Abstract
Background
DNA damage response (DDR) plays pivotal roles in maintaining genome integrity and stability. An effective DDR requires the involvement of hundreds of genes that compose a complicated network. Because DDR is highly conserved in evolution, studies in lower eukaryotes can provide valuable information to elucidate the mechanism in higher organisms. Fission yeast (Schizosaccharomyces pombe) has emerged as an excellent model for DDR research in recent years. To identify novel genes involved in DDR, we screened a genome-wide S. pombe haploid deletion library against six different DNA damage reagents. The library covered 90.5% of the nonessential genes of S. pombe.
Results
We have identified 52 genes that were actively involved in DDR. Among the 52 genes, 20 genes were linked to DDR for the first time. Flow cytometry analysis of the repair defective mutants revealed that most of them exhibited a defect in cell cycle progression, and some caused genome instability. Microarray analysis and genetic complementation assays were carried out to characterize 6 of the novel DDR genes in more detail. Data suggested that SPBC2A9.02 and SPAC27D7.08c were required for efficient DNA replication initiation because they interacted genetically with DNA replication initiation proteins Abp1 and Abp2. In addition, deletion of sgf73+, meu29+, sec65+ or pab1+ caused improper cytokinesis and DNA re-replication, which contributed to the diploidization in the mutants.
Conclusions
A genome-wide screen of genes involved in DDR emphasized the key role of cell cycle control in the DDR network. Characterization of novel genes identified in the screen helps to elucidate the mechanism of the DDR network and provides valuable clues for understanding genome stability in higher eukaryotes.
Background
Genomes are under constant threat of damage from exogenous factors and endogenous processes that result in DNA lesions. Correspondingly, cells have evolved elaborate DNA damage response (DDR) mechanisms to maintain genome integrity and stability [1]. DDR integrates the DNA-repair process with the cell cycle regulation, chromatin dynamics and programmed cell death, requiring delicate coordination of hundreds of genes [2]. Because DNA damage underlies the onset of cancer, aging, immune deficiencies, and other degenerative diseases, urgent needs of public health have made DDR a major target of study for decades [3].
DDR is highly conserved during evolution. Essential components of the DDR network, including ATM/ATR pathway, non-homologous ends joining (NHEJ) and homologous recombination (HR) repair, share homologues among almost all the eukaryotes [4]. Therefore, studies of the DDR in lower eukaryotes can provide valuable information to elucidate the mechanism in higher organisms. Because of their experimental amenabilities, budding yeast (Saccharomyces cerevisiae) and fission yeast (Schizosaccharomyces pombe) have become excellent models for DDR research [5,6]. Fission yeast separated from budding yeast about 1,000 million years ago during evolution. S. pombe contains about 150 metazoan-homologous genes which can’t be found in S. cerevisiae, and a similar number is seen when this comparison is made for S. cerevisiae. This emphasizes the advantage of using both yeasts for basic studies [7]. With the completion of the Saccharomyces Genome Deletion Project in 1999, genome-wide screens using a deletion library have become an effective way to identify novel genes involved in DDR [8]. Using such systematic screens, scientists have discovered 40 genes required for repairing DNA lesions caused by MMS [9], 31 genes involved in DDR to UV [10], and 107 new loci that influence sensitivity to γ radiation [11]. A haploid deletion library of S. pombe was created by Korea Research Institute of Biotechnology and Bioscience and supplied by Bioneer Corporation (http://pombe.bioneer.co.kr/). This commercial library facilitates the genome-wide screen in fission yeast. By using this library, colleagues identified 229 genes relevant to DDR, among which 23 genes were previously uncharacterized [12]. Following, an upgraded library was applied to investigate the global fitness of deletions after different kinds of DNA damage by barcode sequencing [13]. Both studies made impressive progress to gain a better understanding of DDR. However, the deletion libraries applied in these studies only covered around 70% of non-essential S. pombe genes. In this sense, screening a deletion library with a higher coverage of genes seemed worthwhile in order to build a more comprehensive DDR network.
In this study, we screened a S. pombe haploid deletion library, containing 3,235 deletions, against six different DNA damage reagents. The library represented approximately 90.5% of non-essential genes in the genome. 52 genes were identified to be closely related with DDR, 20 of which were reported for the first time. We characterized six novel DDR genes by flow cytometry and microarray analysis. Data suggest these genes might function in DNA replication and cytokinesis, providing a basis for further characterization of their roles in DDR.
Results
Genome-wide screen of DNA damage sensitive mutants
Six chemical reagents that can cause different kinds of DNA damage were chosen for the screen. Hydroxyurea (HU) inhibits ribonucleotide reductase, depletes nucleotides pool and thus leads to an S-phase arrest [14]. Bleomycin (BLM), a mimetic of gamma irradiation, causes double-strand breaks [15]. Methyl methanesulfonate (MMS), an alkylating agent, primarily methylates DNA on N7-deoxyguanine and N3-deoxyadenine, leading to DNA synthesis defects [9]. Camptothecin (CPT) locks topoisomerase I covalently onto the DNA and thus causes strand breaks during S phase [16]. Ultraviolent radiation (UV) results in an abnormal covalent bond between adjacent pyrimidine bases [17]. Thiabendazole (TBZ) depolymerizes the microtubule and was used to check the integrity of the spindle checkpoint [18]. Before the screen was performed, the growth of WT cells with different concentrations of DNA damaging agents were monitored. The highest concentration that did not affect the growth of WT cells was chosen for large scale screen. By using this concentration, it was easier to compare the growth with WT cells and to pick the sensitive mutants.
The screen was carried out in three rounds. First, 3,235 deletions were exposed to each DNA damage reagent in 96-well microtiter plates. 630 mutants showing sensitivities to at least one reagent were picked to create a sub-library. In the second round, mutants from the sub-library were grown in test-tubes to repeat the sensitivity assays, and 322 sensitive deletions were obtained (Additional file 1: Table S1). In the last round of the screen, 322 deletions were subjected to spot assays to quantify the sensitivities. We found that deletion of 52 genes caused viability to decrease by 25 fold or more upon treatment of at least one reagent, suggesting those genes play important roles in DDR (Table 1 and Additional file 1: Figure S1).
Table 1.
