Abstract
Many ion channels are modulated by multiple stimuli, which allow them to integrate a variety of cellular signals and precisely respond to physiological needs. Understanding how these different signaling pathways interact has been a challenge in part because of the complexity of underlying models. In this study, we analyzed the energetic relationships in polymodal ion channels using linkage principles. We first show that in proteins dually modulated by voltage and ligand, the net free-energy change can be obtained by measuring the charge-voltage (Q-V) relationship in zero ligand condition and the ligand binding curve at highly depolarizing membrane voltages. Next, we show that the voltage-dependent changes in ligand occupancy of the protein can be directly obtained by measuring the Q-V curves at multiple ligand concentrations. When a single reference ligand binding curve is available, this relationship allows us to reconstruct ligand binding curves at different voltages. More significantly, we establish that the shift of the Q-V curve between zero and saturating ligand concentration is a direct estimate of the interaction energy between the ligand- and voltage-dependent pathway. These free-energy relationships were tested by numerical simulations of a detailed gating model of the BK channel. Furthermore, as a proof of principle, we estimate the interaction energy between the ligand binding and voltage-dependent pathways for HCN2 channels whose ligand binding curves at various voltages are available. These emerging principles will be useful for high-throughput mutagenesis studies aimed at identifying interaction pathways between various regulatory domains in a polymodal ion channel.
INTRODUCTION
Voltage-gated ion channels constitute a superfamily of ion channels that have evolved over millions of years and are known to mediate a variety of physiological processes ranging from electrical and chemical signaling (Hille, 2001) to gene expression (Dolmetsch, 2003) and cell division (DeCoursey et al., 1984; Day et al., 1993). Among the various protein superfamilies involved in signal transduction, they are one of the largest. They share a common minimal architecture of a central pore domain surrounded by four voltage-sensing domains (Long et al., 2005; Moiseenkova-Bell et al., 2008; Wang and Sigworth, 2009; Payandeh et al., 2011). In addition to membrane potential, some members of this superfamily are regulated by other physical and chemical stimuli, which sometimes involve additional regulatory domains. For instance, the BK channels are also regulated by free calcium (Marty, 1981), whereas the activity of members of the transient receptor potential (TRP) channel family is regulated by heat, ligands, and/or mechanical stretching (Caterina et al., 1997; Clapham, 2003; Voets et al., 2004). This polymodal behavior of many members of the voltage-gated ion channel superfamily makes them outstanding models to study the fundamental basis of signal propagation in membrane proteins (Horrigan and Aldrich, 2002).
To gain insight into the mechanisms of how information is transferred between domains, it is necessary to map the network of residues involved in mediating such crosstalk. This appears seemingly straightforward because all it would require is to measure the effect of site-specific perturbation on interaction energies. In practice, however, domain-level interaction energies cannot be directly estimated and require development of detailed discrete state models to fully explain the voltage- and ligand-dependent properties of these ion channels. This can be an enormous challenge because even a simple model of a dually regulated tetrameric ion channel requires a minimum of 50 states (Magleby, 2003). Symmetry arguments would reduce the number of free equilibrium parameters to a minimum of six, but even this is by no means straightforward. These practical considerations restrict the analyses of interaction networks to a limited set of well-defined sites and to systems where detailed kinetic models have been developed.
In this study, we examine the free-energy relationships between ligand- and voltage-gating pathways of polymodal ion channels. Our aim is to develop a theoretical framework that will allow us to extract the various free energies without requiring a detailed knowledge of kinetic models. Our approach is, to a degree, an extension of Wyman’s linkage principles to electrochemical equilibria (Wyman and Gill, 1990). A direct consequence of these ideas is that the total chemical free-energy change of a voltage- or ligand-dependent system is directly estimable by extracting the median voltage of activation or the median ligand concentration from the gating-charge displacement (charge-voltage; Q-V) or the ligand binding curve, respectively (Wyman, 1967; Chowdhury and Chanda, 2012). Here, we show that the ligand binding curve at any voltage can be obtained by measuring the Q-V curves at multiple ligand concentrations and a single reference ligand binding curve. This information can be further transformed to obtain the net free-energy change of a bimodal system. Finally, we show that the net displacement of the Q-V curve on the voltage axis between zero and fully saturating ligand concentrations is linearly related to the total interaction energy of the ligand binding sites with the pore and the voltage sensors. We apply these theoretical propositions on BK channels and hyperpolarization-activated hyperpolarization activated cyclic nucleotide-gated (HCN) channels, both of which are regulated by voltage and ligand, to estimate the interaction energies between the two regulatory domains. Finally, we discuss the potential of applicability of our theoretical framework to systems of varying size and complexity and even to nonallosteric systems.
THEORY
Free-energy component for the ligand- or voltage-dependent pathway
As shown by Wyman, the work done to saturate a macromolecule with a ligand can be directly estimated from the ligand binding curve, by extracting the median ligand concentration (Wyman, 1967; Wyman and Gill, 1990). In his original derivation, however, it was assumed that the free energy of the macromolecule does not change upon ligand binding (Wyman, 1964). Here, we show that such an assumption is not essential to deduce energetic information from the binding curves.
Consider a protein that can bind a ligand (X). The protein is presumed to exist in N different conformational states. Each state is associated with a variable ni, which depicts the number of molecules of ligand X bound to the conformational state. The probability of occurrence of each state is assumed to follow a Boltzmann distribution such that
where β = 1/kBT, Ei is the energy of the state, and Z is the partition function of the system (). The Boltzmann weight of each state, Bi (= ), is assumed to have an overall functional form as:
| (1) |
In Eq. 1, x represents the ligand concentration and is related to the chemical energy of the state (which includes contributions from the intrinsic energies of the different component structural units and the interactions between them). The probability distribution of the conformational states will depend on the ligand concentration. The average number of ligands bound to the protein is:
| (2) |
The free energy of the system is:
| (3) |
For a small change in ligand concentration, the change in the free-energy of system can be obtained through partial differentiation of Eq. 3 and thus:
| (4) |
Using Eqs. 2 and 3, Eq. 4 can be converted to:
| (5) |
To obtain the net change in free-energy of the system due to a process completely driven by the ligand, we integrate Eq. 5 from zero to a specified ligand concentration:
| (6) |
Thus the free energy associated with a ligand binding process is related to the area under a ligand binding curve ( vs. ln x). Integrating the right side of Eq. 6, by parts, gives:
| (7) |
In the above integral equation (Eq. 7), we have used the boundary condition that at zero ligand concentration, = 0. At saturating ligand concentration, approaches Nmax, which is the maximum number of ligands that each protein molecule can bind. Using these limits in Eq. 7, the net free-energy change associated with full-scale transformation of the protein, associated with binding of ligand, is:
| (8) |
The free-energy expression in Eq. 8 comprises of two terms: the first is a nonsaturating component that increases in magnitude with increasing ligand concentration; the second component is a saturating component that equals the area between the ligand binding curve and the ordinate axis, as shown in Fig. 1.
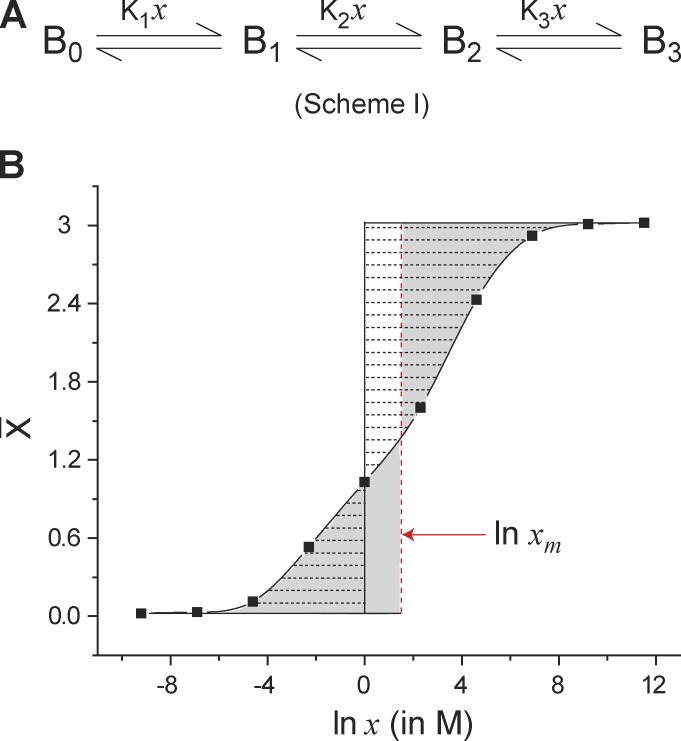
Figure 1.
Median ligand activity of an arbitrary three-step ligand binding process. (A) A three-step sequential ligand binding process (Nmax = 3) where Bi represents the state of the protein with “i” ligands bound (i = 0, 1, 2, and 3) and Kj is the equilibrium constant for the jth step of the process (j = 1, 2, or 3). In each step, a single molecule of the ligand binds to the protein, B. (B) The ligand binding curve for Scheme I generated using arbitrary values of Kj (K1 = 0.01 M−1, K2 = 0.1 M−1, K3 = 10 M−1). Demarcated by horizontal broken lines is the area between the binding curve and the ln x = 0 axis (solid black vertical line), which represents the integral . By virtue of the integration, the area on the right of the ln x = 0 axis is negative and that on its right is positive. The sum of these areas (taking their signs into consideration) is a measure of total free energy associated with saturating the protein with the ligands (, where ). The vertical red dashed line is the median ligand activity axis. By its definition, the two shaded regions on either side of the ln xm axis are equal in area. Eq. 11 implies that the region demarcated by the horizontal dashed lines and the rectangle defined by the ln x = 0, ln xm, = 0, and = Nmax lines are equal in area and that both give a measure of .
What do these individual components mean? We take a look at the overall “reaction” that is occurring here:
[P]and represent the completely unbound and bound states of the protein. The overall free-energy change of this process can be represented as:
| (9) |
μ of each component of the reaction is its chemical potential. The chemical potential of the free/unbound ligand in turn has two components, with the standard chemical potential (μ0) superposed on a secondary term, kBT ln x, which originates from “mixing” (entropic factors) of the free ligand in solution (Depaula and Atkins, 2009). Comparing Eqs. 8 and 9, therefore, tells us that the nonsaturating term in Eq. 8 is the mixing term, whereas the second term is associated with the full-scale binding of the ligand to the protein. The difference in our derivation from Wyman’s principally results in the appearance of the nonsaturating component in the free-energy change expression (Eq. 8).
