Abstract
Objectives
To summarize existing evidence about whether the presence of mutant or upregulated p53 is a prognostic factor for patients presenting with squamous cell carcinoma arising from the larynx, oropharynx, hypopharynx, or oral cavity.
Method
Relevant articles were identified using strict criteria for systematic searches. Associations between mutant or upregulated p53 versus wild-type or low/undetectable p53 in relation to overall survival and DFS were summarized by extracting or deriving hazard ratio (HR) estimates. Random-effects meta-analyses were used to account for between-study heterogeneity and to summarize the effect of p53 across studies.
Results
The meta-analyses gave a statistically significant pooled HR for overall survival in oral cavity [pooled HR, 1.48; 95% confidence interval, (95% CI), 1.03-2.11], and for disease-free survival in oral cavity (pooled HR, 1.47; 95% CI, 1.12-1.93) and in oropharynx (pooled HR, 0.45; 95% CI, 0.27-0.73). Despite attempts to limit it, between-study heterogeneity was large in the majority of meta-analyses and the prognostic value of p53 was generally inconsistent and inconclusive across studies.
Conclusion
The meta-analysis results highlight that current evidence about the prognostic value of p53 in patients with squamous cell carcinaoma of the head and neck is inconclusive. Large heterogeneity exists across studies in study-level and patient-level characteristics, making it difficult to ascertain a clear picture. Future studies are required in which p53 expression is investigated in a more standardized and biologically informative manner. In particular, prospectively planned individual patient data meta-analyses are needed to establish the prognostic importance of p53 for specific subgroups of patients undergoing specific treatments.
Introduction
Loss of function of the p53 tumor suppressor gene plays an important role in the development of cancer (1). p53 regulates the activity of pathways, which lead variously to cell cycle arrest, DNA repair, senescence, or apoptosis following exposure of cells to endogenous or exogenous cellular stresses (2). With respect to cancer, those stresses that cause DNA damage are of particular interest. p53 has been extensively investigated across the whole spectrum of human malignancy, with mutations of the p53 gene (TP53) found in over 50% of cancers (3). p53 gene status in tumors such as carcinomas of the bladder affects upon disease outcome—mutations are linked with poorer patient survival, and this is likely due, in part, to reduced susceptibility of such cells to apoptosis (4).
Head and neck cancer accounts for 3% of incident cancer in the United Kingdom, with squamous cell carcinoma (SCC) being the commonest histologic type. The larynx, oral cavity, oropharynx, and hypopharynx comprise the four main anatomic sites within the head and neck, accounting for 90% of SCC cases (5). Smoking, alcohol, diet, and presence of the human papilloma virus (HPV) have all been identified as etiologic factors in the development of SCC of the head and neck (SCCHN; refs. 6, 7). Whatever the etiology, p53 overexpression (which is usually indicative of the presence of mutant p53) is also commonly seen in SCCHN, which is to be expected given an overall cited UK mutation prevalence of 50.7%.5 However, despite this knowledge and despite the fact that the presence of p53 expression and its effect on prognosis in SCCHN has been widely investigated in the published literature, no consensus exists about its true clinical role in determining key parameters such as treatment responses or prognostic value. Existing studies are often small, heterogeneous (e.g., with respect to the primary tumor site and outcomes assessed), and often conflicting in their results, making it difficult to assess the clinical benefit of p53. Moreover, in many studies, the method used (immunohistochemistry) is often performed in a manner that is inherently ineffective at interrogating p53 function. This is disappointing because superior strategies based on the same simple technology have been widely known for several years (8).
The potential limitations of undertaking such a review were previously shown by Kyzas et al. in 2005 (9). They showed that the association between p53 status and mortality varied widely depending on whether indexed, published, or unpublished data were included in the meta-analysis. Such variation ranged from a risk ratio (RR) of 1.38 [95% confidence interval (CI), 1.13-1.67; P = 0.001], to 1.23 (95% CI, 1.03-1.47; P = 0.02), and to 1.16 (95% CI, 0.99-1.35; P = 0.06) when indexed, published, or unpublished data were considered, respectively. These authors examined reports of p53 status in SCCHN tumors, and their meta-analysis of the data highlighted some important limitations and potential problems in such studies of p53 status compared with clinical outcomes. Although Kyzas et al. (9) reported results for tumors of the larynx and oropharynx separately, they did not include tumors originating from the other two main anatomic subsites from which SCCHN can arise, namely the oral cavity and hypopharynx. Due to the heterogeneous biological behavior and response to treatment of tumors originating from different subsites (10), it is essential to consider tumors originating from each site separately to avoid such confounding factors in this kind of meta-analysis. Furthermore, Kyzas et al. (9) gave priority to data from immunohistochemical (IHC) analyses when both IHC and genetic data (from PCR/single-strand conformational polymorphism) were available and used 2-year overall survival (OS) as their outcome indicator. Although IHC is the most commonly used method to examine such phenotype/outcome questions and has the merit of permitting the largest number of studies to be compared using a single method for p53 analysis, when performed for p53 alone—as it routinely is—it is not a reliable indicator of p53 status (as discussed below). Similarly, although OS at 2 years has the advantage of being readily obtainable information, disease-free survival (DFS) is arguably a more relevant outcome in this context.
In this article, we report our results from a systematic review and meta-analysis of existing studies specifically to investigate whether p53 is a prognostic factor in SCCHN in relation to OS and DFS for each of the major primary tumor sites. This allows us to produce evidence-based results for the prognostic value of p53 in SCCHN, to consider how heterogeneity across studies affects the observed prognostic effect of p53, and to identify further research needs.
Materials and Methods
Search Strategy
Medline (1966-2007), the Cochrane Central Register of Controlled Trials (1993-2007), and Embase (1974-2007) were systematically searched for relevant articles. The search strategy can be seen in Table 1. Conference abstracts, dissertations, and theses were reviewed for unpublished material; these include the American Society of Clinical Oncology (2000-2007), ISI Proceedings (1990 to present), and the ProQuest Dissertations & Theses Database (1637 to present). The bibliographies of retrieved articles were also reviewed for any additional studies. Search terms used were as follows:
p53
Tumour suppressor protein p53
Palatal neoplasm, cancer, or carcinoma
Tongue neoplasm, cancer, or carcinoma
Laryngeal neoplasm, cancer, or carcinoma
Hypopharyngeal neoplasm, cancer, or carcinoma
Oropharyngeal neoplasm, cancer, or carcinoma
Tonsillar neoplasm, cancer, or carcinoma
Oral cavity neoplasm, cancer, or carcinoma
Mouth neoplasm, cancer, or carcinoma
Head and neck squamous cell carcinoma
Neck metastasis
Table 1. Medline and Embase search strategy.
