Abstract
Studies of bacteriophage Mu transposition paved the way for understanding retroviral integration and V(D)J recombination as well as many other DNA transposition reactions. Here we report the structure of Mu transposase (MuA) in complex with bacteriophage DNA ends and target DNA, determined from data that extend anisotropically to 5.2/5.2/3.7Å resolution, in conjunction with previously-determined structures of individual domains. The highly intertwined structure illustrates why chemical activity depends on formation of the synaptic complex, and reveals that individual domains play different roles when bound to different sites. It also suggests explanations for the increased stability of the final product complex and for its preferential recognition by the ATP-dependent unfoldase ClpX. Although MuA and many other recombinases share a structurally conserved “DDE” catalytic domain, comparisons among the limited set of available complex structures suggest that some conserved features, such as catalysis in trans and target DNA bending, arose through convergent evolution because they are important for function.
Mobile DNA elements are important in many aspects of biology, such as disease, evolution, and the spread of antibiotic resistance, and the recombinases they encode, including MuA, are useful genetic tools1,2. The DNA transposition system of bacteriophage Mu was the first to be developed in vitro3. MuA, many other DNA transposases, and retroviral integrases share a conserved RNaseH-like or “DDE” catalytic domain, named for the 3 Mg++-binding carboxylates in their active sites4. Structural studies have lagged behind biochemical ones: only 3 family members have been cocrystallized in active, DNA-bound complexes, and only one with target DNA5–7. Despite mechanistic similarities, only the catalytic domain is conserved among all of these, and their overall architectures are completely different4. More examples are needed to understand the diverse ways in which these enzymes harness a common catalytic domain to accomplish transposition. The richness of the known biochemistry for Mu, from assembly of the initial complex to targeted disassembly of the product complex, makes it a particularly informative example for structural studies.
The first steps of Mu transposition (Fig. 1) are common to many other DNA transposition systems as well as retroviral integration: (1) pairing of the mobile element’s ends by the recombinase to form a “transpososome” or “intasome,” (2) hydrolytic nicking at the phage-host junction, and (3) attack of the newly freed 3′ hydroxyls on a target DNA (“strand transfer”), creating a new connectivity. Bacteriophage Mu uses this mechanism to form a lysogen, and to replicate when it becomes lytic. During the lytic phase, host enzymes are recruited to convert the branched product into replication forks, resulting in duplication of the entire phage genome. However, during initial lysogen formation, the “flanking host” DNA (gray in Fig. 1) consists only of extra sequences appended during headful packaging. In this case, a poorly understood signal causes the transposase to cleave both strands at each genome end, leading to a simple insertion without replication8,9.
Figure 1.

Transposition pathway and structure determination. a) Cartoon of transposition. The transposase (MuA) pairs the phage genome ends (blue and red). At each end, the same active site catalyzes the attack of H2O at the phage-host junction and then the direct attack of the phage 3′-OH on target DNA (“strand transfer”). Target binding is nonspecific, and there is a 5 bp stagger between the sites of attack. Host and target DNAs may be entire circular replicons. After the ATP-dependent unfoldase ClpX disassembles the final strand transfer complex, the 3′ hydroxyls are used as replication primers, resulting in duplication of the phage genome. Our crystals contain the strand transfer product (3rd panel). b) Domain structure of MuA. c) Experimental electron density map after phase improvement with Parrot superimposed on the model (contours are 1.2 and 2σ).
Note: part a was originally drawn in Illustrator, and an editable pdf can be supplied if needed. Part b was drawn in Word.
MuA is chemically active only when incorporated into transpososomes that pair the two ends of the phage genome, and thus assembly of this complex is a regulatory step. Mu transpososomes become increasingly stable as the reaction progresses10,11. After strand transfer the complex is so stable that the “enzyme” MuA does not actually turn over. The strand transfer reaction can only be reversed if the complex is disrupted, for instance by heating to 75C12,13. This may be a thermodynamic necessity for a reaction in which the 1st step (hydrolysis) is committed, yet the 2nd step (strand transfer) is chemically isoenergetic, with no net change in the number of phosphodiester bonds. In vivo, Mu transpososomes must be disassembled by the ATP-dependent unfoldase ClpX before DNA replication can proceed14–17. Our new transpososome structure suggests explanations for the increased stability of the final complex and its preferential recognition by ClpX.
Crystallization of the strand transfer complex was facilitated by two observations. First, despite a lack of target sequence specificity, MuA attacks mismatch-containing target DNA with single-nucleotide precision (Fig. S1)1. Second, the natural transpososome assembly pathway (described below) can be simplified under permissive conditions in vitro. The resulting active complexes contain 4 copies of the MuA protein and 2 copies of a ~50bp DNA derived from the phage genome’s right end, each carrying two MuA binding sites (termed R1 and R2)18,19. Modeling based on the structure of these complexes suggests that transpososomes formed on full left and right ends are quite similar.
Overall Architecture
Viewed in isolation, the 5 domains of each subunit resemble beads on a string (Fig. S2). However, when all four subunits are assembled on the DNA, they intertwine to form a network of protein-protein and protein-DNA interactions (Fig.2 and movie S2). The overall transpososome resembles a pair of scissors, with the phage end DNAs forming the handles and the sharply bent target DNA the blades. A 34Å resolution EM reconstruction of Mu transpososomes in the absence of target DNA found a similar V shape, although the arms were shorter and the accompanying electron spectroscopic imaging predicted a more contorted path for the DNA20. Within the transpososome, most of the individual protein domains perform different roles in the R1- vs. R2-bound subunits. Where a system encoded by a larger genome might evolve two separate polypeptides, phage Mu cleverly re-uses the same sequence to perform different functions within the complex.
Figure 2.
