Abstract
Members of the tripartite interaction motif (TRIM) family of E3 ligases are emerging as critical regulators of innate immunity. To identify new regulators, we carried out a screen of 43 human TRIM proteins for the ability to activate NF-κB, AP-1, and interferon, hallmarks of many innate immune signaling pathways. We identified 16 TRIM proteins that induced NF-κB and/or AP-1. We found that one of these, TRIM62, functions in the TRIF branch of the TLR4 signaling pathway. Knockdown of TRIM62 in primary macrophages led to a defect in TRIF-mediated late NF-κB, AP-1, and interferon production after lipopolysaccharide challenge. We also discovered a role for TRIM15 in the RIG-I-mediated interferon pathway upstream of MAVS. Knockdown of TRIM15 limited virus/RIG-I ligand-induced interferon production and enhanced vesicular stomatitis virus replication. In addition, most TRIM proteins previously identified to inhibit murine leukemia virus (MLV) demonstrated an ability to induce NF-κB/AP-1. Interfering with the NF-κB and AP-1 signaling induced by the antiretroviral TRIM1 and TRIM62 proteins rescued MLV release. In contrast, human immunodeficiency virus type 1 (HIV-1) gene expression was increased by TRIM proteins that induce NF-κB. HIV-1 resistance to inflammatory TRIM proteins mapped to the NF-κB sites in the HIV-1 long terminal repeat (LTR) U3 and could be transferred to MLV. Thus, our work identifies new TRIM proteins involved in innate immune signaling and reinforces the striking ability of HIV-1 to exploit innate immune signaling for the purpose of viral replication.
INTRODUCTION
Tripartite interaction motif (TRIM) proteins are RING-type E3 ligases that share an N-terminal structure consisting of a RING domain, one or two B boxes, a putative coiled-coil domain, and a variable C terminus (1). Based on the variable C terminus, the ∼100 members of the TRIM family can be classified into 11 subgroups (2, 3). Despite their common domain structure, TRIM proteins function in distinct cellular processes that include cell differentiation, transcriptional regulation, cytoskeleton remodeling, intracellular trafficking, membrane repair, and oncogenesis (4–6). Several TRIM proteins display a broad range of antiviral activities, and their RNA levels are upregulated by type I interferons (IFNs) (1, 7–13). TRIM5 is well known for its ability to restrict various retroviruses, such as human immunodeficiency virus type 1 (HIV-1) and N-tropic murine leukemia virus (MLV) (14–17). TRIM19 can restrict diverse viruses, such as influenza virus, vesicular stomatitis virus (VSV), herpes simplex virus, cytomegalovirus, and HIV-1 (18). TRIM22 inhibits HIV-1 long terminal repeat (LTR)-driven transcription and hepatitis B RNA synthesis, whereas TRIM28 functions as a corepressor of retroviral LTRs in embryonic stem cells (19–23). TRIM21/Ro52 is an intracellular IgG receptor that degrades and neutralizes viruses in the cytoplasm by binding to antibody-coated virions (24). TRIM56 restricts bovine viral diarrhea virus replication, while TRIM79α mediates degradation of the viral polymerase to inhibit replication of tick-borne encephalitis virus (25, 26). Moreover, a systematic screen of 55 human and mouse TRIM proteins for antiretroviral activities targeting HIV-1 and MLV revealed ∼20 TRIMs capable of inhibiting early and late stages of retrovirus replication (13).
In addition to antiviral activities, TRIM proteins regulate the pattern recognition receptor (PRR)-driven pathways that shape the immune response to viruses, bacteria, and parasites (4, 27, 28). PRR pathways regulated by TRIMs include the Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), and DNA sensors. Mouse TRIM30 induces the lysosomal degradation of TAB2 and TAB3, thereby negatively regulating NF-κB induction in the lipopolysaccharide (LPS)-triggered TLR4 signaling pathway (29). TRIM5 and TRIM8 activate transforming growth factor beta (TGF-β)-activated kinase 1 (TAK1), a common downstream kinase utilized by TLRs and RLRs to induce NF-κB and MAP kinase (MAPK) signaling (30–32). TRIM21 negatively regulates TLR3, -4, -7, and -9 and RLR signaling pathways by modulating the activities of IκB kinase complexes (IKKs) and interferon regulatory factors (IRFs) (33). TRIM27 targets all IKKs and negatively regulates the PRR pathways (34). CARD domain ubiquitination by TRIM25 is essential for RIG-I-mediated type I interferon induction (35). TRIM23/ARD1 plays a role in NF-κB and IRF3 activation downstream of RLRs and TLR3 by promoting K27-linked ubiquitination of NEMO (36, 37). TRIM56 facilitates double-stranded DNA-stimulated interferon induction by ubiquitination of STING (stimulator of interferon genes) (38).
To identify new regulators of innate immune signaling pathways, in this study, we carried out a transient-expression-based screen of 43 human TRIM proteins for the ability to induce NF-κB-, AP-1-, and interferon-related luciferase reporters. We identified 16 TRIM proteins that induce NF-κB and/or AP-1, 5 of which have not been previously reported to signal. Among these was TRIM62, which we show plays a critical role in the TRIF branch of the TLR4 signaling pathway. In addition, we discovered the role of TRIM15 as a regulator in the RIG-I sensing pathway. Moreover, we noticed a striking correlation between NF-κB/AP-1 induction by TRIM proteins and antiviral activity targeting MLV (13). MLV, in contrast to HIV-1, was particularly vulnerable to the expression of many TRIM proteins. The differential susceptibility of MLV and HIV-1 to inflammatory TRIM proteins mapped to the U3 promoter of the viral LTR. Thus, our work identified new TRIM proteins involved in inflammatory and innate immune signaling and reinforced the striking ability of HIV-1 to efficiently replicate despite a strong innate immune response.
MATERIALS AND METHODS
Cell lines, viruses, plasmids, antibodies, and reagents.
A549 cells were obtained from the American Type Culture Collection (Manassas, VA). HEK293 and DFJ8 cells were described previously (13). BHK21 cells were obtained from John Rose (Yale University). The interferon reporter cell line HL116, which carries the luciferase gene under the control of the IFN-inducible 6-16 promoter, was a kind gift from Olivier Schwartz, Institut Pasteur. HL116 cells were grown in medium supplemented with hypoxanthine-aminopterin-thymidine (HAT) (Sigma-Aldrich). All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies) supplemented with 10% fetal calf serum (FCS) (Gemini Biotech) at 5% CO2.
Vesicular stomatitis virus expressing cytoplasmic enhanced green fluorescent protein (GFP) (VSV-eGFP) was a kind gift from John Rose (Yale University), Newcastle disease virus (NDV) expressing GFP was obtained from Peter Palese (Mount Sinai School of Medicine, New York, NY), and Sendai virus (SeV) strain Cantell was from Charles River Laboratories (Wilmington, MA).
Most TRIM expression plasmids have been described previously (13). TRIM51, TRIM61, and TRIM68 were gifts from Sebastien Nisole (CNRS, Paris, France). pcDNA3.1-based constructs expressing TRIM22, TRIM25, and TRIM34 were generated by amplifying the gene from a cDNA library prepared from PRR agonist-stimulated THP-1, primary mononucleocyte-derived macrophages, and dendritic cells. All the TRIM proteins used in the study were full-length isoforms. DNA sequencing of clones matched the reference sequence listed in Table S1 in the supplemental material. Other expression plasmids and their sources used in the current study were as follows: IKK beta superactivator (IKKβ SA), IκB superrepressor (dominant-negative IκB [dnIκB]), c-jun, dominant-negative Fos (dnFos), and IKK-ε were from Sankar Ghosh (Columbia University, NY); human TLR4, MD2, and IRF3 were from Ruslan Medzhitov (Yale University); human MyD88, TRIF, and TRAM were from Addgene (Boston, MA; deposited by Doug Golenbock, University of Massachusetts, Worcester, MA); TIRAP was from OpenBiosystems; and RIG-I, MDA5, and STING were from John MacMicking (Yale University). Deletions of individual domains from TRIMs were generated using overlap PCR, and the mutated TRIM genes were cloned into XhoI and EcoRI sites of the pEYFP C1 vector. E3 mutants of the TRIMs described were created by replacing two active-site cysteines with alanines in the RING domain using QuikChange site-directed mutagenesis (Agilent Technologies). MLV and HIV-1 LTR luciferase constructs were generated by PCR amplification using Friend MLV pLRB303 and pNL4-3, respectively, as templates and cloning into the pHTS-MCS vector (Biomyx, San Diego, CA). Overlap PCR was used to generate a chimeric LTR (MLV with HIV-1 U3), which was subsequently cloned into a luciferase vector or the pLRB303 vector to generate replication-competent MLV. The two NF-κB sites (AGGGACTTTCC) in the HIV LTR were disrupted with a QuikChange site-directed mutagenesis kit to ACTCACTTTCC based on previous studies (39).
The antibodies used and their sources were as follows: anti-GFP (Life Technologies); anti-actin and anti-beta-tubulin (Sigma); anti-hemagglutinin (HA) (Covance); anti-IκB, -phosphorylated p38, -p-IRF3, -TAK1, -MyD88,- RIP1, and -TRAF6 (Cell Signaling); anti-MLV gag p30 (Quality Biotech, Camden, NJ, and ATCC); anti-TRIM15, -TRIM31, and -TRIM62 (Proteintech); and anti-TRIM13 and -TRIM25 (Abnova). The reagents used in the study and their sources were as follows: Ultrapure LPS from Salmonella enterica serovar Minnesota and 5′triphosphorylated-double-stranded RNA (dsRNA) RIG-I ligand (Invivogen, San Diego, CA), tumor necrosis factor alpha (TNF-α) (Sigma, St. Louis, MO), and puromycin (Gemini Bio-Products, Sacramento, CA).
Dual-luciferase assay.
HEK293 cells seeded at 5 × 104 per well of a 48-well plate were transfected using Fugene6 (3 μl per μg DNA; Roche, Indianapolis, IN, or Promega Inc.) with 5 to 25 ng of DNA expressing the indicated TRIM proteins or bona fide inducers/repressors of immune signaling pathways and with 25 ng of plasmid DNA expressing firefly luciferase under the control of various transcriptional response element or viral LTR constructs. In addition, each transfection mixture contained 5 ng of internal control reporter plasmid pRL-TK (Promega), which expresses Renilla luciferase for normalization of transfection efficiencies. The total amount of DNA was equalized to 250 ng for each transfection using pcDNA3.1. Luciferase assays conducted with A549 cells were carried out at a density of 4 × 105 cells per well of 48-well plates. Transfections were carried out using 3 μl of Transfectin (Bio-Rad, Hercules, CA) per microgram of DNA. Transfection mixtures contained 100 ng of plasmid expressing bona fide inducers or pcDNA 3.1, 80 ng of beta interferon (IFN-β)-luciferase, and 20 ng of pRL-TK plasmids. Cells were lysed 24 to 48 h posttransfection with passive lysis buffer (Promega), and the luciferase activity was measured using a Dual-Glo luciferase assay system (Promega, WI) in a Berthold multiwell luminometer. Firefly luciferase data were normalized to Renilla luciferase readings in each well, and the data were represented as the fold change compared to empty pcDNA (and standard deviation) for at least two triplicate experiments carried out on separate days. The significance cutoff value or threshold for each transcriptional response element was calculated by adding 2 standard deviations to the mean luciferase values obtained for empty-vector transfections (the 95% confidence interval value of the control) from experiments carried out on different days. Any value above this threshold was considered positive. The following firefly luciferase plasmids were used: NF-κB–luciferase was from Promega, the AP-1–luciferase construct was from Ruslan Medzhitov (Yale School of Medicine), human beta 1 interferon (IFN-β1–luc) was from Jürg Tschopp (University of Lausanne); interferon-stimulated response element (ISRE) and IRF3-luc were from Akiko Iwasaki (Yale School of Medicine), and gamma interferon activation sequence (GAS)-luciferase was from Agilent Technologies (Santa Clara, CA).
Viral infectivity assay.