| 1. | SEARCH: | P53.TI,AB. |
| 2. | SEARCH: | PROTEIN-P53.DE. OR TUMOUR-SUPPRESSOR PROTEIN-P53.DE |
| 3. | SEARCH: | LARYNX-CANCER.DE. OR LARYNX-CARCINOMA.DE. |
| 4. | SEARCH: | LARYNX ADJ CANCER.TI,AB. |
| 5. | SEARCH: | LARYNX NEAR CANCER.TI,AB. |
| 6. | SEARCH: | LARYNX NEAR NEOPLASM.TI,AB. |
| 7. | SEARCH: | OROPHARYNX-CANCER.DE. OR OROPHARYNX-CARCINOMA.DE. |
| 8. | SEARCH: | OROPHARYNX NEAR CANCER.TI,AB. |
| 9. | SEARCH: | OROPHARYNX NEAR NEOPLASM.TI,AB. |
| 10. | SEARCH: | HYPOPHARYNX ADJ CANCER.TI,AB. |
| 11. | SEARCH: | HYPOPHARYNX-CANCER.DE. OR HYPOPHARYNX-CARCINOMA.DE. |
| 12. | SEARCH: | HYPOPHARYNX NEAR CANCER.TI,AB. |
| 13. | SEARCH: | HYPOPHARYNX NEAR NEOPLASM.TI,AB. |
| 14. | SEARCH: | ORAL ADJ CAVITY ADJ CANCER.TI,AB. |
| 15. | SEARCH: | ORAL ADJ CAVITY NEAR CANCER.TI,AB. |
| 16. | SEARCH: | ORAL ADJ CAVITY NEAR NEOPLASM.TI,AB. |
| 17. | SEARCH: | ORAL ADJ CAVITY-CANCER.DE. OR ORAL ADJ CAVITY-CARCINOMA.DE. |
| 18. | SEARCH: | MOUTH-CANCER.DE. OR MOUTH-CARCINOMA.DE |
| 19. | SEARCH: | MOUTH ADJ CANCER.TI,AB. |
| 20. | SEARCH: | MOUTH NEAR CANCER.TI,AB. |
| 21. | SEARCH: | MOUTH NEAR NEOPLASM.TI,AB. |
| 22. | SEARCH: | TONGUE ADJ CANCER.TI,AB. |
| 23. | SEARCH: | TONGUE-CANCER.DE. OR TONGUE-CARCINOMA.DE. |
| 24. | SEARCH: | TONGUE NEAR CANCER.TI,AB. |
| 25. | SEARCH: | TONGUE NEAR NEOPLASM.TI,AB. |
| 26. | SEARCH: | TONSIL ADJ CANCER.TI,AB. |
| 27. | SEARCH: | TONSIL-CANCER.DE. OR TONSIL-CARCINOMA.DE. |
| 28. | SEARCH: | TONSIL NEAR CANCER.TI,AB. |
| 29. | SEARCH: | TONSIL NEAR NEOPLASM.TI,AB. |
| 30. | SEARCH: | NECK ADJ CANCER.TI,AB. |
| 31. | SEARCH: | NECK-CANCER.DE. |
| 32. | SEARCH: | NECK NEAR CANCER.TI,AB. |
| 33. | SEARCH: | NECK NEAR NEOPLASM.TI,AB. |
| 34. | SEARCH: | HEAD-AND-NECK-SQUAMOUS-CELL-CARCINOMA.DE. |
| 35. | SEARCH: | NECK-METASTASIS.DE. |
| 36. | SEARCH: | 1 OR 2 |
| 37. | SEARCH: | 3 OR 4 OR 5 OR 6 OR 7 OR 8 OR 9 OR 10 OR 11 OR 12 OR 13 OR 14 OR 15 OR 16 |
| 38. | SEARCH: | 17 OR 18 OR 19 OR 20 OR 21 OR 22 OR 23 OR 24 OR 25 OR 26 OR 27 OR 28 OR 29 OR 30 OR 31 OR 32 OR 33 OR 34 |
| 39. | SEARCH: | 37 OR 38 |
| 40. | SEARCH: | 36 AND 39 |
Inclusion and Exclusion Criteria
The results from these searches were reviewed by title and abstract by two independent reviewers. Studies not written in English were excluded. If studies seemed appropriate, the full manuscript was scrutinized and the study deemed “relevant” if it met the following inclusion criteria:
Primary tumors were biopsy-proven nontreated SCCHN from the larynx, oropharynx, hypopharynx, or oral cavity (cutaneous and lip SCC, nasopharyngeal carcinoma, and cases of tumor recurrence were excluded).
Treatment modalities used included radiotherapy, chemotherapy, surgery, or a combination of these.
Studies assessed p53 status in pretreatment tumor samples and compared this with survival outcomes, and sufficient data were available to allow meaningful statistical analysis.
Survival outcomes were of a minimum 2-y follow-up after primary treatment.
These strict inclusion criteria were used to limit heterogeneity across studies and thus facilitate more clinically meaningful meta-analysis results. Between-study heterogeneity is a known problem for meta-analysis of prognostic marker studies (11), and we wanted to limit this problem from the outset by focusing on clinically similar studies with good follow-up that measured p53 at the same pretreatment baseline point and allowed primary tumor sites to be distinguished.
Outcomes
The primary outcome measures used were OS and DFS. As defined in our study, OS is the measure of the proportion of people within a treatment group who are expected to be alive after a specified time. It takes into account death due to any cause—both related and unrelated to the cancer in question. DFS, on the other hand, measures the proportion of people among those treated for a cancer that will remain free of disease at a specified time after treatment.
Data Extraction
Data were extracted from the full-published article for each study and these were collated in a Microsoft Excel worksheet. Site and stage of tumor, primary treatment, and number of cases analyzed for p53 were all recorded. The method of p53 analysis used was categorized as IHC or mutational analysis. IHC data recorded included the antibody used for analysis and the cutoff level used to dichotomize p53-positive (high expression) versus p53 negative (low or undetectable expression) cases. Mutational analysis data included method of analysis—single-strand conformational polymorphism with direct sequencing—and the specific exons investigated. Authors of trials were contacted and asked to supply further data wherever necessary.
The recommended summary statistics for meta-analysis of time-to-event data (OS and DFS) are the log(hazard ratio) and its variance, which account for both the time it takes for an event to occur, as well as censoring. As many trials do not report this information directly (12), appropriate data such as log-rank test P values were extracted to allow estimation of the log(hazard ratio) and its variance using previously reported methods (13). For the studies (n = 6) that reported P values as less than a given significance (e.g., P < 0.05) rather than reporting the exact P value, the value reported as less than was used to calculate the estimated hazard ratio (HR) for that study group. If only Kaplan Meier graphs were published, the “Mouseyes” image digitization and linear separation program (14) was used to extract data directly from the survival curves (15). For the purposes of this review, only HRs from unadjusted analyses were included; we did not use results from adjusted analyses (that is, those that assessed p53 in a model that adjusted for other prognostic factors) because there was clearly a large heterogeneity across studies in the sets of adjustment factors that were used, and thus, meta-analysis would have been more difficult to interpret. Where relevant extractable data were not present (e.g., an unadjusted HR), the primary author was contacted requesting this for the purposes of our analysis. Results from studies by the same authors were closely scrutinized to avoid repetition of data in the meta-analysis. If results for distinct patient groups were presented in the same article, for example when results were separated by stage or treatment type, these groups were each included in the meta-analysis as separate entries. Three authors verified the accuracy of data extraction from included articles independently. We used a software (version 3.0; 28 Sept, 2004) developed by Matthew Sydes and Jayne Tierney of the MRC Clinical Trials Unit, London, to undertake calculations to estimate the trial level log(hazard ratio) and variance based on the summary data extracted from publications.
Meta-analysis
Primary Analyses
Trial level log(hazard ratios) and their variances were pooled using an inverse variance-weighted random effects meta-analysis and results presented graphically in the form of a Forest Plot with pooled HR and 95% CI. The meta-analysis was undertaken using Review Manager (RevMan) version 5.0 (Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2008). A random-effects meta-analysis here estimates and accounts for any between-study heterogeneity, estimates the average (“pooled”) HR across studies and its CI, and provides a prediction interval for the HR in a future p53 study (16). It thus provides an important summary of the current evidence base for p53, and we were able to apply it to both outcomes (OS and DFS) separately for the primary tumor sites of larynx, oral cavity, and oropharynx, and for just the OS outcome for the hypopharynx.
In addition, in which sufficient data were available for each primary site individually, meta-analysis for OS and DFS for early and advanced stage disease was calculated for each site. The effect of between-study heterogeneity in our meta-analyses was assessed by the I2 statistic (17). I2 describes the proportion of total variation in meta-analysis estimates, which is due to interstudy heterogeneity rather than sampling error and is measured from 0% to 100%, with increasing I2 values indicating a larger effect of between-study heterogeneity in the meta-analysis.
Subgroup Analyses
To investigate the effect of study design and stage of disease on the p53 results, we also performed subgroup analyses grouping studies that (a) prospectively or retrospectively collected data, and (b) included patients with advanced stage only compared with early stage only. A test for subgroup differences was calculated as described by Deeks et al. (18).
Publication Bias
For those meta-analyses including 10 or more studies, we assessed the possibility of publication bias by visually assessing a funnel plot for asymmetry and by performing Egger’s test for such asymmetry with a 10% significance level due to the low power of this test (19).
Results
Search Results
Our search strategy identified 1,548 articles from Embase, 948 articles from Medline, 16 articles from the Cochrane library of randomized control trials, 3 articles from conference proceedings, and 109 articles from the bibliographies of retrieved articles. Following deduplication, two reviewers independently screened the titles and abstracts identified. They subsequently agreed that 269 studies should be retrieved for detailed review. Fortyone relevant studies were eligible for inclusion (the other 228 were excluded for several reasons; Fig. 1) and the characteristics of these studies can be seen below. No unpublished work was included.