Following Wyman’s elegant mathematical analysis, the saturating component of Eq. 8 () can be shown to be related to the overall chemical free-energy change of the protein–ligand system using the median ligand activity (xm; Wyman, 1964). The latter is defined as follows (Fig. 1):
| (10) |
Using this definition, is:
| (11) |
The thermodynamic equations describing a ligand binding process are strictly analogous to those of a voltage-driven process. We have previously shown (Chowdhury and Chanda, 2012) that for a purely voltage-driven process, the net change in the chemical free energy of the system can be estimated directly via the measurement of the gating-charge displacement versus voltage curves as:
| (12) |
where V is the voltage, is the mean gating-charge of the ensemble, Qmax is the maximum number of gating-charge translocated in the voltage-dependent activation of the protein, and VM is the median voltage of activation.1 VM is defined in a manner similar to Eq. 10:
| (13) |
The physical significance of the median transformation of the ligand binding and gating-charge displacement curves is discussed in more detail in Appendix 1.
The preceding thermodynamic relations, although esoteric in construction, are simply the consequence of a fundamental thermodynamic proposition; the work done by a system undergoing a reversible change equals the decrease in the free energy of the system. Thus the free-energy change of a system caused by a change in an intensive environmental variable (such as voltage, pressure, temperature, ligand concentration, etc.) is directly estimable through the measurement of the conjugate extensive (charge, volume, entropy, bound ligands, etc.). The median measure (median voltage, median ligand concentration) is simply a convenient transformation of the free-energy change, enabling straightforward comparisons to gauge the energetic perturbation of the system. The physical implication of the median transformation is further discussed in Appendix 1.
Net free-energy change for combined ligand and voltage-dependent activation
So far we have seen that the Q-V and the ligand binding curve give an accurate estimate of the total free-energy change associated with the full-scale activation of a voltage-dependent and a ligand-dependent process, respectively. How does one estimate the net free-energy change when the channels are regulated by both voltage and ligand?
Consider a protein with dual sensitivity to voltage and a ligand (X). It exists in N different conformational states, each of which has an associated charge (or valence), qi, as well as a variable ni, which depicts the number of molecules of ligand X bound to the conformational state. The probability distribution of states, and thus the free energy of the system, , will now be sensitive to voltage as well as ligand. is the mean gating charge of the entire ensemble and is equal to , where Pi (V, x) is the occupancy of the ith state at voltage V and ligand concentration, x. Similarly, is the average number of bound ligands.
The differential change in free energy in such a system will be:
| (14) |
In Eq. 14, the subscript in each partial differential indicates the parameter held constant during the differentiation. Using our previous descriptions in Eq. 14, it can be shown that:
| (15) |
Suppose we measure the Q-V curve at a particular ligand concentration. Integrating the area under the curve will yield a measure of the net free-energy change that has occurred:
| (16a) |
This free-energy change is the work done to change the distribution of states by changing only voltage, whereas ligand concentration is maintained as a constant. A similar relationship can be derived for a transformation caused by a change in ligand concentration alone:
| (16b) |
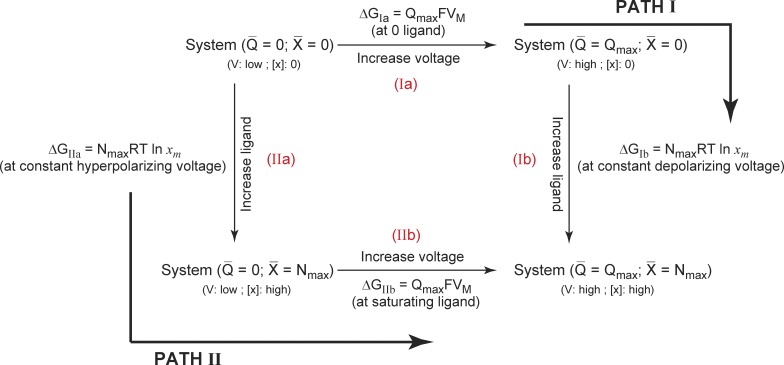
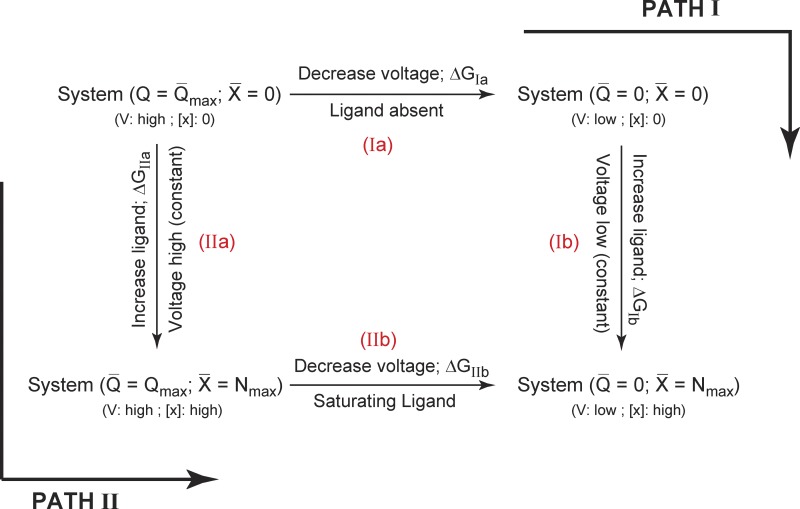
To deconstruct the free-energy changes associated with charge movement and ligand binding in such a system, we look at the thermodynamic cycle shown in Fig. 2. Our initial reference state (Sref) is the one where all the gating charges are in the resting state with no bound ligands. The final state of the system (Sfinal) is where all the gating charges have translocated to activated state and the channel is fully liganded. We are specifically interested in evaluating the free-energy difference between Sfinal and Sref, i.e., . The net free energy of activation of this system can be calculated in two different ways. In the first pathway, all the gating charges are moved (step I a), and subsequently the ligands are allowed to bind (step I b), whereas in the second pathway, we first saturate the system with ligands (step II a) and then allow the translocation of the gating-charges (step II b). Each of these steps are associated with a free-energy change and, therefore, the net free-energy change associated with the full-scale activation of the protein is ΔGIa + ΔGIb = ΔGIIa + ΔGIIb = .
Figure 2.
A thermodynamic cycle for a channel dually modulated by voltage and ligand. The reference state of the system is that in which all gating charges are in their resting position and all ligand binding sites are unoccupied (at hyperpolarizing voltages and zero ligand concentration). The final state of the system is where all gating charges have moved and all ligand binding sites are occupied by the ligand (depolarizing voltages and saturating ligand concentrations). The free-energy difference between these two states can be calculated by two different ways. In Path I, Q-V is measured at zero ligand concentration followed by the ligand binding curve at highly depolarized voltages. In Path II, the ligand binding curve is measured at hyperpolarizing voltages, whereas the Q-V curve is measured at saturating ligand concentration. The free energy expressions for each of the steps are shown alongside.
We first consider the free-energy estimation strategy along Path I. From our previous discussion, ΔGIa is estimable from the Q-V curve for the channel at zero ligand concentration, whereas ΔGIb can be calculated from the ligand binding curve obtained at highly depolarizing voltages. Thus the total free-energy change in the system, ΔGnet, can be obtained as:
| (17) |
The first integral in Eq. 17 is the area under the Q-V curve at zero ligand concentration, whereas the second is the area under the ligand binding curve at highly depolarized voltages, when all charge has moved. Similarly the free-energy estimation along the second pathway can be accomplished as:
In the preceding equation, the first integral is the area under the ligand binding curve at highly hyperpolarized voltages, whereas the second integral is the area under the Q-V curve at saturating ligand concentrations. The chemical component of this free-energy change, , can be extracted by calculation of median variables (Eqs. 10–13). In summary,
| (18) |
Voltage dependence of ligand occupancy from Q-V curves
As shown in the previous section, if we measure the ligand binding curves at various voltages and the Q-V curves at various ligand concentrations, we can directly map the free-energy landscape for dual regulation by ligand and voltage (Eq. 16). Although the Q-V curves can, in most cases, be measured at different ligand concentrations, estimating the ligand binding curve at very high or very low voltages is not straightforward. In this section, we describe how this information can be extracted from thermodynamic principles.
Because the free energy, , is a state function, the differential, , (from Eqs. 14 and 15) is exact. The result is the relation:
| (19a) |
or
| (19b) |
The differentials depicted in Eqs. 19a and 19b are reminiscent of Maxwell’s thermodynamic relations. We multiply both sides of Eq. 19b with dV and perform the integration, i.e.:
| (20a) |
This integral equation can be rewritten as:
| (20b) |
Eq. 20b tells us that the gradient of median voltage of activation with respect to the natural logarithm of the ligand concentration is linearly related to the change in the number of ligands bound to protein, because of a change in voltage. Note that in Eq. 20b it has been assumed that Qmax is not affected by the ligand.
Although it may be experimentally challenging to obtain ligand binding curves at different voltages, obtaining a single ligand binding curve in the absence of membrane potential (0 mV) is relatively simpler. By using this reference binding curve and Q-V curves at different ligand concentrations, it is possible to obtain the entire ligand binding surface. Integrating Eq. 20a and within finite limits:
| (21) |
The right-hand side of Eq. 21 is the change in the ligand occupancy caused by a change in voltage from Vref to V. This can be obtained by computing the gradient of the area under the Q-V curve, between Vref and V, with respect to ln x. Because V is arbitrary and (the ligand binding curve at a reference voltage) is available, we can potentially generate the entire ligand binding surface. Similarly, the Q(V, x) surface can also be obtained once a reference Q-V curve and ligand binding curves at different voltages are generated.
