Abstract
Metastatic renal cell carcinoma (RCC) is one of the most treatment-resistant malignancies, and patients have a dismal prognosis, with a <10% five-year survival rate. The identification of markers that can predict the potential for metastases will have a great effect in improving patient outcomes. In this study, we used differential proteomics with isobaric tags for relative and absolute quantitation (iTRAQ) labeling and LC-MS/MS analysis to identify proteins that are differentially expressed in metastatic and primary RCC. We identified 1256 non-redundant proteins, and 456 of these were quantified. Further analysis identified 29 proteins that were differentially expressed (12 overexpressed and 17 underexpressed) in metastatic and primary RCC. Dysregulated protein expressions of profilin-1 (Pfn1), 14–3-3 zeta/delta (14–3-3ζ), and galectin-1 (Gal-1) were verified on two independent sets of tissues by means of Western blot and immunohistochemical analysis. Hierarchical clustering analysis showed that the protein expression profile specific for metastatic RCC can distinguish between aggressive and non-aggressive RCC. Pathway analysis showed that dysregulated proteins are involved in cellular processes related to tumor progression and metastasis. Furthermore, preliminary analysis using a small set of tumors showed that increased expression of Pfn1 is associated with poor outcome and is a potential prognostic marker in RCC. In addition, 14–3-3ζ and Gal-1 also showed higher expression in tumors with poor prognosis than in those with good prognosis. Dysregulated proteins in metastatic RCC represent potential prognostic markers for kidney cancer patients, and a greater understanding of their involved biological pathways can serve as the foundation of the development of novel targeted therapies for metastatic RCC.
Renal cell carcinoma (RCC)1 is the most common neoplasm of the adult kidney. Worldwide incidence and mortality rates of RCC are rising each decade (1). Seventy-five percent of kidney tumors are of the clear cell (ccRCC) subtype (2). Although modern imaging techniques for abdominal screening have led to increased incidental detection of renal tumors (3), unfortunately ∼25% to 30% of patients still have metastases at presentation.
The prognosis with RCC is quite variable. The greatest risk of recurrence following nephrectomy is within the first 3 to 5 years (4). The ability to predict which tumors will metastasize would have a significant effect on patient outcomes, because the likelihood of a favorable response to treatment is greater when the metastatic burden is limited, and surgical resection of a single or limited number of metastases can result in longer survival (5). Furthermore, ∼3% of patients will develop a second primary renal tumor, either synchronous or metachronous. Currently, patient prognosis is assessed based on histological parameters and a multivariate analysis developed at Memorial Sloan Kettering (6), but neither is sufficiently accurate. A more accurate assessment of prognosis is urgently needed to better guide patient management.
Although surgery can be curative for localized disease, many patients eventually relapse. Metastatic RCC is one of the most treatment-resistant malignancies, with chemotherapy and radiotherapy having limited effect. The five-year survival rate for metastatic RCC is ≤10% (7). Although there has been much progress in RCC treatment with the new era of antiangiogenic therapy, the majority of patients ultimately suffer a relapse and die from progression of the cancer. A more in-depth understanding of the pathogenesis of metastasis will be a cornerstone in the development of new targeted therapies. A number of prognostic markers have previously been identified based on comparative analysis of primary and metastatic tumors, including C-reactive protein, tetraspanin 7, hypoxia-inducible factor 1 α, phos-S6, U3 small nucleolar ribonucleoprotein protein, carbonic anhydrase IX, and microvascular density (8–14). However, no biomarker has yet had an established clinical role independent of stage (15). Differential protein expression between primary RCC and normal tissues was previously studied (16–18). Also, differential expression between primary and metastatic kidney disease has been investigated at the microRNA level (19, 20). Molecular analyses hold the promise of providing a better understanding of the pathogenesis of kidney cancer (21).
In this study, we aimed to elucidate the pathogenesis of RCC metastasis through proteomic analysis and to identify potential prognostic markers for kidney cancer. We performed quantitative proteomic analysis using isobaric tags for relative and absolute quantitation (iTRAQ) labeling and LC-MS/MS to identify proteins that were dysregulated in metastatic RCC relative to primary RCC. Differential expressions of selected biologically interesting proteins—profilin-1 (Pfn1), 14–3-3 zeta/delta (14–3-3ζ), and galectin-1 (Gal-1)—were validated on two independent sets of tumors by means of western blot (WB) analysis and immunohistochemistry (IHC). Hierarchical clustering analysis showed that differential protein expression can distinguish between aggressive and non-aggressive tumors. In order to explore the role of these dysregulated proteins in tumor progression, we performed Gene Ontology (GO) and pathway analyses. In addition, we carried out a preliminary analysis to assess the potential of Pfn1, 14–3-3ζ, and Gal-1 as prognostic markers in RCC.
EXPERIMENTAL PROCEDURES
Patients and Specimens
Primary ccRCC tissues and matched normal kidney tissues from the same patient were obtained from nephrectomy specimens at St. Michael's Hospital and the Ontario Tumor Bank, Toronto, Ontario, Canada. We also collected unmatched metastatic RCC tissues. Specimens were collected immediately following nephrectomy and flash frozen in liquid nitrogen in 2-ml cryogenic tubes. As RCC is known to arise from the proximal tubules (22), the kidney cortex is considered a suitable representation of normal kidney tissue (23). All specimens were histologically confirmed by a pathologist. The study was approved by the Research Ethics Boards of York University, St. Michael's Hospital and the Ontario Cancer Institute. Relevant clinical information on the patients is shown in supplemental Table S1.
