Abstract
G88R emerged as a compensatory mutation in matrix protein 1 (M1) of influenza virus A/WSN/33 when its nuclear localization signal (NLS) was disrupted by R101S and R105S substitutions. The resultant M1 triple mutant M(NLS-88R) regained replication efficiency in vitro while remaining attenuated in vivo with the potential of being a live vaccine candidate. To understand why G88R was favored by the virus as a compensatory change for the NLS loss and resultant replication deficiency, three more M1 triple mutants with an alternative G88K, G88V, or G88E change in addition to R101S and R105S substitutions in the NLS were generated. Unlike the other M1 triple mutants, M(NLS-88R) replicated more efficiently in vitro and in vivo. The G88R compensatory mutation not only restored normal functions of M1 in the presence of a disrupted NLS but also resulted in a strong association of M1 with viral ribonucleoprotein. Under a transmission electron microscope, only the M1 layer of the M(NLS-88R) virion exhibited discontinuous fingerprint-like patterns with average thicknesses close to that of wild-type A/WSN/33. Computational modeling suggested that the compensatory G88R change could reestablish the integrity of the M1 layer through new salt bridges between adjacent M1 subunits when the original interactions were interrupted by simultaneous R101S and R105S replacements in the NLS. Our results suggested that restoring the normal functions of M1 was crucial for efficient virus replication.
INTRODUCTION
The viral ribonucleoprotein (vRNP) core of influenza A viruses consists of viral RNA (vRNA), nucleoprotein (NP), and RNA polymerases, which are surrounded by matrix protein 1 (M1) (1, 2). As the major structural component, M1 lies beneath the lipid bilayer of viral particles, forming a shell to connect vRNP with the membrane proteins hemagglutinin (HA), neuraminidase (NA), and matrix protein 2 (3, 4). Following infection, M1 is dissociated from vRNP to allow the latter to enter into the host nucleus for vRNA synthesis. Later, M1 also assists the nuclear export of newly formed vRNP and transports it to the cellular membrane for virion assembly and budding (5–7). Thus, M1 possesses multiple functions in support of virus replication cycles.
The multibasic motif 101RKLKR105 of M1 is a nuclear localization signal (NLS) and is involved in binding of vRNA and vRNP (3, 8). Disrupting 101RKLKR105 with R101S and R105S was reported to affect virus replication negatively and render the virus sensitive to high temperatures (9, 10). However, this R101S-R105S double mutant in an A/WSN/33 (WSN, H1N1) background was unstable and the phenotypes changed after two passages in eggs (11). During our process of regenerating an R101S-R105S mutant, a third mutation with Arg substituted for a Gly at amino acid (aa) position 88 emerged in addition to R101S and R105S in M1. In fact, G88R is also a spontaneous mutation that naturally occurs in M1 of wild-type WSN (wt-WSN) independent of other changes, which by itself has no negative effect on virus replication (unpublished observation). We thus intentionally introduced G88R along with R101S and R105S into wt-WSN M1. The resultant M1 triple mutant, M(NLS-88R), was found not only to replicate efficiently in vitro but also to be attenuated in mice exhibiting high vaccine potential with broad cross-protection against influenza infections (11).
The highly promising nature of M(NLS-88R) invokes the fundamental question of why G88R was favored by the virus as a compensatory change for the NLS loss and resultant replication deficiency (9–11). A structural model based on partially crystallized M1 (aa 1 to 164) indicates that M1 monomers interact with each other through charge-charge interactions (1). Hence, we hypothesized that a positive charge at position 88 was required to restore the normal functions of M1 when the NLS was disrupted. We thus generated three more M1 triple mutants in the wt-WSN background with an alternative G88K, G88E, or G88V change in addition to R101S and R105S in the 101RKLKR105 motif to prove this hypothesis.
MATERIALS AND METHODS
Viruses.
Unless stated otherwise, all of the plasmids expressing 8 gene segments of wt-WSN were in the pHW2000 backbone (9, 10). The mutation (i) G88R, (ii) G88K, (iii) G88E, or (iv) G88V, in addition to R101S and R105S, was introduced into the plasmid expressing the full-length M cDNA of wt-WSN using a QuikChange II site-directed mutagenesis kit (Agilent Technologies). Reverse genetics was performed in cocultured 293T and MDCK cells as described previously (11, 12). The rescued M1 triple mutants with different substitutions at position 88 were designated M(NLS-88R), M(NLS-88K), M(NLS-88E), and M(NLS-88V), respectively. All recombinant viruses, including reverse genetic-generated wt-WSN (11) and mutant recombinant WSN (mu-rWSN) (13), were propagated twice in 9-day-old embryonated eggs at 33°C, and aliquots were stored at −70°C until use. All of the mutants were confirmed by full-length DNA sequencing (CBER/FDA core facility) and were titrated on MDCK cells by plaque assay (14).
In vitro replication.
MDCK cells were infected with M1 triple mutants (multiplicity of infection [MOI] of 0.002) at 33°C, 5% CO2 for up to 72 h. Infectious particles sampled at different time points were titrated by plaque assay. The temperature sensitivity (ts) was determined by comparing virus replication in MDCK cells at 33°C to that at 39°C.
Confocal microscopy.
Infected MDCK cells were fixed and permeabilized before being probed with biotin-conjugated rabbit anti-M1 and mouse anti-NP antibodies (Abcam). NorthernLights streptavidin NL557 and NorthernLights 493 fluorochrome-labeled donkey anti-mouse IgG (R&D Systems) were used as the secondary antibodies. Nuclei were counterstained by 4′,6-diamidino-2-phenylindole (DAPI). Images were acquired under an LSM 510 confocal laser scanning microscope (Zeiss).
M1-NP peptide binding assay.
