Abstract
Developing neural tissue undergoes a period of neurogenesis followed by a period of gliogenesis. The lineage relationships among glial cell types have not been defined for most areas of the nervous system. Here we use retroviruses to label clones of glial cells in the chick retina. We found that almost every clone had both astrocytes and oligodendrocytes. In addition, we discovered a novel glial cell type, with features intermediate between those of astrocytes and oligodendrocytes, which we have named the diacyte. Diacytes also share a progenitor cell with both astrocytes and oligodendrocytes.
Introduction
It is currently unknown whether progenitor cells in the developing CNS are able to give rise to both astrocytes and oligodendrocytes. Studies of cultures derived from dissociated rat optic nerve suggested that glial progenitor cells, the O2A cells, give rise to both type 2 astrocytes and oligodendrocytes (Raff et al., 1983a, 1984; Temple and Raff, 1985). However, two groups transplanted labeled O2A cells into the developing brain and found that only oligodendrocytes were produced, suggesting that there are distinct progenitors for astrocytes and oligodendrocytes (Espinosa de los Monteros et al., 1993; Groves et al., 1993). Subsequent studies in various systems led to the proposal that there are distinct domains for astrocyte and oligodendrocyte production, supporting the notion of distinct progenitors for the two glial types (Rowitch, 2004).
Recently, the distinction between astrocyte-producing and oligodendrocyte-producing regions has become blurred. The dorsal domains of the spinal cord and telencephalon, which were thought to give rise exclusively to astrocytes, have been found to also produce oligodendrocytes (Cai et al., 2005; Fogarty et al., 2005; Vallstedt et al., 2005, Kessaris et al., 2006; Richardson et al., 2006). Furthermore, one study reported that glia that express Ng2, which have been shown previously to be progenitor cells for glia during development, in the adult mouse are capable of producing both oligodendrocytes and protoplasmic astrocytes in vivo (Zhu et al., 2008). However, two independent studies found that adult Ng2-positive (Ng2+) glia gave rise to oligodendrocytes and neurons, but not astrocytes, in vivo (Menn et al., 2006; Rivers et al., 2008). These studies all rely on fate mapping to irreversibly label large numbers of cells. Thus, these studies cannot distinguish between a single multipotent progenitor cell giving rise to more than one cell type and a progenitor pool with distinct progenitor cells that give rise to only one cell type or a limited subset of cell types.
One set of studies, however, did use clonal lineage tracing to determine the lineage relationships of glial cells. Levison and Goldman found that 15% of clones arising from retroviral infections of the postnatal day 0 (P0) subventricular zone had both astrocytes and oligodendrocytes (Levison and Goldman, 1993; Zerlin et al., 2004). However, two studies from the Luskin group, which used a similar viral marking strategy, did not observe this multipotency (Luskin et al., 1993; Luskin and McDermott, 1994).
In this study, we used retrovirus-mediated clonal analysis to determine whether chick retinal astrocytes and oligodendrocytes are produced by multipotent progenitor cells. We found that glial clones radiated into the retina from the optic nerve head in patterns suggestive of migration directed toward the periphery, with little deviation from this direction of migration. Almost every clone (>97%) exhibited both astrocytes and oligodendrocytes. In addition, we discovered a novel glial cell type, which we have named the diacyte, that was present in almost every clone. These data demonstrate that the glial cell types of the inner retina derive from a common, multipotent progenitor cell.
Materials and Methods
Viral construction and production.
The membrane-bound GFP (mGFP) and tdTomato (tandem dimer Tomato) genes were cloned into the pQXIX retroviral vector (Clontech). Viral production, concentration, and titering were done by standard methods (Cepko and Pear, 1997). Briefly, pQmGFP or pQtdTomato and the vesicular stomatitis virus-glycoprotein-encoding plasmid (Naldini et al., 1996) were transiently transfected with FUGENE6 (Roche) onto the PLAT-E cell line, which expresses the necessary gag and pol genes for viral production (Morita et al., 2000).
Chick injections and tissue processing.
Embryonic day 3 (E3) to E4 chicks (Hamburger and Hamilton stage 17–22) were injected with an ∼1:1 mixture of the two viruses in the anterior neural tube, taking care not to inject into the optic cup. Injections were done as described previously (Fekete and Cepko, 1993). Harvesting, preparation of whole mounts, and immunohistochemical staining of chick retinas were conducted as described previously (Rompani and Cepko, 2008). Each batch of retinas sharing the same experiment number were injected with a particular viral mix, with the exception of 248 and 251, which shared the same viral mix.
Antibodies used were chicken anti-GFP at 1:2000 (Abcam AB13970), rabbit anti-red fluorescent protein at 1:500 (discontinued; Millipore Bioscience Research Reagents), rabbit anti-Olig2 (AB9610; Millipore Bioscience Research Reagents), mouse anti-Olig1 at 1:1000 (MAB 5540; Millipore Bioscience Research Reagents), rabbit anti-Ng2 (AB5320; Millipore Bioscience Research Reagents), rabbit anti-GFAP at 1:500 (G9269; Sigma), mouse anti-S100B (S2657; Sigma), mouse anti-myelin/oligodendrocyte specific protein (MOSP) (MAB328; Millipore Bioscience Research Reagents), mouse anti-myelin-associated glycoprotein (MAG) (MAB1567; Millipore Bioscience Research Reagents), and rat anti-proteolipid protein (PLP) [courtesy of Ben Barres (Stanford University, Stanford, CA) and Kazuhiro Ikenaka (National Institute for Physiological Sciences, Okazaki, Japan)]. Secondary antibodies were Alexa488 goat anti-chicken at 1:500 (A-11039; Invitrogen), cyanine 3 (Cy3) goat anti-rabbit at 1:500 (111-165-144; Jackson ImmunoResearch), and Cy3 goat anti-mouse at 1:500 (115-165-146; Jackson ImmunoResearch).
Image acquisition and processing.
Retinal whole mounts became between 50 and 100 μm thick, depending on the degree of flattening induced by the mounting. However, as we reported previously (Rompani and Cepko, 2008), even in the thinnest samples, processes did not seem altered. Confocal images were taken on a Leica DMRXE laser-scanning confocal microscope. Tiled images were taken with an Olympus Fluoview LSM with an automated stage. Maximum intensity projection (MIP), stitching, and cell counting were done with custom-made software produced in Labview (National Instruments). Cell counting for supplemental Figure 6 (available at www.jneurosci.org as supplemental material) was conducted with the Imaris Spot Detector (Bitplane). The number of Olig2+ cells in Figure 8 was quantified using custom Labview software that stitched together all of the confocal images and then automatically counted the number of Olig2+ cells in 150 × 150 μm squares. The value of each square was then outputted into a single pixel of an output image file and pseudocolored such that red was the highest value and green was the lowest.
Figure 8.
