Abstract
The genes for DNA uptake and recombination in Bacilli are commonly regulated by the transcriptional factor ComK. We have identified a ComK homologue in Bacillus coagulans, an industrial relevant organism that is recalcitrant for transformation. Introduction of B. coagulans comK gene under its own promoter region into Bacillus subtilis comK strain results in low transcriptional induction of the late competence gene comGA, but lacking bistable expression. The promoter regions of B. coagulans comK and the comGA genes are recognized in B. subtilis and expression from these promoters is activated by B. subtilis ComK. Purified ComK protein of B. coagulans showed DNA-binding ability in gel retardation assays with B. subtilis- and B. coagulans-derived probes. These experiments suggest that the function of B. coagulans ComK is similar to that of ComK of B. subtilis. When its own comK is overexpressed in B. coagulans the comGA gene expression increases 40-fold, while the expression of another late competence gene, comC is not elevated and no reproducible DNA-uptake could be observed under these conditions. Our results demonstrate that B. coagulans ComK can recognize several B. subtilis comK-responsive elements, and vice versa, but indicate that the activation of the transcription of complete sets of genes coding for a putative DNA uptake apparatus in B. coagulans might differ from that of B. subtilis.
Introduction
The ability to take-up DNA from the environment is widely spread among eubacteria, including Gram-positive and Gram-negative species [1]. It allows the exchange of genetic material, possibly contributing to the survival of bacteria under harsh growth conditions [2]–[4]. Cells that activate the expression of genes coding for a DNA uptake and recombination apparatus can benefit from foreign DNA after it recombines into the genome. Due to the need for homologous sequences for recombination it is proposed that DNA is utilized more efficiently from closely related species [5]. The induction of the competence genes has been studied in various bacteria [2], [6]. In Gram-positives, a global transcription factor or sigma factor coordinate the expression of genes required for efficient DNA uptake and recombination, the so-called late competence genes. In Streptococci, the conserved ComX sigma factor activates the late competence genes [5], while the global transcription factor ComK has been identified in various Bacilli to activate gene expression of genes related to DNA uptake [7]. As the induction of functional DNA uptake can be a useful tool for molecular biotechnological applications, numerous studies aim to better characterize the regulators involved in competence and try to achieve highly transformable strains [8]–[14].
The genes coding for DNA uptake and recombination are conserved among Bacilli [7]. The functional uptake of exogenously provided genomic DNA has been shown in various strains of B. subtilis [8], [12], [15], and also in other Bacilli, like B. licheniformis [10], [16], B. amyloliquefaciens [17], and B. cereus [11]. The regulation and function of late competence genes have been mainly studied in B. subtilis [18]. The 7 genes containing comG operon encodes a type IV pilus that facilitates DNA to pass the cell wall and reach the cell membrane [19]. The maturation of the pilin like proteins is facilitated by the ComC prepilin protease [20]. DNA is bound and transported across the membrane in a single stranded form by the ComEA protein and ComEC permease, respectively, with the aid of ComFA and NucA proteins [18]. The single-stranded DNA is then integrated via recombination by a protein complex containing among others RecA, SsbB, DprA and YjbF [21].
The late competence genes are scattered around the B. subtilis chromosome. To coordinate the expression of these genes and operons, B subtilis utilizes the global transcription factor ComK. If the protein level of ComK increases in the cells, ComK directly or indirectly activates more than 100 genes [6], [22]–[24]. ComK binds to the so-called K-boxes, that contains two AT-boxes (AAAA-N5-TTTT) separated by a spacer of a discrete number of helical turns [25]–[27]. To ensure that competence develops only under particular conditions, the expression of the comK gene and the protein level of ComK are tightly regulated. Transcription of comK is repressed by AbrB, CodY, and Rok and activated by the DegU protein [6], while the ComK protein is trapped by the adaptor protein MecA, and targeted to proteolysis by ClpCP [28]. At high cell densities, the expression of the comS gene, embedded in the srfA operon, is activated in a quorum sensing dependent manner [29]. ComS protein hijacks the MecA protein and prevents ComK degradation [28]. The increase of ComK amounts in the cells leads to a positive feedback loop and the protein level further increases. However, this enhanced level of ComK is only developed in a subpopulation of cells [30], [31]. The occurrence of two subpopulations of cells with a distinct expression state is called bistability [32] and has not been only described for competence, but also for other phenotypes of B. subtilis, like sporulation, motility, biofilm formation, and protease production [33]–[37].
In this study we characterized the function of the Bacillus coagulans ComK homologue in B. coagulans and in B. subtilis. B. coagulans is a spore forming, microaerophilic, lactic acid producing species of the Bacillus genus. It is frequently isolated as food spoilage organism [38], while its propitious features are used in probiotics [39]. It can be applied as a lactic acid production organism in biotechnological procedures and various molecular tools have been developed recently [40]–[42]. The genome sequences of several strains have been determined that facilitate genomics studies in this group of organisms [43]–[45]. As there is no study published on DNA uptake in B. coagulans, our aim was to better characterize the ComK homologue from B. coagulans DSM 1. First, we assayed the B. coagulans comK gene (denoted as comKBco) and promoter regions of comKBco and comGBco in the heterologous host, B. subtilis. In vitro studies further supported the conserved role of ComKBco as a DNA binding protein. Finally, we assayed the effect of comKBco overexpression in B. coagulans.
Results
Identification of comK Homologue in B. coagulans
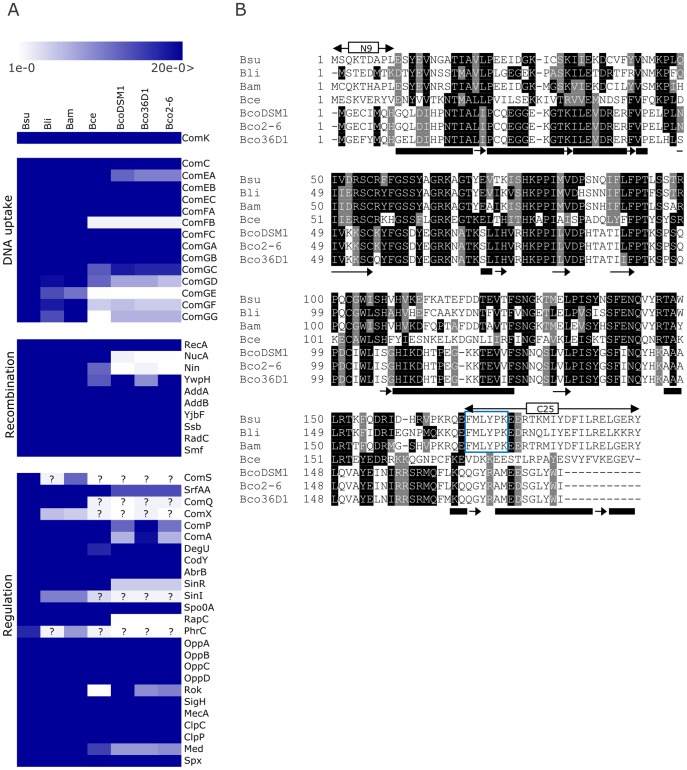
Genomic inspection of various B. coagulans strains, including DSM 1 (unpublished data), 36D1 and 2–6 showed that several genes and operons can be identified with high sequence similarity to Bacillus genes that code for the late competence genes in B. subtilis and their homologues in other Bacillus species (Fig. 1A). BLAST analyses revealed the presence of many orthologous genes putatively involved in DNA uptake and recombination, which we visualized with Genesis software. As in the case of many Bacillus species [7], the comFB gene is missing in the comF operon. While the putative ComGEFG proteins lack high similarity to the corresponding proteins of B. subtilis, the number of genes in the comG operon is conserved and the coded proteins show higher similarity to the corresponding proteins of B. cereus, where functional DNA uptake has been shown [11]. Interestingly, the genomes of all B. coagulans species also lack the nucA-nin operon that is required for the DNA cleavage during transformation in B. subtilis [46]. The absence of these genes reduces transformation efficiency to 8–15% of the wild type in B. subtilis. Still, the genomic analysis of competence genes shows that homologues of the majority of genes coding for the B. subtilis DNA-uptake and recombination machinery are present and conserved in all B. coagulans strains.