The following M1 peptides were synthesized with reverse-phase high-performance liquid chromatography (RP-HPLC) purification (CBER/FDA core facility): wt 88G-101R-105R, 87NGDPNNMDKAVKLYRKLKRE106; 88G-101S-105S, 87NGDPNNMDKAVKLYSKLKSE106; 88R-101S-105S, 87NRDPNNMDKAVKLYSKLKSE106; 88K-101S-105S, 87NKDPNNMDKAVKLYSKLKSE106; 88E-101S-105S, 87NEDPNNMDKAVKLYSKLKSE106; and 88V-101S-105S, 87NVDPNNMDKAVKLYSKLKSE106, where the underlining indicates amino acids different from the wt. All of the peptides contained a 6C linker with biotinylation at the N terminus. Total vRNA was extracted from wt-WSN using a QIAamp viral RNA minikit (Qiagen). Immulon 2HB 96-well enzyme-linked immunosorbent assay (ELISA) plates were coated with 10 μg/ml of seasonal NP peptide (ProSci Incorporated) in phosphate-buffered saline (PBS) (pH 7.2) at 4°C overnight. M1 peptides were 2-fold diluted in PBS (pH 7.2) and were added into NP-precoated plates with or without freshly purified vRNA (100 to 120 ng/ml). The plates were incubated at 37°C for 2 to 4 h, followed by color development using streptavidin-horseradish peroxidase (HRP). The plates were read at 450 nm on a Victor V multilabel plate reader (PerkinElmer).
M1-vRNP dissociation.
M1/vRNP complexes were obtained by incubating viruses in disruption buffer (10 mM Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.05 M NaCl, and 1.25 mM dithiothreitol) with or without 0.5 M NaCl at room temperature for 40 min (9). The disrupted protein complexes were centrifuged through a 20% sucrose cushion at 10,000 × g for 30 min. Purified pellets were subjected to electrophoresis under nonreducing conditions. The M1/vRNP ratios were estimated by densitometric determination of M1/NP ratios relative to the M1/NP ratios of intact wt-WSN virion using a Gel Doc-It system (UVP LLC).
Cellular membrane binding assay.
Cellular membrane-associated M1 was assessed as described previously, with minor modifications (15). Briefly, infected MDCK cells (MOI, 2) were dissociated in hypotonic buffer containing 1 mM Tris-HCl (pH 7.4) and 0.1 mM MgCl2 on ice for 60 min. After being passed through a 20-gauge needle at least 20 times, cell homogenates were centrifuged at 1,000 × g at 4°C for 10 min. The postnuclear supernatants were subjected to ultracentrifugation at 100,000 × g at 4°C for another 60 min. The pellets containing the membranes were collected and analyzed by Western blotting directly. The supernatants (cytosol) after ultracentrifugation were also collected, and proteins were precipitated with ice-cold acetone before Western blotting. The sheep anti-HA of A/Puerto Rico/8/34, biotin-conjugated rabbit anti-M1 (Abcam), and mouse anti-β-actin antibody (Santa Cruz) were used for probing, followed by IRDye-680LT-labeled donkey anti-sheep, IRDye-680LT-labeled streptavidin, and IRDye-800CW-labeled donkey anti-mouse (LI-COR). The blots were imaged and analyzed using an Odyssey imaging system (LI-COR).
TEM.
Egg-amplified viruses were subjected to ultracentrifugation (30,000 rpm for 90 min, 4°C), followed by a discontinuous 15 to 30 to 60% sucrose gradient purification (16). Purified viruses were fixed with 2% paraformaldehyde and 2% glutaraldehyde in PBS at room temperature overnight, followed by postfixation with 1% osmium tetroxide for another 1 h. After dehydration and infiltration, fixed viruses were embedded in epoxy resin. Ultrathin sections were stained with uranyl acetate and lead citrate and were examined under a Zeiss EM 912 transmission electron microscope (TEM) equipped with a Keenview digital camera (Olympus). The thickness of the M1 layer was measured clockwise at 3, 6, 9, and 12 o'clock and was averaged for each virion. Ten representative virions of each virus were measured.
Animal study.
Four- to 6-week-old specific-pathogen-free female BALB/c mice (Taconic Farm) were infected intranasally under light isoflurane anesthesia. Lungs harvested on day 3 postinfection (p.i.) were homogenized and titrated by plaque assay. HA inhibition (HAI) titers of mouse sera collected at 4 weeks p.i. were determined using 0.5% turkey red blood cells (11). The animal study was approved by the IACUC of CBER/FDA.
Statistical analysis.
One-way analysis of variance (ANOVA) was performed to determine significant differences (P < 0.05) between treatments/groups.
Computational modeling of M1.
The dimeric crystal structure of influenza A M1 (residues 1 to 164) at neutral pH was retrieved from the Protein Data Bank (PDB; code 1EA3) (1) and used to model the outer M1 layer of the virion. A tetramer that involved expansion of the M1 dimer in the x direction was created using ZDOCK (http://zlab.umassmed.edu/zdock/software/). PROCHECK (http://www.doe-mbi.ucla.edu/Software/PROCHECK.html) was used to assess the stereochemical fitness of the protein backbone. Unless otherwise noted, all molecular modeling tasks were performed using SYBYL 8.1 (Tripos Inc.).
RESULTS
In vitro and in vivo characterization.
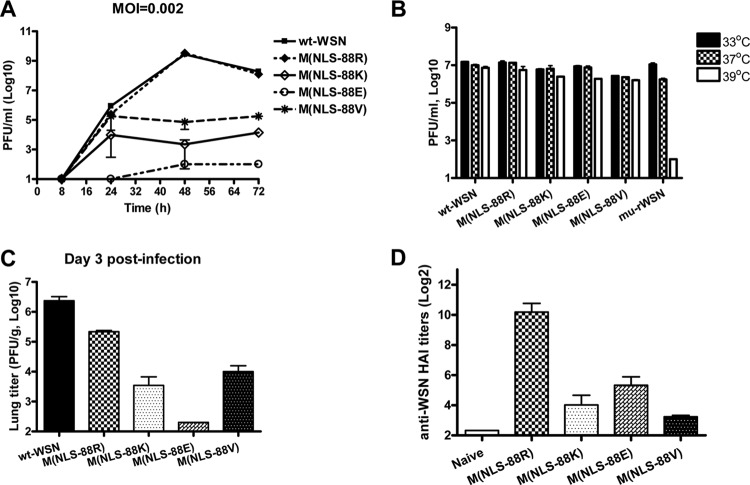
Among all of the triple mutants, M(NLS-88R) replicated most efficiently in MDCK cells, at a rate similar to that of wt-WSN (Fig. 1A). M(NLS-88V) was the second most efficient, followed by M(NLS-88K) and M(NLS-88E) (Fig. 1A). Unlike a ts mu-rWSN showing ≥2 log reduction in viral titers when temperature rose from 33 to 39°C, all four M1 triple mutants replicated equally efficiently at both temperatures (Fig. 1B), indicating none of them was ts.