Awwad; mixed stage larynx, 40 cases; oral cavity, 14 cases; oropharynx, 14 cases treated with radiotherapy; retrospective IHC study, cutoff is any staining; OS and DFS (20)
Buyukbayram; mixed stage larynx, 60 cases; retrospective IHC study, 20% cutoff; OS and DFS (21)
Caminero; mixed stage oropharynx, 106 cases treated with surgery with or without radiotherapy; retrospective IHC study, 10% cutoff; OS (22)
Chien; mixed stage hypopharynx, 58 cases treated with surgery; retrospective IHC study, 50% cutoff; OS (23)
Cho; stage I/II oral cavity, 33 cases treated with surgery with or without radiotherapy; prospective IHC study, 50% cutoff; DFS (24)
Chomchai; mixed stage larynx, 45 cases mixed treatment; retrospective mutation analysis exons (5-8); OS & DFS (25)
Costa; oral cavity, 51 cases treated with chemoradiation or surgery; prospective IHC study, cutoff not given; DFS (26)
de Aguiar; mixed stage oral cavity, 81 cases treated with surgery ± radiotherapy; retrospective IHC study, 5% cutoff; OS (27)
Fan; mixed stage larynx, 109 cases mixed treatment; retrospective IHC study, 10% cutoff; OS and DFS (28)
Frank; mixed stage hypopharynx, 43 cases mixed treatment; retrospective IHC study, strong (10%) cutoff; OS (29)
Friedman; stage III/IV larynx, 69 cases mixed treatment; retrospective IHC study, 10% cutoff; OS (30)
Friesland; stage III/IV oropharynx, 37 cases mixed treatment; retrospective IHC study, 5% cutoff; OS (31)
Grabenbauer; mixed stage oropharynx, 84 cases treated with surgery; prospective IHC study, 10% cutoff; OS and DFS (32)
Hiranuma; mixed stage oral cavity, 45 cases mixed treatment; prospective mutation analysis exons (5-8); OS (33)
Hirvikoski; mixed stage larynx, 99 cases mixed treatment; retrospective IHC study, 20% cutoff; OS (34)
Homma; mixed stage larynx, 59 cases treated with chemoradiation; prospective IHC study, 10% cutoff; OS (35)
Jackel; mixed stage larynx, 88 cases treated with surgery with or without radiotherapy; retrospective IHC study, cutoff any staining; OS (36)
Jayasurya; mixed stage oral cavity, 316 cases mixed treatment; prospective IHC study, cutoff median staining; OS and DFS (37)
Jeannon; mixed stage larynx, 60 cases treated with radiotherapy; retrospective IHC study, 25% cutoff; OS (38)
Kozomara; mixed stage oral cavity, 42 cases treated with surgery & radiotherapy; prospective mutation analysis exons 4-8; OS (39)
Miyahara; mixed stage hypopharynx, 28 cases; IHC study, 5% cutoff; OS (40)
Nadal; mixed stage larynx, 89 cases treated with surgery; prospective IHC study, cutoff any staining; OS & DFS (41)
Nakashima; mixed stage larynx, 92 cases; IHC study. 1% cutoff; OS (42)
Narayana; stage I larynx, 102 cases treated with radiotherapy; retrospective IHC study, 10% cutoff; DFS (43)
Nibu; stage III/IV hypopharynx, 53 cases treated with radiotherapy and surgery; retrospective IHC study, 10% cutoff; OS (44)
Obata; mixed stage oropharynx, 33 cases mixed treatment; IHC study, 10% cutoff; OS (45)
Otero; stage III/IV oropharynx, 34 cases mixed treatment, retrospective IHC study, 10% cutoff; OS and DFS (46)
Pai; stage I/II larynx, 86 cases treated with radiotherapy; retrospective IHC study, cutoff any staining; DFS (47)
Parikh; stage I/II larynx, 111 cases treated with radiotherapy; retrospective IHC study, 20% cutoff; OS and DFS (48)
Pruneri; mixed stage larynx, 82 cases treated with surgery; mutation analysis exons 5-8; OS & DFS (49)
Pulkkinen; mixed stage larynx, 62 cases mixed treatment; retrospective IHC study, cutoff intermediate staining; OS and DFS (50)
Rowley; mixed stage larynx, 35 cases treated with radiotherapy; retrospective IHC study, cutoff any staining; OS (51)
Russo; stage III/IV larynx, 81 cases treated with surgery; prospective mutation analysis exons 5 to 8; OS and DFS (52)
Shigyo; mixed stage larynx, 32 cases treated with radiotherapy or surgery; prospective IHC study, 10% cutoff; DFS (53)
Siegelmann; oral cavity, 45 cases; retrospective IHC study, 20% cutoff; OS and DFS (54)
Sittel; stage I/II larynx, 47 cases treated with laser surgery; retrospective IHC study, 10% cutoff; DFS (55)
Tan; mixed larynx, 90 cases treated with radiotherapy; retrospective IHC study, cutoff is strong staining; OS (56)
Tatemoto; mixed stage oral cavity, 150 cases treated with chemoradiation with or without surgery; IHC study, 10% cutoff; OS (57)
Tsuji; oral cavity, 50 cases; IHC study, cutoff any staining; OS (58)
Vielba; mixed surgery larynx, 62 cases mixed treatment, retrospective IHC study, 5% cutoff; OS and DFS (59)
Yanamoto; mixed stage oral cavity, 69 cases treated with surgery, IHC study, 10% cutoff; OS (60)
Figure 1.
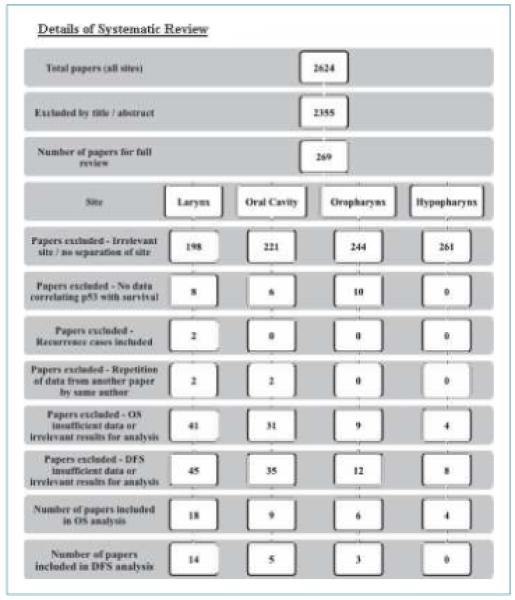
Details of systematic review.
Primary Meta-analysis Results
Forrest plots of the primary meta-analyses for the individual primary tumor sites can be seen in Figs. 2 and 3. Table 2 reports the average (pooled) HR and its 95% CI for each of the meta-analyses. HR values of >1 indicate a poorer prognosis for overexpressing or mutant p53 tumors. There was some evidence from the meta-analyses that p53 status may provide prognostic information. The pooled HR was statistically significant for OS (pooled HR, 1.48; 95% CI, 1.03-2.11) and DFS (pooled HR, 1.47; 95% CI, 1.12-1.93) in relation to oral cavity, and for DFS in relation to oropharynx (pooled HR, 0.45; 95% CI, 0.27-0.73).
Figure 2.

A, hypopharynx overall survival forrest plot. B, larynx overall survival forrest plot. C, oral cavity overall survival forrest plot. D, oropharynx overall survival forrest plot.
Figure 3.
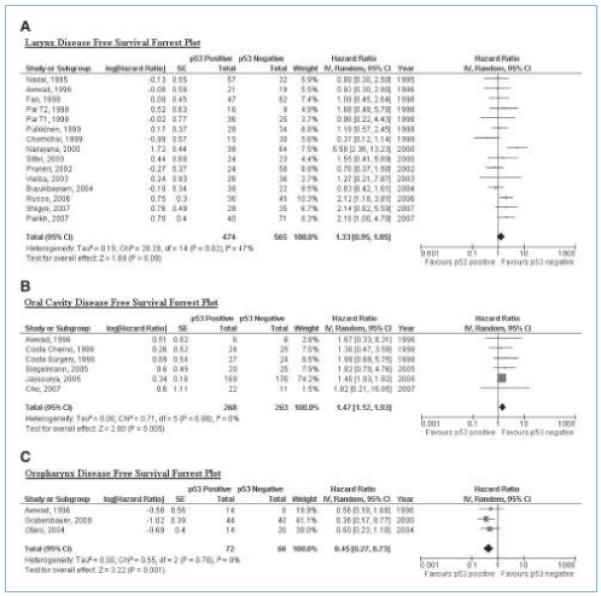
A, larynx disease free survival forrest plot. B, oral cavity disease free survival forrest plot. C, oropharynx disease free survival forrest plot.