Figure 1. Survey on the presence of competence genes and the alignment of ComK protein sequences from various Bacillus strains.
(A) Results of BLAST searches were visualized with Genesis 1.6 software: white is absent (with E-value of E–0), dark blue is present (E-value<E–20). BLAST analysis was performed with B. subtilis protein sequences against translated protein database of a given genome. Protein names are indicated on the right. Bsu, B. subtilis; Bli, B. licheniformis, Bam, B. amyloliquefaciens, Bce, B. cereus Bco, B. coagulans. Question marks denote small ORFs where identification is uncertain using the available bioinformatic tools that can miss homologues. (B) Multiple alignment of ComK homologues. Black background represents conserved amino acids and grey background represents similar amino acids. Alignment was performed using ClustalW [59], and presented using Boxshade 3.21 program. The N- and C-terminal deletions analyzed by Susanna et al [47] are marked (ΔN9 and ΔC25, respectively). Boxed amino acid residues indicate the residues involved in interaction with MecA [60]. Alpha-helices and beta-sheets of B. subtilis ComK protein are indicated with rectangles and arrows under the alignment, respectively.
Homologues of the comK gene are present in all B. coagulans species. Although the comK homologue is not annotated in the complete genome of B. coagulans 2–6, a gene that codes for a putative ComK homologue can be identified between nucleotides 860419 and 860961 of the B. coagulans 2–6 chromosome (NCBI reference sequence NC_015634.1). The B. coagulans ComK homologues are somewhat shorter than the ComK protein of B. subtilis (13 aminoacids shorter compared to ComK of B. subtilis), but most regions are conserved (Fig. 1B). The C-terminal region of B. coagulans ComK proteins is truncated by 11 amino acids. Previous studies have shown that a 25–35 amino acid C-terminal truncation is incapable of transcriptional induction of comG operon [47]. The ComK proteins of B. coagulans strains have half of this C-terminal part. Interestingly, as shown in many Bacillus species, the region recognized by the adaptor protein MecA is not conserved in any of the B. coagulans species suggesting that the interaction site is different or that the ComK level is not controlled by a MecA homologue in B. coagulans species, while putative MecA homologues are present in all B. coagulans strains (Fig. 1A).
Examination of the presence of the early regulatory competence genes suggest that pleiotropic regulators (DegU, CodY, AbrB, and Spo0A) that directly or indirectly control comK transcription in B. subtilis are present in B. coagulans, while SinR and the Rap-Phr signaling systems seem to be less conserved or absent in B. coagulans (Fig. 1A). Interestingly, rok can be identified in B. coagulans, while it was previously described to be present only in the B. subtilis/amyloliquefaciens/pumilus/licheniformis group [7].
Introduction of comKBco into B. subtilis Results in Activation of Gene Expression from PcomGABsu
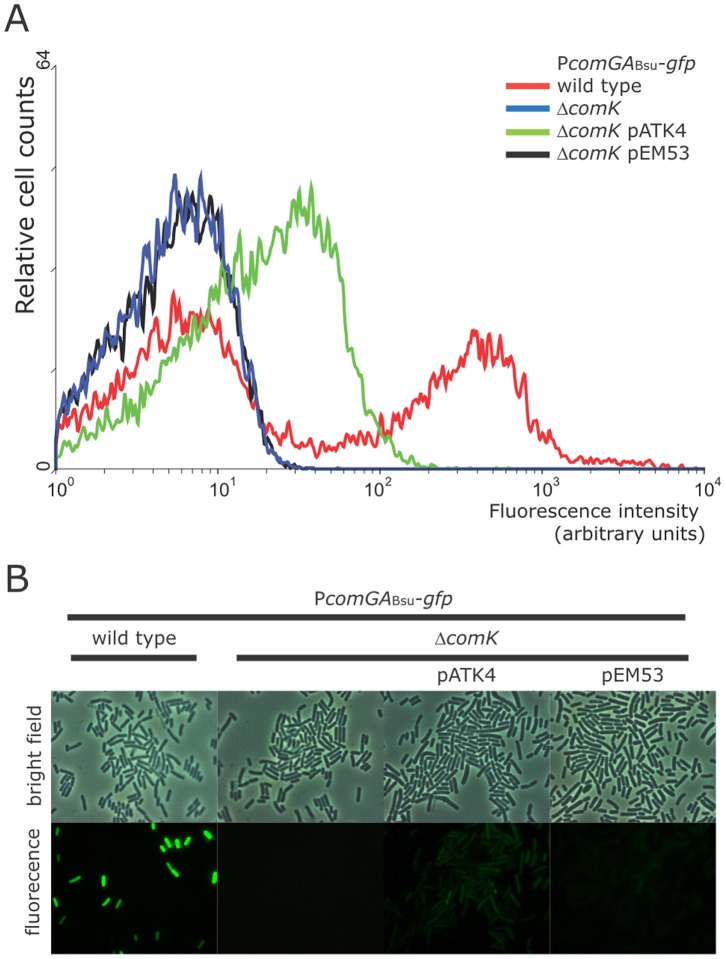
On the basis of its protein sequence analysis, the comKBco gene of B. coagulans DSM1 appears to code for another member of the ComK family. Therefore, we wanted to test if the product of the comK gene can activate transcription. For this we first introduced the comKBco gene (cloned in pATK4) into B. subtilis harboring a PcomGABsu-gfp reporter that enables us to monitor the activation of gene expression. The expression of comKBco is driven by its own promoter region. Subsequently, we deleted the endogenous comKBsu gene in this strain so we can solely monitor the effect of comKBco. In this synthetic background, the reporter activity observed depends on the presence of ComKBco. As depicted in Fig. 2, we observed reporter activity from the comGABsu promoter when the comKBco gene was introduced, but not when the empty plasmid was present. The gene expression was detected using both flow cytometry (Fig. 2A) and fluorescence microscopy (Fig. 2B). The expression from PcomGABsu was low compared to the strain in which wild type comKBsu was present, and expression of comGABsu was not bistable in contrast to the expression observed in the wild type B. subtilis strain. However, the lack of bistable gene expression of the reporter gene could also originate from a low expression level from the comKBco promoter in B. subtilis. These experiments suggest that comKBco is able to affect gene expression in Bacilli. Introduction of comKBco into B. subtilis resulted in low comGABsu expression, which suggests the lack of complete functional complementation of comKBsu deletion under the tested conditions. Accordingly, no natural transformation was observed in the complemented B. subtilis strain (data not shown).
Figure 2. Single cell analysis of PcomGABsu-gfp in the presence of comKBco in B. subtilis.
Samples were taken 2 hours after the transition point between the exponential and stationary growth phase. (A) Flow cytometric analyses of comGABsu expression in wild type (red line), ΔcomK mutant (blue line), ΔcomK strain with the comKBco containing plasmid pATK4 (green line), and ΔcomK strain with the empty plasmid (black line). The relative numbers of cells are indicated on the y axis, and their relative fluorescence levels are indicated on the x axis on a logarithmic scale. For each experiment at least 20,000 cells were analyzed. The graph is the representative of at least three independent experiments. (B) Light-microscopic phase-contrast picture (top row) and fluorescence image (bottom row) of cells. Strains used from left to right are wild type, ΔcomK mutant, ΔcomK with pATK4, and ΔcomK with pEM53, respectively.