Free energy of interaction between voltage- and ligand-dependent pathways
In the bimodal systems considered here, a change in the Q-V curve caused by changes in ligand concentration is caused by energetic coupling of the voltage-dependent and ligand-dependent pathways. How can the strength of this energetic linkage be estimated? Under zero ligand conditions, the area under the Q-V curve is the work done to move all the charges when all the ligand binding sites are unoccupied (ΔGIa). Under saturating ligand conditions, the area bound by the Q-V curve is the work done to move all the charges when all the ligand binding sites are occupied (ΔGIIb). The difference between the two energy measures is the energetic connectivity between the voltage-dependent and the ligand-dependent processes. Mathematically, the net free energy of interaction, , between the ligand-dependent and voltage-dependent pathways is:
| (22) |
Thus, the change in the areas under the Q-V curve between fully saturating and zero ligand concentrations gives a measure of the energetic connectivity between the voltage- and ligand-dependent pathways. Specifically, if the maximum charge translocated in the voltage-dependent activation pathway is unchanged due to the ligand, Eq. 22 can be rewritten as:
| (23a) |
This principle is illustrated for a hypothetical channel gated by voltage and ligand (Fig. 3 A). As shown in Fig. 3 B, the area between the Q-V curves measured at zero and fully saturating ligand concentrations equals the area of the rectangle bound by the median voltage axes of the two Q-V curves. This area is the energetic facilitation afforded by ligand binding on the voltage-dependent activation pathway of the protein.
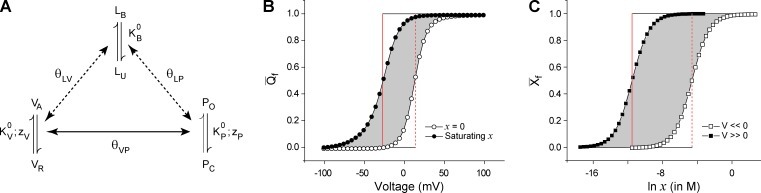
Figure 3.
Energetic linkage between the voltage-dependent and ligand-dependent pathways. (A) An allosteric model of a hypothetical ion channel with a single voltage-sensing and ligand binding domain. The voltage sensor exists in two states (VR and VA) and its activation energetics are governed by its intrinsic equilibrium constant, , and its voltage dependence, zV. The pore unit has an intrinsic activation constant and intrinsic voltage dependence given by and zP, respectively. The ligand binding domain has an intrinsic binding affinity of . All the three “structural units” are allosterically coupled to each other via interactions represented by the terms θVP, θLV, and θLP. The model parameters are = 0.1, = 0.01, = 100 M−1, θVP = 100, θLV = 1, θLP = 1,000, zV = 2.5, and zP = 1.5. (B) Simulated fractional gating charge displacement (Qf vs. V) curves for this system at saturating (closed circles) and zero (open circles) ligand concentrations. The red vertical line is the VM axis for each of the curves and the shaded area is equal to the area of the rectangle bound the two VM axes. This area, when scaled by Qmax of the system (zP + zV), gives the net energetic facilitation of the voltage-dependent pathway by the ligand and is equal to −kBTln(θLVθLP). (C) The fractional ligand binding curves (Xf vs. ln x) at very low (open circles) and very high voltages (closed circles) are shown, and the red vertical lines are the corresponding ln xm axes. The area of the shaded region is equal to area of the rectangle bound the two ln xm axes, which when scaled by Nmax of the system (equal to 1 in this case) and kBT, gives the net energetic facilitation of the ligand-dependent pathway by voltage and is equal to −kBTln(θLVθLP). Note that the two shaded in areas in B and C after appropriate scaling will be equal.
A similar relation can be derived for the ligand binding curve as well, wherein is estimated from the difference in the area’s ligand binding curves obtained at highly depolarized (all gating charge has moved) and hyperpolarized (all charges in resting configuration) voltages (Fig. 3 C). This is a consequence of the thermodynamic cycle in Fig. 2: ΔGIIb − ΔGIa = ΔGIb − ΔGIIa. Thus, in terms of the ligand binding curves, will be:
| (23b) |
Eqs. 22, 23a, and 23b have an important historical context to understand, with which we transform Eq. 22 as:
| (24) |
The partial differential under the double integral on the left of Eq. 24 is an example of hetero-tropic linkage functions (Wyman, 1984; Gill et al., 1985; Wyman and Gill, 1990), which have been the cornerstone of Wyman’s linkage theory (Wyman, 1964) and have been used to understand the cross-regulation of enzyme activity by multiple modulators. Our derivation that the double integral of the linkage function is an energetic measure of cooperativity (or linkage) in the system is one example of its utility. In general, the reason why such a mathematical formulation reflects cooperativity in a system can be understood by evaluating the partial differential in terms of the microscopic state/system variables:
| (25) |
Noting the functional expression on the right of Eq. 25, one can see that the linkage function is actually a measure of the covariance between gating-charge movement and ligand binding. If the two processes are strongly interdependent, the covariance will be higher, and in the case of independent processes, the covariance will be zero.
RESULTS
Ligand binding curves as an estimate of total free-energy change in ligand gated ion channels
The first major conclusion of our thermodynamic treatment is that total free-energy change associated with saturating a macromolecule with a ligand is estimable from the ligand binding curve, via extraction of the median ligand activity (ln xm) and the maximum number of ligands that can bind to the macromolecule. This is fundamentally different from the Hill equation based metrics to characterize the free-energy responses in ligand-gated ion channels (Nache et al., 2008; Gleitsman et al., 2009). We consider two ligand-gated channels, the cyclic nucleotide-gated (CNG) channels and the HCN channels, both of whose direct ligand binding curves have been measured using a fluorescent ligand (Biskup et al., 2007; Kusch et al., 2010, 2012). Kinetic models of activation of these channels have been developed using constraints from direct ligand binding and ionic current measurements. We compared the free-energy estimate obtained directly from the kinetic model parameters and the median metric, extracted from the simulated ligand binding curves.
In both these channels, there are four ligand binding sites. The cooperativity in the different ligand binding steps are asymmetric: the second and fourth steps of ligand binding show positive coupling, whereas the third step shows negative coupling. The ligand-dependent activation of these channels can be described by a 10-state model with five closed and five open states, each of the five states being unique in the number of bound ligands (Fig. 4 A; Biskup et al., 2007; Kusch et al., 2012). Our initial reference state for both the channels is the closed state with zero ligands bound, and the final state of the channel is the open state with all the ligand binding sites occupied with ligands. Using the equilibrium model parameters, we simulated the binding curves of both channels and extracted their median ligand concentrations (Fig. 4 B). For the CNG channel, the median metric comes out to be ∼−29.4 kcal/mol, which is nearly identical to the kinetic model based on the free-energy change estimate from state C0 to O4. This estimate is very different from the estimates derived by fitting the Hill equation to the ligand binding curve (or the simulated dose response curve; Table 1). Similarly, for the HCN channel, the median metric and the kinetic model yield matching estimates of the total free-energy change for ligand-dependent activation of the channel. The simulations clearly demonstrate the utility of the median metric and its applicability even when ligand binding steps are energetically asymmetric.
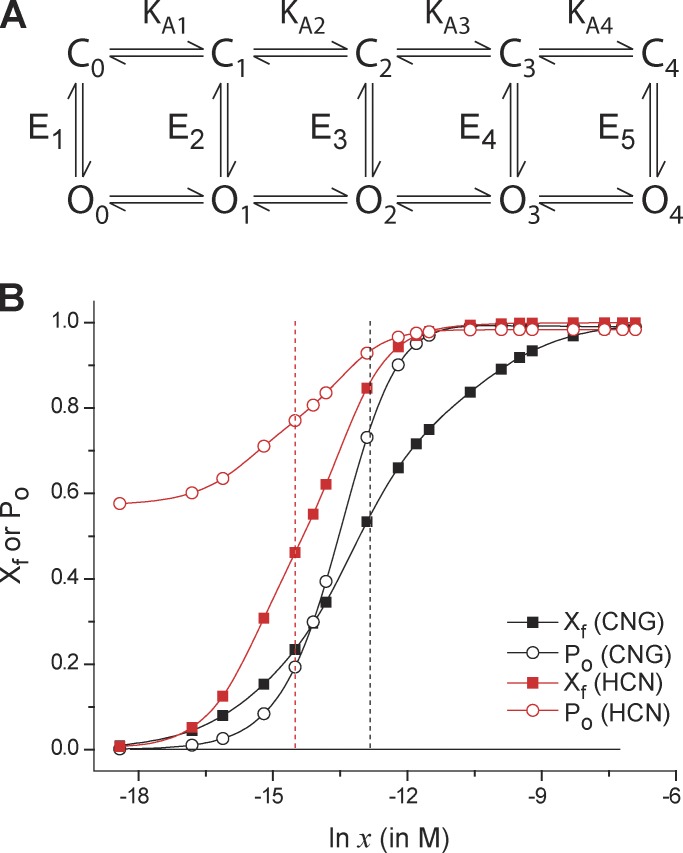
Figure 4.
Comparison of median estimates of free-energy change in CNG and HCN channels. (A) The proposed model of the ligand-induced activation pathway of the CNG and HCN channels. There are five open and closed states (labeled Oi and Ci, (I = 0, 1, 2 …, 4), where “i” represents the number of ligands bound to state). KAi (i = 1, 2, 3, or 4) indicates the step-wise binding affinities of the ligand in the closed state. Ei (i = 1, 2, …, 5) represents the closed-open equilibrium constant for a unique ligation state of the channel. For the CNG channel, each state is considered to have a multiplicity of 4!/(4 − i)!i! (i = 0, 1, 2, …, 4), whereas for the HCN channel, each of the states are considered to be unique (multiplicity = 1). (B) Using the model parameters provided in Biskup et al. (2007; for the CNG channel) and Kusch et al. (2012; for the HCN channel), the steady-state open probability curves (open symbols) and the fractional binding curves (closed symbols) were simulated. The vertical broken lines represent the median ligand concentration axes (for the binding curves) for the CNG (black) and HCN (red) channels. For the HCN channels, these curves represent the channel response at −130 mV.
Table 1.
Comparison of free-energy estimates of CNG and HCN channel obtained from dose–response curves, binding curves and kinetic models
| Response | Free-energy estimation method | Free-energy estimate | ||||
| CNG | HCN | |||||
| Fitting function | Parameters | Parameter values | Parameter values | |||
| kcal/mol | kcal/mol | |||||
| DRCa | 1.65 µM | −7.7 | 0.54 µM | −8.3 | ||
| H | 1.9 | 1.3 | ||||
| BCb | 2.3 µM | −7.5 | 0.54 µM | −8.4 | ||
| H | 0.75 | 1.14 | ||||
| BCc | Nmax RT ln xm + CF | xm | 2.42 µM | −29.4 (−30d) | 0.45 µM | −33.6 (−33.8d) |
| Nmax | 4 | 4 | ||||
| CF | 0 | −0.5 kcal/mol | ||||
The table lists the estimates of free-energy change of ligand-dependent activation of CNG channel and HCN channel (at −130 mV) calculated by different methods.