Fig 1.
In vitro and in vivo characterization of M1 triple mutants. The replication kinetics (MOI, 0.002) (A) and temperature-sensitive (ts) phenotype (B) were determined in MDCK cells by plaque assay. A cold-adapted A/WSN/34 mutant (mu-rWSN) was used as a ts control. BALB/c mice were inoculated i.n. with wild-type A/WSN/33 (wt-WSN) or M1 triple mutants at 5 × 104 PFU/50 μl/mouse. (C) Lung virus titers (n = 3 mice/group) were determined by plaque assay on day 3 postinfection (p.i.). (D) wt-WSN-specific serum HA inhibition (HAI) titers were determined at 4 weeks p.i. (n = 8 mice/group). Because all wt-WSN-infected mice died within 2 weeks of infection, no HAI titers were available for this group.
Similar to in vitro replication, M(NLS-88R) replicated more efficiently than the other triple mutants in mouse lungs, and M(NLS-88E) replicated least efficiently (Fig. 1C). Unlike wt-WSN, which was pathogenic in mice, all M1 triple mutants were attenuated in vivo with no mortality observed at a dose of up to 106 PFU/mouse (data not shown). Compared to the other triple mutants, M(NLS-88R) elicited a stronger HAI response with a geometric mean titer (GMT) of 698 against wt-WSN virus at 4 weeks p.i. (Fig. 1D). In contrast, none of the groups infected with M(NLS-88K), M(NLS-88E), or M(NLS-88V) had an HAI GMT of >20 against wt-WSN (Fig. 1D).
M1-NP and M1-vRNP interactions.
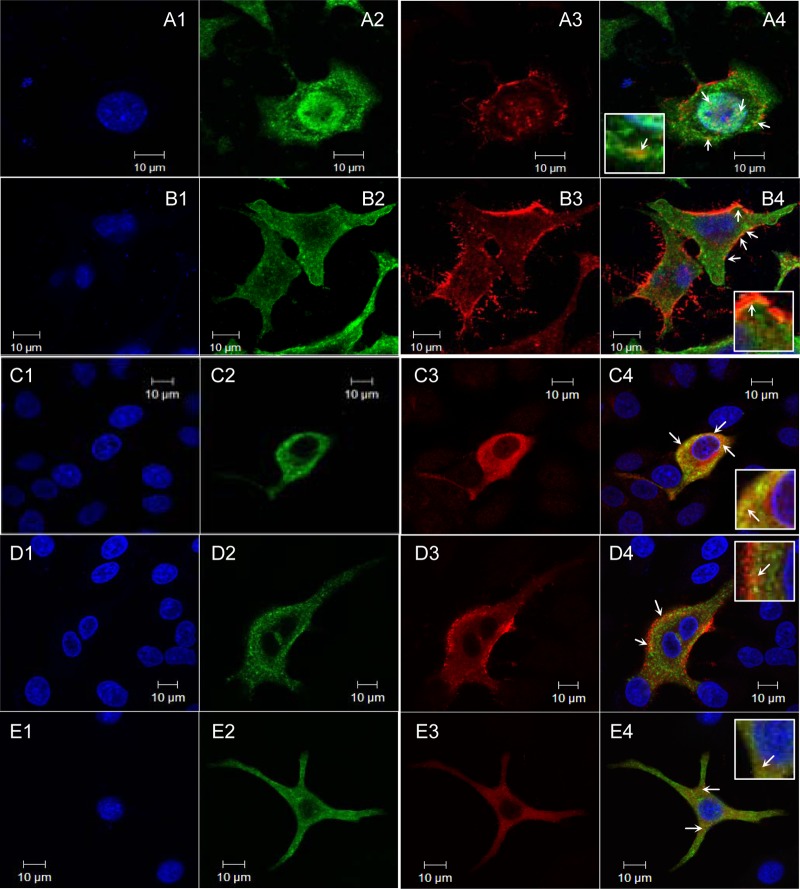
In infected cells, newly synthesized wt-WSN M1 was observed in the cytoplasm and nucleus as well as near the cellular membrane (Fig. 2A3). Unlike M1 of R101S-R105S, the distribution of which was limited to the cytoplasm (9), newly synthesized M1 of M(NLS-88R) and M(NLS-88K) but not M(NLS-88E) or M(NLS-88V) was also observed in the nuclei, although the nuclear-localized M1 appeared to be less obvious in M(NLS-88K)-infected cells (Fig. 2B3 to E3). Among all four triple mutants, the M1 cellular distribution pattern of M(NLS-88R) most resembled that of wt-WSN (Fig. 2A3 and B3). Unlike M1, there appeared to be no big differences in the cellular distribution of NP among wt-WSN and the M1 triple mutants (Fig. 2A2, B2, C2, D2, E2, and data not shown). In wt-WSN-infected cells, colocalized M1 and NP were observed in the cytoplasm and nucleus and near the membrane (Fig. 2A4). Infection with M(NLS-88R) or M(NLS-88K) also resulted in strong M1-NP colocalization, as indicated by bright orange to yellow color along the cellular membrane or in the cytoplasm (Fig. 2B4 and C4). In contrast, the M1 and NP of M(NLS-88E) or M(NLS-88V) showed a weak colocalization in infected cells (Fig. 2D4 and E4).
Fig 2.
