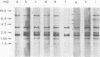
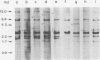
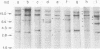
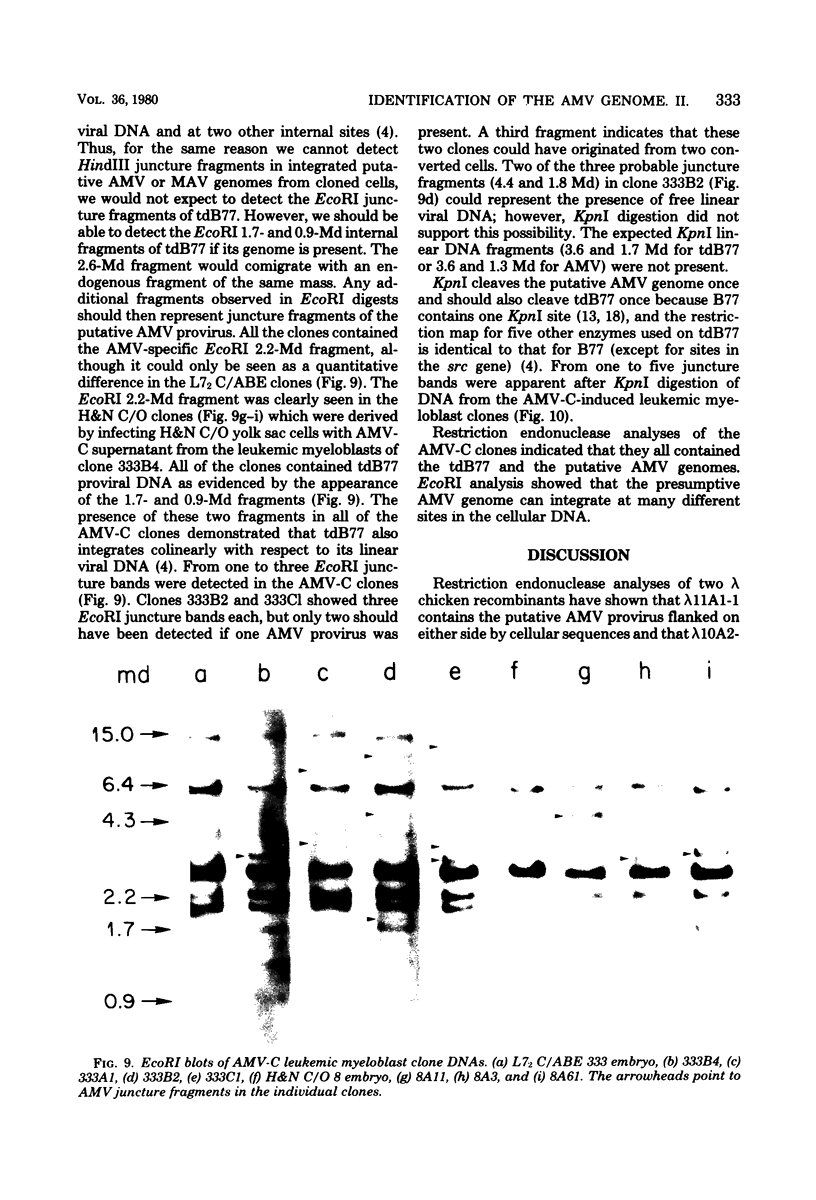
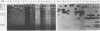Abstract
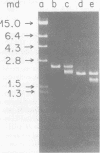
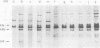
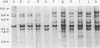
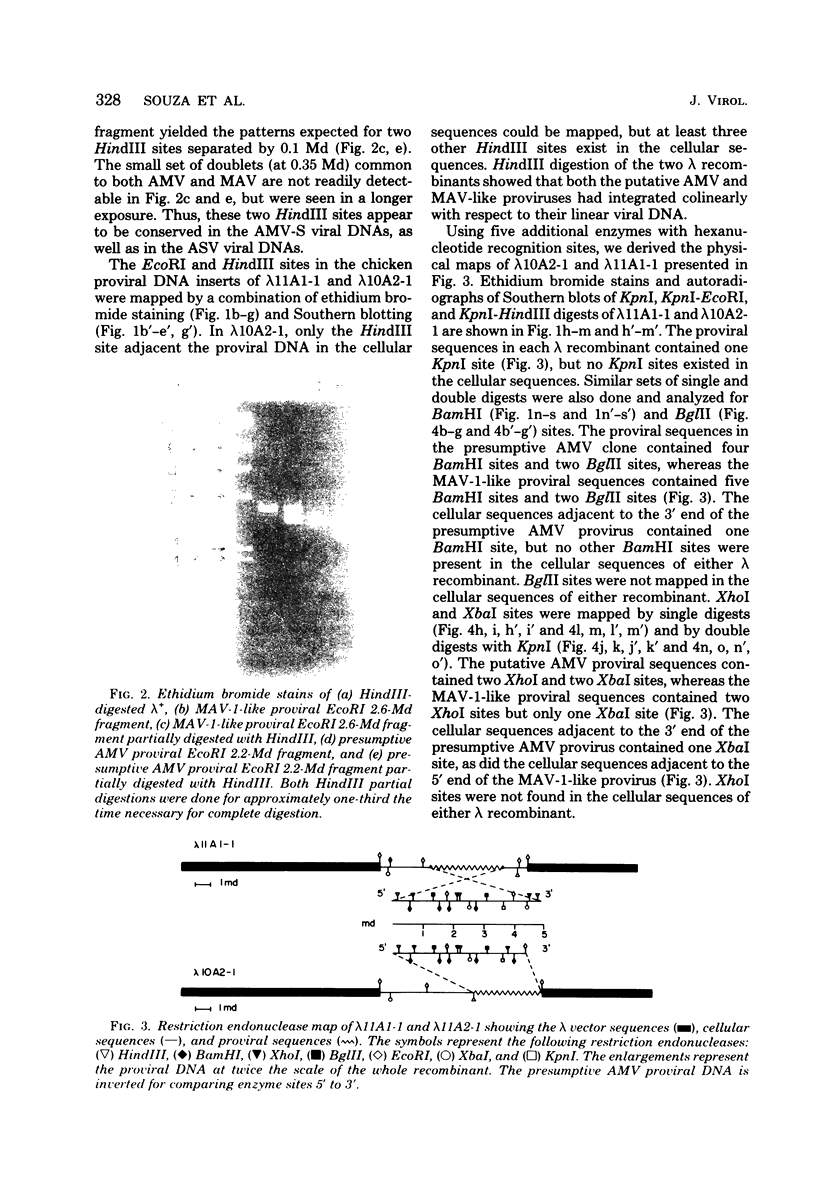
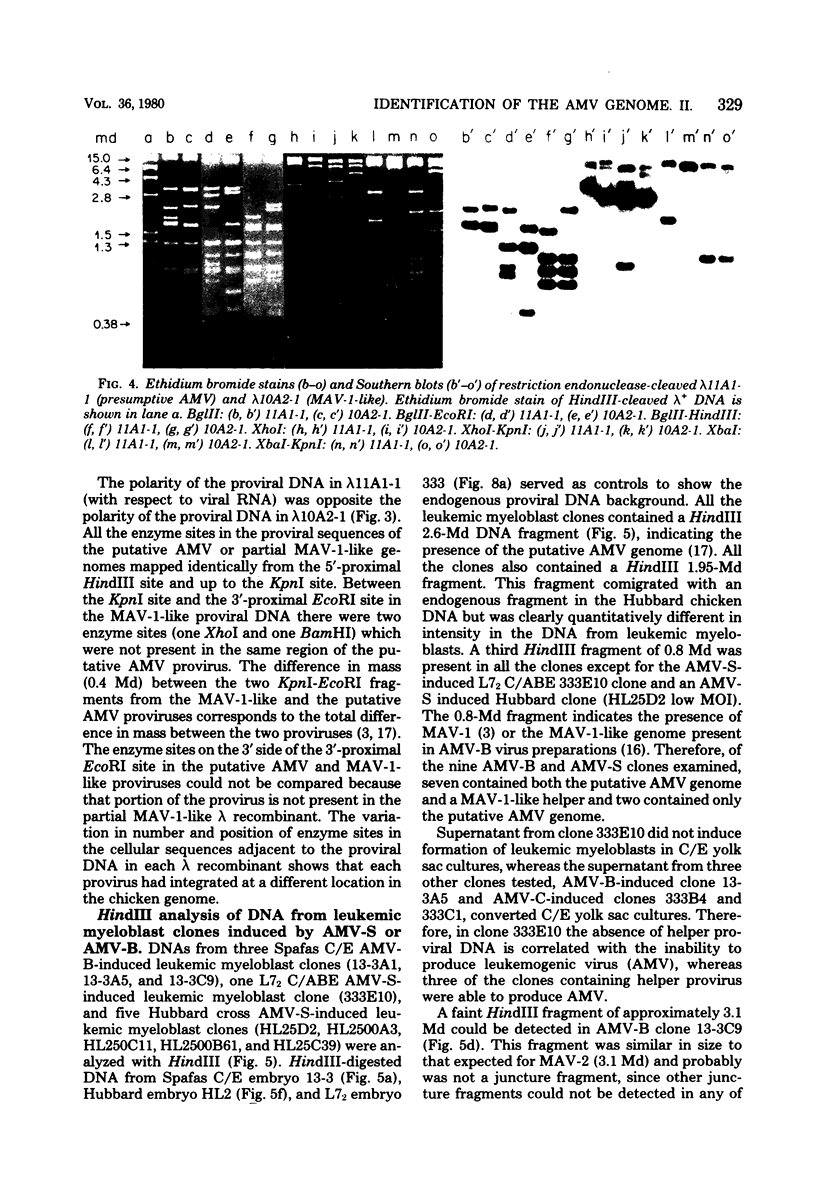
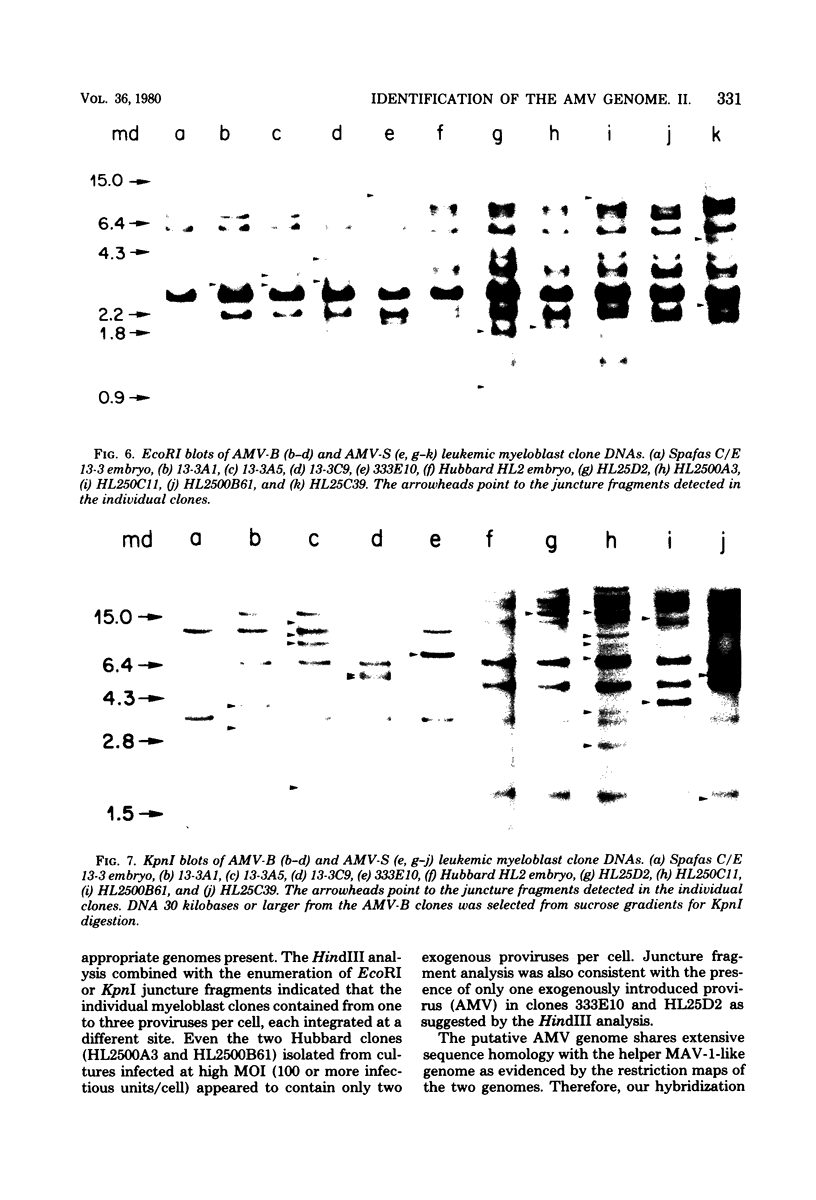