The simulated dose-response curve (DRC, relative open probability curve) is fitted with a Hill equation, normalized to account for nonzero open probabilities at zero ligand concentration, i.e., (and non-unity open probabilities at saturating ligand concentrations, ). is calculated as RT lnx1/2.
The simulated binding curve (BC) is fitted with the Hill equation, and ΔGC is calculated as RT lnx1/2.
From the simulated binding curve (BC), the median ligand concentration is extracted. CF is the correction factor in Eq. 26 (informed by and ).
ΔGC estimates of the net free-energy difference between the first closed state (C0) and the last open state (O5) () obtained from the kinetic model parameters (Fig. 4 A).
It is important to consider the limiting states of the ensemble while inferring energetics from these measurements. For the CNG channel model, this is not a complication because PO at zero ligand is very small, whereas it is approximately one under saturating ligand concentration. However, for HCN channels, the minimum PO in absence of ligand is 0.55 (from kinetic models). Because the C-to-O transition is ligand independent, even in complete absence of ligand all the channels are not in C0 state. As described in Chowdhury and Chanda (2012), we can apply equivalent correction factors to estimate the complete free energy of activation of the ligand-dependent pathway. Thus, the net expression for the work done in saturating the channel with the ligand is:
| (26) |
where is the open probability at saturating ligand concentration and is the open probability at zero ligand concentration. The first of the two correction factors () accounts for the fact that at saturating ligand concentrations, there might be some closed states with relatively large occupancy, whereas the second () accounts for the fact that at zero ligand concentration, there might be substantial occupancy of the open states of the channel. For the case of the wild-type HCN channel model, this correction factor amounts to ∼0.5 kcal/mol, which is much smaller than the median metric estimate alone. However, this might be an important consideration to accurately describe the activation energies of deleterious mutations.
Generating ligand binding curves at different voltages in allosteric ion channels modulated by voltage and ligand
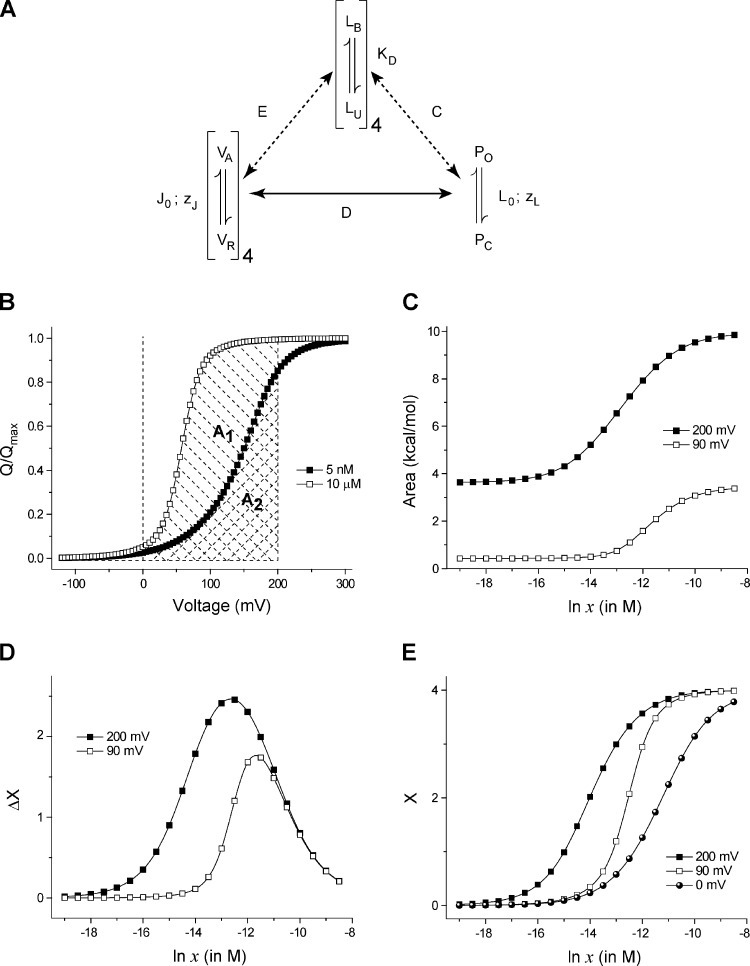
Another important utility of the linkage principles described in the theory section is the ability to generate ligand binding curves at different voltages by measuring the Q-V curves at multiple ligand concentrations and one reference ligand binding curve (at zero voltage). Here, we test this idea using the BK channel for which well-described kinetic models are available (Rothberg and Magleby, 1998; Horrigan and Aldrich, 1999; Horrigan et al., 1999; Rothberg and Magleby, 2000; Horrigan and Aldrich, 2002; Niu and Magleby, 2002). We consider the two-tier Horrigan and Aldrich (HA) model (Fig. 5 A) with four identical voltage-sensing domains, each of which have an intrinsic activation constant J0 and a voltage-dependence zJ. The pore has an intrinsic activation constant L0 and an intrinsic voltage-dependence zL. The equilibrium constant of activation of voltage sensor (J) and that of the pore (L) are assumed to have an exponential voltage dependence (I = I0 exp[zIFV/RT], where I = J or L). In the absence of calcium, the voltage-dependent activation of the channel can be described by a 10-state Monod-Wyman-Changeux (MWC)-type allosteric scheme (Cox et al., 1997; Cui et al., 1997; Talukder and Aldrich, 2000), wherein the pore is capable of opening but with reduced potency at very low voltages, and depolarization-induced activation of the voltage-sensing domains progressively facilitates channel opening. This facilitation is described by an interaction parameter, D. To explain the calcium regulation, it is assumed that there exists a single calcium-binding site in each of the subunits, each with a dissociation constant KD. The binding equilibrium constant is described as: x/KD. Calcium binding can facilitate the opening of the pore via an interaction described by the parameter, C. It could also facilitate activation of the voltage-sensing domain (within the same subunit) via an interaction parameter, E. This model of channel activation has been shown to accurately describe the steady-state and kinetic properties of channel gating over a wide range of voltages and ligand concentrations, and hence will be the primary model on which we test our derivations.
Figure 5.
Generating ligand binding curves at different voltages using Q-V curves at different ligand concentrations. (A) The allosteric model proposed for the BK channel (Horrigan and Aldrich, 2002). There are four voltage-sensing domains, four ligand binding sites (one per subunit), and a single pore. Each of the voltage sensors and the ligand binding sites are allosterically linked to the pore. Each ligand binding site is coupled to a single voltage sensor (in the same subunit). J0 and L0 are the voltage-independent components of the equilibrium constants of activation of the voltage sensor and the pore, respectively. zJ and zL reflect the voltage dependence of activation of each voltage sensor and the pore. KD is the dissociation constant of each of the ligand binding sites. The voltage sensor and pore coupling is described by the cooperativity parameter, D. E and C depict the cooperativity parameters describing coupling of the ligand binding sites with the voltage sensors and pore, respectively. The best fit estimates of these parameters (Fit B; Horrigan and Aldrich, 2002) are: J0 = 0.0319, KD = 0.000011, L0 = 9.8 × 10−7, C = 8, D = 25, E = 2.4, zJ = 0.58, and zL = 0.3. (B) Q-V curves at two ligand concentrations, for the model shown in A. The cross-hashed area (A2) is the area under the Q-V curve at 5 nM ligand concentration, between 0 and 200 mV. The area under the Q-V curve at 10 µM ligand concentration, between 0 and 200 mV, is the total shaded area (A1 + A2, A1 being the area between the two curves). (C) The area under the Q-V curve between 0 and 200 (or 90) mV is plotted against ligand concentration. As the ligand concentration increases, the Q-V curve shifts leftward and thus this area increases as depicted in B. (D) The change in ligand occupancy of the protein caused by increasing the voltage from 0 to 200 (or 90) mV (derived as the gradient of the curves in C) is plotted against different ligand concentrations. In the intermediate ligand concentrations (ln x between −15 and −10), changing voltage brings about a large change in the mean number of ligands bound to the protein, even though the ligand concentration is unchanged. (E) The linkage equation–derived ligand binding curve at 200 and 90 mV, obtained by adding ΔX versus ln x curves for individual voltages to the reference ligand binding curve. In this case, the reference binding curve was simulated from the model parameters.
To estimate the ligand binding curve at a particular voltage (say 200 mV), we first generate a set of QV curves at different ligand concentrations. For each QV curve at a particular concentration, we evaluate the area bound the curve, the V = 200 mV axis, and the reference voltage axis (Vref = 0 mV; Fig. 5 B), and plot these areas with respect to ln x (Fig. 5 C). The gradient of this plot with respect to ln x gives a measure of the change in the ligand occupancy of the protein due to change in voltage (Fig. 5 D). A linear addition of this curve with the reference-binding curve (at 0 mV) generates the binding curve at 200 mV (Fig. 5 E).
Using such a method of estimation, the total free-energy change associated with full-scale activation of the BK channel was estimated as −24 kcal/mol. This value is nearly identical to that estimated from the model parameters (Table 2). It is to be noted that the reference binding curve used in Fig. 5 E is inferred directly from the kinetic model; direct calcium binding curves for the BK channel have not yet been measured, although calcium-induced conformational changes in purified preparations of calcium-binding domains have been spectroscopically monitored (Yusifov et al., 2010).
Table 2.
Energetic parameters for the BK channel gating model
| Energetic parameter | Value |
| VM (x = 0)a | 138.6 mV |
| VM (x → ∞)a | 30.7 mV |
| xm (V → −∞)b | 14.7 µM |
| xm (V → ∞)b | 0.83 µM |
| c | −24.1 kcal/mol |
| d | −23.9 kcal/mol |
| ΔGmodele | −24.6 kcal/mol |
| f | −6.7 kcal/mol |
| g | −6.5 kcal/mol |
| (model)h | −6.8 kcal/mol |
Vm values were extracted from Q-V curves simulated in the absence and saturation concentrations of ligand (x = 75 µM) using parameters of Fit B of the HA model.
The ligand binding curves at very high (300 mV) and very low (−150 mV) voltages were deduced using the linkage method (Eq. 21), using the binding curve at 0 mV (directly simulated from the model) as the reference curve.
The free-energy change along Path I of thermodynamic cycle, ΔGIa + ΔGIb.
The free-energy change along Path II of thermodynamic cycle, ΔGIIa + ΔGIIb.
