Abstract
A monoclonal antibody against prostate stem cell antigen (PSCA) has emerged as a novel cancer therapy currently being tested in clinical trials for prostate and pancreatic cancers, but this treatment is likely to be efficient only in patients with PSCA-expressing tumors. The present study demonstrates that a genetic variant (rs2294008) discovered by bladder cancer genome-wide association studies is a strong predictor of PSCA protein expression in bladder tumors, as measured by two-sided multivariable linear regression (P = 6.46×10−11; n = 278). The association pattern is similar in non-muscle-invasive tumors, stages Ta (P = 3.10×10−5; n = 173) and T1 (P = 2.64×10−5; n = 60), and muscle-invasive tumors, stages T2 (P =.01; n = 23) and T3/4 (P =.03; n = 22). The study suggests that anti-PSCA immunotherapy might be beneficial for bladder cancer patients with high tumor PSCA expression, which is statistically significantly associated with the presence of CT and TT genotypes of a common genetic variant, rs2294008. Future clinical studies will be needed to validate PSCA as a therapeutic target for bladder cancer.
Genome-wide association studies have discovered genetic variants associated with many cancer types (1). It is anticipated that these findings will help improve understanding of disease mechanisms and lead to novel translational applications. Bladder cancer genome-wide association studies identified a single nucleotide polymorphism, rs2294008, within the PSCA gene (2,3). PSCA is a cell-surface, glycosylphosphatidylinositol-anchored protein expressed in various cancers (4–14); however, its biological role in normal and cancer conditions remains unclear. PSCA antibody-based immunotherapy is currently being used in clinical trials for prostate and pancreatic cancers (15–20). In this article, we suggest PSCA as a candidate drug target for bladder cancer as well.
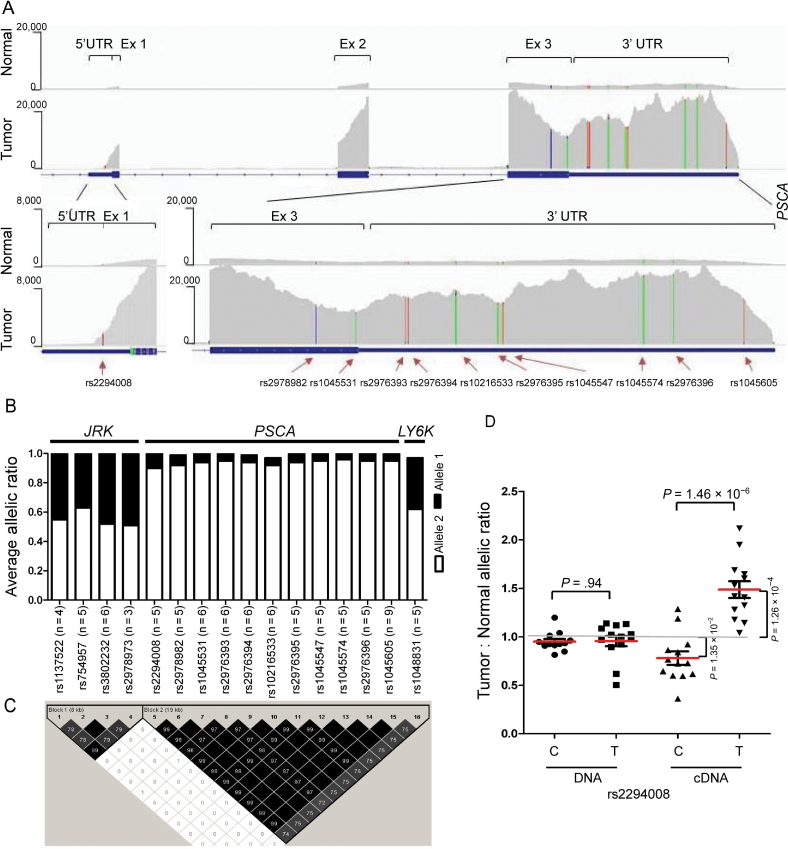
Previously, we found increased PSCA mRNA expression in bladder tumors and, specifically, in the presence of the risk T allele of rs2294008 (21). Now we have discovered an additional effect associated with rs2294008—an allelic expression imbalance, which is a deviation from an expected 50%:50% allelic ratio in heterozygous transcribed single nucleotide polymorphisms. We performed RNA sequencing of six normal and six tumor bladder tissues, and validation studies in 14 normal and 13 tumor bladder tissue samples heterozygous for rs2294008 (Figure 1, A–D; Supplementary Methods, available online). We quantified rs2294008 T and C alleles in DNA and cDNA samples and calculated an average tumor:normal (T:N) ratio. In DNA samples the T:N ratio was close to 1.0 and similar for both alleles (Figure 1, D; Supplementary Table 1, available online), suggesting no difference in DNA copy number variation between normal and tumor tissues within the PSCA gene. This is an important