Transpososome structure. The complex sits on a crystallographic twofold (vertical) that relates the blue and red halves. The pale- and dark-colored subunits adopt different conformations within the homotetramer. DNA colors match Figure 1. a) Cartoon. Catalytic sites are marked as yellow and tan stars (facing the viewer or the background, respectively) and domains of the blue subunits are labeled. b) Ribbon drawing, with the scissile phosphate groups shown as yellow spheres. c) Same drawing as in B, rotated ~90° about a vertical axis.
Catalysis and Mu DNA end binding in trans
The catalytic sites lie within domain IIα. In agreement with biochemical studies, only the R1-bound subunits’ active sites engage with DNA, and they do so in trans: e.g. the dark blue subunit binds the blue Mu DNA via domains Iβ and γ while its active site domain docks at the red Mu DNA – target junction (the interdomain linkers are too short for any other connectivity) (Fig. 3). First characterized for Mu transposition21,22, such trans catalysis is a recurring theme in DNA transposition, and helps render chemical activity dependent on full complex assembly4. Domains IIαβ of the R2 subunits bridge the two Mu end DNAs and play a primarily structural role. IIα of each R2 subunit interacts with DBD Iβ of the subunit bound to the R1 site of the same DNA segment, whereas IIβ of each R2 subunit binds to the opposite DNA segment. Biochemical studies had predicted domain IIβ to interact with the target DNA, which does occur in the R1 subunits23.
Figure 3.
This is a stereo pair.
Stereo close-up of interactions near the Mu DNA-target junction. Colors are the same as in Figure 2. A segment of DNA from a symmetry-related complex (yellow) binds the positively-charged domain IIIα of the R2-bound subunit (cyan). If the red Mu end DNA were extended to include flanking host DNA, it could lie where the yellow DNA does. The yellow sphere marks the phosphate group at the Mu-target DNA junction, and the main chains of the two active site D’s are also yellow (a third active site residue lies on a helix that could not be modeled). The loop that extends from domain IIα aa 410–430) to interact with the black target DNA is circled on the red subunit.
Domains Iβ and Iγ recognize the specific binding sites on the phage DNA ends and their positions agree with footprinting and mutagenesis data24,25. The closest structural match to MuA’s tandem DNA binding domains is the centromere binding protein CENP-B, which probably evolved from an ancestral transposase26. The eukaryotic mariner family Mos1 and Tc3 transposase structures also include tandem DBDs7,27. In both, contacts between the DBDs equivalent to MuA’s Iβ mediate synapsis of the two transposon ends. In the Mu transpososome, only the R1-bound DBDs mediate protein-protein contacts: Iβ as described above, and Iγ to both IIα of the other Mu end’s R1 subunit and IIIα of same Mu end’s R2 subunit. The importance of these interactions is underscored by the high sequence conservation within the interaction surfaces (Fig. S3).
Although the resolution precludes detailed analysis of DNA bending, the path of the backbone is clear. The R2 site is bent by ~28°, largely through compression of the major groove around domain Iβ, which agrees with DNaseI hypersensitive sites, and the CENP-B – DNA structure10,25,28. The R1 site is less bent (~17°). Stronger bending there would cause a steric clash between domain Iγ of the R1-bound subunit and the β barrel (IIβ) of the adjacent R2 subunit. We propose that the R1 site straightens somewhat upon transpososome formation. The formation of favorable protein-DNA and protein-protein contacts within the transpososome could offset the cost of weakening contacts between DNA and domain Iβ.
Larger DNA conformational changes may occur on transpososome formation, as implied by solution experiments that found ~90° bends in monomeric MuA-DNA complexes29. In monomeric MuA, domain IIα could interact in cis with its own Iβ and additional DNA bending might be induced by electrostatic interactions with IIβ and IIIα. A different but stable monomer conformation would raise the energy barrier to spontaneous tetramer formation, making it more amenable to regulation. It could also prevent premature encounters between the active site and the DNA.
Target DNA
The target DNA is bent through a total of ~140°. Protein-DNA contacts are mediated by the R1 subunits’ domains IIα, IIβ and IIIα and extend to all but the outermost base pairs of the target DNA, as predicted by footprinting30. A long loop (residues 410 to 430) extends from each catalytic domain (IIα) underneath the target. This loop could be fit onto experimental electron density by a rigid-body rotation from its position in the 2.8Å domain IIαβ structure (PDB ID 1bcm)31. The positively charged β barrel, domain IIβ, interacts loosely with the outer end of the target. Although poorly ordered, its position is defined by the SeMet signal from two adjacent Met residues. The relative orientation between domains IIα and IIβ of the R1 subunit is shifted slightly from that in the unbound protein and in the R2 subunit. This alters the connecting loop, which also lies near the target, but could not be modeled. Finally, the R1 subunits’ domain IIIαs pair to form a coiled coil on the concave side of the bent target DNA.
Two roles for domain IIIα
Domain IIIα, the final ~45 residues in our structure, also plays different roles in the R1 vs. R2 subunits. It is highly positively charged and binds DNA as an isolated peptide32. The R1 subunits’ domain IIIαs appear to stabilize the bent target DNA in two ways: (1) alleviating electrostatic repulsion between the sides of the U-shaped target, and (2) physically trapping the target DNA within the complex. In the absence of target DNA, they must be either mobile or in a different location. Since the C-terminus of MuA contains a ClpX binding tag, rearrangement of the R1 subunits’ domain IIIs upon target binding could allow ClpX to preferentially recognize the final strand transfer complex for ATP-dependent disassembly. Furthermore, it is these endmost subunits that ClpX preferentially unfolds33,34.