ΔGmodel is directly obtained from the model parameters:
Energetic connectivity calculated as: ΔGIIb − ΔGIa.
Energetic connectivity calculated as: GIb − ΔGIIa.
Energetic connectivity evaluated from the model parameters: −RT ln(CE)4.
The principle utility of the formulation shown in Fig. 5 is that it allows us to estimate the total free energy of the activation of the channel. It is important to remember that standard biochemical assays of ligand binding are performed over time scales much longer than those of electrophysiological recordings. This may allow the proteins to populate slowly accessible long-lived desensitized states (Colquhoun, 1998) that are not normally detectable during the time scale of electrophysiology measurements. The median metric extracted from such binding curves would incorporate the free-energy contributions of such transitions, along with the energetic changes occurring during the early channel activation transitions. Thus, to estimate the free energy of activation (i.e., the transition from the first closed state, where all voltage sensors are resting and all ligand binding sites are unoccupied to the final open, conducting state, where all voltage sensors are activated and all ligand bind sites are occupied), one needs to incorporate a correction factor related to the maximum open probability at saturating ligand concentrations and very high voltages, measured at very long time scales. The final free-energy equations in this scenario will be:
| (27a) |
and
| (27b) |
where, t → ∞ indicates that the measurements are made on a very long time scale. It is to be noted that for the kind of systems to which this discussion directly pertains to (e.g., BK channels), the existence of such deep desensitized states has not yet been reported. This issue however may be more pertinent for the cys loop receptor channels, which are not directly voltage dependent and are not the systems toward which our current discussion is geared toward.
Energetic interdependence of voltage- and ligand-induced transitions in bimodal allosteric ion channels
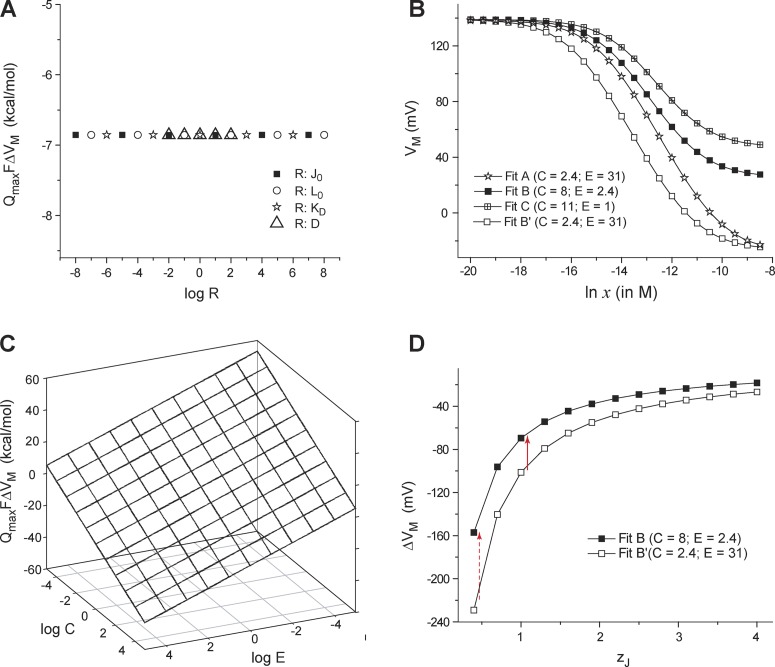
To obtain the interaction energy between the ligand- and voltage-dependent pathways, we generated Q-V curves at zero and fully saturating calcium concentrations using the parameters described by the HA model (Horrigan and Aldrich, 2002). Thereafter, each of the parameters were varied over several orders of magnitude, and in each case we extracted the median voltage at VM at zero calcium (VM (x = 0)) and at saturating calcium concentrations (VM (xsat)) from the simulated Q-V curves. For this system, Qmax = 4zJ + zL. We plotted QmaxFΔVM (ΔVM = VM(xsat) – VM(x = 0)) against different values of KD, J0, L0, and D (Fig. 6 A). The plot shows that this free-energy metric, QmaxFΔVM, is completely independent of the ligand binding affinity, the intrinsic stability of the voltage-sensing and pore domains, and the voltage sensor and pore coupling.
Figure 6.
Dependence of the shift of Q-V curves due to ligand concentration on different parameters of an allosteric model. (A) QmaxFΔVM is plotted against the (logarithm of) a subset of parameter values for the allosteric model of the BK channel (Fig. 5 A). In each case, a single parameter of the model was changed, whereas the others were maintained at the control (Fit B) values. Symbols correspond to different parameters that were varied. For all the four equilibrium parameters tested (J0, L0, KD, and D), QmaxFΔVM remains constant at −6.8 kcal/mol. (B) VM of the simulated Q-V curves is plotted against ligand concentrations for four sets of the BK channel model parameters as described previously (Horrigan and Aldrich, 2002): for Fit A, J0 = 0.03807, KD = 0.000032, L0 = 2 × 10−6, C = 2.4, D = 18.4, E = 31, zJ = 0.55, and zL = 0.4; for Fit B, J0 = 0.0319, KD = 0.000011, L0 = 9.8 × 10−7, C = 8, D = 25, E = 2.4, zJ = 0.58, and zL = 0.3; for Fit C, J0 = 0.0319, KD = 0.000011, L0 = 9.8 × 10−7, C = 11, D = 25, E = 1, zJ = 0.58, and zL = 0.3; for Fit B′, same as Fit B except the coupling terms C = 2.4 and E = 31. (C) QmaxFΔVM was evaluated through simulations for values of C and E, varying over several orders of magnitude, and is plotted against log C and log E. (D) A plot of ΔVM with respect to zJ for Fits B and B′. ΔVM for Fits B and B′ are more significantly different for low values of zJ (red broken arrow) than higher values (red solid arrow).
Next, we simulated the Q-V curves for three characteristic model parameters provided by Horrigan and Aldrich (2002; Fits A, B, and C) at multiple calcium concentrations and plotted the VM values at those ligand concentrations (Fig. 6 B). Fit B was chosen as the best fit by Horrigan and Aldrich based on multiple tests. Interestingly, the VM versus ln x plots show that each of the fits generate characteristically different features. When only the interaction terms of the ligand binding domains (parameters C and E) were modified for Fit B to match those in Fit A, a different curve was generated. Strikingly, the VM values converge at limiting ligand concentrations as long as the interaction terms between the two sets of model parameters are the same. Next, we plotted QmaxFΔVM against different values of the parameters C and E (Fig. 6 C). The planar 3D surface generated shows that QmaxFΔVM is linearly related to interaction energies of the ligand binding domain with the voltage sensor and the pore even when they are varied over several orders of magnitude.
Are these numerical simulations consistent with our theoretical proposition? To understand this, we first write the partition function for this system:
| (28) |
Under zero calcium conditions, K = 0 and Eq. 28 reduce to:
QmaxFVM (x = 0) is the measure of the nonelectrical free-energy change associated with full-scale voltage-dependent activation of the channel. Thus:
At saturating calcium conditions, Eq. 28 reduces to:
The median metric will now yield the following free-energy measure:
From this analysis, it is easily seen that the net change in the median voltage of charge movement will be given by:
| (29) |
The conclusion is that the net displacement of the Q-V curve upon ligand binding is solely determined by the interaction of the ligand binding domain with the voltage sensor and the pore.
These simulations illustrate our theoretical results and highlight the crucial utility of Q-V curve measurements in understanding ligand-dependent gating of polymodal systems such as the BK channels. The model of the BK channel considered here, to some extent, is restrictive because it assumes that there exists a single ligand binding site per subunit of the channel that is allowed to interact with the voltage sensor only of the same subunit (Bao et al., 2002; Shi et al., 2002; Xia et al., 2002; Magleby, 2003). In Appendix 2 we show that our overall relationship is valid even when there is more than a single binding site per subunit of the protein.
A striking outcome of Eq. 29 is that for a constant value of C and E, Qmax and ΔVM will be inversely related. For two sets of parameters (Fits B and B′), we plotted ΔVM with respect to the intrinsic voltage-dependence of voltage sensors, zJ (Fig. 6 D). The curves clearly show that the net sensitivity of the Q-V curve to ligand reduces as zJ increases. Furthermore, if zJ is large, the Q-V curves are less sensitive to coupling interactions between the two pathways. This can be mathematically proven by differentiating Eq. 29 with respect to zJ for a constant value of C and E:
| (30) |
Eq. 30 demonstrates that the total gating charge on the channel can influence its sensitivity to ligands. Significantly, the estimated total gating charge of BK channels is ∼2.5 eo (Ma et al., 2006), which is much lower than that of a canonical voltage-gated ion channels such as the Shaker Kv channel (Schoppa et al., 1992; Aggarwal and MacKinnon, 1996; Seoh et al., 1996).
Linkage relationships in a hyperpolarization- and ligand-activated channel with a voltage-independent pore
In this section, we consider the hyperpolarization-activated, cyclic nucleotide gated HCN channels in which the pore is believed to be intrinsically voltage-independent and negative coupling with the four identical voltage-sensing domains causes it to close as the voltage sensors activate (Altomare et al., 2001; Chen et al., 2007). However, ligand binding favors channel opening like the BK channel.
To address such a system, we modified the thermodynamic cycle (Fig. 7). We chose the closed channel state as a reference state, which in this case is predominant at high voltages and zero ligand concentration. This was necessitated by the fact that the ligand binding domain and voltage sensors are oppositely coupled to the pore, unlike BK channels. The final state of the system is the open state, where all ligand binding sites are saturated with ligand and all voltage sensors are deactivated. As shown in Fig. 7, this transformation can be achieved via two paths. Applying the principles that we have detailed in the previous sections on this thermodynamic cycle, our primary energy-linkage equation is:
Figure 7.
Thermodynamic cycle for a hyperpolarization- and ligand-activated ion channels. In this case, our reference state is where all the gating charges are in activated conformation and all the ligand binding sites are unoccupied (at depolarizing voltages and zero ligand concentration). The choice of reference state is based on the considerations of type of coupling between voltage- and ligand-dependent pathways. The final state of the system is where all the gating charges are in the resting conformation and ligand binding sites are saturated. This state will be preferred when the voltage is low and the ligand concentration is high. As in the previous thermodynamic cycle, the net free energy of activation of this channel can be calculated by two alternate paths.