List of genes whose deletions exhibited strong sensitivities to DNA damage reagents
| Systematic ID | Gene name | Description of the gene products | DNA damaging agents | Flow cytometry phenotypea | Referenceb |
|---|---|---|---|---|---|
| SPAC17A5.07c |
ulp2+ |
SUMO deconjugating cysteine peptidase |
HU, BLM, MMS, TBZ, UV |
1C |
[19] |
| SPAC1952.07 |
rad1+ |
checkpoint clamp complex protein |
HU, BLM, MMS, CPT, UV |
1C |
[12,13,20] |
| SPAC23C11.15 |
pst2+ |
Clr6 histone deacetylase complex subunit |
HU, BLM, MMS, TBZ, UV |
2C |
[21] |
| SPAC3C7.03c |
rhp55+ |
RecA family ATPase |
HU, BLM, MMS, TBZ, UV |
2C |
[12,22] |
| SPBC1D7.04 |
mlo3+ |
RNA annealing factor |
HU, BLM, MMS, CPT, UV |
W4C |
[13] |
| SPAC1952.05 |
gcn5+ |
SAGA complex histone acetyltransferase catalytic subunit |
HU, MMS, CPT, TBZ |
2C |
[12,13,23] |
| SPAC227.07c |
pab1+ |
protein phosphatase regulatory subunit |
HU, MMS, TBZ, UV |
S4C |
[12,13,24] |
| SPAC4D7.10c |
spt20+ |
histone acetyltransferase SAGA complex subunit |
HU, CPT, TBZ, UV |
S4C |
[23] |
| SPAC6G9.10c |
sen1+ |
ATP-dependent 5' to 3' DNA/RNA helicase |
HU, BLM, TBZ, UV |
2C |
[25] |
| SPBC146.13c |
myo1+ |
myosin type I |
HU, BLM, TBZ, UV |
2C |
[12,26] |
| SPBC2F12.11c |
rep2+ |
transcriptional activator |
HU, BLM, MMS, UV |
1C |
[13,27] |
| SPBC3D6.04c |
mad1+ |
mitotic spindle checkpoint protein |
HU, BLM, TBZ, UV |
NCc |
[13,28] |
| SPBC342.05 |
crb2+ |
DNA repair protein RAD9 homolog |
HU, BLM, MMS, UV |
NC |
[12,13,29] |
| SPBC409.15 |
|
rRNA processing protein |
HU, MMS, TBZ, UV |
S4C |
[13] |
| SPCC1393.05 |
ers1+ |
RNA-silencing factor |
HU, BLM, TBZ, UV |
2C |
[13] |
| SPCC306.04c |
set1+ |
histone lysine methyltransferase |
HU, BLM, MMS, TBZ |
2C |
[13,30] |
| SPCC417.02 |
dad5+ |
DASH complex subunit |
HU, BLM, TBZ, UV |
NC |
[12,13,31] |
| SPAC16E8.09 |
scd1+ |
Rho guanine nucleotide exchange factor |
HU, BLM, TBZ |
2C |
[32] |
| SPAC4H3.05 |
srs2+ |
ATP-dependent DNA helicase |
BLM, MMS, UV |
1C |
[12,33] |
| SPAC6F6.01 |
cch1+ |
calcium channel |
HU, MMS, UV |
2C |
[12,13] |
| SPAC664.07c |
rad9+ |
checkpoint clamp complex protein |
HU, BLM, MMS |
NC |
[12,13,20] |
| SPBC11B10.10c |
pht1+ |
histone H2A variant |
BLM, MMS, UV |
NC |
[34] |
| SPBC13G1.08c |
ash2+ |
Ash2-trithorax family protein, Set1 complex component |
HU, BLM, TBZ |
2C |
[12,30] |
| SPBC428.08c |
clr4+ |
histone H3 methyltransferase |
HU, MMS ,TBZ |
1C |
[12,13,35] |
| SPBC660.11 |
tcg1+ |
single-stranded telomeric binding protein |
HU, BLM, TBZ |
2C |
[13] |
| SPBC800.05c |
atb2+ |
tubulin alpha 2 |
HU, BLM, TBZ |
W4C |
[13,36] |
| SPCC126.04c |
sgf73+ |
histone acetyltransferase SAGA complex subunit |
HU, MMS, TBZ |
W4C |
[12,23] |
| SPCC126.15c |
sec65+ |
signal recognition particle subunit |
BLM, TBZ, UV |
S4C |
[12,13] |
| SPCC162.12 |
tco89+ |
TORC1 subunit |
HU, BLM, UV |
NC |
[37] |
| SPAC2F7.08c |
snf5+ |
chromatin remodeling complex subunit |
HU, MMS |
1C |
[38] |
| SPAC664.01c |
swi6+ |
chromodomain protein |
HU, TBZ |
NC |
[12,13,35] |
| SPAC19A8.11c |
|
recombination protein |
BLM |
NC |
[13] |
| SPAC3F10.02c |
trk1+ |
potassium ion transporter |
HU, BLM, CPT, TBZ, UV |
2C |
This study |
| SPAC1486.04c |
alm1+ |
medial ring protein |
HU, BLM, MMS, UV |
NC |
This study |
| SPAC17G6.06d |
rps2401+ |
40S ribosomal protein |
HU, BLM, MMS, UV |
2C |
This study |
| SPBC2A9.02e |
|
NAD dependent epimerase/dehydratase family protein |
HU, BLM, MMS, UV |
1C |
This study |
| SPCC63.02c |
aah3+ |
alpha-amylase homolog |
HU, BLM, TBZ, UV |
W4C |
This study |
| SPAC14C4.05cd |
mug61+ |
LEM domain protein, Sad1 interacting factor |
HU, BLM, UV |
2C |
This study |
| SPAC1556.06f |
meu1+ |
meiotic expression up-regulated protein |
HU, BLM, UV |
NC |
This study |
| SPAC22E12.11c |
set3+ |
histone lysine methyltransferase |
BLM, MMS, UV |
W4C |
This study |
| SPAC25H1.05f |
meu29+ |
meiotic expression up-regulated protein 29 precursor |
HU, BLM, TBZ |
W4C |
This study |
| SPAC27D7.05c |
apc14+ |
anaphase-promoting complex subunit |
HU, TBZ, UV |
NC |
This study |
| SPAC3G6.01 |
hrp3+ |
ATP-dependent DNA helicase |
BLM, TBZ, UV |
2C |
This study |
| SPBP8B7.13 |
vac7+ |
Vac7 ortholog |
HU, MMS, TBZ |
W4C |
This study |
| SPCC830.06d |
|
calcineurin regulatory subunit |
HU, BLM, TBZ |
W4C |
This study |
| SPAC27D7.08ce |
|
DUF890 family protein |
HU, BLM |
1C |
This study |
| SPAC3F10.17d |
|
ribosome biogenesis protein |
HU, BLM |
2C |
This study |
| SPBC29A10.02 |
mug12+ |
meiotic RNA-binding protein |
HU, BLM |
NC |
This study |
| SPBC31E1.02c |
pmr1+ |
P-type ATPase, calcium transporting |
HU, UV |
NC |
This study |
| SPBC577.13d |
syj2+ |
inositol polyphosphate phosphatase |
HU, TBZ |
NC |
This study |
| SPCC1494.03 |
arz1+ |
Zfs1 target number 1 |
MMS, UV |
1C |
This study |
| SPBC20F10.10d | psl1+ | cyclin pho85 family | MMS | NC | This study |
a See text for detailed descriptions of flow cytometry phenotypes.
b Reference that reported involvement of gene in DDR.
c NC, no change.
d role inferred from homology.
e conserved hypothetical.
f sequence orphan.
Among these 52 genes, 24 genes (46%) were identified in previous large-scale screens [12,13], and 32 genes (62%) in total have been reported to be related with DDR, which validates the accuracy of our screen (Table 1). For example, genes directly involved in sensing and repairing DNA damage were identified. Proteins encoded by these genes include: Rad1 and Rad9, two subunits of a checkpoint complex (9-1-1) [20]; Crb2, Rep2 and Ulp2, proteins required for cell cycle control [19,27,29]; Rhp55, Sen1 and Srs2, proteins involved in DNA double strand break (DSB) and single strand break (SSB) repair [22,25,33]. As expected, deletions of these genes were sensitive to a broad range of DNA damage reagents (Table 1). Genes involved in spindle assembly and cytokinesis were also obtained, including dad5+, atb2+, mad1+, pab1+, myo1+ and scd1+[24,26,28,31,32,36]. As expected, deletions of these genes exhibited sensitivity to TBZ, a microtubule depolymerizing agent (Table 1). Chromatin controls the accessibility of the DNA repair machinery, and thus it was not surprised to identify genes related to the dynamics of chromatin structure. Such proteins included Set1 and Ash2, subunits of a histone H3K4 methyltransferase complex [30]; Clr4 and Swi6, subunits of an H3K9 methyltransferase [35]; Gcn5, Sgf73 and Spt20, subunits of the SAGA histone acetylase complex [23]; Pst2, a component of Clr6 deacetylase complex [21]; Snf5, a subunit of the Swi/Snf remodeling complex [38]; Pht1, a histone H2A variant [34]. These results stress the importance of histone modification and chromatin remodeling in DDR. SPBC409.15, sec65+, tcg1+, cch1+ and SPAC19A8.11c were identified previously during other genome-wide screens [12,13]. Identification by our screen confirmed the relevance of these genes to DDR. However, several known DDR genes identified in the previous large scale screens, including ctp1+, rhp51+, rad32+, rad26+, pnk1+, rad3+, hus1+, rad17+, rad24+, rhp57+[12,13], were not screened out in this study. This might be caused by different screen strategy, different choice of DNA damaging agents and their working concentrations. Besides, the commercial library we used contained errors. We checked the mutants of several known DDR genes and found rhp51Δ, rad26Δ, rad3Δ were wrong. Therefore, the quality of the library also affected the results of our screen.
On the other hand, another 20 genes were found to be linked with DDR for the first time in this study, and the identities of corresponding mutants have been double checked. Among 20 genes, 10 genes have been already identified to function in different biological processes, including biosynthesis, RNA processing, stress response, transport and chromatin modification. Notably, deletion of trk1+, a gene encoding the potassium ion transporter, caused strong sensitivity to almost all the DNA damage reagents used in our assay [39]. There was no assigned function for the remaining 10 genes; they were classified as “sequence orphan”, “conserved hypothetical” or “role inferred from homolog”. Our data provided novel functional annotations for these unknown genes. Interestingly, deletion of psl1+ and SPAC19A8.11c caused sensitivity to only one reagent, suggesting these genes are required for repairing a specific DNA lesion.
Among these 20 novel DDR genes, 11 genes have homologues in S. cerevisiae. Notably, deletion of 5 homologous genes are sensitive to DNA damage reagents in S. cerevisiae (Table 2), which reflects the functional conservation of these DDR genes in fungi [40-44].
Table 2.