Tissue Preparation and Protein Extraction
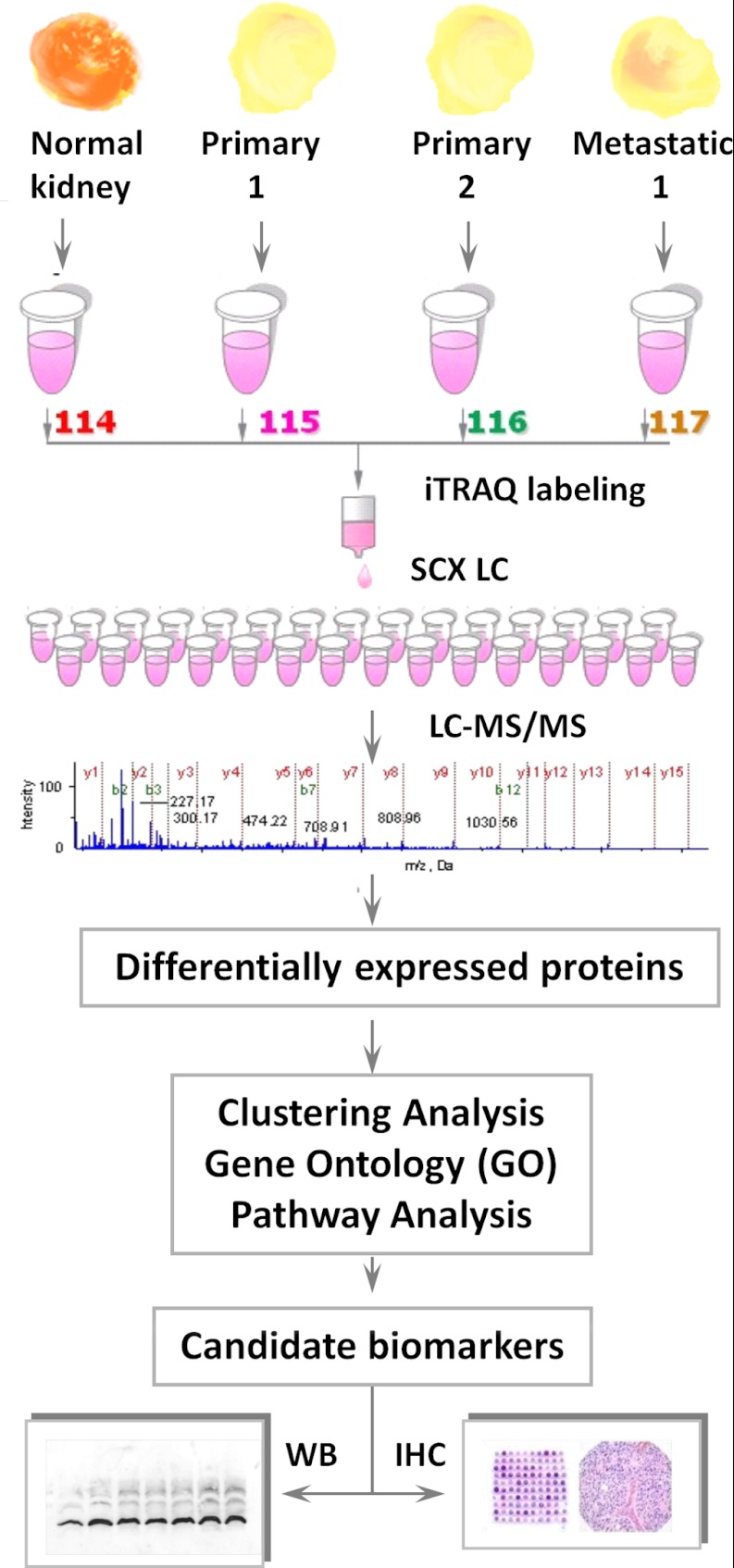
A schematic of the work flow is shown in Fig. 1. Tissues were prepared as described elsewhere (24–26). Briefly, tissues were washed three times in ice-cold phosphate buffered saline (PBS) and homogenized using a hand-held homogenizer in a protease-inhibitor mixture (Roche, Laval, Quebec, Canada). Cell debris was then removed via centrifugation at 4 °C for 30 min at 14,000 rpm. The clarified supernatant was transferred to a fresh 1.5-ml tube. A reference sample was prepared from a pool of six combined normal kidney tissues. Protein concentrations were determined using the Bradford assay (Sigma-Aldrich, St. Louis, MO) (24, 26). Equal amounts of protein from each tissue type were digested with trypsin, labeled with iTRAQ, and combined. Samples were then separated via off-line strong cation exchange (SCX) liquid chromatography and analyzed via reverse phase (RP) LC-MS/MS. Proteins were identified and quantified using Protein Pilot and then subjected to additional characterization, including verification via WB, clustering, GO analysis, pathway analysis, and IHC.
Fig. 1.
Work flow for quantitative proteomic analysis. Six pairs of primary ccRCC and normal matched kidney tissues from the same patient, as well as six metastatic RCC tissues, were analyzed. The reference sample consisted of a homogenate of the six normal kidney tissues. Each sample was digested individually with trypsin and labeled with the appropriate iTRAQ tag. The labeled digests were then pooled and separated via off-line SCX LC. Each fraction was analyzed in triplicate by on-line RP nano-LC-MS/MS. Exclusion lists were used to minimize redundancy. MS data were analyzed by Protein Pilot to identify and quantify proteins using a cut-off of 5% local FDR. Dysregulated proteins were validated by means of WB and IHC and further analyzed in clustering, GO, and pathway analyses.
iTRAQ Sample Labeling
For iTRAQ LC-MS/MS analysis, 100 μg of clarified supernatants were denatured for 1 h at 60 °C, disulfide bonds were reduced, and the cysteine residues were blocked as described in the iTRAQ protocol (Applied Biosystems, Foster City, CA). Supernatants were then divided into sets of four, each containing one aliquot of the reference sample and three ccRCC malignant or individual non-malignant kidney samples. Each sample was then digested with trypsin and labeled with the iTRAQ tags (labeling details are shown in supplemental Table S2). Labeling of the reference sample was randomized for each set to eliminate any potential for bias that might be associated with a particular iTRAQ reporter tag. The iTRAQ-labeled samples were then combined according to the specified set and transferred into fresh 1.5-ml tubes. Each iTRAQ set was then dried using a vacuum centrifuge (Thermo Savant SC110 A, Holbrook, NY).
SCX Chromatography
The iTRAQ sets were dissolved in 1.7 ml of Buffer A (10 mm H3PO4/KH2PO4 in an aqueous solution of 25% acetonitrile and acidified to a pH of 3.0 with phosphoric acid) and filtered using a 0.45-μm syringe filter (Millipore, Cambridge, Ontario, Canada). Each set was then separated via off-line SCX chromatography using an HP1050 HPLC instrument (Agilent, Palo Alto, CA) with a 2.1-mm internal diameter × 100-mm length PolyLC Polysulfoethyl A column packed with 5-μm beads with 300- Å pores (The Nest Group, Southborough, MA) as described elsewhere (27). Separation was performed using a linear binary gradient over 1 h (see details in supplemental Table S3) of Buffer A and Buffer B; Buffer B was composed of Buffer A and 350 mm potassium chloride. Buffer C was composed of Buffer A and 1 m potassium chloride and was used to strip the column after the run. A total of 30 SCX fractions were collected per iTRAQ set. These fractions were dried using a vacuum centrifuge as before.
RP LC-MS/MS
The SCX fractions were analyzed in triplicate using a nanobore LC system (LC Packings, Amsterdam, The Netherlands) and a QSTAR Pulsar mass spectrometer (Applied Biosystems/MDS SCIEX, Foster City, CA) in positive ion mode, externally calibrated with tryptic peptides from bovine serum albumin. The first five fractions were not analyzed because they consisted of the void volume, which contained unreacted iTRAQ labels and byproducts that would compromise the RP column. Fractions 6–17 were redissolved in 16 μl of eluant A (consisting of 94.9% deionized water, 5.0% methanol, and 0.1% formic acid (pH 3)). For subsequent fractions, the amount of eluant A was incremented by 2 μl over the preceding fraction to accommodate the increase in the amount of KCl. A 1-μl aliquot of the sample (∼1 μg of total peptides) was loaded onto a C18 RP pre-column (LC Packings: 300 μm × 5 mm) and desalted before separation on an RP analytical column (75 μm × 150 mm packed in-house with 3-μm Kromasil C18 beads with 100 Å pores; The Nest Group, Southborough, MA). Eluant A, consisting of 94.9% deionized water, 5.0% methanol, and 0.1% formic acid (pH 3), was used to load the sample onto the C18 pre-column at a flow rate of 25 μl min−1. After 4 min, the C18 pre-column was switched in-line with the RP analytical column. Separation was performed at 100 nL min−1 using a nonlinear binary gradient (see gradient below) starting with Eluant A and transitioning to Eluant B, which consisted of 5.0% deionized water, 94.9% methanol, and 0.1% formic acid.