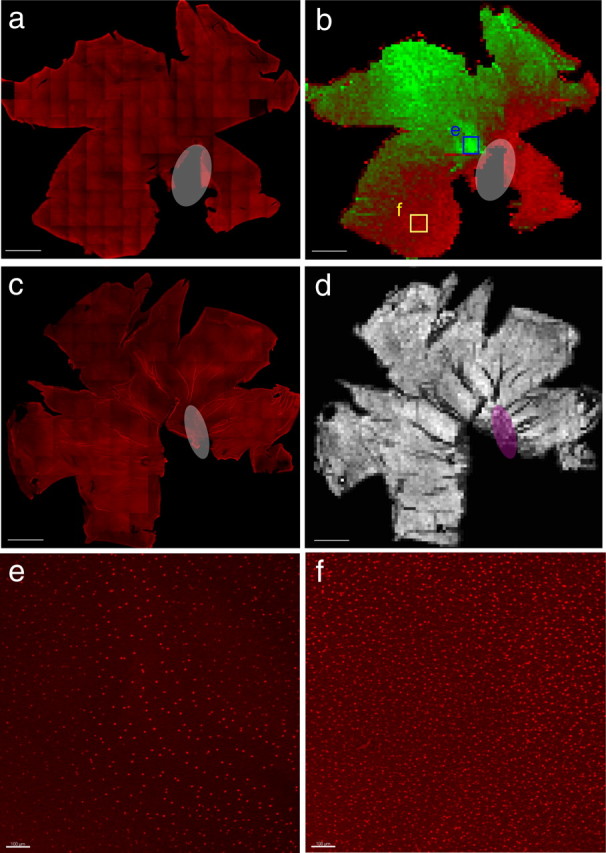
Quantification of Olig2 and neurofilament staining in the P8/9 chick retina. a, Tiled confocal image of the chick P8/9 retina stained for Olig2 (red). b, Pseudocoloring of the automated quantification of Olig2+ cells on the vitreal surface of the retina. Red indicates relatively higher Olig2+ cell density, and green indicates relatively lower cell density. Note the ventral high Olig2 density and dorsal low Olig2 density as well as the spot (outline) in the central retina of very low Olig2 density. The red rim surrounding the retina is an automated quantification artifact. Each pixel corresponds to a single quantified bin and is equivalent to an ∼150 × 150 μm area of the retina. c, Tiled confocal image of the chick P8 retina stained for neurofilament to mark retinal ganglion cell axons (red). d, Quantification of the number of filaments on the vitreal surface of the retina shown with black being low and white being high, indicating no significant difference in the number of axonal fascicles throughout the retina. Each pixel is also equivalent to an ∼150 × 150 μm area of the retina and corresponds to a single quantification bin. Scale bar, 2500 μm. White and magenta ovals indicate ventral fissure. e, MIP corresponding to blue box in b. Scale bar, 100 μm. f, MIP corresponding to yellow box in b. Scale bar, 100 μm.
Results
Glial clones in the chick retina follow patterns of restricted migration
Unlike many mammalian retinas, which have only astrocytes, the chick retina has both astrocytes and oligodendrocytes (Meyer, 1977). Retinal glia have been shown to arise from progenitor cells originating within the floor of the third ventricle (Kim et al., 2006). The glial progenitor cells are induced to produce astrocytes and oligodendrocytes by sonic hedgehog from retinal ganglion cell axons (Gao and Miller, 2006). These glial progenitor cells infiltrate the optic chiasm and migrate down the optic nerve before migrating out of the optic nerve head and onto the inner surface of the retina (Watanabe and Raff, 1988; Fu and Qui, 2001). To target these glial progenitor cells, we infected the E3 chick neural tube in ovo with a mixture of two viruses, one expressing mGFP and another expressing tdTomato. A low number of viral particles was used, such that only a few clones of glia in the retina would be labeled. When the mature tissue was examined, coherent clusters of cells that were either red or green were observed, suggesting that each cluster was a clone. If clusters of labeled cells were not clones but were generated by infection of more than one progenitor cell, then at least some clusters would have a mixture of tdTomato+ cells and GFP+ cells.
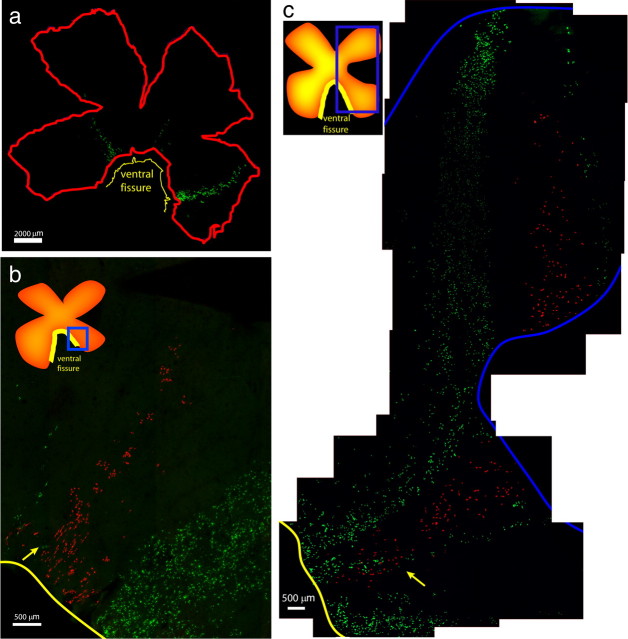
Infected chick retinas were harvested between E20 and P9, and whole mounts were immunohistochemically processed for both GFP and tdTomato. Retinal glia exhibited a distinct pattern of cellular distribution across the retina (Fig. 1), with several features of note. The infected glial cells appeared to radiate from the nerve head and migrate toward the periphery (Fig. 1a). There appeared to be little migration away from this trajectory, because the width of each group of cells was similar throughout the path of migration (Fig. 1). As mentioned above, in general, there was little mixing of GFP- and tdTomato-labeled cells within one of these groupings (Fig. 1b,c). However, there was one region in which some mixing was observed, in the area closest to the optic nerve head (Fig. 1b,c, yellow arrows). Even in these cases, however, the migration paths of clones appeared to diverge because, nearer to the periphery, there was almost no overlap between GFP+ and tdTomato+ cells (Fig. 1b,c). An example of such divergence is shown in Figure 1c, where GFP+ cells near the ventral fissure were intermingled with tdTomato+ cells, but then some members of the presumptive GFP+ clone appear to have crossed over the path of the tdTomato cells, to follow a distinct trajectory out to the periphery. In general, we have scored such discrete groupings of a single color as clones. When there were ambiguities in the assignment of clonal boundaries, we erred on the side of splitting errors, i.e., we considered two groups of GFP+ cells as two separate clones if they shared a common origin in the optic nerve head but had two different migration paths as they left the optic nerve head. Using this method, we likely, in some cases, incorrectly split a single clone into two clones. This was done to avoid lumping errors in the cases in which clonal boundaries were more difficult to assign. Splitting errors, given the findings described below, would not change the conclusions regarding potency.
Figure 1.
Chick retinal glial clones. a, Whole-mount view of an E3 infected retina (231-2R) harvested at E20, showing glial clones of different sizes and shapes. Scale bar, 2000 μm. b, Retina (231-2R) showing a small tdTomato+ clone flanked by two GFP+ clones, with small degree of overlap in which the cells have exited the ventral fissure (yellow arrow). Scale bar, 500 μm. c, Retina (230-2R) showing two large GFP+ clones flanking a single tdTomato+ clone that overlaps with one of the GFP+ clones, close to the ventral fissure (yellow arrow). Scale bar, 500 μm. Insets show diagrams of the retina, with boxed areas indicating the areas occupied by the glial clones, which were imaged at higher resolution. Yellow line indicates ventral fissure.
Retinal glial clones were often seen in retinas that did not have labeled neuronal cells in the retina. This is consistent with the aforementioned origin of retinal glia from the third ventricle and the targeted site of inoculation to that area. The progenitor cells that give rise to retinal neurons line the developing optic vesicle before E3, and, by E3, the age of injection, the optic vesicle had already formed the optic cup. Previous studies in which the optic vesicle and cup, but not the brain or optic nerve, were targeted by viral infection have demonstrated that retinal progenitor cells give rise to all neuronal cell types, as well as Mueller glia, but not to astrocytes and oligodendrocytes (Turner and Cepko, 1987; Turner et al., 1990; Fekete and Cepko 1993). Furthermore, the majority of chick retinal neurons are produced by E8 (Kahn, 1974; Mishima and Fujita, 1978; Dütting et al., 1983; Morris and Cowan, 1995), whereas astrocytes and oligodendrocytes do not start migrating into the retina until E10 (Ono et al., 1998; Fu and Qui, 2001).
