEBRA and Rta (Fig. 2) (22, 57). Therefore, association of Rta with oriLyt and its capacity to interact with replication proteins suggests that the protein might play a role in the assembly or the recruitment of the replication machinery to oriLyt.
A fourth possible contribution of Rta in replication of the EBV genome involves its capacity to localize to replication compartments. Together with other replication proteins, such as BMRF1 and BALF2, Rta localizes to replication compartments in EBV-positive cells undergoing viral replication (26, 27). Thus, Rta might recruit various components of the replication machinery to subnuclear sites at which synthesis of viral DNA takes place. In summary, Rta might alter the chromatin structure at oriLyt, activate transcription of oriLyt flanking genes, stabilize the replication complex, or recruit replication proteins to oriLyt and replication compartments. However, it is likely that these potential functions of Rta in viral replication are not mutually exclusive; Rta might perform multiple roles in replication.
Possible indirect roles of Rta in replication.
Rta might contribute to activation of oriLyt indirectly by inducing transcription and promoting expression of proteins that are necessary for the process of viral DNA replication. Rennekamp and Lieberman recently reported that the BHLF1 or BHRF1 transcripts are required for replication of an oriLyt reporter plasmid (23). The BHLF1 promoter overlaps almost completely the upstream region of oriLyt; this region contains four ZEBRA response elements. Mutational analysis of individual ZREs in the BHLF1 promoter/oriLyt upstream region demonstrated that the same sites are necessary for oriLyt-mediated replication and transcription of BHLF1 (12). Using plasmid reporter assays, it was previously shown that ZEBRA alone can activate the BHLF1 promoter (12). However, in our experiments we found that the BHLF1 promoter is a synergistic target for both Z(S186A) and Rta (Fig. 10), neither of which alone could activate this transcript. This result might explain differences in the requirement of Rta for replication of a plasmid containing oriLyt versus replication of the endogenous viral genome. Thus, Rta might contribute to activation of oriLyt by inducing transcription through the BHLF1 or BHRF1 promoters. In addition, Rta might activate transcription of early viral proteins that enhance the process of viral DNA replication. These might include BMLF1, EBV-encoded dUTPase, ribonucleotide reductase, and thymidine kinase.
Previously, Rta was found to be dispensable for replication of an oriLyt-containing plasmid, although Rta expression augmented the replication efficiency of such a plasmid (7). The mechanism by which Rta exerted its stimulatory effect on replication of the oriLyt plasmid is not understood but presumably was not related to consequences that Rta might have on epigenetic modifications. Contrary to the previously proposed auxiliary role of Rta in the plasmid replication assay, our findings argue for an essential role of Rta in amplification of the endogenous viral genome. In addition to activating expression of genes encoding replication proteins (Fig. 11A), Rta is likely to have distinct and direct roles in the process of viral DNA replication (Fig. 11B). A stimulatory role that is observed in a transient-replication assay could be attributed to a function of Rta in stabilizing the replication complex. Other essential roles, manifested only when activation of the endogenous origin is examined, could be attributed to the ability of Rta to modify the epigenetics of oriLyt and to activate transcription of adjacent genes. It is important to note that our conclusion about the essential function of Rta in replication of the endogenous viral genome does not require an experimental system using the S186A or S173A ZEBRA mutant. In 293 cells carrying an EBV bacmid with inactivation of both BZLF1 and BRLF1, the genes encoding ZEBRA and Rta proteins, respectively, expression of wt ZEBRA together with a mixture of replication proteins failed to support viral DNA replication. However, coexpression of ZEBRA plus Rta restored viral DNA replication (unpublished data). This finding represents an additional proof for the indispensable role of Rta in EBV lytic DNA replication from the endogenous viral genome.
