Abstract
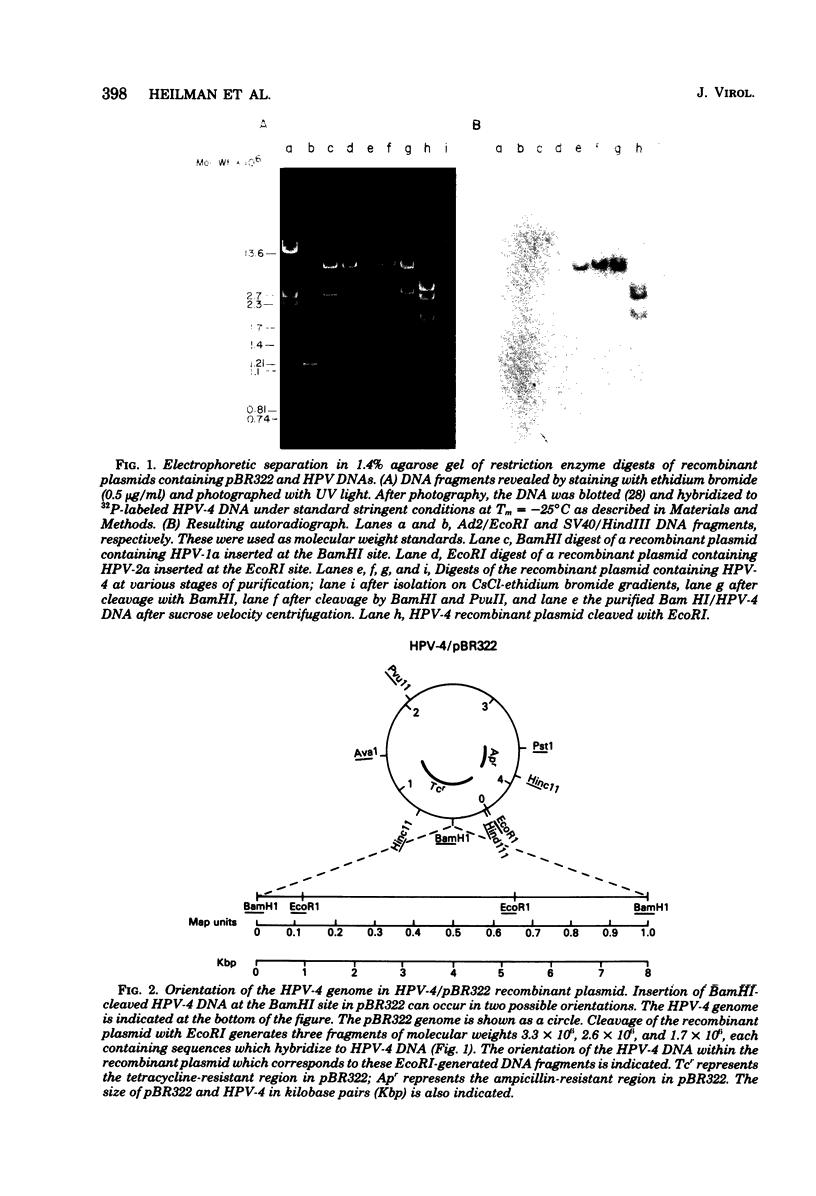
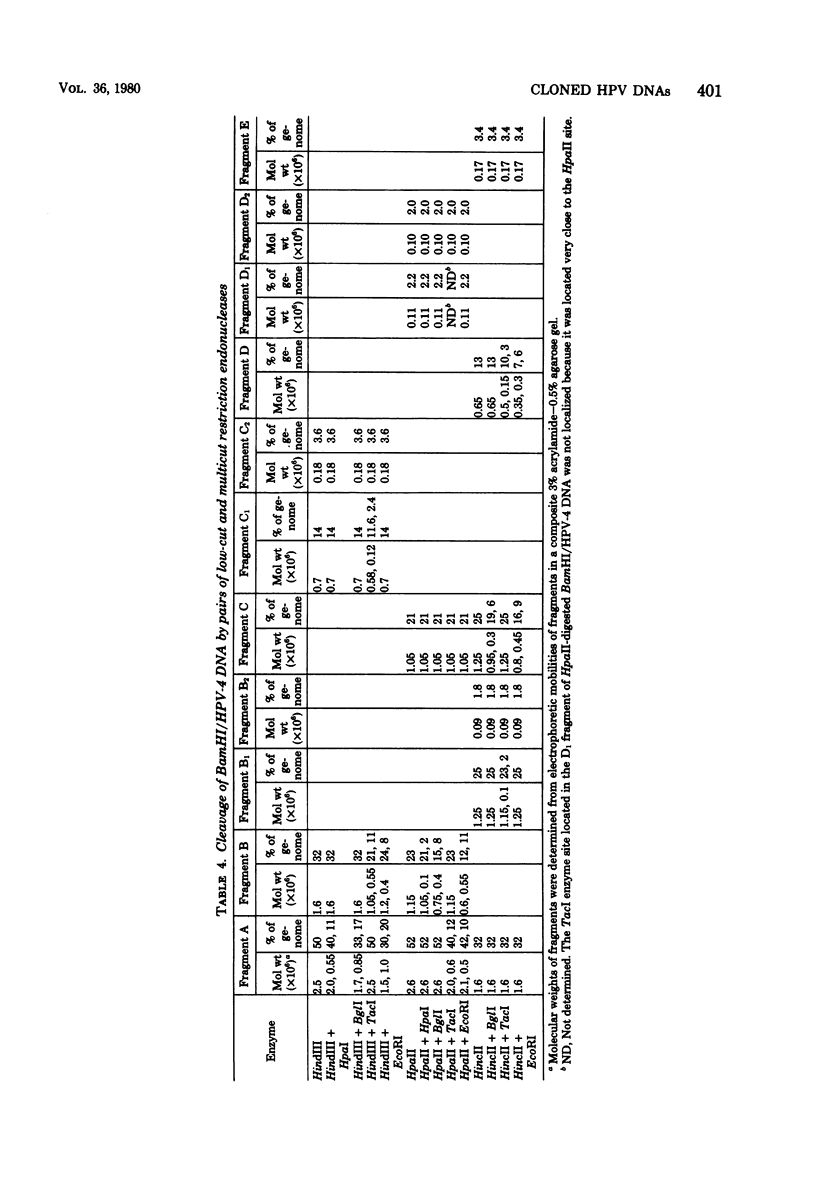
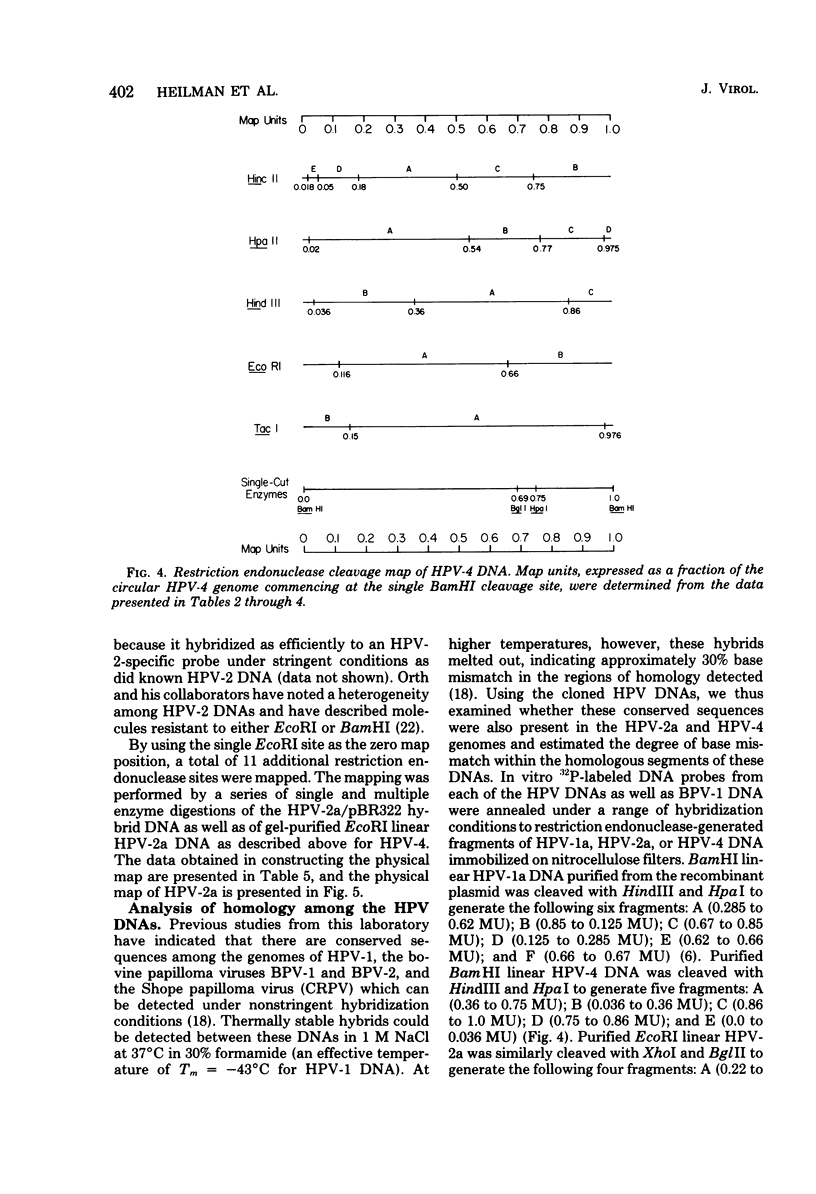
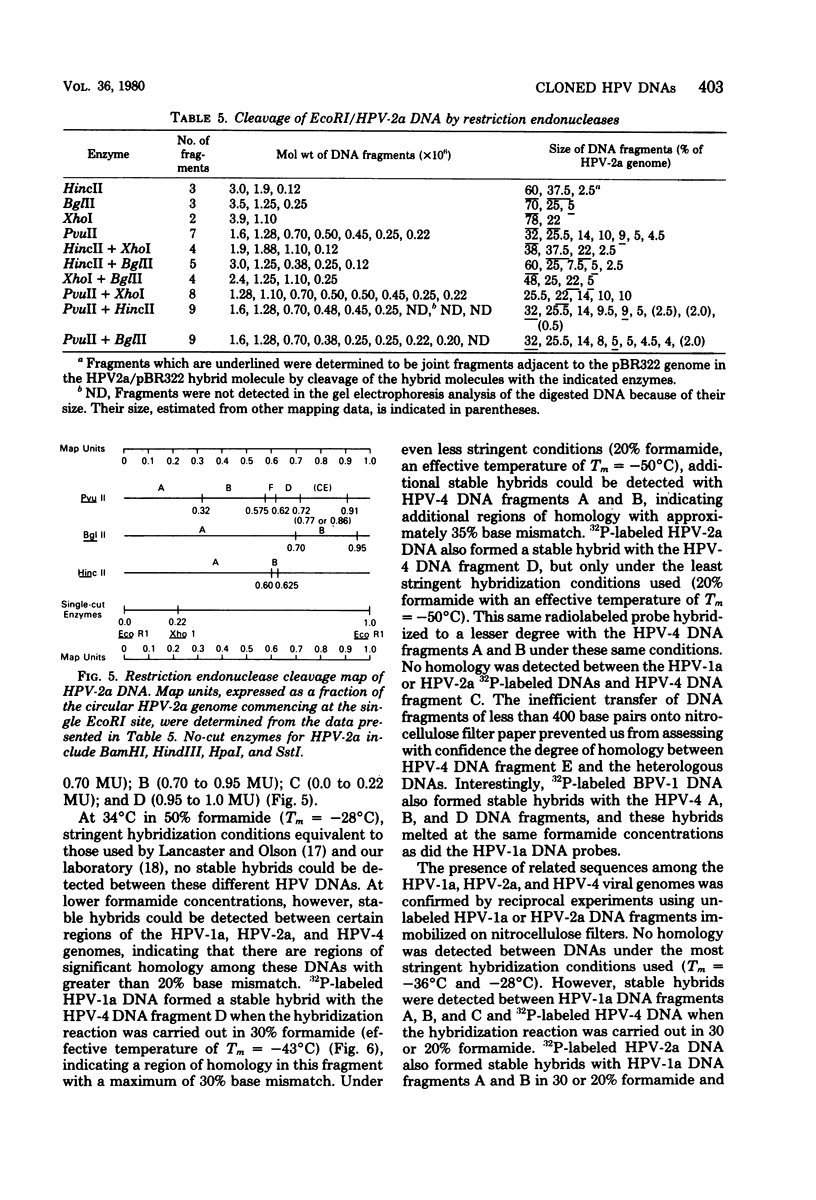
The complete DNA genomes of four distinct human papilloma viruses (human papilloma virus subtype 1a [HPV-1a], HPV-1b, HPV-2a, and HPV-4) were molecularly cloned in Escherichia coli, using the certified plasmid vector pBR322. The restriction endonuclease patterns of the cloned HPV-1a and HPV-1b DNAs were similar to those already published for uncloned DNAs. Physical maps were constructed for HPV-2a DNA and HPV-4 DNA, since these viral DNAs had not been previously mapped. By using the cloned DNAs, the genomes of HPV-1a, HPV-2a, and HPV-4 were analyzed for nucleotide sequence homology. Under standard hybridization conditions (Tm = --28 degrees C), no homology was detectable among the genomes of these papilloma viruses, in agreement with previous reports. However, under less stringent conditions (i.e., Tm = --50 degrees C), stable DNA hybrids could be detected between these viral DNAs, indicating homologous segments in the genomes with approximately 30% base mismatch. By using specific DNA fragments immobilized on nitrocellulose filters, these regions of homology were mapped. Hybridization experiments between radiolabeled bovine papilloma virus type 1 (BPV-1) DNA and the unlabeled HPV-1a, HPV-2a, or HPV-4 DNA restriction fragments under low-stringency conditions indicated that the regions of homology among the HPV DNAs are also conserved in the BPV-1 genome with approximately the same degree of base mismatch.