Table 2.
Meta-analysis results
| Site | No. of included articles |
No. of cases |
Pooled HR (95% CI) |
I2 % | 95% prediction interval for the underlying HR in a future study |
Test for subgroup differences (χ2, df, P) |
|---|---|---|---|---|---|---|
| Primary analyses: | ||||||
| OS | ||||||
| Larynx | 18 | 1,333 | 0.88 (0.65-1.21) | 52 | 0.31-2.52 | — |
| Oral cavity | 9 | 811 | 1.48 (1.03-2.11) | 50 | 0.57-3.86 | — |
| Oropharynx | 6 | 304 | 1.09 (0.56-2.12) | 77 | 0.12-9.53 | — |
| Hypopharynx | 4 | 182 | 2.10 (0.63-7.04) | 87 | 0.01-578 | — |
| DFS | ||||||
| Larynx | 14 | 1,039 | 1.33 (0.95-1.85) | 47 | 0.49-3.63 | — |
| Oral cavity | 5 | 531 | 1.47 (1.12-1.93) | 0 | NA | — |
| Oropharynx | 3 | 140 | 0.45 (0.27-0.73) | 0 | NA | — |
| Subgroup analyses: | ||||||
| Larynx OS | ||||||
| Prospective | 3 | 229 | 1.87 (1.00-3.51) | 25 | 0.01-439 | 12.26 (df = 1) |
| Retrospective | 13 | 930 | 0.74 (0.53-1.02) | 40 | 0.31-1.79 | P = 0.0005 |
| Early stage only | 1 | 111 | 0.54 (0.28-1.06) | NA | Not estimable | 8.61 (df = 1) |
| Advanced stage only | 2 | 261 | 1.74 (0.65-4.65) | 69 | Not estimable | P = 0.003 |
| Larynx DFS | ||||||
| Prospective | 3 | 233 | 1.80 (1.12-2.89) | 6 | 0.06-51.94 | 2.07 (df = 1) |
| Retrospective | 11 | 806 | 1.24 (0.83-1.84) | 50 | 0.38-3.98 | P = 0.15 |
| Early stage only | 4 | 346 | 2.32 (1.30-4.15) | 31 | 0.51-10.48 | NA |
| Advanced stage only | 0 | — | — | — | — | — |
| Oral cavity OS | ||||||
| Prospective | 3 | 402 | 1.41 (0.66-3.03) | 69 | 0-8102 | 0.95 (df = 1) |
| Retrospective | 3 | 140 | 1.29 (0.56-2.95) | 52 | 0-6844 | P = 0.33 |
| Oral cavity DFS | ||||||
| Prospective | 3 | 472 | 1.44 (1.08-1.91) | 0 | NA | 0.23 (df = 1) |
| Retrospective | 2 | 59 | 1.78 (1.09-1.94) | 0 | NA | P = 0.63 |
| Oropharynx DFS | ||||||
| Early stage only | 0 | — | — | — | — | NA |
| Advanced stage only | 2 | 59 | 0.69 (0.32-1.48) | 18 | Not estimable | — |
NOTE: A HR of <1 indicates survival advantage for positive p53 status. A HR of >1 indicates survival advantage for negative p53 status.
Abbreviations: df, degrees of freedom; NA, not applicable—the between-study heterogeneity was estimated as zero so the pooled HR and its 95% CI alone are a suitable summary here.
However, despite our attempts to limit between-study heterogeneity through our strict inclusion criteria and by separating results by tumor site, there was still a large between-study heterogeneity in the effect of p53 status on the majority of meta-analyses, with I2 generally toward 50% or above (Table 2). This means that the underlying HR for upregulated/mutant p53 versus wild-type or low/undetectable p53 actually varies across studies. Only in the oropharynx and oral cavity DFS meta-analysis was I2 equal to zero, such that the between-study variance was estimated as zero. Therefore, only in these latter analyses can the pooled HR and its 95% CI be considered a reasonably reliable summary of the evidence.
In all the other meta-analyses that exhibit heterogeneity, it is clinically and statistically more informative to focus on the estimated 95% prediction interval for the underlying HR in a future study (16); this tells us the range of possible HR values in a new study that chooses its own particular set of study characteristics (e.g., cutoff level, treatment, etc.). All the calculated 95% prediction intervals in these meta-analyses contained the null value of 1 (Table 2), indicating that there is no significant evidence that p53 status maintains prognostic value regardless of the between-study heterogeneity. For example, the prediction interval for OS in the oral cavity site is from 0.57 to 3.86, indicating that given the heterogeneity, we are not sure whether upregulated/mutant or wild-type, low/undetectable p53 values indicate a worse prognosis, or indeed whether p53 has prognostic value at all. Further research is clearly needed to establish more precisely how study-level and patient-level characteristics are causing heterogeneity, and how they are modifying the prognostic importance of p53 (see Discussion).
Publication Bias
Due to the small number of studies in most meta-analyses, it was only sensible to examine the potential for publication bias in the larynx OS and DFS meta-analyses, which both contained >10 studies. There was no clear evidence of funnel plot asymmetry for either OS or DFS, either visually or from Egger’s test (OS: P = 0.95; DFS: P = 0.61), and thus, there was no clear evidence of publication bias.
Subgroup Analyses for Retrospective versus Prospective Studies
Only for the larynx and oral cavity tumor sites were sufficient articles available to compare OS and DFS between retrospective and prospective studies (Supplementary Fig. S1; Table 2). The subgroup analyses for OS excluded two studies from the larynx analysis and three from the oral cavity analysis due to lack of information to classify studies as retrospective or prospective. A statistically significant difference between the pooled results of prospective and retrospective studies was identified for the larynx OS analysis (P = 0.0005) with pooled results from three prospective studies, suggesting increased length of survival for p53-negative status (HR, 1.87; 95% CI, 1.00, 3.51), and pooled results from 13 retrospective studies, suggesting an opposite effect in favor of p53-positive status (HR, 0.74; 95% CI, 0.53-1.02). However, heterogeneity between study results still remains within each subgroup and results should be interpreted cautiously. Statistically significant differences between the two subgroup pooled results were not identified for the other analyses (Table 2).
Subgroup Analyses for Advanced versus Early Stage
Only for the larynx tumor site were sufficient articles available to compare OS between studies with early and advanced stage patients (Supplementary Fig. S2; Table 2). However, this subgroup analysis only includes 3 of the original 18 studies with the OS data available and should be interpreted very cautiously. A statistically significant difference between the pooled results of advanced and early-stage tumors was identified for the larynx OS analysis (P = 0.003) with pooled results from two advanced stage studies, suggesting no significant difference in length of survival for p53-negative status (HR, 1.74; 95% CI, 0.65-4.65), and results from one study in early-stage tumors, suggesting an effect (nonsignificant) in favor of p53-positive status (HR, 0.54; 95% CI, 0.28-1.06). Data were also available from two advanced stage studies (OS oropharynx) and early early-stage studies (DFS Larynx), and are summarized in Table 2 and Supplementary Figs. S3 and S4, respectively.
Discussion
The prognostic value of p53 in SCCHN has long been debated. When considering SCCHN, there is ample evidence of differing biological behavior and response to treatment depending on the primary site (10). Consequently, in this article, we have systematically reviewed whether p53 status is a prognostic factor for DFS and OS in patients presenting with carcinomas of the four main primary head and neck tumor sites, and ultimately included 41 studies in our review. In doing so, it is important to note that we excluded a large number of articles, which reported p53 status with survival outcomes, but without separation of the individual tumor sites. One such recent study was a prospective, multicenter review of p53 status and survival data for SCCHN treated surgically (61). This group extracted DNA from 420 cases (from all head and neck sites) to perform mutational analysis of exons 2 to 11 for the presence of disruptive or nondisruptive mutations. They found evidence of an association between p53 mutation and survival, showing that p53 mutations are significantly associated with a shorter OS in SCCHN compared with wild-type cases. Furthermore, they found that disruptive p53 mutations have a stronger association with shortened survival than nondisruptive mutations. This study contained significant results from a well-designed analysis of p53, but did not comply with the inclusion criteria of our review as it combined tumors from all sites as well as recurrent tumors.
