Abstract
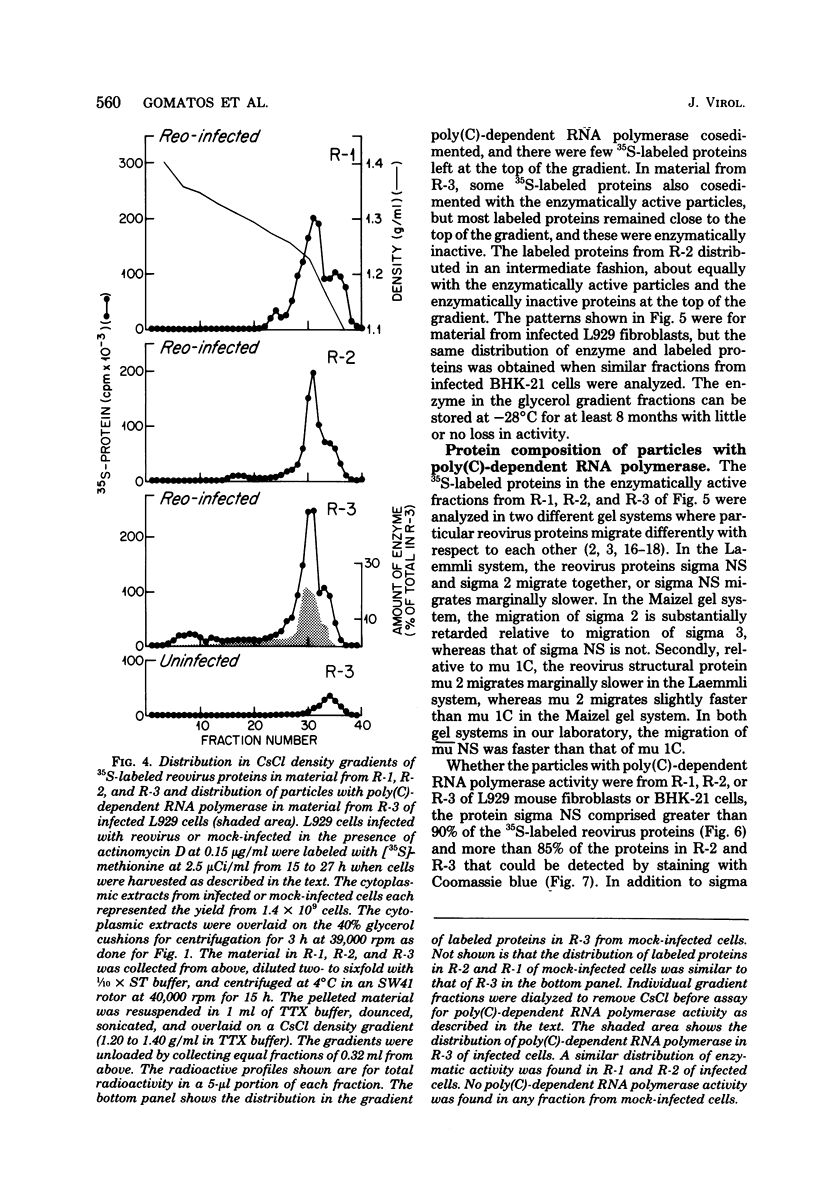
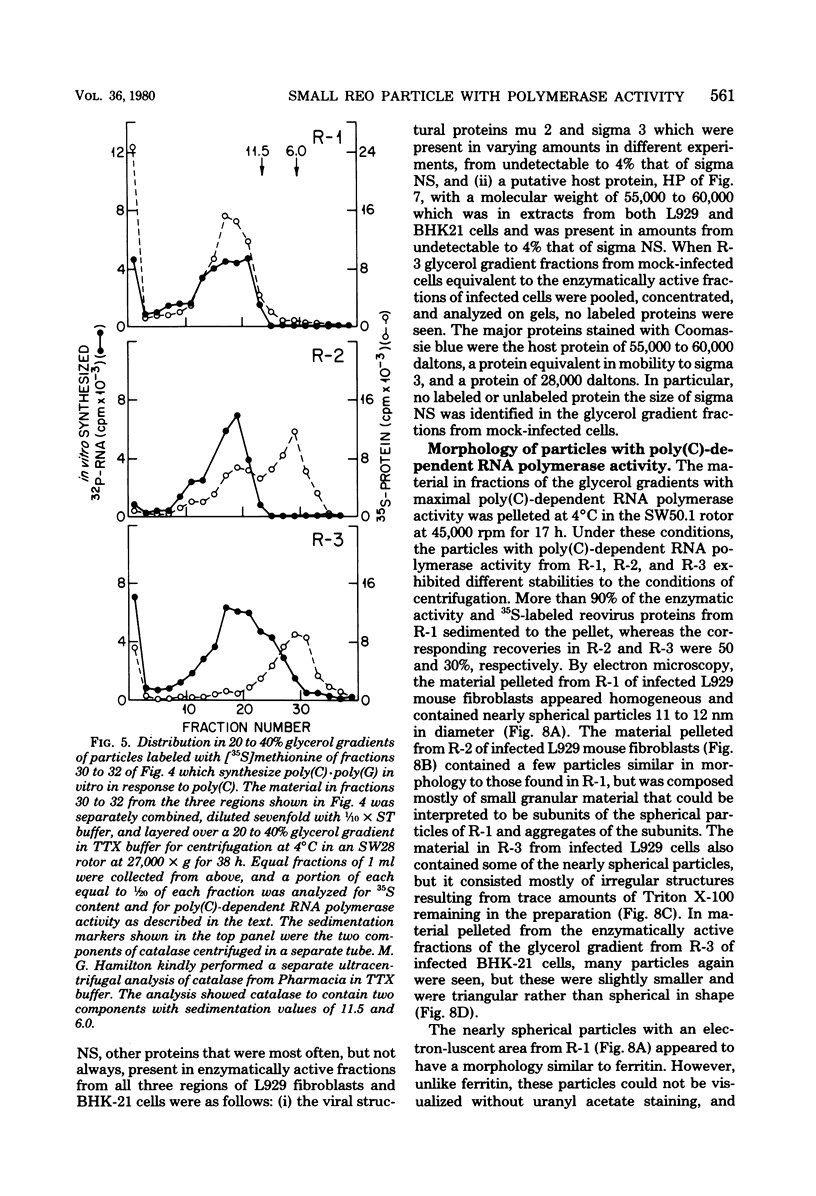
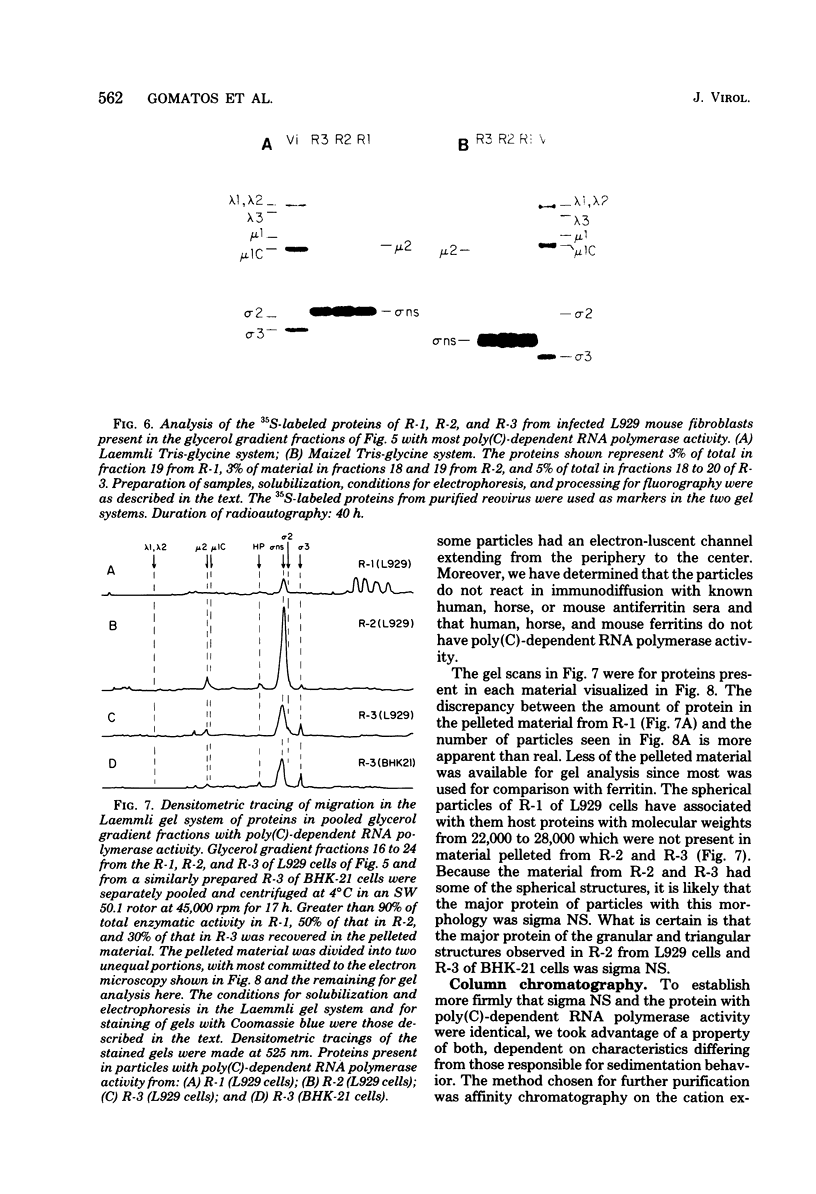
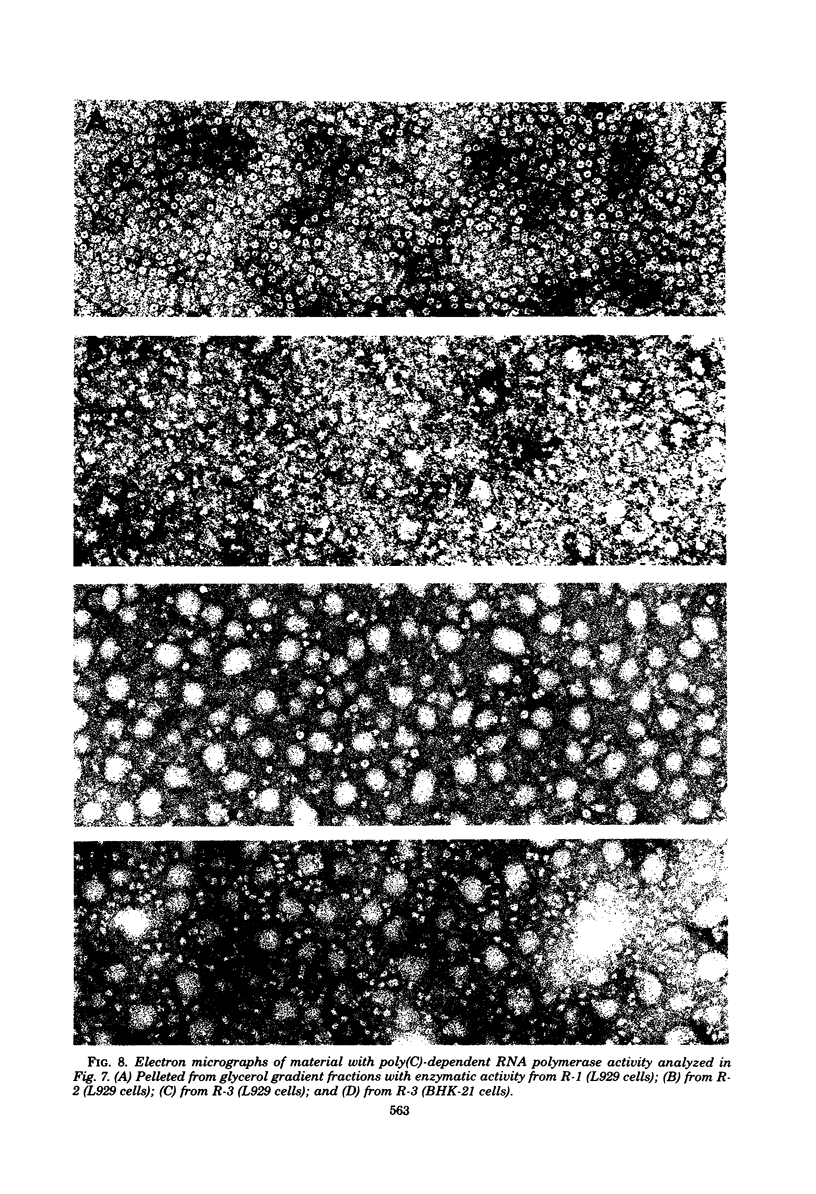
We previously reported that virus-specific particles with polycytidylate [poly(C)]-dependent RNA polymerase activity accumulated at 30 degrees C in reovirus-infected cells. These particles sedimented heterogeneously from 300 to 550S and traversed through a 40% glycerol cushion to the pellet in 3 h at 190,000 x g. In the present report, we found that smaller particles with poly(C)-dependent RNA polymerase activity remained in the glycerol cushion. These smaller, enzymatically active particles, when purified, sedimented at 15 to 1S. They were spherical or triangular with a diameter of 11 to 12 nm. They were comprised mostly, and likely solely, of one reovirus protein, sigma NS. No particles with poly(C)-dependent RNA polymerase activity were found in mock-infected cells. Chromatography on the cation exchanger, CM-Sephadex, ascertained that sigma NS was the poly(C)-dependent RNA polymerase and showed its existence in two forms. In one form, it was enzymatically active and eluted from the column at 0.5 M KCl. In the