Initial transpososome assembly requires domain IIIα on the R2- but not the R1-bound subunits22,35,36. The structure suggests that the R2 subunit’s IIIα stabilizes the complex by wrapping around the other subunits near the active site. It may also anchor the flanking host DNA (Figs. 1 and 3). The construct crystallized included minimal flanking host DNA, but if extended, it could bind the R2 subunit’s IIIα, occupying a spot where symmetry-related DNA interacts in the crystal. This model agrees with footprinting data that predicted a large distortion, and would prevent steric clashes between the flanking host and target DNAs10,30. Domain IIIα was reported to have cryptic nuclease activity that might cleave the flanking DNA flap after the initial insertion reaction8,32. Alternatively, movement of domain IIIα triggered by an unknown signal) might deliver the uncleaved strand to the DDE site for hydrolysis.
Transpososomes with full left and right ends
Although the complex that we crystallized is highly active in vitro, its assembly requires high protein and DNA concentrations or “permissive” solvent conditions18. The natural system is more complicated and provides an interesting example of templated complex assembly. The two ends of phage Mu genome carry different arrays of 3 MuA binding sites each, and the left end also binds the DNA bending protein HU (Fig. 4). Conversion of an initial pairing of right and left ends to an active complex is stimulated by transient binding of the N-terminal domains of several of MuA subunits to an internal enhancer element37,38. However, if complexes assembled this way are treated with a high salt wash, an active complex remains that contains only four subunits of MuA, contacting only the R1, R2 and L1 DNA sites10,19. Thus the other binding sites are important for assembly but not for the final activity.
Figure 4.
Model for a transpososome assembled on full left (reddish) and right (blue) bacteriophage ends. The N-terminus of each domain Iβ is marked with a red sphere to show the approximate position of domain Iα, which transiently binds the enhancer. Domains discussed in the text are labeled. Inset: cartoon of the bacteriophage Mu genome ends and internal enhancer element.
Modeling showed that the functional part of the tetrameric assembly, where Mu ends join to target DNA, can be identical in the crystallized (R1R2)2 complex and in the full left + right complex (Fig. 4). Subunits R1, R2, L1, and domains II and III of the L2 subunit were modeled directly from the crystal structure. The HU-induced bend allows a single protomer of MuA to bind the L2 site via domain Iβ while its domains II and III form part of the core complex. Domain Iγ acts as a linker, which explains why L2 is the only site where Iγ doesn’t footprint25.
Additional interactions involving the L3 and R3-bound subunits may temporarily hold the components together while the intertwining of protein and DNA needed to form a transpososome at the Mu-host junction occurs. We modeled interactions between the L3 subunit’s domain IIα and R2’s Iβ based on those seen in the crystal between the R2 and R1-bound subunits. The R3 subunit’s role is unclear, but if the HU-induced bend were relaxed, similar cross-end interactions might occur between the L3 and R3 subunits.
Topological studies predicted two right-handed superhelical crossings within the transpososome39,40. One such crossing occurs in the crystal structure, near the junction with target DNA (R1 over L1 in Fig. 4). A 2nd crossing (L3 over R2) results from the severe bend induced by HU in the model, in conjunction with a smaller bend in the L2 site. Restraint of two supercoils within such short segments of DNA helps is consistent observations that supercoiling stimulates transpososome assembly41.
During assembly on intact phage DNA, domain Iα transiently binds an internal enhancer, which contains two clusters of MuA binding sites termed O1 and O2. There are too many degrees of freedom to add the enhancer to our model. However, our model does agree with data showing that O1-bound proteins interact with both L3 and R1 and that the somewhat longer O2 bridged to both R3 and L142,43. The multiple protein-protein interaction surfaces of MuA may help stabilize an initial pairing of the phage ends, but interactions between the wrong partners could slow down the transition to a final, cleavage-ready complex. The enhancer may stimulate this transition by preventing unproductive interactions among L- and R- bound subunits as well as by aligning them for productive ones.
Such a baroque assembly process is not limited to Mu: many other mobile elements also require seemingly “extra” recombinase subunits44. Although the details vary among these systems, they may all be using the same fundamental strategy of using additional subunits to temporarily stabilize pairing of the element end DNAs while a complicated, intertwined structure forms at the 3′ ends. The additional complexity may also provide additional opportunities for regulation. Finally, for mobile elements that are present in high copy, it may help ensure that the two ends paired in a single transpososome belong to the same copy of the mobile element.
Convergent and divergent evolution
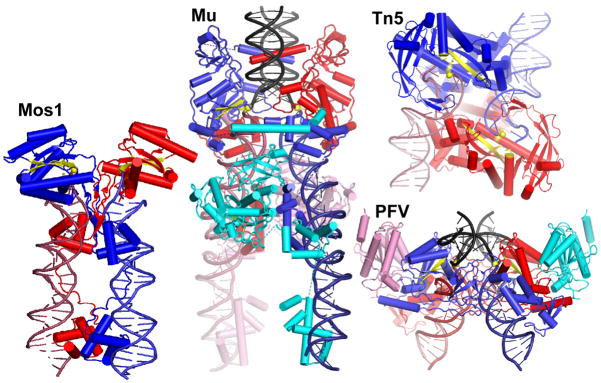
Many DNA transposases and retroviral integrases share a structurally conserved “DDE” or RNaseH-like catalytic domain, suggesting divergence from a common ancestor. However, this is the only domain conserved among these diverse recombinases. Comparison of the four reported structures of DDE recombinases in complex with substrate DNAs shows that other recurring features may reflect convergent evolution for functional reasons (Fig. 5). All four complexes are held together by intertwined networks of protein-protein and protein-DNA contacts, although different domains mediate those contacts4. Mos1 and MuA do have structurally related bipartite DNA binding domains, but even those domains form different protein-protein contacts in their respective transpososomes7.
Figure 5.
Comparison of DDE recombinase-DNA complexes. The mobile element ends are red and blue and target DNA (where included) is black. Subunits that carry out the chemical reactions are red and blue; additional subunits pink and cyan. Active site residues, scissile phosphate groups, and the two β strands of the conserved catalytic domain that carry the catalytic D’s are in yellow. Mos1 is a Tc1/mariner family eukaryotic DNA transposon; Tn5 is a bacterial DNA transposon, and Prototype Foamy Virus (PFV) is a mammalian retrovirus5–7. Mos1 and Tn5 require only a dimer for activity, whereas Mu transposase and PFV integrase require tetramers. In the PFV structure, only the catalytic domains of the additional subunits were visible (pink and cyan).