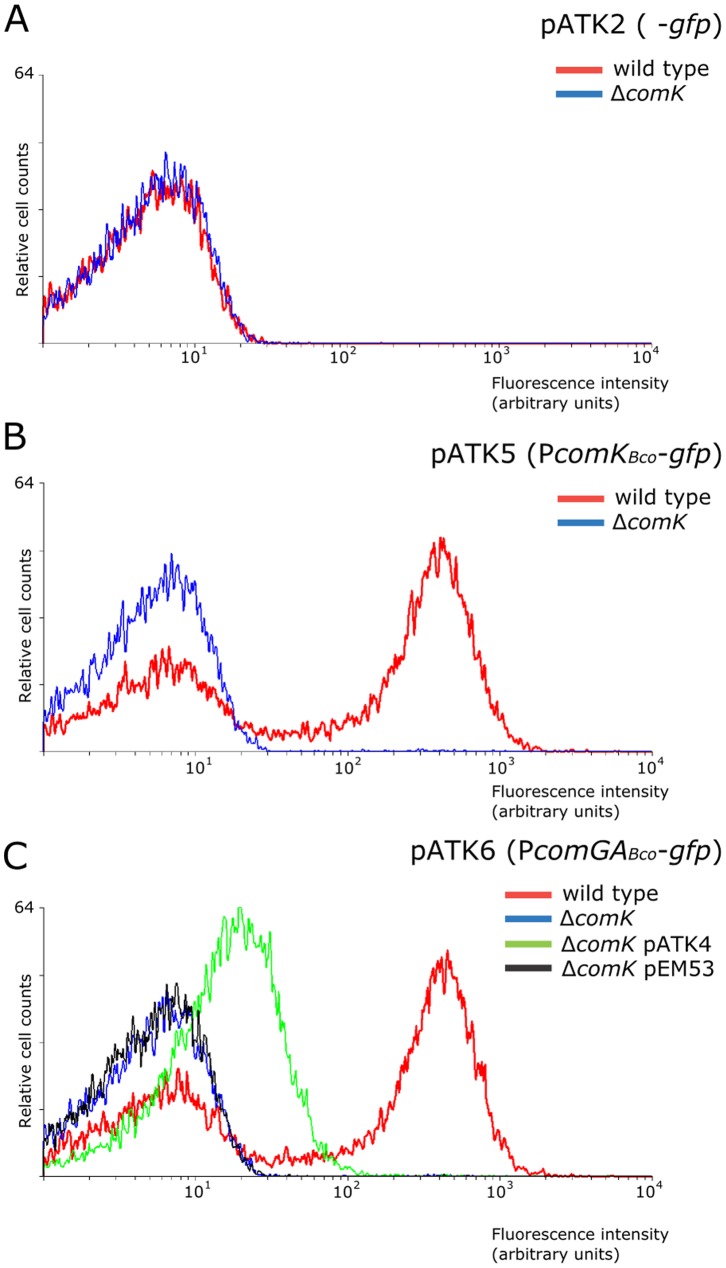
ComKBsu Activates Transcription from the Promoter Regions of comKBco and comGABco
ComKBco can activate gene expression in the heterologous host B. subtilis. The transcription activation by ComK proteins depends on the promoter sequences they bind and their relative amount, and they either activate gene expression (e.g. comGABsu promoter [26]) or relieve transcription repression (e.g. comKBsu promoter [48]). To test whether the elements of the comGABco and comKBco promoters are functionally conserved, we assayed the effect of the ComKBsu protein on these promoter fragments in the heterologous host, B. subtilis. For this we introduced the promoter-gfp constructs pATK5 and pATK6 into B. subtilis and subsequently also assayed the effect of the comKBsu mutation on the expression from these promoters. Expression of a reporter gene from both the comKBco and comGABco promoters was observed in B. subtilis (Fig. 3). This expression was dependent on the presence of the ComKBsu protein. The activation of gene expression from the introduced promoters showed a bimodal expression pattern that could originate from the bimodal level of ComKBsu protein in B. subtilis or due to use of plasmid based system to monitor gene expression. However, we can conclude that the comGABco and comKBco promoters are recognized in B. subtilis in a comKBsu-dependent manner. Introduction of comKBco (pATK4) into the ΔcomKBsu strain containing the pATK6 plasmid showed that ComKBco can activate gene expression from the promoter of comGABco (Fig. 3C, pATK6, ΔcomKBsu with pATK4) similarly to that observed for the comGABsu promoter (Fig. 2A, PcomGABsu-gfp, ΔcomKBsu with pATK4).
Figure 3. Expression from the comKBco and comGABco promoters in B. subtilis.
Single cell analysis of B. subtilis strains containing plasmids with promoter-less gfp (A), with PcomKBco-gfp fusion (B), and the PcomGABco-gfp reporter (C). Samples were taken at the indicated time points given in hours relative to the transition point between the exponential and stationary growth phase (T0). The single cell expression pattern in the wild type strain is indicated with light grey, the ΔcomK mutant is designated with dark grey, and the ΔcomK strain with the comKBco containing plasmid pATK4 is shown in white. The relative numbers of cells are indicated on the y axis, and their relative fluorescence levels are indicated on the x axis on a logarithmic scale. For each experiment at least 20,000 cells were analyzed. The graph is the representative of at least three independent experiments.
ComKBco is a DNA Binding Protein
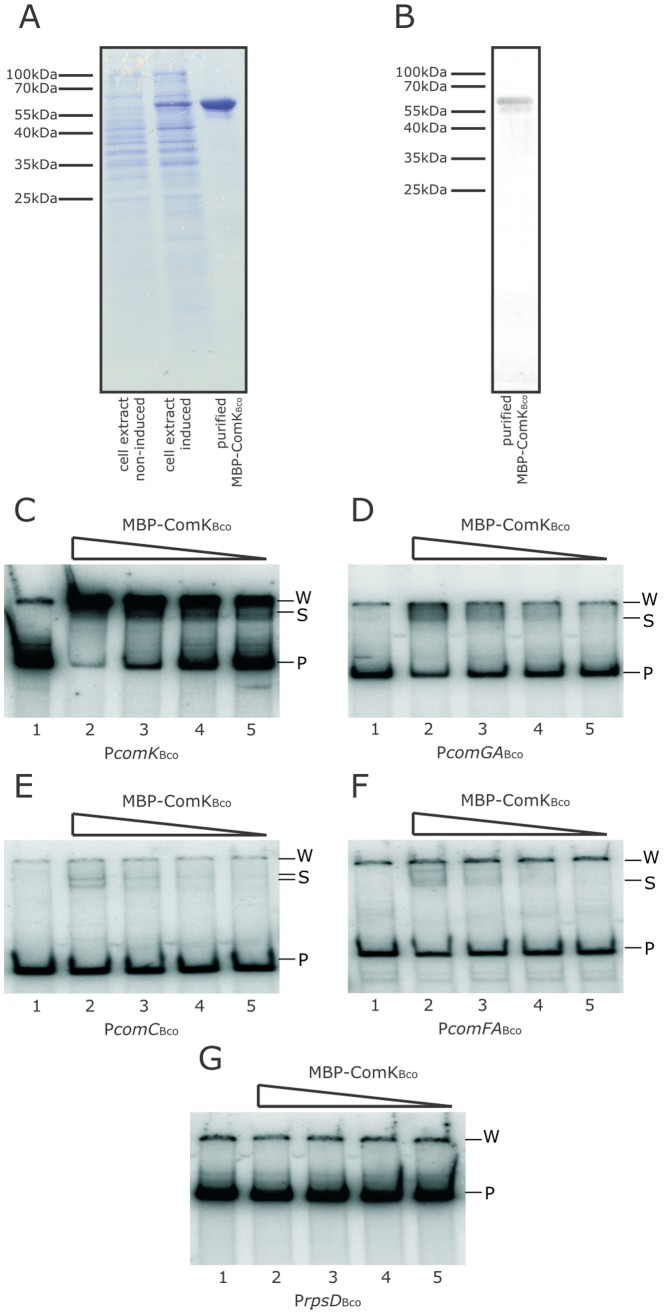
Experiments presented above show that ComKBco affects expression from the comGABsu promoter and that the comGABco and comKBco promoters are also recognized by ComKBsu. To test this in more details, we examined the in vitro DNA binding ability of ComKBco. We overexpressed a malE-comKBco fusion construct in Escherichia coli and purified the ComKBco protein with the aid of the maltose binding protein (MBP) tag (Fig. 4A). MBP fusion tag is generally used to purify DNA binding proteins and assay the in vitro DNA binding ability of target proteins. The MBP tag did not alter the binding ability of ComKBsu protein in previous studies [25]. As a control, we also obtained the MBP-ComKBsu protein using the same purification procedure. The MBP-ComKBco protein was overexpressed in Escherichia coli and purified as described in the Methods section (Fig. 4A). The integrity of the purified MBP-ComKBco protein was also verified using antibodies developed against the ComKBsu protein (Fig. 4B). A smaller protein band was copurified and recognized by the ComKBsu-antibody. The purified MBP-ComKBco clearly bound to the DNA fragment containing the comKBco and comGABco promoter regions in gel retardation assay (Fig. 4C and D). We also observed a weaker DNA binding of MBP-ComKBco to the comCBco and comFABco promoters (Fig. 4E and F). The MBP-ComKBco showed no binding towards the B. coagulans rpsD promoter fragment that is used as a non-specific control in our experiments (Fig. 4G).