To further investigate the migration of glial cells into the retina, infected retinas were harvested at E12 (Fig. 2). In these retinas, cells were observed with a migratory, bipolar morphology close to the optic nerve head, consistent with the morphology of migrating, immature glia (Fig. 2b). These cells were positive for staining with anti-Olig2, consistent with a glial identity (Fig. 2b). Because glial cells are known to divide as they migrate (Nakazawa et al., 1993; Chan-Ling et al., 2009), infected E12 retinas were explanted for 1 d in vitro in the presence of bromodeoxyuridine (BrdU). The GFP+ cells with a migratory posture were found to be labeled with BrdU and thus had undergone an S phase, consistent with being dividing cells (Fig. 2c). Retinas at different stages also were stained for Olig2 to investigate the time course of glial migration into the retina. There were no Olig2+ cells observed in the ganglion cell layer (GCL) at E9 (data not shown). At E11, we found that the extent of Olig2 migration was extremely variable, even within the same retina (supplemental Fig. 1a–e, available at www.jneurosci.org as supplemental material). For example, in a single E11 retina, one group of Olig2 cells extended 13,600 μm from the optic nerve head, whereas another extended only 1700 μm from the optic nerve head (supplemental Fig. 1a, available at www.jneurosci.org as supplemental material). By E13, the Olig2+ cells in the GCL extended mostly throughout the central retina but were not in the peripheral retina, although by E15, the Olig2+ cells extended to the periphery (supplemental Fig. 1f,g, available at www.jneurosci.org as supplemental material). Furthermore, Olig2+ cells could be seen in the outer nuclear layer (ONL) throughout the E9 retina (supplemental Fig. 1h, available at www.jneurosci.org as supplemental material). However, the GCL became increasingly populated with Olig2+ cells migrating in from the optic nerve head, while the Olig2+ cells in the ONL decreased in frequency and were absent in the E15 retina (data not shown).
Figure 2.
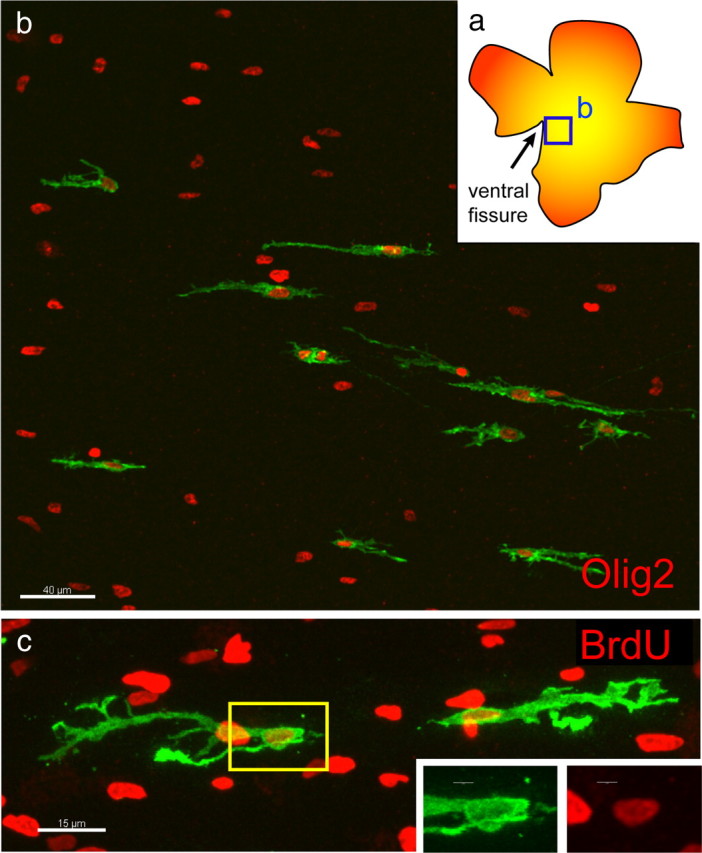
Early glial cell migration and cell division. a, Diagram of retina imaged in b showing region close to the optic nerve head in which GFP+ cells could be seen. b, GFP+ cells derived from an E3 infection, harvested at E12, expressed Olig2 (red). Scale bar, 40 μm. c, E3 infected chick retina explanted for 1 d in vitro at E12 in the presence of BrdU (red), indicating that glial cells underwent S phase as they migrated. Yellow box indicates inset area. Scale bars: c, 15 μm; inset, 3 μm.
Glial clones were composed of astrocytes, oligodendrocytes, and a third glial cell type
The cells within the infected glial clones were characterized by their morphology and gene expression. Three cellular morphologies were seen at E20. One morphology comprised long, thick processes in the fiber layer that were parallel to the ganglion cell axons, consistent with the morphology of an oligodendrocyte (Fig. 3b,c,f). A second morphology was of short, irregular processes in the fiber layer and exuberant processes extending into the inner plexiform layer (IPL), consistent with the morphology of an astrocyte (Fig. 3b,d,g). The third morphology was intermediate to those of oligodendrocytes and astrocytes, with thin, wispy processes in the fiber layer, parallel to the ganglion cell axons, and very small, thin processes in the IPL (Fig. 3b,e,h).
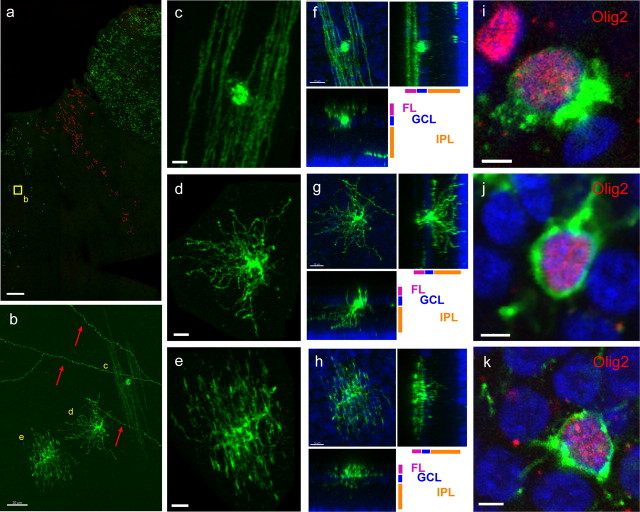
Figure 3.
Morphology and Olig2 expression of GFP+ cells in glial clones. a, Low-magnification view of three glial clones, two expressing GFP and one expressing tdTomato. Yellow box indicates location of cells shown in b. Retina infected at E3 and harvested at E20. Scale bar, 500 μm. b, Examples of the three morphologically distinct types of cells in the inner retina at E20, shown in more detail in c–e. Red arrows indicate IPL processes from infected brain neurons that project axons into the retina. Scale bars, 20 μm. c, GFP+ cell exhibiting oligodendrocytic morphology. d, GFP+ cell exhibiting astrocytic morphology. e, GFP+ cell exhibiting morphology that is intermediate to those of an astrocyte and an oligodendrocyte. f, Oligodendrocyte-like GFP+ cell at P8/9, together with orthogonal projections showing two side views. g, Astrocyte-like GFP+ cell at P8/9 with orthogonal projections showing two side views. h, GFP+ cell with intermediate morphology, with orthogonal projections showing two side views. i, Oligodendrocyte-like GFP cell positive for Olig2 (red). j, Astrocyte-like GFP cell positive for Olig2 (red). k, GFP cell with intermediate morphology, positive for Olig2 (red). Scale bars: c–h, 10 μm; i–k, 3 μm. Green indicates GFP.
