Abstract
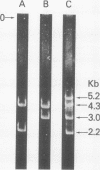
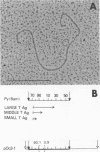
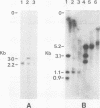
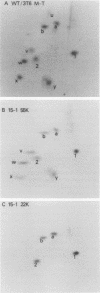
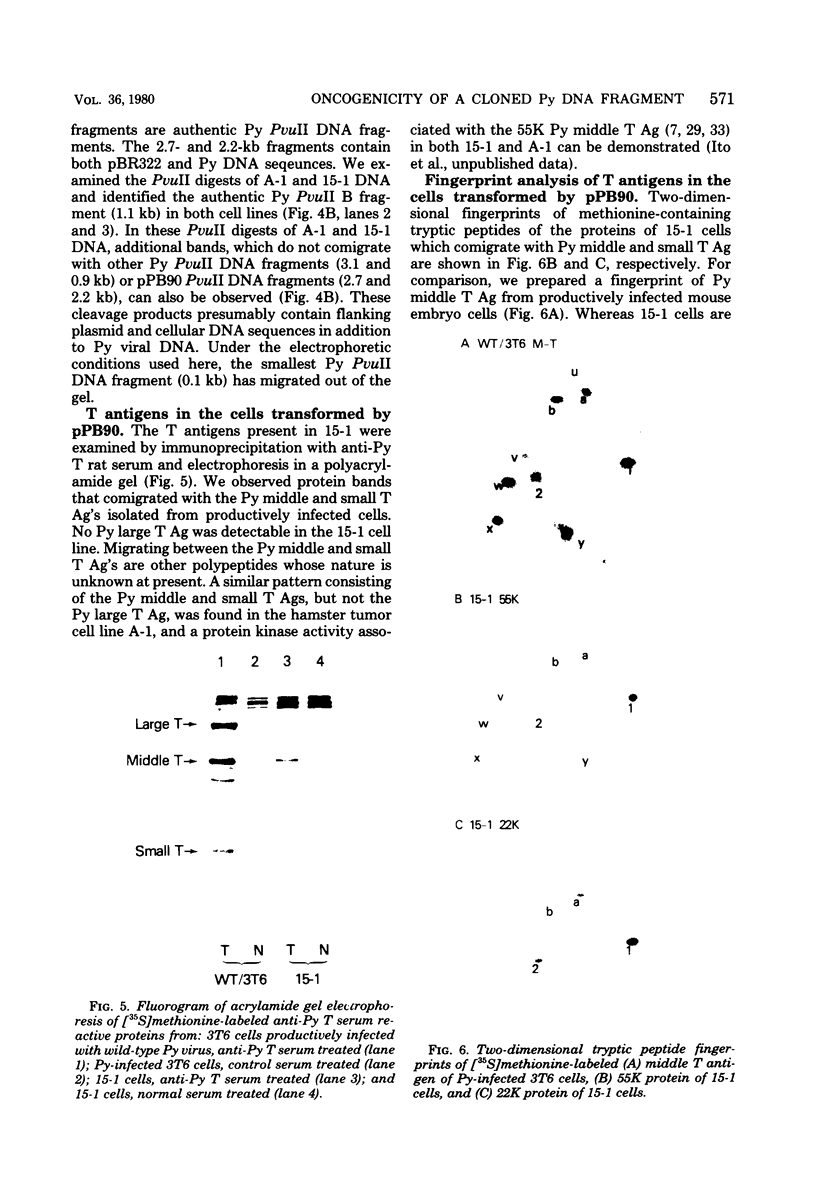
The two polyoma DNA fragments generated by cleavage with BamHI and EcoRI were cloned in pBR322, and their oncogenic potential was tested in vivo and in vitro. Only recombinant plasmid DNA containing a polyoma DNA fragment which extends clockwise from 58 to 0 map units and include approximately the 5'-proximal half of the early gene region produced tumors in newborn hamsters and transformed rat embryo cells in tissue culture. Southern blotting analysis indicated that the entire 2.2-kilobase polyoma BamHI-EcoRI fragment was intact in both a tumor cell line and a cell line transformed in culture which we examined. The presence of polyoma middle and small T antigen in these lines was demonstrated by immunoprecipitation and tryptic peptide mapping. DNA from a recombinant plasmid containing a polyoma genome deleted between 90 and 4 map units failed to induce tumors or transform cells.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bendig M. M., Thomas T., Folk W. R. Viable deletion mutant in the medium and large T-antigen-coding sequences of the polyoma virus genome. J Virol. 1980 Mar;33(3):1215–1220. doi: 10.1128/jvi.33.3.1215-1220.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin T. L. Host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1970 Sep;67(1):394–399. doi: 10.1073/pnas.67.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birg F., Dulbecco R., Fried M., Kamen R. State and organization of polyoma virus DNA sequences in transformed rat cell lines. J Virol. 1979 Feb;29(2):633–648. doi: 10.1128/jvi.29.2.633-648.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- Di Mayorca G., Callender J., Marin G., Giordano R. Temperature-sensitive mutants of polyoma virus. Virology. 1969 May;38(1):126–133. doi: 10.1016/0042-6822(69)90134-2. [DOI] [PubMed] [Google Scholar]
- Eckhart W., Hutchinson M. A., Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979 Dec;18(4):925–933. doi: 10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- Eckhart W. Properties of temperature-sensitive mutants of polyoma virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):37–40. doi: 10.1101/sqb.1974.039.01.007. [DOI] [PubMed] [Google Scholar]
- FRIED M. CELL-TRANSFORMING ABILITY OF A TEMPERATURE-SENSITIVE MUTANT OF POLYOMA VIRUS. Proc Natl Acad Sci U S A. 1965 Mar;53:486–491. doi: 10.1073/pnas.53.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann T., Esty A., LaPorte P., Deininger P. The nucleotide sequence and genome organization of the polyoma early region: extensive nucleotide and amino acid homology with SV40. Cell. 1979 Jul;17(3):715–724. doi: 10.1016/0092-8674(79)90278-2. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Maddock C. New classes of viable deletion mutants in the early region of polyoma virus. J Virol. 1979 Sep;31(3):645–656. doi: 10.1128/jvi.31.3.645-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntheroth W. G. Cardiopulmonary changes in kittens during sleep. Science. 1979 Sep 7;205(4410):1040–1041. [PubMed] [Google Scholar]