List of homologues of novel DDR genes in S. cerevisiae
| Systematic ID in S. pombe | Gene name in S. pombe | Systematic ID in S. cerevisiae | Gene name in S. cerevisiae | DNA damaging agents* | Reference |
|---|---|---|---|---|---|
| SPAC3F10.02c |
trk1+ |
YJL129C |
TRK1 |
- |
|
| SPAC1486.04c |
alm1+ |
YKR095W |
MLP1 |
BLM, MMS, UV |
[40-42] |
| SPAC17G6.06 |
rps2401+ |
YER074W |
RPS24A |
HU |
[43] |
| SPBC2A9.02 |
|
YLL056C |
|
- |
|
| SPAC22E12.11c |
set3+ |
YPL181W |
CTI6 |
- |
|
| SPAC3G6.01 |
hrp3+ |
YER164W |
CHD1 |
HU |
[43] |
| SPCC830.06 |
|
YKL190W |
CNB1 |
- |
|
| SPAC3F10.17 |
|
YKL143W |
LTV1 |
- |
|
| SPBC31E1.02c |
pmr1+ |
YGL167C |
PMR1 |
HU, TBZ |
[43,44] |
| SPBC577.13 |
syj2+ |
YOR109W |
INP53 |
BLM |
[43] |
| SPBC20F10.10 | psl1+ | YIL050W | PCL7 | - |
* Listed are the DNA damage reagents that deletion of S. cerevisiae genes are sensitive to.
Cell cycle analysis of DNA damage sensitive mutants
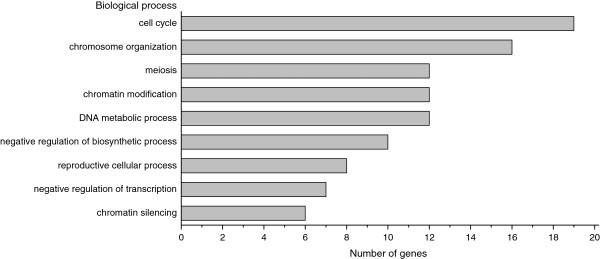
S. pombe genome is extensively annotated using terms from the Gene Ontology Consortium (http://www.geneontology.org), with 98.3% of its genes having at least one GO (Gene Ontology) annotation [45]. The GO term classification of 52 genes was carried out with a significance level smaller than 0.05 (Additional file 1: Table S2), and representative GO terms were shown in Figure 1. This analysis revealed that the 52 genes were significantly enriched in cell cycle and chromatin related processes. As the most over-represented GO term, “cell cycle” was annotated to 36.5% of genes (19/52). Cell cycle control is one of the essential components of the DDR network [46,47]. After DNA damage, the cell cycle is delayed by checkpoint to provide an opportunity for repair. To monitor the cell cycle change in the deletions upon DNA damage, the DNA content of 52 mutants was analyzed by flow cytometry (Additional file 1: Table S3 and Figures S2-S5).
Figure 1.
Representative GO terms of 52 genes. GO terms shown in the chart are: cell cycle (GO:0007049), chromosome organization (GO:0051276), meiosis (GO:0007126), chromatin modification (GO:0016568), DNA metabolic process (GO:0006259), negative regulation of biosynthetic process (GO:0009890), reproductive cellular process (GO:0048610), negative regulation of transcription (GO:0016481) and chromatin silencing (GO:0006342). Complete list of GO terms is shown in Additional file 1: Table S2.
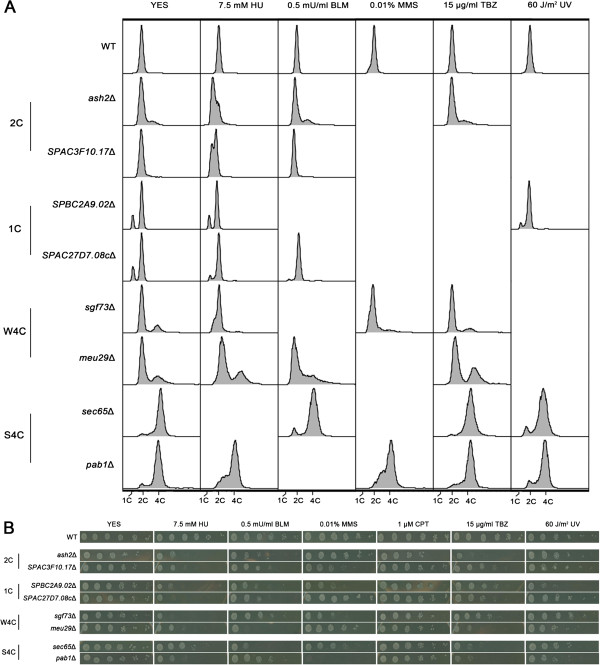
As expected, 37 deletions exhibited abnormal cell cycle profiles after DNA damage. No change was observed for the remaining 15 mutants, probably due to insufficient time for treatment. Based on flow cytometry phenotypes without reagent treatment, the 37 mutants could be divided into four groups which were designated as “2C”, “1C”, “W4C” and “S4C”, respectively (Table 1). Representative cytometry data of each group are shown in Figure 2A. “2C” stands for 2C DNA content. Members of this group, 16 deletions in total, exhibited DNA content peaks at 2C without reagent treatment, the same as WT cells. However, peaks moved towards 1C upon DNA damage caused by HU or MMS, suggesting that these deletions can cause replication arrest in response to damage (Additional file 1: Figure S2). The concentration of HU was the critical concentration that did not cause replication arrest of WT cells (Figure 2A). In the “1C” group, including 9 members, DNA content peaks moved towards 1C without treatment (Additional file 1: Figure S3). This result suggested that these deletions might have a defect in DNA replication [48,49]. Eight mutants in the “W4C” group and 4 mutants in the “S4C” group exhibited peaks of 4C DNA content (Additional file 1: Figure S4-S5) where “W” stands for “Weak”, as the 4C content was less than 35% and “S” represents “Strong”, because the 4C content was above 80%. Cytometry phenotypes suggested members of both groups had undergone diploidization, and the situation was much more severe in the “S4C” group. Genome duplication could be caused by DNA re-replication, a chromosome segregation defect, or improper cytokinesis [50]. Possible reasons for diploidization in the deletions will be discussed in the following section. Quantifications of the 1C, 2C and 4C DNA contents in 37 mutants are listed in Additional file 1: Table S3.
Figure 2.
Flow cytometry analysis and spot assays of eight representative mutants. (A) Flow cytometry analysis of eight mutants. Cells were grown to the logarithmic phase and treated with a DNA damage reagent for 2 h. For UV sensitivity assay, cells were exposed to 60 J/m2 radiation and then grown for 2 h. After treatment cells were harvested and subjected to cytometry analysis. (B) Sensitivity to different DNA damage reagents was quantified by spot assays. Exponentially growing cells, WT or deletions, were harvested and 5-fold serial dilutions were spotted on plates supplemented with DNA damage reagents. The plates were photographed after 3~4 days of incubation at 32°C.
Gene expression profiling of mutants
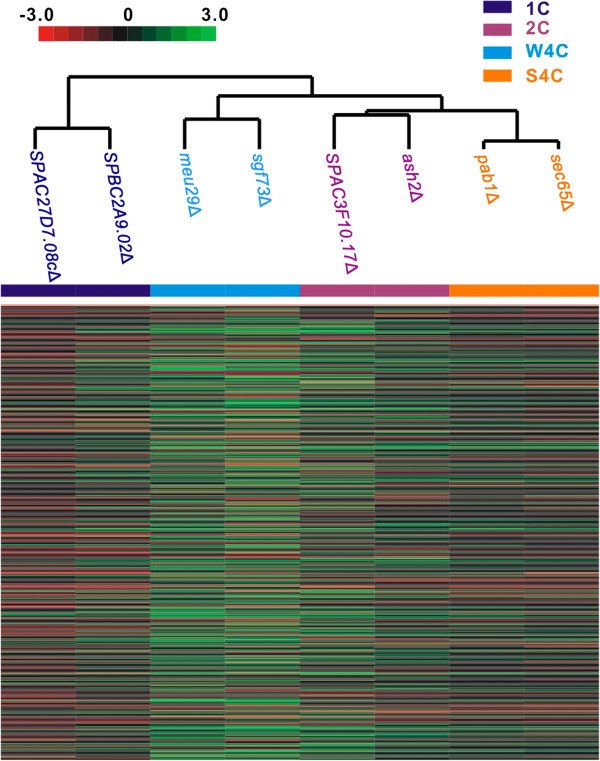
We selected 2 typical mutants from each cytometry phenotype group for further characterization (Figure 2A). All deletions showed strong sensitivity to at least two different DNA damage reagents (Figure 2B). SPAC3F10.17, SPBC2A9.02, SPAC27D7.08c and meu29+ were uncharacterized DDR genes. ash2+, sgf73+, sec65+ and pab1+ were identified during a previous global screen, but their detailed roles in DDR had not been identified yet [12,13]. For a better understanding of the gene function, we performed a DNA microarray assay to analyze the gene expression profiles of these eight deletions [51]. Transcription levels of hundreds of genes changed by 2-fold or more in the mutants. Notably, differentially regulated genes were enriched in the process related to DNA replication and cytokinesis. Representative genes are listed in Table 3. Analysis of microarray data by hierarchical clustering clustered 8 mutants into 4 groups (Figure 3). Notably, clustering perfectly matched the classification based on the flow cytometry phenotypes. It suggested that both genes from each group might function in the same pathway to regulate DDR and cell cycle progression.