To further characterize cells with these three morphologies, the retinas were processed for immunohistochemistry for the glial marker, Olig2. Cells with all three morphologies were positive for Olig2 (Fig. 3i–k). To investigate whether these glial morphologies changed during development, the tissue was harvested at different stages after the glia started migrating into the retina. All glial cells were bipolar at E12 but became shorter and thicker by E14, and, by E16, some of the cells had a more mature morphology, with many elaborated processes, whereas many remained bipolar (supplemental Fig. 2a–d, available at www.jneurosci.org as supplemental material). By E18, most of the cells had a mature morphology, albeit with shorter processes, but some cells with a more bipolar shape could still be seen (supplemental Fig. 2e,f, available at www.jneurosci.org as supplemental material). The immature cells were equally intermixed with cells of more mature morphology. At E20, all three cell types obtained their final morphology, with very few immature cells observed in even the most highly infected retinas. We also harvested retinas between postnatal day 8 or 9 (P8/9). This time point was more than 1 week after the chick hatched, and chicks have a functional visual system at hatch, which suggests that all cell types have developed into a functional morphology by this stage. We found that, although cell processes for all three cell types were longer and thicker at P9 than at E20, they had generally unchanged gross morphology (data not shown). The only noticeable difference in the later harvest was that the vast majority astrocyte cell bodies could now be found within the IPL itself, in addition to the GCL (Fig. 4a).
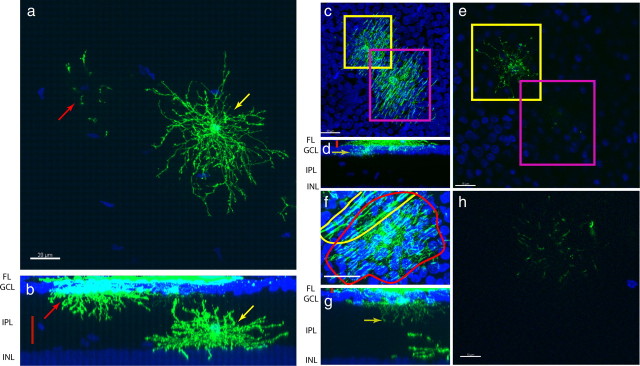
Figure 4.
Post-hatch migration of astrocytes into the IPL and variable diacyte projection into the IPL. a, MIP of an IPL astrocyte (yellow arrow) and processes of an astrocyte with a cell body in the GCL (red arrow). b, Cross-sectional MIP showing GCL astrocyte (red arrow), IPL astrocyte (yellow arrow), and area shown in a (red bar). Scale bar, 20 μm. c, MIP of two diacytes observed from the fiber layer, highlighted in the yellow and magenta boxes. d, Cross-sectional MIP showing the area imaged in c, red bar. Yellow arrow indicates the single confocal slice shown in e. e, Single confocal slice showing one of the cells extending very short processes into the IPL (the yellow-boxed cell in c), whereas the other cell did not extend any processes into the IPL (the magenta-boxed cell). f, MIP of a diacyte (red outline) and an oligodendrocyte (yellow outline). g, Cross-sectional MIP of a diacyte extending processes deep into the IPL. Red bar indicates area depicted in f. Yellow arrow indicates slice shown in h. h, Single confocal slice deep in the IPL showing extensive diacyte processes in the IPL. Scale bars: a, d, 20 μm; c, 15 μm; f, 10 μm. INL, Inner nuclear layer.
We named the GFP+ cells with the third morphology diacytes, given the intermediate morphology between those of retinal astrocytes and oligodendrocytes. The diacyte had processes in the fiber layer (FL), as do oligodendrocytes, but the diacyte processes were more numerous and thinner than those of oligodendrocytes (Fig. 3, compare f, h). Furthermore, the diacyte was able to send projections into the IPL, much like astrocytes (Fig. 4c–e). Unlike astrocytes, however, the diacyte projections into the IPL were short, and diacytes were frequently found with only very short IPL projections or none at all (Fig. 4f–h). Quantification of process length, process diameter, cell body position, nucleus size, and process shape at P8/9 showed that diacytes are significantly different from both astrocytes and oligodendrocytes (Fig. 5) (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Diacytes had an IPL length to width ratio that was similar to astrocytes but had a smaller FL length to width ratio to oligodendrocytes. They were unique in that they had both IPL and FL processes (Fig. 5a,e top left). Furthermore, diacytes had shorter IPL and FL processes than astrocytes and oligodendrocytes, respectively (Fig. 5b,e, top right). Diacytes had IPL process diameters similar to that of astrocytes, but their FL process diameters were significantly smaller than that of oligodendrocytes (Fig. 5c,e, bottom left). Whereas diacytes were located at the GCL together with oligodendrocytes, astrocytes were generally very deep within the IPL (Fig. 5d,e, bottom right). The nucleus of all three cell types were similarly sized (Fig. 5d,e, bottom right).
Figure 5.
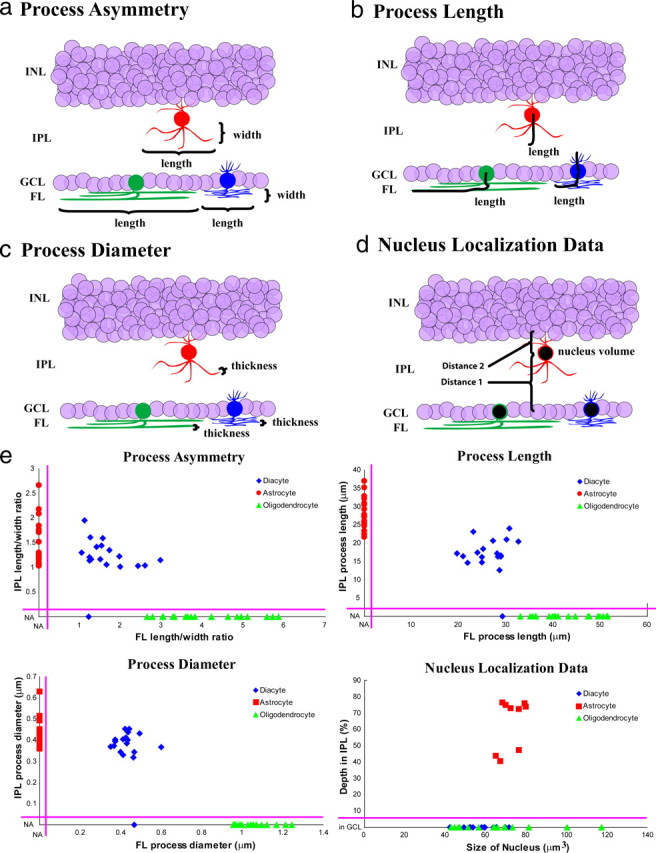
Quantification of glial characteristics. a, Diagram demonstrating process asymmetry (length divided by width), which was measured for processes in the IPL or FL for each cell type. b, Process length for each cell type for their processes in either the IPL or FL. c, Process diameter for each cell type for processes in the IPL or FL. d, Nucleus localization data for each cell type showing depth of the nucleus in the IPL [distance 1/(distance 1 + distance 2)] as well as nucleus size in cubic micrometers. e, Two-dimensional scatter plots of quantified cell morphology. Top left, IPL length/width ratio plotted against FL length/width ratio. Top right, IPL process length plotted against FL process length. Bottom left, IPL process diameter plotted against FL process diameter. Bottom right, Percentage depth in IPL plotted against size of nucleus. Red, Astrocytes; green, oligodendrocytes; blue, diacytes; NA, not applicable (i.e., FL process width for cells without FL processes). INL, Inner nuclear layer.