HEK293 cells (2.5 × 105/well) were transfected in 24-well format using Fugene6 with 200 ng of plasmids carrying MLV or MLV (HIVU3) carrying the full-length Friend 57 MLV genome with a GFP insertion into the envelope protein and 0 to 50 ng of empty vector or plasmid expressing TRIM and 0 to 20 ng of plasmid expressing IKKβ SA, dnIκB, or dnFos. The amount of DNA per transfection was equalized to 500 ng using pcDNA3.1 vector. Forty-eight hours after transfection, the released virus infectivity was measured by applying culture supernatants to DFJ8 target cells in the presence of 5 μg/ml of Polybrene. GFP-positive cells were enumerated after an additional 36 to 48 h by fluorescence-activated cell sorting (FACS). In parallel experiments, the producer cells and culture supernatants from triplicate wells were pooled and sedimented. Gag protein in cells and released Gag were analyzed by Western blot analyses as described previously (13).
For experiments with VSV, target A549 cells and primary human macrophages were infected with VSV-eGFP at multiplicities of infection (MOIs) of 0.25 and 10, respectively, in medium containing 1% fetal bovine serum (FBS) and allowed to attach at 37°C for 1 h. After replacing the medium with complete medium, with or without the indicated amounts of LPS, the culture supernatants were harvested 24 to 30 h postinfection. Ten-fold serial dilutions of the culture supernatants were applied to BHK21 cells, overlaid with a 1:1 mixture of 2× complete DMEM and 2% methyl cellulose, and allowed to form plaques for 48 h. The cells were fixed with 4% paraformaldehyde, and the GFP-positive plaques were counted using a fluorescence microscope or after staining with crystal violet solution.
Culture of primary macrophages, siRNA knockdown, and LPS induction.
Peripheral blood mononuclear cells (PBMCs) were isolated from fresh blood drawn from healthy anonymous donors using Ficoll-Paque Plus (GE Healthcare). CD14+ cells (monocytes) were enriched from PBMCs by allowing the cells to adhere to petri plates in RPMI without serum at 37°C for 2 h. The nonadherent cells were removed by three sequential washes with warm medium. The enriched monocytes were allowed to differentiate into macrophages for a week in macrophage medium (RPMI 1640 medium supplemented with 10% pooled human AB serum [Atlanta Biologicals, Lawrenceville, GA], 20 mmol/liter HEPES, 1× minimal essential medium nonessential amino acids, 1× GlutaMax-I (Life Technologies), 10 μM β-mercaptoethanol, and 50 ng/ml recombinant human macrophage colony-stimulating factor [e-bioscience]). For small interfering RNA (siRNA) knockdown experiments, the differentiated macrophages were detached using trypsin-EDTA, washed, and plated at a density of 4 × 105 cells per well of a 48-well plate in macrophage medium. Twelve hours after plating, the cells were transfected with AllStars negative-control siRNA (Qiagen) or with TRIM62- or TRIM15-specific siRNA at a final concentration of 100 nM using 1 μl Interferin reagent (Polyplus Transfection, France). The medium was replaced with fresh macrophage medium 5 h posttransfection, and the cells were incubated for 3 days to allow gene silencing. The knockdown efficiency was monitored from extracted RNA using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA), followed by real-time PCR using gene-specific primers (obtained from SA Biosciences) and a SYBR Fast Bio-Rad iCycler quantitative PCR (qPCR) kit (Kapa Biosystems, Woburn, MA). PCR primers specific for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were used to normalize the data, and the knockdown of TRIM62 and TRIM15 RNAs compared to control cells was assessed to be in the range of 76 to 82% and 72 to 80%, respectively, for separate experiments. For LPS induction experiments, macrophages were treated with 100 ng/ml UltraPure S. Minnesota LPS in Optimem. The cells were then harvested at the indicated time points by phosphate-buffered saline (PBS) wash, followed by lysis in the well using RIPA buffer (Boston Bioproducts, MA) supplemented with protease and phosphatase inhibitor mixture (Sigma). The cell lysates were clarified by sedimentation at 20,000 × g and subjected to SDS-PAGE, followed by Western blotting using the indicated antibodies. Acquisition of Western blot signal and densitometric quantification of band intensities were carried out using the ImageQuant LAS 4010 digital imaging system and ImageQuant TL 1D gel analysis software from GE Healthcare Life Sciences.
siRNA and short hairpin RNAs (shRNAs) used in the study.
Silencing of endogenous human TRIM proteins in A549 and HEK293 cells was carried out using On-Targetplus siRNA smart pools (a mixture of 4 siRNAs), or individual siRNAs where indicated, from Dharmacon, Inc., predesigned to reduce off-target effects by up to 90%, as described previously (13). siRNA transfections were carried out using 0.75 μl Interferin reagent (Polyplus Transfection, VWR International, LLC) for 50 nM siRNA per well in a 48-well format. We routinely obtained 70 to 90% knockdown of specific TRIM proteins, as assessed by monitoring the levels of the transiently expressed GFP- or the HA-tagged version by Western blotting or endogenous levels by quantitative real-time RT-PCR (Q-RT-PCR) 48 to 96 h posttransfection. The primer pairs for quantitative PCR were purchased from SA Biosciences. PCR primers specific for GAPDH were used to normalize the data. The sense sequences of siRNAs targeting (i) TRIM1 were number 1, GCAAAUGACGCUGUCAAAUU, number 2, GGUCAACAGUCCUCAUCAAUU, number 3, AAUCAAAUGGCCAAACUAAUU, and number 4, GCGCAACAGCGAACUAGAAUU; (ii) TRIM13 were number 1, UAAAACAGCCGAUUUCAUAUU, number 2, GACACUGGCACAUUCAUUAUU, number 3, GAGACCAGCUCCAUUCAAGUU, and number 4, GAAGGGAGUGUGCGGAAUUUU; (iii) TRIM14 were number 1, CAGAUUACUACUUGACGAA, number 2, GCUAAUGCAGAGUCAAGUA, number 3, UCCAGAGGCUUCAGGCAUA, and number 4, CAACAUAACCCAGAUAGAA; (iv) TRIM27 were number 1, GAGCAGGGCUGAAAGAAUC, number 2, UAAGAGAGGCUCAGUUAUA, number 3, GCUGAACUCUUUGAGCCUAA, and number 4, GAAGAUUGUUUGGGAGUUU; (v) TRIM47 were number 1, CAUCAAGAGUGCAGCCGUA, number 2, GCAUAUCCGUGCUGAAGAG, number 3, GAACCAAAGGUGUCAAGAG, and number 4, GUACGGGACGGCAAGAUGA; (vi) RIG-I were number 1, GCACAGAAGUGUAUAUUGGUU, number 2, CCACAACACUAGUAAACAAUU, number 3, CGGAUUAGCGACAAAUUUAUU, and number 4, UCGAUGAGAUUGAGCAAGAUU; and (vii) MAVS were number 1, CCGUUUGCUGAAGACAAGA, number 2, CCACCUUGAUGCCUGUGAA, number 3, CAGAGGAGAAUGAGUAUAA, and number 4, UUGCUGAAGACAAGACCUAUA. The sequences for the siRNA Dharmacon Smartpool for human TRIM5, TRIM15, and TRIM62 were as previously reported (13). The sequences for siRNAs that were most effective in knocking down gene expression for TRIM15 and TRIM62 were CCCUGAAGGUGGUCCAUGA and CGCCAAAGCGCUUCGAUGU, respectively. These siRNAs were used for gene-silencing experiments carried out in macrophages.
Stable knockdown of gene expression was carried out using lentivirus-based shRNA systems after standard viral transduction, followed by puromycin selection in HEK293 cells. Constructs expressing shRNA against TAK1 (target sequence, AGCGCCCTTCAATGGAGGAAAT) and TRAF6 (number 1, AGGGTACAATACGCCTTACAAT; number 2, CGAAGAGATAATGGATGCCAAC) were described in previous studies (31, 40). Constructs expressing shRNAs targeting multiple sequences in TRIM62, RIP1, MyD88, and TRIF were obtained from Open Biosystems. For TRIM62, the efficacy of the knockdown was tested with each shRNA using quantitative RT-PCR, and the most effective target sequence, CATGAGACCAACCTCACATAT (shRNA number 1), was used in the functional experiments. In addition to monitoring the levels of transiently expressed GFP-tagged versions, multiple shRNAs for RIP1, MyD88, and TRIF were subjected to functional analyses to determine the best target sequence. RIP1 knockdown was assessed using RIP1-dependent TNF-α-triggered induction of NF-κb. TIRAP- and TRAM-induced NF-κb were used to evaluate the knockdown efficiency of MyD88 and TRIF. The best target sequences for (i) RIP1 (GIPZ based) were number 1, AGAGTAAACTCCAAGACGA, and number 2, GCGAGATGGACTGAAAGAA; (ii) MyD88 (pLKO.1 based) was GCAGAGCAAGGAATGTGACTT; and (iii) TRIF (pLKO.1 based) was ATCTTCTACAGAAAGTTGGAG. Scrambled shRNA, also called nontargeting shRNA (NTshRNA), was obtained from Open Biosystems.
IFN-β secretion and induction assays in siRNA-treated cells.
A549 cells (4 × 105/well) in 48-well plates were transfected with the indicated siRNAs. Sixty hours posttransfection, the cells were transfected with 250 ng RIG-I-specific ligand (Invivogen) using 1 μl of Interferin reagent. The medium was replaced 5 h posttransfection, and the cells were incubated further for 24 h to allow secretion of type I interferons. The culture supernatant was applied to interferon reporter HL116 cells, and 24 h later, the cells were lysed for measurement of firefly luciferase activity. For interferon induction assays, 60 h post-siRNA treatment, the cells were transfected with 100 ng of plasmid expressing the indicated proteins or pcDNA, along with 80 ng of IFN-β luciferase reporter and 20 ng of pRLTK vector. If virus infections were carried out, they were done 5 h after transfection. Cells were lysed 24 h later, and luciferase activity was measured as described above.
Statistical analysis.
Student's t test, available in GraphPad Prism v5.5, was used to determine the statistical significance of the data.
RESULTS
TRIM proteins induce inflammatory and innate immune signaling.