Time (min) 0.1 5 10 70 85 95 98 135
B (%) 5 5 15 35 80 80 5 Stop
MS data were acquired in information-dependent acquisition mode using Analyst QS 1.1 software (Applied Biosystems/MDS SCIEX, Foster City, CA). The LC-MS/MS analysis was performed using a 1-s TOF-MS survey scan from 400 to 1500 Da, followed by four 2-s product-ion scans, from 80 to 2000 Da, of the four most abundant ion peaks in the survey scan. The collision energy was automatically controlled by the information-dependent acquisition collision energy parameter script. Switching criteria were set for ions with m/z ≥ 400 and ≤ 1500, charge states of +2 to +4, and abundances of ≥10 counts. Using Analyst QS 1.1 controlled dynamic exclusion, former target ions were excluded for 30 s, and ions within a 100-ppm window were ignored. Precursor ion exclusion (PIE) lists were used to minimize redundancy.
Bioinformatics Analysis
Protein Identification by Protein Pilot
LC-MS/MS data for each fraction were used to identify proteins by searching a concatenated Swissprot/Panther database of 66,082 distinct human protein entries (version June 2, 2010). The database was searched using Protein Pilot software, version 2.0.1 (AB SCIEX, Foster City, CA), which uses the Paragon algorithm (28). Protein identification was performed with methylmethanethiosulfonate selected for cysteine modification, with the search option “emphasis on biological modifications,” and with “PSPEP” (Proteomics System Performance Evaluation Pipeline Software) analysis checked. Peptide and protein summaries and false discovery rate (FDR) reports were generated. Only proteins identified with local FDR ≤ 5% were considered for further analysis (29).
Iterative Runs with PIE
To minimize redundancy in subsequent iterations, a PIE list was added to the acquisition method after each iteration as described elsewhere (30, 31). PIE lists were generated using an Excel template developed in-house. To generate the list for each iteration, the peptide summary of a fraction, obtained after the previous iteration, was imported into an Excel template in which (1) the m/z values and elution times of peptides identified with >95% confidence were extracted, (2) alternative charge states (only +2, +3, and +4 were considered) of the peptides were calculated, and (3) the next three higher isotopic m/z values of extracted and calculated peptides were determined. The resulting m/z ratios from all three of these considerations constituted the PIE list. This list was saved and imported into the acquisition method for the next iteration. The list used for each iteration was cumulative of all the m/z values and elution times derived from all previous iterations for the fraction. Tolerance windows for exclusion were set at 100 ppm for m/z and 360 s for elution time. The template is available in the supplementary data (PIE Template).
iTRAQ Ratio Re-calculation and Identification of Differentially Expressed Proteins
To identify non-redundant proteins, data acquired for all 25 fractions from each iTRAQ set injected in triplicate were searched using Protein Pilot software. Proteins identified in five iTRAQ sets were compiled and matched by accession numbers using a script written in Matlab (version 7.7.0.471). Redundant proteins and peptides and proteins identified in reverse sequence were removed from the list. To improve the confidence of protein quantitation, the mean expression iTRAQ ratios of the proteins were re-calculated based on the criteria that the protein must be identified by a minimum of three peptides, with ≥95% confidence, and with an expression ratio error factor of <11.1%. To enhance confidence in the protein quantitation even more, we included only 95% of all quantified proteins with the lowest computed error factor (which corresponds to a confidence > 0.05 in supplemental Table S4) for further consideration. Proteins were considered as differentially expressed if iTRAQ ratios were ≥1.5 or ≤0.67 in ≥50% in metastatic relative to primary ccRCC samples.
GO Analysis
Proteins were classified into groups according to biological processes (e.g. metabolic process), molecular function (e.g. protein binding), and subcellular compartmentalization (e.g. cytoplasm, organelle, etc.) using the GO Consortium databases.
Clustering Analysis
To determine whether differentially expressed proteins can discriminate between metastatic and primary RCC samples, the samples were hierarchically clustered based on quantified proteins. Proteins were included in the analysis if quantification was available in at least 50% of the samples. The average iTRAQ ratios were logarithmically transformed for hierarchical clustering via the City-block distance method. Hierarchical clustering analysis was performed using the Cluster 3.0 software, and the result was visualized using TreeView software (Stanford University, Palo Alto, CA), both of which were developed by Eisen et al. (32).
WB Analysis
Dysregulated protein expression in metastatic RCC samples was verified via WB analysis. Briefly, 30 μg of total protein were electrophoretically separated on a 10% SDS-PAGE gel. Proteins were then transferred to a PVDF membrane and probed with the following polyclonal antibodies: anti-Gal-1, anti-Pfn1 (both from Abcam, Cambridge, MA), and anti-14–3-3ζ (Santa Cruz Biotechnologies, Santa Cruz, CA). β-actin (Cell Signaling Technology, Danvers, MA) was used as a loading control. Membranes were incubated with primary antibodies overnight at 4 °C. Protein expression was visualized after incubation with secondary anti-rabbit antibodies conjugated with horseradish peroxidase and enhanced chemoluminescence reagent (Amersham Biosciences, Piscataway, NJ). The intensity of protein staining was determined using ImageJ, a publicly available Java-based image processing program. Average protein expression was calculated based on two independent WB analyses. Primary ccRCC samples were compared with non-malignant kidney samples using the paired sample two-tailed t test. Metastatic RCC samples were compared with primary ccRCC samples using the Mann-Whitney two-tailed test. p ≤ 0.05 was considered as significant.
Tissue Microarray Construction and Immunohistochemistry
Appropriate areas from normal kidney tissue, primary ccRCC, and metastatic RCC were selected and circled from donor blocks by a pathologist. Tissue microarray (TMA) blocks containing duplicate 1.0-mm cores from each specimen were constructed with a manual tissue microarrayer (Beecher Instruments, Sun Prairie, WI). The TMAs contained 22 cases of primary ccRCC and matched normal kidney tissues from the same patient, 12 cases of primary ccRCC from patients who later developed metastasis, and 26 metastatic RCC tissues. In addition, each block contained two marker cores for TMA orientation.