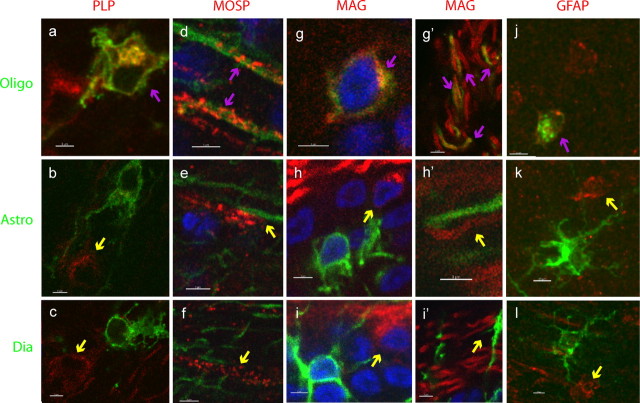
To further confirm the identity of some of these cells as astrocytes and oligodendrocytes, immunohistochemistry for glial markers was conducted. The cell type exhibiting oligodendrocyte-like morphology, with long, parallel processes in the FL, was positive for the markers PLP, MOSP, and MAG (Fig. 6, top row). In addition, this cell type was positive for GFAP, which may be attributable to cross reactivity (see Discussion) (Fig. 6j). The cells with the astrocyte morphology or the diacyte morphology were negative for these markers (Fig. 6, middle and bottom rows). Software-assisted quantification of colocalization of PLP or MAG with GFP+ cells with each type of morphology was performed to confirm these observations for a large number of cells (data not shown). This quantification showed that colocalization of MAG with GFP was 39% for cells with oligodendrocyte morphology but only about 6% for cells with the morphologies of astrocytes or diacytes. Meanwhile, colocalization of PLP with GFP was 86% for cells with oligodendrocyte morphology and <27% for cells with morphologies of astrocytes and diacytes. MOSP was not quantified because it had too punctate a distribution for accurate colocalization with GFP. In addition, antibodies to S100β, ALDH1L1, GalC, Id3, Aldolase, Ng2, MBP, Canary Brain antigen, O1, O4, and Olig1, which have been shown to work well in mouse and human tissue, were used. A variety of antigen retrieval and tissue processing conditions were used on the chick tissue without success.
Figure 6.
Markers expressed by retinal glial cells. Expression of glial markers in the three morphological types of glial cells, marked by GFP. Oligodendrocyte morphology (a, d, g, g′, j), astrocyte morphology (b, e, h, h′, k), and intermediate morphology (diacyte) (c, f, i, i′, l). Antigens are indicated in column headings and are shown in red. Magenta arrows indicate colocalized signals, and yellow arrows indicate non-colocalization. Scale bar, 3 μm.
The composition of the clones and their distribution across the retina were also analyzed. Of the 182 glial clones from 32 retinas, 179 had all three cell types (Fig. 7a) (supplemental Table 1, available at www.jneurosci.org as supplemental material). Three clones contained relatively few cells, namely <20, and were composed entirely of astrocytes. The three cell types were approximately distributed in a 1:1:1 distribution in the majority of the glial clones. For analysis of the distribution of clones across the retina, the retina was divided into four domains. The domains were drawn to encompass the origin of the clones within the optic nerve head and to respect the general distribution patterns of the clones. The number of glial clones within each domain was then quantified. When the number of virally labeled clones was normalized to the surface area of each domain, the clones were seen to be evenly distributed (χ2 test, p = 0.491) (Fig. 7d). The sizes of virally labeled glial clones were roughly quantified and generally were very large (>200 cells), although several were small (<50 cells) and some were intermediate in size (supplemental Table 1, available at www.jneurosci.org as supplemental material). Glial clones could be found in a variety of shapes, including ones that were short, narrowing slightly as they reached the periphery (supplemental Fig. 4a, available at www.jneurosci.org as supplemental material). Others were composed of discontinuous groups of cells (supplemental Fig. 4b, available at www.jneurosci.org as supplemental material), and others were long and uniformly wide across the extent of their path from the optic nerve head (supplemental Fig. 4c, red arrow, available at www.jneurosci.org as supplemental material). However, there was more or less a continuum of shapes throughout the set of clones.
Figure 7.
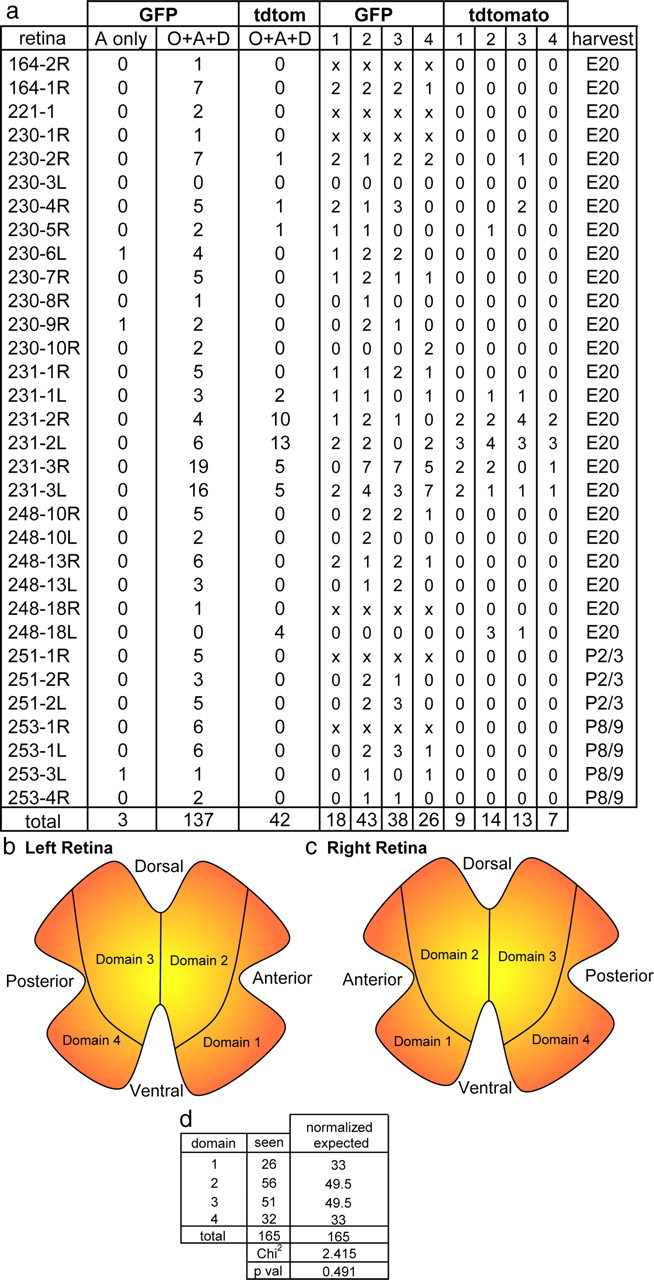
Quantification of glial clones. a, Column labeled “retina” lists the experiment number and retina number. The numbers of GFP and tdTomato clones in each retina are listed as astrocyte-only clones or clones with all three glial cell types (clones with other combinations of cell types were not observed). Also shown is the number of clones per domain of the retina (key in b and c) and age of harvest. Retinas whose domains were unable to be determined because of fragmentation of the retinal whole mounts are denoted by “x” on each domain. All retinas were derived from E3 infections. b, c, Delineation of the four domains for the left (b) and right (c) retina, with domains drawn to mimic shapes of clones. d, Statistical analysis of number of glial clones per domain in the retina, normalized for expected clone distribution based on domains 2 and 3 being 50% larger than domains 1 and 4. O, Oligodendrocyte; A, astrocyte; D, diacyte.