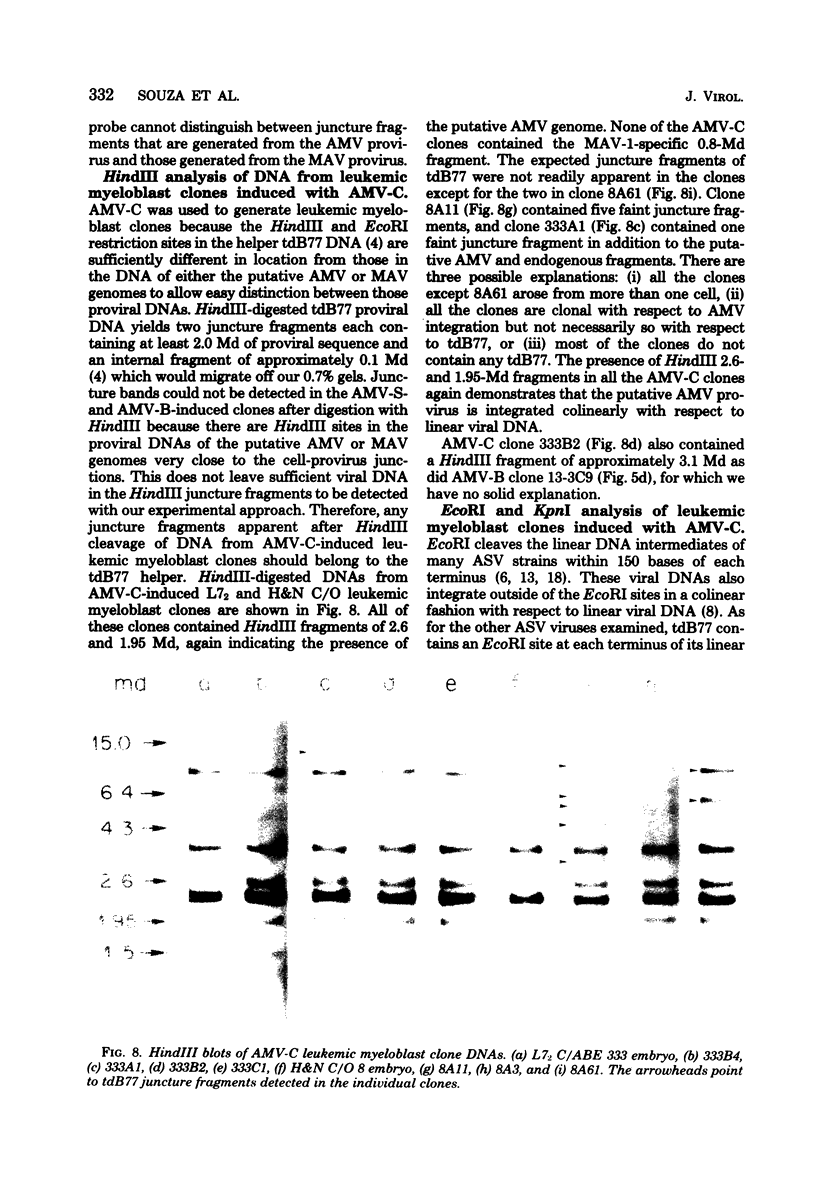
Two lambda proviral DNA recombinants were characterized with a number of restriction endonucleases. One recombinant contained a complete presumptive avian myeloblastosis virus (AMV) provirus flanked by cellular sequences on either side, and the second recombinant contained 85% of a myeloblastosis-associated virus type 1 (MAV-1)-like provirus with cellular sequences adjacent to the 5' end of the provirus. Comparing the restriction maps for the proviral DNAs contained in each lambda hybrid showed that the putative AMV and MAV-1-like genomes shared identical enzyme sites for 3.6 megadaltons beginning at the 5' termini of the proviruses with respect to viral RNA. Two enzyme sites near the 3'-end of the MAV-1-like provirus were not present in the putative AMV genome. We also examined a number of leukemic myeloblast clones for proviral content and cell-provirus integration sites. The presumptive AMV provirus was present in all the leukemic myeloblast clones regardless of the endogenous proviral content of the target cells or the AMV pseudotype used for conversion. Multiple cellular sites were suitable for integration of the putative AMV genome and the helper genomes. The proviral genomes were all integrated colinearly with respect to linear viral DNA.