Figure 4. Purification of ComKBco protein and its DNA binding ability.
(A) SDS-PAGE analysis of overexpression and purification of ComKBco protein from E. coli. Non-induced and 0.1 mmol l−1 IPTG induced cell extracts are loaded on the first and second lanes, respectively, while purified MBP-ComKBco is run in the third lane. (B) Immunoblot analysis of the purified MBP-ComKBco protein using α-ComKBsu antibodies. Marker sizes are indicated on the left of the blot. (C–G) Gel retardation assay with the purified MBP-ComKBco protein on the comKBco (C), comGABco (D), comCBco (E), and comFABco (F) promoter fragments. Lane 1 contains no protein, lanes 2–5 contain 2.2 µmol l−1 to 275 nmol l−1 purified MBP-ComKBco at 2 fold dilutions, respectively. The promoter fragment of rpsDBco is used as negative control with no apparent binding by MBP-ComKBco (G). Free probes (P), shifted bands (S), and signal specific to the wells of the gel (W) are indicated.
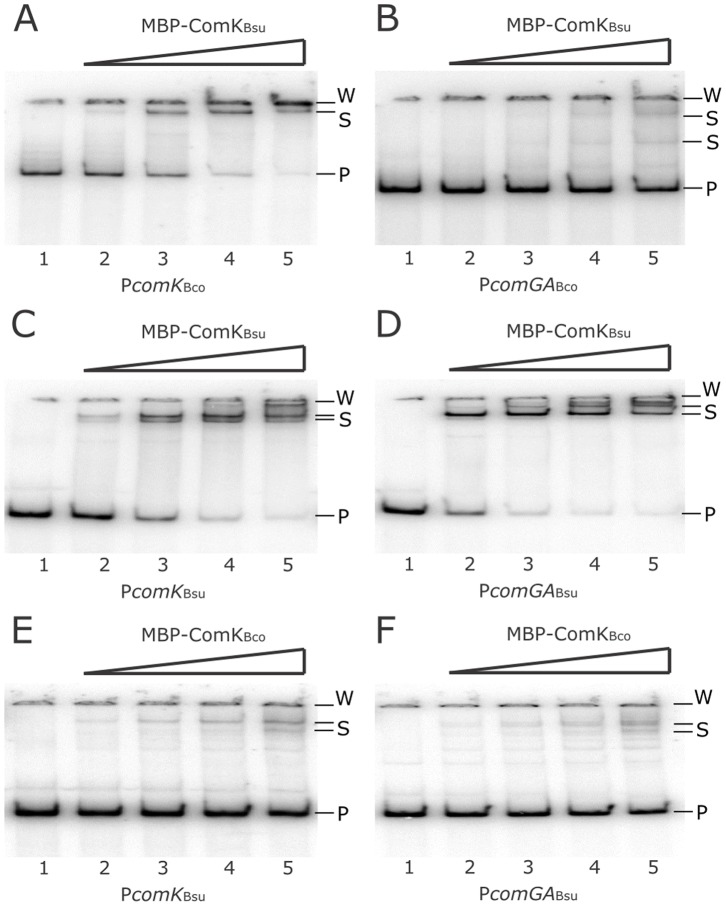
Experiments performed in B. subtilis suggest that the ComK proteins can activate gene expression on the heterologous comGA promoters (see above). To examine if this effect of ComK proteins is achieved by direct binding and transcription activation, we examined the in vitro binding of various ComK proteins on the B. coagulans and B. subtilis promoters of the comGA and comK genes. Results depicted in Fig. 5 show that the ComK proteins bind to the heterologous promoters, although the affinity of the ComK proteins was different in the case of different promoters. As our gel retardation experiments were not controlled in competition experiments with a cold probe, we can only judge the presence of DNA binding, but no indisputable conclusion can be drawn on the affinity differences. However, the binding of ComK proteins of B. coagulans and B. subtilis to the heterologous promoter fragments is in agreement with the in vivo experiments done in B. subtilis. Taken together the in vivo and in vitro experiments all suggest that B. coagulans possesses a functional ComK homologue that is presumably able to activate the transcription of several late competence genes in B. coagulans (see also below).
Figure 5. Gel retardation assay with ComKBsu and ComKBco.
The binding of MBP-ComKBsu (A–D) and MBP-ComKBco (E–F) was assayed at a doubling concentration of the proteins from 125 nmol l−1 to 1 µmol l−1 (lanes 2 to 5, respectively). Lane 1 of each picture lacks any added protein. DNA binding was detected on promoters of comKBco (A), comGABco (B), comKBsu (C and E), and comGABsu (D and F) genes. Free probes (P), shifted bands (S), and signal specific to the wells of the gel (W) are indicated.
ComKBsu activates transcription by binding K-boxes that are composed of two AT-boxes with a consensus sequence AAAA-N5-TTTT. The boxes are separated of a discrete number of helical turns (8-, 18- or 31-bp between the two AT-boxes), which places them on the same side of the DNA-helix [25]–[27]. The analysis of the promoter region of putative competence related genes in B. coagulans showed the presence of several AT-boxes (allowing maximum 3 mismatches to the consensus AT-box), however, K-boxes could be only found in the promoter regions of comKBco and comCBco (Figure S1). Interestingly, the promoter regions of comCBco, comEABco and comFABco contain an overrepresented GCC-N8-TGC motif (identified 1, 2, and 3 times, respectively). This motif is not found within the promoter regions of the comGABco and comKBco genes. However, due to the low number of analyzed promoters, we cannot conclude whether the K-boxes or this latter overrepresented motif are functional in B. coagulans and their role requires additional functional characterization.
Overexpression of comKBco in B. coagulans Results in Elevated comGABco Expression
In our final experiments, we assayed the effect of comKBco overexpression in B. coagulans DSM 1. For this, we cloned the comKBco gene under control of the IPTG (isopropyl-β-d-thiogalactopyranoside) inducible spac promoter, resulting in plasmid pATK10. We introduced this construct into B. coagulans DSM 1 by electroporation and assayed whether the level of ComK protein is enhanced in B. coagulans containing pATK10 upon induction. An increased level of ComK protein was detected in Western blot analysis using antibodies against ComKBsu (Figure S2). Next, we monitored the expression of late competence genes using quantitative RT-PCR. As expected, the expression level of comGABco gene was increased (ratio of 30.5±3.7) in the strain where comKBco expression was induced with 1 mM IPTG compared to the wild type strain that lacks the plasmid. However, the expression level of another late competence gene (i.e. comCBco) showed no significant change (ratio of 0.85±0.3) in the comKBco-overexpression strain compared to the plasmid-free strain under this given condition. Other late competence genes (comEA-CBco and comFACBco) also lacked the increased expression in the comKBco overexpression strain (data not shown). Overexpression of the comKBsu gene using the previously published pNWcomKBsu plasmid [11] resulted in slightly increased comGABco expression (ratio of 3.2±1.2) and unaltered comCBco transcription (ratio of 1.3±0.5) compared to the plasmid free wild type strain. These experiments demonstrate that ComKBco can activate gene expression in B. coagulans in line with previous observations presented above.
To test if the increased expression of comGABco by comKBco overexpression is sufficient to observe a functional DNA uptake in B. coagulans, we tested the uptake of genomic DNA (e.g. chromosomal DNA of DSM1 ΔsigF::Cmr described in [40]) or plasmid DNA (e.g. pNW33N). The expression of comKBco was induced at mid-exponential phase and DNA was supplied at different time points (1–4 hours) after induction. Cells were plated on medium containing chloramphenicol. We could not observe reproducible DNA uptake under the above presented comKBco overexpressing conditions in B. coagulans, suggesting the lack of a fully functional DNA uptake machinery under these specific conditions. Similarly, DNA uptake was not detected in B. coagulans when comKBsu was overexpressed, in contrast to the experiments with B. cereus [11]. Since an increased level of ComKBco is detected by Western blot analysis when comKBco is overexpressed in B. coagulans (Figure S2) and the comGABco gene expression was induced roughly 30 times, it may be that the resulting level of ComKBco is not high enough to activate the whole DNA-uptake and recombination apparatus.