- Hassell J. A., Topp W. C., Rifkin D. B., Moreau P. E. Transformation of rat embryo fibroblasts by cloned polyoma virus DNA fragments containing only part of the early region. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3978–3982. doi: 10.1073/pnas.77.7.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hutchinson M. A., Hunter T., Eckhart W. Characterization of T antigens in polyoma-infected and transformed cells. Cell. 1978 Sep;15(1):65–77. doi: 10.1016/0092-8674(78)90083-1. [DOI] [PubMed] [Google Scholar]
- Israel M. A., Chan H. W., Hourihan S. L., Rowe W. P., Martin M. A. Biological activity of polyoma viral DNA in mice and hamsters. J Virol. 1979 Mar;29(3):990–996. doi: 10.1128/jvi.29.3.990-996.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel M. A., Chan H. W., Rowe W. P., Martin M. A. Molecular cloning of polyoma virus DNA in Escherichia coli: plasmid vector system. Science. 1979 Mar 2;203(4383):883–887. doi: 10.1126/science.217087. [DOI] [PubMed] [Google Scholar]
- Israel M. A., Chowdhury K., Ramseur J., Chandrasekaran K., Vanderryn D. F., Martin M. A. Tumorigenicity of polyoma virus in hamsters. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):591–596. doi: 10.1101/sqb.1980.044.01.061. [DOI] [PubMed] [Google Scholar]
- Israel M. A., Simmons D. T., Hourihan S. L., Rowe W. P., Martin M. A. Interrupting the early region of polyoma virus DNA enhances tumorigenicity. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3713–3716. doi: 10.1073/pnas.76.8.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel M. A., Vanderryn D. F., Meltzer M. L., Martin M. A. Characterization of polyoma viral DNA sequences in polyoma-induced hamster tumor cell lines. J Biol Chem. 1980 Apr 25;255(8):3798–3805. [PubMed] [Google Scholar]
- Ito Y. Polyoma virus-specific 55K protein isolated from plasma membrane of productively infected cells is virus-coded and important for cell transformation. Virology. 1979 Oct 15;98(1):261–266. doi: 10.1016/0042-6822(79)90545-2. [DOI] [PubMed] [Google Scholar]
- Ito Y., Spurr N., Dulbecco R. Characterization of polyoma virus T antigen. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1259–1263. doi: 10.1073/pnas.74.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Spurr N., Griffin B. E. Middle T antigen as primary inducer of full expression of the phenotype of transformation by polyoma virus. J Virol. 1980 Jul;35(1):219–232. doi: 10.1128/jvi.35.1.219-232.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Spurr N. Polyoma virus T antigens expressed in transformed cells: significance of middle T antigen in transformation. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):149–157. doi: 10.1101/sqb.1980.044.01.017. [DOI] [PubMed] [Google Scholar]