Despite the diversity of these complexes, catalysis is always in trans: the subunit that catalyzes DNA cleavage and joining on one mobile element end binds to specific sequences on the other end. This feature ensures that the chemical reactions at the two element ends are coordinated, because the complex requires proper pairing of the ends for assembly.
Another recurring feature is strong bending of the target DNA. Target DNA bound the PFV intasome is also bent, although not quite as severely as that in the Mu transpososome6. The Mos1 transpososome was crystallized without target DNA, but additional end DNAs found in the crystal bind where target is expected to, and in a way that requires target bending7. Modeling of target DNA onto the Tn5 transpososome structure also requires bending, which agrees with biochemical data for the related Tn10 system5,45. Outside of the catalytic domain, contacts to the target DNA vary widely among these structures. Why then have they all evolved to strongly bend the target DNA? As noted for the PFV structure, target bending may help render strand transfer irreversible by straining the DNA conformation such that the ends snap away from the active site after strand transfer. This may be a source of the product binding energy that drives forward the otherwise-isoenergetic strand transfer reaction. The overall conformation of the target DNA in the Mu transpososome resembles that bound by IHF. In that case, a nick at the kink does enhance affinity by allowing the ends to spring apart46.
The DDE catalytic domain is thus a conserved module that has been co-opted by numerous mobile elements to perform similar chemical reactions. However, other similarities in the way that it has been harnessed to mobilize these elements appear to reflect convergent evolution to satisfy functional requirements.
FULL METHODS
Overview
We determined the structure of the final strand transfer complex, which contains a tetramer of MuA, 2 copies of the phage end DNA, and one target DNA (Fig. 2 and movie S1). Crystallizations used a slightly truncated protein, MuA 77-605, which is active in vitro and lacks only the N-terminal enhancer-binding domain and the C-terminal domain that interacts with ClpX and MuB, a second phage-encoded protein that helps deliver an appropriate target DNA under non-permissive conditions47. The phage end DNA mimics precleaved right ends, and the 35bp target DNA contains a central G:G mismatch. Although MuA displays little sequence specificity for target DNA, it attacks mismatch-containing DNA with single-nucleotide precision1. This feature facilitated production of a homogenous sample for crystallization.
Phases were determined by MIRAS using 3 derivatives. The crystals diffracted anisotropically to 5.2Å, 5.2Å and 3.7Å along the 3 principal axes. Model building was possible despite the low resolution because >90% of the protein structure had been previously determined as isolated domains31,48,49. Placement of the protein domains was verified by SeMet data, and the DNA sequence register by an additional data set collected from crystals where every T on one strand had been substituted with 5-Br-dU (Fig. S4). No prior structure was available for domain IIIα, which comprises ~45aa that are strongly predicted to form one long helix followed by a short one. Although density for these helices was visible, the sequence register is uncertain. The complex lies on a crystallographic twofold axis such that the asymmetric unit contains half a transpososome. After highly restrained refinement R and Rfree were 39.3 and 43.7%, well within the range expected for a low-resolution structure.
Expression and Purification of the MuA transposase
The pMK599 plasmid, a pET3c derivative that contains the MUA open reading frame coding for residues 77 to 605, was a gift from the Mizuuchi lab50. This plasmid was transformed into E. coli Rosetta pLysS strain (EMD Biosciences) for protein overexpression. After plating transformants, a starter culture was prepared by inoculating multiple colonies into LB media (with 100 ug/mL ampicillin) and growing at 37 °C until the OD600 was ~0.7. Typically, 100 ml of starter culture was prepared per liter of final culture. After the addition of starter culture to fresh ampicillin-containing LB media, cells were grown to an OD600 of ~0.8, then protein expression was induced with IPTG (added to a final concentration of 0.5 mM). Cells were harvested 2 hours after induction by centrifugation at ~8000g for 10 minutes, and cell pellets were stored at −80 °C for later use.
Cell pellets were resuspended in a lysis buffer (25 mM HEPES [pH 7.5], 1 mM EDTA, 1 M NaCl, 10% sucrose, 10% glycerol, 5 mM DTT, 200 ug/ml lysozyme, protease inhibitor cocktail from Roche Diagnostics), sonicated, and centrifuged at 40,000g for 1 hour (18,000 rpm in a SS-34 rotor). Ammonium sulfate was added to the supernatant to 30% saturation in order to precipitate the protein. The pellet was collected by centrifugation, and redissolved in buffer A (20 mM MES [pH 5.5], 0.5 mM EDTA, 5% glycerol, 0.2 M NaCl, and 1 mM DTT). The protein sample was filtered before loading onto a heparin affinity column (GE Healthcare). Proteins were eluted with salt gradient from 0.2 M to 2.0 M NaCl. To improve the purity, MuA-containing fractions were rechromatographed on heparin after dialysis into buffer A. The protein was then dialyzed into buffer A again and loaded onto a Mono-S column (GE Healthcare). A gradient similar to that from the heparin affinity purification was applied. Fractions containing MuA were pooled and dialyzed at 4 °C into 20 mM HEPES [pH 7.5], 0.5 mM EDTA, 0.2 M ammonium sulfate, 20% glycerol, and 1 mM DTT. The protein was concentrated to approximately 10 mg/ml, and stored at −80 °C. Minimal nuclease contamination was detected when samples (0.5 mg/ml final concentration) were incubated for 2 hours at 37°C in 10 mM HEPES pH 7.5 with supercoiled plasmid DNA, 50 mM NaCl, and 10 mM MgCl2.