In the HCN channels, the PO does not approach unity at hyperpolarizing conditions in the absence of ligand. This means that an ensemble of states is populated under these conditions. To account for this multiplicity of states, we add a correction term to the interaction energy for HCN channels,
| (31) |
The correction factor used in Eq. 31 is equivalent to that shown in Eq. 26 for the ligand-dependent activation. In the wild-type channel, the PO value is zero or unity under all other limiting conditions of voltage and ligand. However, many mutations compromise the channel’s ability to fully close or fully open and thus the PO values at each of the four limiting conditions have to be taken into account to calculate the net interaction energy for the voltage- and ligand-dependent pathway. Thus, extending Eq. 31 for the thermodynamic cycle shown in Fig. 7, we can write a general formula from of net interaction energy between the two pathways:
| (32) |
where Vmin and Vmax are the highly hyperpolarizing and depolarizing limits, respectively, and xmin and xmax are the zero and saturating ligand concentrations, respectively.
For HCN2 channels, ligand binding curves have been measured at multiple voltages using confocal microscopy combined with voltage clamp fluorometry. However, to the best of our knowledge, the Q-V curves for the same subtype are not available (although Q-V curves for spHCN channels have been reported; Männikkö et al., 2002, 2005; Ryu et al., 2012). Using the binding curves reported in Kusch et al. (2010), our thermodynamic relationships (Eqs. 23a, 23b, and 32) allow us to predict that the net interaction energy between the ligand binding domain and other domains for HCN2 channels is nearly −3 kcal/mol (Table 3). Furthermore, from the kinetic model for ligand-induced activation of the HCN2 channel, the net interaction between the ligand binding domains and the pore can be estimated to be ∼−2.2 kcal/mol. This implies that the net energetic coupling of the voltage sensors and the ligand binding domains is weak (∼−0.8 kcal/mol). This is consistent with the notion that cAMP binding to the HCN channels does not affect the voltage-dependent transitions but modifies the final, voltage-independent closed–open transition (Craven and Zagotta, 2004; Shin et al., 2004). It is to be noted however that the coupling associated with ligand binding domains in the HCN2 channels is not completely symmetrical (Ulens and Siegelbaum, 2003); some steps (the second and fourth) show positive coupling whereas the others (the third) show negative coupling (Kusch et al., 2012). Hence the relative small interaction energy estimates may be the result of the net balance of interaction energies. This prediction can be further tested by measuring the Q-V curves at different ligand concentrations and Eqs. 23a and 23b.
Table 3.
Energetic parameters for the HCN channel
| Parameter | Parameter value |
| xm (V → −∞) (V = −160 mV)a | 0.54 µM |
| xm (V → ∞) (V = −30 mV)a | 1.72 µM |
| b | −3.0 kcal/mol |
| ΔGint (ligand pore)c | −2.2 kcal/mol |
The median ligand concentrations at very low and high voltages were approximated to the EC50 values of the ligand binding curves reported in Kusch et al. (2010).
The interaction energy between the ligand binding domain and the rest of the protein and CF is the correction factor (Eq. 31). The following limiting values were used in calculating CF: PO(Vmin, xmin) = 0.58; PO(Vmin, xmax) = 1; PO(Vmax, xmax) = PO(Vmax, xmin) = 0.
The interaction between the ligand binding domains is the pore domain calculated from the HCN channel model parameters proposed in Kusch et al. (2012; equal to ; Fig. 4 A).
DISCUSSION
In a recent study (Chowdhury and Chanda, 2012), we have shown that the net free energy associated with voltage-dependent activation of an ion channel can be estimated by extracting the median voltage of activation from the Q-V curve using the relation:
This relationship is analogous to Wyman’s thermodynamic description of ligand binding equilibrium (Wyman, 1967), which when applied to ligand-dependent ion channels, yields the relation:
Thus for ligand-gated ion channels, the total work done to drive the channels to activated conformation can be obtained by measuring the median ligand activity and the maximum number of bound ligands.
Here, we extend these thermodynamic principles to develop the free-energy relationships in ion channels dually modulated by voltage and ligand. For these systems, the net free-energy change for activation by both ligand and voltage is:
Thus, using the Q-V curves at zero ligand concentrations and ligand binding curves at depolarizing conditions, we can determine the net free energy for activation of a voltage- and ligand-activated channel. The thermodynamic cycle (Fig. 2) dictates that we can also obtain the same information by measuring the Q-V curves at saturating ligand concentration and ligand binding curves at hyperpolarizing conditions.
Although Q-V curves at zero and saturating ligand concentrations can be obtained, measuring ligand binding curves at various holding potentials is not straightforward. Recent technological developments have allowed us to directly estimate the ligand binding curves for certain ion channels, but this approach has not been extended to many other voltage- and ligand-activated ion channels. Here, we derive another thermodynamic relationship, which allows us to estimate the ligand curves at various potentials by using a single reference ligand binding curve. Accordingly, the ligand occupancy at various voltages and ligand concentrations is:
If we obtain ligand occupancy at a particular voltage and ligand concentration, , then the Q-V curves for the channel at various ligand concentrations will allow us to generate the entire ligand binding surface. This formulation is further illustrated in Fig. 5. Typically, ligand binding curves are measured biochemically using detergent solubilized protein preparations that lack a voltage gradient (Szallasi et al., 1999; Jordt and Julius, 2002; Picollo et al., 2009; Piscitelli et al., 2010; Yusifov et al., 2010). In such cases, Vref is 0 mV, and this ligand binding curve can serve as a reference. Similarly, using multiple ligand binding curves at different voltages, we can also obtain Q-V curves at multiple ligand concentrations as long as a single reference Q-V curve is available.
These thermodynamic relationships need some qualifications. The median metric of net free-energy change of polymodal channel (Eq. 18) incorporates all the sources of free-energy change in the system including the (true) binding energies of the ligands and the relative stabilities of the voltage sensors and the pore, as well as interactions between them. In case the channel undergoes inactivation/desensitization, its free-energy contribution is also linearly incorporated in the median metric. This free-energy metric allows us to characterize the net energetic effect of a site-specific perturbation (which includes its effect on the free energies of all transitions accompanying the “full-scale” activation of the protein) without any assumptions about the nature and connectivity of intermediate states. Under conditions, correction factors have to be considered that can be obtained by using open probability measurements.
Possibly the most important thermodynamic relationship that we propose here is that the difference between VM of the Q-V curves, measured at saturating and zero ligand concentrations, directly yields an estimate of the interaction energy between the voltage-dependent and the ligand-dependent pathways of the channel. This interaction energy can also be estimated from the shift in the ligand binding curve between hyperpolarizing and depolarizing voltages:
This relationship is independent of the number of possible intermediates and the nature of connectivity between them. This is an important experimental advantage, as identification of allosteric pathways in the complex polymodal ion channels most often requires tedious and complex model fitting procedures. For instance, in the HA model for BK channels, ligand binding involves four identical and independent binding sites. However, additional ligand binding sites have been described, some of which also interact with each other (Bao et al., 2002; Shi et al., 2002; Xia et al., 2002; Magleby, 2003; Sweet and Cox, 2008; Zhou et al., 2012). By measuring the ΔVM value and total gating charge, we can estimate the interaction energies of the binding sites with the voltage-dependent pathways, as was shown for BK and HCN channels in Tables 2 and 3. This approach can be also used to identify sites that mediate coupling interactions between voltage- and ligand-dependent pathways. Note that in case the channel undergoes slowly developing desensitization transitions during binding measurements (not observed in the time scale of gating current measurements), correction factors, related to the limiting PO values (as described in Eq. 27), need to be incorporated with the median ligand concentration measures.
It is important to note that this energy metric is a de facto measure of interaction energy only in systems that are allosteric in nature. For instance, in BK channels, voltage sensor activation, ligand binding, and pore opening are independent equilibria, but they interact with each other allosterically. To ascertain whether such an allosteric framework is applicable, two conditions have to be satisfied. First, Qmax of the protein is independent of ligand concentration such that irrespective of ligand concentration, all the charges can be moved if a sufficiently large voltage is applied. Second, the shift in the Q-V curve saturates at both high and low ligand concentrations. If either condition is not fulfilled, it would imply that certain transitions are under the obligatory control of the ligand and occur only in the presence of ligand. Similar conditions apply to ligand binding phenomenon wherein the maximum possible occupancy is independent of voltage and the shifts in ligand binding curves saturate at limiting voltages. Additionally, in allosteric systems where ligand binding processes are directly voltage dependent because of the interaction of the ligand with the membrane electric field, Qmax for the voltage-dependent transitions might change with ligand concentration, and the effect of ligand on Qmax (in addition to VM) must be taken into consideration in the calculations described.
The median method of interaction energy has the advantage in that it can provide an estimate of interaction energy in a relatively straightforward manner. Other methods such as the χ-value analysis (Chowdhury and Chanda, 2010) can also provide an estimate of interaction energies, but these measurements require limiting conditions where the signal-to-noise ratio is low. The BK channels, for instance, have a large single channel conductance, which makes it possible to obtain these estimates at potentials where the PO values are <10−8. Most other ion channels have ∼10-fold lower single channel conductance, which makes such measurements under limiting conditions a challenging proposition. To eschew potential incorrect experimental application of our thermodynamic relationships, it must be emphasized that these relationships may be applied directly using only Q-V curves or the direct ligand binding curves and not the more commonly used conductance–voltage or dose-response curves. Channel open probability measurements sample only the states that are conducting and thus cannot be used directly as the conjugate extensives to extract the energetic details of the system, as described here.
Allosteric voltage-dependent ion channels such as the BK, HCN, and KCNQ channels are known to be acutely sensitive to ligands, such as calcium, cyclic nucleotides, PIP2, etc. However, all of these channels are believed to have a much lower Qmax than the Shaker KV channels (Craven and Zagotta, 2006; Ma et al., 2006; Miceli et al., 2012), despite having a large number of positively charged residues on their S4 segments. Additionally, experimental evidence has been reported wherein mutation of a gating charge residue in the S4 segment of BK channel increased the channel’s sensitivity to calcium (Cui and Aldrich, 2000). This mutual reciprocity may initially seem to be obvious, when considering a simplified two-state process where the single transition is directly influenced by voltage and ligand. Nevertheless it is unclear why this inverse correlation should hold in multistate allosteric systems where voltage and ligand act through different pathways. Furthermore, quantification of such correlations, using the simplified two-state process–based calculations, is at best empirical. Our generalized thermodynamic analysis shows that if the interaction energy remains unchanged, changes in ligand concentration will cause greater shifts in the Q-V curves if the gating charge is reduced. In other words, even if the interaction energy between two pathways is modest, the voltage-dependent activation will appear to be very sensitive to ligand concentration as long as the total gating charge is small. Similarly, decreasing the total number of ligand binding sites will increase the magnitude of voltage-dependent shifts in ligand binding curves. This implies that for proper physiological function of these polymodal channels, both the charge of the channel and interaction energies associated with the ligand binding domains have to be optimized. Thus, a low value of Qmax observed in many polymodal allosteric channels may provide an evolutionary advantage, which renders channel gating highly sensitive to specific ligands.