Table 3.
Differentially regulated genes in eight deletions
|
Systematic ID |
Gene name |
Description of gene products |
2C |
1C |
W4C |
S4C |
Gene ontology |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ash2Δ | SPAC3F10.17Δ | SPBC2A9.02Δ | SPAC27D7.08cΔ | sgf73Δ | meu29Δ | sec65Δ | pab1Δ | ||||
| SPAC14C4.09 |
agn1+ |
glucan endo-1,3-alpha-glucosidase |
2.7 |
2.0 |
2.4 |
1.9 |
2.6 |
1.9 |
2.9 |
3.1 |
cell septum edging catabolic process |
| SPAC6G10.12c |
ace2+ |
transcription factor |
4.1 |
3.8 |
1.3 |
1.4 |
2.4 |
1.6 |
3.9 |
3.4 |
cytokinetic cell separation |
| SPAC821.09 |
eng1+ |
endo-1,3-beta-glucanase |
5.7 |
4.1 |
1.5 |
1.1 |
3.1 |
4.2 |
6.3 |
5.9 |
primary cell septum disassembly |
| SPBC83.18c |
fic1+ |
C2 domain protein |
1.8 |
1.6 |
1.3 |
1.3 |
2.3 |
1.9 |
1.3 |
1.3 |
cell cycle cytokinesis |
| SPCC320.13c |
ark1+ |
aurora-B kinase |
1.4 |
1.4 |
1.2 |
1.5 |
2.7 |
2.3 |
1.5 |
1.3 |
cell cycle cytokinesis |
| SPAC17H9.19c |
cdt2+ |
WD repeat protein |
2.6 |
1.4 |
3.3 |
1.6 |
2.5 |
1.2 |
2.2 |
3.0 |
DNA replication checkpoint |
| SPAC1F7.05 |
cdc22+ |
ribonucleoside reductase large subunit |
2.7 |
2.3 |
2.1 |
1.6 |
3.3 |
2.3 |
2.5 |
2.8 |
regulation of DNA-dependent DNA replication |
| SPAC27E2.10c |
rfc3+ |
DNA replication factor C complex subunit |
1.7 |
1.2 |
1.1 |
1.1 |
1.6 |
2.1 |
1.3 |
1.3 |
DNA-dependent DNA replication |
| SPAC3G6.06c |
rad2+ |
FEN-1 endonuclease |
0.98 |
1.8 |
1.6 |
1.6 |
2.1 |
2.5 |
1.7 |
1.7 |
DNA replication, removal of RNA primer |
| SPAC821.08c |
slp1+ |
sleepy homolog |
2.6 |
2.2 |
1.2 |
1.2 |
3.6 |
2.0 |
2.5 |
2.2 |
DNA replication checkpoint |
| SPBC1105.04c |
abp1+ |
CENP-B homolog |
0.64 |
0.69 |
0.43 |
0.40 |
0.41 |
0.46 |
0.57 |
0.52 |
DNA-dependent DNA replication initiation |
| SPBC12D12.02c |
cdm1+ |
DNA polymerase delta subunit |
1.9 |
2.2 |
1.5 |
1.9 |
1.9 |
1.3 |
2.8 |
3.1 |
DNA strand elongation involved in DNA replication |
| SPBC14C8.07c |
cdc18+ |
MCM loader |
1.9 |
1.4 |
1.4 |
1.0 |
2.3 |
1.2 |
2.0 |
1.9 |
DNA replication checkpoint |
| SPBC1861.02 |
abp2+ |
ARS binding protein |
1.5 |
1.2 |
0.28 |
0.50 |
0.31 |
0.70 |
0.91 |
0.80 |
DNA-dependent DNA replication initiation |
| SPBC428.18 |
cdt1+ |
replication licensing factor |
3.0 |
2.8 |
2.3 |
1.7 |
2.2 |
2.2 |
3.5 |
3.3 |
DNA replication checkpoint |
| SPBC660.14 |
mik1+ |
mitotic inhibitor kinase |
2.5 |
3.2 |
1.5 |
1.5 |
3.4 |
1.7 |
2.3 |
2.4 |
DNA replication checkpoint |
| SPCC1672.02c |
sap1+ |
switch-activating protein |
1.2 |
0.75 |
1.6 |
1.0 |
2.3 |
1.2 |
1.4 |
1.4 |
replication fork arrest at rDNA repeats |
| SPCC23B6.05c |
ssb3+ |
DNA replication factor A subunit |
2.1 |
1.8 |
1.1 |
1.6 |
1.3 |
1.4 |
1.2 |
1.3 |
DNA-dependent DNA replication |
| SPCC970.10c | brl2+ | ubiquitin-protein ligase E3 | 2.6 | 1.3 | 1.1 | 0.89 | 1.1 | 1.2 | 1.1 | 1.1 | DNA replication-independent nucleosome assembly |
Differentially regulated genes in the deletions comparing to WT were analyzed using an Affymetrix microarray. Genes relevant to DNA replication and cytokinesis are listed. The number represents the ratio of mRNA level in the deletion relative to that in WT.
Figure 3.
Clustering analysis of eight mutants. Hierarchical clustering matrix of eight mutants with microarray data for S pombe gene transcription. Color panel indicates relative increase (red) and decrease (green) to the median (black) of eight mutants for each transcript.
abp1+ and abp2+ function downstream of SPBC2A9.02 and SPAC27D7.08c to initiate DNA replication
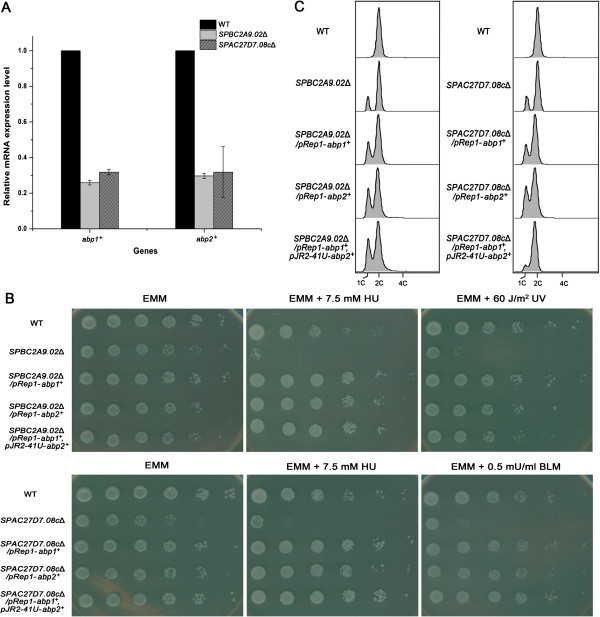
As members of the “1C” group, SPBC2A9.02Δ or SPAC27D7.08cΔ exhibited a discrete 1C DNA peak, suggesting G1 arrest and a defect in replication initiation [52]. Consistently, both mutants displayed a growth defect on EMM plates (Figure 4B). Both microarray and real-time PCR analysis revealed that the expression levels of abp1+ and abp2+ were simultaneously down-regulated by more than 2-fold in both deletions (Table 3 and Figure 4A). Abp1 and Abp2 are ARS (autonomously replicating sequence) binding proteins and are required for initiation of DNA replication [53,54]. It is possible that down-regulation of abp1+ and abp2+ contributed to the replication defects observed in SPBC2A9.02Δ and SPAC27D7.08cΔ. To check this possibility, we overexpressed abp1+ and abp2+ in the deletions. Without DNA damage, the growth defects of SPBC2A9.02Δ and SPAC27D7.08cΔ were partially rescued by overexpression of abp1+ and abp2+ (Figure 4B). The improvement was more obvious in the case of SPAC27D7.08cΔ, and was relatively mild, nevertheless, observable in the case of SPBC2A9.02Δ. In face of DNA damage, overexpressing either abp1+ and abp2+ could significantly improve the growth of SPBC2A9.02Δ and SPAC27D7.08cΔ (Figure 4B). Correspondingly, G1-arrest in SPAC27D7.08cΔ could also be reproducibly relieved by overexpression of both abp1+ and abp2+ (Figure 4C). The data suggested that abp1+ and abp2+ function downstream of SPBC2A9.02 and SPAC27D7.08c to ensure the proper initiation of DNA replication under normal circumstances or after DNA damage.