SeMet-labeled MuA 77-605 was prepared similarly except that cells were grown differently51: Instead of using LB, the cells were inoculated in M9 media plus 0.4% glucose, 10 mM NaCl, 0.1 mM CaCl2, 2 mM MgSO4, and 100 mg/ml ampicillin until OD600 reached ~ 0.5. An amino acid cocktail containing L-isoleucine, L-leucine, L-lysine, L-phenylalanine, L-threonine, and L-valine was then added to a final concentration of 100 mg of each amino acid per liter. Seleno-DL-methione (Sigma) was also added to a final concentration of 60 mg/liter. The culture was grown for 15 more minutes before 0.5 mM IPTG was added. Cells were harvested after 3 hours of induction.
Preparation of Mu end and target DNA
Mu end DNA duplexes were designed to contain the R1 and R2 binding sites for MuA. Each duplex was prepared by mixing four single strands in equal molar amounts. The oligonucleotides used for the structure determination are listed below:
TL: 5′-GCTTGAAGCGGCGCACGAAAAACGCG,
TR: 5′-AAAGCGTTTCACGATAAATGCGAAAAC,
BL: 5′-AACGCTTTCGCGTTTTTCGTGCGCCGCTTCA
BR: 5′-CGGTTTTCGCATTTATCGTGA.
These strands were heated at 80°C for 20 minutes, and annealed by slow cooling to room temperature. The final concentration of the duplex DNA is 0.2 mM in TEN buffer (10mM Tris-HCl and 0.5mM EDTA, 100 mM NaCl, pH 8.0). The resulting DNA mimics the product of initial DNA cleavage by MuA, and has a three-nt 5′-overhang on the uncleaved strand, and a two-nt 5′-overhang on the other end. Each strand of the resulting duplex is nicked at a position that does not interfere with transpososome assembly.
The target DNA contains a central mismatch and was designed to be asymmetric to avoid hairpin formation during annealing. The target DNA was prepared in a manner similar to that of the Mu end DNA using the following oligonucleotides:
5′-TATCGCAACAACACATCGGATAACCATAAGTAATA
5′-TATTACTTATGGTTATCGGATGTGTTGTTGCGATA.
All unmodified oligonucleotides were obtained from IDT Technologies. Brominated oligonucleotides (discussed in the later section) that were used in this study to validate the sequence of the donor DNA and the location of the target DNA were obtained from Yale University’s Keck Facility. Since brominated oligonucleotides are photolabile, they were handled in the dark as much as possible.
Crystallization and Data collection
Strand-transfer complexes were assembled by mixing the target DNA, Mu end DNA, and MuA protein in 1: 1.4 : 3.7 molar ratios in a solution containing 10 mM MgCl2, 25 mM Hepes [pH 7.5], 10 mM DTT, 0.02% Zwittergent, 14% glycerol, and 0.2 M (NH4)2SO4. This was incubated for at least an hour at room temperature to ensure completeness of the strand transfer reaction. Although DMSO is usually added to stimulate assembly of transpososomes with two right end DNAs, we found that it was not necessary at the high protein and DNA concentrations used for crystallographic work18. Crystallization trials were then performed using hanging-drop vapor diffusion method: drops contained a 1:1 mixture of complex stock solution (~2 mg/ml) and well, and were incubated at 19°C. Crystals appeared in 22–28% (v/v) PEG400, 0.1 M Hepes [pH 7.5], and 0.2 M MgCl2, and grew to their full size in two to four weeks. Tantalum bromide derivatives were obtained by soaking the crystals of the strand-transfer complex for 1 to 8 days with 0.4 mM [Ta6Br12]2+ cluster (Jena Bioscience) in a solution that mimicked the condition of the drop. For derivatization with mercury, crystals were soaked in 32% PEG400, 0.1 M Hepes [pH 7.5], 0.2 M MgCl2, and 0.1 mM mersalyl acid (Sigma) for 1 to 2 days. For derivatization with selenium, crystallization setups were done with SeMet-protein. And for that with bromine, the brominated donor DNA where every thymine on the T-rich strand (oligos BL & BR) was replaced with 5-bromo-dU was used. All crystals were frozen in liquid N2 directly from the drop.
Numerous data sets were collected from several different beamlines at the Advanced Photon Source in Argonne, IL. Many crystals were screened at BIO-CARS 14-BM. All data sets used for the final phasing and refinement were collected at SBC-CAT 19-ID beamline at 100K temperature. For the SeMet data, data sets collected from two different crystals were merged to improve completeness of data, especially at the low resolution shells. X-ray data collected from the native and Ta-derivative crystals were integrated and scaled with HKL3000 suite and the others with HKL200052. A summary of the data collection statistics is shown in Table S1.
Structure Determination and Refinement
The toehold in solving the structure of the Mu transpososome was a single Tantalum bromide cluster (Table S1). This cluster was initially found using direct methods in SHELXD53 from a 5-day-soaked Ta-dataset, and was consistent with the anomalous difference Patterson maps54 generated from other Ta-datasets where crystals were soaked for 1, 3, 5, 7, and 8 days. SIRAS phases from this one cluster were generated using MLPHARE and were utilized in anomalous difference Fourier methods to determine the substructure for the mercury derivative55. We used SIRAS phases calculated from the Hg derivative to independently confirm the Ta site. Following several rounds of difference Fourier calculation, we were able to locate the rest of the heavy atoms. Final MIRAS phases were generated from 4 [Ta6Br12]2+, 3 Hg, and 17 Se sites. Reasonable figures of merit were obtained prior to density modification: 0.41 for centric and 0.25 for acentric reflections. With 77% solvent in the crystal, further phase improvement was achieved by density modification using Parrot56. Electron density maps generated showed clear density for the DNA as well as tubular densities that represent protein helices. The protein structure was initially modeled by docking previously determined structural domains (Iβ, Iγ, and catalytic domains) of the MuA transposase into the density using Se peaks as markers. The bromine sites, despite not being included in calculating phases due to the low resolution of that data set, were particularly useful in guiding the model building for the donor DNA. We also have a low resolution dataset from a crystal that contains a symmetric brominated target DNA:
5′-TATCGCAACAACACATCGGATGTGTTGTTGCGATA.