APPENDIX 1
The median measure is able to extract the free-energy estimates associated with voltage-dependent and ligand-dependent transitions, but the reason “why it works” needs some more clarification. Such a clarification does not have any direct bearing on the overall utility of the median transformation, but is necessary to show that the median transformation is not just an arbitrary transformation, but has a core thermodynamic principle behind it.
To understand this principle, we first consider a simple binary voltage-dependent transition between two states: Si and Sf. Si is the state of the channel with zero gating charge/valence and Sf is the state of the channel with all translocated gating charge. In such a binary system, monitoring the occupancy of any one of the states (say Sf) gives completely thermodynamic information about the entire system. At a particular voltage, when the two states are equally populated, the chemical energy difference between the two states is counterbalanced by the electrical energy difference between the two states. This voltage, the V1/2, is frequently measured by monitoring the occupancy of the open state of the channel (through conductance measurements). If the charge associated with the binary transition is known, then the electrical energy difference between the two states at V1/2 can be easily calculated, which in turn yields the chemical free-energy difference between the two states.
In the more general case, where voltage-dependent activation pathways are multistate, there would be several intermediate states between Si and Sf. Also, in channels that undergo inactivation, Sf might not be the final “open” state, but rather simply the final state (whether conducting or not). The parameter of interest is still the free-energy difference between Si and Sf. In such situations, it has been shown that Si (the state with zero gating charge) and Sf (the state with maximum gating-charge) are equally populated, not necessarily at V1/2, but at VM, the median voltage. This implies that at the median voltage, the electrical energy difference between the two states balances the chemical component of the free-energy difference between the two states, and thus the total change in chemical free energy of the system can be directly estimated using VM and Qmax. The same logic would apply to ligand-dependent cases as well where at the median ligand activity, the completely unbound, and the fully bound forms are equally populated.
To understand the principle of median response better, we take a different approach and consider the slope of the Q-V curve (generally referred to as the slope function) in a voltage-dependent channel. The total gating-charge translocated over a voltage range can be written as:
| (A1) |
Similarly we write Eq. 14 as:
| (A2) |
We divide both Eqs. A1 and A2 with Qmax, noting that fractional gating charge moved, , is . As a result, we end up with the following pair of equations:
| (A3) |
and
| (A4) |
The important consequence of Eqs. A3 and A4, is that, because is continuous and differentiable, and its infinite limit integration over voltage is unity, it represents a probability density function (pdf), and the median voltage is mean of the pdf. More interestingly, it can be realized that the V1/2 (the voltage at which half of all the gating-charges have moved) is
| (A5) |
which is the definition of the median of a pdf. Thus (by Eq. A5), V1/2 of the Q-V curve is the median of the pdf, and VM of the Q-V curve (by Eq. A4) is the mean of the pdf. In a similar manner, ln xm can be shown to be the mean of the pdf described by the slope of the fractional ligand binding curve, and ln x1/2 is the median of the pdf. These pdfs (the slope functions) encapsulate several thermodynamic details, for example about cooperativity (Di Cera et al., 1988; Di Cera and Chen, 1993), and will be left for a more detailed exposition later.
APPENDIX 2
We consider here the case of a channel with a single pore domain that is allosterically coupled to “m” identical voltage sensors (one in each subunit). There are “n” binding sites per subunit (in different subunits, the ith binding sites have the same ligand binding affinities, but within the same subunit, the different sites have different binding affinities). Each of the voltage sensors have an equilibrium constant J0 and an intrinsic voltage-dependence zJ. The pore domain has an intrinsic activation constant L0 and an intrinsic voltage dependence zL. The equilibrium constant of activation of voltage sensor (J) and that of the pore (L) are assumed to have an exponential voltage dependence (as described for the other models considered in this study). The voltage-dependent activation of the channel in zero calcium conditions is described by a 2× m-state MWC-type allosteric scheme. Activation of each voltage sensor facilitates the opening of the pore through interactions described by D. The ith binding site of the channel has a binding affinity Ki (i = 1, 2, … n). The ith binding site interacts with the jth voltage sensor, the interaction being described by Eij (j = 1, 2, … m), whereas its interaction with the pore is described by the parameter Ci. The binding sites within the same subunit do not necessarily interact equally with the pore or the voltage sensors, but the same binding site in different subunits interact equally with the pore and the voltage sensors (i.e., Ci and Eij are the same for the ith binding site in different subunits, but Ci may or may not equal Ck, and Eij may or may not equal Ekj).
For this system, at zero calcium, the median metric would be related to the overall free-energy change of the system as
| (A6) |
where Qmax is mzJ + zL. Under saturating calcium concentration, at very low voltages, all the ligand binding sites will be occupied to start with. Upon full translocation of the gating charge, the activated voltage sensors and the open pore begin to interact with the ligand binding sites. These interactions along with the change in energy caused by the voltage-dependent pathway will govern the free-energy change associated with voltage-dependent activation under saturating ligand concentrations. Thus, the median metric at very high calcium ligand concentrations will yield the following equation:
| (A7) |
Hence the displacement of the Q-V curve along the voltage axis, according to Eqs. A6 and A7, will be:
| (A8) |
This equation imposes a constraint on the maximum gating charges transferred, maximum number of ligand binding sites (mn), ΔVM, and the change in the median ligand concentrations caused by an increase in voltage. The linkage relations that we describe also remain valid in this situation.
Acknowledgments
The project was supported by funds from the National Institutes of Health (grant GM084140) and a Shaw Scientist award to B. Chanda.
Christopher Miller served as editor.
Footnotes
In this paper, we designate the median voltage of activation by VM, which is different than what was we had previously used (Chowdhury and Chanda, 2012). This was done to avoid confusion with membrane potential that is routinely designated by Vm (our previously used symbol for median voltage). However, for the median ligand concentration, we retain the designation xm, originally used by Wyman (Wyman, 1964, 1967).
Abbreviations used in this paper:
- CNG
- cyclic nucleotide-gated
- HA
- Horrigan and Aldrich
- HCN
- hyperpolarization activated cyclic nucleotide-gated
- probability density function
- Q-V
- charge-voltage
References
- Aggarwal S.K., MacKinnon R. 1996. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 16:1169–1177 10.1016/S0896-6273(00)80143-9 [DOI] [PubMed] [Google Scholar]
- Altomare C., Bucchi A., Camatini E., Baruscotti M., Viscomi C., Moroni A., DiFrancesco D. 2001. Integrated allosteric model of voltage gating of HCN channels. J. Gen. Physiol. 117:519–532 10.1085/jgp.117.6.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Rapin A.M., Holmstrand E.C., Cox D.H. 2002. Elimination of the BK(Ca) channel’s high-affinity Ca(2+) sensitivity. J. Gen. Physiol. 120:173–189 10.1085/jgp.20028627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskup C., Kusch J., Schulz E., Nache V., Schwede F., Lehmann F., Hagen V., Benndorf K. 2007. Relating ligand binding to activation gating in CNGA2 channels. Nature. 446:440–443 10.1038/nature05596 [DOI] [PubMed] [Google Scholar]
- Caterina M.J., Schumacher M.A., Tominaga M., Rosen T.A., Levine J.D., Julius D. 1997. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 389:816–824 10.1038/39807 [DOI] [PubMed] [Google Scholar]
- Chen S., Wang J., Zhou L., George M.S., Siegelbaum S.A. 2007. Voltage sensor movement and cAMP binding allosterically regulate an inherently voltage-independent closed-open transition in HCN channels. J. Gen. Physiol. 129:175–188 10.1085/jgp.200609585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Chanda B. 2010. Deconstructing thermodynamic parameters of a coupled system from site-specific observables. Proc. Natl. Acad. Sci. USA. 107:18856–18861 10.1073/pnas.1003609107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Chanda B. 2012. Estimating the voltage-dependent free energy change of ion channels using the median voltage for activation. J. Gen. Physiol. 139:3–17 10.1085/jgp.201110722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham D.E. 2003. TRP channels as cellular sensors. Nature. 426:517–524 10.1038/nature02196 [DOI] [PubMed] [Google Scholar]
- Colquhoun D. 1998. Binding, gating, affinity and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. Br. J. Pharmacol. 125:924–947 10.1038/sj.bjp.0702164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D.H., Cui J., Aldrich R.W. 1997. Allosteric gating of a large conductance Ca-activated K+ channel. J. Gen. Physiol. 110:257–281 10.1085/jgp.110.3.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven K.B., Zagotta W.N. 2004. Salt bridges and gating in the COOH-terminal region of HCN2 and CNGA1 channels. J. Gen. Physiol. 124:663–677 10.1085/jgp.200409178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven K.B., Zagotta W.N. 2006. CNG and HCN channels: two peas, one pod. Annu. Rev. Physiol. 68:375–401 10.1146/annurev.physiol.68.040104.134728 [DOI] [PubMed] [Google Scholar]
- Cui J., Aldrich R.W. 2000. Allosteric linkage between voltage and Ca(2+)-dependent activation of BK-type mslo1 K(+) channels. Biochemistry. 39:15612–15619 10.1021/bi001509+ [DOI] [PubMed] [Google Scholar]
