Abstract
Extracellular matrix (ECM) has many effects beyond providing structural support. ECM proteins typically comprise multiple, independently folded domains whose sequences and arrangement are highly conserved. Some of these domains bind adhesion receptors such as integrins that mediate cell-matrix adhesion and also transduce signals into cells. ECM proteins also bind soluble growth factors and regulate their distribution, activation and presentation to cells and are capable of integrating complex, multivalent signals to cells in a spatially organized and regulated fashion. These properties need to be incorporated into considerations of the roles of ECM and ECM proteins in phenomena as diverse as developmental patterning, stem cell niches, cancer and genetic diseases.
Introduction
All cells are in close contact with extracellular matrix, either continuously or at important phases of their development (e.g., as stem or progenitor cells or during cell migration and invasion). The extracellular matrix (ECM) is well known for its ability to provide structural support for organs and tissues, for cell layers in the form of basement membranes, and for individual cells as substrates for cell motility. The role of ECM in cell adhesion and in signaling to cells through adhesion receptors such as integrins has received much attention1–3 and, more recently, the idea has been developed that mechanical characteristics of the matrix (stiffness, deformability) also provide inputs into cell behavior4,5. Thus, it is clear that ECM proteins and structures play vital roles in the determination, differentiation, proliferation, survival, polarity and migration of cells – ECM signals are arguably at least as important as soluble signals in governing these processes and probably more so. That work has been well summarized elsewhere and there is not space to review it here. Instead, I want to emphasize somewhat different aspects of the contributions of ECM and ECM proteins to cell and tissue behavior, namely their roles in binding, presenting and integrating growth factor signals to cells.
The complex domain structures of ECM proteins
There are hundreds of ECM proteins encoded in vertebrate genomes. Many of the genes are ancient, such as those comprising the basement membrane toolkit (type IV collagens, laminins, nidogen, perlecan, type XV/XVIII collagen), which are found in most metazoa, and one can argue that basement membranes were crucial to the evolution of multilayered organisms6. However, many vertebrate ECM proteins/genes have evolved much more recently, during the evolution of the deuterostome lineage, and that expansion includes not only elaboration of preexisting families (e.g., laminins, collagens, etc.) but also novel proteins (e.g., fibronectins, tenascins etc.). What purposes are served by this proliferation of ECM proteins? Almost universal properties of ECM proteins are that they are large and complex, with multiple distinct domains, and that those domains are highly conserved among different taxa (Figure 1). It is not necessary for proteins to be large or complex in order to generate strong, stable fibrils – intermediate filament proteins and type I collagen provide notable examples to the contrary. So, why are most ECM proteins so large, complex and conserved? Many ECM proteins have dozens of individually folded domains but in most cases we do not understand the functions of more than a few of them. What are the rest there for? The conserved domains are arranged in specific juxtapositions with one another, sometimes controlled by highly regulated alternative splicing. The clear implication is that the specific domains and architectures of ECM proteins contain information of biological significance and evolutionary value. This article will explore that hypothesis in light of recent discoveries concerning the structure, functions and interactions of representative ECM proteins.
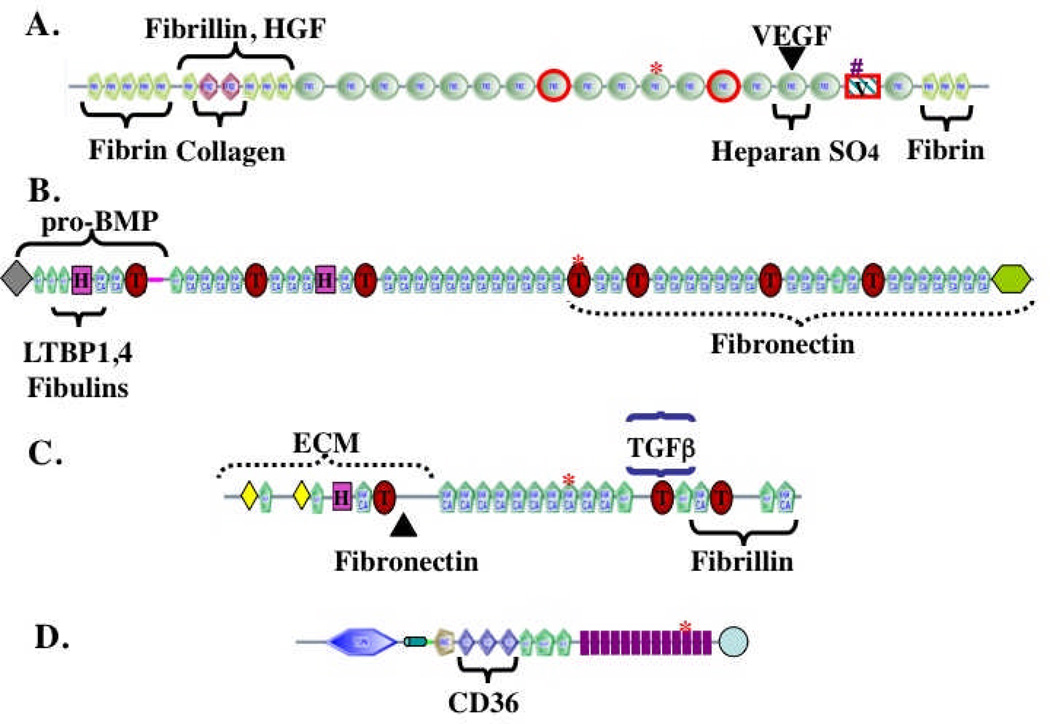
Figure 1. The complex domain structures of ECM proteins.