conclusion because PSCA is located in 8q24.3 region and relatively close (12Mb) to MYC oncogene in 8q24.21 region, which is reported to be amplified in many cancers (22–24). Thus, our results do not support the involvement of PSCA in differential genomic amplification in bladder tumors. In cDNA of tumors compared with normal tissue, we detected a notable increase of T allele expression (T:N ratio = 1.48; P = 1.26×10−4), accompanied by decrease of C allele expression (T:N ratio = 0.78; P = 1.35×10−2), resulting in an overall difference of T:N ratio for C and T alleles (P = 1.46×10−6) in cDNA but not in DNA from the same samples (Figure 1, D; Supplementary Table 2, available online). Importantly, the observed allelic expression imbalance in bladder tumors could result from decreased expression of transcripts with nonrisk C allele and/or from increased expression of transcripts with risk T allele of rs2294008.
Figure 1.
Allelic expression imbalance (AEI) in bladder tissue samples (see Supplementary Methods, available online). A) RNA sequencing analysis of PSCA expression in representative unpaired normal (n = 1) and tumor (n = 1) bladder tissue samples heterozygous for rs2294008. PSCA exon structure, level of mRNA expression, and location of transcribed heterozygous genetic variants are shown. B) Summary of AEI analysis in RNA sequencing of normal (n = 6) and tumor (n = 6) bladder tissue samples. Numbers of samples heterozygous for each of the transcribed single nucleotide polymorphisms (SNPs) within PSCA and surrounding genes, JRK and LY6K, are indicated in parenthesis. Strong AEI is suggested by the 90%:10% allelic ratio for 11 heterozygous transcribed variants within PSCA but not for variants within JRK and LY6K, which showed the expected 50%:50% allelic ratio. C) Linkage disequilibrium plot for 16 SNPs from JRK, PSCA, and Ly6K genes included in the AEI analysis. The PSCA variants with strong AEI are strongly correlated, as indicated by high pair-wise r 2 values (black shading on plot), calculated based on 3532 case patients and 5120 control subjects of European ancestry from the bladder cancer genome-wide association studies (21). D) Different sample types (DNA, cDNA, C and T alleles of rs2294008) are marked by circles, squares and triangles. Red bars mark mean values within each sample group. A grey line at the T:N allelic ratio “1” indicates allelic expression balance, whereas significant deviations from this line indicate allelic expression imbalance. Results of AEI measured by a TaqMan allele-specific expression/genotyping assay in DNA and cDNA from 14 normal and 13 tumor bladder tissue samples heterozygous for rs2294008. The analysis shows that AEI is detected in cDNA but not DNA samples. The Tumor:Normal expression ratio indicates increased expression of risk T allele, whereas decreased expression on nonrisk C allele in tumors compared with normal tissue samples.