Discussion
Genetic engineering of microorganisms allows to improve them or introduce alternative biochemical reactions and thereby to develop improved or novel strains or products. However, genetic engineering can be time consuming for recalcitrant bacteria. The use of competence for DNA uptake and recombination improves the engineering process by allowing or enhancing genetic accessibility. Competence has been described for many laboratory type strains of Bacilli [15]–[17] [12]. The genes coding for functional DNA uptake and recombination are widely conserved in Bacilli suggesting that natural competence exists in more species than described before [7]. However, highly efficient DNA uptake is not identified under laboratory conditions in many species. Different strains of the same species might also differ in their degrees of competence. Natural isolates of B. subtilis show a low DNA uptake efficiency that can be improved by induction of the late competence genes through overexpression of the comK gene [12].
In this study, we present the genomic conservation of genes coding for putative homologues for DNA uptake and the recombination apparatus in B. coagulans. Further characterization of the comK homologue in B. coagulans DSM 1 indicates that comKBco codes for a DNA binding transcriptional activator. Introduction of comKBco into a synthetic B. subtilis background that lacks its own comKBsu gene results in gene expression activation from the promoter regions of comG operons of B. subtilis and B. coagulans. These experiments clearly suggest a conserved role of ComK homologues in Bacilli, although the set of target genes might vary. This is also supported by the induction of functional DNA uptake in B. cereus by the ComKBsu protein [11]. However, overexpression of either or both comK genes of B. cereus into B. subtilis does not result in a similar induction of comG expression (unpublished observation, AM Mironczuk and ÁT Kovács). Previous studies on the binding site of ComKBsu described K-boxes, where the distance between the two AT-boxes is important for its function [25], [26]. While AT-boxes can be identified in several promoter regions of late competence genes in various Bacilli, properly spaced K-boxes are found only in the promoter regions of comKBco and comCBco genes. In contrast with these in silico observations, purified ComKBsu protein binds in vitro to the promoter regions of late competence genes of B. coagulans (Fig. 5) and B. cereus [11] and overexpression of comKBsu results in enhanced comG expression in vivo (RT-qPCR results and [11]). This suggests that the recognition and transcriptional activation by ComK proteins might not be so stringent in the heterologous hosts. Alternatively, ComK proteins of B. coagulans and B. cereus could act on deviating binding sites or their effect is indirect on the comG promoter.
Overexpression of various comK genes in different Bacilli results in increased transcription from the comG promoter. In the present and previous studies, we used the fusion between the comG promoter and the reporter gene gfp, for general use of comGBsu expression as a reporter of activation of competence in B. subtilis [30], [31], [49]. However, microarray analysis and RT-qPCR experiments in B. cereus showed that while expression of comK genes increases comG transcription, the transcript levels of other late competence genes are not induced equally [11]. In B. coagulans, when the ComKBco protein level is increased to a certain level that results in roughly 30 times induction of comGABco, the expression of comCBco is not changed. We can hypothesize that the produced ComK protein level is not high enough to activate gene expression from these promoters or one or more additional regulatory mechanisms act on the late competence genes. In vitro transcription assays using these promoter regions and purified ComK protein could show us whether this is the case.
While overexpression of comK genes in B. coagulans results in increased comG expression similar to the experiments in B. cereus [11], we did not detect functional uptake of DNA under these conditions. Our survey on the presence of late competence genes in B. coagulans also points to the absence of genes that are required for high efficiency DNA uptake in B. subtilis (e.g. nucA-nin genes). However, our study clearly shows that ComKBco is a DNA-binding protein that is capable of activating gene expression. Therefore, it presents an important element of future research for better understanding of late competence gene induction in B. coagulans.
Methods
Bacterial Strains, Growth Conditions and Transformation
The strains and plasmids used in this study are listed in Table 1. B. coagulans strains were grown in BC medium at 50°C, 120 rpm [40]. BC medium contains per liter: 10 g yeast-extract (Difco), 2 g (NH4)2HPO4, 3.5 g (NH4)2SO4, 10 g Bis-Tris (bis[2-hydroxymethyl]iminotris[hydroxymethyl]-methane), 5 mg MgCl2 · 6 H2O, 3 mg CaCl2 · 2 H2O, 1 ml of filter sterilized trace elements (containing per liter 0.05 g ZnCl2, 0.03 g MnCl2 · 4 H2O, 0.3 g H3BO3, 0.2 g CoCl2 · 6 H2O, 0.01 g CuCl2 · 2 H2O, 0.02 g NiSO4 · 6 H2O, and 0.03 g Na2MoO4 · 2 H2O), pH 6.7. B. subtilis strains were grown in minimal medium [15]. For cloning, Escherichia coli DH5α and Lactococcus lactis MG1363 were grown in TY and GM17 (37.5 g M17 broth (Difco) per liter supplemented with 0.5% glucose) medium, respectively, grown at 30°C or 37°C. Antibiotics were used at a concentration of 5 µg ml−1 for chloramphenicol, 6 µg ml−1 for tetracycline, and 100 µg ml−1 for ampicillin. Transformation of L. lactis and B. coagulans was performed by electroporation as previously described [40], [50]. Transformation of E. coli was performed by heat-shock [51]. DNA was introduced into B. subtilis strains using natural competence [52].
Table 1. Strains, plasmids used in this study.
| Properties | Reference | |
| Strain | ||
| B. coagulans DSM 1 | wild type strain | DSMZ collection |
| B. subtilis 168 | wild type strain | laboratory strain |
| B. subtilis ΔcomK | comK::Kmr mutant | [30] |
| B. subtilis PcomG-gfp | PcomG-gfp fusion in B. subtilis 168 strain (Cmr) | [30] |
| L. lactis MG1363 | lac− prt−; plasmid-free derivative of NCDO712 | [61] |
| E. coli DH5α | endA1 hsdR17 supE44 thi-1 λ − recA1 gyrA96 relA1 ΔlacU169 (φ80dlacZΔM15) | Bethesda Research Laboratories |
| Plasmids | ||
| pNW33N | 4.2 kb, Cmr, Geobacillus-E. coli shuttle vector | Bacillus Genetic Stock Centre |
| pEM53 | 5.6 kb, Tcr, pNZ124-based cloning vector | [40] |
| pDG148 | 8.3 kb, Ampr, Kmr,Pspac, lacI integration vector | [62] |
| pMALc2X | 6.6 kb, Ampr, overexpression vector for MalE fusion | New England Biolabs |
| pSG1151 | 4.6 kb, Ampr, Cmr, gfpmut1 harboring plasmid | [63] |
| pATK2 | 5.0 kb, Cmr, gfpmut1 cloned into pNW33N | This study |
| pATK4 | 4.9 kb, Tcr, comKBco gene and promoter region in pEM53 | This study |
| pATK8 | 4.5 kb, Tcr, Pspac-comKBco in pEM53 | This study |
| pATK10 | 5.8 kb, Tcr, Pspac-comKBco overexpression construct and lacI in pEM53 | This study |
| pATK5 | 5.6 kb, Cmr, PcomGABco-gfp fusion | This study |
| pATK6 | 5.5 kb, Cmr, PcomKBco-gfp fusion | This study |
| pMALcomKBco | 7.3 kb, Ampr, MAL-ComK overproduction vector | This study |
CmR, chloramphenicol resistant;
TcR, tetracycline resistant,
KmR, kanamycine resistant,
AmpR, ampicillin resistant.