M1-NP colocalization. Following 20 h of infection (MOI, 2), MDCK cells were fixed and permeabilized before being probed with biotin-conjugated rabbit anti-M1 (red) and mouse anti-NP (green) monoclonal antibodies followed by NorthernLights streptavidin NL557 and NorthernLights 493 fluorochrome-labeled donkey anti-mouse IgG. Nucleus was counterstained by DAPI (blue). Images were acquired with a 63×, 1.4-numeric-aperture oil differential interference contrast objective lens under an LSM 510 confocal laser scanning microscope. (A1 to A4) wt-WSN; (B1 to B4) M(NLS-88R); (C1 to C4) M(NLS-88K); (D1 to D4) M(NLS-88E); (E1 to E4) M(NLS-88V). M1-NP colocalization (yellow or orange spots) is indicated by a white arrow.
Fig 3.
M1-NP peptide binding and M1-vRNP complex association. Biotinylated M1 peptides were 2-fold diluted and were added into NP peptide-precoated plates with or without freshly purified vRNA. Each M1 peptide concentration was tested in duplicate. The optical density readings at 450 nm were plotted against corresponding M1 peptide concentrations. The average results from the three representative experiments are shown. (A) wt 88G-101R-105R; (B) 88G-101S-105S; (C) 88R-101S-105S; (D) 88K-101S-105S; (E) 88E-101S-105S; and (F) 88V-101S-105S. Purified M1/vRNP complexes were incubated in the disruption buffer with or without 0.5 M NaCl at room temperature for 40 min. The disrupted protein complexes were centrifuged through a 20% sucrose cushion at 10,000 × g for 30 min. The pellets were subjected to electrophoresis in 10% NuPAGE Bis-Tris minigels under nonreducing conditions. The M1/vRNP ratios were estimated by densitometric determination of M1/NP ratios relative to M1/NP ratios of intact wt-WSN virion. (G) M1/vRNP ratios under different conditions from 3 to 4 independent experiments. wt, wt-WSN; 88R, M(NLS-88R); 88K, M(NLS-88K); 88E, M(NLS-88E); 88V, M(NLS-88V). *, P < 0.05 by one-way ANOVA.
Fig 4.
Transmission electron images of M1 triple mutants. The epoxy resin-embedded M1 triple mutants were cut by an ultramicrotome, and ultrathin sections were stained with uranyl acetate and lead citrate. Images were acquired under a Zeiss EM 912 transmission electron microscope equipped with a Keenview digital camera. The inset in each transmission electron image shows an enlarged portion of the M1 layer. White arrows indicate the lipid bilayers. The inner part between two black arrows is the M1 layer. (A) wt-WSN (wt); (B) M(NLS-88R) (88R); (C) M(NLS-88K) (88K); (D) M(NLS-88E) (88E); (F) M(NLS-88V) (88V).
We next simulated M1-NP binding in ELISA. As shown in Fig. 3A, wt 88G-101R-105R peptide showed a dose-dependent binding to immobilized NP peptide with and without vRNA. Substitution of Arg for Ser at positions 101 and 105 dramatically impaired the resultant M1 peptide binding to immobilized NP peptide (Fig. 3B), suggesting that the 101RKLKR105 motif is important in direct M1-NP binding. The presence of vRNA did not improve the binding of 88G-101S-105S to NP significantly (Fig. 3B), further indicating that the 101RKLKR105 motif is involved in direct M1-NP binding. Additional G88R (Fig. 3C) or G88K (Fig. 3D) but not G88E (Fig. 3E) or G88V (Fig. 3F) alteration improved the overall binding of 101S-105S to immobilized NP peptide in a concentration-dependent manner, suggesting that a positively charged amino acid at position 88 is beneficial for direct M1-NP binding after the NLS was partially depolarized. Interestingly, the binding of 88R-101S-105S but not 88K-101S-105S to immobilized NP peptide was obviously improved in the presence of vRNA (Fig. 3C and D), implying that a positive charge at position 88 is not essential for vRNP binding after the partial charge loss of NLS.
However, this M1-NP binding ELISA is not a functional assay, and the demonstrated physical interactions between M1 and NP peptides may not reflect the real scenario in assembled virions. As shown in Fig. 3G, the intact viral particles of all M1 mutants had M1/vRNP ratios similar to that of wt-WSN M1 particles. Detergent disruption had no obvious impact on the M1-vRNP association (Fig. 3G). Under high-salt conditions, however, all M1 triple mutants except M(NLS-88R) had significantly less M1 remaining in the M1/vRNP complexes than those isolated from intact viral particles (Fig. 3G), indicating a weak association between M1 and vRNP in these mutants.
TEM of purified virions.
The mature virions of all M1 triple mutants had HA spikes well arranged on the surface with morphology indistinguishable from that of wt-WSN (Fig. 4A to E). Under TEM, the M1 layer was seen as a dark line (the inner part between two black arrows) lying immediately underneath the lipid bilayer (the white line, as indicated by white arrows) (Fig. 4A to E). In mature virions of wt-WSN and the M(NLS-88R) mutant, M1 appeared as a bilayer arranged in well-organized, discontinuous fingerprint-like pattern (Fig. 4A and B) (17, 18). The M1 thickness of wt-WSN or M(NLS-88R) virions was relatively uniform, with an average value of 13 and 11 nm, respectively (Table 1). In contrast, M1 from the other triple mutants was displayed as a disrupted single layer with uneven thickness that ranged from 4 to 9 nm on average, respectively (Fig. 4C to E and Table 1). The statistical analysis also showed that the M1 layer from M(NLS-88K), M(NLS-88E), or M(NLS-88V) was significantly thinner than that of wt-WSN and/or M(NLS-88R) (Table 1).
Table 1.
Measurement of the M1 layer in mature virionsf
| Wild type or mutant | Thickness of M1 layer (nm) of virionc: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Avgb | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| wt-WSNa | 13 | 10 (9–11) | 14 (11–16) | 13 (11–17) | 12 (10–16) | 16 (14–18) | 18 (13–22) | 18 (12–22) | 11 (9–12) | 12 (9–12) | 10 (9–10) |
| M(NLS-88R) | 11 | 8 (6–10) | 11 (9–13) | 11 (9–12) | 11 (8–17) | 14 (11–21) | 10 (8–11) | 12 (10–13) | 12 (11–14) | 10 (7–12) | 11 (7–13) |
| M(NLS-88K) | 9d | 5 (4–6) | 10 (8–13) | 9 (8–11) | 9 (7–10) | 7 (5–10) | 13 (9–15) | 10 (8–11) | 11 (8–15) | 5 (3–6) | 7 (6–8) |
| M(NLS-88E) | 4d,e | 4 (2–6) | 4 (3–5) | 4 (2–4) | 4 (3–5) | 4 (3–4) | 5 (4–5) | 4 (3–5) | 5 (4–5) | 4 (2–6) | 4 (3–4) |
| M(NLS-88V) | 7d,e | 5 (4–6) | 7 (5–11) | 9 (7–11) | 6 (4–8) | 7 (5–9) | 7 (5–8) | 7 (5–9) | 7 (5–8) | 7 (6–8) | 7 (7–8) |
wt-WSN contains 88G in M1.