Inconclusive Evidence
Our meta-analysis results highlight that the current evidence base about the prognostic value of p53 is inconclusive, and there is large heterogeneity observed across studies. For all but two of our meta-analysis, due to the heterogeneity, we could not be sure whether positive or negative p53 values indicate a worse prognosis, or indeed that p53 has prognostic value at all; the prediction intervals for the HR in a future study are all wide (Table 2) and contain the null value of 1. Only for DFS in relation to oropharynx and oral cavity were there no estimated heterogeneity and a significant meta-analysis result. Why there was no heterogeneity in these particular settings is unclear, but caution is perhaps advised, as there were only three and five studies in the analyses. Interestingly, for oropharynx, the meta-analysis indicates that p53 over-expression/mutation confers a survival advantage; in contrast for oral cavity, the meta-analysis indicates that p53 overexpression/mutation confers a survival disadvantage. Such differences might derive from underlying biological differences or from sampling issues as outlined above due to the small numbers involved in these studies. However, the relatively high incidence of HPV infection in oropharyngeal SCC (40-70% of oropharynx tumors are HPV positive; refs. 62, 63) provides the most obvious potential biological source of altered p53 outcome associations because HPV E6 inactivates p53 by promoting MDM2-independent degradation of p53 (64). However, studies of oropharyngeal cancers have found the presence of HPV to be associated with good outcomes (65, 66) and thus it is not trivial to explain why HPV infection might contribute to reduced survival in the p53-low/negative population relative to the survival for p53-positive patients as the apparent HR association suggests. However, the relationship between HPV presence and p53 expression is complex as evidenced in a recent study of oropharynx cancers in which it was found that HPV DNA was present in 16 of 19 cases displaying wild-type p53 expression as confirmed by analysis of exons 4 to 9 (67) in these tumors. Therefore, the distribution of p53-positive samples in an HPV-positive cohort seems abnormal with respect to p53 mutational status and may well contribute to the results obtained from our meta-analysis. Clearly, even when prognostic significance is evident, the direction of effect is not consistent across tumor subtypes, and thus, we obtain an apparently confusing picture about how p53 status is associated with outcome. Nevertheless, such results provide further support for the need to perform separate analyses of individual tumor sites.
The Problem of Heterogeneity
It is clear that heterogeneity across studies here is severely “muddying the water.” We tried to limit such heterogeneity at the onset of our review by focusing on particular tumor sites, including just those studies with an adequate follow-up (2 or more years), ensuring p53 was measured in the pretreatment tumor tissue and, where possible, looking at advanced and early stage of disease separately. Yet despite this, much heterogeneity remained, and this is likely caused by other factors such as type of treatment, p53 cutoff levels, and method of measuring p53. It was very difficult to examine or explain this heterogeneity in our meta-analyses due to the generally small number of studies, the poor reporting of information, the lack of patient-level data, as well as the variability in clinical characteristics across patients within studies.
For example, although all included articles have reported results clearly for a single primary site, the majority have grouped together tumors of all stages, which will increase heterogeneity as the two most important prognostic factors for survival in SCCHN are primary tumor size (T stage) and nodal status (N stage; ref. 68). We attempted to control for this by performing subgroup analysis for early (stage I and II) and advanced (stage III and IV) disease whenever adequate data were reported; however, this was only possible on two occasions (Table 2). In addition, fewer than half of the included articles reported a single mode of treatment for all patients; the remainder include patients treated heterogeneously using combinations of radiotherapy, chemotherapy, or surgery. Although in general it is reasonable to accept that on a stage by stage basis for the four primary sites, the differing treatment types carry similar survival outcomes, we have been unable to control for the inevitable effect of this treatment variability on survival outcome. Other patient demographics such as age, smoking, and alcohol use may also increase outcome variability.
In addition, the method of analysis of p53 status varies considerably between articles. Although IHC is the commonest method reported in the included articles, the primary antibody used to detect p53 expression was not consistent. Because it is not possible to distinguish wild-type from mutant p53 by IHC, the commonest antibodies used (DO7, DO1, and Pab 1801) are unable to differentiate tumor in which there is a primary p53 mutation from those in which wild-type p53 is upregulated in response to genotoxic stress. Thus, although p53 expression is commonly assumed to be a surrogate for the detection of mutant p53, this is evidently not always the case (8). Similarly, although the absence of p53 staining by IHC is considered synonymous with a wild-type phenotype, such a staining pattern could also be explained by gene deletion (8). A further issue is that the reported cutoff point chosen to dichotomize p53 positivity versus p53 negativity varied from the presence of one positively stained cell to 50% positivity. Both extremes are difficult to justify from a biological perspective. Occasional positive nuclei might result from the normal upregulation of wild-type p53 in response to localized stress. Heterogeneous upregulation of demonstrably mutant p53 has frequently been shown (69), and thus, using a cutoff of 50% will exclude many mutant samples that display lower levels of p53 positivity. The result of using such cutoffs would be, on one hand, to include wild-type samples in a presumed mutant population and, on the other, to place mutant samples with the wild-type group. Consequently, both of these scenarios would be expected to result in diminishing differences between p53 “wild-type” and “mutant” groups.
Other Problems Encountered During Our Meta-analysis Approach
Other problems beside heterogeneity limited our systematic review and meta-analysis; one problem in particular is the poor reporting of individual studies. This has also been highlighted elsewhere for prognostic markers in neuroblastoma (12). Despite our best efforts, we were unable to obtain all the published and unpublished pertinent data necessary for a complete review of p53 in SCCHN. For example, several articles discussed the prognostic value of p53 without presenting any useable data (despite our attempts to contact corresponding authors, these data were not made available to us). Of those studies that were included, the majority did not explicitly report their outcomes as a HR, necessitating the indirect estimation of HR using the techniques previously described (13, 15). On occasions, we used the 5-year survival rate to estimate the number of events, as the actual number was not directly reported; however, we are aware that if there was a lot of censoring, this may be an overestimate of the true number of events. This issue highlights the problems we found when extracting results from the literature for our meta-analysis. In addition, the use of inexact P values to calculate HR—as was necessary in the case of several articles—is a conservative approach and is likely to under estimate the true value of the HR. Results were only included from articles in which unadjusted results were available; published multivariate results were adjusted for different factors across individual studies, including other tumor markers, T stage, and N stage. These results were poorly presented—those factors not found to be significant in the model used were not reported. As a consequence of this, synthesis of these studies into a pooled analysis would be problematic and hence was not undertaken. Selective reporting in studies of prognostic indicators, particularly the presentation of data, outcomes, and analyses, which give a statistically significant and favorable result, is also a concern. As explained previously, this problem has been highlighted by Kyzas et al. (70) who analyzed 1,915 articles on cancer prognostic markers and found that 90% to 96% of published articles gave a positive prognosis for the factor being investigated. Future primary studies should adhere to the REMARK guidelines (71), which provide details of good practice for reporting prognostic tumor marker studies. We did not assess the quality of the primary studies in our review, although Hayden et al. (72) have recently provided some suggestions on how to do this.
Future Research
Given the heterogeneity and the difficulty in extracting suitable results from individual studies, it is thus not surprising that the prognostic value of p53 remains unresolved in SCCHN. Similar conclusions have been found in meta-analyses of p53 in bladder cancer, in which it was concluded that “after 10 years of research (including over 10,000 patients), evidence is not sufficient to conclude whether changes in p53 act as markers of outcome in patients with bladder cancer” (73), and “from this analysis, it becomes evident that further retrospective investigations will not contribute to the solution of the problem and thus are obsolete. There is an obvious need for standardization of the assay procedure and the assessment of the specimens as well as for the initiation of a prospective multicenter trial to provide definite answers” (74). We strongly echo these statements for the future analysis of p53 in SCCHN.