Full text
PDF

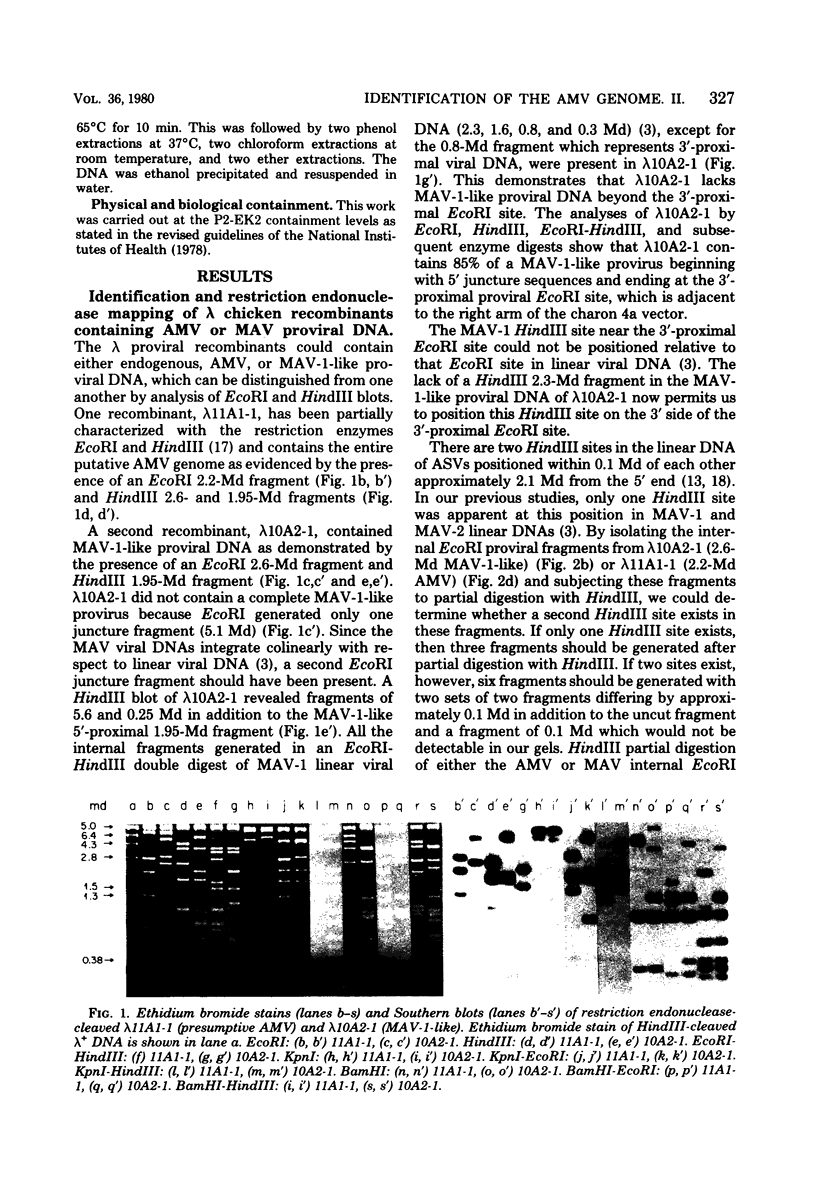




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Astrin S. M. Endogenous viral genes of the White Leghorn chicken: common site of residence and sites associated with specific phenotypes of viral gene expression. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5941–5945. doi: 10.1073/pnas.75.12.5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALUDA M. A., GOETZ I. E. Morphological conversion of cell cultures by avian myeloblastosis virus. Virology. 1961 Oct;15:185–199. doi: 10.1016/0042-6822(61)90234-3. [DOI] [PubMed] [Google Scholar]
- Bergmann D. G., Souza L. M., Baluda M. A. Characterization of avian myeloblastosis-associated virus DNA intermediates. J Virol. 1980 May;34(2):366–372. doi: 10.1128/jvi.34.2.366-372.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmer T. M., Parsons J. T. Analysis of cellular integration sites in avian sarcoma virus infected duck embryo cells. J Virol. 1979 Dec;32(3):762–769. doi: 10.1128/jvi.32.3.762-769.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Hsu T. W., Sabran J. L., Mark G. E., Guntaka R. V., Taylor J. M. Analysis of unintegrated avian RNA tumor virus double-stranded DNA intermediates. J Virol. 1978 Dec;28(3):810–818. doi: 10.1128/jvi.28.3.810-818.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. H., Payvar F., Spector D., Schimke R. T., Robinson H. L., Payne G. S., Bishop J. M., Varmus H. E. Heterogeneity of genetic loci in chickens: analysis of endogenous viral and nonviral genes by cleavage of DNA with restriction endonucleases. Cell. 1979 Oct;18(2):347–359. doi: 10.1016/0092-8674(79)90054-0. [DOI] [PubMed] [Google Scholar]