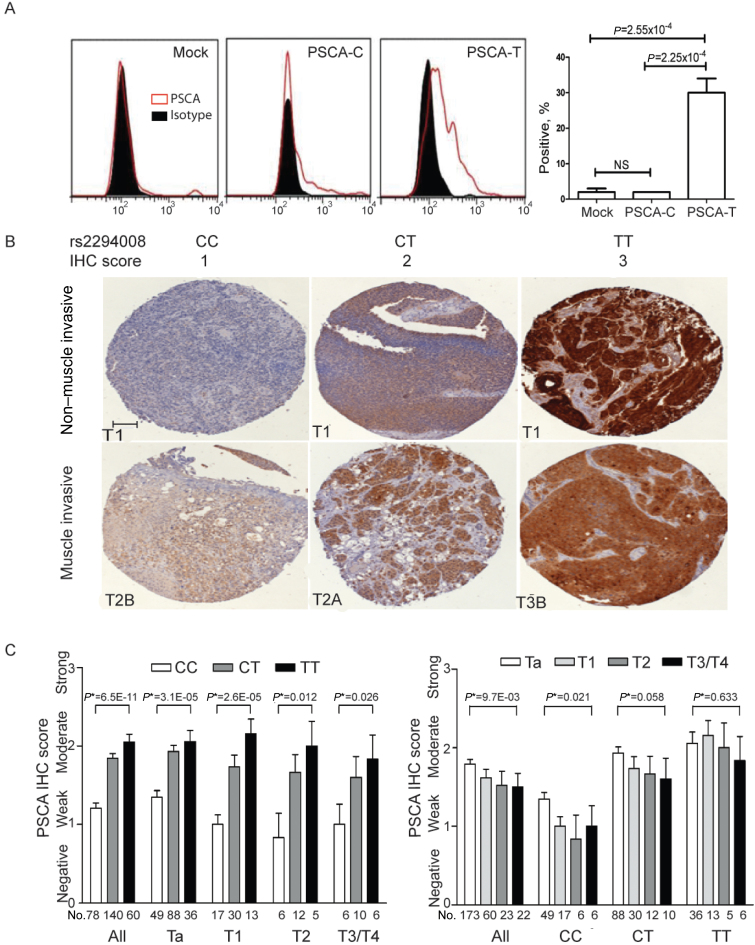
Next, we evaluated the effect of rs2294008 on PSCA protein expression. The risk T allele of rs2294008 creates a novel alternative translation start site, extending the leader peptide from 11 to 20 amino acids, which might affect the efficiency of PSCA post-translational processing and cell surface expression. To test this hypothesis, we performed fluorescence activated cell sorter analysis in HeLa cells transfected with the allelic PSCA expression constructs (Supplementary Methods, available online). We observed stronger surface PSCA expression in cells transfected with PSCA-T construct compared with those transfected with PSCA-C construct (P = 2.25×10−4) and with a mock-control (P = 2.55×10−4) (Figure 2, A).
Figure 2.