TMA sections were cut 5 μm thick and placed on charged slides. Slides were deparaffinized in xylene, hydrated in gradient ethanol, and pre-treated in a microwave oven for 20 min at 800 W in 1 l of citrate buffer (0.01 m, pH 6.0) for antigen retrieval. Sections were then incubated with hydrogen peroxide (0.3% v/v) in PBS for 15 min to quench the endogenous peroxidase activity, followed by blocking with 10% fetal bovine serum in PBS to preclude nonspecific binding. Thereafter, the slides were incubated with primary antibodies Gal-1, Pfn1, or 14–3-3ζ overnight at 4 °C. Protein expression was detected using the streptavidin-biotin complex with the Dako LSAB+ kit (Dako Cytomation, Glostrup, Denmark) and diaminobenzidine as the chromogen. All procedures were carried out at room temperature unless otherwise specified. Slides were washed with 0.025% Triton X 100 in PBS (0.1 m, pH = 7.3) three times after each step. Finally, sections were counterstained with Mayer's hematoxylin and mounted with DPX mountant. In the negative control tissue sections, the primary antibody was replaced by isotype-specific non-immune mouse/rabbit IgG.
Immunoexpression of each protein was evaluated by a pathologist. Quantification in tumor sections was classified into four categories: (A) moderate to strong membrane, cytoplasmic, and nuclear staining in greater than 50% of tumor cells; (B) moderate to strong cytoplasmic staining in >50% of either the cytoplasm or nuclei, but not both; (C) overall weak staining in the cytoplasm and/or nuclei; and (D) no staining.
RESULTS
Identification of Differentially Expressed Proteins in Primary and Metastatic RCC
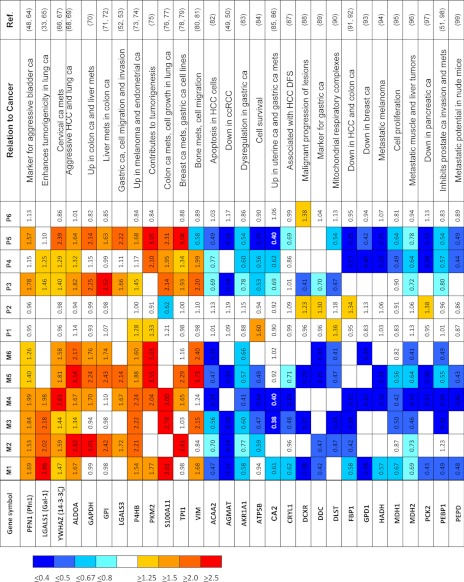
Using Protein Pilot, we identified a total of 1256 non-redundant proteins with local FDR ≤ 5% (supplemental Table S4); 456 of these proteins were reliably quantified (supplemental Table S5). Twenty-nine proteins met our definition of differential expression (see “Experimental Procedures”) in a comparison between metastatic and primary RCC: 12 were overexpressed (iTRAQ ratios of ≥1.5), and 17 were underexpressed (iTRAQ ratios of ≤0.67). Table I and supplemental Table S6 show a heat map of the 29 differentially expressed proteins. A literature search showed that all 29 proteins had previously been associated with other malignancies. For example, Gal-1 has previously been reported to be associated with cell migration and invasion in a metastatic murine lung cancer model (33). Gal-1 was also shown to have prognostic significance in epithelial ovarian cancer (34).
Table I. Heat map showing expression of 29 proteins that are dysregulated in metastatic compared to primary renal cell carcinoma.
ca, cancer; DFS, disease-free survival; HCC, hepatocellular carcinoma; mets, metastasis. For “protein name” we used the gene name according to UniProtKB. For a full name of the protein and its accession number, see supplementary Table S6.
Clustering Analysis Indicated that Differential Protein Expression Can Discriminate between Metastatic and Primary RCC
In order to examine the hypothesis that metastatic potential was present at least in part of the primary tumor, cluster analysis was performed on 456 proteins for which quantitative information was available. The samples were clustered into two main groups: one that contained the primary RCC samples, P1, P2, and P6, and a second that had the six metastatic cases plus the other three primary RCC samples, P3, P4, and P5 (data not shown). Clustering of the last three primary RCC samples with the six metastatic samples becomes less puzzling after data from clinical follow-up are examined: one of the primary RCC patients developed subsequent metastasis to the liver and had a history of colon cancer, a second patient also had an earlier cancer, and the third patient had no reported metastasis. The other group of primary RCC patients, P1, P2, and P6, did not develop metastasis for five years. If validated on a larger tumor set, these data then strongly suggest that RCCs have a unique protein expression pattern that is required for metastasis and that differentially expressed proteins can discriminate between aggressive and non-aggressive RCCs.
Validation of Dysregulated Protein Expression
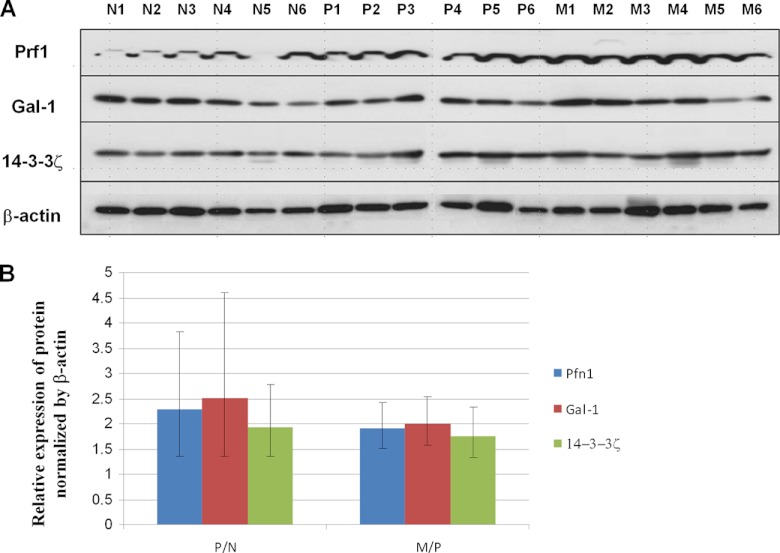
To proceed with the first steps in validating our MS analysis, we confirmed the differential expression of three dysregulated proteins via WB analysis using samples from the same RCC patient cohort. The proteins Gal-1, Pfn1, and 14–3-3ζ were selected for validation based on their interesting biology and potential significance in RCC tumorigenesis (35–40). The expression of Gal-1, Pfn1, and 14–3-3ζ was found to be elevated in primary ccRCC relative to normal kidney tissue (a typical set of results is shown in Fig. 2A) when analyzed via WB analysis. Additionally, all three proteins were further up-regulated in metastatic than in primary ccRCC. Densitometry showed that 14–3-3ζ, Pfn1, and Gal-1 were up-regulated 1.93-fold (p < 0.05), 2.28-fold (p < 0.05), and 2.50-fold (p < 0.1), respectively, in primary ccRCC relative to normal kidney tissue (Fig. 2B). When we compared protein expression in metastatic and primary tissues, 14–3-3ζ and Pfn1 showed significantly increased expression (1.77-fold change, p < 0.05, and 1.92-fold change, p < 0.01, respectively); Gal-1 showed a 2.5-times increase in expression, but this finding did not reach 95% statistical significance (p < 0.1).
Fig. 2.