Indeed, a gold-standard approach would be to now initiate large, prospectively planned meta-analyses of individual patient data. Such an approach has many scientific advantages to a situation in which large (and small) centers conduct studies on their own patients, but with variations in inclusion criteria, measurement techniques, analysis strategies, etc. Riley et al. (75) discuss the benefits of IPD and a prospectively planned IPD meta-analysis. These include the following:
Consistency in the inclusion and exclusion criteria across studies
Use of up-to-date follow-up information—potentially longer-term than that used in the study publication
An ability to estimate for missing or poorly reported outcomes and summary statistics across studies—thus potentially reducing the problem of selective within-study reporting
More direct estimation of the HR in which previously only indirect estimates were available
Standardization of statistical analysis across studies
The production of adjusted estimates in relation to a consistent set of adjustment factors, that is, an assessment of the value of p53 in relation to other existing prognostic markers
The use of a consistent cutoff point across studies
The assessment of the benefits of using combinations of markers
The assessment of specific subgroups of patients across studies
A proper investigation of the causes of study-level and patient-level heterogeneity in the prognostic effect of p53
The identification of those studies that contain the same or overlapping sets of patients
McShane et al. (76) state that “More importantly, the necessity of large, definitive prospective studies or prospectively planned meta-analyses for tumor marker research must be recognized.” To achieve this, research groups need to collaborate and the focus should be on prospective, large, coherent, and high-quality research. “Cultural changes will be required” (76), but the necessity of this type of collaboration has been recognized before by clinical trialists and the epidemiologic community, and is achievable. With respect to SCCHN, research groups should particularly endeavor to examine the role of p53 in tumors from distinct primary sites, in strictly defined patient groups in which tumor stage and the treatment modality used are not only taken into consideration but are also controlled for. In addition, it is essential that consensus about the optimum methods of p53 detection as well as the scoring parameters chosen be agreed. For p53, there is simply no biological justification for using cutoffs for positivity of single nuclei, or 50% of cells, nor is the median a sensible criterion for dichotomization of this biomarker. Combinations of prognostic markers (e.g., p53 status and MDM2 and p21) should be investigated, as they can have increased prognostic power that exceeds that of the individual markers themselves (8, 77). It is relatively simple in this manner to improve upon the typically low specificity for p53 status determination by IHC and at the same time to increase the sensitivity of this process. For example, Nenutil et al. (8) proposed a simple strategy that combines highly sensitive detection of p53, with monitoring of p53 target gene expression (MDM2 and/or p21). Such an approach will greatly diminish the false positive and false negative mutation inference rate (inevitable with cutoffs of 1 positive nuclei and 50% positivity, respectively) and is amenable to standardization in scoring. For example, cells expressing p53 but not expressing MDM2 and/or p21, regardless of the relative levels, likely harbor mutant p53, whereas coexpression of p53 and MDM2 would be likely to indicate the retention of a wild-type p53 allele. Although such analyses are not as precise as direct analysis of DNA, the use of biologically justifiable combinations of markers can, for relatively little extra effort and using a low-tech approach (that is, IHC), provide a great deal of information about pathway function, and because it is a pathway function that determines cellular behavior, it is this that future studies must aim to interrogate.
Supplementary Material
Acknowledgments
We thank Professor Graham Ogden, Professor Saman Warnakulasuriya, Dr. Gloria Niehans, Dr. Maria Grazia Daidone, Dr. Angela Flavia Logullo, Dr. Ruben Cabanillas, and Dr. Rahul Parikh for generously providing background data from their published studies.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Cancer Epidemiology, Biomarkers and Prevention Online (http://cebp.aacrjournals.org/).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1.Soussi T. p53 alterations in human cancer: more questions than answers. Oncogene. 2007;26:2145–56. doi: 10.1038/sj.onc.1210280. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH. Outcomes of p53 activation-spoilt for choice. J Cell Sci. 2006;119:5015–20. doi: 10.1242/jcs.03293. [DOI] [PubMed] [Google Scholar]
- 3.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–9. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 4.Lu ML, Wikman F, Orntoft TF, et al. Impact of alterations affecting the p53 pathway in bladder cancer on clinical outcome, assessed by conventional and array-based methods. Clin Cancer Res. 2002;8:171–9. [PubMed] [Google Scholar]
- 5.Wilson JA, editor. Effective head and neck cancer management. British Association of Otorhinolaryngologists Head and Neck Surgeons; London: 2002. Third consensus document. [Google Scholar]
- 6.Rafferty MA, Fenton JE, Jones AS. The history, aetiology and epidemiology of laryngeal carcinoma. Clin Otolaryngol Allied Sci. 2001;26:442–6. doi: 10.1046/j.1365-2273.2001.00507.x. [DOI] [PubMed] [Google Scholar]
- 7.zur Hausen H. Papillomaviruses in the causation of human cancers—a brief historical account. Virology. 2009;384:260–5. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 8.Nenutil R, Smardova J, Pavlova S, et al. Discriminating functional and non-functional p53 in human tumours by p53 and MDM2 immunohistochemistry. J Pathol. 2005;207:251–9. doi: 10.1002/path.1838. [DOI] [PubMed] [Google Scholar]
- 9.Kyzas PA, Loizou KT, Ioannidis JP. Selective reporting biases in cancer prognostic factor studies. J Natl Cancer Inst. 2005;97:1043–55. doi: 10.1093/jnci/dji184. [DOI] [PubMed] [Google Scholar]
- 10.Timar J, Csuka O, Remenar E, Repassy G, Kasler M. Progression of head and neck squamous cell cancer. Cancer Metastasis Rev. 2005;24:107–27. doi: 10.1007/s10555-005-5051-5. [DOI] [PubMed] [Google Scholar]
- 11.Altman DG, Riley RD. Primer: an evidence-based approach to prognostic markers. Nat Clin Pract Oncol. 2005;2:466–72. doi: 10.1038/ncponc0287. [DOI] [PubMed] [Google Scholar]
- 12.Riley RD, Abrams KR, Sutton AJ, et al. Reporting of prognostic markers: current problems and development of guidelines for evidence-based practice in the future. Br J Cancer. 2003;88:1191–8. doi: 10.1038/sj.bjc.6600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson PR, Smith CT, Hutton JL, Marson AG. Aggregate data meta-analysis with time-to-event outcomes. Stat Med. 2002;21:3337–51. doi: 10.1002/sim.1303. [DOI] [PubMed] [Google Scholar]
- 14.Taylor RJ. “Mouseyes”: an aid to wound measurement using a computer. J Wound Care. 1997;6:123–6. doi: 10.12968/jowc.1997.6.3.123. [DOI] [PubMed] [Google Scholar]
- 15.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins JP, Thompson SG, Spiegelhalter DJ. A re-evaluation of random-effects meta-analysis. J R Stat Soc Ser A Stat Soc. 2009;172:137–59. doi: 10.1111/j.1467-985X.2008.00552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 18.Deeks JJ, Higgins JPT, Altman DG. Analysing data and undertaking meta-analyses. In: Higgins JPT, Green S, editors. Cochrane Handbook for Systematic Reviews of Interventions. John Wiley & Sons Ltd.; Chichester, Engand: 2008. pp. 243–36. [Google Scholar]