The mean measurement of 10 representative virions.
The average measurement of each representative virion with the range of thickness in parentheses.
P < 0.001 compared to wt-WSN by one-way ANOVA with Tukey's multiple-comparison test.
P < 0.001 compared to M(NLS-88R) by one-way ANOVA with Tukey's multiple-comparison test.
Purified wild-type A/WSN/33 (wt-WSN) and the M1 mutants with R101S and R105S alterations in the nuclear localization signal (NLS) motif 101RKLKR105 and an additional change in amino acid position 88 were fixed and embedded in epoxy resin. Ultrathin sections were stained with uranyl acetate and lead citrate and were examined under a Zeiss EM 912 transmission electron microscope equipped with a Keenview digital camera (Olympus). The thickness of the M1 layer of each virion was measured clockwise at 3, 6, 9, and 12 o'clock and was averaged. Ten representative virions of each virus were measured.
M1 association with cellular membrane.
To assess cellular membrane-associated M1, β-actin was used as the loading control (Fig. 5A and B) because M1 interacts with the cellular membrane at least partly through host cytoskeleton (19). At 24 h p.i., the majority of newly synthesized M1 from wt-WSN and M(NLS-88R) was precipitated with the cellular membrane with only a small fraction remaining in the cytosol (Fig. 5A). Due to slow replication, the level of M1 of M(NLS-88K), M(NLS-88E), or M(NLS-88V) was below the detection limit in both membrane and cytosol fractions at 24 h p.i., with some surface HA0 protein present in the cytosol fraction (Fig. 5A). By 44 h p.i., newly synthesized M1 from all four triple mutants, regardless of replication speeds, had a cellular membrane binding pattern similar to that of wt-WSN (Fig. 5B), suggesting that the disruption of the 101RKLKR105 motif did not interrupt M1 association with the host cellular membrane. However, cellular membrane-associated M1 in proportion to HA0 was significantly less in M(NLS-88K)-, M(NLS-88E)-, and M(NLS-88V)-infected cells than those of M(NLS-88R)- or wt-WSN-infected MDCK (Fig. 5B).
Fig 5.
Cellular membrane association of newly synthesized M1 protein. At 24 (A) or 44 (B) h postinfection (MOI, 4), MDCK cells were dissociated in hypotonic buffer, followed by low-speed centrifugation at 1,000 × g at 4°C for 10 min. The postnuclear supernatants were subjected to ultracentrifugation at 100,000 × g at 4°C for another 60 min. The pellets containing the membranes (m) were analyzed directly by Western blotting. The ultracentrifugation supernatants containing cytosol (c) were subjected to protein precipitation with ice-cold acetone before Western blotting. An equal amount of total protein was loaded per lane for each preparation of membrane and cytosol fractions. The antibody pairs included (i) sheep antiserum against HA of A/Puerto Rico/8/34 and IRDye-680LT-labeled donkey anti-sheep, (ii) biotin-conjugated rabbit anti-M1 and IRDye-680LT-labeled streptavidin, and (iii) β-actin-specific mouse monoclonal antibody and IRDye-800CW-labeled donkey anti-mouse. The blots were imaged and analyzed using an Odyssey imaging system. The M1/HA0 ratio of each virus in the corresponding membrane fraction was estimated by densitometry and is shown at the bottom of the images. wt, wt-WSN; 88R, M(NLS-88R); 88K, M(NLS-88K); 88E, M(NLS-88E); 88V, M(NLS-88V).
Computational modeling of M1.
The dimeric crystal structure of influenza A M1 (aa 1 to 164) at neutral pH contains approximately brick-like monomers, with the 101RKLKR105 motif on one monomeric corner surrounded by strongly basic residues and a negatively charged region on the opposite side (1, 20, 21). A large electrostatic dipole of dimeric M1 thus was possible according to this organization (Fig. 6A). A ZDOCK-based complementarity fit showed that these oppositely charged monomeric faces steered the M1 dimeric formation of 40- by 40- by 60-Å3 dimensions (Fig. 6B) (1, 22). Dimers were then computationally docked in both the x- and y-directions using ZDOCK to simulate an extended M1 layer (Fig. 6B). The model predicted that the M1 dimer-dimer association forming a tetramer showed a slight 30° tilt, which was accommodated well in the formation of an octamer (Fig. 6B). Interestingly, ZDOCK predicted that subunit A/B of wt M1 in this octamer model had the R105 side chain form salt bridges with D38, E40, and E44 of adjacent subunit E/F; meanwhile, subunit B/F had the R101 side chain interact similarly with E152 and D156 of the adjacent subunit D/H (Fig. 6C and Table 2). Along with the other predicted interactions (Table 2), these salt bridges could impart stability to the noncovalent associations, which were expected to be lost in R101S-R105S. Replacing Gly with Arg at position 88 potentially introduced three new salt bridges with E6, E114, or E152 in the diagonally opposite subunit D (Fig. 6D), thereby compensating for the loss resulting from R101S and R105S.
Fig 6.