Figure 4.
abp1+ and abp2+ function downstream of SPBC2A9.02 and SPAC27D7.08c to initiate DNA replication. (A) Reduced expression levels of abp1+ and abp2+ in SPBC2A9.02Δ and SPAC27D7.08cΔ. The mRNA levels were quantified by real time PCR and those of act1+ served as an internal control (n=3). The relative level in WT was designated as arbitrary unit 1. (B) Overexpression of abp1+ and abp2+ partially rescued the growth defect of SPBC2A9.02Δ and SPAC27D7.08cΔ. pREP1-abp1+ or pREP1-abp2+ were transformed into each deletion separately. pREP1-abp1+ and pJR2-41U-abp2+ were co-transformed into SPBC2A9.02Δ or SPAC27D7.08cΔ. Transformants were harvested and 5-fold serial dilutions were spotted on plates supplemented with DNA damage reagents. Plates were photographed after 3 days of incubation at 32°C. (C) Overexpression of abp1+ or abp2+ partially relieved the G1-arrest in SPBC2A9.02Δ and SPAC27D7.08cΔ. Transformants described in Figure 4B were grown to logarithmic phase and harvested for flow cytometry analysis. Reproducible results were obtained in three independent experiments.
Members of “W4C” and “S4C” groups exhibited defects in cytokinesis and replication
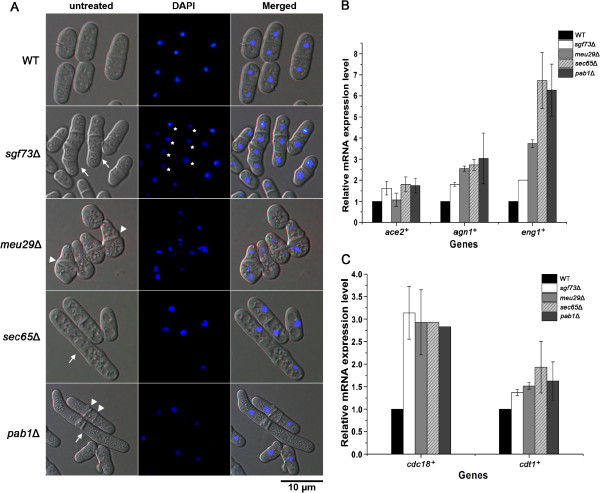
Deletions from the “W4C” and “S4C” groups exhibited discrete peaks of 4C DNA content, suggesting the mutants underwent diploidization. Diploidization in S. pombe is commonly caused by a defect in cytokinesis. Correspondingly, microscopic analysis revealed abnormal morphological changes in these mutants (Figure 5A). WT cells were rod-shaped and contained a single nucleus, or double nuclei separated by a sharp septum. In contrast, mutant cells exhibited elongated cell length (sgf73Δ, sec65Δ and pab1Δ), multiple nuclei (sgf73Δ), thick septum (meu29Δ) or multiple septa (pab1Δ), resembling typical defects in cytokinesis [55]. As expected, all 4 deletions displayed strong sensitivity to TBZ, a microtubule depolymerizing agent [56]. Microarray and real-time PCR analysis showed that the expressions of several cytokinesis related genes were up-regulated in the mutants, including those of ace2+, agn1+ and eng1+ (Table 3 and Figure 5B). Ace2 is a transcription factor that controls the expression of genes required for cell separation, while eng1+ and agn1+ are both targets of Ace2. Eng1, a β-glucanase, degrades the primary division septum between the new ends of daughter cells. Agn1, an α-glucanase, hydrolyses the old cell wall surrounding the septum and leads to full separation of daughter cells [57,58]. The data suggest that deletion of sgf73+, meu29+, sec65+ or pab1+ delays proper progression of cytokinesis, while a ruptured cell wall constitutively generates a signal to activate the Ace2 pathway and up-regulate target genes [58].
Figure 5.
sgf73+, meu29+, sec65+ and pab1+ function in cytokinesis and DNA replication. (A) Microscopic analysis of deletions. Cells were stained with DAPI to visualize the nuclei. Cells exhibiting elongated cell length are indicated by arrows, multiple nuclei by asterisk, and abnormal septum by arrow heads. The bar represents 10 μm. (B) Increased expression levels of ace2+, agn1+ and eng1+ in the deletions. The mRNA levels were quantified by real time PCR and those of act1+ served as internal controls (n=3). The relative level in WT was designated as arbitrary unit 1. (C) Increased expression levels of cdc18+ and cdt1+ in deletions. Real time PCR was performed as described in Figure 5B (n=3).
On the other hand, diploidization could also result from DNA re-replication during one cell cycle. Consistent with this idea, expression levels of cdc18+ and cdt1+ were up-regulated in all 4 mutants (Table 3 and Figure 5C). Presence of Cdc18 and Cdt1 at pre-RCs (pre-replicative complexes) is important for efficient DNA replication initiation, and inactivation of these proteins after initiation is crucial to ensure only one round of DNA replication in each cell cycle. Overexpression of cdc18+ and cdt1+ in fission yeast causes replication origins to re-fire, and drive re-replication of DNA sequences genome-wide [59,60]. Therefore, up-regulation of cdc18+ and cdt1+ in sgf73Δ, meu29Δ, sec65Δ and pab1Δ might lead to DNA re-replication, and that contributes to the observed diploidization. Meanwhile, disturbed replication initiation probably compromises DDR during early S phase. Correspondingly, all the members in “W4C” and “S4C” groups showed strong sensitivities to HU.
Discussion
In this study, six reagents were applied in the screen and they can cause different kinds of DNA damage. The screen revealed six genes whose deletions displayed strong sensitivities to five reagents, including rad1+, rhp55+, ulp2+, pst2+, mlo3+ and trk1+ (Table 1). Broad sensitivities to different DNA damage reagents suggest that these genes function in the universal pathway of DDR, for example, in the conserved ATM/ATR pathway [2]. As expected, rad1+ plays a role in the ATR pathway, and rhp55+ in the ATM pathway [2,22]. ulp2+, encoding a SUMO protease, is required for cell division after termination of the DNA damage checkpoint [61]. The high sensitivity of ulp2Δ to a broad range of DNA damage reagents emphasizes the importance of silencing of the DNA damage checkpoint and restart of the cell cycle. pst2+ encodes a subunit of the Clr6 histone deacetylase. Deletion of pst2+ could lead to hyperacetylation of histones and down-regulation of histone H3 S10 phosphorylation, resulting in abnormal chromosome condensation and a defect in DNA damage repair [62]. Identification of pst2+ during the screen indicates the importance of chromatin condensation and decondensation in DDR. The protein encoded by mlo3+ was required for export and quality control of mRNA [63], suggesting DDR is related to the level and quality of mRNA. The screen has revealed the novel link between DDR and trk1+, gene encoding a potassium ion transporter [39]. Two calcium transporter genes, cch1+, and pmr1+, have also been identified in this study. cch1+, along with other ion transporter genes, including zrg17+, fep1+, ctr4+ and zhf1+, were identified during previous global screens for DDR genes [12,13].These results imply a close connection between ion transport and DDR. Ion transport controls several crucial physiological parameters, including membrane potential and ion balance [64]. It will be intriguing to uncover the mechanism how ion transport influences the DDR in future studies.
The screen also identified genes whose deletion exhibited sensitivity to only one kind of DNA damage reagent. Characterization of these genes will help to elucidate the specific DDR for a certain DNA lesion. For example, deletion of psl1+ displayed specific sensitivity to MMS. Previous screens have identified similar genes, including cac2+, mag1+, rev3+ and slx4+[9]. These genes, along with psl1+, might work together to remove the damage caused by alkylated DNA. SPAC19A8.11cΔ caused exclusive sensitivity to BLM. BLM abstracts a hydrogen atom from DNA deoxyribose and causes alkali-labile sites in DNA [15]. Genomic screen in budding yeast identified 23 genes exhibiting specific sensitivity to BLM [11]. SPAC19A8.11c might be an additional gene needed to repair lesions caused by BLM.
Cell cycle is delayed by checkpoints in response to DNA damage, thus providing a chance to repair DNA lesions. Several DNA damage checkpoints have been described in S. pombe, including G2-M, intra-S, S-M, G1-M and G1-S checkpoints [46,65-68]. Among the 52 deletion identified in this study, 37 deletions were found to affect cell cycle progression. Notably, 16 deletions in the “2C” group caused replication arrest upon treatment with HU or MMS. It suggested that these genes might be involved in DNA damage repair in S phase. Failures of repairing lesions in the deletions might persist intra-S checkpoint and slow the replication. Another member of “2C”, myo1Δ caused a 4C peak of DNA content after treatment of TBZ, indicating the diploidization of the genome. Since Myo1 regulates the assembly of actin and contributes to proper septation, observed diploidiation might be caused by a cytokinesis defect in myo1Δ [26].