Effect of rs2294008 on prostate stem cell antigen (PSCA) protein expression (see Supplementary Methods, available online). A) Fluorescence activated cell sorting (FACS) analysis of cell surface PSCA expression in HeLa cells. The cells were transiently transfected with an empty vector (mock) or PSCA expression constructs with C and T alleles of rs2294008, encoding PSCA proteins with leader peptides of 11 (C allele) or 20 amino acids (T allele). The staining with anti-PSCA antibody 1G8 (red curves) is compared with isotype control (black area). Percentage of positive cells (y axis) is plotted against fluorescence intensity in logarithmic scale (x axis). The graphic representation of FACS results (right panel) shows mean values of three biological replicates with standard errors of the mean; P values are from a two-sided unpaired t test. Cell surface PSCA expression is statistically significantly increased in cells transfected with an expression construct carrying the risk T allele of rs2294008 compared with the construct with a nonrisk C allele of rs2294008 and mock control, which also shows negligible endogenous level of PSCA expression in HeLa cells. NS = not statistically significant. B) Immunohistochemistry (IHC) analysis of PSCA expression using bladder tumor tissue microarrays. Representative images are from groups of samples stratified by rs2294008 genotypes (CC, CT, TT) and tumor stages—non-muscle-invasive tumors (stages Ta and T1) and muscle-invasive tumors (stages T2–T4). PSCA expression was detected with 1G8 anti-PSCA monoclonal antibody (brown staining) and scored as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong), based on overall intensity. Scale bar corresponds to 0.1mm, and all images are presented in the same scale. C) Association between PSCA IHC scores and rs2294008 genotypes, stratified by tumor stage (left panel) or, for tumor stages, stratified by rs2294008 genotypes (right panel). PSCA IHC scores are shown as mean values with standard errors; two-sided P* values were estimated from multivariable linear models, assuming additive genetic effect of rs2294008, adjusted for age, sex, study site, and smoking status (ever or never). Single nucleotide polymorphism rs2294008 was found to be the strongest predictor for PSCA expression, regardless of all other factors tested, such as age, sex, smoking status, and tumor grade and stage.
An immunohistochemistry analysis in a pilot set of matching pairs of bladder tumor-normal tissue samples (5 pairs with TT and 5 pairs with CC genotype of rs2294008) supported our RNA sequencing and allelic expression imbalance results. In samples with risk TT genotypes, PSCA was expressed even in normal tissues, whereas in samples with nonrisk CC genotypes, there was low or no PSCA expression even in tumors (Supplementary Figure 1, A, available online). A validation immunohistochemistry analysis was performed in an additional set of 278 bladder tumors obtained from patients enrolled in the New England Bladder Cancer study (Figure 2, B). Genotype of rs2294008 was strongly associated with PSCA protein expression, as measured by two-sided multivariable linear regression (P = 6.46×10−11) (Figure 2, C; Supplementary Table 3, available online). The pattern for rs2294008 association was similar in non-muscle-invasive (stages Ta [P = 3.10×10−5; n = 173] and T1 [P = 2.64×10−5; n = 60]), and muscle-invasive (stages T2 [P =.01; n = 23] and T3/4 [P =.03; n = 22]) tumors (25) (Figure 2, C); this association could not be explained by any other factors (Supplementary Table 4, available online). Stratification by tumor grades (low and high grade) (26) or a stage and grade combination provided similar results (Supplementary Figure 1, B, available online). Surprisingly, PSCA expression was decreased with higher tumor stage (P = 9.69×10−3) (Figure 2, C). We found that this effect was due to decreased expression of non-risk C allele (P =.02 in CC group and P =.06 in CT group), whereas PSCA expression of risk T allele was not associated with tumor staging (P =.63) (Figure 2, C). Together with allelic expression imbalance data in mRNA samples, these results reiterate functional changes on PSCA mRNA and protein expression reflected in both gain of T allele and loss of C allele of rs2294008 in bladder tumors compared with normal tissue and during progression through tumor stages.
The PSCA function and the mechanism of the anti-PSCA antibody remain unclear. PSCA might be functionally involved in oncogenic development (5,27–32), cause differential immune response (14), or mark some specific cell populations. Although there are several reports on genetic association of rs2294008 with bladder, gastric, duodenal, and breast cancers (2,3,14,21,33–40), little data is available for relationships between rs2294008 and PSCA expression in different tissue types (21,39). Our study is a unique and detailed investigation of the allele-specific effects of rs2294008 on PSCA mRNA and protein expression in bladder tissue.
Up to 75% of bladder cancer patients of European descent are estimated to carry the risk rs2294008 CT and TT genotypes. Possibly, if used at early cancer stages (Ta and T1), anti-PSCA immunotherapy might prevent progression to advanced muscle-invasive cancer, a devastating disease that is treated by radical bladder cystectomy together with removal of parts of surrounding organs and lymph nodes to prevent metastasis.