In a few retinas, clones containing exclusively cells of unusual and irregular morphologies were seen. Because there was occasional nicking of blood vessels during the injections, potentially causing virus to enter the vasculature, it was possible that these cells might have been derived from a hematopoietic progenitor cell that produced macrophages (Kezic and McMenamin, 2008). Macrophages have been shown to enter the quail retina in a central to peripheral gradient starting at E7 (Navascués et al., 1995). To investigate whether infection of blood vessels might generate cells with such irregular morphologies within the retina, E3 embryos were injected with virus directly into the heart. Cells with the irregular morphologies and the previously observed distribution were observed (supplemental Fig. 5, available at www.jneurosci.org as supplemental material). Of the two dozen animals injected in the heart, approximately half had these clones, with several clones seen per retina. These cells were Olig2 negative, and no typical glial clones, or neuronal retinal clones, were observed after the heart injections (data not shown).
Glial distribution in the post-hatch chick retina
To investigate how many glial clones might be needed to carpet any region of the retina, the total number of Olig2+ cells within single 375 × 375 μm regions of either large, dense clones or small, sparse clones were quantified. Dense clones accounted for 6–10% of the Olig2+ cells within the area subserved by such clones (supplemental Fig. 6a,b, available at www.jneurosci.org as supplemental material), whereas sparse clones accounted for 2.5–5% of the Olig2+ cells within their boundaries (supplemental Fig. 6c,d, available at www.jneurosci.org as supplemental material). Thus, because there are 57 small clones, 21 medium clones, and 82 large clones in our dataset, we can estimate that 35% of the clones in the retina are small, 13% are medium, and 51% are large. From these data and the estimated coverage of each sized clone of the retina (i.e., that large clones spread across ∼5% of the surface of the chick retina but, in that patch, only accounted for 6–10% of the Olig2+ cells in that area), we estimate that there are ∼1600 clones carpeting the chick retina, approximately composed of 800 large clones, 200 medium clones, and 600 small clones.
To examine the total distribution of glia in the mature chick retina, post-hatch chick retinas were stained with Olig2, and the entire 22 × 22 mm surface area of the retina was captured at high resolution using a confocal microscope (Fig. 8a). Glial cell density was found to have a more than fivefold density change across the retina. The highest cell density was in the ventral retina, up to 37.8 cells per 100 × 100 μm, and the lowest cell density was observed in the dorsal retina, with as few as 6.8 cells per 100 × 100 μm (Fig. 8). In addition to the steep dorsoventral gradient, the central retina had a spot of very low glial cell density (Fig. 8b). Because clones contained an ∼1:1:1 ratio of the three cell types, each 100 × 100 μm region of the ventral retina would have 12.6 of each of the cell types, whereas the dorsal retina would have 2.27 of each of the cell types. Because the surface area of the chick retina is ∼160,000 μm2, there are ∼13,000 of each cell type in the chick retina, after correcting for regional differences.
To determine whether the central spot of reduced glial density might correlate with the decreased ganglion cell axon density within the central retina, which occurs in some mammals, anti-neurofilament immunohistochemistry was conducted (Fig. 8c). Using analytic methods similar to that used to quantify Olig2 staining, no significant change in the density of axons was seen in the central retina (Fig. 8d) (supplemental Fig. 6, available at www.jneurosci.org as supplemental material).
Discussion
Patterned glial cell migration into the chick retina
Infections of the chick neural tube generated groups of cells distributed in distinct patterns across the inner surface of the chick retina. The cells appeared to enter the retina, migrate across the retina, and differentiate during the time period described previously for glia of the chick retina (Fu and Qui, 2001). They appeared within the ventral fissure (the avian optic nerve head) at approximately E10 and migrated toward the periphery, while dividing and maintaining a bipolar shape, as reported previously (Nakazawa et al., 1993). The groups of cells were interpreted to be clones because of the general lack of mixing of cells infected with two different viruses encoding two different fluorescent proteins.
The pattern of distribution of clonally related cells from the optic nerve head to the periphery of the retina suggested a fairly restricted path of migration. Cells appeared to head more or less straight to the periphery once they entered the retina, with little lateral migration. Moreover, and rather surprisingly, the migration paths appeared to be clonally based. Even when GFP+ and tdTomato+ cells were intermingled at the optic nerve head, the GFP+ cells and the tdTomato+ cells sorted themselves out such that they were not intermingled at the periphery. A good example of this is shown in Figure 1c, where GFP+ cells appeared to have crossed over the path of the tdTomato+ cells but to not have joined the path of the tdTomato+ cells. In fact, we used this observation of migration path divergence in our assignment of clonal boundaries, i.e., we traced the boundaries of a clone by examining the peripheral distribution and then followed it back to the ventral fissure. An additional feature of note regarding clones was revealed by an analysis of the ratio of GFP+ to tdTomato+ clones (Fig. 7). Individual retinas within a group of retinas (e.g., experiment 231) infected with a given mixture of viruses showed quite different ratios of GFP+ to tdTomato+ clones (e.g., 4:10 in 231-2R vs 16:5 in 231-3L) (Fig. 7). This could be attributable to random variations in the infection of progenitor cells with the two types of viruses. Alternatively, it is possible that a single, or a few, infected progenitor cells in the third ventricle gave rise to subclones, each of which we scored as a clone, attributable to the fact that they created the aforementioned distinct patterns as they entered the retina. In such a case, one might observe a skewed ratio of GFP+ to tdTomato+ clones, because each retina might then reflect very few events that occurred in the ventricle, which were magnified by the formation of subclones.
What might these migration patterns suggest in terms of mechanisms of cellular distribution? First, the fairly direct trajectory away from the ventral fissure toward the periphery might suggest that glial cells follow an attractive cue located in the periphery of the retina or arrayed in a peripheral to central gradient across the retina. They may also be repulsed by a negative signal from the ventral fissure. However, if there were purely repulsive and/or purely attractive cues, one might expect cells to fan out a bit more as they migrated rather than maintaining a straight path toward the periphery. Very little spreading beyond the straight path to the periphery was seen. This behavior may be explained by a migration along axons. However, this mechanism alone would not explain the divergence of clonal paths, discussed above, in which GFP+ and tdTomato+ cells were intermingled at the start, and then sorted themselves out toward the periphery. This latter observation suggests some type of clonal recognition of cues or paths, for which there is no known basis. Whatever the mechanism is that creates the clonal patterns of migration, it is not specific to retinal glial cells. Presumptive macrophages, which originated from infections in the heart, also showed this same overall pattern of migration. Thus, the observed migration patterns may reflect a shared mechanism through which all optic nerve-derived cells cover the inner surface of the retina.
We were curious as to the distribution of glial cells across the inner retinal surface, which has not been investigated in detail in the chick. We found a relatively steep gradient of glial cell density along the dorsoventral axis. This gradient matches the gradient of rods, with rods being higher ventrally (Bruhn and Cepko, 1996). This correlation is strengthened by the observations that there is a central spot of very low density of Olig2+ cells, which likely occurs within the rod-free zone of the central retina. Furthermore, the dorsal region also contains a domain of lower glial density, and this again matches an area of lower rod density (Bruhn and Cepko, 1996). The reason for higher glial cell density in areas of high rod density is not clear but might reflect some underlying difference in metabolic activity (Okawa et al., 2008) or signaling between rods and cones and/or between ganglion cells that somehow might reflect such differences.