enzymatically inactive state, it did not bind to the column. Our results suggest that the enzymatically active form of sigma NS carries a greater net positive charge than the inactive form. They also suggest that both forms of sigma NS are associated with a particle which has poly(C)-dependent RNA polymerase activity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Both G. W., Lavi S., Shatkin A. J. Synthesis of all the gene products of the reovirus genome in vivo and in vitro. Cell. 1975 Feb;4(2):173–180. doi: 10.1016/0092-8674(75)90124-5. [DOI] [PubMed] [Google Scholar]
- Cross R. K., Fields B. N. Reovirus-specific polypeptides: analysis using discontinuous gel electrophoresis. J Virol. 1976 Jul;19(1):162–173. doi: 10.1128/jvi.19.1.162-173.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DULBECCO R., VOGT M. Plaque formation and isolation of pure lines with poliomyelitis viruses. J Exp Med. 1954 Feb;99(2):167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EAGLE H. Amino acid metabolism in mammalian cell cultures. Science. 1959 Aug 21;130(3373):432–437. doi: 10.1126/science.130.3373.432. [DOI] [PubMed] [Google Scholar]
- Fields B. N., Raine C. S., Baum S. G. Temperature-sensitive mutants of reovirus type 3: defects in viral maturation as studied by immunofluorescence and electron microscopy. Virology. 1971 Mar;43(3):569–578. doi: 10.1016/0042-6822(71)90282-0. [DOI] [PubMed] [Google Scholar]
- GOMATOS P. J., TAMM I., DALES S., FRANKLIN R. M. Reovirus type 3: physical characteristics and interaction with L cells. Virology. 1962 Jul;17:441–454. doi: 10.1016/0042-6822(62)90139-3. [DOI] [PubMed] [Google Scholar]
- Gomatos P. J., Kuechenthal I. Reovirus-specific enzyme(s) associated with subviral particles responds in vitro to polyribocytidylate to yield double-stranded polyribocytidylate-polyriboguanylate. J Virol. 1977 Jul;23(1):80–90. doi: 10.1128/jvi.23.1.80-90.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomatos P. J. Reovirus-specific, single-stranded RNA's synthesized in vitro with enzyme purified from reovirus-infected cells. J Mol Biol. 1968 Nov 14;37(3):423–439. doi: 10.1016/0022-2836(68)90112-5. [DOI] [PubMed] [Google Scholar]
- Huismans H., Joklik W. K. Reovirus-coded polypeptides in infected cells: isolation of two native monomeric polypeptides with affinity for single-stranded and double-stranded RNA, respectively. Virology. 1976 Apr;70(2):411–424. doi: 10.1016/0042-6822(76)90282-8. [DOI] [PubMed] [Google Scholar]