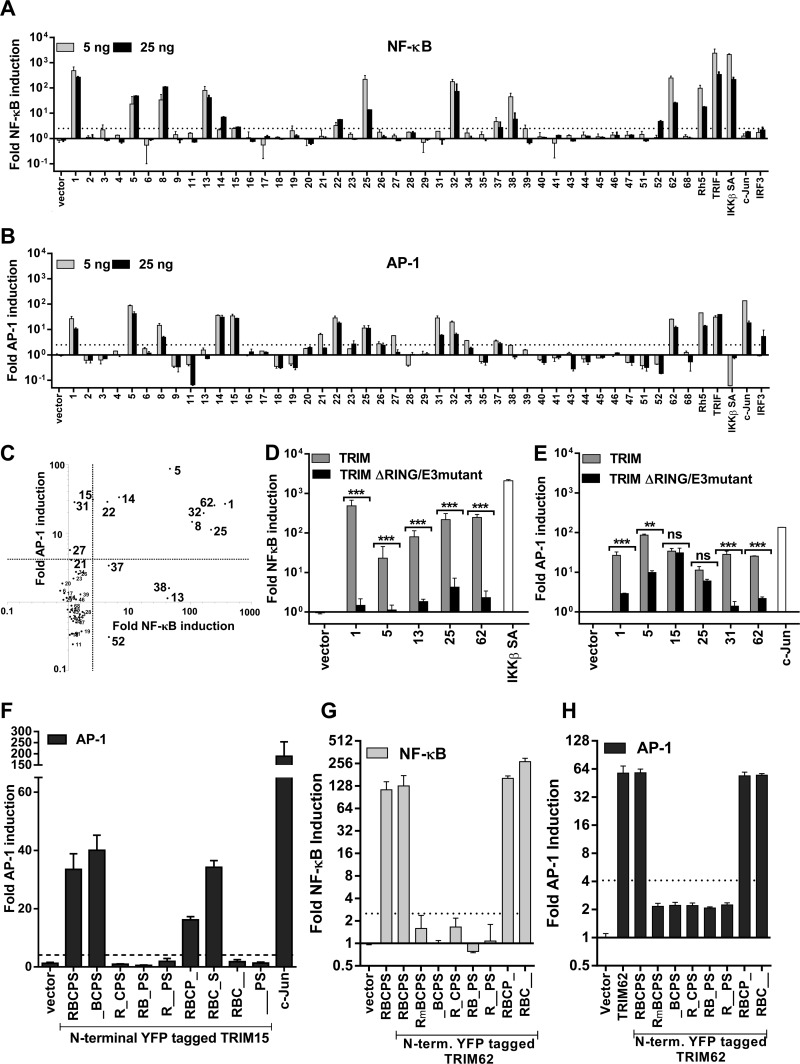
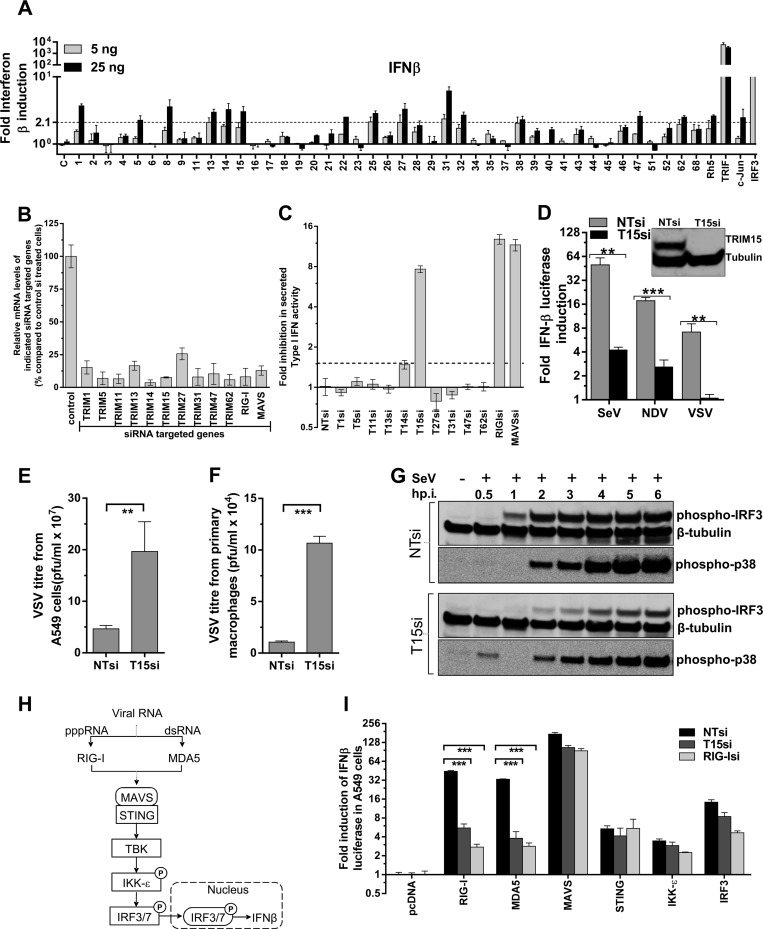
To identify TRIM proteins capable of inducing inflammatory and innate immune signaling, we cotransfected 5 or 25 ng of plasmids expressing 43 human TRIM proteins with NF-κB or AP-1 firefly luciferase reporter constructs in HEK293 cells (Fig. 1A and B). Constitutively active thymidine kinase promoter-driven Renilla luciferase was used for normalization and estimation of fold induction. The empty vectors pcDNA and pCMVSPORT6 were used as negative controls, and constructs expressing TRIF, IKKβ SA (IKKβ kinase with phosphomimetic mutations [S177E/S181E] that is a constitutive superactivator of NF-κB [41]), c-jun, and IRF-3 were used as specificity controls. A scatter plot of AP-1 versus NF-κB induction by the TRIM proteins showed that 16 out of the 43 proteins were able to induce signal transduction (Fig. 1A to C) based on the threshold calculated for each response element (see Materials and Methods). TRIM13, -37, -38, and -52 activated only NF-κB, while TRIM15, -21, -27, and -31 induced only AP-1 signaling (Fig. 1C). TRIM1, -5α, -8, -14, -22, -25, -32, and -62 induced NF-κB, as well as AP-1 (Fig. 1C). Importantly, we identified five proteins, TRIM1, -14, -15, -52, and -62, whose signaling properties were not previously described.
Fig 1.
TRIM proteins induce NF-κB and AP-1. (A and B) HEK293 cells were transfected with 5 or 25 ng of plasmids expressing the indicated TRIM proteins (see Table S1 in the supplemental material) or bona fide inducer controls, with Renilla luciferase and NF-κB (A) or AP-1 (B) firefly luciferase reporter constructs. Luciferase activity was measured at 24 and 48 h posttransfection, respectively. Firefly luciferase was normalized to Renilla luciferase readings in each well, and the data were plotted as fold change compared to empty pcDNA/vector (with standard deviation) for at least two triplicate experiments carried out on separate days. (C) Scatter plot of the TRIM-induced NF-κB versus AP-1 activities shown in panels A and B. (D and E) Effects of RING domain deletion on NF-κB (D) and AP-1 (E) induction by TRIM proteins selected based on their signaling activities. (F to H) Normalized luciferase activity in HEK293 cells transfected with NF-κB or AP-1 luciferase and pRLTK reporter constructs, along with empty vector or 10 ng of plasmids expressing untagged TRIM62 or N-terminal YFP-tagged versions of TRIM62 or TRIM15. R, B, C, P, and S represent the RING, B-box, coiled-coil, PRY, and SPRY domains. The E3 ligase inactive mutant (C11A/C14A) is denoted as E3mutant or Rm. The normalized reporter activity in cells transfected with empty vector was set to 1 to determine fold induction. A statistically significance cutoff value (dotted lines; 2.5 for NF-κB; 4.1 for AP-1) was obtained by adding 2 standard deviations to the mean value obtained for empty-vector samples for each response element. Statistical significance values: ***, P < 0.001; **, P < 0.01; *, P < 0.05, respectively; ns, not significant.
In order to test if the ability to signal required a functional E3 ligase activity, we generated RING domain deletions or point-mutated versions of TRIMs that disrupt the catalytic sites. All RING domain mutant constructs were expressed at levels similar to that of the wild-type (wt) protein (see Fig. S1A in the supplemental material). Interfering with the E3 ligase function of TRIM1, -5, -13, -31, and -62 significantly reduced their observed signaling activities (Fig. 1D and E). In the case of TRIM25, E3 ligase activity was necessary for NF-κB induction, but not for AP-1 signaling (Fig. 1D and E). E3 ligase activity of TRIM15 was dispensable for AP-1 signaling (Fig. 1E). Deletion of individual domains revealed that, in addition to the B box and coiled coil, the PRY-SPRY domains were required for AP-1-inducing activity of TRIM15 (Fig. 1F; see Fig. S1B in the supplemental material). In contrast, deletion of the PRY-SPRY domains of TRIM62 did not compromise its signal transduction properties. Rather, in addition to the RING domain, both B-box and coiled-coil domains were required for NF-κB and AP-1 induction by TRIM62 (Fig. 1G and H; see Fig. S1C in the supplemental material). Thus, the B-box and/or coiled-coil domains that confer oligomerization/dimerization properties (42) are indispensable for both TRIM15- and -62-mediated signaling.
TRIM proteins use the TAK1-dependent canonical NF-κB pathway for signaling.
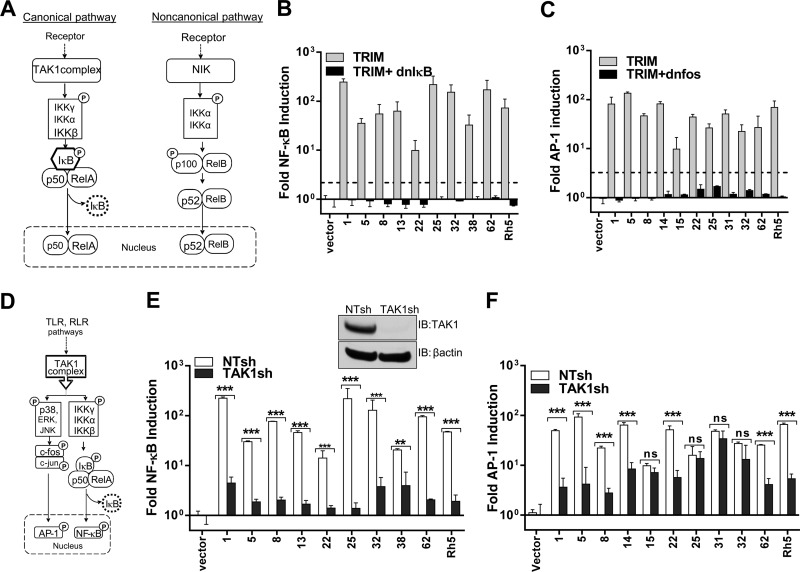
The activation of NF-κB by various stimuli can result in generation of active RelA-p50 and RelB-p52 dimers via the canonical (IKKβ- and IκB-dependent) and the noncanonical (IKKα- and p100/δ-dependent) pathways, respectively (Fig. 2A) (43). To differentiate the two pathways, we tested NF-κB induction by TRIMs when coexpressed with dnIκB, which inhibits only the canonical RelA-p50 activation (43, 44). dnIκB, also designated “superrepressor,” is a mutant of IκB that cannot be phosphorylated (S32R/S36R) and consequently cannot be ubiquitinated and degraded (45). Coexpression of dnIκB inhibited NF-κB activation by all the NF-κB-inducing TRIMs tested, indicating that they function upstream of IκB in the canonical pathway (Fig. 2B). These data indicate that expression of the TRIM proteins indeed triggered the signaling pathway and ruled out their involvement as components of transcription factors. A similar inhibition in AP-1 transcriptional activation was seen when AP-1-inducing TRIMs were coexpressed with dnFos (Fig. 2C). dnFos inhibits the DNA binding activity of the Fos and Jun heterodimer (46).
Fig 2.
TRIM proteins use the TAK1-dependent canonical NF-κB pathway for signaling. (A) Schematic of canonical and noncanonical NF-κB signaling pathways (41, 43). The canonical pathway involves activation of the IKK complex by TAK1 and IKK-mediated IκB phosphorylation followed by degradation, resulting in nuclear translocation of the NF-κB heterodimer p50-RelA. The noncanonical NF-κB pathway is independent of IκB and relies on NIK (NF-κB-inducing kinase) for phosphorylation of IKKα to induce p100 processing. This results in nuclear translocation of the p52-RelB complex. (B and C) NF-κB and AP-1 luciferase activity was determined as for Fig. 1A after transfection with NF-κB luciferase or AP-1 luciferase and pRLTK reporter constructs, along with empty vector (vector) or 5 ng of plasmids expressing the indicated TRIMs in the absence or presence of 10 ng plasmid expressing dnIκB or dnFos. (D) Schematic depicting the critical role of TAK1 as the common IKK-activating kinase engaged by activated TLR and RLR for induction of NF-κB and AP-1 (47). (E and F) The indicated TRIM-induced NF-κB and AP-1 luciferase reporter activity was determined in NT (open bars) and TAK1sh (filled bars) expressing HEK293 cells as for Fig. 1A and B. The error bars denote standard deviations between triplicate samples. The inset in panel E shows Western blot (IB, immunoblot) analysis of HEK293 cell lysates stably expressing nontargeting shRNA (NTsh) or shRNA targeting TAK1 (TAK1sh) probed with antibodies to TAK1 and β-actin as a loading control. Statistical significance values: ***, P < 0.001; **, P < 0.01; *, P < 0.05, respectively; ns, not significant.