Cloning of comK Bco Gene
To facilitate the purification of MBP-ComKBco, the comKBco gene was PCR amplified from the genome of B. coagulans DSM 1 using oligos oATK26 and oATK14 (for the sequences of oligos, see Table 2) containing BamHI and SalI sites, respectively. The construct pMALcomK was created by ligating the BamHI and SalI digested PCR into the corresponding site of the pMAL-c2 (New England Biolabs). The comK Bco gene harboring its own promoter was PCR amplified with oligos oATK1 and oATK2, and cloned into the ScaI site of pEM53 vector, resulting pATK4. The cloned fragment contains the comKBco gene and the 732 bp upstream region. Vector pEM53 is derived from pNZ124 by replacing the chloramphenicol resistance gene cat by the tetracycline resistance gene tetK amplified from pGhost8::ISS1 [40]. To overexpress comKBco in B. coagulans, the comKBco gene was cloned after the Pspac promoter. For this, the comKBco containing PCR fragment was obtained with oATK13 and oATK14 oligonucleotides (Table 2), digested with HindIII-SalI enzymes, and ligated together with the Pspac containing EcoRI-HindIII fragment from pDG148 and the EcoRI-XhoI digested vector, resulting in pATK8. Subsequently, the lacI gene was introduced from pDG148 (1294 bp BamHI-SwaI fragment) into the BamHI-ScaI digested pATK8 vector, resulting pATK10. The resulting vectors were validated using restriction analysis and inserts were verified by sequencing.
Table 2. Oligonucleotides used in this study.
| Oligo name | target | Sequence (5′ - 3′) | Restriction site |
| oATK1 | comKBco | GGACCGTTACGCCGTAGAGA | |
| oATK2 | comKBco | GGACTTGCAGTTCGCAATGT | |
| oATK5 | comKBco | GGTACCTCCGCATGCTGGAAGAAT | KpnI |
| oATK6 | comKBco | GGGCCCCAATTGCCCATGTTGCATAA | ApaI |
| oATK7 | comGABco | GGTACCTTCCTGGACGGATACTTC | KpnI |
| oATK8 | comGABco | GGGCCCTTCTACCGACATAATCCATC | ApaI |
| oATK13 | comKBco | GCAAAGCTTAGAGAGTGGATCATGAGATA | HindIII |
| oATK14 | comKBco | CAGGTCGACGGACTTGCAGTTCGCAATGT | SalI |
| oATK16 | rpsDBco | GGGTACCAATCCAGTAAACGGGACTTAT | KpnI |
| oATK17 | rpsDBco | GGGGCCCTTTCCAGCTTGGACCTGTAT | ApaI |
| oATK26 | comKBco | TGGGATCCATGGGGGAATGCATTATGCAA | BamHI |
| oATK48 | comCBco | ACGGGGCCCCGCAAAATAAGCTGTCCATA | KpnI |
| oATK49 | comCBco | ACGGGTACCATTTGCCGGAAATCGACGTG | ApaI |
| oATK50 | comFABco | ACGGGGCCCCTGTTCGGAGAAAACAGAAG | KpnI |
| oATK51 | comFABco | ACGGGTACCTGCCTGGATGCTGAAATAAG | ApaI |
| oATK83 | rpiABco | AATAGCAGACTTGAACGACAC | |
| oATK84 | rpiABco | CACCAAATGCTTGTATCCGA | |
| oATK87 | comGABco | AAGCAGGCATTACTTATAGCAC | |
| oATK88 | comGABco | GGACAACGCAATGTAATCAG | |
| oATK89 | comCBco | CCTCCTCTATCTCATTGCCT | |
| oATK90 | comCBco | GAAACGCAAATACATCCCGA | |
| comG1 | comGABsu | CCGGAATTCATGGTGACCATGTCTGCT | |
| comG2 | comGABsu | CGCGGATCCCTCTCCTTTCAACGC | |
| pK-F | comKBsu | AATCTATCGACATATCCTGCAA | |
| KFPr | comKBsu | GGAATTCTTGCGCCGTTCACTTCATAC |
Construction of Promoter-gfp Reporter Plasmids
The gfp gene was first obtained from pSG1151 using KpnI-XbaI restriction enzymes and ligated into the corresponding sites of the broad host range pNW33N vector, resulting pATK2. pATK2 was digested with KpnI and ApaI and used to ligate the promoter fragments of comKBco and comGABco obtained with PCR reaction using oligonucleotides oATK5 and oATK6 (for comKBco) and oATK7 and oATK8 (for comGBco) and digested with the same restriction enzyme pairs. The integrity of cloned fragments was verified by sequencing.
Protein Overexpression and Purification
1 liter culture of cells containing the pMALcomKBco or pMALcomK [53] was grown for 2 hours at 37°C and induced with 0.1 mmol l−1 of IPTG at 0.8 of OD600. Cells were harvested by centrifugation (10 min, 4°C, 6500×g). Pellets were washed with a buffer containing 1.17% NaCl, 25 mmol l−1 EDTA, 10 mmol l−1 NaN3, 0.15% DTT, 50 mmol l−1 Tris-HCl pH 7.4. Cells were lysed by sonification (15×10 s at 10 kHz with 30 s intervals), and the sonicated fractions were centrifuged (20 min, 4°C, 9000×g) to obtain a supernatant that contains the MBP-ComK. The fraction with the MBP-ComK has been loaded on an amylose column which had been equilibrated with a buffer containing 0.5 mM DTT, 20 mM Tris-HCl pH 8.0. Elution was performed with the same buffer, now containing 10 mM maltose. The fractions were stored immediately at 4°C (after analysis the fractions were pooled and stored at −80°C). The purity of MBP-ComKBco and MBP-ComKBsu was verified on SDS-PAGE and the purified proteins were also validated using Western hybridization with antibody raised against ComKBsu as described in [53]. Sample preparation and Western hybridization on the B. coagulans samples were performed as described previously for B. cereus [11]. B. coagulans wild-type and comKBco overexpression strains were grown in BC medium until 0.8 of OD600 and induced with 0.1 mmol l−1 IPTG. Three hours after induction, samples were harvested by centrifugation (10.397×g, 1 min, 4°C), disrupted using lysozyme treatment. The ComKBco protein level was detected after SDS-PAGE using Western hybridization with ComKBsu– specific antibody [53].
Gel Retardation Assay
Gel retardation assays were carried out essentially as described by Susanna et al. [26]. The promoter regions of B. coagulans putative competence genes comK, comG, comC and comFA were obtained by PCR using oligos oATK5-oATK6, oATK7-oATK8, oATK48-oATK49, and oATK50-oATK51, respectively (Table 2). The B. subtilis comK and comG promoter fragments were obtained using oligos pK-F – KFPr [54] and comG1-comG2 [26], respectively. The B. coagulans rpsD promoter region was used as negative control. The resulting fragments were end-labeled with [γ-33P] ATP using T4 polynucleotide kinase (Roche Nederland B.V., The Netherlands). Purified MBP-ComKBco and MBP-ComKBsu proteins and probes were premixed on ice in binding buffer. Reaction mixtures contained poly (dI-dC) that is known to eliminate non-specific DNA binding of ComK [25]. Samples were incubated at 30°C, and were loaded on a 6% polyacrylamide gel after 20 min incubation. Gels were run in 1× TBE buffer (0,089 mmol l−1 Tris, 0,089 mmol l−1 Boric Acid, 0,022 mmol l−1 EDTA) at 90 V for 60 minutes, dried in a vacuum dryer and autoradiographed using phosphoscreens and a Cyclone PhosphorImager (Packard Instruments, Meridien, CT).
Quantitative RT-PCR
B. coagulans wild type and comKBco overexpression strains were grown in BC medium until 0.8 of OD600 and induced with 0.1 mmol l−1 IPTG. Two hours after induction, samples were harvested by centrifugation (10.397×g, 1 min, 4°C). A total of three independent biological replicates were included. RNA preparation of quantitative PCR was performed as described before [55], [56]. The pellets were immediately frozen in liquid nitrogen and stored at −80°C. RNA extraction was performed with the Macaloid/Roche protocol [57]. Samples were treated with RNase-free DNase I (Fermentas, St. Leon-Rot, Germany) for 60 min at 37°C in DNaseI buffer (10 mmol l−1 Tris·HCl (pH 7.5), 2.5 mmol l−1 MgCl2, 0.1 mmol l−1 CaCl2), and re-purified with the Roche RNA isolation Kit. RNA concentration and purity was assessed using NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific). Reverse transcription was performed with 50 pmol random nonamers on 4 µg of total RNA using RevertAid™ H Minus M-MuLV Reverse Transcriptase (Fermentas, St. Leon-Rot, Germany). Quantification of cDNA was performed on an CFX96 Real-Time PCR System (BioRad, Hercules, CA) using Maxima SYBR Green qPCR Master Mix (Fermentas, St. Leon-Rot, Germany). The following oligos were used: for comGA, oATK87 and oATK88, for comC, oATK89 and oATK90 and for rpiA gene of B. coagulans, oATK83 and oATK84 (oligo sequences are listed in Table 2). The amount of comGA and comC cDNA levels was normalized to the level of rpiA cDNA using the 2−ΔΔCt method [58].