Molecular modeling of the putative M1 self association. The dimeric (consisting of subunits A and B) crystal structure of influenza A M1 (residues 1 to 164) at neutral pH was retrieved from the Protein Data Bank (code 1EA3) and used to model the outer M1 layer of the virion. The molecular modeling was performed using SYBYL 8.1. (A) Connolly surface of an M1 dimer onto which the electrostatic potential has been mapped. (B) Cartoon representation of the putative M1 self-associated layer model in which the oppositely charged faces interact with one another. The nuclear localization signal (NLS) 101RKLKR105 motif and G88, indicated by pink rectangles and red circles, respectively, are located in oppositely charged faces and cannot be viewed directly from the front. (C and D) Potential ionic interactions that basic residues of the NLS motif may make with nearby acidic residues of adjacent subunits in the putative M1 self-associated layer model. Rendering styles: 101RKLKR105 motif, ball and stick (orange carbon atoms); adjacent acidic residues, ball and stick; G88, yellow spheres; R88, ball and stick (green carbon atoms). Subunit color codes are the following: A, dark blue; B, light blue; C, dark red; D, light red; E, dark purple; F, light purple; G, dark green; H, light green. (C) Interactions between subunits B and D of wt M1. (D) Interactions between subunits A, B, and D of the M(NLS-88R) triple mutant M1.
Table 2.
Predicted interactions in the interfacial regions between adjacent M1 subunitsa
| Interaction | Subunits | Cα-Cα distance (Å) | Type |
|---|---|---|---|
| R76-E8 | B-D/F-H | 11.3 | wt (G88/R101/R105) |
| R76-E29 | B-D/F-H | 10.2 | wt |
| R78-E29 | B-D/F-H | 13.6 | wt |
| K98-D156 | B-D/F-H | 6.7 | wt |
| R101-E152◀ | B-D/F-H | 9.2 | wt |
| R101-D156◀ | B-D/F-H | 8.7 | wt |
| K104-E152◀ | B-D/F-H | 11.5 | wt |
| R134-E6 | B-D/F-H | 7.3 | wt |
| R134-E8 | B-D/F-H | 9.4 | wt |
| R134-E152 | B-D/F-H | 11.7 | wt |
| R88-E6 | A-D | 10.8 | G88R/R101S/R105S |
| R88-E114 | A-D | 8.5 | G88R/R101S/R105S |
| R88-E152 | A-D | 13.6 | G88R/R101S/R105S |
| K102-E44◀ | A-E/B-F | 7.7 | wt |
| R105-D38◀ | A-E/B-F | 11.0 | wt |
| R105-E40◀ | A-E/B-F | 8.4 | wt |
| R105-E44◀ | A-E/B-F | 8.4 | wt |
| E114-R27 | A-E/B-F | 13.9 | wt |
| E114-R49 | A-E/B-F | 10.4 | wt |
| D94-R72 | A-F | 6.8 | wt |
| K95-D38 | A-F | 8.8 | wt |
| K95-E40 | A-F | 10.6 | wt |
| R76-E152 | A-C/E-G | 9.6 | wt |
| R134-E152 | A-C/E-G | 7.5 | wt |
| R134-D156 | A-C/E-G | 8.1 | wt |
| D94-R127 | C-G/D-H | 9.9 | wt |
| K95-E23 | C-G/D-H | 10.3 | wt |
| K98-D30 | C-G/D-H | 8.7 | wt |
| K98-D38 | C-G/D-H | 9.1 | wt |
| R101-D38◀ | C-G/D-H | 11.5 | wt |
The dimeric (consisting of subunits A and B) crystal structure of the M1 protein (residues 1 to 164) of influenza A virus at neutral pH was retrieved from the Protein Data Bank (codes 1EA3 and 1AA7) and used to model the outer M1 layer of the virion. The molecular modeling was performed using SYBYL 8.1. The new putative M1 layer model, in which the positively and negatively charged surfaces face one another in the interfacial region between M1 subunits, was constructed by conceptually defining the M1 layer to be (locally) coincident with the x-y plane, with the z axis being normal (perpendicular to the surface of the layer). A small section of the M1 layer was generated from the dimer by first expanding in the x direction to generate a tetramer (i.e., a dimer of dimers) and then in the y direction to generate an octamer using the ZDOCK server (http://zdock.bu.edu/). Of the highest scoring M1 octamer models proposed by ZDOCK, one model (ZSCORE = 53.02), displaying excellent dimer-packing characteristics, was selected for further refinement and analysis. The distance between residues on adjacent subunits was measured between the backbone Cα atoms and not the usual hydrogen-bonded heavy atoms of the side chains, because a given side chain may interact with multiple partners, though not necessarily at the same time. Interactions involving the nuclear localization signal (NLS) motif 101RKLKR105 are indicated with a triangle (◀).
DISCUSSION
G88R is a compensatory mutation that emerged during our attempts to regenerate the R101S-R105S double mutant. The resultant M(NLS-88R) triple mutant not only replicated efficiently in vitro and in vivo but also exhibited robust immunogenicity in mice. These results suggest that 101RKLKR105 and its adjacent region influence virus replication. Since a structural model suggests that M1 monomers interact with each other via charge-charge interactions (1), G88R favored by the virus could be reasoned to compensate for the charge loss of NLS resulting from R101S and R105S alterations. If this hypothesis was correct, then M(NLS-88K) with a positively charged Lys at position 88 should have behaved similarly to M(NLS-88R). To our surprise, however, M(NLS-88K), like M(NLS-88E) and M(NLS-88V), had significantly retarded replication compared to M(NLS-88R) and wt-WSN. It clearly indicates that G88R was not randomly picked by the virus based solely on amino acid charges.
A simulated peptide binding assay was performed to examine M1-NP physical interactions. The direct M1-NP binding without vRNA was interrupted by replacing Arg with Ser at aa 101 and 105 of the corresponding M1 peptide, and a positive charge (R or K) at position 88 improved the binding of resultant M1 peptides to immobilized NP peptide in a concentration-dependent manner. This is consistent with a previous report that the middle domain (aa 87 to 165) of M1 is responsible for binding to NP without requiring vRNA (23). However, some vRNA binding activity still remained in the 88G-101S-105S peptide, since most of the positive charges in M1 NSL had to be decreased in order to completely inhibit its vRNA binding (3). In vRNP, M1-NP interactions are likely to be dominant over M1-vRNA, since the affinity of NP for vRNA is approximately 10-fold higher than that of M1 for vRNA (2, 3, 22). Interestingly, G88R, but not G88K, of mutant M1 peptide showed substantially enhanced binding to NP peptide with addition of vRNA, implying that the M1-vRNP interactions are not completely oriented by positive-negative charge interactions. The functional M1-vRNP dissociation assay showed that vRNP purified from M(NLS-88R) viral particles but not from other triple mutants also exhibited a strong association with M1 under high-salt conditions. This result clearly indicates that M1-vRNP association in assembled virions is more complicated than positive-negative charge interactions of M1-NP, as demonstrated by the direct peptide binding ELISA.