The majority of innate immune-sensing pathways activated by Toll- and RIG-I-like receptors induce NF-κB and/or AP1 by utilizing TAK1 for activation of the IKK complex (Fig. 2D) (47). TRIM5 and -8 have been previously shown to activate TAK1 (30, 31). Therefore, we generated HEK293 cells stably expressing NTshRNA and shRNA targeting TAK1 to determine the requirement for TAK1 in TRIM-induced signaling (Fig. 2E, inset). Knockdown of TAK1 largely abolished the ability of the TRIM proteins to induce NF-κB (Fig. 2E). AP-1 signaling by a number of TRIM proteins, including TRIM1, -5, -8, -14, -22, and -62, was also significantly reduced in cells lacking TAK1 (Fig. 2F). However, TAK1 was not needed for AP-1 induction by TRIM15, -25, -31, and -32 (Fig. 2F). Thus, TAK1 was essential for NF-κB induction by TRIM proteins, but its requirement for AP-1 signaling was variable.
TRIM62 functions in the TLR4 signaling pathway.
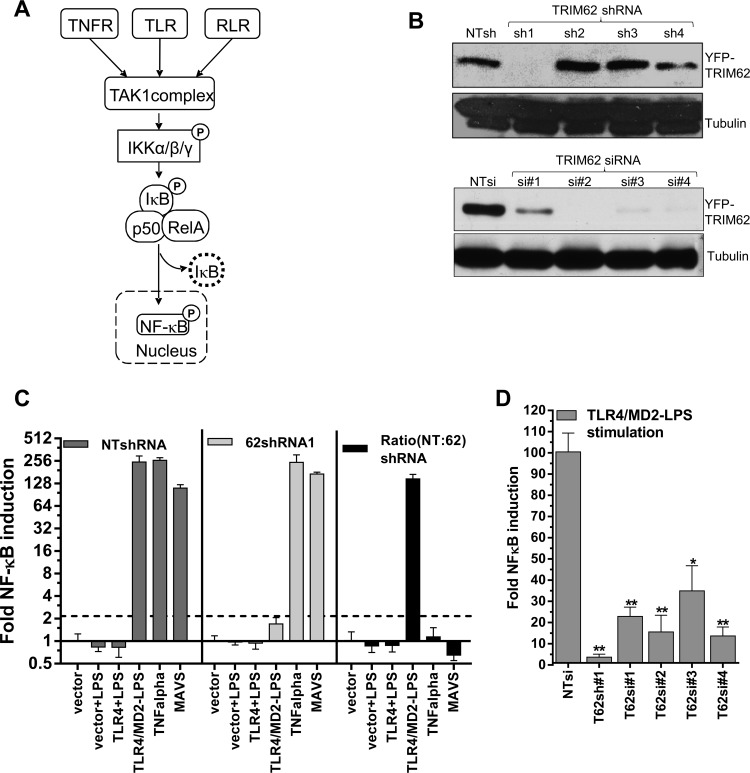
We next focused our study on the previously uncharacterized signal transduction induced by TRIM62. TRIM62 potently induced both NF-κB and AP-1 in a TAK1-dependent manner, a hallmark of TNF, TLR, and RLR signaling pathways (Fig. 3A). To identify the specific signaling pathway utilized by TRIM62, we first silenced TRIM62 expression using different shRNAs and siRNAs in HEK293 cells (Fig. 3B). The knockdown efficiency for shRNA1 and all four siRNAs ranged between 75 and 89% when endogenous mRNA levels were measured and was also confirmed at the protein level using transiently expressed yellow fluorescent protein (YFP)-tagged TRIM62 expression constructs (Fig. 3B). We then measured NF-κB induction in HEK293 cells expressing NTshRNA or TRIM62 shRNA1 in response to TNF-α, LPS and by expression of MAVS to study TNFR, TLR4, and RLR pathways (Fig. 3C). HEK293 cells endogenously express the TNF-α receptor. To achieve LPS responsiveness in HEK293 cells, we had to express the receptor TLR4, together with its cofactor, MD2 (Fig. 3C) (48). Interestingly, LPS-stimulated TLR4 signaling was strongly affected in TRIM62-silenced cells, suggesting that TRIM62 functions in the TLR4 pathway (Fig. 3C). The requirement for TRIM62 in TLR4-mediated NF-κB signaling was also confirmed by silencing TRIM62 using four different siRNAs (Fig. 3D).
Fig 3.
TRIM62 is a novel protein involved in TLR4 signaling. (A) Schematic depicting the possible scenarios for activation of TAK1-dependent NF-κB and AP-1 induction involving stimulation of tumor necrosis factor alpha-, Toll-like, and RIG-I-like receptors (47). (B) HEK293 cells stably expressing nontargeting shRNA (NTsh) or shRNAs 1 to 4 targeting TRIM62 (top) or treated for 24 h with control (NT) or siRNAs 1 to 4 targeting TRIM62 (bottom) were transfected with YFP-tagged TRIM62, and Western blot analysis of cell lysates was carried out 48 h posttransfection using antibodies to GFP and tubulin as a loading control. (C) Normalized NF-κB reporter luciferase activity induced by 100 ng/ml LPS, 5 ng/ml TNF-α, and transfection of empty vector or 5 ng plasmid encoding MAVS protein in HEK293 cells stably expressing NTshRNA or shRNA targeting TRIM62. LPS-treated cells were transfected with 25 ng plasmids expressing TLR4 alone or together with cofactor MD2 5 h prior to treatment. The ratio depicting the fold inhibition in reporter activity observed in cells expressing TRIM62sh1 compared to NTsh (NT:62) is shown by the black bars. (D) Fold induction of NF-κB luciferase activity compared to empty vector by transfection of 25 ng plasmid encoding TLR4 and MD2 protein into HEK293 cells. These cells either were stably expressing shRNA targeting TRIM62 or were transfected with control nontargeting siRNA or four different siRNAs targeting TRIM62. The cells treated with siRNAs were transfected with NF-κB inducers after a 72-h period to allow gene silencing. The cells were triggered to induce NF-κB by addition of LPS as in panel C. The reporter activity induced by empty vector was set to 1 for each condition to determine the fold induction. A statistical significance cutoff value (dashed line; 2.5 for NF-κB; 4.1 for AP-1) was obtained by adding 2 standard deviations to the mean value obtained for empty-vector samples for each response element. The error bars denote standard deviations between triplicate samples. Statistical significance values: **, P < 0.01; *, P < 0.05.
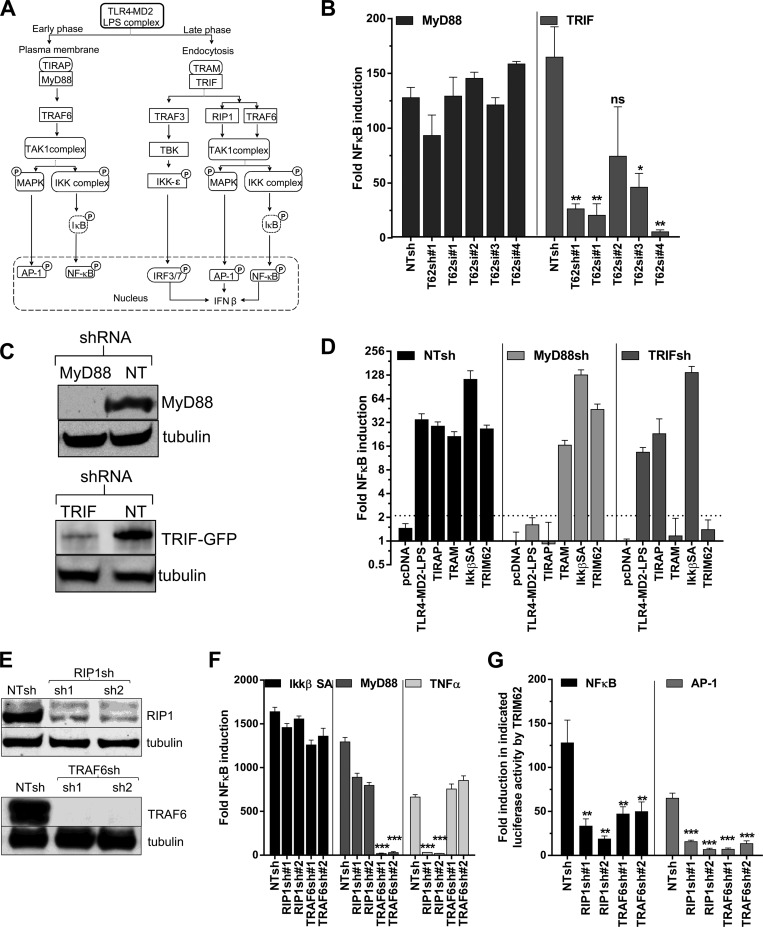
The LPS-activated TLR4 pathway utilizes MyD88 and TRIF as adaptors to trigger NF-κB. TIRAP acts as an adaptor to recruit MyD88 at the plasma membrane to induce early NF-κB and AP-1 signaling (Fig. 4A) (49). TRAM recruits TRIF to the endocytosed LPS-TLR4 complex to trigger the late wave of NF-κB and AP-1 signaling (Fig. 4A) (50, 51). TRIF also activates IRF3/7, which together with NF-κB and AP-1 stimulate transcription from the IFN-β promoter (50–52). TRIM62 silencing using shRNA1 and 3 different siRNAs in HEK293 cells significantly compromised TRIF-mediated, but not MyD88-mediated, NF-κB induction (Fig. 4B). To substantiate the requirement for TRIM62 in the TLR4 signaling branches, we generated MyD88- and TRIF-silenced cells using shRNAs (Fig. 4C). Specific knockdown was functionally evident, as NF-κB induction by the MyD88-proximal adaptor TIRAP was compromised in MyD88 and not in cells where TRIF expression was silenced (Fig. 4D). Similarly, NF-κB induction by the TRIF adaptor protein TRAM was specifically reduced in TRIF- and not MyD88-silenced cells (Fig. 4D). Importantly, TRIM62-mediated NF-κB induction was abrogated in cells where TRIF, but not MyD88, expression was silenced (Fig. 4D).
Fig 4.