The figure shows representative ECM proteins (out of hundreds encoded in the genome). The proteins are built from multiple, independently folded domains, which occur in different combinations in different ECM proteins as a consequence of exon shuffling during evolution. The domain structures were generated from the SMART web site (http://smart.embl-heidelberg.de/) and edited in light of specific knowledge about individual proteins.
A. Fibronectin. Encoded by a single gene but alternatively spliced at three regions (boxed in red) to generate 12 proteins in rodents and 20 in humans. FN3 domains are widespread in ECM proteins. Binding sites for other matrix proteins are marked. The heparan-sulfate-binding site can interact with proteoglyans (PGs) or with syndecan, an integral membrane PG. The RGD (arg-gly-asp) integrin-binding site is marked by a red asterisk and a second LDV (leu-asp-val) integrin-binding site is marked by a pound sign. Fibronectin is a proangiogenic molecule, whose function is compromised by elimination of the RGD site or of the two alternatively spliced FN3 domains36,37. FN also binds the proangiogenic growth factors VEGF and HGF16,17.
B. Fibrillin-1. A member of a three-gene family. Fibrillins are made up of EGF-like domains, which are found in many ECM proteins, as well as TB (TGFβ-binding, marked by T) and hybrid (H) domains that are both specific to fibrillins and LTBPs21,22. Known binding sites within fibrillin-1 for other matrix proteins and growth factors are marked.
C. LTBP-1. A member of a four-gene family with structure related to that of fibrillins. Known binding sites for TGFβ/LAP latent complex and for fibrillin and fibronectin are marked.
D. Thrombospondin-1. Member of a five-gene family38. Thrombospondins 1 and 2 have the structure shown and both are antiangiogenic. Antiangiogenic activity lies in the TSP1 repeats, which bind to the CD36 receptor. TSP1 repeats are also found in other ECM proteins. Thrombospondins also contain EGF-like repeats and a VWC domain, known in other proteins to bind BMPs. The 13 TSP3 repeats (purple) and C-terminal domain are unique to thrombospondins and bind multiple Ca++ ions.
In all proteins, the asterisks mark RGD (arg-gly-asp) tripeptide sequences that may bind to integrins, as is well documented for the similar motif in fibronectin (see A).
ECM proteins and growth factor signaling
One long-standing idea is that ECM binds growth factors and that is certainly true. Many growth factors (e.g., FGFs, VEGFs) bind avidly to heparin and to heparan sulfate, a component of many ECM proteoglycans. So, a generally held view is that heparan sulfate proteoglycans (PGs) act as a sink or reservoir of growth factors and may assist in establishing stable gradients of growth factors bound to the ECM; such gradients of morphogens play vital roles in patterning developmental processes. It is also often proposed, and sometimes even demonstrated, that growth factors can be released from ECM by degradation of ECM proteins or of the glycosaminoglycan components of PGs. Those models place ECM in a distal role, acting as localized reservoirs for soluble growth factors that will be released from the solid phase to function as traditional, soluble ligands. However, some growth factors actually bind to their signaling receptors using heparan sulfate as a cofactor. The binding of FGF to FGFR depends on a heparan sulfate chain binding at the same time7 and TGFβ ligands bind first to integral membrane proteoglycans (e.g., endoglin, betaglycan) and that binding and “presentation” play key roles in signaling by these ligands8 – in a sense, they are acting as solid-phase ligands. Such phenomena may well be much more widespread than the few, well studied examples known. Less well known are examples of growth factors binding to ECM proteins themselves without involvement of glycosaminoglycans but there are increasing numbers of such documented interactions and I will argue here that presentation of growth factor signals by ECM proteins is an important part of ECM function.