Verification of Gal-1, Pfn1, and 14–3-3ζ overexpression in metastasis via Western blot analysis. A, representative blots showing the expression of Gal-1, Pfn1, and 14–3-3ζ in normal kidney tissues (N) and in primary (P) and metastatic (M) ccRCC. For each of these proteins, expression was increased in primary tumor tissues relative to normal kidney tissue; as well, the expression of all proteins was increased in metastatic tissues relative to primary ccRCCs. β-actin was used as a loading control. B, graphical representation of the average fold change in expression of the three proteins between six primary ccRCCs and matched normal specimens (P/N) and that between six primary ccRCCs and six unmatched metastatic cases.
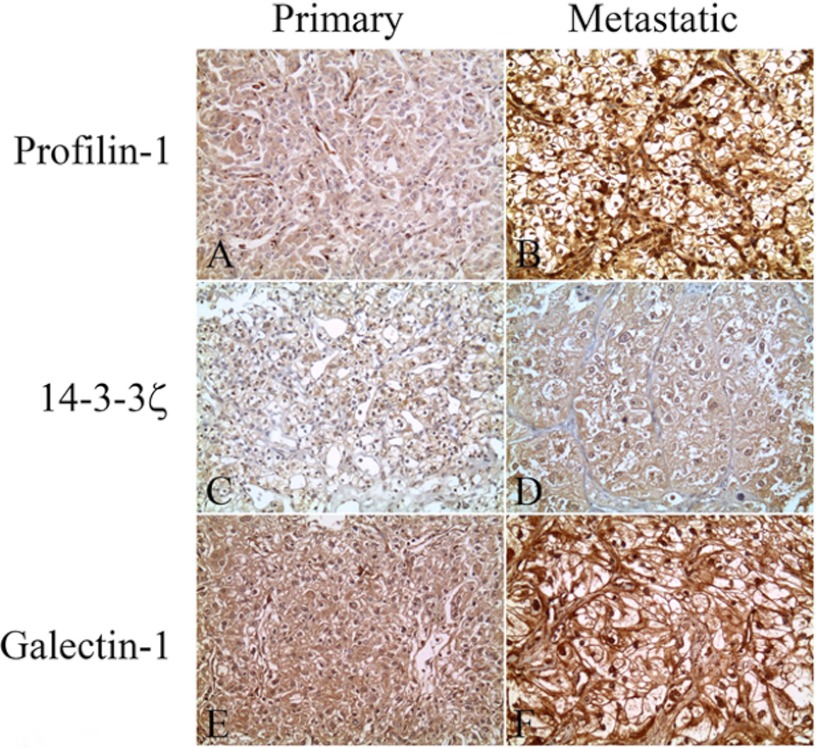
We additionally verified the overexpression of Pfn1, Gal-1, and 14–3-3ζ in metastatic relative to primary RCC in an independent cohort of patients by means of IHC using TMAs consisting of 22 cases of primary ccRCC and 26 metastatic RCC tissues (Fig. 3 shows a typical set of results). We quantified immunoexpression using the four-tier scale described under “Experimental Procedures” (ranging from A, which was the highest, to D, with no staining). For Pfn1 expression, we found increased expression in metastatic relative to primary ccRCC. Most of the primary tissues (19/22, 86%) showed the B expression level (Fig. 3A). There were also two (9%) tumor samples that exhibited the A expression level and one (5%) tumor sample that showed the C level. For metastasis, we found that the majority (18/23, 78%; three cases were omitted because the samples were washed off the slide) of samples showed the A level of expression (Fig. 3B) and five (22%) showed the B level, indicating overall increased Pfn1 expression in metastatic relative to primary tissues.
Fig. 3.
Verification of Gal-1, Pfn1, and 14–3-3ζ overexpression in metastatic ccRCCs via immunohistochemical analysis. Representative photomicrographs show the overexpression of Gal-1, Pfn1, and 14–3-3ζ in metastatic relative to primary RCC tissue in IHC studies. Pfn1 shows higher intensity and percent positivity in metastatic (B) than in primary tumors (A). 14–3-3ζ shows more intense cytoplasmic and nuclear staining in metastatic (D) than in primary (C) RCCs. Similarly, Gal-1 staining is more intense in metastatic (F) than in primary (E) tumor samples. Original magnification × 200.
We then examined the expression of 14–3-3ζ in this independent tumor set. Similar to the MS results, 14–3-3ζ showed increased expression in metastatic relative to primary ccRCC. 28% (6/22) of primary ccRCC tumor samples had the A level of expression (Fig. 3C), 36% (8/22) exhibited the B level, and 36% (8/22) exhibited the C level. By contrast, in the 24 metastatic tissues that were available for the examination of 14–3-3ζ expression, we found that 67% (16/24; two cases were omitted because the samples were washed off the slide) showed the A level of expression (Fig. 3D), 29% (7/24) showed the B level, and 4% (1/24) showed the C level, again indicating an overall increase of 14–3-3ζ expression in metastatic relative to primary RCC. The differences in Gal-1 expression between primary and metastatic RCC were not statistically significant (Figs. 3E and 3F). This may be attributed to the high background staining that does not allow accurate quantification. A summary of our IHC analyses is shown in Table II.
Table II. Immunohistochemical expression of profilin-1, galectin-1, and 14–3-3ζ in metastatic and primary ccRCC.
| Protein Name | Expression Level | 22 Primary Paired ccRCC | 26 Metastatic ccRCC |
|---|---|---|---|
| Pfn1 | A | 2 (9%) | 18 (78%)a |
| B | 19 (86%) | 5 (22%) | |
| C | 1 (5%) | ||
| Gal-1 | A | 12 (55%) | 13 (57%)a |
| B | 8 (36%) | 8 (35%) | |
| C | 2 (9%) | 1 (4%) | |
| D | 1 (4%) | ||
| 14–3-3ζ | A | 6 (28%) | 16 (67%)b |
| B | 8 (36%) | 7 (29%) | |
| C | 8 (36%) | 1 (4%) |
A, moderate to strong membrane, cytoplasmic, and nuclear staining in >50% tumor cells; B, moderate to strong cytoplasmic staining in >50% of either the cytoplasm or nuclei, but not both; C, overall weak staining in the cytoplasm and/or nuclei; D, no staining.
a Three cases were omitted because the samples were washed off the slide.
b Two cases were omitted because the samples were washed off the slide.