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- CRAWFORD L. V., CRAWFORD E. M. A COMPARATIVE STUDY OF POLYOMA AND PAPILLOMA VIRUSES. Virology. 1963 Oct;21:258–263. doi: 10.1016/0042-6822(63)90265-4. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Favre M., Orth G., Croissant O., Yaniv M. Human papillomavirus DNA: physical map. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4810–4814. doi: 10.1073/pnas.72.12.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissmann L., Pfister H., Zur Hausen H. Human papilloma viruses (HPV): characterization of four different isolates. Virology. 1977 Feb;76(2):569–580. doi: 10.1016/0042-6822(77)90239-2. [DOI] [PubMed] [Google Scholar]
- Gissmann L., zur Hausen H. Human papilloma virus DNA: physical mapping and genetic heterogeneity. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1310–1313. doi: 10.1073/pnas.73.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howley P. M., Israel M. A., Law M. F., Martin M. A. A rapid method for detecting and mapping homology between heterologous DNAs. Evaluation of polyomavirus genomes. J Biol Chem. 1979 Jun 10;254(11):4876–4883. [PubMed] [Google Scholar]
- Howley P. M., Khoury G., Byrne J. C., Takemoto K. K., Martin M. A. Physical map of the BK virus genome. J Virol. 1975 Oct;16(4):959–973. doi: 10.1128/jvi.16.4.959-973.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel M. A., Chan H. W., Rowe W. P., Martin M. A. Molecular cloning of polyoma virus DNA in Escherichia coli: plasmid vector system. Science. 1979 Mar 2;203(4383):883–887. doi: 10.1126/science.217087. [DOI] [PubMed] [Google Scholar]
- Jarrett W. F., McNeil P. E., Grimshaw W. T., Selman I. E., McIntyre W. I. High incidence area of cattle cancer with a possible interaction between an environmental carcinogen and a papilloma virus. Nature. 1978 Jul 20;274(5668):215–217. doi: 10.1038/274215a0. [DOI] [PubMed] [Google Scholar]
- Ketner G., Kelly T. J., Jr Integrated simian virus 40 sequences in transformed cell DNA: analysis using restriction endonucleases. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1102–1106. doi: 10.1073/pnas.73.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster W. D., Olson C. Demonstration of two distinct classes of bovine papilloma virus. Virology. 1978 Sep;89(2):372–379. doi: 10.1016/0042-6822(78)90179-4. [DOI] [PubMed] [Google Scholar]
- Law M. F., Lancaster W. D., Howley P. M. Conserved polynucleotide sequences among the genomes of papillomaviruses. J Virol. 1979 Oct;32(1):199–207. doi: 10.1128/jvi.32.1.199-207.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M. F., Martin J. D., Takemoto K. K., Howley P. M. The colinear alignment of the genomes of papovaviruses JC, BK, and SV40. Virology. 1979 Jul 30;96(2):576–587. doi: 10.1016/0042-6822(79)90113-2. [DOI] [PubMed] [Google Scholar]