Flow Cytometric Analyses and Microscopy
B. subtilis wild type and ΔcomK strains carrying either pATK5 or pATK6 were grown ON in minimal medium supplemented with chloramphenicol (5 µg ml−1). For the flow cytometric analyses, cultures were inoculated into fresh minimal medium. Samples were taken after transition to stationer phase every hour. Cells were diluted 10 fold in minimal salts and analyzed on a Coulter Epics XL-MCL flow cytometer (Beckman Coulter Mijdrecht, NL) operating an argon laser at 488 nm. Green fluorescent protein (GFP) signals were collected through an FITC filter with the photomultiplier voltage set between 700 and 800 V. Date were obtained using EXPO32 software (Beckman Coulter) and further analyzed using WinMDI 2.8 (The Scripps Research Institute). Figures were prepared using WinMDI 2.8 and Adobe CS4 Illustrator.
The fluorescence of the GFP reporter protein was visualized with a Zeiss Axiophot microscope, using filter set 09 (excitation, 450 to 490; emission, >520 nm). Imaging of PcomGABsu-gfp in individual cells using fluorescence microscopy was performed as described by Smits et al. [30] using AxioVs20 software (Zeiss) for image capturing and figures were prepared for publication using Adobe CS4 Illustrator.
Nucleotide Sequence Accession Numbers
Sequences used in this study have been deposited in GenBank under accession numbers JX518619 (comKBco), JX518620 (comGABco), JX518621 (comCBco), JX518622 (comFABco), JX518623 (rpsDBco), JX518624 (rpiABco).
Supporting Information
A. Schematic presentation of the promoter region of putative competence related genes. Filled boxes indicate putative AT-boxes (maximum 3 mismatches to the consensus AAAA-N5-TTTT), open boxes indicate upstream open reading frames and com genes, numbers denote spacing between AT-boxes resulting in a so called K-box (8 bp and 31 bp in the case of comKBco and comCBco, respectively). B. Sequences of B. coagulans DSM1 promoter regions related to competence. Bold letters indicate putative AT-boxes. The putative open reading frames, com genes are indicated below the sequence.
(PDF)
Detection of the ComKBco protein. Equal amounts of proteins were loaded in each lane, Samples were taken from induced (+) or non-induced (−) cultures. Cells were centrifuged, lysed and analysed by Western blotting using ComKBsu-specific antibodies. The arrows indicate ComK specific signal (K) and non-specific signal (NP). Positions of molecular weight marker bands are indicated on the right side of the gel.
(TIF)
Funding Statement
Part of this work was financially supported by a SenterNovem subsidy IS044081. This project was carried out within the research programme of the Kluyver Centre for Genomics of Industrial Fermentation which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lorenz MG, Wackernagel W (1994) Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev 58(3): 563–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Claverys JP, Prudhomme M, Martin B (2006) Induction of competence regulons as a general response to stress in Gram-positive bacteria. Annu Rev Microbiol 60: 451–475. [DOI] [PubMed] [Google Scholar]
- 3. Finkel SE, Kolter R (2001) DNA as a nutrient: Novel role for bacterial competence gene homologs. J Bacteriol 183(21): 6288–6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palchevskiy V, Finkel SE (2006) Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J Bacteriol 188(11): 3902–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martin B, Quentin Y, Fichant G, Claverys JP (2006) Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol 14(8): 339–345. [DOI] [PubMed] [Google Scholar]
- 6. Hamoen LW, Venema G, Kuipers OP (2003) Controlling competence in Bacillus subtilis: Shared use of regulators. Microbiology 149(Pt 1): 9–17. [DOI] [PubMed] [Google Scholar]
- 7. Kovacs AT, Smits WK, Mironczuk AM, Kuipers OP (2009) Ubiquitous late competence genes in Bacillus species indicate the presence of functional DNA uptake machineries. Environ Microbiol 11(8): 1911–1922. [DOI] [PubMed] [Google Scholar]
- 8. Ashikaga S, Nanamiya H, Ohashi Y, Kawamura F (2000) Natural genetic competence in Bacillus subtilis Natto OK2. J Bacteriol 182(9): 2411–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blomqvist T, Steinmoen H, Havarstein LS (2006) Natural genetic transformation: A novel tool for efficient genetic engineering of the dairy bacterium Streptococcus thermophilus . Appl Environ Microbiol 72(10): 6751–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoffmann K, Wollherr A, Larsen M, Rachinger M, Liesegang H, et al. (2010) Facilitation of direct conditional knockout of essential genes in Bacillus licheniformis DSM13 by comparative genetic analysis and manipulation of genetic competence. Appl Environ Microbiol 76(15): 5046–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mironczuk AM, Kovacs AT, Kuipers OP (2008) Induction of natural competence in Bacillus cereus ATCC14579. Microb Biotechnol 1(3): 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nijland R, Burgess JG, Errington J, Veening JW (2010) Transformation of environmental Bacillus subtilis isolates by transiently inducing genetic competence. PLoS One 5(3): e9724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Woodbury RL, Wang X, Moran CP Jr (2006) Sigma X induces competence gene expression in Streptococcus pyogenes . Res Microbiol 157(9): 851–856. [DOI] [PubMed] [Google Scholar]
- 14. Wydau S, Dervyn R, Anba J, Dusko Ehrlich S, Maguin E (2006) Conservation of key elements of natural competence in Lactococcus lactis ssp. FEMS Microbiol Lett 257(1): 32–42. [DOI] [PubMed] [Google Scholar]
- 15. Venema G, Pritchard RH, Venema-Schroeder T (1965) Fate of transforming deoxyribonucleic acid in Bacillus subtilis . J Bacteriol 89: 1250–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thorne CB, Stull HB (1966) Factors affecting transformation of Bacillus licheniformis . J Bacteriol 91(3): 1012–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, et al. (2004) Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol 186(4): 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen I, Dubnau D (2004) DNA uptake during bacterial transformation. Nat Rev Microbiol 2(3): 241–249. [DOI] [PubMed] [Google Scholar]
- 19. Chen I, Provvedi R, Dubnau D (2006) A macromolecular complex formed by a pilin-like protein in competent Bacillus subtilis . J Biol Chem 281(31): 21720–21727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chung YS, Dubnau D (1995) ComC is required for the processing and translocation of comGC, a pilin-like competence protein of Bacillus subtilis . Mol Microbiol 15(3): 543–551. [DOI] [PubMed] [Google Scholar]
- 21. Kramer N, Hahn J, Dubnau D (2007) Multiple interactions among the competence proteins of Bacillus subtilis . Mol Microbiol 65(2): 454–464. [DOI] [PubMed] [Google Scholar]
- 22. Berka RM, Hahn J, Albano M, Draskovic I, Persuh M, et al. (2002) Microarray analysis of the Bacillus subtilis K-state: Genome-wide expression changes dependent on ComK. Mol Microbiol 43(5): 1331–1345. [DOI] [PubMed] [Google Scholar]
- 23. Hamoen LW, Smits WK, de Jong A, Holsappel S, Kuipers OP (2002) Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res 30(24): 5517–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ogura M, Yamaguchi H, Kobayashi K, Ogasawara N, Fujita Y, et al. (2002) Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J Bacteriol 184(9): 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hamoen LW, Van Werkhoven AF, Bijlsma JJ, Dubnau D, Venema G (1998) The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix. Genes Dev 12(10): 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Susanna KA, van der Werff AF, den Hengst CD, Calles B, Salas M, et al. (2004) Mechanism of transcription activation at the comG promoter by the competence transcription factor ComK of Bacillus subtilis . J Bacteriol 186(4): 1120–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Susanna KA, Mironczuk AM, Smits WK, Hamoen LW, Kuipers OP (2007) A single, specific thymine mutation in the ComK-binding site severely decreases binding and transcription activation by the competence transcription factor ComK of Bacillus subtilis . J Bacteriol 189(13): 4718–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Turgay K, Hahn J, Burghoorn J, Dubnau D (1998) Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J 17(22): 6730–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hamoen LW, Eshuis H, Jongbloed J, Venema G, van Sinderen D (1995) A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis . Mol Microbiol 15(1): 55–63. [DOI] [PubMed] [Google Scholar]