GO Analysis
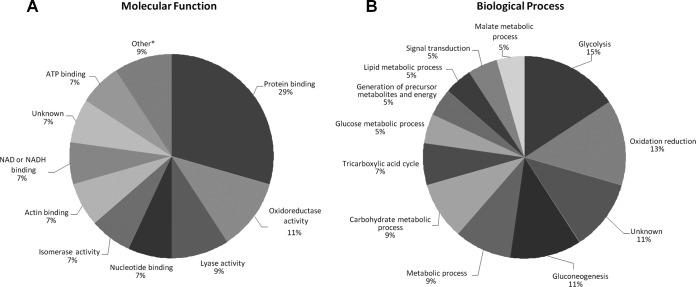
We subjected the 29 identified proteins that were differentially expressed in metastatic and primary ccRCC to GO analysis and categorized them according to molecular function, biological processes, and cellular components. When we analyzed dysregulated proteins for molecular function, we found that over one-third of proteins were grouped under the GO term “catalytic activity” (GO:0003824, p = 0.0016, Fig. 4A), which included proteins involved in isomerase activity such as glucose-6-phosphate isomerase, triosephosphate isomerase 1, and protein disulfide isomerase, among others. In addition, we found that 69% of proteins analyzed were grouped under the GO term “protein binding” (GO:0005515, p < 0.001). This category includes actin binding (GO:0003779) in which both Pfn1 and alcohol dehydrogenase A fall under.
Fig. 4.
Gene Ontology (GO) analysis. PIE charts showing the results of GO analysis. The 29 dysregulated proteins were analyzed for (A) molecular function and (B) biological process. Significance values for each function and process are shown in the figure.
We also grouped dysregulated proteins based on biological processes. We found that a significant number of proteins were grouped under the “multicellular organismal process” (GO:0032501, p = 0.00441) and “biological regulation” (GO:0065007; p < 0.001, Fig. 4B) headings, including the proteins Gal-1 and Pfn1. In addition, when the 29 identified dysregulated proteins were analyzed for their cellular localization, we found that 76% of proteins were located intracellularly (p = 0.03016), including proteins associated with both plasma and organelle membranes (p = 0.01703). There was also a significant association with the extracellular region (p < 0.001, details not shown).
Differentially Expressed Proteins in Metastatic RCC Are Involved in a Number of Pathways Related to Tumor Progression and Metastasis
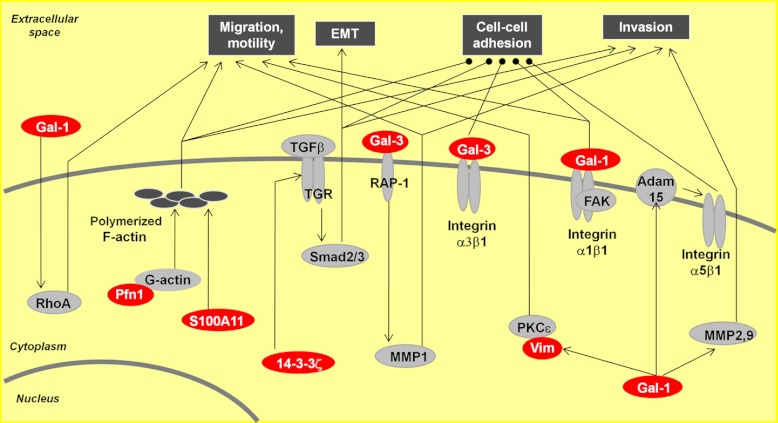
We performed pathway analysis on the 29 dysregulated proteins. A subset of proteins identified in our study are involved in cell migration and invasion, cell–cell adhesion, and epithelial to mesenchymal transition (Fig. 5). Gal-1 has been reported to be involved in a number of pathways that can contribute to tumor progression and metastasis. It can promote tumor invasion by upregulating matrix metalloproteinase (MMP)-9 and MMP-2 and by reorganizing the actin cytoskeleton in lung adenocarcinoma (40). In addition, Gal-1 enhances the activation of Cdc42, increasing the number and length of filopodia on tumor cells (40). Dysregulated Pfn1 has been previously reported to be involved in the restoration of adherent junctions in breast cancer cells (41), and galectin-3 (Gal-3) has been shown to be involved in breast cancer cell adhesion (42). The identified proteins also have an effect on cellular migration, as S100A11 has been shown to mediate hypoxia-induced mitogenic factor-induced smooth muscle cell migration (43), and both 14–3-3ζ (44) and vimentin (45) have been shown to be involved in cellular migration.
Fig. 5.
The involvement of a subgroup of differentially expressed proteins in cell–cell adhesion, migration, and invasion. Gal-1 is involved in cell migration through interaction with the α1β1 integrin, modulates cell adhesion and cell motility via Gal-1-induced expression of RhoA and alteration of the polymerization of the actin cytoskeleton, and promotes tumor invasion by reorganizing the actin cytoskeleton and upregulating matrix metalloproteinase (MMP)-9 and MMP-2. 14–3-3ζ cooperates with ErbB2 to promote cell motility and migration via the activation of Src and to induce epithelial-mesenchymal transition (EMT) by activating the TGFβ pathway to reduce cell adhesion. Pfn1 plays a role in cell adhesion and motility through interaction with G-actin. Pathways involved in tumor progression and metastasis are shown in white lettering on a black rectangle, differentially expressed proteins are denoted by white lettering on a red oval, and other proteins involved in pathways are represented by black letters on a gray oval. Lines with arrowheads represent documented interactions.
The Potential Clinical Significance of Proteins Dysregulated in Metastatic RCC
In order to determine whether the dysregulated proteins identified herein had potential as prognostic markers or not, we performed additional preliminary analyses using IHC on an expanded set of primary RCC samples. We analyzed the expression of Pfn1, Gal-1, and 14–3-3ζ in a total of 34 primary ccRCCs: 12 cases with poor prognosis (developed metastasis within three years) and 22 cases with good prognosis (no metastasis within five years; same set used in the comparison with metastatic ccRCC; Table II). We quantified immunoexpression in terms of the four categories described above. A summary of the expression levels of the three proteins in patients with poor versus good prognosis is shown in Fig. 6.
Fig. 6.
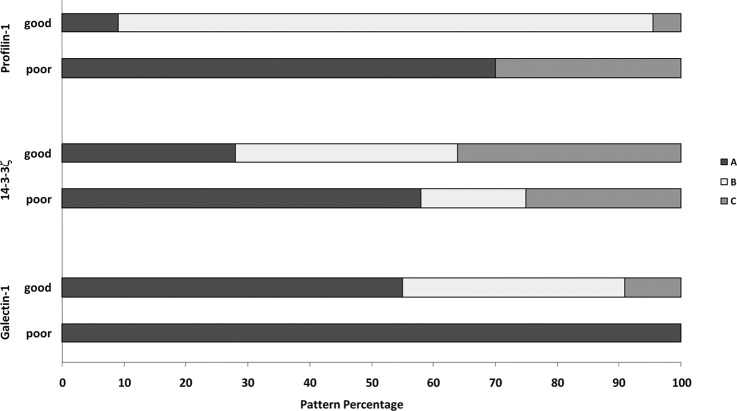
Stacked bar graphs showing differential staining patterns of Pfn1, Gal-1, and 14–3-3ζ in patients with good prognosis and those with poor prognosis. Higher expression levels were associated with tumors with poor prognosis. Expression was quantified in a four-tier scale (from A with the highest expression to D with no expression, as described in the text).