TRIM62 functions in the TRIF branch of the TLR4 signaling pathway. (A) Schematic of an LPS-triggered TLR4 signaling pathway that uses plasma membrane-localized TIRAP/MyD88 as an adaptor to induce the early phase of TRAF6- and TAK1-dependent NF-κB and AP-1. After endocytosis of the LPS-TLR4 complex, TRAM and TRIF are recruited, triggering RIP1-, TRAF6-, and TAK1-dependent late NF-κB and AP-1 induction. TRIF also engages TBK, resulting in activation of IRF3 and -7. AP-1, IRF3/7, and NF-κB together bind the positive regulatory domain elements in the IFN-β promoter to activate robust transcription of IFN-β (53). Phosphorylation of signaling components are denoted by circled P. (B) Normalized NF-κB reporter luciferase activity induced by transfection of empty vector or 2.5 ng plasmid expressing MyD88 or TRIF in HEK293 cells stably expressing NTshRNA, shRNA targeting TRIM62, or cells treated with four different siRNAs targeting TRIM62. The cells treated with siRNAs were transfected with NF-κB inducers after a 72-h period to allow gene silencing. The reporter activity induced by empty vector was set to 1 for each condition to determine the fold induction. (C) Western blot analysis of HEK293 cell lysates stably expressing control NTshRNA or shRNA against MyD88 or TRIF using antibodies to GFP, endogenous MyD88, and the loading control tubulin. A TRIF-GFP expression construct (50 ng) was transfected 48 h prior to harvesting the cells for testing the knockdown efficiency in TRIF shRNA-expressing cells. (D) Normalized NF-κB reporter luciferase activity induced by transfection of empty vector (pcDNA) or a plasmid encoding TRIM62 (10 ng), TIRAP (50 ng), TRAM (50 ng), or IKKβ SA (2.5 ng) in HEK293 cells stably expressing NTshRNA or shRNA targeting MyD88 or TRIF. (E) Western blot analysis of HEK293 cell lysates stably expressing control NTshRNA or shRNA against RIP1 or TRAF6 using antibodies to endogenous RIP1, TRAF6, and the loading control tubulin. (F) HEK293 cells stably expressing NTshRNA or two shRNAs targeting RIP1 or TRAF6 were transfected with NF-κB luciferase and pRLTK reporter constructs, along with either empty vector or 2.5 ng of plasmid encoding IKKβ SA or MyD88 or treated with 5 ng/ml of TNF-α. Normalized reporter activity induced by the empty vector was set to 1 for each cell line to determine the fold induction. (G) Normalized NF-κB and AP-1 reporter luciferase activity induced by 10 ng of plasmid expressing TRIM62 in HEK293 cells stably expressing NTshRNA or two different shRNAs targeting RIP1 and TRAF6. The dotted lines denote the statistical significance cutoff values (2.5 for NF-κB; 4.1 for AP-1) obtained by adding 2 standard deviations to the mean value obtained for empty-vector samples for each response element. The error bars denote standard deviations of experiments carried out in triplicate. Statistical significance values: ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
RIP1 and TRAF6 function as a kinase and a nondegradative E3 ubiquitin ligase, respectively, in the TLR4 pathway (47, 53). MyD88 recruits TRAF6, whereas TRIF utilizes both RIP1 and TRAF6 to activate TAK1 (Fig. 4A). We therefore explored the signaling activity of TRIM62 in cells expressing shRNAs targeting RIP1 and TRAF6. The knockdown was confirmed by monitoring the endogenous protein levels of RIP1 and TRAF6 (Fig. 4E). The knockdown was also functionally validated by measuring the NF-κB induction triggered by TNF-α, which uses RIP1 as the main proximal kinase, and MyD88, which mainly utilizes TRAF6 (Fig. 4F). NF-κB induction by a downstream component of IKKβ SA was not significantly affected in RIP1 and TRAF6 shRNA-expressing cells (Fig. 4F). Importantly, induction of both NF-κB and AP-1 by TRIM62 was significantly reduced in cells expressing shRNAs targeting RIP1 and TRAF6 compared to cells expressing control NTshRNA (Fig. 4G). These data suggest that TRIM62, like TRIF, uses both TRAF6 and RIP1 to activate TAK1.
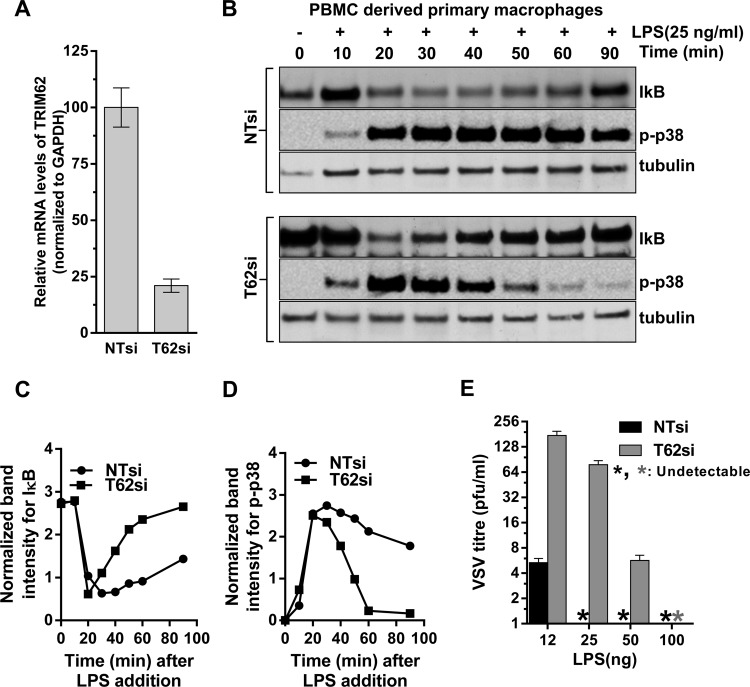
To confirm the role of TRIM62 in the TRIF branch of the TLR4 signaling pathway in primary cells, we tested the effect of TRIM62 silencing on LPS signaling in PBMC-derived primary human macrophages. The knockdown efficiency of TRIM62 was ∼80% in TRIM62 siRNA-treated macrophages compared to the control (Fig. 5A). We then monitored the temporal progression of NF-κB and AP-1 signaling in these macrophages in response to LPS stimulus (Fig. 5B). The induction of NF-κB signaling was analyzed by measuring IκB degradation, and AP-1 induction was examined by monitoring the levels of phosphorylated MAPK p38 using Western blotting followed by densitometry (Fig. 5B to D). In control siRNA-treated macrophages, LPS triggered a prolonged degradation of IκB from 20 to 60 min, as both MyD88-dependent early and TRIF-dependent late NF-κB signaling were functional (Fig. 5B and C) (52). This was further corroborated by the sustained levels of active phosphorylated p38 in the same period (Fig. 5B and D). In contrast, LPS-stimulated TRIM62-silenced macrophages displayed only a short burst of IκB degradation, followed by a quick recovery to steady-state levels (Fig. 5B and C). This was associated with activation of p38 only at early time points after LPS addition (20 to 30 min) (Fig. 5B and D). These data were consistent with intact early MyD88-dependent signaling but a compromised late wave of TRIF-dependent signaling in TRIM62-silenced primary human macrophages.
Fig 5.
TRIM62 is required for TRIF signaling in primary macrophages. (A) Quantitative real-time PCR analysis of TRIM62 mRNA levels normalized to GAPDH in primary macrophages 72 h after siRNA treatment. TRIM62 mRNA levels in control NTsiRNA-treated cells were set to 100%. (B) Western blot analyses of cell lysates from PBMC-derived macrophages treated with negative-control siRNA (NTsi) or siRNA targeting TRIM62 (T62si) at the indicated times after LPS addition. The blots were probed with antibodies to the indicated proteins. Tubulin was used as a loading control. (C and D) Plots of IκB and phosphorylated p38 normalized band intensities shown in panel B as a function of time. The band intensities were normalized to tubulin levels using densitometric quantification. (E) VSV PFU released per ml in culture supernatants 30 h postinfection of PBMC-derived macrophages treated with negative-control siRNA (NTsi) or siRNA against TRIM62 (T62si) and infected with VSV at an MOI of 10, along with the indicated amounts of LPS. The error bars represent standard deviations of three experiments carried out in triplicate.
TRIF signaling also activates IRF3/7 via TANK binding kinase (TBK) (Fig. 4A) (54). The activation of all three transcription factors, NF-κB, AP-1, and IRF3/7, by TRIF results in the synthesis of IFN-β to establish an antiviral state (Fig. 4A) (52). We therefore tested if TRIM62 is required for the LPS-stimulated induction of an antiviral state in the primary human macrophages. Indeed, these experiments revealed that the VSV titer in the culture supernatant 30 h postinfection was increased up to 20-fold in TRIM62 siRNA-treated cells (Fig. 5E). This effect is more evident at lower concentrations of LPS, as MyD88-dependent signaling could potentially compensate for the TRIM62 defect at higher concentrations. These data suggest that in primary human macrophages, silencing of TRIM62 also curtailed LPS stimulation of an antiviral state. Thus, collectively, we conclude that TRIM62 functions in the TRIF branch of the TLR4 pathway.
TRIM15 functions in the RLR-induced interferon pathway.
In addition to the induction of NF-κB and AP-1, we also tested if TRIM protein expression stimulated interferon reporters, as activation of some innate immunity pathways culminates in synthesis of inflammatory cytokines, such as interferons. Unlike with NF-κB/AP-1, TRIM expression induced only weak activation (less than 7-fold over background) of interferon-related reporters, including IFN-β, IRF3, GAS, and ISRE response elements (Fig. 6A; see Fig. S2 in the supplemental material). Because of the limitations associated with an expression screen for interferon in HEK293 cells, we decided to specifically ask if TRIM proteins are required for RIG-I ligand-induced type I interferon in more relevant cell types. We chose A549 lung epithelial cells, as they responded strongly to RIG-I ligand and secreted measurable type I interferons. We conducted a small-scale siRNA screen for 10 TRIM proteins in A549 cells, most of which mildly induced IFN-β reporter luciferase in HEK293 cells (Fig. 6A). Real-time PCR analysis for relative mRNA levels of targeted genes revealed that the knockdown efficiencies ranged between 70 and 90% (Fig. 6B). Interestingly, the RIG-I ligand-induced secretion of type I interferon was strongly dependent on TRIM15 (Fig. 6C). The extent of inhibition was similar to that seen in cells silenced for RIG-I or MAVS expression. TRIM15 knockdown also significantly diminished SeV-, NDV-, and VSV-induced interferon induction in A549 cells compared to controls (Fig. 6D). Accordingly, the VSV titer in culture supernatants from TRIM15-silenced A549 cells, as well as PBMC-derived primary macrophages, was enhanced 4- to 10-fold compared to control cells (Fig. 6E and F). A temporal analysis of SeV-induced interferon signaling revealed that TRIM15 silencing significantly diminished phosphorylation of IRF3 and MAPK p38 compared to control cells (Fig. 6G). These data suggested that TRIM15 functions upstream of IRF3 and MAPK in the RLR pathway (Fig. 6H). We next compared the levels of interferon induction by various components of the RLR pathway in cells treated with siRNA to the control, TRIM15, and RIG-I. We detected a significant 9-fold reduction in RIG-I- and MDA5-induced interferon luciferase activity upon silencing of TRIM15 compared to control cells, suggesting that TRIM15 functions in a process that is common to both pathways (Fig. 6G and H). Both RIG-I and MDA5 use MAVS as a common adaptor for downstream signaling (Fig. 6H). Interferon induction by MAVS and the downstream factors STING, IKK-ε, and IRF3, however, was intact in cells silenced for TRIM15 expression (Fig. 6H and I). Thus, our data suggest that TRIM15 must function upstream or at the level of MAVS in the RLR pathway.
Fig 6.
TRIM15 is required for RIG-I-mediated interferon induction. (A) HEK293 cells were transfected with 5 or 25 ng of plasmids expressing the indicated TRIM proteins, bona fide inducers, Renilla luciferase, and human IFN-β–firefly luciferase reporter construct, and luciferase activities were measured at 48 h posttransfection. The normalized luciferase activities were plotted as the fold change compared to empty pcDNA/vector (with standard deviation) for at least two triplicate experiments carried out on separate days. A statistical significance cutoff value (dashed line) of 2.1 for the IFN-β luciferase assay was obtained by adding 2 standard deviations to the mean value obtained for empty-vector samples. (B) Quantitative real-time PCR analyses for the indicated genes 72 h post-siRNA treatment in A549 cells. Relative mRNA levels were calculated by normalizing to GAPDH. The normalized mRNA level for each gene was set to 100% in control NTsiRNA-treated cells. (C) A549 cells were treated with control siRNA (NTsi) or siRNAs targeting the indicated genes. Sixty hours posttreatment, they were activated by transfecting 250 ng of RIG-I ligand, and the activity of type I interferons in the culture supernatants was measured using the HL116 reporter cell line after 24 h. The fold inhibition in IFN-β activity was plotted using NTsi-treated cells as a control and set to 1. (D) Control (NTsi) or TRIM15 (T15si)-treated A549 cells were transfected 60 h posttreatment with IFN-β–luc and pRLTK vector. The cells were then infected with SeV (25 HA units/ml), NDV-GFP (MOI = 0.25), or VSV-eGFP (MOI = 0.25), and dual-luciferase activities were measured 24 h postinfection, normalized, and plotted. (E and F) VSV titers in culture supernatants of A549 cells and PBMC-derived macrophages treated with either control or TRIM15si. A549 cells and macrophages were infected with VSV at MOIs of 0.25 and 10 for 24 and 30 h, respectively. (G) Western blot analyses of cell lysates from A549 cells treated with negative-control siRNA (NTsi) or siRNA targeting TRIM15 (T15si) at the indicated times after infection (p.i.) with SeV (25 HA units/ml). The blots were probed with antibodies to the indicated proteins. β-Tubulin was used as a loading control. (H) Signal transduction pathway for cytoplasmic viral RNA sensors RIG-I and MDA5 that sense short triphosphate and long double-stranded RNA, respectively. Both sensors use MAVS as the common signaling adaptor for activation of IRF3/7 via STING, TBK, and IKK-ε (27). Phosphorylation of signaling components are denoted by circled P. (I) A549 cells pretreated for 60 h with NT, TRIM15, and RIG-I siRNA were transfected with IFN-β–luc and pRLTK vector, along with pcDNA or plasmids expressing the indicated proteins. Dual-luciferase activity was measured 24 h later, and normalized values were plotted, with the activity in pcDNA-transfected cells set to 1. The error bars in panels B to F and I represent standard deviations from experiments carried out in triplicate. Statistical significance values: ***, P < 0.001; **, P < 0.01.