Bromine peaks obtained from this particular crystal was useful in confirming the location of our target DNA (Figure S4).
The transpososome lies on a crystallographic twofold axis such that the asymmetric unit comprises half of a transpososome: two MuA protomers, one Mu end DNA, and ½ target DNA. The initial model revealed possible loose contacts between two crystallographically-related copies of domain IIβ (a β-barrel). To improve the diffraction of our crystals, we engineered three sets of mutations (Quikchange, Stratagene) in that region of contact: a single M521W mutation and two double mutations, M521W/N525L and M521L/N525L. The latter led to an improvement of the resolution along the best diffracting axis of our “native dataset” from 4.2 to 3.7 Å. The structure was modeled in COOT57, and refined using PHENIX58. Density was visible for several sections that unfortunately could not be modeled due to the resolution: e.g. the linker between domains Iβ and Iγ lies in the minor groove as seen for Mos1 and CENP-B, and the region around the 3rd active site residue of the R1-bound subunits, which clearly changes conformation from the inactive form seen in isolated domain II structures. Multiple restraints were employed during refinement due to the low resolution of the data. These include H-bond restraints on the DNA base pairs, secondary structure restraints, model restraints where models of the individual domains of the MuA transposase were obtained from the PDB, NCS restraints, and Ramachandran restraints. Nine TLS groups were used: 1) R1β and the DNA with which it is interacting, (2) R2β and DNA, (3) R1γ and DNA, (4) R2γ and DNA plus R2-domain IIIα, (5) R2-domain II including the β-barrel, (6) R1-domain II without the β-barrel, (7) R1-β-barrel, (8) R1-domain IIIα, and (9) target DNA including the sequences after the CA step in the donor DNA (that do not interact directly with the DNA binding domains). As categorized by PROCHECK, the % of residues in the following regions of the Ramachandran plot were: favored/allowed/generous/disallowed = 93.8/5.4/0.2/0.5. Several variations on this protocol were tried. Simply removing the Ramachandran restraints made very little difference, probably because most of the model was already restrained to previously-determined domain structures. The unrestrained Ramachandran plot statistics were: favored/allowed/generous/disallowed = 90.1/9.0/0.4/0.5, and Rwork/Rfree were 40.1/43.6% (as opposed to 39.3/43.7% with restraints). Superimposing the two structures (refined +/− Rama restraints) revealed some slight differences in residues 347 to 356. This is the region where Ramachandran outliers were observed. However, upon inspection of the experimental map and the difference maps, it was difficult to discern which was more correct. Hence, we are choosing to report a structure that has better geometry. We also tried refinement with DEN59, but it only improved the Rfree by 0.3% and greatly degraded the Ramachandran plot. Our structure may be an unusual test case for DEN because of the low resolution of our data and the high quality of our individual domain models.
During the initial rounds of refinement, progress stalled when Rfree was ~49%. However, after ellipsoidal truncation and anisotropic scaling were performed on the native dataset using the Diffraction Anisotropy server60, the Rfree considerably improved to ~44%. The server truncated the dataset to 3.7 Å along c* and 5.2 Å along a* and b*. The final refined structure has an Rwork and Rfree of 39.30 and 43.70, respectively.
Modeling the full transpososome
To model the full complex, additional model B-form DNA coordinates was created using the W3DNA server61. DNA and protein coordinates were manipulated in both pymol and coot. Subunits R1, R2, L1, and domains II and III of the L2 subunit in the model could be taken directly from the crystal structure. To model the other subunits, we docked domain Iβ of the R2 subunit and the DNA segment it binds onto the appropriate site in the modeled DNA. The L1 and L2 binding sites are separated by an ~80bp segment where the DNA bending protein HU binds. Modeling of the HU-induced bend was based the structure of a closely-related IHF-DNA complex and on footprinting data for HU synergistically bound within this loop62,63. Modeled DNA for the L end was broken and appropriate sections abutted to the ends of the DNA in the IHF-DNA structure. We justified some additional bending of the DNA on the L2 end of the HU site based on the symmetry-related DNA in the IHF structure, and the fact that IHF- and HU-induced bends are known to be flexible. Bending of the model DNA in the L2 binding site was based on bending seen crystallographically in the R2 site. In modeling the R3-bound subunit, the other domains simply followed Iβ as a rigid body, which gives only a rough placement of domain II. For the L3-bound subunit, we modeled an interaction between its domain II and the R2 subunit’s domain Iβ based on the II – Iβ interactions seen in the crystal.
Figures were prepared using Pymol (The PyMOL Molecular Graphics System, Version 1.3 Schrödinger, LLC.)
Supplementary Material
Acknowledgments
We thank Kiyoshi Mizuuchi for initiating this project, Kerren K. Swinger and Beata Vertessy for early crystallization efforts, and Xiaojing Yang and the staff of APS beamlines 14, 19, and 21 for assistance with data collection. This work was funded in part by NIH grant GM086826 (to PAR).
Footnotes
Author Contributions: SPM carried out most of the crystallographic work, YZP grew the first diffracting transpososome crystals and assisted with all other aspects of the project, and PAR designed the project and assisted in computational work and interpretation of the results.
Author Information:
Coordinates and structure factors were deposited at the PDB, with ID 4fcy. Reprints and permissions information is available at www.nature.com/reprints. The authors have no competing financial interests.