For the expression of Pfn1 in the 10 primary ccRCCs with poor prognosis (samples from two cases were washed off during IHC handling), seven (70%) samples showed the A level of expression, and three (30%) showed the C level. By contrast, only 2/22 cases (9%) in the good prognostic group showed the A expression level; 19/22 (86%) showed the B level and 1/22 (5%) the C level of expression. Significantly, the percentage of tumor cases displaying the A level of expression (70% versus 5%, respectively) was drastically different between the poor and the good prognosis cases and resembled that in metastatic RCCs, suggesting that Pfn1 might have prognostic value for RCC patients.
We also investigated the potential of 14–3-3ζ as a prognostic marker. In the tumor cases with poor prognosis, 58% (7/12) exhibited the A level of expression. The B level was seen in 17% (2/12) and the C level in 25% (3/12) of cases. This is contrasted with the expression in 22 tumor cases with good prognosis; we found that only 6/22 (28%) of the samples showed the A expression level. The B and C levels were each seen in 36%. Again, the expression of 14–3-3ζ in ccRCC of poor prognosis resembled more closely that of metastatic ccRCC, which strongly suggests that 14–3-3ζ might have clinical significance for prognostication in kidney cancer patients.
Finally, for Gal-1 we found that 12/12 (100%) tumors from patients with poor prognosis exhibited the A level of expression. When we examined the patients with good prognosis, we found that only 12/22 (55%) exhibited the A level, with 8/22 (36%) showing the B level and 2/22 (9%) showing the C level of expression.
DISCUSSION
Despite the many recent advances in metastatic RCC treatment through targeted therapies, the survival rate with metastatic RCC is extremely low (five-year survival rate of <10%). Regrettably, there are no prognostic molecular markers that would predict whether a tumor will behave aggressively or remain indolent. It is abundantly clear that tumor biology plays a significant role in resultant tumor behavior (15). Unfortunately, RCC primary tumors that are placed in the same prognostic category based on currently used parameters might behave differently. It is our hypothesis that the underlying biology of these tumors and differences in its details will determine a particular tumor's potential for metastasis. In addition, we can use these biological differences to identify novel molecular markers that might be useful for diagnostic, prognostic, or predictive purposes, the success of which would pave the road to a new era of personalized medicine in kidney cancer (46, 47).
In this study, we performed quantitative proteomic profiling to identify differential protein expression between metastatic and primary RCC and to identify potential prognostic markers for RCC patients. We identified 29 proteins that were significantly differentially expressed in metastatic and primary RCC. Interestingly, all 29 proteins had previously been reported to be involved in tumor progression and metastasis (Table I and supplemental Table S6). For example, two proteins that we identified as dysregulated in our study, Pfn1 and agmatinase, were also reported to have increased (48) or decreased (49, 50) expression in RCC. In addition, many of the proteins we reported had been shown to be involved in cellular migration and invasion. For example, phosphatidylethanolamine-binding protein 1/Raf kinase inhibitor protein, which was downregulated in our study, was shown to inhibit the migration and invasive ability of prostate cancer cells through the extracellular matrix (51). Gal-3, which had increased expression in our study, was reported to facilitate cell migration and invasion in vitro and induce metastasis in vivo (52, 53). Furthermore, we identified a subgroup (5/12) of overexpressed enzymes that are involved in glycolysis. These include fructose-bisphosphate aldolase A, glucose-6-phosphate isomerase, glyceraldehyde-3-phosphate dehydrogenase, pyruvate kinase M1/M2, and triosephosphate isomerase. Interestingly, proteins involved in the glycolytic pathway have also been reported to be involved in the metastasis of other cancers such as melanoma (54, 55).
Hierarchical cluster analysis based on the quantified proteins resulted in patients' being clustered into two distinct groups. One group contained all the patients with metastatic RCC and three of the six primary RCCs, and the other group had the remaining three primaries. A review of the clinical conditions for the three primaries in the metastatic group showed that two of these patients had histories of cancer and one had a subsequent RCC recurrence, suggesting that the protein expression profile of primary tumors might correlate with tumor aggressiveness. These data agree with those of Ramaswamy et al. (56), who hypothesized that the metastatic potential of tumors is encoded in the bulk of the primary tumor. This implies that there exists a biological difference between primary tumors that will metastasize and those that will remain indolent. The clinical implication is that exploitation of the protein expression profile might enable prediction of which patients will likely develop metastasis. This will encourage more intense follow-up of these patients and likely lead to earlier detection of any new tumors, the result of which is a more aggressive course of treatment and an overall increase in survival. Dissimilar tumor biology between more and less aggressive tumors might be the underlying factor in different responses to treatment. This can be advantageous, as clinicians will have the ability to administer the optimal treatment for patients rather than alternative treatments that will not be effective and which might cause negative side effects. A better understanding of the tumor biology will facilitate the realization of personalized medicine for RCC patients.
Pathway analysis predicted that a number of the identified dysregulated proteins are involved in similar biological signaling pathways (Fig. 6), suggesting that their dysregulation might be a cooperative effect imposed by the malignancy. For example, Gal-1 interacts with the α1β1 integrin subunit, inducing the phosphorylation of focal adhesion kinase, which modulates cell migration (57). Binding of Gal-1 to integrin is involved in cell adhesion (37, 58). Gal-1 has also been found to be involved in cell motility (35, 37) and cellular invasion (40). 14–3-3ζ is an isoform in a family of evolutionally highly conserved acidic proteins expressed in all eukaryotic organisms (59) and has been shown to be involved in tumor progression and metastasis. 14–3-3ζ, in cooperation with ErbB2, was found to drive breast cancer metastasis (36). In addition, Pfn1, an actin-binding protein, plays a critical role in cell migration by regulating the actin-cytoskeleton pathway (38, 39). Pfn1 remodels the actin cytoskeleton by regulating actin polymerization via the regeneration of actin monomers from disassembling filament networks through G-actin. Interestingly, our study showed that G-actin was overexpressed in all metastatic samples, which is in agreement with this mechanism (details shown in supplemental Table S5).
An interesting point with respect to Pfn1 expression is similar to what Minamida et al. (48) reported: we found Pfn1 overexpression in RCC tissues and cell lines. In contrast, Pfn1 has been found to be underexpressed in most, if not all, aggressive adenocarcinomas. For example, Pfn1 was reported to be downregulated in human breast cancer tissue and cell lines (60), pancreatic (60) and hepatic (61) carcinoma cells, squamous cell carcinoma (62), and nasopharyngeal cancer cell lines (63). These differences suggest that Pfn1 might be involved in different tumorigenic mechanisms in different tissue types.
In order to assess the potential prognostic value of our dysregulated proteins, we examined the expressions of Pfn1, Gal-1, and 14–3-3ζ in a small set of RCC patients who had either good or poor prognoses. These proteins were chosen based on their increased expression in metastatic tumors relative to primary tumors and their previously reported involvement as cancer markers (62–64). Our analysis indicated that of the three proteins, Pfn1 has the most promise as a prognostic marker. Patients who had poor prognosis had high Pfn1 expression (A level of expression in 70% of the tumors), whereas only 1% of patients with a good prognosis had this level of expression. Success in this preliminary study is limited because of the small number of cases examined, but the encouraging results certainly warrant follow-up and validation in a larger cohort of patients.