M1 regulates intracellular trafficking of vRNP for subsequent virion assembly (6, 24–26). Unlike M1 of R101S-R105S, which showed altered cellular distribution (9), the triple mutants with G88R or G88K but not G88E or G88V had M1 observed in both cytoplasm and nuclei of infected cells. This observation indicates that a positively charged amino acid at position 88 of M1 is important in restoring the normal cellular distribution of M1. However, M1 from M(NLS-88R) appeared to have a stronger nuclear localization than M(NLS-88K), implying that an Arg, but not Lys, at position 88 is more beneficial for vRNP export. Additionally, M(NLS-88E) and M(NLS-88V) had impaired M1-NP colocalization, in contrast to M(NLS-88R) and M(NLS-88K). This result is consistent with those from M1-NP peptide binding assays and confirms that M1-NP binding is mainly dependent on the positive-negative charge.
In addition to NLS, M1 also has several regions rich in basic amino acid residues. Recently it was reported that replacing Arg with Ala or Asp on the basic amino acid stretch 76RRR78 of M1 resulted in defective virus assembly and replication, whereas substitution(s) of Arg for Lys in 76RRR78 led to similar or even higher rates of virus replication than wt-WSN (5). In contrast, M(NLS-88K), similar to M(NLS-88E) and M(NLS-88V), had significantly retarded growth compared to M(NLS-88R) or wt-WSN. These results indicate that 101RKLKR105 and its adjacent region affect virus replication through different mechanism(s) than 76RRR78, implying that M1-NP interactions are not as critical for virus replication as expected.
To prepare for virion assembly and budding, M1 needs to be associated with the host cellular membrane mainly through its N terminus (4, 20, 27, 28). An NLS knockout M1 mutant was reported to have altered membrane association (20). However, the impaired growth of M(NLS-88K), M(NLS-88E), or M(NLS-88V) was not due to defective cellular membrane association of M1. There may be multiple membrane binding domains within M1, and NLS is not essential for M1 association with the cellular membrane (15, 22, 28). However, the cellular membrane-associated M1 in proportion to viral surface protein HA was significantly reduced in M(NLS-88K)-, M(NLS-88E)-, or M(NLS-88V)-infected cells compared that in M(NLS-88R)- or wt-WSN-infected MDCK, implying that less M1 is incorporated into assembled virions of these mutants.
M1 in assembled virions functions as a bridge to interact with vRNP and the cytoplasmic tails of viral surface HA and NA (4, 5, 17, 28, 29). Consistent with the observation from the cellular membrane association experiment, the virions of M(NLS-88K), M(NLS-88E), and M(NLS-88V) showed a disrupted thin M1 layer under TEM. Only the mutant with G88R had the formation and thickness of M1 layer that closely resemble those of wt-WSN. Our TEM results along with the M1-vRNP complex association data imply that a disrupted M1 layer leads to a weak association with vRNP in assembled virions. Our data support that reduced M1 incorporation affects vRNP recruitment and changes virion structure (4), which could be the reasons for the retarded growth of M(NLS-88K) despite its restored M1-NP interactions.
An octamer model that is consistent with published biophysical data of M1 (1, 30–32) was developed to better understand why G88R was beneficial for regaining replication competency. The excellent complementarity of the polybasic and polyacidic domains of M1 highlighted key salt bridges between selected acidic and basic residues as the origin for self-association in generating rod-like M1 structures. The self association of the M1 brick likely results in an M1 layer that displays two leaves, as shown under TEM (Fig. 4A and B). Several key salt bridges that facilitate interactions between adjacent M1 subunits will be absent from the R101S-R105S mutant, ostensibly resulting in a significant reduction in the stability and integrity of the M1 layer, which is expected to negatively impact virus replication. Nature likely uses multiple salt bridges to protect against high mutation rates (33), and eliminating some salt bridges may not completely obviate replication but may negatively affect its rate. The M(NLS-88R) octamer modeling predicts reengineering of several new salt bridges (Fig. 6D) that likely regenerate the stability of the M1 layer and consequently improve virus replication. This may explain why nature prefers spontaneous G88R change even without NLS loss.
Taken together, we have demonstrated that in addition to restored M1-NP and M1-vRNP interactions, maintaining the M1 integrity in assembled virion is also crucial in regaining replication competency of influenza viruses with a disrupted NLS in M1.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of Center for Biologics Evaluation and Research, U.S. Food and Drug Administration.
We are grateful to Zhiping Ye and Teresa Liu of CBER/FDA for providing the R101S-R105S double mutant and wt-WSN plasmids. We thank Nga Yen Nguyen of the Core Facility, CBER/FDA, and Yangqing Zhao of DVP, CBER/FDA, for technical assistance. We also thank Qibing Zhou of VCU and Mingjie Zhang and Vladimir Lugovtsev of CBER/FDA for critical review of the manuscript.