Chick retinal glial astrocytes and oligodendrocytes are derived from multipotent progenitors
Three cell types were observed in almost every clone of retinal glia. Of the three cell types, two had morphologies consistent with either astrocytes or oligodendrocytes (Seo et al., 2001). We were able to label the cells with oligodendrocyte morphology using antibodies to several oligodendrocyte-specific markers: PLP, MAG, and MOSP. These cells also were labeled with a GFAP polyclonal antibody, which should label astrocytes and not oligodendrocytes. Curiously, this antibody, in addition to several others that we tried, did not label cells whose morphology was consistent with that of astrocytes. It is possible that the anti-GFAP antibody that did stain the oligodendrocytes had reactivity with other proteins, e.g., intermediate filament proteins, as has been reported previously for other polyclonal anti-GFAP antibodies that react strongly to human GFAP (Vaughan et al., 1990; Dolman et al., 2004).
Antibodies to astrocyte-specific genes, such as S100B, Aldh1L1, Id3, and ALDOH, that reliably stain astrocytes in the mouse and human did not work in chick tissue (data not shown). Furthermore, in situ hybridization for S100B and Aldh1L1 failed to produce signal in the GCL. This might suggest that these genes are expressed not at all or at a level below detection (Elliott et al., 2008) However, given that cells with astrocytic morphology expressed Olig2, which has been shown to be expressed in astrocytes as well as oligodendrocytes (Cai et al., 2007), and the fact that cells with astrocytic morphology were clearly negative for all oligodendrocyte markers, we conclude that cells with the morphology of astrocytes were in fact astrocytes.
Of the 182 retinal glial clones, 179 contained astrocytes and oligodendrocytes, as well as the newly described diacyte, indicating that >98% of the infected progenitor cells were able to make all three types of glia. This is in keeping with the previous work on the O2A progenitor cell reported in the 1980s, in which cultures derived from the rat optic nerve were shown to have a progenitor cell capable of making both type 2 astrocytes and oligodendrocytes (Raff et al., 1983a,b, 1984). This previous work also showed a progenitor cell that only made type 1 astrocytes (Raff et al., 1984). In addition to clones with both astrocytes and oligodendrocytes, we did observe three small retinal glial clones with only astrocytes. Although this may support the existence of two types of glial progenitor cells, with one being restricted to making only one type of astrocyte, we were unable to notice any morphological differences among chick retinal astrocytes in the mixed clones versus the astrocyte-only clones. It is possible that, in the chick retina, all glia share common progenitor cells and there is no independent source of a “type 1” astrocyte. However, the rat does not have retinal oligodendrocytes, and we cannot directly compare our data with those from the rat optic nerve. In addition, because the chick astrocytes do not share markers or morphologies with the rat type 2 and type 1 astrocytes, we cannot conclude that indeed the chick has an “O2A” progenitor. However, the data presented here are supportive of the interpretations of the rat optic nerve cultures regarding a multipotent glial progenitor cell. Furthermore, it is thought that tectal glia are derived from both local tectal progenitor cells and progenitor cells within the floor of the third ventricle (Kim et al., 2006; Seo et al., 2008). Thus, the progenitor cells that give rise to retinal glia also may give rise to tectal glia because at least some of them likely are members of the same progenitor pool.
Novel glial cell type in the chick retina
In all of the clones containing astrocytes and oligodendrocytes, a third cell type was seen that was distinct from astrocytes and oligodendrocytes, which we have named the diacyte. This cell type had processes in the FL that resembled those of astrocytes. However, in contrast to what can be extensive astrocytic processes in the IPL, diacytes exhibited thin and short processes in the IPL. In addition, the cell bodies of diacytes were never found within the IPL, whereas astrocyte cell bodies were often observed within the IPL. The diacyte contained processes in the FL, as do oligodendrocytes, but lacked staining for any oligodendrocyte marker. It also differs from the oligodendrocyte in that its processes are much finer than the thick oligodendrocytic processes, which are at least 1 μm thick to accommodate axonal bundles. Thus, it is unlikely the diacyte is an oligodendrocyte. The diacyte is most likely not an immature glial cell, because its morphology remained distinct until P9. At this later stage, quantification of several morphological features compared the diacyte with astrocytes and oligodendrocytes. The features of process asymmetry, length, and diameter were plotted comparing FL and IPL values, and, in such plots, diacytes were consistently intermediate between astrocytes and oligodendrocytes, further supporting the classification of the diacyte as a distinct cell type. It is also unlikely that the diacyte is an Ng2+ glial cell type, because in situ hybridization for Ng2 in the postnatal chick retina showed staining in the photoreceptor layer but not the GCL (data not shown). Furthermore, Ng2+ cells are known to also express PLP, a marker that this cell type did not express (Ye et al., 2003). The function of this cell type clearly requires additional study.
Footnotes
This study was funded by the Howard Hughes Medical Institute. We thank Ben Barres (Stanford University, Stanford, CA) and Kazuhiro Ikenaka (National Institute for Physiological Sciences, Okazaki, Japan) for the PLP antibody and Ben Barres for helpful suggestions of glial markers.
References
- Bruhn SL, Cepko CL. Development of the pattern of photoreceptors in the chick retina. J Neurosci. 1996;16:1430–1439. doi: 10.1523/JNEUROSCI.16-04-01430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45:41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, Parada LF, Lu QR. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134:1887–1899. doi: 10.1242/dev.02847. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Pear WS. In: Current protocols in molecular biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Hoboken, NJ: Greene; 1997. pp. 9.9–9.14. [Google Scholar]
- Chan-Ling T, Chu Y, Baxter L, Weible Ii M, Hughes S. In vivo characterization of astrocyte precursor cells (APCs) and astrocytes in developing rat retinae: differentiation, proliferation, and apoptosis. Glia. 2009;57:39–53. doi: 10.1002/glia.20733. [DOI] [PubMed] [Google Scholar]
- Dolman DE, Lawrenson JG, Abbott NJ. Cross reactivity of polyclonal GFAP antiserum: implications for the in-vitro characterisation of brain endothelium. Brain Res. 2004;1012:185–186. doi: 10.1016/j.brainres.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Dütting D, Gierer A, Hansmann G. Self-renewal of stem cells and differentiation of nerve cells in the developing chick retina. Brain Res. 1983;312:21–32. doi: 10.1016/0165-3806(83)90117-7. [DOI] [PubMed] [Google Scholar]
- Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60:26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Espinosa de los Monteros A, Zhang M, De Vellis J. O2A progenitor cells transplanted into the neonatal rat brain develop into oligodendrocytes but not astrocytes. Proc Natl Acad Sci U S A. 1993;90:50–54. doi: 10.1073/pnas.90.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete DM, Cepko CL. Replication-competent retroviral vectors encoding alkaline phosphatase reveal spatial restriction of viral gene expression/transduction in the chick embryo. Mol Cell Biol. 1993;13:2604–2613. doi: 10.1128/mcb.13.4.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M, Richardson WD, Kessaris N. A subset of oligodendrocytes generated from radial glia in the dorsal spinal cord. Development. 2005;132:1951–1959. doi: 10.1242/dev.01777. [DOI] [PubMed] [Google Scholar]