Inflammatory signaling induced by TRIM proteins contributes to their anti-MLV activity.
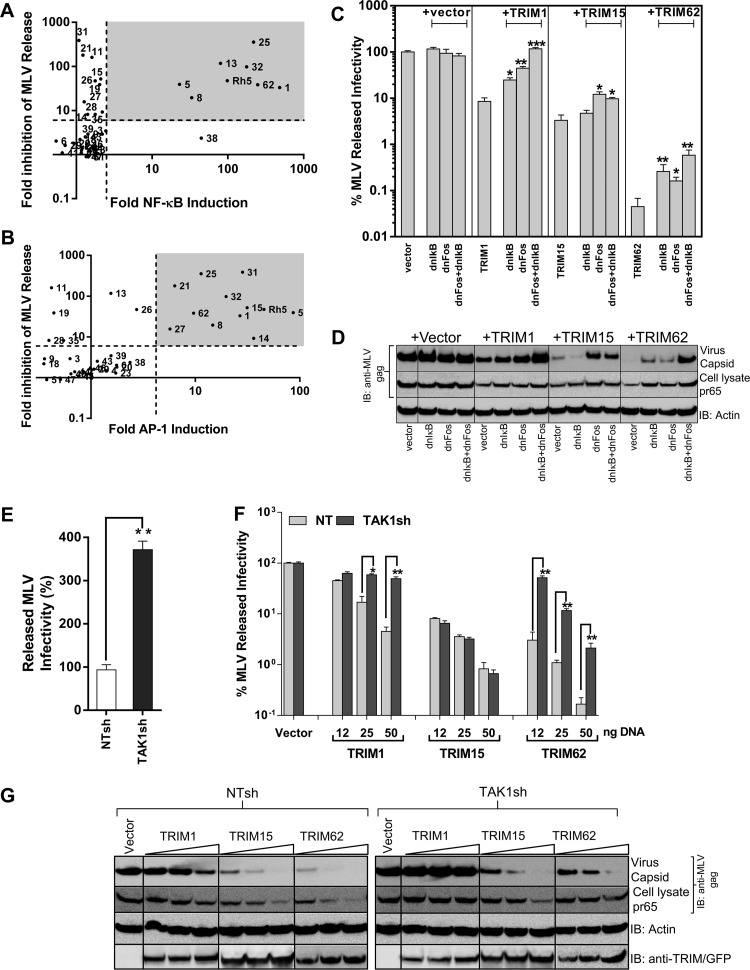
Our laboratory had previously carried out a screen for antiretroviral activities of TRIM proteins and identified many family members with antiretroviral activity (13). In light of the innate immune signaling cascades induced by TRIM proteins, we asked if these features were potentially associated with their previously observed antiviral activities. To this end, we plotted the fold induction in NF-κB/AP-1 activities of TRIM proteins against the previously observed inhibition in MLV released infectivity. Out of the 17 TRIMs that inhibited MLV released infectivity, 12 induced NF-κB and/or AP-1 signaling, raising the possibility of an association between signaling and inhibitory activities of TRIM proteins (Fig. 7A and B). To functionally test if inflammatory signaling plays a role in the antiviral effects, we expressed the dominant-negative proteins dnIκB and/or dnFos that suppressed NF-κB or AP-1 induction by the TRIM proteins (Fig. 2B and C). The infectivity inhibited by NF-κB and AP-1 inducing TRIM1 and -62 was partially rescued by both dnIκB and dnFos (Fig. 7C). There was up to 11- and 37-fold rescue of MLV release by a combination of the dominant-negative proteins from the inhibitory effects of TRIM1 and -62, respectively, by restoring MLV gene expression and virus release (Fig. 7C and D). The antiviral activities of AP-1 inducing TRIM15 were rescued to a lesser extent (∼4-fold) in a statistically significant manner by dnFos, but not by dnIκB (Fig. 7C and D). We next asked if interfering with TAK1 signaling could rescue TRIM-mediated inhibition of MLV assembly and release. Interestingly, TAK1 silencing alone resulted in ∼4-fold enhancement of released MLV infectivity, suggesting that TAK1-mediated signaling restricts MLV replication in HEK293 cells (Fig. 7E). TAK1 silencing rescued released MLV infectivity from the antiviral effects of TRIM1 and -62 by up to 10- and 16-fold, respectively (Fig. 7F and G). The rescue of infectivity correlated well with restoration of MLV gene expression and virus release in TAK1-silenced cells (Fig. 7G). In contrast, the anti-MLV properties of TAK1-independent TRIM15 were largely unaffected (Fig. 7F and G). Thus, the antiviral activities exhibited by TRIM1 and -62 are to a significant extent due to the ability of both proteins to induce inflammatory and innate immune signaling cascades in a TAK1-dependent manner.
Fig 7.
Inhibition of MLV by TRIM proteins correlates with their signaling activities. (A and B) Scatter plot analyses of fold inhibition in MLV released infectivity by TRIM proteins described previously (13) versus their NF-κB- and AP-1-inducing activities. The grey areas highlight TRIM proteins that inhibit MLV release in addition to inducing NF-κB/AP-1. (C) Friend MLV env-GFP released infectivity from HEK293 cells determined 48 h posttransfection using MLV permissive DFJ8 target cells in the presence of empty vector or the indicated TRIM protein expression construct either alone or by transfecting 5 and 10 ng of plasmids expressing dnIκB and/or dnFos, respectively. MLV infectivity released in the presence of vector was set to 100%. (D) Western blot analyses of sedimented virus from culture supernatants and HEK293 cell lysates for the experiment shown in panel C using antibodies to MLV gag and actin. (E and F) Amounts of MLV infectivity released from NT and TAK1 shRNA-expressing HEK293 cells transfected with 200 ng of plasmid encoding full-length MLV together with empty vector or the indicated amounts of TRIM-expressing plasmids. The released MLV infectivity in the presence of empty vector for cell lines expressing NT and TAK1 shRNAs was set to 100% in panel F. The error bars represent standard deviations for triplicate experiments. (G) Western blot analyses of sedimented virus from culture supernatants and HEK293 cell lysates for the experiment shown in panel F using antibodies to MLV gag p30, actin, GFP (for detecting TRIM1), TRIM15, and TRIM62. Statistical significance values: ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Resistance of HIV-1 to TRIM-induced signaling is due to the NF-κB binding sites in its LTR and can be transferred to MLV.
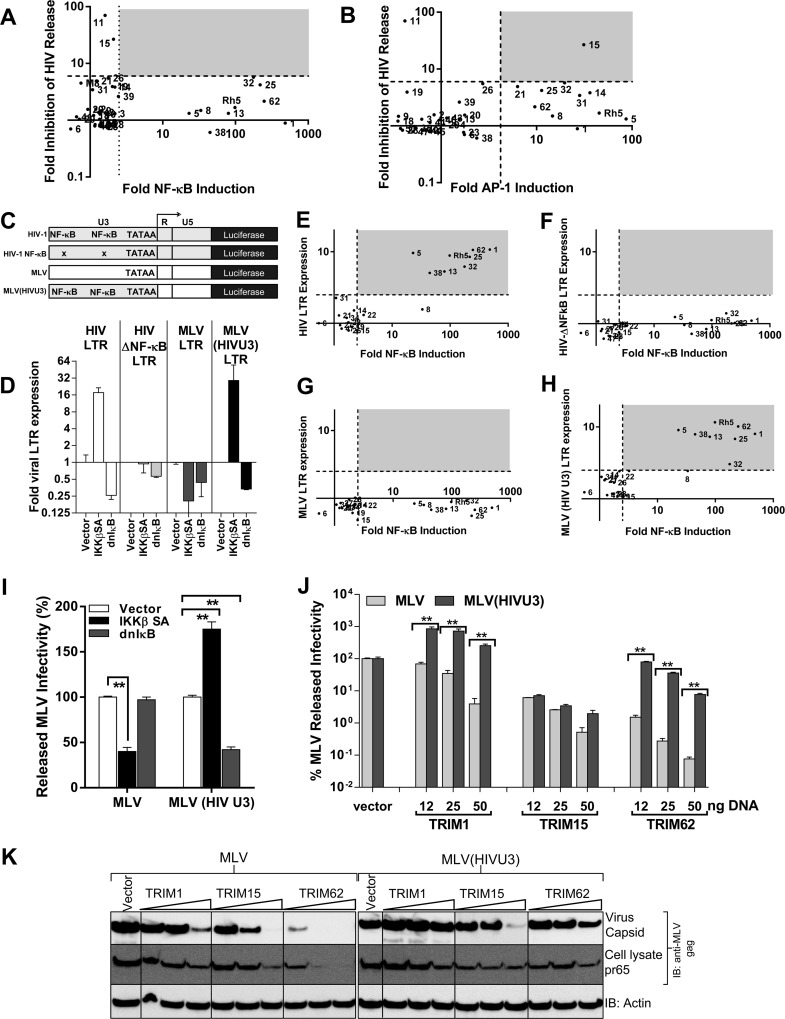
A scatter plot analysis of TRIM signaling activities against the previously observed inhibition in HIV released infectivity revealed that, in contrast to MLV, HIV-1 assembly and release were resistant to most of the NF-κB/AP-1-inducing TRIM proteins, except TRIM15 (Fig. 8A and B). In contrast to MLV, HIV-1 contains two NF-κB binding sites in the U3 promoter region of the LTR (39). To investigate the different effects of NF-κB signaling on HIV-1- and MLV LTR-driven expression, we used HIV-1 LTR with wild-type or mutant NF-κB sites and MLV LTR-driven luciferase constructs (Fig. 8C). We also generated an MLV LTR that carried the U3 promoter region of HIV-1 [MLV (HIV U3) LTR] (Fig. 8C). The luciferase expression from viral LTR constructs was analyzed when coexpressed with IKKβ SA and dnIκB. Expectedly, luciferase expression correlated strongly with the presence of NF-κB sites in the LTR (Fig. 8D). Importantly, MLV LTR activity was rendered responsive to NF-κB once it carried the U3 promoter of HIV-1 (Fig. 8D). We next tested the effect of TRIM protein expression on viral LTR-driven luciferase activity. There was good association between the NF-κB induction by TRIMs and enhanced expression from viral LTRs that contained the NF-κB binding sites (Fig. 8E to H). We next asked if the resistance of HIV-1 to TRIM-induced inflammatory signaling can be transferred to MLV in the context of full-length virus. In contrast to wt MLV, coexpression with IKKβ SA enhanced released infectivity of MLV (HIVU3) (Fig. 8I), suggesting that it was no longer sensitive to innate immune signaling. We next tested the effect of TRIM protein expression on MLV release. MLV and MLV (HIVU3) were equally susceptible to TRIM15-mediated inhibition. In contrast, expression of NF-κB-inducing TRIM1 enhanced MLV (HIVU3) released infectivity by 10-fold, making it completely resistant to TRIM1 inhibition compared to wt MLV (Fig. 8J and K). MLV (HIVU3) was also significantly resistant to the inhibitory effects of TRIM62 (Fig. 8J and K). Thus, as with HIV-1, we were able to render MLV resistant to TRIM-mediated inhibition by incorporating the HIV-1 U3 promoter. Therefore, taken together, our data demonstrated that the different vulnerabilities of MLV and HIV-1 to inflammatory TRIM proteins mapped to the U3 promoter of the viral LTRs.