In contrast to the “2C” group, deletions in the “1C” group caused G1 or S phase arrest without DNA damage. The data suggest these genes are required for cell cycle progression. These deletions interfere with cell cycle regulation in response to DNA damage, thus leading to high sensitivity to damage reagents. Further investigation revealed that SPBC2A9.02 and SPAC27D7.08c might function in the initiation of DNA replication through initiation factors, Abp1 and Abp2. Since deletion of SPBC2A9.02 and SPAC27D7.08c share a similar cytometry phenotype and gene expression profiling, it is likely both genes work in the same pathway. SPAC27D7.08c contains a methyltransferase 10 domain, harboring potential SAM-dependent methyltransferase activity (http://www.rcsb.org/pdb/explore.do?structureId=2H00). It suggests that SPAC27D7.08c might regulate replication by methylating downstream proteins.
Flow cytometry analysis indicated that the members of “S4C” and “W4C” groups underwent diploidization. Gene expression and microscopic analysis of sgf73Δ, meu29Δ, sec65Δ and pab1Δ suggested diploidization might be caused by a cytokinesis defect and DNA re-replication. It is possible that proteins encoded by these genes function as subunits of large complexes, involved in the regulations of different processes, including replication, chromosome segregation and cytokinesis. A similar case was reported for a subunit of the Orc complex, Orc6 [69]. Consistent with this idea, Sgf73 is a subunit of the SAGA complex, a conserved multifunctional co-activator [70]. SAGA complex is known to regulate transcriptional activation, transcription elongation and mRNA export [71]. However, its roles in DNA re-replication and cytokinesis are yet to be identified. Recently, Pab1 has been revealed to be a novel component of the septation initiation network (SIN) complex [24]. SIN plays an important role in cytokinesis [72]. Whether the SIN complex also contributes to the replication initiation needs further characterization. Notably, pab1+, along with other 3 genes from the “W4C” group (SPCC830.06, set3+, atb2+), is conserved from S. pombe to mammals. Thus, further characterization of these genes is expected to provide valuable information for studies of genome stability and DDR in higher eukaryotes, especially in human.
Conclusions
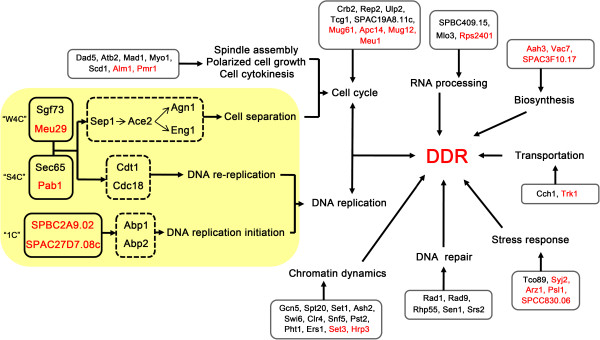
Genome-wide screening is a fast and efficient way to explore unknown genes, clarify signaling pathways, and to ultimately build a comprehensive gene network. In this study, we performed a systematic screen of the S. pombe deletion library to uncover genes involved in DDR. 52 genes were characterized, among which 20 genes were linked to DDR for the first time. Most of the genes take part in cell cycle control, DNA repair, chromatin dynamics and DNA replication, all of which are well-known components of DDR [2]. In addition, many novel genes functioning in biosynthesis, transport, RNA processing and stress response were uncovered, suggesting their substantial contributions to DDR. Further characterizations suggested 6 novel genes might function in DDR through DNA replication and cytokinesis. Our study introduces new members to the long list of DDR genes and provides new clues to clarify the dynamic DDR network (Figure 6).
Figure 6.
Graphic presentation of the involvement of 52 genes in DDR network. Proteins encoded by 52 DDR genes are shown in the solid line box and previously uncharacterized proteins are labelled in red. Proteins are categorized into different biological processes based on the GO analysis. Possible pathways that link 6 novel DDR proteins are shown in orange background.
Methods
Genome-wide haploid deletion library
The S. pombe haploid deletion library used in this study was bought from Bioneer (http://pombe.bioneer.co.kr/). It contains 3,235 haploid deletion strains covering 65.8% of the 4,914 protein coding open reading frames (ORFs) based on the annotated genome sequence (http://www.genedb.org/genedb/pombe). As 3,576 genes are nonessential [73], this library represents approximately 90.5% of the nonessential S. pombe genes. Fission yeast were cultured in YES or EMM medium at 32°C as described before [74].
Screen of deletions sensitive to DNA damage
The screen was performed in three rounds. In the first round, deletion strains from the Bioneer library were grown in YES medium till saturation. 20 μl culture from each strain was diluted into 180 μl liquid YES medium containing different DNA damage reagents in 96-well microtiter plates. As a control, cells were also diluted into medium without any reagent. Concentrations of reagents were: 7.5 mM hydroxyurea (HU) (SIGMA, H8627), 0.5 mU/ml bleomycin (BLM) (NIPPON KAYAKU, 480890), 0.01% methyl methanesulfonate (MMS) (ACROS ORGANICS, 156890050), 1 μM camptothecin (CPT) (SIGMA, C9911), 15 μg/ml thiabendazole (TBZ) (TCI, T0830) and 60 J/m2 ultraviolet radiation (UV). After 24 hours of incubation at 32°C, the optical densities of the cultures were measured at 600 nm (A600) and compared to those of the controls. Deletions with A600 that dropped by 5 fold or more upon reagent treatment were designated as sensitive. Deletion mutants showing sensitivity to at least one reagent were picked to create a sub-library. This round of the screen was repeated once. In the second round, strains from the sub-library were grown in YES medium overnight, and then inoculated into 1 ml YES medium containing different reagents at an A600 of 0.02. After 24 hours of incubation at 32°C, A600 was measured and compared to those of no reagent controls. In the third round, strains showing sensitivity to at least one DNA-damaging agent in the second round were grown in liquid medium to an A600 of 1.0. Cultures were diluted by five-fold for five times, and 2 μl dilutions were spotted onto YES or EMM plates containing DNA damage reagents of indicated concentrations. The growth of the cells was checked after 3~4 days of incubation at 32°C. If the growth of a mutant on the plate containing certain reagent was 2-spot lesser than that on YES plate (25 fold reduction in viability), this mutant was designated as sensitive.
Gene ontology analysis
Gene ontology (GO) classifications were performed at http://amigo.geneontology.org with the database filter set as GeneDB S. pombe. Maximum P-value was 0.05 as the threshold for significance assessment, and minimum number of gene products was 3 in each GO term. GO analysis was based on the biological process classifications in this study [73].
Flow cytometry
1~2×107 exponentially growing cells were treated with DNA damage reagent for 2 h. For the UV sensitivity assay, cells were exposed to 60 J/m2 radiation and then grown for 2 h. Cells were harvested and fixed in 70% (v/v) cold ethanol at 4°C for 1 h. Cells were resuspended in 0.5 ml of 50 mM sodium citrate containing 0.1 mg/ml RNase A and incubated at 37°C for 2 h. Cells were briefly sonicated, and then stained with 4 μg/ml propidium iodide (PI) at room temperature for 15 min. 1~2×104 cells were measured by a FACS Calibur flow cytometer (Becton-Dickinson) and data were analyzed by Flowjo 2.0 [48].
DNA microarray analysis
cDNAs were prepared from the exponentially growing wild type (WT) cells or deletion cells as previously described [75]. cDNA was labeled and hybridized to the Yeast genome 2.0 array according to the manufacturer’s protocol (900553, Affymetrix). Data was analyzed by Shanghai GeneTech Company (Shanghai, China). The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus [76] and are accessible through GEO Series accession number GSE40747 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40747).
Clustering analysis
Hierarchical clustering was carried out by Gene Cluster with differentially regulated genes of eight mutants, using the correlation (uncentered) and centroid linkage clustering method. The clustering results were visualized with Java TreeView.
Real time PCR analysis
Experiments were performed as described before [77]. Briefly, total RNAs were prepared from exponentially growing cells by using TRIzol (Invitrogen) and reverse-transcribed to make first strand cDNAs. cDNAs were used as templates for real time PCR. PCR were performed using SYBR Premix ExTaq TMII (DRR081C, Takara) on an ABI Prism 5700 sequence detection system (Applied Biosystems) according to manufacturer’s protocol. The threshold cycle (CT) of each sample was determined by the ABI system and then normalized to the value for act1+ by the following equation: ΔCT = CT(gene of interest) − CT (act1+). Relative level was calculated as 2-ΔCT. Reaction for each sample was performed in triplicate. Primers are listed in Additional file 1: Table S4.