Before considering the potential roles of ECM proteins in modulating responses to growth factor signals, it is important to address first some related concepts that need to be kept separate in thinking about and analyzing the functions of ECM in signaling to cells. First, it is clear that standard ECM receptors such as integrins and DDR tyrosine kinase receptors are signal transduction receptors in their own right – their ligands are specific domains and motifs embedded in the ECM proteins and ECM-integrin interactions lead to signal transduction responses by cells that are at least as complex and important as those triggered by soluble ligands such as EGF, PDGF and VEGF. That topic has been well reviewed1–3 and I will not discuss it further here. Second, and less clearly, there are numerous reports of “crosstalk” and “synergy” between signaling by integrins and that by various growth factors9. In most cases it is uncertain whether such crosstalk involves [1] membrane-proximal interactions or [2] cooperation in the downstream signal transduction pathways. We will not be interested here in the second case but will return later to the first. Another concept, first suggested by Jurgen Engel 20 years ago, when the modular nature of ECM proteins was first becoming apparent, is that intrinsic domains within ECM proteins might act as ligands for canonical growth factor receptors10. This suggestion arose from the observation that laminin contains multiple copies of EGF-like domains, as do many ECM proteins (e.g., laminins, tenascins, thrombospondins, fibrillins). Engel suggested that they might bind to EGF receptors and signal as solid-phase ligands. It has been demonstrated that EGF-like domains from laminin11,12 or tenascin13,14 presented as soluble ligands can bind to EGFR and modulate its signaling and it is often hypothesized that fragments of ECM proteins can be released by proteolysis (e.g., by matrix metalloproteases) and act as soluble ligands. That model is similar to the idea that matrix-bound growth factors can be released by ECM degradation. In both cases, the ECM acts as a reservoir of growth factors (bound or intrinsic), which can be released as soluble factors to bind their receptors. However, the interesting idea that intrinsic growth factor-like ligands can act from the solid-phase deserves more intensive investigation and careful experimental distinction from alternatives such as release of bound or intrinsic ligands. We will explore this idea and the related concept that ECM proteins bind and present growth factors as organized solid-phase ligands.
Growth factor binding to ECM proteins
As mentioned, it is widely accepted that many growth factors bind to glycosaminoglycan chains attached to ECM and membrane proteins. However, there is increasing evidence for specific binding of growth factors by the proteins themselves. For example, both fibronectin and vitronectin bind to HGF and form complexes of Met (the HGF receptor) and integrins (the ECM receptors), leading to enhanced cell migration15. Similarly VEGF binds to specific FN type III (FN3) domains in both fibronectin (FN) and tenascin-C and these associations promote cell proliferation16,17. Importantly, in the case of the FN-VEGF binding, the effect on proliferation requires the binding sites for integrins and for VEGF to be in the same molecule, indicating a requirement for juxtaposition of the two receptors (integrin α5β1and VEGFR2), rather than some downstream crosstalk16. It is worth noting that FN3 domains are prevalent in many ECM proteins and membrane receptors and their potential for binding soluble factors needs further investigation.
There are other examples of widely distributed ECM domains that bind and present growth factors. For example, in Drosophila, collagen IV binds Dpp (a BMP homologue) and enhances its interactions with BMP receptors; this collagen/BMP interaction is crucial in regulating the dorsoventral axis and the numbers of germinal stem cells in the ovary, both processes dependent on gradients of Dpp18. Collagen IV is a universal constituent of basement membranes and the key Dpp-binding motif identified in the C-terminal domains of the two Drosophila collagen IV subunits is highly conserved across phyla, suggesting that this interaction may be important in many other contexts18. Collagen II, the major collagen of cartilage, provides another instructive example. This collagen contains a chordin-like VWC domain near its N-terminus, which binds TGF-β1 and BMP-2, two chondrogenic growth factors. The VWC domain is alternatively spliced, being included in prechondrogenic mesoderm and early developing cartilage but excluded in mature cartilage19. The VWC/chordin domain is found in many ECM proteins as well as in known regulators of BMPs and it typically acts as a negative regulator of their functions20. These two examples illustrate the capacity of conserved elements of ECM proteins to regulate, either positively or negatively, the functions of diffusible growth factors/morphogens of the BMP family.