- 19.Sterne JAC, Egger M, Moher D. Addressing reporting biases. In: Higgins JPT, Green S, editors. Cochrane Handbook for Systematic Reviews of Interventions. John Wiley & Sons Ltd.; Chichester, Engand: 2008. pp. 297–334. [Google Scholar]
- 20.Awwad S, Jaros E, Somes J, Lunec J. P53 overexpression in head and neck carcinoma and radiotherapy results. Int J Radiat Oncol Biol Phys. 1996;34:323–32. doi: 10.1016/0360-3016(95)02108-6. [DOI] [PubMed] [Google Scholar]
- 21.Buyukbayram H, Cureoglu S, Arslan A, Isikakdogan AR. Prognostic value of PCNA and mutant p53 expression in laryngeal squamous cell carcinoma. Cancer Invest. 2004;22:195–202. doi: 10.1081/cnv-120030207. [DOI] [PubMed] [Google Scholar]
- 22.Caminero MJ, Nunez F, Suarez C, Ablanedo P, Riera JR, Dominguez F. Detection of p53 protein in oropharyngeal carcinoma. Prognostic implications. Arch Otolaryngol Head Neck Surg. 1996;122:769–72. doi: 10.1001/archotol.1996.01890190065015. [DOI] [PubMed] [Google Scholar]
- 23.Chien CY, Huang CC, Cheng JT, Chen CM, Hwang CF, Su CY. The clinicopathological significance of p53 and p21 expression in squamous cell carcinoma of hypopharyngeal cancer. Cancer Lett. 2003;201:217–23. doi: 10.1016/s0304-3835(03)00484-1. [DOI] [PubMed] [Google Scholar]
- 24.Cho JH, Kim HS, Park CS, et al. Maspin expression in early oral tongue cancer and its relation to expression of mutant-type p53 and vascular endothelial growth factor (VEGF) Oral Oncol. 2007;43:272–7. doi: 10.1016/j.oraloncology.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 25.Chomchai JS, Du W, Sarkar FH, et al. Prognostic significance of p53 gene mutations in laryngeal cancer. Laryngoscope. 1999;109:455–9. doi: 10.1097/00005537-199903000-00021. [DOI] [PubMed] [Google Scholar]
- 26.Costa A, Licitra L, Veneroni S, et al. Biological markers as indicators of pathological response to primary chemotherapy in oral-cavity cancers. Int J Cancer. 1998;79:619–23. doi: 10.1002/(sici)1097-0215(19981218)79:6<619::aid-ijc11>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 27.de Aguiar AF, Jr., Kowalski LP, de Almeida OP. Clinicopathological and immunohistochemical evaluation of oral squamous cell carcinoma in patients with early local recurrence. Oral Oncol. 2007;43:593–601. doi: 10.1016/j.oraloncology.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Fan GK, Fujieda S, Sunaga H, Tsuzuki H, Ito N, Saito H. Expression of protein p27 is associated with progression and prognosis in laryngeal cancer. Laryngoscope. 1999;109:815–20. doi: 10.1097/00005537-199905000-00025. [DOI] [PubMed] [Google Scholar]
- 29.Frank JL, Bur ME, Garb JL, et al. p53 tumor suppressor oncogene expression in squamous cell carcinoma of the hypopharynx. Cancer. 1994;73:181–6. doi: 10.1002/1097-0142(19940101)73:1<181::aid-cncr2820730131>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 30.Friedman M, Lim JW, Manders E, et al. Prognostic significance of Bcl-2 and p53 expression in advanced laryngeal squamous cell carcinoma. Head Neck. 2001;23:280–5. doi: 10.1002/hed.1031. [DOI] [PubMed] [Google Scholar]
- 31.Friesland S, Mellin H, Munck-Wikland E, et al. Human papilloma virus (HPV) and p53 immunostaining in advanced tonsillar carcinoma-relation to radiotherapy response and survival. Anticancer Res. 2001;21:529–34. [PubMed] [Google Scholar]
- 32.Grabenbauer GG, Muhlfriedel C, Rodel F, et al. Squamous cell carcinoma of the oropharynx: Ki-67 and p53 can identify patients at high risk for local recurrence after surgery and postoperative radiotherapy. Int J Radiat Oncol Biol Phys. 2000;48:1041–50. doi: 10.1016/s0360-3016(00)00737-9. [DOI] [PubMed] [Google Scholar]
- 33.Hiranuma H, Jikko A, Maeda T, et al. An analysis of the prognostic significance of p53 status for squamous cell carcinoma of the oral cavity treated by radiotherapy. Oral Oncol. 1998;34:513–8. doi: 10.1016/s1368-8375(98)00058-x. [DOI] [PubMed] [Google Scholar]
- 34.Hirvikoski P, Kumpulainen E, Virtaniemi J, et al. p53 expression and cell proliferation as prognostic factors in laryngeal squamous cell carcinoma. J Clin Oncol. 1997;15:3111–20. doi: 10.1200/JCO.1997.15.9.3111. [DOI] [PubMed] [Google Scholar]
- 35.Homma A, Furuta Y, Oridate N, et al. Correlation of clinicopathological parameters and biological markers related to apoptosis and proliferative activity with a clinical outcome in squamous cell carcinoma of the larynx treated with concurrent chemoradiotherapy. Auris Nasus Larynx. 2001;28(Suppl):S87–94. doi: 10.1016/s0385-8146(01)00067-0. [DOI] [PubMed] [Google Scholar]
- 36.Jackel MC, Sellmann L, Dorudian MA, Youssef S, Fuzesi L. Prognostic significance of p53/bcl-2 co-expression in patients with laryngeal squamous cell carcinoma. Laryngoscope. 2000;110:1339–45. doi: 10.1097/00005537-200008000-00022. [DOI] [PubMed] [Google Scholar]
- 37.Jayasurya R, Sathyan KM, Lakshminarayanan K, et al. Phenotypic alterations in Rb pathway have more prognostic influence than p53 pathway proteins in oral carcinoma. Mod Pathol. 2005;18:1056–66. doi: 10.1038/modpathol.3800387. [DOI] [PubMed] [Google Scholar]
- 38.Jeannon JP, Soames J, Lunec J, Awwad S, Ashton V, Wilson JA. Expression of cyclin-dependent kinase inhibitor p21(WAF1) and p53 tumour suppressor gene in laryngeal cancer. Clin Otolaryngol Allied Sci. 2000;25:23–7. doi: 10.1046/j.1365-2273.2000.00318.x. [DOI] [PubMed] [Google Scholar]
- 39.Kozomara R, Jovic N, Magic Z, Brankovic-Magic M, Minic V. p53 mutations and human papillomavirus infection in oral squamous cell carcinomas: correlation with overall survival. J Craniomaxillofac Surg. 2005;33:342–8. doi: 10.1016/j.jcms.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 40.Miyahara H, Yane K, Naitoh H, et al. p53 tumor suppressor gene and ras oncogene mutations in hypopharyngeal squamous cell carcinomas. Int J Oncol. 1997;11:133–7. doi: 10.3892/ijo.11.1.133. [DOI] [PubMed] [Google Scholar]
- 41.Nadal A, Campo E, Pinto J, et al. p53 expression in normal, dysplastic, and neoplastic laryngeal epithelium. Absence of a correlation with prognostic factors. J Pathol. 1995;175:181–8. doi: 10.1002/path.1711750205. [DOI] [PubMed] [Google Scholar]
- 42.Nakashima T, Wang XF, Masuda M, Inokuchi A, Komiyama S. Over-expression of p53 nuclear protein in premalignant and malignant laryngeal lesions. Eur Arch Otorhinolaryngol. 1999;256(Suppl 1):S56–9. doi: 10.1007/pl00014155. [DOI] [PubMed] [Google Scholar]
- 43.Narayana A, Vaughan AT, Kathuria S, Fisher SG, Walter SA, Reddy SP. P53 overexpression is associated with bulky tumor and poor local control in T1 glottic cancer. Int J Radiat Oncol Biol Phys. 2000;46:21–6. doi: 10.1016/s0360-3016(99)00348-x. [DOI] [PubMed] [Google Scholar]
- 44.Nibu KI, Yanagisawa A, Nakamizo M, et al. Clinical role of p53 and p21WAF1/CIP1 in squamous cell carcinoma of the pyriform sinus. Acta Otolaryngol. 1998;118:432–7. doi: 10.1080/00016489850183575. [DOI] [PubMed] [Google Scholar]
- 45.Obata A, Eura M, Sasaki J, et al. Clinical significance of p53 functional loss in squamous cell carcinoma of the oropharynx. Int J Cancer. 2000;89:187–93. doi: 10.1002/(sici)1097-0215(20000320)89:2<187::aid-ijc14>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 46.Otero-Garcia JE, Youssef E, Enamorado II, et al. Prognostic significance of p53 and FHIT in advanced oropharyngeal carcinoma. Am J Otolaryngol. 2004;25:231–9. doi: 10.1016/j.amjoto.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Pai HH, Rochon L, Clark B, Black M, Shenouda G. Overexpression of p53 protein does not predict local-regional control or survival in patients with early-stage squamous cell carcinoma of the glottic larynx treated with radiotherapy. Int J Radiat Oncol Biol Phys. 1998;41:37–42. doi: 10.1016/s0360-3016(98)00025-x. [DOI] [PubMed] [Google Scholar]