- Ito Y., Joklik W. K. Temperature-sensitive mutants of reovirus. I. Patterns of gene expression by mutants of groups C, D, and E. Virology. 1972 Oct;50(1):189–201. doi: 10.1016/0042-6822(72)90359-5. [DOI] [PubMed] [Google Scholar]
- Krug R. M., Gomatos P. J. Absence of adenine-rich ribonucleic acid from purified infectious reovirus 3. J Virol. 1969 Nov;4(5):642–650. doi: 10.1128/jvi.4.5.642-650.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lau R. Y., Van Alstyne D., Berckmans R., Graham A. F. Synthesis of reovirus-specific polypeptides in cells pretreated with cycloheximide. J Virol. 1975 Sep;16(3):470–478. doi: 10.1128/jvi.16.3.470-478.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrae M. A., Joklik W. K. The nature of the polypeptide encoded by each of the 10 double-stranded RNA segments of reovirus type 3. Virology. 1978 Sep;89(2):578–593. doi: 10.1016/0042-6822(78)90199-x. [DOI] [PubMed] [Google Scholar]
- Mustoe T. A., Ramig R. F., Sharpe A. H., Fields B. N. Genetics of reovirus: identification of the ds RNA segments encoding the polypeptides of the mu and sigma size classes. Virology. 1978 Sep;89(2):594–604. doi: 10.1016/0042-6822(78)90200-3. [DOI] [PubMed] [Google Scholar]