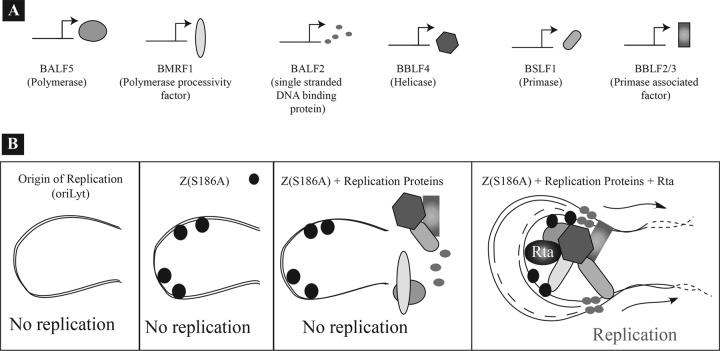
Fig 11.
Models for the role of Rta in lytic EBV replication. Rta has at least two roles in viral DNA replication. Rta activates expression of genes encoding replication proteins (A); in addition, Rta plays a direct role in activation of oriLyt from the endogenous genome (B). In the experiments described here, we found that Z(S186A) has the capacity to bind oriLyt. Provision of Z(S186A) as an origin binding protein and of all the replication proteins in trans failed to support viral DNA replication. EBV genome amplification was restored only when Z(S186A), Rta, and a mixture of the six replication proteins were ectopically expressed. Rta interacted with oriLyt; ZEBRA significantly enhanced this interaction. Rta is required for synthesis of the BHLF1 transcript, which facilitates strand separation and recruitment of replication proteins to oriLyt during replication (23). While the model shows that replication proteins do not bind to oriLyt in the absence of Rta, additional experiments are necessary to explore this postulate.
In conclusion, we have described an essential role for Rta in amplification of the EBV genome during the viral lytic cycle. Further experiments on the function of Rta in replication will provide new insights into the complex process of lytic EBV DNA replication, particularly origin recognition, tethering of replication proteins to oriLyt, and the mechanisms required to activate oriLyt.
ACKNOWLEDGMENTS
The research reported in this paper was supported by NIH grants CA16038 and CA12055 to G.M.
Footnotes
Published ahead of print 17 October 2012
REFERENCES
- 1. Countryman J, Miller G. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. U. S. A. 82:4085–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hardwick JM, Lieberman PM, Hayward SD. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Manet E, Gruffat H, Trescol-Biemont MC, Moreno N, Chambard P, Giot JF, Sergeant A. 1989. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 8:1819–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seibl R, Motz M, Wolf H. 1986. Strain-specific transcription and translation of the BamHI Z area of Epstein-Barr virus. J. Virol. 60:902–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Countryman J, Jenson H, Seibl R, Wolf H, Miller G. 1987. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J. Virol. 61:3672–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feederle R, Kost M, Baumann M, Janz A, Drouet E, Hammerschmidt W, Delecluse HJ. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fixman ED, Hayward GS, Hayward SD. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kenney S, Holley-Guthrie E, Mar EC, Smith M. 1989. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J. Virol. 63:3878–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lieberman PM, Hardwick JM, Sample J, Hayward GS, Hayward SD. 1990. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quinlivan EB, Holley-Guthrie EA, Norris M, Gutsch D, Bachenheimer SL, Kenney SC. 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 21:1999–2007 (Corrected and republished with original paging.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ragoczy T, Heston L, Miller G. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978–7984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schepers A, Pich D, Hammerschmidt W. 1996. Activation of oriLyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology 220:367–376 [DOI] [PubMed] [Google Scholar]
- 13. Schepers A, Pich D, Hammerschmidt W. 1993. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein-Barr virus. EMBO J. 12:3921–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zalani S, Holley-Guthrie E, Kenney S. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. U. S. A. 93:9194–9199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hammerschmidt W, Sugden B. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427–433 [DOI] [PubMed] [Google Scholar]
- 16. Cho MS, Bornkamm GW, Hzur Hausen 1984. Structure of defective DNA molecules in Epstein-Barr virus preparations from P3HR-1 cells. J. Virol. 51:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raab-Traub N, Dambaugh T, Kieff E. 1980. DNA of Epstein-Barr virus. VIII. B95-8, the previous prototype, is an unusual deletion derivative. Cell 22:257–267 [DOI] [PubMed] [Google Scholar]