TGF-β regulation by ECM binding
The best developed story about growth factors and ECM concerns the roles of diverse ECM proteins and their receptors in binding and regulation of TGF-β. There are three genes encoding the precursors of TGF-β isoforms 1–3. Each precursor is cleaved by a furin protease to the mature TGF-β and its propeptide, known as LAP (latency-associated peptide). The LAP and TGF-β remain non-covalently associated in a complex called the small latency complex (SLC) and in this form TGF-βs are inactive21,22. The LAPs are then S-S-bonded to one of the latent TGF-β-binding proteins (LTBPs) to form large latent complexes (LLCs) and many cells secrete TGF-βs already assembled into such complexes. The LTBPs bind in turn to other ECM proteins (including fibrillins and fibronectins) thereby incorporating the different TGF-β isoforms into extracellular matrices in latent form (see Figures 1 and 2A). LTBP-mediated incorporation into ECM is necessary for subsequent effective activation of TGF-βs. There are several mechanisms for activation (see Figure 2B); they include degradation of ECM proteins such as fibrillin or LTBPs. Activation can also occur by cleavage or conformational change in LAP, exposing or releasing the TGF-βs so that they can bind and activate their receptors21,22. Another ECM protein, thrombospondin, can activate TGF-βs by binding and disassociating LAP or by activating metalloproteases; mice lacking thrombospondin-1 develop pneumonia because of reduced levels of active TGF-β in their lungs23. Yet another mechanism for activation of TGF-βs involves αvβ6 and αvβ8 integrins, which bind to RGD sequences in LAP1 and LAP324,25. αvβ8 integrin appears to cooperate with metalloproteases to release TGF-β. However, αvβ6 integrin activates TGF-β without any requirement for proteolysis. Instead, it binds to LAP and, in the presence of mechanical strain between the cells expressing the integrin and the ECM to which the SLC is attached, deforms LAP to expose the associated TGF-β (see Figure 2B). The activated TGF-β is not released in soluble, diffusible form but appears to act only at short range, perhaps as a bound solid-phase ligand. Thus, the binding, sequestration in latent form and subsequent activation of TGF-βs all intimately involve a variety of ECM proteins; LTBPs and fibrillins act to sequester TGF-β/LAP complexes, thrombospondin can act to activate TGF-β and integrin-based mechanical strain, which requires LTBP, fibrillin and fibronectin, is an important mechanism for activation (see Figure 2). The whole assemblage acts like a regulated machine incorporating both negative and positive regulation; incorporation of TGF-β into the matrix anchors and localizes the growth factor in a latent form, which can subsequently be activated by proteolysis or by mechanical strain21–25. Mutations in many of the ECM proteins, integrins and the RGD sites in the LAPs confirm the relevance of these interactions in vivo.
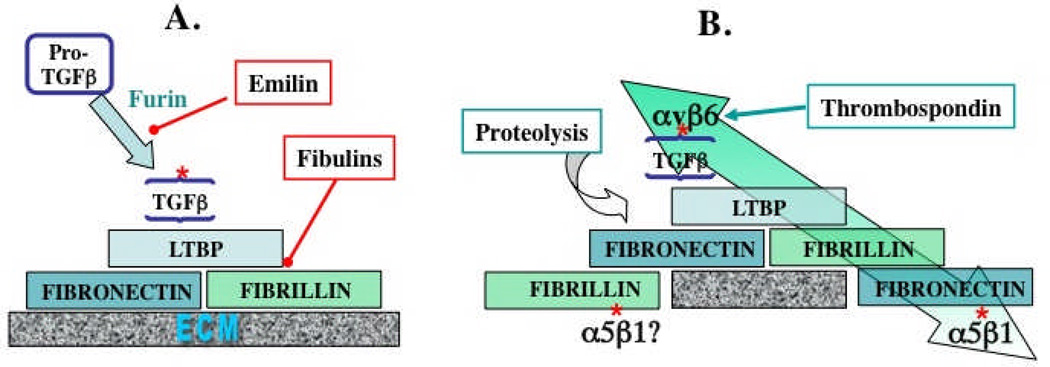
Figure 2. ECM interactions regulating TGFβ.