- Hughes S. H., Shank P. R., Spector D. H., Kung H. J., Bishop J. M., Varmus H. E., Vogt P. K., Breitman M. L. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978 Dec;15(4):1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- McClements W., Hanafusa H., Tilghman S., Skalka A. Structural studies on oncornavirus-related sequences in chicken genomic DNA: two-step analyses of EcoRI and Bgl I restriction digests and tentative mapping of a ubiquitous endogenous provirus digests and tentative mapping of a ubiquitous endogenous provirus. Proc Natl Acad Sci U S A. 1979 May;76(5):2165–2169. doi: 10.1073/pnas.76.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscovici C., Zanetti M. Studies on single foci of hematopoietic cells transformed by avian myeloblastosis virus. Virology. 1970 Sep;42(1):61–67. doi: 10.1016/0042-6822(70)90238-2. [DOI] [PubMed] [Google Scholar]
- Sabran J. L., Hsu T. W., Yeater C., Kaji A., Mason W. S., Taylor J. M. Analysis of integrated avian RNA tumor virus DNA in transformed chicken, duck and quail fibroblasts. J Virol. 1979 Jan;29(1):170–178. doi: 10.1128/jvi.29.1.170-178.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Hughes S. H., Kung H. J., Majors J. E., Quintrell N., Guntaka R. V., Bishop J. M., Varmus H. E. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell. 1978 Dec;15(4):1383–1395. doi: 10.1016/0092-8674(78)90063-6. [DOI] [PubMed] [Google Scholar]
- Souza L. M., Baluda M. A. Identification of the avian myeloblastosis virus genome. I. Identification of restriction endonuclease fragments associated with acute myeloblastic leukemia. J Virol. 1980 Nov;36(2):317–324. doi: 10.1128/jvi.36.2.317-324.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza L. M., Komaromy M. C., Baluda M. A. Identification of a proviral genome associated with avian myeloblastic leukemia. Proc Natl Acad Sci U S A. 1980 May;77(5):3004–3008. doi: 10.1073/pnas.77.5.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M., Hsu T. W., Lai M. M. Restriction enzyme sites on the avian RNA tumor virus genome. J Virol. 1978 May;26(2):479–484. doi: 10.1128/jvi.26.2.479-484.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]