- Ramig R. F., Cross R. K., Fields B. N. Genome RNAs and polypeptides of reovirus serotypes 1, 2, and 3. J Virol. 1977 Jun;22(3):726–733. doi: 10.1128/jvi.22.3.726-733.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramig R. F., Mustoe T. A., Sharpe A. H., Fields B. N. A genetic map of reovirus. II. Assignment of the double-stranded RNA-negative mutant groups C, D, and E to genome segments. Virology. 1978 Apr;85(2):531–534. doi: 10.1016/0042-6822(78)90459-2. [DOI] [PubMed] [Google Scholar]
- Sarkar N. H., Moore D. H. Surface structure of mouse mammary tumor virus. Virology. 1974 Sep;61(1):38–55. doi: 10.1016/0042-6822(74)90240-2. [DOI] [PubMed] [Google Scholar]
- Spandidos D. A., Graham A. F. Nonpermissive infection of L cells by an avian reovirus: restricted transcription of the viral genome. J Virol. 1976 Sep;19(3):977–984. doi: 10.1128/jvi.19.3.977-984.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Millward S., Graham A. F. Regulation of transcription of the Reovirus genome. J Mol Biol. 1968 Aug 28;36(1):107–123. doi: 10.1016/0022-2836(68)90223-4. [DOI] [PubMed] [Google Scholar]