Footnotes
Published ahead of print 17 October 2012
REFERENCES
- 1. Arzt S, Baudin F, Barge A, Timmins P, Burmeister WP, Ruigrok RW. 2001. Combined results from solution studies on intact influenza virus M1 protein and from a new crystal form of its N-terminal domain show that M1 is an elongated monomer. Virology 279(2):439–446 [DOI] [PubMed] [Google Scholar]
- 2. Baudin F, Bach C, Cusack S, Ruigrok RW. 1994. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 13(13):3158–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Elster C, Larsen K, Gagnon J, Ruigrok RW, Baudin F. 1997. Influenza virus M1 protein binds to RNA through its nuclear localization signal. J. Gen. Virol. 78(Pt 7):1589–1596 [DOI] [PubMed] [Google Scholar]
- 4. Rossman JS, Lamb RA. 2011. Influenza virus assembly and budding. Virology 411(2):229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Das SC, Watanabe S, Hatta M, Noda T, Neumann G, Ozawa M, Kawaoka Y. 2012. The highly conserved arginine residues at positions 76 through 78 of influenza A virus matrix protein M1 play an important role in viral replication by affecting the intracellular localization of M1. J. Virol. 86(3):1522–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin K, Helenius A. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67(1):117–130 [DOI] [PubMed] [Google Scholar]
- 7. Sakaguchi A, Hirayama E, Hiraki A, Ishida Y, Kim J. 2003. Nuclear export of influenza viral ribonucleoprotein is temperature-dependently inhibited by dissociation of viral matrix protein. Virology 306(2):244–253 [DOI] [PubMed] [Google Scholar]
- 8. Ye Z, Robinson D, Wagner RR. 1995. Nucleus-targeting domain of the matrix protein (M1) of influenza virus. J. Virol. 69(3):1964–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu T, Ye Z. 2004. Introduction of a temperature-sensitive phenotype into influenza A/WSN/33 virus by altering the basic amino acid domain of influenza virus matrix protein. J. Virol. 78(18):9585–9591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu T, Ye Z. 2005. Attenuating mutations of the matrix gene of influenza A/WSN/33 virus. J. Virol. 79(3):1918–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xie H, Liu TM, Lu X, Wu Z, Belser JA, Katz JM, Tumpey TM, Ye Z. 2009. A live attenuated H1N1 M1 mutant provides broad cross-protection against influenza A viruses, including highly pathogenic A/Vietnam/1203/2004, in mice. J. Infect. Dis. 200(12):1874–1883 [DOI] [PubMed] [Google Scholar]
- 12. Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97(11):6108–61131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qi L, Carbone KM, Ye Z, Liu T, Ovanesov M, Pletnikov M, Sauder C, Rubin SA. 2008. Genetic contributions to influenza virus attenuation in the rat brain. J. Neurovirol. 14(2):136–142 [DOI] [PubMed] [Google Scholar]
- 14. Xie H, Liu T, Chen H, Huang X, Ye Z. 2007. Evaluating the vaccine potential of an influenza A viral hemagglutinin and matrix double insertion DNA plasmid. Vaccine 25(44):7649–7655 [DOI] [PubMed] [Google Scholar]
- 15. Thaa B, Herrmann A, Veit M. 2009. The polybasic region is not essential for membrane binding of the matrix protein M1 of influenza virus. Virology 383(1):150–155 [DOI] [PubMed] [Google Scholar]
- 16. Adamo JE, Liu T, Schmeisser F, Ye Z. 2009. Optimizing viral protein yield of influenza virus strain A/Vietnam/1203/2004 by modification of the neuraminidase gene. J. Virol. 83(9):4023–4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burleigh LM, Calder LJ, Skehel JJ, Steinhauer DA. 2005. Influenza a viruses with mutations in the m1 helix six domain display a wide variety of morphological phenotypes. J. Virol. 79(2):1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruigrok RW, Calder LJ, Wharton SA. 1989. Electron microscopy of the influenza virus submembranal structure. Virology 173(1):311–316 [DOI] [PubMed] [Google Scholar]
- 19. Zhang J, Lamb RA. 1996. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology 225(2):255–266 [DOI] [PubMed] [Google Scholar]
- 20. Arzt S, Petit I, Burmeister WP, Ruigrok RW, Baudin F. 2004. Structure of a knockout mutant of influenza virus M1 protein that has altered activities in membrane binding, oligomerisation and binding to NEP (NS2). Virus Res. 99(2):115–119 [DOI] [PubMed] [Google Scholar]
- 21. Harris A, Forouhar F, Qiu S, Sha B, Luo M. 2001. The crystal structure of the influenza matrix protein M1 at neutral pH: M1-M1 protein interfaces can rotate in the oligomeric structures of M1. Virology 289(1):34–44 [DOI] [PubMed] [Google Scholar]
- 22. Ruigrok RW, Barge A, Durrer P, Brunner J, Ma K, Whittaker GR. 2000. Membrane interaction of influenza virus M1 protein. Virology 267(2):289–298 [DOI] [PubMed] [Google Scholar]
- 23. Noton SL, Medcalf E, Fisher D, Mullin AE, Elton D, Digard P. 2007. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J. Gen. Virol. 88(Pt 8):2280–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boulo S, Akarsu H, Ruigrok RW, Baudin F. 2007. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res. 124(1–2):12–21 [DOI] [PubMed] [Google Scholar]
- 25. Bui M, Wills EG, Helenius A, Whittaker GR. 2000. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J. Virol. 74(4):1781–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whittaker G, Bui M, Helenius A. 1996. The role of nuclear import and export in influenza virus infection. Trends Cell Biol. 6(2):67–71 [DOI] [PubMed] [Google Scholar]
- 27. Baudin F, Petit I, Weissenhorn W, Ruigrok RW. 2001. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology 281(1):102–108 [DOI] [PubMed] [Google Scholar]
- 28. Kretzschmar E, Bui M, Rose JK. 1996. Membrane association of influenza virus matrix protein does not require specific hydrophobic domains or the viral glycoproteins. Virology 220(1):37–45 [DOI] [PubMed] [Google Scholar]
- 29. Ali A, Avalos RT, Ponimaskin E, Nayak DP. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 74(18):8709–8719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. 2010. Structural organization of a filamentous influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 107(23):10685–10690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sha B, Luo M. 1997. Crystallization and preliminary X-ray crystallographic studies of type A influenza virus matrix protein M1. Acta Crystallogr. D Biol. Crystallogr. 53(Pt 4):458–460 [DOI] [PubMed] [Google Scholar]
- 32. Sha B, Luo M. 1997. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat. Struct. Biol. 4(3):239–244 [DOI] [PubMed] [Google Scholar]
- 33. Elena SF, Carrasco P, Daros JA, Sanjuan R. 2006. Mechanisms of genetic robustness in RNA viruses. EMBO. Rep. 7(2):168–173 [DOI] [PMC free article] [PubMed] [Google Scholar]