A. Incorporation into the ECM.
Cleavage by furin protease of Pro-TGFβ to the small latent complex (SLC) comprising TGFβ and LAP is inhibited by emilin, an ECM protein. The SLC binds to LTBP, via S-S bonding to a TB domain, to form the large latent complex (LLC), in which form the TGFβ is inactive21,22. LTBP then binds to fibrillin and to fibronectin (see Figure 1 for specific interaction domains). Fibulins compete for LTBP binding to fibrillin39. Fibrillin binds to preexisting fibronectin fibrils or assembles into microfibrils and both fibrillin and fibronectin undergo further homomeric and heteromeric interactions within the ECM.
B. Activation of ECM-bound latent TGFβ.
TGFβ can be activated by proteolysis of the ECM proteins and/or of LAP or directly by thrombospondin (see text). TGFβ can also be activated by mechanical strain (large green arrow). This strain arises from cytoskeletal force applied through αvβ6 integrin, which binds to an RGD site in LAP and requires attachment of the TGFβ/LAP complex through LTBP to the fibronectin-rich matrix, which is, in turn, attached via α5β1 integrin to other cells. Fibrillin might also be attached to cells via integrins.
Further analyses of LAPs, LTBPs and fibrillins have uncovered the molecular details of the interactions. The TGF-β/LAP complex binds to LTBP-1 through a specific TGF-binding (TB) domain and adjacent EGF domains (see Figure 1). TB domains, as well as hybrid domains, which are hybrids of TB and EGF domains, are unique to fibrillins and LTBPs and there are several in each of those proteins, suggesting that they may be able to bind other BMP family members (Figure 1). Indeed it is known that proBMP-7 can bind to fibrillin-1 in an N-terminal region containing a hybrid and a TB domain26. Furthermore, fibrillin-2 and BMP-7 mutations show genetic interactions in causing syndactyly and polydactyly in mice27 and a related human disease, congenital contractual arachnodactyly, arises from mutations in fibrillin-2. So it seems very likely that other functionally important interactions between members of the TGF/BMP and LTBP/fibrillin families remain to be discovered. The interactions of different LTBPs and fibrillins with diverse TGF/BMP family members have the potential to target different signals to different locations.
The implications of this ECM-based regulation of TGF-β function for human disease have recently become abundantly clear in the case of Marfan’s syndrome, a genetic disease resulting from mutations in the gene for fibrillin-128,29. Like many other genetic diseases whose target genes encode ECM proteins, this disease is associated with defective assembly of extracellular matrix components, in this case the microfibrils of which fibrillins are components. The phenotype was originally attributed to mechanical consequences of these structural defects of the ECM. However, the known associations of fibrillins with LTBPs suggested that activation of TGF-βs might also play a role and it was shown in mouse models of Marfan’s syndrome that activation of TGF-β was markedly increased and that many of the phenotypic consequences of mutations in fibrillin-1 could be ameliorated by TGF-β antagonists28,29. These insights are already having clinical applications.
Extracellular matrix proteins as localized, multivalent signal integrators
The examples discussed illustrate the roles of discrete domains in ECM proteins in binding and regulating the functions of canonical growth factors. Many of these domains are found in multiple ECM proteins in different combinations and arrangements and it is a reasonable proposition that many more such ECM/growth factor interactions remain to be discovered.
Other domains and motifs in these ECM proteins have the potential to bind directly to cell-surface adhesion receptors such as integrins. At the very least, the coexistence in the same ECM proteins of sites for cell adhesion and binding sites for growth factors concentrates the growth factors close to their own cell-surface receptors. Therefore, localization of growth factors at the cellular level by binding to ECM can localize their signaling and this concept underlies the idea that binding of growth factors to ECM contributes to establishment of stable gradients. According to this model, morphogen gradients are composed jointly of soluble, diffusible factors and ECM – both are necessary. ECM-bound growth factors could be released locally or could be presented as complexes still bound to the ECM proteins and, as mentioned, there is also the potential (as yet unproven) for specific intrinsic domains in ECM proteins (e.g., EGF-like domains) to bind directly to growth factor receptors.