- 18. Schepers A, Pich D, Mankertz J, Hammerschmidt W. 1993. cis-acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J. Virol. 67:4237–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang J, Liao G, Chen H, Wu FY, Hutt-Fletcher L, Hayward GS, Hayward SD. 2006. Contribution of C/EBP proteins to Epstein-Barr virus lytic gene expression and replication in epithelial cells. J. Virol. 80:1098–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baumann M, Feederle R, Kremmer E, Hammerschmidt W. 1999. Cellular transcription factors recruit viral replication proteins to activate the Epstein-Barr virus origin of lytic DNA replication, oriLyt. EMBO J. 18:6095–6105 (Erratum, 19:315, 2000.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gruffat H, Renner O, Pich D, Hammerschmidt W. 1995. Cellular proteins bind to the downstream component of the lytic origin of DNA replication of Epstein-Barr virus. J. Virol. 69:1878–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liao G, Huang J, Fixman ED, Hayward SD. 2005. The Epstein-Barr virus replication protein BBLF2/3 provides an origin-tethering function through interaction with the zinc finger DNA binding protein ZBRK1 and the KAP-1 corepressor. J. Virol. 79:245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rennekamp AJ, Lieberman PM. 2011. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. J. Virol. 85:2837–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manet E, Rigolet A, Gruffat H, Giot JF, Sergeant A. 1991. Domains of the Epstein-Barr virus (EBV) transcription factor R required for dimerization, DNA binding and activation. Nucleic Acids Res. 19:2661–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen LW, Raghavan V, Chang PJ, Shedd D, Heston L, Delecluse HJ, Miller G. 2009. Two phenylalanines in the C-terminus of Epstein-Barr virus Rta protein reciprocally modulate its DNA binding and transactivation function. Virology 386:448–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Daikoku T, Kudoh A, Fujita M, Sugaya Y, Isomura H, Shirata N, Tsurumi T. 2005. Architecture of replication compartments formed during Epstein-Barr virus lytic replication. J. Virol. 79:3409–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park R, Heston L, Shedd D, Delecluse HJ, Miller G. 2008. Mutations of amino acids in the DNA-recognition domain of Epstein-Barr virus ZEBRA protein alter its sub-nuclear localization and affect formation of replication compartments. Virology 382:145–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sarisky RT, Gao Z, Lieberman PM, Fixman ED, Hayward GS, Hayward SD. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y, Tang Q, Maul GG, Yuan Y. 2006. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: dual role of replication and transcription activator. J. Virol. 80:12171–12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Graham FL, Smiley J, Russell WC, Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59–74 [DOI] [PubMed] [Google Scholar]
- 31. Shaw G, Morse S, Ararat M, Graham FL. 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16:869–871 [DOI] [PubMed] [Google Scholar]
- 32. El-Guindy A, Heston L, Delecluse HJ, Miller G. 2007. Phosphoacceptor site S173 in the regulatory domain of Epstein-Barr Virus ZEBRA protein is required for lytic DNA replication but not for activation of viral early genes. J. Virol. 81:3303–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. El-Guindy A, Heston L, Miller G. 2010. A subset of replication proteins enhances origin recognition and lytic replication by the Epstein-Barr virus ZEBRA protein. PLoS Pathog. 6:e1001054 doi:10.1371/journal.ppat.1001054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Francis AL, Gradoville L, Miller G. 1997. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J. Virol. 71:3054–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen LW, Chang PJ, Delecluse HJ, Miller G. 2005. Marked variation in response of consensus binding elements for the Rta protein of Epstein-Barr virus. J. Virol. 79:9635–9650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pearson GR, Vroman B, Chase B, Sculley T, Hummel M, Kieff E. 1983. Identification of polypeptide components of the Epstein-Barr virus early antigen complex with monoclonal antibodies. J. Virol. 47:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Francis A, Ragoczy T, Gradoville L, El-Guindy A, Miller G. 1999. Amino acid substitutions reveal distinct functions of serine 186 of the ZEBRA protein in activation of lytic cycle genes and synergy with the EBV Rta transactivator. J. Virol. 73:4543–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bhende PM, Seaman WT, Delecluse HJ, Kenney SC. 2005. BZLF1 activation of the methylated form of the BRLF1 immediate-early promoter is regulated by BZLF1 residue 186. J. Virol. 79:7338–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bhende PM, Seaman WT, Delecluse HJ, Kenney SC. 2004. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 36:1099–1104 [DOI] [PubMed] [Google Scholar]