- Cui J., Cox D.H., Aldrich R.W. 1997. Intrinsic voltage dependence and Ca2+ regulation of mslo large conductance Ca-activated K+ channels. J. Gen. Physiol. 109:647–673 10.1085/jgp.109.5.647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day M.L., Pickering S.J., Johnson M.H., Cook D.I. 1993. Cell-cycle control of a large-conductance K+ channel in mouse early embryos. Nature. 365:560–562 10.1038/365560a0 [DOI] [PubMed] [Google Scholar]
- DeCoursey T.E., Chandy K.G., Gupta S., Cahalan M.D. 1984. Voltage-gated K+ channels in human T lymphocytes: a role in mitogenesis? Nature. 307:465–468 10.1038/307465a0 [DOI] [PubMed] [Google Scholar]
- Depaula J., Atkins P.W. 2009. Physical Chemistry. Ninth edition W.H. Freeman & Company; 1060 pp [Google Scholar]
- Di Cera E., Chen Z.Q. 1993. The binding capacity is a probability density function. Biophys. J. 65:164–170 10.1016/S0006-3495(93)81033-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cera E., Gill S.J., Wyman J. 1988. Binding capacity: cooperativity and buffering in biopolymers. Proc. Natl. Acad. Sci. USA. 85:449–452 10.1073/pnas.85.2.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch R. 2003. Excitation-transcription coupling: signaling by ion channels to the nucleus. Sci. STKE. 2003:PE4 10.1126/stke.2003.166.pe4 [DOI] [PubMed] [Google Scholar]
- Gill S.J., Richey B., Bishop G., Wyman J. 1985. Generalized binding phenomena in an allosteric macromolecule. Biophys. Chem. 21:1–14 10.1016/0301-4622(85)85001-8 [DOI] [PubMed] [Google Scholar]
- Gleitsman K.R., Shanata J.A., Frazier S.J., Lester H.A., Dougherty D.A. 2009. Long-range coupling in an allosteric receptor revealed by mutant cycle analysis. Biophys. J. 96:3168–3178 10.1016/j.bpj.2008.12.3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. 2001. Ion Channels of Excitable Membranes. Sinauer Associates, Sunderland, UK: 814 pp [Google Scholar]
- Horrigan F.T., Aldrich R.W. 1999. Allosteric voltage gating of potassium channels II. Mslo channel gating charge movement in the absence of Ca(2+). J. Gen. Physiol. 114:305–336 10.1085/jgp.114.2.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrigan F.T., Aldrich R.W. 2002. Coupling between voltage sensor activation, Ca2+ binding and channel opening in large conductance (BK) potassium channels. J. Gen. Physiol. 120:267–305 10.1085/jgp.20028605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrigan F.T., Cui J., Aldrich R.W. 1999. Allosteric voltage gating of potassium channels I. Mslo ionic currents in the absence of Ca(2+). J. Gen. Physiol. 114:277–304 10.1085/jgp.114.2.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordt S.E., Julius D. 2002. Molecular basis for species-specific sensitivity to “hot” chili peppers. Cell. 108:421–430 10.1016/S0092-8674(02)00637-2 [DOI] [PubMed] [Google Scholar]
- Kusch J., Biskup C., Thon S., Schulz E., Nache V., Zimmer T., Schwede F., Benndorf K. 2010. Interdependence of receptor activation and ligand binding in HCN2 pacemaker channels. Neuron. 67:75–85 10.1016/j.neuron.2010.05.022 [DOI] [PubMed] [Google Scholar]
- Kusch J., Thon S., Schulz E., Biskup C., Nache V., Zimmer T., Seifert R., Schwede F., Benndorf K. 2012. How subunits cooperate in cAMP-induced activation of homotetrameric HCN2 channels. Nat. Chem. Biol. 8:162–169 10.1038/nchembio.747 [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., Mackinnon R. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- Ma Z., Lou X.J., Horrigan F.T. 2006. Role of charged residues in the S1-S4 voltage sensor of BK channels. J. Gen. Physiol. 127:309–328 10.1085/jgp.200509421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magleby K.L. 2003. Gating mechanism of BK (Slo1) channels: so near, yet so far. J. Gen. Physiol. 121:81–96 10.1085/jgp.20028721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Männikkö R., Elinder F., Larsson H.P. 2002. Voltage-sensing mechanism is conserved among ion channels gated by opposite voltages. Nature. 419:837–841 10.1038/nature01038 [DOI] [PubMed] [Google Scholar]
- Männikkö R., Pandey S., Larsson H.P., Elinder F. 2005. Hysteresis in the voltage dependence of HCN channels: conversion between two modes affects pacemaker properties. J. Gen. Physiol. 125:305–326 10.1085/jgp.200409130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty A. 1981. Ca-dependent K channels with large unitary conductance in chromaffin cell membranes. Nature. 291:497–500 10.1038/291497a0 [DOI] [PubMed] [Google Scholar]
- Miceli F., Vargas E., Bezanilla F., Taglialatela M. 2012. Gating currents from Kv7 channels carrying neuronal hyperexcitability mutations in the voltage-sensing domain. Biophys. J. 102:1372–1382 10.1016/j.bpj.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseenkova-Bell V.Y., Stanciu L.A., Serysheva I.I., Tobe B.J., Wensel T.G. 2008. Structure of TRPV1 channel revealed by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA. 105:7451–7455 10.1073/pnas.0711835105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nache V., Kusch J., Biskup C., Schulz E., Zimmer T., Hagen V., Benndorf K. 2008. Thermodynamics of activation gating in olfactory-type cyclic nucleotide-gated (CNGA2) channels. Biophys. J. 95:2750–2758 10.1529/biophysj.108.129718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X., Magleby K.L. 2002. Stepwise contribution of each subunit to the cooperative activation of BK channels by Ca2+. Proc. Natl. Acad. Sci. USA. 99:11441–11446 10.1073/pnas.172254699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N., Catterall W.A. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picollo A., Malvezzi M., Houtman J.C., Accardi A. 2009. Basis of substrate binding and conservation of selectivity in the CLC family of channels and transporters. Nat. Struct. Mol. Biol. 16:1294–1301 10.1038/nsmb.1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscitelli C.L., Krishnamurthy H., Gouaux E. 2010. Neurotransmitter/sodium symporter orthologue LeuT has a single high-affinity substrate site. Nature. 468:1129–1132 10.1038/nature09581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg B.S., Magleby K.L. 1998. Kinetic structure of large-conductance Ca2+-activated K+ channels suggests that the gating includes transitions through intermediate or secondary states. A mechanism for flickers. J. Gen. Physiol. 111:751–780 10.1085/jgp.111.6.751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg B.S., Magleby K.L. 2000. Voltage and Ca2+ activation of single large-conductance Ca2+-activated K+ channels described by a two-tiered allosteric gating mechanism. J. Gen. Physiol. 116:75–99 10.1085/jgp.116.1.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S., Yellen G. 2012. Charge movement in gating-locked HCN channels reveals weak coupling of voltage sensors and gate. J. Gen. Physiol. 140:469–479 10.1085/jgp.201210850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppa N.E., McCormack K., Tanouye M.A., Sigworth F.J. 1992. The size of gating charge in wild-type and mutant Shaker potassium channels. Science. 255:1712–1715 10.1126/science.1553560 [DOI] [PubMed] [Google Scholar]
- Seoh S.A., Sigg D., Papazian D.M., Bezanilla F. 1996. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 16:1159–1167 10.1016/S0896-6273(00)80142-7 [DOI] [PubMed] [Google Scholar]
- Shi J., Krishnamoorthy G., Yang Y., Hu L., Chaturvedi N., Harilal D., Qin J., Cui J. 2002. Mechanism of magnesium activation of calcium-activated potassium channels. Nature. 418:876–880 10.1038/nature00941 [DOI] [PubMed] [Google Scholar]
- Shin K.S., Maertens C., Proenza C., Rothberg B.S., Yellen G. 2004. Inactivation in HCN channels results from reclosure of the activation gate: desensitization to voltage. Neuron. 41:737–744 10.1016/S0896-6273(04)00083-2 [DOI] [PubMed] [Google Scholar]
- Sweet T.B., Cox D.H. 2008. Measurements of the BKCa channel’s high-affinity Ca2+ binding constants: effects of membrane voltage. J. Gen. Physiol. 132:491–505 10.1085/jgp.200810094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szallasi A., Blumberg P.M., Annicelli L.L., Krause J.E., Cortright D.N. 1999. The cloned rat vanilloid receptor VR1 mediates both R-type binding and C-type calcium response in dorsal root ganglion neurons. Mol. Pharmacol. 56:581–587 [DOI] [PubMed] [Google Scholar]
- Talukder G., Aldrich R.W. 2000. Complex voltage-dependent behavior of single unliganded calcium-sensitive potassium channels. Biophys. J. 78:761–772 10.1016/S0006-3495(00)76634-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulens C., Siegelbaum S.A. 2003. Regulation of hyperpolarization-activated HCN channels by cAMP through a gating switch in binding domain symmetry. Neuron. 40:959–970 10.1016/S0896-6273(03)00753-0 [DOI] [PubMed] [Google Scholar]
- Voets T., Droogmans G., Wissenbach U., Janssens A., Flockerzi V., Nilius B. 2004. The principle of temperature-dependent gating in cold- and heat-sensitive TRP channels. Nature. 430:748–754 10.1038/nature02732 [DOI] [PubMed] [Google Scholar]
- Wang L., Sigworth F.J. 2009. Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy. Nature. 461:292–295 10.1038/nature08291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman J.J. 1964. Linked functions and reciprocal effects in hemoglobin: A second look. In Advances in Protein Chemistry 223–286 [DOI] [PubMed] [Google Scholar]
- Wyman J. 1967. Allosteric linkage. J. Am. Chem. Soc. 89:2202–2218 10.1021/ja00985a037 [DOI] [Google Scholar]
- Wyman J. 1984. Linkage graphs: a study in the thermodynamics of macromolecules. Q. Rev. Biophys. 17:453–488 10.1017/S0033583500004881 [DOI] [PubMed] [Google Scholar]
- Wyman J., Gill S.J. 1990. Binding and linkage: functional chemistry of biological macromolecules. University Science Books, Mill Valley, CA: 330 pp [Google Scholar]
- Xia X.M., Zeng X., Lingle C.J. 2002. Multiple regulatory sites in large-conductance calcium-activated potassium channels. Nature. 418:880–884 10.1038/nature00956 [DOI] [PubMed] [Google Scholar]
- Yusifov T., Javaherian A.D., Pantazis A., Gandhi C.S., Olcese R. 2010. The RCK1 domain of the human BKCa channel transduces Ca2+ binding into structural rearrangements. J. Gen. Physiol. 136:189–202 10.1085/jgp.200910374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zeng X.H., Lingle C.J. 2012. Barium ions selectively activate BK channels via the Ca2+-bowl site. Proc. Natl. Acad. Sci. USA. 109:11413–11418 10.1073/pnas.1204444109 [DOI] [PMC free article] [PubMed] [Google Scholar]