ECM proteins are highly conserved, not only in the sequences of specific domains but also in the arrangements of those domains within the proteins. It is also of interest that specific domains are often inserted or omitted by highly regulated alternative splicing, thus changing the complement of domains. This could alter the binding of specific growth factors, as in the case of the VWC domain in type II collagen19, or the interactions with cell surface receptors, as in the case of agrin. In agrin, inclusion of two small exons (A/y and B/z) confers on agrin the ability to bind to heparan sulfate and dystroglycan, respectively, and greatly enhances the clustering of acetylcholine receptors30. I also mentioned the extensive body of data suggesting that ECM proteins can synergize with growth factors in affecting cell proliferation and migration9. While such synergy does not in principle require juxtaposition, experiments such as those concerning VEGF binding by fibronectin (FN) show that the synergy requires the binding sites for integrins and for VEGF to be coupled in the same molecule – presenting them as two separate, substrate-bound fragments of FN does not suffice16. This sort of result suggests that proximity is important and raises the hypothesis that ECM molecules, by virtue of their ordered domain organization, act to organize complexes of receptors in the plane of the membrane. Such complexes could enhance membrane-proximal regulation among the receptors and promote integration of the signals transduced (Figure 3). An instructive parallel can be found in the clustering of immunoregulatory receptors in immunological synapses (which also involve crosstalk among integrins and other receptors)31,32. Immunological synapses have substructure – different receptors occupy different zones within the synapse. ECM-mediated clusters could have highly detailed substructure, the juxtaposition of different receptors could be driven by the arrangement of domains in the ECM protein at a resolution of several nanometers. One could think of ECM proteins and their associated partners (growth factors and other ECM proteins) as solid-phase growth factors metaphorically playing tunes, in contrast with soluble growth factors that one could view as playing single notes (Figure 3).
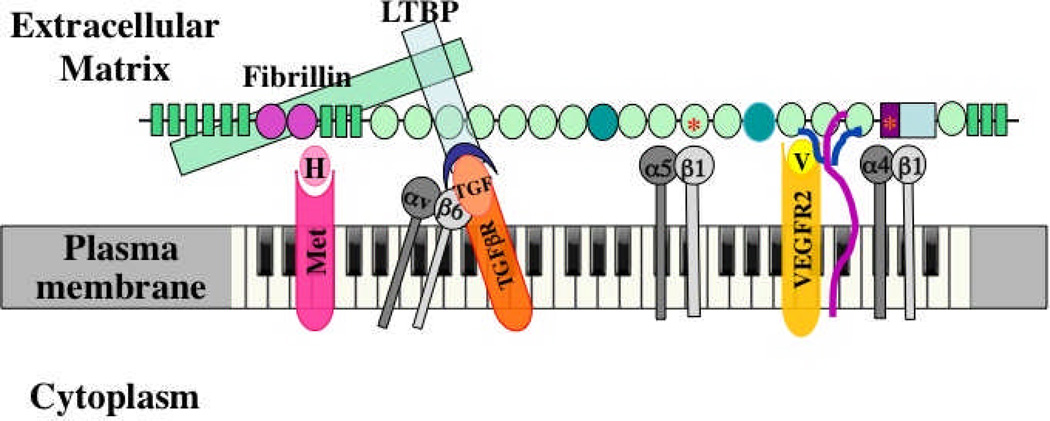
Figure 3. Multidomain interactions of ECM proteins with cells.
The example shown is fibronectin40. Multiple domains are known to bind to integrins, to other ECM proteins and to growth factors, as shown. Integrins α5β1 and α4β1 bind, respectively, to RGD and LDV motifs; heparan sulfate chains of syndecan (purple/blue) bind to FN3-13 as does VEGF. Evidence suggests that VEGF (V, yellow) signals through its own receptor (VEGFR2) more effectively when bound to fibronectin16. The same is proposed here for HGF (H) and its receptor (Met, pink). As shown in Figures 1 and 2, fibrillin (green) binds to an N-terminal region of fibronectin and in turn binds LTBP (blue), which recruits TGFβ in a latent complex with LAP (blue crescent). αvβ6 integrin can bind an RGD site in LAP, activating TGFβ, so that it can bind its own receptors (orange). The proposal is that fibronectin organizes and integrates all these signals at two levels. First, by recruiting growth factors to the ECM, fibronectin localizes those signals at the cellular level. Second, the close juxtaposition of the domains in fibronectin brings the different receptors together into an organized submicron patch in the cell surface membrane. Each domain is 2–4 nm in diameter and the entire fibronectin subunit shown is 60–70 nm long, so the receptors will be brought into close apposition such that their signals provide complex, integrated information to the cell – metaphorically generating a melodies and chords in contrast with the “single notes” generated by each receptor. Fibronectin is essential for angiogenesis, and most of the bound receptors and ligands have been shown to play roles in angiogenesis. This model suggests that fibronectin and its associated ECM proteins orchestrate and integrate these signals. In addition, alternatively spliced domains of fibronectin (darker green ovals) are also necessary for proper vascular development and it is a reasonable hypothesis that they introduce additional ligands and/or receptors into the mix.