References
- 1.Yanagihara K, Mizuuchi K. Mismatch-targeted transposition of Mu: a new strategy to map genetic polymorphism. Proc Natl Acad Sci U S A. 2002;99:11317–11321. doi: 10.1073/pnas.132403399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haapa S, Taira S, Heikkinen E, Savilahti H. An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res. 1999;27:2777–2784. doi: 10.1093/nar/27.13.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizuuchi K. In vitro transposition of bacteriophage Mu: a biochemical approach to a novel replication reaction. Cell. 1983;35:785–794. doi: 10.1016/0092-8674(83)90111-3. [DOI] [PubMed] [Google Scholar]
- 4.Montano SP, Rice PA. Moving DNA around: DNA transposition and retroviral integration. Curr Opin Struct Biol. 2011;21:370–378. doi: 10.1016/j.sbi.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 6.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson JM, Colloms SD, Finnegan DJ, Walkinshaw MD. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell. 2009;138:1096–1108. doi: 10.1016/j.cell.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi W, Harshey RM. DNA repair by the cryptic endonuclease activity of Mu transposase. Proc Natl Acad Sci U S A. 2010;107:10014–10019. doi: 10.1073/pnas.0912615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaconas G, Kennedy DL, Evans D. Predominant integration end products of infecting bacteriophage Mu DNA are simple insertions with no preference for integration of either Mu DNA strand. Virology. 1983;128:48–59. doi: 10.1016/0042-6822(83)90317-3. [DOI] [PubMed] [Google Scholar]
- 10.Lavoie BD, Chan BS, Allison RG, Chaconas G. Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J. 1991;10:3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Surette MG, Buch SJ, Chaconas G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987;49:253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- 12.Au TK, Pathania S, Harshey RM. True reversal of Mu integration. EMBO J. 2004;23:3408–3420. doi: 10.1038/sj.emboj.7600344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuuchi M, Rice PA, Wardle SJ, Haniford DB, Mizuuchi K. Control of transposase activity within a transpososome by the configuration of the flanking DNA segment of the transposon. Proc Natl Acad Sci U S A. 2007;104:14622–14627. doi: 10.1073/pnas.0706556104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kruklitis R, Welty DJ, Nakai H. ClpX protein of Escherichia coli activates bacteriophage Mu transposase in the strand transfer complex for initiation of Mu DNA synthesis. EMBO J. 1996;15:935–944. [PMC free article] [PubMed] [Google Scholar]
- 15.Levchenko I, Luo L, Baker TA. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 16.Mhammedi-Alaoui A, Pato M, Gama MJ, Toussaint A. A new component of bacteriophage Mu replicative transposition machinery: the Escherichia coli ClpX protein. Mol Microbiol. 1994;11:1109–1116. doi: 10.1111/j.1365-2958.1994.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 17.Abdelhakim AH, Oakes EC, Sauer RT, Baker TA. Unique contacts direct high-priority recognition of the tetrameric Mu transposase-DNA complex by the AAA+ unfoldase ClpX. Mol Cell. 2008;30:39–50. doi: 10.1016/j.molcel.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savilahti H, Rice PA, Mizuuchi K. The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J. 1995;14:4893–4903. doi: 10.1002/j.1460-2075.1995.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker TA, Mizuuchi K. DNA-promoted assembly of the active tetramer of the Mu transposase. Genes Dev. 1992;6:2221–2232. doi: 10.1101/gad.6.11.2221. [DOI] [PubMed] [Google Scholar]
- 20.Yuan JF, Beniac DR, Chaconas G, Ottensmeyer FP. 3D reconstruction of the Mu transposase and the Type 1 transpososome: a structural framework for Mu DNA transposition. Genes Dev. 2005;19:840–852. doi: 10.1101/gad.1291405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savilahti H, Mizuuchi K. Mu transpositional recombination: donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell. 1996;85:271–280. doi: 10.1016/s0092-8674(00)81103-4. [DOI] [PubMed] [Google Scholar]
- 22.Aldaz H, Schuster E, Baker TA. The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell. 1996;85:257–269. doi: 10.1016/s0092-8674(00)81102-2. [DOI] [PubMed] [Google Scholar]
- 23.Krementsova E, Giffin MJ, Pincus D, Baker TA. Mutational analysis of the Mu transposase. Contributions of two distinct regions of domain II to recombination. J Biol Chem. 1998;273:31358–31365. doi: 10.1074/jbc.273.47.31358. [DOI] [PubMed] [Google Scholar]
- 24.Namgoong SY, Sankaralingam S, Harshey RM. Altering the DNA-binding specificity of Mu transposase in vitro. Nucleic Acids Res. 1998;26:3521–3527. doi: 10.1093/nar/26.15.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou AH, Leung PC, Harshey RM. Transposase contacts with mu DNA ends. J Biol Chem. 1991;266:20476–20482. [PubMed] [Google Scholar]
- 26.Tanaka Y, et al. Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 2001;20:6612–6618. doi: 10.1093/emboj/20.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watkins S, van Pouderoyen G, Sixma TK. Structural analysis of the bipartite DNA-binding domain of Tc3 transposase bound to transposon DNA. Nucleic Acids Res. 2004;32:4306–4312. doi: 10.1093/nar/gkh770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craigie R, Mizuuchi M, Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984;39:387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- 29.Kuo CF, Zou AH, Jayaram M, Getzoff E, Harshey R. DNA-protein complexes during attachment-site synapsis in Mu DNA transposition. EMBO J. 1991;10:1585–1591. doi: 10.1002/j.1460-2075.1991.tb07679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizuuchi M, Baker TA, Mizuuchi K. DNase protection analysis of the stable synaptic complexes involved in Mu transposition. Proc Natl Acad Sci U S A. 1991;88:9031–9035. doi: 10.1073/pnas.88.20.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice P, Mizuuchi K. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell. 1995;82:209–220. doi: 10.1016/0092-8674(95)90308-9. [DOI] [PubMed] [Google Scholar]