- Lowy D. R., Dvoretzky I., Shober R., Law M. F., Engel L., Howley P. M. In vitro tumorigenic transformation by a defined sub-genomic fragment of bovine papilloma virus DNA. Nature. 1980 Sep 4;287(5777):72–74. doi: 10.1038/287072a0. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Morrison J. M., Keir H. M., Subak-Sharpe H., Crawford L. V. Nearest neighbour base sequence analysis of the deoxyribonucleic acids of a further three mammalian viruses: Simian virus 40, human papilloma virus and adenovirus type 2. J Gen Virol. 1967 Jan;1(1):101–108. doi: 10.1099/0022-1317-1-1-101. [DOI] [PubMed] [Google Scholar]
- Orth G., Favre M., Croissant O. Characterization of a new type of human papillomavirus that causes skin warts. J Virol. 1977 Oct;24(1):108–120. doi: 10.1128/jvi.24.1.108-120.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth G., Jablonska S., Favre M., Croissant O., Jarzabek-Chorzelska M., Rzesa G. Characterization of two types of human papillomaviruses in lesions of epidermodysplasia verruciformis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1537–1541. doi: 10.1073/pnas.75.3.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- SYVERTON J. T. The pathogenesis of the rabbit papilloma-to-carcinoma sequence. Ann N Y Acad Sci. 1952 Jul 10;54(6):1126–1140. doi: 10.1111/j.1749-6632.1952.tb39983.x. [DOI] [PubMed] [Google Scholar]
- Schildkraut C. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3(2):195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]