Such models also have other implications. The very nature of ECM imposes spatial context on the signaling. Cells are often polarized by their associations with ECM – the basement membrane to which epithelial sheets attach defines the base and polarity of the cell and confers ability to respond to soluble growth factors such as EGF. There is good evidence that the deformability of the ECM affects the responses of cells to it2–4,33,34. ECM molecules are flexible and extendable and mechanical tension can uncover cryptic sites within them35. Such mechanically exposed cryptic sites could bind additional cell-surface receptors or growth factors. Mechanical extension or the inclusion or exclusion of alternatively spliced domains could also alter the physical relationships among other domains, thus affecting the composition and spatial arrangement of the hypothesized organized patches of receptors.
Implications for future research
The ideas explored in this brief review need further experimental investigation. There are only relatively few well documented examples of specific growth factor binding by domains in ECM proteins but it would be straightforward to investigate this possibility more extensively. There are even fewer cases where it is clear whether ECM-bound growth factors need to be released to soluble form or can act as solid-phase ligands and the proposition that intrinsic domains of ECM proteins can directly affect canonical growth factor receptors, either as solid-phase ligands or as locally released soluble ligands, needs more study. The idea that specific arrangements of domains confer important information can be tested. The possible effects of mechanical strain on exposure of cryptic binding sites for growth factors, receptors or other ECM proteins are just beginning to be explored. The nature of ECM-induced receptor complexes in the membrane can be investigated by methods such as FRET, FLIM, high resolution EM and crosslinking. The effects of regulated alternative splicing of ECM proteins on all of these questions and the implications of the diversity within families of proteins such as LTBPs and fibrillins need to be investigated.
The ECM is a fundamental component of the microenvironment of cells and has been significantly expanded during the evolution of vertebrates. Some of that elaboration has contributed to structural components such as bones and teeth but it is evident that this is only one role of ECM. The abundant evidence that ECM provides much more than mechanical support and a locus for cell adhesion and migration should be incorporated into our thinking about the potential roles of ECM in basement membranes, stem cell niches and tumors. All epithelial cells are in association with basement membranes for at least part of their lives and many stem cell niches have ECM within them. ECM composition and organization undergo radical alterations in cancer and could affect survival, proliferation and other properties of both tumor and stromal cells. Ever since McKusick’s initial recognition and cataloguing of a diverse set of genetic diseases affecting extracellular matrix over 50 years ago, it has been implicitly assumed that the pathological consequences were a direct result of defects in ECM assembly. While those defects do exist and no doubt contribute, in Marfan’s syndrome and related diseases, it is now clear that many phenotypic consequences are indirect effects of dysregulation of TGFβ signaling consequent on the ECM defects. Structural defects are difficult to treat, absent gene therapy or stem cell therapies, but growth factor signaling offers simpler and more accessible targets for intervention. One can hope that further investigations of the roles of ECM proteins in regulating signaling events will yield additional leads of this sort.
Acknowledgements
I thank Alexandra Naba and Kaan Certel for constructive criticisms of the text and figures.
I gratefully acknowledge support from the Howard Hughes Medical Institute and from the National Institutes of Health.