A limitation of our study is that we only studied samples from patients of European ancestry. Similar studies should be performed in other population groups. The systemic delivery of this treatment in non-muscle-invasive tumors might be limited because of lower vascularization of these tumors. Alternative approaches may include intravesical administration of the anti-PSCA antibody and/or its conjugation with other bioactive molecules to enhance treatment efficiency and facilitate bladder tumor-specific drug delivery.
This is one of the first studies to show direct translational implications of a genetic variant identified through genome-wide association studies for common cancers. Clinical trials are warranted in order to validate PSCA as a therapeutic target in bladder cancer and rs2294008 as a predictive marker for treatment response.
Funding
This work was supported in whole or in part with federal funds from the Intramural Research Program of the National Institutes of Health, National Cancer Institute (contract HHSN261200800001E). The New England Bladder Cancer study, the National Cancer Institute bladder cancer genome-wide association studies, and follow-up studies are supported by the Division of Cancer Epidemiology and Genetics, Intramural Research Program of the National Institutes of Health, National Cancer Institute. The New England Bladder Cancer study is also supported by a National Cancer Institute contract (NCI N02-CP-01037). This work was supported by a National Cancer Institute Director’s Career Development Award (to Y-PF).
Supplementary Material
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
We are grateful to all the patients who have provided samples for this study. We thank Dr. Robert Reiter (UCLA) for providing monoclonal anti-PSCA antibody 1G8 and for critical reading of the manuscript, Dr. Dimitra Bourmpoulia (NCI) for providing help with fluorescence activated cell sorter analysis, and Anne Taylor (IMS, Rockville) for help with data management.
References
- 1. Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Hum Genet. 2011; 130(1):59–78 [DOI] [PubMed] [Google Scholar]
- 2. Rothman N, Garcia-Closas M, Chatterjee N, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010; 42(11):978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu X, Ye Y, Kiemeney LA, et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet. 2009; 41(9):991–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amara N, Palapattu GS, Schrage M, et al. Prostate stem cell antigen is overexpressed in human transitional cell carcinoma. Cancer Res. 2001; 61(12):4660–4665 [PubMed] [Google Scholar]
- 5. Barbisan F, Mazzucchelli R, Santinelli A, et al. Expression of prostate stem cell antigen in high-grade prostatic intraepithelial neoplasia and prostate cancer. Histopathology. 2010; 57(4):572–579 [DOI] [PubMed] [Google Scholar]
- 6. Cheng L, Reiter RE, Jin Y, et al. Immunocytochemical analysis of prostate stem cell antigen as adjunct marker for detection of urothelial transitional cell carcinoma in voided urine specimens. J Urol. 2003; 169(6): 2094–2100 [DOI] [PubMed] [Google Scholar]
- 7. Elsamman EM, Fukumori T, Tanimoto S, et al. The expression of prostate stem cell antigen in human clear cell renal cell carcinoma: a quantitative reverse transcriptase-polymerase chain reaction analysis. BJU Int. 2006; 98(3):668–673 [DOI] [PubMed] [Google Scholar]
- 8. Geiger KD, Hendruschk S, Rieber EP, Morgenroth A, Weigle B, Juratli T, et al. The prostate stem cell antigen represents a novel glioma-associated antigen. Oncol Rep. 2011; 26(1):13–21 [DOI] [PubMed] [Google Scholar]