- Ketner G., Kelly T. J., Jr Integrated simian virus 40 sequences in transformed cell DNA: analysis using restriction endonucleases. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1102–1106. doi: 10.1073/pnas.73.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura G. Temperature-sensitive growth of cells transformed by ts-a mutant of polyoma virus. Nature. 1975 Feb 20;253(5493):639–641. doi: 10.1038/253639b0. [DOI] [PubMed] [Google Scholar]
- Lania L., Gandini-Attardi D., Griffiths M., Cooke B., De Cicco D., Fried M. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980 Feb;101(1):217–232. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- Magnusson G., Berg P. Construction and analysis of viable deletion mutants of polyoma virus. J Virol. 1979 Nov;32(2):523–529. doi: 10.1128/jvi.32.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. L., Chowdhury K., Martin M. A., Israel M. A. Polyoma large tumor antigen is not required for tumorigenesis mediated by viral DNA. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1336–1340. doi: 10.1073/pnas.77.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak U., Dilworth S. M., Griffin B. E. Coding capacity of a 35 percent fragment of the polyoma virus genome is sufficient to initiate and maintain cellular transformation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3278–3282. doi: 10.1073/pnas.77.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B. S., Benjamin T. L. Phosphorylation of polyoma T antigens. Cell. 1979 Dec;18(4):935–946. doi: 10.1016/0092-8674(79)90206-x. [DOI] [PubMed] [Google Scholar]
- Seif R., Cuzin F. Temperature-sensitive growth regulation in one type of transformed rat cells induced by the tsa mutant of polyoma virus. J Virol. 1977 Dec;24(3):721–728. doi: 10.1128/jvi.24.3.721-728.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons D. T., Chang C., Martin M. A. Multiple forms of polyoma virus tumor antigens from infected and transformed cells. J Virol. 1979 Mar;29(3):881–887. doi: 10.1128/jvi.29.3.881-887.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart J. E., Ito Y. Three species of polyoma virus tumor antigens share common peptides probably near the amino termini of the proteins. Cell. 1978 Dec;15(4):1427–1437. doi: 10.1016/0092-8674(78)90066-1. [DOI] [PubMed] [Google Scholar]
- Smith A. E., Smith R., Griffin B., Fried M. Protein kinase activity associated with polyoma virus middle T antigen in vitro. Cell. 1979 Dec;18(4):915–924. doi: 10.1016/0092-8674(79)90204-6. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGT M., DULBECCO R. Studies on cells rendered neoplastic by polyoma virus: the problem of the presence of virus-related materials. Virology. 1962 Jan;16:41–51. doi: 10.1016/0042-6822(62)90200-3. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978 Jul;14(3):725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]