- 32.Wu Z, Chaconas G. A novel DNA binding and nuclease activity in domain III of Mu transposase: evidence for a catalytic region involved in donor cleavage. EMBO J. 1995;14:3835–3843. doi: 10.1002/j.1460-2075.1995.tb00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdelhakim AH, Sauer RT, Baker TA. The AAA+ ClpX machine unfolds a keystone subunit to remodel the Mu transpososome. Proc Natl Acad Sci U S A. 2010;107:2437–2442. doi: 10.1073/pnas.0910905106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burton BM, Baker TA. Mu transpososome architecture ensures that unfolding by ClpX or proteolysis by ClpXP remodels but does not destroy the complex. Chem Biol. 2003;10:463–472. doi: 10.1016/s1074-5521(03)00102-9. [DOI] [PubMed] [Google Scholar]
- 35.Naigamwalla DZ, Coros CJ, Wu Z, Chaconas G. Mutations in domain III α of the Mu transposase: evidence suggesting an active site component which interacts with the Mu-host junction. J Mol Biol. 1998;282:265–274. doi: 10.1006/jmbi.1998.2011. [DOI] [PubMed] [Google Scholar]
- 36.Yang JY, Kim K, Jayaram M, Harshey RM. A domain sharing model for active site assembly within the Mu A tetramer during transposition: the enhancer may specify domain contributions. EMBO J. 1995;14:2374–2384. doi: 10.1002/j.1460-2075.1995.tb07232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Surette MG, Chaconas G. The Mu transpositional enhancer can function in trans: requirement of the enhancer for synapsis but not strand cleavage. Cell. 1992:68. doi: 10.1016/0092-8674(92)90081-m. [DOI] [PubMed] [Google Scholar]
- 38.Mizuuchi M, Mizuuchi K. Conformational isomerization in phage Mu transpososome assembly: effects of the transpositional enhancer and of MuB. EMBO J. 2001;20:6927–6935. doi: 10.1093/emboj/20.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harshey RM, Jayaram M. The mu transpososome through a topological lens. Crit Rev Biochem Mol Biol. 2006;41:387–405. doi: 10.1080/10409230600946015. [DOI] [PubMed] [Google Scholar]
- 40.Craigie R, Mizuuchi K. Role of DNA topology in Mu transposition: mechanism of sensing the relative orientation of two DNA segments. Cell. 1986;45:793–800. doi: 10.1016/0092-8674(86)90554-4. [DOI] [PubMed] [Google Scholar]
- 41.Surette MG, Chaconas G. A protein factor which reduces the negative supercoiling requirement in the Mu DNA strand transfer reaction is Escherichia coli integration host factor. J Biol Chem. 1989;264:3028–3034. [PubMed] [Google Scholar]
- 42.Allison RG, Chaconas G. Role of the A protein-binding sites in the in vitro transposition of Mu DNA. A complex circuit of interactions involving the Mu ends and the transpositional enhancer. J Biol Chem. 1992;267:19963–19970. [PubMed] [Google Scholar]
- 43.Jiang H, Yang JY, Harshey RM. Criss-crossed interactions between the enhancer and the att sites of phage Mu during DNA transposition. EMBO J. 1999;18:3845–3855. doi: 10.1093/emboj/18.13.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Craig NL. Mobile DNA II. ASM Press; 2002. [Google Scholar]
- 45.Pribil PA, Haniford DB. Target DNA bending is an important specificity determinant in target site selection in Tn10 transposition. J Mol Biol. 2003;330:247–259. doi: 10.1016/s0022-2836(03)00588-6. [DOI] [PubMed] [Google Scholar]
- 46.Swinger KK, Rice PA. Structure-based analysis of HU-DNA binding. J Mol Biol. 2007;365:1005–1016. doi: 10.1016/j.jmb.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levchenko I, Yamauchi M, Baker TA. ClpX and MuB interact with overlapping regions of Mu transposase: implications for control of the transposition pathway. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- 48.Clubb RT, Schumacher S, Mizuuchi K, Gronenborn AM, Clore GM. Solution structure of the Iγ subdomain of the Mu end DNA-binding domain of phage Mu transposase. J Mol Biol. 1997;273:19–25. doi: 10.1006/jmbi.1997.1312. [DOI] [PubMed] [Google Scholar]
- 49.Schumacher S, et al. Solution structure of the Mu end DNA-binding Iβ subdomain of phage Mu transposase: modular DNA recognition by two tethered domains. EMBO J. 1997;16:7532–7541. doi: 10.1093/emboj/16.24.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker TA, Mizuuchi M, Savilahti H, Mizuuchi K. Division of labor among monomers within the Mu transposase tetramer. Cell. 1993;74:723–733. doi: 10.1016/0092-8674(93)90519-v. [DOI] [PubMed] [Google Scholar]
- 51.Ducruix A, Giegg R. In: Preparation of selenomethionyl protein crystals. Dublie S, Carter CWJ, editors. Oxford University Press; 1992. [Google Scholar]
- 52.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter CW Jr, Sweet RM, editors. Vol. 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 53.Sheldrick G. A short history of SHELX. Acta Crystallographica Section A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 54.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 55.CCP4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallographica Section D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 56.Zhang KY, Cowtan K, Main P. Combining constraints for electron-density modification. Methods Enzymol. 1997;277:53–64. doi: 10.1016/s0076-6879(97)77006-x. [DOI] [PubMed] [Google Scholar]
- 57.Emsley P, Lohkamp B, Scott W, Cowtan K. Features and Development of Coot. Acta Crystallographica Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schroder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng G, Lu XJ, Olson WK. Web 3DNA--a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009;37:W240–246. doi: 10.1093/nar/gkp358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lavoie BD, Chaconas G. Site-specific HU binding in the Mu transpososome: conversion of a sequence-independent DNA-binding protein into a chemical nuclease. Genes Dev. 1993;7:2510–2519. doi: 10.1101/gad.7.12b.2510. [DOI] [PubMed] [Google Scholar]
- 63.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr Opin Struct Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.