- 9. Gu Z, Thomas G, Yamashiro J, Shintaku IP, Dorey F, Raitano A, et al. Prostate stem cell antigen (PSCA) expression increases with high gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene. 2000; 19(10):1288–1296 [DOI] [PubMed] [Google Scholar]
- 10. Kawaguchi T SM, Tojo T, Yamato I, Nomi T, Hotta K, Hamada K, Suzaki Y, Sugiura S, Kushibe K, Nakajima Y, Taniguchi S. Clinical significance of prostate stem cell antigen expression in non-small cell lung cancer. Jpn J Clin Oncol. 2010; 40(4):319–326 [DOI] [PubMed] [Google Scholar]
- 11. Liu WK JX, Zhang ZX. Expression of PSCA, PIWIL1, and TBX2 in endometrial adenocarcinoma. Onkologie. 2010; 33(5):241–245 [DOI] [PubMed] [Google Scholar]
- 12. Reiter RE, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, et al. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 1998; 95(4):1735–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saeki N, Gu J, Yoshida T, Wu X. Prostate stem cell antigen: a Jekyll and Hyde molecule? Clin Cancer Res. 2010; 16(14):3533–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanikawa C, Urabe Y, Matsuo K, et al. A genome-wide association study identifies two susceptibility loci for duodenal ulcer in the Japanese population. Nat Genet. 2012; 44(4): 430–434 [DOI] [PubMed] [Google Scholar]
- 15. Antonarakis ES, Carducci MA, Eisenberger MA, et al. Phase I rapid dose-escalation study of AGS-1C4D4, a human anti-PSCA (prostate stem cell antigen) monoclonal antibody, in patients with castration-resistant prostate cancer: a PCCTC trial. Cancer Chemother Pharmacol. 2012; 69(3):763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morris MJ, Eisenberger MA, Pili R, Denmeade SR, Rathkopf D, Slovin SF, et al. A phase I/IIA study of AGS-PSCA for castration-resistant prostate cancer. Ann Oncol. 2012;23(10):2714–2719. .; [DOI] [PMC free article] [PubMed]
- 17. Raff AB, Gray A, Kast WM. Prostate stem cell antigen: a prospective therapeutic and diagnostic target. Cancer Lett. 2009; 277(2):126–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ross S, Spencer SD, Holcomb I, et al. Prostate stem cell antigen as therapy target: tissue expression and in vivo efficacy of an immunoconjugate. Cancer Res. 2002; 62(9):2546–2553 [PubMed] [Google Scholar]
- 19. Wente MN, Jain A, Kono E, et al. Prostate stem cell antigen is a putative target for immunotherapy in pancreatic cancer. Pancreas. 2005; 31(2): 119–125 [DOI] [PubMed] [Google Scholar]
- 20. Wolpin BM, O’Reilly EM, Ko Y, et al. Global, multicenter, open-label, randomized phase II trial comparing gemcitabine (G) with. G plus AGS-1C4D4 (A) in patients (pts) with metastatic pancreatic cancer (mPC). J Clin Oncol. 2011; 29(Suppl):abstrct 4031 [Google Scholar]
- 21. Fu YP, Kohaar I, Rothman N, Earl J, Figueroa JD, Ye Y, et al. Common genetic variants in the PSCA gene influence gene expression and bladder cancer risk. Proc Natl Acad Sci U S A. 2012; 109(13):4974–49799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burbano RR, Assumpcao PP, Leal MF, et al. C-MYC locus amplification as metastasis predictor in intestinal-type gastric adenocarcinomas: CGH study in Brazil. Anticancer Res. 2006; 26(4B):2909–2914 [PubMed] [Google Scholar]
- 23. Reiter RE, Sato I, Thomas G, et al. Coamplification of prostate stem cell antigen (PSCA) and MYC in locally advanced prostate cancer. Genes Chromosomes Cancer. 2000; 27(1):95–103 [DOI] [PubMed] [Google Scholar]
- 24. Zaharieva B, Simon R, Ruiz C, et al. High-throughput tissue microarray analysis of CMYC amplificationin urinary bladder cancer. Int J Cancer. 2005; 117(6):952–956 [DOI] [PubMed] [Google Scholar]
- 25. Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. (Eds.): TNM staging system for bladder cancer. In AJCC Cancer Staging Manual. 7th ed. New York: Springer; 2010:497–505 [Google Scholar]