Microscopic analysis
After overnight incubation at 32°C, cells were washed with phosphate-buffered saline and stained with 1 μg/ml 4’, 6’-diamidino-2-phenylindole (DAPI) to visualize nuclei. Cells were observed and captured by a Zeiss Axioplan microscope equipped with a chilled video charge-coupled device camera (C4742-95; Hamamatsu Photonics, Bridgewater, NJ). Images were analyzed by kinetic image AQM software (Kinetic Imaging, Nottingham, UK).
Abbreviations
The abbreviations used are: DDR: DNA damage response; GO: Gene ontology; SAGA: Spt-Ada-Gcn5 acetyltransferase; HU: Hydroxyurea; BLM: Bleomycin; MMS: Methyl methanesulfonate; CPT: Camptothecin; TBZ: Thiabendazole; UV: Ultraviolet radiation; WT: Wild type; DSB: Double strand breaks; SSB: Single strand breaks.
Competing interests
The authors declare that they have no competing financial interests.
Authors’ contributions
XP carried out the screening work, performed the molecular genetic studies, analyzed the data, and drafted the manuscript. BL participated in the screening work and molecular genetic studies. NZ participated in the molecular genetic studies. BF and WY participated in the screening work. XZ conceived of the study. YY participated in the design of the experiments and helped to draft the manuscript. HL conceived the study, designed and coordinated the work, and critically revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Table S1. List of genes whose deletions exhibited sensitivity to DNA damage reagents during the second round of screen. Table S2. GO profiling of 52 genes whose deletion mutants showed strong sensitivity to DNA damage reagents (P ≤ 0.05). Table S3. Flow cytometry analysis of 37 mutants. Table S4. Primers used for real time PCR analysis in this study. Figure S1. Spot assay of 52 deletions. Exponentially growing cells, WT or deletions, were harvested and 5-fold serial dilutions were spotted on the plates supplemented with DNA damage reagents. The plates were photographed after 3~4 days of incubation at 32°C. Figure S2. Flow cytometry analysis of deletions in “2C” group. Figure S3. Flow cytometry analysis of deletions in “1C” group. Figure S4. Flow cytometry analysis of deletions in “W4C” group. Figure S5. Flow cytometry analysis of deletions in “S4C” group.
Contributor Information
Xian Pan, Email: 081023041@fudan.edu.cn.
Bingkun Lei, Email: 082023052@fudan.edu.cn.
Nan Zhou, Email: 10210700077@fudan.edu.cn.
Biwei Feng, Email: 0570122@fudan.edu.cn.
Wei Yao, Email: 06300700078@fudan.edu.cn.
Xin Zhao, Email: xin.zhao@mcgill.ca.
Yao Yu, Email: steveyuyao@hotmail.com.
Hong Lu, Email: honglu0211@yahoo.com.
Acknowledgements
We thank Rolf Sternglanz from Stony Brook University for his valuable comments on the manuscript. This paper is dedicated to Xuping Fu, dear collegue from school of life sciences, Fudan University. We thank him for great help in bioinformatic analysis during last days of his life, may his soul rest in peace. This work was supported by grants from the National Program on Key Basic Research Project (973 Program, 2009CB825601), the National Natural Science Foundation of China (30771145 and 30671175).
References
- Jeggo PA, Löbrich M. Contribution of DNA repair and cell cycle checkpoint arrest to the maintenance of genomic stability. DNA Repair (Amst) 2006;5:1192–1198. doi: 10.1016/j.dnarep.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- Dudásová Z, Dudás A, Chovanec M. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol Rev. 2004;28:581–601. doi: 10.1016/j.femsre.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Raji H, Hartsuiker E. Double-strand break repair and homologous recombination in Schizosaccharomyces pombe. Yeast. 2006;23:963–976. doi: 10.1002/yea.1414. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S. et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci USA. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrell GW, Giaever G, Chu AM, Davis RW, Brown JM. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc Natl Acad Sci USA. 2001;98:12608–12613. doi: 10.1073/pnas.231366398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. Genes required for ionizing radiation resistance in yeast. Nat Genet. 2001;29:426–434. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- Deshpande GP, Hayles J, Hoe KL, Kim DU, Park HO, Hartsuiker E. Screening a genome-wide S. pombe deletion library identifies novel genes and pathways involved in genome stability maintenance. DNA Repair (Amst) 2009;8:672–679. doi: 10.1016/j.dnarep.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TX, Xu XY, Zhang MJ, Peng X, Du LL. Global fitness profiling of fission yeast deletion strains by barcode sequencing. Genome Biol. 2010;11:R60. doi: 10.1186/gb-2010-11-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch T, Carr AM, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- Levin JD, Demple B. In vitro detection of endonuclease IV-specific DNA damage formed by bleomycin in vivo. Nucleic Acids Res. 1996;24:885–889. doi: 10.1093/nar/24.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S, Capasso H, Walworth NC. The topoisomerase I poison camptothecin generates a Chk1-dependent DNA damage checkpoint signal in fission yeast. Yeast. 1999;15:821–828. doi: 10.1002/(SICI)1097-0061(199907)15:10A<821::AID-YEA422>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Nilssen EA, Synnes M, Kleckner N, Grallert B, Boye E. Intra-G1 arrest in response to UV irradiation in fission yeast. Proc Natl Acad Sci USA. 2003;100:10758–10763. doi: 10.1073/pnas.1833769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Nurse P. Regulation of cell polarity by microtubules in fission yeast. J Cell Biol. 1998;142:457–471. doi: 10.1083/jcb.142.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy A, Calonge TM, Outwin EA, O'Connell MJ. Fission yeast Rnf4 homologs are required for DNA repair. J Biol Chem. 2007;282:20388–20394. doi: 10.1074/jbc.M702652200. [DOI] [PubMed] [Google Scholar]
- Helt CE, Wang W, Keng PC, Bambara RA. Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle. 2005;4:529–532. doi: 10.4161/cc.4.4.1598. [DOI] [PubMed] [Google Scholar]
- Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol. 2007;14:372–380. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- Tsutsui Y, Khasanov FK, Shinagawa H, Iwasaki H, Bashkirov VI. Multiple interactions among the components of the recombinational DNA repair system in Schizosaccharomyces pombe. Genetics. 2001;159:91–105. doi: 10.1093/genetics/159.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Marguerat S, Villén J, Gygi SP, Bähler J, Winston F. The S. pombe SAGA complex controls the switch from proliferation to sexual differentiation through the opposing roles of its subunits Gcn5 and Spt8. Genes Dev. 2008;22:3184–3195. doi: 10.1101/gad.1719908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahoz A, Alcaide-Gavilán M, Daga RR, Jimenez J. Antagonistic roles of PP2A-Pab1 and Etd1 in the control of cytokinesis in fission yeast. Genetics. 2010;186:1261–1270. doi: 10.1534/genetics.110.121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic D, Chinchilla K, Finkel JS, Culbertson MR. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res. 2004;32:2441–2452. doi: 10.1093/nar/gkh561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Bezanilla M, Pollard TD. Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J Cell Biol. 2000;151:789–800. doi: 10.1083/jcb.151.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N, Tanaka K, Sturm S, Okayama H. Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle 'start' function of Res2-Cdc10. EMBO J. 1995;14:4794–4802. doi: 10.1002/j.1460-2075.1995.tb00161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G, Zachariae W, Schleiffer A, Nasmyth K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Moser BA, Du LL, Russell P. Cooperative control of Crb2 by ATM family and Cdc2 kinases is essential for the DNA damage checkpoint in fission yeast. Mol Cell Biol. 2005;25:10721–10730. doi: 10.1128/MCB.25.24.10721-10730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Aasland R, Stewart AF. High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J Biol Chem. 2003;278:8487–8493. doi: 10.1074/jbc.M209562200. [DOI] [PubMed] [Google Scholar]