- 48.Parikh RR, Yang Q, Haffty BG. Prognostic significance of vascular endothelial growth factor protein levels in T1-2 N0 laryngeal cancer treated with primary radiation therapy. Cancer. 2007;109:566–73. doi: 10.1002/cncr.22432. [DOI] [PubMed] [Google Scholar]
- 49.Pruneri G, Pignataro L, Manzotti M, et al. p63 in laryngeal squamous cell carcinoma: evidence for a role of TA-p63 down-regulation in tumorigenesis and lack of prognostic implications of p63 immunoreactivity. Lab Invest. 2002;82:1327–34. doi: 10.1097/01.lab.0000032373.99569.73. [DOI] [PubMed] [Google Scholar]
- 50.Pulkkinen JO, Klemi P, Martikainen P, Grenman R. Apoptosis in situ, p53, bcl-2 and AgNOR counts as prognostic factors in laryngeal carcinoma. Anticancer Res. 1999;19:703–7. [PubMed] [Google Scholar]
- 51.Rowley H, Roland NJ, Helliwell TR, Caslin A, Kinsella AR, Jones AS. p53 protein expression in tumours from head and neck subsites, larynx and hypopharynx, and differences in relationship to survival. Clin Otolaryngol Allied Sci. 1998;23:57–62. doi: 10.1046/j.1365-2273.1998.00087.x. [DOI] [PubMed] [Google Scholar]
- 52.Russo A, Corsale S, Agnese V, et al. TP53 mutations and S-phase fraction but not DNA-ploidy are independent prognostic indicators in laryngeal squamous cell carcinoma. J Cell Physiol. 2006;206:181–8. doi: 10.1002/jcp.20447. [DOI] [PubMed] [Google Scholar]
- 53.Shigyo H, Nonaka S, Katada A, et al. Inducible nitric oxide synthase expression in various laryngeal lesions in relation to carcinogenesis, angiogenesis, and patients’ prognosis. Acta Otolaryngol. 2007;127:970–9. doi: 10.1080/00016480601089382. [DOI] [PubMed] [Google Scholar]
- 54.Siegelmann-Danieli N, Ben-Izhack O, Hanlon A, et al. P53 alteration in oral tongue cancer is not significantly associated with age at diagnosis or tobacco exposure. Tumori. 2005;91:346–50. doi: 10.1177/030089160509100412. [DOI] [PubMed] [Google Scholar]
- 55.Sittel C, Eckel HE, Damm M, von Pritzbuer E, Kvasnicka HM. Ki-67 (MIB1), p53, and Lewis-X (LeuM1) as prognostic factors of recurrence in T1 and T2 laryngeal carcinoma. Laryngoscope. 2000;110:1012–7. doi: 10.1097/00005537-200006000-00024. [DOI] [PubMed] [Google Scholar]
- 56.Tan LK, Ogden GR. p53 over-expression in laryngeal carcinoma is not predictive of response to radiotherapy. Oral Oncol. 1997;33:177–81. doi: 10.1016/s0964-1955(96)00082-6. [DOI] [PubMed] [Google Scholar]
- 57.Tatemoto Y, Osaki T, Yoneda K, Yamamoto T, Ueta E, Kimura T. Expression of p53 and p21 proteins in oral squamous cell carcinoma: correlation with lymph node metastasis and response to chemoradiotherapy. Pathol Res Pract. 1998;194:821–30. doi: 10.1016/S0344-0338(98)80084-8. [DOI] [PubMed] [Google Scholar]
- 58.Tsuji T, Mimura Y, Wen S, et al. The significance of PCNA and p53 protein in some oral tumors. Int J Oral Maxillofac Surg. 1995;24:221–5. doi: 10.1016/s0901-5027(06)80132-3. [DOI] [PubMed] [Google Scholar]
- 59.Vielba R, Bilbao J, Ispizua A, et al. p53 and cyclin D1 as prognostic factors in squamous cell carcinoma of the larynx. Laryngoscope. 2003;113:167–72. doi: 10.1097/00005537-200301000-00031. [DOI] [PubMed] [Google Scholar]
- 60.Yanamoto S, Kawasaki G, Yoshitomi I, Mizuno A. p53, mdm2, and p21 expression in oral squamous cell carcinomas: relationship with clinicopathologic factors. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2002;94:593–600. doi: 10.1067/moe.2002.127404. [DOI] [PubMed] [Google Scholar]
- 61.Poeta ML, Manola J, Goldwasser MA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–61. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiest T, Schwarz E, Enders C, Flechtenmacher C, Bosch FX. Involvement of intact HPV16 E6/E7 gene expression in head and neck cancers with unaltered p53 status and perturbed pRb cell cycle control. Oncogene. 2002;21:1510–7. doi: 10.1038/sj.onc.1205214. [DOI] [PubMed] [Google Scholar]
- 63.Hafkamp HC, Speel EJ, Haesevoets A, et al. A subset of head and neck squamous cell carcinomas exhibits integration of HPV 16/18 DNA and overexpression of p16INK4A and p53 in the absence of mutations in p53 exons 5-8. Int J Cancer. 2003;107:394–400. doi: 10.1002/ijc.11389. [DOI] [PubMed] [Google Scholar]
- 64.Hubbert NL, Sedman SA, Schiller JT. Human papillomavirus type 16 E6 increases the degradation rate of p53 in human keratinocytes. J Virol. 1992;66:6237–41. doi: 10.1128/jvi.66.10.6237-6241.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hafkamp HC, Manni JJ, Haesevoets A, et al. Marked differences in survival rate between smokers and nonsmokers with HPV 16-associated tonsillar carcinomas. Int J Cancer. 2008;122:2656–64. doi: 10.1002/ijc.23458. [DOI] [PubMed] [Google Scholar]
- 66.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–9. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 67.Kumar B, Cordell KG, Lee JS, et al. EGFR, p16, HPV Titer, Bcl-xL and p53, sex, and smoking as indicators of response to therapy and survival in oropharyngeal cancer. J Clin Oncol. 2008;26:3128–37. doi: 10.1200/JCO.2007.12.7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carvalho AL, Nishimoto IN, Califano JA, Kowalski LP. Trends in incidence and prognosis for head and neck cancer in the United States: a site-specific analysis of the SEER database. Int J Cancer. 2005;114:806–16. doi: 10.1002/ijc.20740. [DOI] [PubMed] [Google Scholar]
- 69.Tandon S, Beer H, Jones TM, Birchall M, Boyd M, Helliwell TR. Immunohistochemical expression of p53 and MDM2 within head and neck squamous cell carcinoma. Clin Otolaryngol Allied Sci. 2006;31:247. [Google Scholar]
- 70.Kyzas PA, Denaxa-Kyza D, Ioannidis JP. Almost all articles on cancer prognostic markers report statistically significant results. Eur J Cancer. 2007;43:2559–79. doi: 10.1016/j.ejca.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 71.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–4. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 72.Hayden JA, Cote P, Bombardier C. Evaluation of the quality of prognosis studies in systematic reviews. Ann Intern Med. 2006;144:427–37. doi: 10.7326/0003-4819-144-6-200603210-00010. [DOI] [PubMed] [Google Scholar]
- 73.Malats N, Bustos A, Nascimento CM, et al. P53 as a prognostic marker for bladder cancer: a meta-analysis and review. Lancet Oncol. 2005;6:678–86. doi: 10.1016/S1470-2045(05)70315-6. [DOI] [PubMed] [Google Scholar]
- 74.Schmitz-Drager BJ, Goebell PJ, Ebert T, Fradet Y. p53 immunohistochemistry as a prognostic marker in bladder cancer. Playground for urology scientists? Eur Urol. 2000;38:691–9. doi: 10.1159/000020364. discussion 700. [DOI] [PubMed] [Google Scholar]
- 75.Riley RD, Sauerbrei W, Altman DG. Prognostic markers in cancer: the evolution of evidence from single studies to meta-analysis, and beyond. Br J Cancer. 2009;100:1219–29. doi: 10.1038/sj.bjc.6604999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McShane LM, Altman DG, Sauerbrei W. Identification of clinically useful cancer prognostic factors: what are we missing? J Natl Cancer Inst. 2005;97:1023–5. doi: 10.1093/jnci/dji193. [DOI] [PubMed] [Google Scholar]
- 77.Boyd MT, Vlatkovic N. p53: a molecular marker for the detection of cancer. Exp Opin Med Diagn. 2008;2:1013–24. doi: 10.1517/17530059.2.9.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