- 26. Epstein JI, Amin MB, Reuter VR, Mostofi FK. The World Health Organization/International Society of Urological Pathology consensus classification of urothelial (transitional cell) neoplasms of the urinary bladder. Bladder Consensus Conference Committee. Am J Surg Pathol. 1998; 22(12):1435–1448 [DOI] [PubMed] [Google Scholar]
- 27. Gu Z, Yamashiro J, Kono E, Reiter RE. Anti-prostate stem cell antigen monoclonal antibody 1G8 induces cell death in vitro and inhibits tumor growth in vivo via a Fc-independent mechanism. Cancer Res. 2005; 65(20):9495–9500 [DOI] [PubMed] [Google Scholar]
- 28. Katz BZ, Eshel R, Sagi-Assif O, Witz IP. An association between high Ly-6A/E expression on tumor cells and a highly malignant phenotype. Int J Cancer. 1994;59(5):684–691 [DOI] [PubMed] [Google Scholar]
- 29. Marra E, Uva P, Viti V, et al. Growth delay of human bladder cancer cells by prostate stem cell antigen downregulation is associated with activation of immune signaling pathways. BMC Cancer. 2010; 10:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saffran DC, Raitano AB, Hubert RS, Witte ON, Reiter RE, Jakobovits A. Anti-PSCA mAbs inhibit tumor growth and metastasis formation and prolong the survival of mice bearing human prostate cancer xenografts. Proc Natl Acad Sci U S A. 2001; 98(5):2658–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao Z, Ma W, Zeng G, Qi D, Ou L, Liang Y. Small interference RNA-mediated silencing of prostate stem cell antigen attenuates growth, reduces migration and invasion of human prostate cancer PC-3M cells. Urol Oncol.2004. Article in press; http://dx.doi.org/10.1016/j.urolonc.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 32. Ryu B, Jones J, Blades NJ, et al. Relationships and differentially expressed genes among pancreatic cancers examined by large-scale serial analysis of gene expression. Cancer Res. 2002; 62(3):819–826 [PubMed] [Google Scholar]
- 33. Golka K, Selinski S, Lehmann ML, et al. Genetic variants in urinary bladder cancer: collective power of the “wimp SNPs.”. Arch Toxicol. 2011; 85(6):539–554 [DOI] [PubMed] [Google Scholar]
- 34. Lu Y, Chen J, Ding Y, et al. Genetic variation of PSCA gene is associated with the risk of both diffuse- and intestinal-type gastric cancer in a Chinese population. Int J Cancer. 2010; 127(9):2183–2189 [DOI] [PubMed] [Google Scholar]
- 35. Matsuo K, Tajima K, Suzuki T, et al. Association of prostate stem cell antigen gene polymorphisms with the risk of stomach cancer in Japanese. Int J Cancer. 2009; 125(8):1961–1964 [DOI] [PubMed] [Google Scholar]
- 36. Qiao L, Feng Y. Genetic variations of prostate stem cell antigen (PSCA) contribute to the risk of gastric cancer for Eastern Asians: a meta-analysis based on 16792 individuals. Gene. 2012; 493(1):83–91 [DOI] [PubMed] [Google Scholar]
- 37. Sakamoto H, Yoshimura K, Saeki N, et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008; 40(6):730–740 [DOI] [PubMed] [Google Scholar]
- 38. Shi D, Wang S, Gu D, et al. The PSCA polymorphisms derived from genome-wide association study are associated with risk of gastric cancer: a meta-analysis. J Cancer Res Clin Oncol. 2012; 138(8):1339–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang S, Tang J, Wang M, Yuan L, Zhang Z. Genetic variation in PSCA and bladder cancer susceptibility in a Chinese population. Carcinogenesis.. 2010; 31(4):621–624 [DOI] [PubMed] [Google Scholar]
- 40. Wang T, Zhang L, Li H, Wang B, Chen K. Prostate stem cell antigen polymorphisms and susceptibility to gastric cancer: a systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012; 21(5):843–850 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.