References
- 1.Hynes RO. Cell. 2002;110:673. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 2.Berrier AL, Yamada KM. J. Cell Physiol. 2007;213:565. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 3.Legate KR, Wickstrom SA, Fassler R. Genes Dev. 2009;23:397. doi: 10.1101/gad.1758709. [DOI] [PubMed] [Google Scholar]
- 4.Discher DE, Mooney DJ, Zandstra PW. Science. 2009;324:1673. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geiger B, Spatz JP, Bershadsky AD. Nat. Rev. Mol. Cell Biol. 2009;10:21. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 6.Whittaker CA, et al. Dev. Biol. 2006;300:252. doi: 10.1016/j.ydbio.2006.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammadi M, Olsen SK, Goetz R. Curr. Opin. Cell Biol. 2005;15:506. doi: 10.1016/j.sbi.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Shi Y, Massague J. Cell. 2003;113:685. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 9.Alam N, et al. J. Cell Physiol. 2007;213:649. doi: 10.1002/jcp.21278. [DOI] [PubMed] [Google Scholar]
- 10.Engel J. FEBS Letters. 1989;251:1. doi: 10.1016/0014-5793(89)81417-6. [DOI] [PubMed] [Google Scholar]
- 11.Panayotou G, et al. Cell. 1989;56:93. doi: 10.1016/0092-8674(89)90987-2. [DOI] [PubMed] [Google Scholar]
- 12.Shenk S, et al. J. Cell Biol. 2003;161:197. doi: 10.1083/jcb.200208145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shenk CS, et al. J. Cell Biol. 2001;154:459. [Google Scholar]
- 14.Krishnan AKV, Tran KT, Griffith L, Wells A. J. Cell Physiol. 2007;214:504. doi: 10.1002/jcp.21232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahman S, et al. BMC Cell Biol. 2005;6:8. doi: 10.1186/1471-2121-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wielath ES, et al. Circ. Res. 2006;99:853. doi: 10.1161/01.RES.0000246849.17887.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishitsuka T, Ikuta T, Ariga H, Matsumoto KI. Biol. Pharm. Bull. 2009;32:1004. doi: 10.1248/bpb.32.1004. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Harris RE, Bayston LJ, Ashe HL. Nature. 2008;455:72. doi: 10.1038/nature07214. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Y, Oganesian A, Keene DR, Sandell LJ. J. Cell Biol. 1999;144:1069. doi: 10.1083/jcb.144.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abreu JG, et al. Gene. 2002;287:39. [Google Scholar]
- 21.Rifkin DB. J. Biol. Chem. 2005;280:7409. doi: 10.1074/jbc.R400029200. [DOI] [PubMed] [Google Scholar]
- 22.ten Dyke P, Arthur HM. Nat. Rev. Mol. Cell Biol. 2007;8:857. doi: 10.1038/nrm2262. [DOI] [PubMed] [Google Scholar]
- 23.Lawler J, et al. J. Clin. Invest. 1998;101:982. doi: 10.1172/JCI1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheppard D. Cancer Metast. Rev. 2005;24:395. doi: 10.1007/s10555-005-5131-6. [DOI] [PubMed] [Google Scholar]
- 25.Wipff PJ, Hinz B. Eur. J. Cell Biol. 2008;87:601. doi: 10.1016/j.ejcb.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Gregory KE, et al. J. Biol. Chem. 2005;280:27970. doi: 10.1074/jbc.M504270200. [DOI] [PubMed] [Google Scholar]
- 27.Arteaga-Solis E, et al. J. Cell Biol. 2001;154:275. doi: 10.1083/jcb.200105046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez F, Sakai LY, Rifkin DB, Dietz HC. Cell. Mol. Life Sci. 2007;64:2437. doi: 10.1007/s00018-007-7166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez F, Dietz HC. J. Biol. Chem. 2009;284:14677. doi: 10.1074/jbc.R900004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campanelli JT, Gayer GG, Scheller RH. Development. 1996;122:1663. doi: 10.1242/dev.122.5.1663. [DOI] [PubMed] [Google Scholar]
- 31.Kupfer A, Kupfer H. Semin. Immunol. 2003;15:295. doi: 10.1016/j.smim.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Dustin ML. Annu. Rev. Cell Dev. Biol. 2008;24:577. doi: 10.1146/annurev.cellbio.24.110707.175226. [DOI] [PubMed] [Google Scholar]
- 33.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 34.Vogel V, Sheetz MP. Curr. Opin. Cell Biol. 2009;21:38. doi: 10.1016/j.ceb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogel V. Annu. Rev. Biophys. Biomol. Struct. 2006;35:459. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi S, et al. J. Cell Biol. 2007;178:167. doi: 10.1083/jcb.200703021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Astrof S, Crowley D, Hynes RO. Dev. Biol. 2007;311:11. doi: 10.1016/j.ydbio.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams JC, Lawler J. Int. J. Biochem. Cell Bio. 2004;36:961. doi: 10.1016/j.biocel.2004.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ono RN, et al. J. Biol. Chem. 2009;284:16872. doi: 10.1074/jbc.M809348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hynes RO. Fibronectins. New York: Springer-Verlag; 1990. [Google Scholar]