Fig 8.
Incorporation of HIV-1 U3 promoter rescues MLV from TRIM signaling-induced inhibition. (A and B) Scatter plot analyses of fold inhibition in HIV released infectivity by TRIM proteins described previously (13) versus their NF-κB- and AP-1-inducing activities. The grey areas highlight TRIM proteins that inhibit HIV release in addition to inducing NF-κB/AP-1. (C) Schematic of viral luciferase constructs used in the study derived from HIV-1 LTR, HIV-1 LTR with mutant NF-κB-responsive sites (HIV ΔNF-κB), MLV LTR, and MLV LTR containing the U3 region of HIV-1 [MLV (HIV U3) LTR]. (D) Effect of NF-κB signaling on LTR-driven luciferase activity shown in panel A when cotransfected with empty vector or plasmid expressing IKKβ SA or IκB superrepressor (dnIκB), which results in activation or inhibition of NF-κB signaling, respectively. The error bars represent standard deviations in normalized luciferase values from triplicate experiments. (E to H) Viral LTR-driven luciferase activity was determined in the presence or absence of TRIM protein expression as for panel A. The data were plotted as scatter plots against TRIM-protein-induced NF-κB shown in FIG 1A. The dashed lines represent significance cutoff values (∼2.5 for NF-κB and viral LTRs) obtained by adding 2 standard deviations to the mean value obtained for empty-vector samples for each reporter construct from experiments carried out on separate days. The grey areas highlight TRIM proteins that induce luciferase expression from indicated viral LTRs in addition to inducing NF-κB/AP-1. (I and J) Amounts of viral infectivity released from HEK293 cells transfected with 200 ng of plasmids encoding either full-length MLV or MLV carrying HIV U3 [MLV (HIVU3] and cotransfected with either empty vector, plasmid expressing IKKβ SA, dnIκB (I), or the indicated amount of TRIM-expressing plasmids (J). The released infectivity for each virus in the presence of empty vector was set to 100%. The error bars represent standard deviations from triplicate experiments. (K) Western blot analyses of sedimented virus from culture supernatants and HEK293 cell lysates for the experiment shown in panel J using antibodies to MLV gag p30 and actin. Statistical significance values: **, P < 0.05.
DISCUSSION
In this study, we screened 43 human TRIM proteins for the ability to induce innate immune signaling pathways to identify 16 TRIM proteins that induce NF-κB and/or AP-1 signaling. The signaling properties of five TRIM proteins, TRIM1, -14, -15, -52, and -62, were not previously described. Thus, our work reinforces the emerging importance of TRIM proteins in innate immune signaling pathways (27). The observed signaling properties were not restricted to a single subgroup of TRIM proteins, as half (8 of 16) belonged to subgroup 4 containing PRY and SPRY domains at the variable C terminus (domain structure, RBCPS; R, RING; B, B box; C, coiled-coil; P, PRY; S, SPRY), while the remainder were distributed to other subgroups (Fig. 1C; see Table S1 in the supplemental material). Out of the 16 signaling, active TRIM proteins, 8 induced both NF-κB and AP-1, suggesting that they function upstream of the signaling bifurcation or activate different pathways. Consistently, the majority of TRIM proteins activated NF-κB and AP-1 in a TAK1-dependent manner, a major bifurcation point for many innate immune pathways. The few observed TAK1-independent activities, like the AP-1 signaling induced by TRIM25, may not be so surprising, given that TRIM25 also functions in the cell cycle by regulating levels of 14-3-3-σ (55). Similarly, the TAK1-independent AP-1 signaling activity seen for TRIM32 is likely due to its role in multiple cellular activities, such as cell differentiation, microRNA regulation, degradation of c-myc, and ubiquitination of STING and actin (6, 56, 57).
We studied TRIM62 in more detail and found that it induces NF-κB/AP-1 through the TRIF branch of the TLR4 signaling pathway. Stable knockdown of TAK1, TRAF6, and RIP1 diminished TRIM62-mediated NF-κB induction, suggesting that it acts upstream of these factors in the signaling cascade. TRIM62 most probably acts at the level of TRIF, as NF-κB induction by both proteins required each to be expressed. Future investigations are required to (i) determine the mechanism by which TRIM62 mediates its effect on TRIF-induced signaling; (ii) test if TRIM62 also functions in other TRIF-dependent pathways, such as TLR3; and (iii) identify molecular interactors and the E3 ligase substrate of TRIM62.
In addition to playing a role in innate immune signaling as documented here, missense mutations and homozygous deletion that abrogate TRIM62 expression have been associated with early onset of breast cancer (58). TRIM62, also known as DEAR1, is required for proper acinar morphogenesis in the mammary gland. Three-dimensional (3D) culture of breast epithelial cells with TRIM62 expression silenced did not form a bona fide lumen due to a failure in apoptosis exemplified by reduced caspase 3 activation (58). Interestingly, TLR activation and subsequent TRIF signaling can result in the induction of apoptosis (59, 60). TRIF recruits RIP1/3 via the RIP homotypic interaction motif (RHIM) and Fas/Apo-1-associated death domain protein (FADD) to activate caspase 8/3 (59). The RHIM present in the C terminus of TRIF is also required for induction of NF-κB (53). These data potentially link the role of TRIM62 in innate immune signaling with apoptotic signaling observed in cell differentiation and oncogenesis.
Our previous study had identified TRIM15 as a protein with antiviral activities against both MLV and HIV-1 (13). Importantly, silencing of TRIM15 expression in HeLa cells increased the assembly and release of both viruses, indicating that the endogenous protein contributed to the suppression (13). In the current study, we identify TRIM15 as a critical component of the RLR signaling pathway. Our analyses suggest that TRIM15 functions upstream or at the level of MAVS in the RLR sensing pathway. We have failed to detect interaction of TRIM15 with RIG-I, MDA5, or MAVS. Whether TRIM15 regulates these factors directly by transient interaction(s) or functions indirectly remains to be resolved. Interestingly, retroviral RNAs have been shown to be sensed by RIG-I (61, 62). It is therefore possible that TRIM15-mediated RIG-I sensing of retroviruses contributes to the establishment of an antiviral state.
The unifying feature of the deletion analyses for TRIM15 and TRIM62 were that both required the B box and coiled coil for signal transduction. B-box and coiled-coil domains have been observed to confer oligomerization properties on TRIM proteins (42, 63). Our data support the hypothesis that both TRIM15 and -62 may act as scaffolding proteins to recruit components of signal transduction cascades, similar to the role played by p62/sequestosome (64).
Our identification of inflammatory and innate immune signaling activities induced by multiple TRIM proteins raised the question of whether these activities contributed to the previously observed antiviral activities against MLV (13). Indeed, both activities correlated well, and we addressed this aspect using NF-κB-inducing TRIM 1 and -62 proteins. TRIM1 was previously shown to specifically restrict N-tropic MLV and not B-tropic MLV, a finding that was later confirmed by a number of laboratories, including ours (13, 17, 65). In addition to its capsid-specific restriction activity, our work showed that TRIM1 can also inhibit late stages of MLV replication, probably by inhibiting viral gene expression (13). Our current study demonstrates that TRIM1-induced signaling contributed to downregulation of MLV gene expression. Interference with TRIM1 signaling activities by dnIκB/dnFos and TAK1 gene silencing led to a significant rescue of MLV released infectivity by restoration of viral gene expression. In addition, there was complete rescue from TRIM1-mediated inhibition, together with enhancement of MLV gene expression and release, when the MLV U3 promoter region was replaced with that of HIV. These data suggest that the major mechanism for TRIM1-mediated reduction of MLV released infectivity is the inhibition of viral gene expression. TRIM62-mediated inhibition was partially rescued by the dominant-negative proteins and TAK1 gene silencing, suggesting that, in addition to signaling and subsequent reduction in MLV gene expression, another mechanism(s) also plays a role in virus inhibition. Interestingly, TAK1 silencing enhanced MLV released infectivity even in the absence of any TRIM protein expression, suggesting that MLV is sensed in human cells by inflammatory signaling that utilizes TAK1. Together, these results highlight the sensitivity of MLV to inflammatory and innate immune signaling. This likely explains why MLV does not induce an interferon response despite robust replication in mice (66). HIV-1 replication, on the other hand, induces a striking cascade of cytokines and chemokines, including interferons (67). As documented for simian immunodeficiency virus (SIV), an inflammatory response, especially early in the infection stage, likely also allows HIV-1 to establish itself, as bacterial infections expand the repertoire of founder HIV-1 strains in the host (68–70). In addition, there is chronic activation of the immune system due to translocation of microbial products, such as LPS, from the gut and HIV-1-activated interferon-producing plasmacytoid dendritic cells (pDCs) (71, 72). Interferon, though found to inhibit HIV-1 in vitro, is not necessarily protective in vivo in most individuals (73, 74). Overactivated interferon-producing pDCs inhibit antiviral T cell responses and contribute to T cell death and HIV-1 immunopathogenesis (75). Such chronic activation is a hallmark of progressive HIV-1 infection and has been shown to be a reliable predictor of disease outcome (76). Incorporation of the NF-κB sites in the LTR, in addition to the accessory proteins, certainly contributes to the ability of HIV-1 to replicate under potent inflammatory and innate immune responses. Thus, by screening the different effects of signaling TRIM proteins on MLV and HIV-1, we were able to revisit the fundamentally different replication and immune evasion strategies used by these two retroviruses.
Supplementary Material
ACKNOWLEDGMENTS
The research was supported by grants R21-AI087467 and R01-CA098727 from the National Institutes of Health to W.M.
We thank Xaver Sewald for help and guidance in culturing primary cells and Anasuya Chattopadhyay for help with VSV titration.
We declare that no competing interests exist.
Footnotes
Published ahead of print 17 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01804-12.