- Liu X, McLeod I, Anderson S, Yates JR 3rd. He X: Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 2005;24:2919–2930. doi: 10.1038/sj.emboj.7600762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chang EC. Schizosaccharomyces pombe Ras1 effector, Scd1, interacts with Klp5 and Klp6 kinesins to mediate cytokinesis. Genetics. 2003;165:477–488. doi: 10.1093/genetics/165.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Whitby MC. The involvement of Srs2 in post-replication repair and homologous recombination in fission yeast. Nucleic Acids Res. 2004;32:1480–1491. doi: 10.1093/nar/gkh317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Vanoosthuyse V, Fillingham J, Roguev A, Watt S, Kislinger T, Treyer A, Carpenter LR, Bennett CS, Emili A. et al. An acetylated form of histone H2A.Z regulates chromosome architecture in Schizosaccharomyces pombe. Nat Struct Mol Biol. 2009;16:1286–1293. doi: 10.1038/nsmb.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar S, Saini A, Nanda JS, Saini S, Singh J. Role of Swi6/HP1 self-association-mediated recruitment of Clr4/Suv39 in establishment and maintenance of heterochromatin in fission yeast. J Biol Chem. 2011;286:9308–9320. doi: 10.1074/jbc.M110.143198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Kume K, Kanai M, Goshima T, Miyahara K, Dhut S, Tee WW, Hirata D, Toda T. The V260I mutation in fission yeast alpha-tubulin Atb2 affects microtubule dynamics and EB1-Mal3 localization and activates the Bub1 branch of the spindle checkpoint. Mol Biol Cell. 2006;17:1421–1435. doi: 10.1091/mbc.E05-08-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Lancaster CS, Shi B, Guo H, Thimmaiah P, Bjornsti MA. TOR signaling is a determinant of cell survival in response to DNA damage. Mol Cell Biol. 2007;27:7007–7017. doi: 10.1128/MCB.00290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan BJ, Villén J, Marguerat S, Bähler J, Gygi SP, Winston F. Fission yeast SWI/SNF and RSC complexes show compositional and functional differences from budding yeast. Nat Struct Mol Biol. 2008;15:873–880. doi: 10.1038/nsmb.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero F, Gómez N, Ariño J, Ramos J. Trk1 and Trk2 define the major K(+) transport system in fission yeast. J Bacteriol. 2000;182:394–399. doi: 10.1128/JB.182.2.394-399.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F, Dubrana K, Gasser SM. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J Struct Biol. 2002;140:79–91. doi: 10.1016/S1047-8477(02)00533-6. [DOI] [PubMed] [Google Scholar]
- Svensson JP, Pesudo LQ, Fry RC, Adeleye YA, Carmichael P, Samson LD. Genomic phenotyping of the essential and non-essential yeast genome detects novel pathways for alkylation resistance. BMC Syst Biol. 2011;5:157. doi: 10.1186/1752-0509-5-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolling R, Nguyen T, Chen EY, Botstein D. A new yeast gene with a myosin-like heptad repeat structure. Mol Gen Genet. 1993;237:359–369. doi: 10.1007/BF00279439. [DOI] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D. et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan PW, Klein F, Leach DR. Novel roles for selected genes in meiotic DNA processing. PLoS Genet. 2007;3:e222. doi: 10.1371/journal.pgen.0030222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslett M, Wood V. Gene Ontology annotation status of the fission yeast genome: preliminary coverage approaches 100% Yeast. 2006;23:913–919. doi: 10.1002/yea.1420. [DOI] [PubMed] [Google Scholar]
- Humphrey T. DNA damage and cell cycle control in Schizosaccharomyces pombe. Mutat Res. 2000;451:211–226. doi: 10.1016/S0027-5107(00)00051-8. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Kumar S, Huberman JA. On the slowing of S phase in response to DNA damage in fission yeast. J Biol Chem. 2004;279:43574–43580. doi: 10.1074/jbc.M407819200. [DOI] [PubMed] [Google Scholar]
- Sabatinos SA, Forsburg SL. Measuring DNA content by flow cytometry in fission yeast. Methods Mol Biol. 2009;521:449–461. doi: 10.1007/978-1-60327-815-7_25. [DOI] [PubMed] [Google Scholar]
- Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD. et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/S0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Muzi Falconi M, Brown GW, Kelly TJ. cdc18+ regulates initiation of DNA replication in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1996;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locovei AM, Spiga MG, Tanaka K, Murakami Y, D'Urso G. The CENP-B homolog, Abp1, interacts with the initiation protein Cdc23 (MCM10) and is required for efficient DNA replication in fission yeast. Cell Div. 2006;1:27. doi: 10.1186/1747-1028-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JP, Murakami Y, Huberman JA, Hurwitz J. Isolation, characterization, and molecular cloning of a protein (Abp2) that binds to a Schizosaccharomyces pombe origin of replication (ars3002) Mol Cell Biol. 1998;18:1670–1681. doi: 10.1128/mcb.18.3.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Grallert B, Ribar B, Sipiczki M. Coordination of initiation of nuclear division and initiation of cell division in Schizosaccharomyces pombe: genetic interactions of mutations. J Bacteriol. 1998;180:892–900. doi: 10.1128/jb.180.4.892-900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz MJ, Petersen J, Valovin S, Hagan IM. MTOC formation during mitotic exit in fission yeast. J Cell Sci. 2001;114:4521–4532. doi: 10.1242/jcs.114.24.4521. [DOI] [PubMed] [Google Scholar]
- Bähler J. A transcriptional pathway for cell separation in fission yeast. Cell Cycle. 2005;4:39–41. doi: 10.4161/cc.4.1.1336. [DOI] [PubMed] [Google Scholar]
- Sipiczki M. Splitting of the fission yeast septum. FEMS Yeast Res. 2007;7:761–770. doi: 10.1111/j.1567-1364.2007.00266.x. [DOI] [PubMed] [Google Scholar]
- Nielsen O, Løbner-Olesen A. Once in a lifetime: strategies for preventing re-replication in prokaryotic and eukaryotic cells. EMBO Rep. 2008;9:151–156. doi: 10.1038/sj.embor.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Kipreos ET. Preventing DNA re-replication–divergent safeguards in yeast and metazoa. Cell Cycle. 2003;2:431–434. [PubMed] [Google Scholar]
- Felberbaum R, Hochstrasser M. Ulp2 and the DNA damage response: desumoylation enables safe passage through mitosis. Cell Cycle. 2008;7:52–56. doi: 10.4161/cc.7.1.5218. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Xiao G, Noma K, Malikzay A, Bjerling P, Ekwall K, Kobayashi R, Grewal SI. Alp13, an MRG family protein, is a component of fission yeast Clr6 histone deacetylase required for genomic integrity. EMBO J. 2003;22:2776–2787. doi: 10.1093/emboj/cdg248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakurta AG, Gopal G, Yoon JH, Kozak L, Dhar R. Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast. EMBO J. 2005;24:2512–2523. doi: 10.1038/sj.emboj.7600713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R, Bortner CD, Cidlowski JA. Potential roles of electrogenic ion transport and plasma membrane depolarization in apoptosis. J Membr Biol. 2006;209:43–58. doi: 10.1007/s00232-005-0837-5. [DOI] [PubMed] [Google Scholar]
- Marchetti MA, Kumar S, Hartsuiker E, Maftahi M, Carr AM, Freyer GA, Burhans WC, Huberman JA. A single unbranched S-phase DNA damage and replication fork blockage checkpoint pathway. Proc Natl Acad Sci USA. 2002;99:7472–7477. doi: 10.1073/pnas.112702399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Toda T, MacCallum R, Harris AL, Norbury C. Cid1, a fission yeast protein required for S-M checkpoint control when DNA polymerase delta or epsilon is inactivated. Mol Cell Biol. 2000;20:3234–3244. doi: 10.1128/MCB.20.9.3234-3244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synnes M, Nilssen EA, Boye E, Grallert B. A novel chk1-dependent G1/M checkpoint in fission yeast. J Cell Sci. 2002;115:3609–3618. doi: 10.1242/jcs.00004. [DOI] [PubMed] [Google Scholar]
- Krohn M, Skjolberg HC, Soltani H, Grallert B, Boye E. The G1-S checkpoint in fission yeast is not a general DNA damage checkpoint. J Cell Sci. 2008;121:4047–4054. doi: 10.1242/jcs.035428. [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Gulli MP, Simanis V. SIN and the art of splitting the fission yeast cell. Curr Biol. 2004;14:R722–730. doi: 10.1016/j.cub.2004.08.049. [DOI] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G. et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bähler J. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics. 2003;4:27. doi: 10.1186/1471-2164-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro N, Esteve-Zarzoso B, González A, Mas A, Guillamón JM. Real-time quantitative PCR (QPCR) and reverse transcription-QPCR for detection and enumeration of total yeasts in wine. Appl Environ Microbiol. 2006;72:7148–7155. doi: 10.1128/AEM.00388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of genes whose deletions exhibited sensitivity to DNA damage reagents during the second round of screen. Table S2. GO profiling of 52 genes whose deletion mutants showed strong sensitivity to DNA damage reagents (P ≤ 0.05). Table S3. Flow cytometry analysis of 37 mutants. Table S4. Primers used for real time PCR analysis in this study. Figure S1. Spot assay of 52 deletions. Exponentially growing cells, WT or deletions, were harvested and 5-fold serial dilutions were spotted on the plates supplemented with DNA damage reagents. The plates were photographed after 3~4 days of incubation at 32°C. Figure S2. Flow cytometry analysis of deletions in “2C” group. Figure S3. Flow cytometry analysis of deletions in “1C” group. Figure S4. Flow cytometry analysis of deletions in “W4C” group. Figure S5. Flow cytometry analysis of deletions in “S4C” group.