- 40. Adamson AL, Kenney SC. 1998. Rescue of the Epstein-Barr virus BZLF1 mutant, Z(S186A), early gene activation defect by the BRLF1 gene product. Virology 251:187–197 [DOI] [PubMed] [Google Scholar]
- 41. Hung CH, Liu ST. 1999. Characterization of the Epstein-Barr virus BALF2 promoter. J. Gen. Virol. 80:2747–2750 [DOI] [PubMed] [Google Scholar]
- 42. Furnari FB, Adams MD, Pagano JS. 1992. Regulation of the Epstein-Barr virus DNA polymerase gene. J. Virol. 66:2837–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu C, Sista ND, Pagano JS. 1996. Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J. Virol. 70:2545–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. El-Guindy AS, Miller G. 2004. Phosphorylation of Epstein-Barr virus ZEBRA protein at its casein kinase 2 sites mediates its ability to repress activation of a viral lytic cycle late gene by Rta. J. Virol. 78:7634–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hardwick JM, Tse L, Applegren N, Nicholas J, Veliuona MA. 1992. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J. Virol. 66:5500–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heston L, El-Guindy A, Countryman J, Dela Cruz C, Delecluse HJ, Miller G. 2006. Amino acids in the basic domain of Epstein-Barr virus ZEBRA protein play distinct roles in DNA binding, activation of early lytic gene exprssion, and promotion of viral DNA replication. J. Virol. 80:9115–9133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang P, Day L, Dheekollu J, Lieberman PM. 2005. A redox-sensitive cysteine in Zta is required for Epstein-Barr virus lytic cycle DNA replication. J. Virol. 79:13298–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. El-Guindy A, Heston L, Endo Y, Cho MS, Miller G. 2002. Disruption of Epstein-Barr virus latency in the absence of phosphorylation of ZEBRA by protein kinase C. J. Virol. 76:11199–11208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heilmann AM, Calderwood MA, Portal D, Lu Y, Johannsen E. 2012. Genome-wide analysis of Epstein-Barr virus Rta DNA binding. J. Virol. 86:5151–5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adamson AL, Darr D, Holley-Guthrie E, Johnson RA, Mauser A, Swenson J, Kenney S. 2000. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J. Virol. 74:1224–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Adamson AL, Kenney S. 1999. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein. J. Virol. 73:6551–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Deng Z, Chen CJ, Chamberlin M, Lu F, Blobel GA, Speicher D, Cirillo LA, Zaret KS, Lieberman PM. 2003. The CBP bromodomain and nucleosome targeting are required for Zta-directed nucleosome acetylation and transcription activation. Mol. Cell. Biol. 23:2633–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Swenson JJ, Holley-Guthrie E, Kenney SC. 2001. Epstein-Barr virus immediate-early protein BRLF1 interacts with CBP, promoting enhanced BRLF1 transactivation. J. Virol. 75:6228–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gwack Y, Baek HJ, Nakamura H, Lee SH, Meisterernst M, Roeder RG, Jung JU. 2003. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell. Biol. 23:2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shimizu N, Sakuma S, Ono Y, Takada K. 1989. Identification of an enhancer-type sequence that is responsive to Z and R trans-activators of Epstein-Barr virus. Virology 172:655–658 [DOI] [PubMed] [Google Scholar]
- 56. Gruffat H, Manet E, Rigolet A, Sergeant A. 1990. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 18:6835–6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gao Z, Krithivas A, Finan JE, Semmes OJ, Zhou S, Wang Y, Hayward SD. 1998. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication proteins. J. Virol. 72:8559–8567 [DOI] [PMC free article] [PubMed] [Google Scholar]