REFERENCES
- 1. Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Han K, Lou DI, Sawyer SL. 2011. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 7:e1002388 doi:10.1371/journal.pgen.1002388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Short KM, Cox TC. 2006. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem. 281:8970–8980 [DOI] [PubMed] [Google Scholar]
- 4. McNab FW, Rajsbaum R, Stoye JP, O'Garra A. 2011. Tripartite-motif proteins and innate immune regulation. Curr. Opin. Immunol. 23:46–56 [DOI] [PubMed] [Google Scholar]
- 5. Sato T, Okumura F, Kano S, Kondo T, Ariga T, Hatakeyama S. 2011. TRIM32 promotes neural differentiation through retinoic acid receptor-mediated transcription. J. Cell Sci. 124:3492–3502 [DOI] [PubMed] [Google Scholar]
- 6. Schwamborn JC, Berezikov E, Knoblich JA. 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136:913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carthagena L, Bergamaschi A, Luna JM, David A, Uchil PD, Margottin-Goguet F, Mothes W, Hazan U, Transy C, Pancino G, Nisole S. 2009. Human TRIM gene expression in response to interferons. PLoS One 4:e4894 doi:10.1371/journal.pone.0004894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. El Bougrini J, Dianoux L, Chelbi-Alix MK. 2011. PML positively regulates interferon gamma signaling. Biochimie 93:389–398 [DOI] [PubMed] [Google Scholar]
- 9. Everett RD, Chelbi-Alix MK. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819–830 [DOI] [PubMed] [Google Scholar]
- 10. Nisole S, Stoye JP, Saib A. 2005. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3:799–808 [DOI] [PubMed] [Google Scholar]
- 11. Rajsbaum R, Stoye JP, O'Garra A. 2008. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur. J. Immunol. 38:619–630 [DOI] [PubMed] [Google Scholar]
- 12. Sayah DM, Sokolskaja E, Berthoux L, Luban J. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569–57315243629 [Google Scholar]
- 13. Uchil PD, Quinlan BD, Chan WT, Luna JM, Mothes W. 2008. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 4:e16 doi:10.1371/journal.ppat.0040016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hatziioannou T, Cowan S, Goff SP, Bieniasz PD, Towers GJ. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee K, KewalRamani VN. 2004. In defense of the cell: TRIM5alpha interception of mammalian retroviruses. Proc. Natl. Acad. Sci. U. S. A. 101:10496–10497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 [DOI] [PubMed] [Google Scholar]
- 17. Yap MW, Nisole S, Lynch C, Stoye JP. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. U. S. A. 101:10786–10791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geoffroy MC, Chelbi-Alix MK. 2011. Role of promyelocytic leukemia protein in host antiviral defense. J. Interferon Cytokine Res. 31:145–158 [DOI] [PubMed] [Google Scholar]
- 19. Barr SD, Smiley JR, Bushman FD. 2008. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 4:e1000007 doi:10.1371/journal.ppat.1000007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao B, Duan Z, Xu W, Xiong S. 2009. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology 50:424–433 [DOI] [PubMed] [Google Scholar]
- 21. Hattlmann C. J., Kelly J. N., Barr S. D. 2012. TRIM22: A diverse and dynamic antiviral protein. Mol. Biol. Int. 2012:153415 doi:10.1155/2012/153415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kajaste-Rudnitski A, Marelli SS, Pultrone C, Pertel T, Uchil PD, Mechti N, Mothes W, Poli G, Luban J, Vicenzi E. 2011. TRIM22 inhibits HIV-1 transcription independently of its E3 ubiquitin ligase activity, Tat, and NF-kappaB-responsive long terminal repeat elements. J. Virol. 85:5183–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolf D, Goff SP. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131:46–57 [DOI] [PubMed] [Google Scholar]
- 24. Keeble AH, Khan Z, Forster A, James LC. 2008. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc. Natl. Acad. Sci. U. S. A. 105:6045–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Taylor RT, Lubick KJ, Robertson SJ, Broughton JP, Bloom ME, Bresnahan WA, Best SM. 2011. TRIM79alpha, an interferon-stimulated gene product, restricts tick-borne encephalitis virus replication by degrading the viral RNA polymerase. Cell Host Microbe 10:185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang J, Liu B, Wang N, Lee YM, Liu C, Li K. 2011. TRIM56 is a virus- and interferon-inducible E3 ubiquitin ligase that restricts pestivirus infection. J. Virol. 85:3733–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kawai T, Akira S. 2011. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol. Med. 3:513–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ozato K, Shin DM, Chang TH, Morse HC., III 2008. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8:849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, Wu X, Tao Z, Li Z, Cai X, Sun S, Xiang C, Sun B. 2008. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat. Immunol. 9:369–377 [DOI] [PubMed] [Google Scholar]
- 30. Li Q, Yan J, Mao AP, Li C, Ran Y, Shu HB, Wang YY. 2011. Tripartite motif 8 (TRIM8) modulates TNFalpha- and IL-1beta-triggered NF-kappaB activation by targeting TAK1 for K63-linked polyubiquitination. Proc. Natl. Acad. Sci. U. S. A. 108:19341–19346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De Castillia C, Mothes W, Pizzato M, Grutter MG, Luban J. 2011. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472:361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tareen SU, Emerman M. 2011. Human Trim5alpha has additional activities that are uncoupled from retroviral capsid recognition. Virology 409:113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oke V, Wahren-Herlenius M. 2012. The immunobiology of Ro52 (TRIM21) in autoimmunity: a critical review. J. Autoimmun. 39:77–82 [DOI] [PubMed] [Google Scholar]
- 34. Zha J, Han KJ, Xu LG, He W, Zhou Q, Chen D, Zhai Z, Shu HB. 2006. The Ret finger protein inhibits signaling mediated by the noncanonical and canonical IkappaB kinase family members. J. Immunol. 176:1072–1080 [DOI] [PubMed] [Google Scholar]
- 35. Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920 [DOI] [PubMed] [Google Scholar]
- 36. Arimoto K, Funami K, Saeki Y, Tanaka K, Okawa K, Takeuchi O, Akira S, Murakami Y, Shimotohno K. 2010. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc. Natl. Acad. Sci. U. S. A. 107:15856–15861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Poole E, Groves I, MacDonald A, Pang Y, Alcami A, Sinclair J. 2009. Identification of TRIM23 as a cofactor involved in the regulation of NF-kappaB by human cytomegalovirus. J. Virol. 83:3581–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. 2010. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33:765–776 [DOI] [PubMed] [Google Scholar]
- 39. Nabel G, Baltimore D. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711–713 [DOI] [PubMed] [Google Scholar]
- 40. Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, Lin HK. 2009. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325:1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. 1997. IKK-1 and IKK-2: Cytokine-activated IκB kinases essential for NF-κB activation. Science 278:860–866 [DOI] [PubMed] [Google Scholar]
- 42. Diaz-Griffero F, Qin XR, Hayashi F, Kigawa T, Finzi A, Sarnak Z, Lienlaf M, Yokoyama S, Sodroski J. 2009. A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 83:10737–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Basak S, Kim H, Kearns JD, Tergaonkar V, O'Dea E, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, Hoffmann A. 2007. A fourth IkappaB protein within the NF-kappaB signaling module. Cell 128:369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Monaco C, Andreakos E, Kiriakidis S, Mauri C, Bicknell C, Foxwell B, Cheshire N, Paleolog E, Feldmann M. 2004. Canonical pathway of nuclear factor kappa B activation selectively regulates proinflammatory and prothrombotic responses in human atherosclerosis. Proc. Natl. Acad. Sci. U. S. A. 101:5634–5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang CY, Mayo MW, Baldwin AS., Jr 1996. TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science 274:784–787 [DOI] [PubMed] [Google Scholar]
- 46. Wick M, Lucibello FC, Muller R. 1992. Inhibition of Fos- and Ras-induced transformation by mutant Fos proteins with structural alterations in functionally different domains. Oncogene 7:859–867 [PubMed] [Google Scholar]
- 47. Wertz I. E., Dixit V. M. 2010. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb. Perspect. Biol. 2:a003350 doi:10.1101/cshperspect.a003350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, Akira S, Kitamura T, Kosugi A, Kimoto M, Miyake K. 2002. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat. Immunol. 3:667–672 [DOI] [PubMed] [Google Scholar]
- 49. Horng T, Barton GM, Flavell RA, Medzhitov R. 2002. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature 420:329–333 [DOI] [PubMed] [Google Scholar]
- 50. Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. 2008. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 9:361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takeda K, Akira S. 2004. TLR signaling pathways. Semin. Immunol. 16:3–9 [DOI] [PubMed] [Google Scholar]
- 52. Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. 2003. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301:640–643 [DOI] [PubMed] [Google Scholar]
- 53. Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11:373–384 [DOI] [PubMed] [Google Scholar]
- 54. Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, Akira S. 2003. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171:4304–4310 [DOI] [PubMed] [Google Scholar]
- 55. Urano T, Saito T, Tsukui T, Fujita M, Hosoi T, Muramatsu M, Ouchi Y, Inoue S. 2002. Efp targets 14-3-3[sigma] for proteolysis and promotes breast tumour growth. Nature 417:871–875 [DOI] [PubMed] [Google Scholar]
- 56. Kano S, Miyajima N, Fukuda S, Hatakeyama S. 2008. Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 68:5572–5580 [DOI] [PubMed] [Google Scholar]
- 57. Zhang J, Hu MM, Wang YY, Shu HB. 2012. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 287:28646–28655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lott ST, Chen N, Chandler DS, Yang Q, Wang L, Rodriguez M, Xie H, Balasenthil S, Buchholz TA, Sahin AA, Chaung K, Zhang B, Olufemi SE, Chen J, Adams H, Band V, El-Naggar AK, Frazier ML, Keyomarsi K, Hunt KK, Sen S, Haffty B, Hewitt SM, Krahe R, Killary AM. 2009. DEAR1 is a dominant regulator of acinar morphogenesis and an independent predictor of local recurrence-free survival in early-onset breast cancer. PLoS Med. 6:e1000068 doi:10.1371/journal.pmed.1000068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Han KJ, Su X, Xu LG, Bin LH, Zhang J, Shu HB. 2004. Mechanisms of the TRIF-induced interferon-stimulated response element and NF-kappaB activation and apoptosis pathways. J. Biol. Chem. 279:15652–15661 [DOI] [PubMed] [Google Scholar]
- 60. Hasan UA, Caux C, Perrot I, Doffin AC, Menetrier-Caux C, Trinchieri G, Tommasino M, Vlach J. 2007. Cell proliferation and survival induced by Toll-like receptors is antagonized by type I IFNs. Proc. Natl. Acad. Sci. U. S. A. 104:8047–8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Berg RK, Melchjorsen J, Rintahaka J, Diget E, Soby S, Horan KA, Gorelick RJ, Matikainen S, Larsen CS, Ostergaard L, Paludan SR, Mogensen TH. 2012. Genomic HIV RNA induces innate immune responses through RIG-I-dependent sensing of secondary-structured RNA. PLoS One 7:e29291 doi:10.1371/journal.pone.0029291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Solis M, Nakhaei P, Jalalirad M, Lacoste J, Douville R, Arguello M, Zhao T, Laughrea M, Wainberg MA, Hiscott J. 2011. RIG-I-mediated antiviral signaling is inhibited in HIV-1 infection by a protease-mediated sequestration of RIG-I. J. Virol. 85:1224–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. 2011. Hexagonal assembly of a restricting TRIM5alpha protein. Proc. Natl. Acad. Sci. U. S. A. 108:534–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moscat J, Diaz-Meco MT, Wooten MW. 2007. Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 32:95–100 [DOI] [PubMed] [Google Scholar]
- 65. Zhang F, Hatziioannou T, Perez-Caballero D, Derse D, Bieniasz PD. 2006. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology 353:396–409 [DOI] [PubMed] [Google Scholar]
- 66. Liberatore RA, Bieniasz PD. 2011. Tetherin is a key effector of the antiretroviral activity of type I interferon in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 108:18097–18101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stacey AR, Norris PJ, Qin L, Haygreen EA, Taylor E, Heitman J, Lebedeva M, DeCamp A, Li D, Grove D, Self SG, Borrow P. 2009. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J. Virol. 83:3719–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Galvin SR, Cohen MS. 2004. The role of sexually transmitted diseases in HIV transmission. Nat. Rev. Microbiol. 2:33–42 [DOI] [PubMed] [Google Scholar]
- 69. Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, Karita E, Manigart O, Mulenga J, Keele BF, Shaw GM, Hahn BH, Allen SA, Derdeyn CA, Hunter E. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 5:e1000274 doi:10.1371/journal.ppat.1000274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li Q, Estes JD, Schlievert PM, Duan L, Brosnahan AJ, Southern PJ, Reilly CS, Peterson ML, Schultz-Darken N, Brunner KG, Nephew KR, Pambuccian S, Lifson JD, Carlis JV, Haase AT. 2009. Glycerol monolaurate prevents mucosal SIV transmission. Nature 458:1034–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12:1365–1371 [DOI] [PubMed] [Google Scholar]
- 72. O'Brien M, Manches O, Sabado RL, Baranda SJ, Wang Y, Marie I, Rolnitzky L, Markowitz M, Margolis DM, Levy D, Bhardwaj N. 2011. Spatiotemporal trafficking of HIV in human plasmacytoid dendritic cells defines a persistently IFN-alpha-producing and partially matured phenotype. J. Clin. Invest. 121:1088–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Goujon C, Malim MH. 2010. Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells. J. Virol. 84:9254–9266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Herbeuval JP, Shearer GM. 2007. HIV-1 immunopathogenesis: how good interferon turns bad. Clin. Immunol. 123:121–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Boasso A, Royle CM, Doumazos S, Aquino VN, Biasin M, Piacentini L, Tavano B, Fuchs D, Mazzotta F, Lo Caputo S, Shearer GM, Clerici M, Graham DR. 2011. Overactivation of plasmacytoid dendritic cells inhibits antiviral T-cell responses: a model for HIV immunopathogenesis. Blood 118:5152–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, Shih R, Lewis J, Wiley DJ, Phair JP, Wolinsky SM, Detels R. 1999. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J. Infect. Dis. 179:859–870 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.