- 30. Smits WK, Eschevins CC, Susanna KA, Bron S, Kuipers OP, et al. (2005) Stripping bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol Microbiol 56(3): 604–614. [DOI] [PubMed] [Google Scholar]
- 31. Maamar H, Dubnau D (2005) Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol 56(3): 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smits WK, Kuipers OP, Veening JW (2006) Phenotypic variation in bacteria: The role of feedback regulation. Nat Rev Microbiol 4(4): 259–271. [DOI] [PubMed] [Google Scholar]
- 33. Veening JW, Hamoen LW, Kuipers OP (2005) Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis . Mol Microbiol 56(6): 1481–1494. [DOI] [PubMed] [Google Scholar]
- 34. Veening JW, Igoshin OA, Eijlander RT, Nijland R, Hamoen LW, et al. (2008) Transient heterogeneity in extracellular protease production by Bacillus subtilis . Mol Syst Biol 4: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kearns DB, Losick R (2005) Cell population heterogeneity during growth of Bacillus subtilis . Genes Dev 19(0890–9369; 0890–9369 24): 3083–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chai Y, Chu F, Kolter R, Losick R (2008) Bistability and biofilm formation in Bacillus subtilis . Mol Microbiol 67(0950-382; 0950-382 2): 254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abee T, Kovacs AT, Kuipers OP, van der Veen S (2011) Biofilm formation and dispersal in gram-positive bacteria. Curr Opin Biotechnol 22(2): 172–179. [DOI] [PubMed] [Google Scholar]
- 38. Oomes SJ, van Zuijlen AC, Hehenkamp JO, Witsenboer H, van der Vossen JM, et al. (2007) The characterisation of Bacillus spores occurring in the manufacturing of (low acid) canned products. Int J Food Microbiol 120(1–2): 85–94. [DOI] [PubMed] [Google Scholar]
- 39. Doron SI, Hibberd PL, Gorbach SL (2008) Probiotics for prevention of antibiotic-associated diarrhea. J Clin Gastroenterol 42 Suppl 2S58–63. [DOI] [PubMed] [Google Scholar]
- 40. Kovacs AT, van Hartskamp M, Kuipers OP, van Kranenburg R (2010) Genetic tool development for a new host for biotechnology, the thermotolerant bacterium Bacillus coagulans . Appl Environ Microbiol 76(12): 4085–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rhee MS, Kim JW, Qian Y, Ingram LO, Shanmugam KT (2007) Development of plasmid vector and electroporation condition for gene transfer in sporogenic lactic acid bacterium, Bacillus coagulans . Plasmid 58(1): 13–22. [DOI] [PubMed] [Google Scholar]
- 42. Wang Q, Ingram LO, Shanmugam KT (2011) Evolution of D-lactate dehydrogenase activity from glycerol dehydrogenase and its utility for D-lactate production from lignocellulose. Proc Natl Acad Sci U S A 108(47): 18920–18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhee M, Moritz B, Xie G, Glavina Del Rio T, Dalin E, et al.. (2011) Complete genome sequence of a thermotolerant sporogenic lactic acid bacterium, Bacillus coagulans strain 36D1. Standards in Genomic Sciences 5(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Su F, Xu K, Zhao B, Tai C, Tao F, et al. (2011) Genome sequence of the thermophilic strain Bacillus coagulans XZL4, an efficient pentose-utilizing producer of chemicals. J Bacteriol 193(22): 6398–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Su F, Yu B, Sun J, Ou HY, Zhao B, et al. (2011) Genome sequence of the thermophilic strain Bacillus coagulans 2–6, an efficient producer of high-optical-purity L-lactic acid. J Bacteriol 193(17): 4563–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Provvedi R, Chen I, Dubnau D (2001) NucA is required for DNA cleavage during transformation of Bacillus subtilis . Mol Microbiol 40(3): 634–644. [DOI] [PubMed] [Google Scholar]
- 47. Susanna KA, Fusetti F, Thunnissen AM, Hamoen LW, Kuipers OP (2006) Functional analysis of the competence transcription factor ComK of Bacillus subtilis by characterization of truncation variants. Microbiology 152(Pt 2): 473–483. [DOI] [PubMed] [Google Scholar]
- 48. Smits WK, Hoa TT, Hamoen LW, Kuipers OP, Dubnau D (2007) Antirepression as a second mechanism of transcriptional activation by a minor groove binding protein. Mol Microbiol 64(2): 368–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Albano M, Hahn J, Dubnau D (1987) Expression of competence genes in Bacillus subtilis . J Bacteriol 169(0021–9193 7): 3110–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Holo H, Nes IF (1989) High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55(12): 3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Russel DW (2001) Molecular cloning: A laboratory manual. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 52. Kunst F, Rapoport G (1995) Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis . J Bacteriol 177(9): 2403–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van Sinderen D, Luttinger A, Kong L, Dubnau D, Venema G, et al. (1995) comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis . Mol Microbiol 15(3): 455–462. [DOI] [PubMed] [Google Scholar]
- 54. Albano M, Smits WK, Ho LT, Kraigher B, Mandic-Mulec I, et al. (2005) The rok protein of Bacillus subtilis represses genes for cell surface and extracellular functions. J Bacteriol 187(6): 2010–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grande Burgos MJ, Kovacs AT, Mironczuk AM, Abriouel H, Galvez A, et al. (2009) Response of Bacillus cereus ATCC 14579 to challenges with sublethal concentrations of enterocin AS-48. BMC Microbiol 9: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mellegard H, Kovacs AT, Lindback T, Christensen BE, Kuipers OP, et al. (2011) Transcriptional responses of Bacillus cereus towards challenges with the polysaccharide chitosan. PLoS One 6(9): e24304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. van Hijum SA, de Jong A, Baerends RJ, Karsens HA, Kramer NE, et al. (2005) A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25(4): 402–408. [DOI] [PubMed] [Google Scholar]
- 59. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22): 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Prepiak P, Dubnau D (2007) A peptide signal for adapter protein-mediated degradation by the AAA+ protease ClpCP. Mol Cell 26(1097–2765 5): 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gasson M (1983) Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 154(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Joseph P, Fantino JR, Herbaud ML, Denizot F (2001) Rapid orientated cloning in a shuttle vector allowing modulated gene expression in Bacillus subtilis . FEMS Microbiol Lett 205(1): 91–97. [DOI] [PubMed] [Google Scholar]
- 63. Lewis PJ, Marston AL (1999) GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis . Gene 227(1): 101–110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Schematic presentation of the promoter region of putative competence related genes. Filled boxes indicate putative AT-boxes (maximum 3 mismatches to the consensus AAAA-N5-TTTT), open boxes indicate upstream open reading frames and com genes, numbers denote spacing between AT-boxes resulting in a so called K-box (8 bp and 31 bp in the case of comKBco and comCBco, respectively). B. Sequences of B. coagulans DSM1 promoter regions related to competence. Bold letters indicate putative AT-boxes. The putative open reading frames, com genes are indicated below the sequence.
(PDF)
Detection of the ComKBco protein. Equal amounts of proteins were loaded in each lane, Samples were taken from induced (+) or non-induced (−) cultures. Cells were centrifuged, lysed and analysed by Western blotting using ComKBsu-specific antibodies. The arrows indicate ComK specific signal (K) and non-specific signal (NP). Positions of molecular weight marker bands are indicated on the right side of the gel.
(TIF)