- Fu H, Qui M. Migration and differentiation of Nkx-2.2+ oligodendrocyte progenitors in embryonic chicken retina. Brain Res Dev Brain Res. 2001;23:115–118. doi: 10.1016/s0165-3806(01)00178-x. [DOI] [PubMed] [Google Scholar]
- Gao L, Miller RH. Specification of optic nerve oligodendrocyte precursors by retinal ganglion cell axons. J Neurosci. 2006;26:7619–7628. doi: 10.1523/JNEUROSCI.0855-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, Barnett SC, Franklin RJ, Crang AJ, Mayer M, Blakemore WF, Noble M. Repair of demyelinated lesions by transplantation of purified O-2A progenitor cells. Nature. 1993;362:453–455. doi: 10.1038/362453a0. [DOI] [PubMed] [Google Scholar]
- Kahn AJ. An autoradiographic analysis of the time of appearance of neurons in the developing chick neural retina. Dev Biol. 1974;38:30–40. doi: 10.1016/0012-1606(74)90256-5. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kezic J, McMenamin PG. Differential turnover rates of monocyte-derived cells in varied ocular tissue microenvironments. J Leukoc Biol. 2008;84:721–729. doi: 10.1189/jlb.0308166. [DOI] [PubMed] [Google Scholar]
- Kim DW, Park SW, Jeon GS, Seo JH, Golden JA, Cho SS. The multiple dorsoventral origins and migratory pathway of tectal oligodendrocytes in the developing chick. Brain Res. 2006;1076:16–24. doi: 10.1016/j.brainres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Levison SW, Goldman JE. Both oligodendrocytes and astrocytes develop from progenitors in the subventricular zone of postnatal rat forebrain. Neuron. 1993;10:201–212. doi: 10.1016/0896-6273(93)90311-e. [DOI] [PubMed] [Google Scholar]
- Luskin MB, McDermott K. Divergent lineages for oligodendrocytes and astrocytes originating in the neonatal forebrain subventricular zone. Glia. 1994;11:211–226. doi: 10.1002/glia.440110302. [DOI] [PubMed] [Google Scholar]
- Luskin MB, Parnavelas JG, Barfield JA. Neurons, astrocytes, and oligodendrocytes of the rat cerebral cortex originate from separate progenitor cells: an ultrastructural analysis of clonally related cells. J Neurosci. 1993;13:1730–1750. doi: 10.1523/JNEUROSCI.13-04-01730.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menn B, Garcia-Verdugo JM, Yaschine C, Gonzalez-Perez O, Rowitch D, Alvarez-Buylla A. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DB. The avian eye and its adaptations. In: Crescitelli F, editor. The visual system in vertebrates. Berlin: Springer; 1977. pp. 549–611. [Google Scholar]
- Mishima H, Fujita H. Studies on the cytodifferentiation of the neuroblasts and visual cells in the chick embryo retina, using the electron-microscopic autoradiography of 3H-thymidine. Albrecht Von Graefes Arch Klin Exp Ophthalmol. 1978;206:1–10. doi: 10.1007/BF00411331. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Morris VB, Cowan R. An analysis of the growth of the retinal cell population in embryonic chicks yielding proliferative ratios, numbers of proliferative and non-proliferative cells and cell-cycle times for successive generations of cell cycles. Cell Prolif. 1995;28:373–391. doi: 10.1111/j.1365-2184.1995.tb00079.x. [DOI] [PubMed] [Google Scholar]
- Nakazawa T, Tachi S, Aikawa E, Ihnuma M. Formation of the myelinated nerve fiber layer in the chicken retina. Glia. 1993;8:114–121. doi: 10.1002/glia.440080207. [DOI] [PubMed] [Google Scholar]
- Naldini L, Blömer U, Gage FH, Trono D, Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci U S A. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navascués J, Moujahid A, Almendros A, Marin-Teva JL, Cuadros MA. Origin of microglia in the quail retina: central-to-peripheral and vitreal-to-scleral migration of microglial precursors during development. J Comp Neurol. 1995;354:209–228. doi: 10.1002/cne.903540205. [DOI] [PubMed] [Google Scholar]
- Okawa H, Sampath AP, Laughlin SB, Fain GL. ATP consumption by mammalian rod photoreceptors in darkness and in light. Curr Biol. 2008;18:1917–1921. doi: 10.1016/j.cub.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Tsumori T, Kishi T, Yokota S, Yasui Y. Developmental appearance of oligodendrocytes in the embryonic chick retina. J Comp Neurol. 1998;398:309–322. [PubMed] [Google Scholar]
- Raff MC, Miller RH, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature. 1983a;303:390–396. doi: 10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- Raff MC, Abney ER, Cohen J, Lindsay R, Noble M. Two types of astrocytes in cultures of developing rat white matter: differences in morphology, surface gangliosides, and growth characteristics. J Neurosci. 1983b;3:1289–1300. doi: 10.1523/JNEUROSCI.03-06-01289.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff MC, Abney ER, Miller RH. Two glial cell lineages diverge prenatally in rat optic nerve. Dev Biol. 1984;106:53–60. doi: 10.1016/0012-1606(84)90060-5. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci. 2008;11:1392–1401. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompani SB, Cepko CL. Retinal progenitor cells can produce restricted subsets of horizontal cells. Proc Natl Acad Sci U S A. 2008;105:192–197. doi: 10.1073/pnas.0709979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Seo JH, Haam YG, Park SW, Kim DW, Jeon GS, Lee C, Hwang DH, Kim YS, Cho SS. Oligodendroglia in the avian retina: immunocytochemical demonstration in the adult bird. J Neurosci Res. 2001;65:173–183. doi: 10.1002/jnr.1140. [DOI] [PubMed] [Google Scholar]
- Seo JH, Chang JH, Song SH, Lee HN, Jeon GS, Kim DW, Chung CK, Cho SS. Spatiotemporal gradient of astrocyte development in the chick optic tectum: evidence for multiple origins and migratory paths of astrocytes. Neurochem Res. 2008;33:1346–1355. doi: 10.1007/s11064-008-9590-3. [DOI] [PubMed] [Google Scholar]
- Temple S, Raff MC. Differentiation of a bipotential glial progenitor cell in a single cell microculture. Nature. 1985;313:223–225. doi: 10.1038/313223a0. [DOI] [PubMed] [Google Scholar]
- Turner DL, Cepko CL. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- Turner DL, Snyder EY, Cepko CL. Lineage-independent determination of cell type in the embryonic mouse retina. Neuron. 1990;4:833–845. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- Vallstedt A, Klos JM, Ericson J. Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron. 2005;45:55–67. doi: 10.1016/j.neuron.2004.12.026. [DOI] [PubMed] [Google Scholar]
- Vaughan DK, Erickson PA, Fisher SK. Glial fibrillary acidic protein (GFAP) immunoreactivity in rabbit retina: effect of fixation. Exp Eye Res. 1990;50:385–392. doi: 10.1016/0014-4835(90)90139-l. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Raff MC. Retinal astrocytes are immigrants from the optic nerve. Nature. 1988;332:834–837. doi: 10.1038/332834a0. [DOI] [PubMed] [Google Scholar]
- Ye P, Bagnell R, D'Ercole AJ. Mouse NG2+ oligodendrocyte precursors express mRNA for proteolipid protein but not its DM-20 variant: a study of laser microdissection-captured NG2+ cells. J Neurosci. 2003;23:4401–4405. doi: 10.1523/JNEUROSCI.23-11-04401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerlin M, Milosevic A, Goldman JE. Glial progenitors of the neonatal subventricular zone differentiate asynchronously, leading to spatial dispersion of glial clones and to the persistence of immature glia in the adult mammalian CNS. Dev Biol. 2004;270:200–213. doi: 10.1016/j.ydbio.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Zhu X, Bergles DE, Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008;135:145–157. doi: 10.1242/dev.004895. [DOI] [PubMed] [Google Scholar]