In short, through quantitative proteomic analysis, we identified differential protein expressions that can distinguish between aggressive and non-aggressive RCC tumors. Many of these proteins are involved in biological pathways pertinent to tumor progression and metastasis. In addition, our preliminary analysis showed that some of these dysregulated proteins might be useful clinical markers. Validation of these markers would greatly improve RCC patient treatment and increase overall survival.
Supplementary Material
Footnotes
* This work was supported by grants from the Canadian Cancer Society (CCS Grant No. 20185), the Ministry of Research and Innovation of the Government of Ontario, the Kidney Foundation of Canada, and the Cancer Research Society.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- 14–3-3ζ
- 14–3-3 protein zeta/delta
- ccRCC
- clear cell renal cell carcinoma
- Gal-1
- galectin-1
- GO
- Gene Ontology
- IHC
- immunohistochemistry
- iTRAQ
- isobaric tags for relative and absolute quantitation
- PIE
- precursor ion exclusion
- Pfn1
- profilin-1
- RCC
- renal cell carcinoma
- RP
- reverse phase
- SCX
- strong cation exchange
- TMA
- tissue microarray
- WB
- Western blot.
REFERENCES
- 1. Hollingsworth J. M., Miller D. C., Daignault S., Hollenbeck B. K. (2006) Rising incidence of small renal masses: a need to reassess treatment effect. J. Natl. Cancer Inst. 98, 1331–1334 [DOI] [PubMed] [Google Scholar]
- 2. Cheville J. C., Lohse C. M., Zincke H., Weaver A. L., Blute M. L. (2003) Comparisons of outcome and prognostic features among histologic subtypes of renal cell carcinoma. Am. J. Surg. Pathol. 27, 612–624 [DOI] [PubMed] [Google Scholar]
- 3. Russo P. (2001) Renal cell carcinoma : clinical features and management. Methods Mol. Med. 53, 3–33 [DOI] [PubMed] [Google Scholar]
- 4. Lam J. S., Shvarts O., Leppert J. T., Pantuck A. J., Figlin R. A., Belldegrun A. S. (2005) Postoperative surveillance protocol for patients with localized and locally advanced renal cell carcinoma based on a validated prognostic nomogram and risk group stratification system. J. Urol. 174, 466–472 [DOI] [PubMed] [Google Scholar]
- 5. Hofmann H. S., Neef H., Krohe K., Andreev P., Silber R. E. (2005) Prognostic factors and survival after pulmonary resection of metastatic renal cell carcinoma. Eur. Urol. 48, 77–81 [DOI] [PubMed] [Google Scholar]
- 6. Motzer R. J., Mazumdar M., Bacik J., Berg W., Amsterdam A., Ferrara J. (1999) Survival and prognostic stratification of 670 patients with advanced renal cell carcinoma. J. Clin. Oncol. 17, 2530–2540 [DOI] [PubMed] [Google Scholar]
- 7. Tsui K. H., Shvarts O., Smith R. B., Figlin R. A., deKernion J. B., Belldegrun A. (2000) Prognostic indicators for renal cell carcinoma: a multivariate analysis of 643 patients using the revised 1997 TNM staging criteria. J. Urol. 163, 1090–1095 [DOI] [PubMed] [Google Scholar]
- 8. Johnson T. V., Young A. N., Force S., Master V. A. (2011) C-reactive protein may represent sensitive measure of renal cell carcinoma metastasis. Urol. Nurs. 31, 181–182, 194 [PubMed] [Google Scholar]
- 9. Dubinski W., Gabril M., Iakovlev V. V., Scorilas A., Youssef Y. M., Faragalla H., Kovacs K., Rotondo F., Metias S., Arsanious A., Plotkin A., Girgis A. H., Streutker C. J., Yousef G. M. (2011) Assessment of the prognostic significance of endoglin (CD105) in clear cell renal cell carcinoma using automated image analysis. Hum. Pathol. 43, 1037–1043 [DOI] [PubMed] [Google Scholar]
- 10. Iakovlev V. V., Gabril M., Dubinski W., Scorilas A., Youssef Y. M., Faragalla H., Kovacs K., Rotondo F., Metias S., Arsanious A., Plotkin A., Girgis A. H., Streutker C. J., Yousef G. M. (2012) Microvascular density as an independent predictor of clinical outcome in renal cell carcinoma: an automated image analysis study. Lab. Invest. 92, 46–56 [DOI] [PubMed] [Google Scholar]
- 11. Wuttig D., Zastrow S., Fussel S., Toma M. I., Meinhardt M., Kalman K., Junker K., Sanjmyatav J., Boll K., Hackermuller J., Rolle A., Grimm M. O., Wirth M. P. (2011) CD31, EDNRB and TSPAN7 are promising prognostic markers in clear-cell renal cell carcinoma revealed by genome-wide expression analyses of primary tumors and metastases. Int. J. Cancer 131, E693–704 [DOI] [PubMed] [Google Scholar]
- 12. Hoffmann N. E., Sheinin Y., Lohse C. M., Parker A. S., Leibovich B. C., Jiang Z., Kwon E. D. (2008) External validation of IMP3 expression as an independent prognostic marker for metastatic progression and death for patients with clear cell renal cell carcinoma. Cancer 112, 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tostain J., Li G., Gentil-Perret A., Gigante M. (2010) Carbonic anhydrase 9 in clear cell renal cell carcinoma: a marker for diagnosis, prognosis and treatment. Eur. J. Cancer 46, 3141–3148 [DOI] [PubMed] [Google Scholar]
- 14. Schultz L., Chaux A., Albadine R., Hicks J., Kim J. J., De Marzo A. M., Allaf M. E., Carducci M. A., Rodriguez R., Hammers H. J., Argani P., Reuter V. E., Netto G. J. (2011) Immunoexpression status and prognostic value of mTOR and hypoxia-induced pathway members in primary and metastatic clear cell renal cell carcinomas. Am. J. Surg. Pathol. 35, 1549–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arsanious A., Bjarnason G. A., Yousef G. M. (2009) From bench to bedside: current and future applications of molecular profiling in renal cell carcinoma. Mol. Cancer 8, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Romaschin A. D., Youssef Y., Chow T. F., Siu K. W., DeSouza L. V., Honey R. J., Stewart R., Pace K. T., Yousef G. M. (2009) Exploring the pathogenesis of renal cell carcinoma: pathway and bioinformatics analysis of dysregulated genes and proteins. Biol. Chem. 390, 125–135 [DOI] [PubMed] [Google Scholar]
- 17. Shi T., Dong F., Liou L. S., Duan Z. H., Novick A. C., DiDonato J. A. (2004) Differential protein profiling in renal-cell carcinoma. Mol. Carcinog. 40, 47–61 [DOI] [PubMed] [Google Scholar]
- 18. Siu K. W., DeSouza L. V., Scorilas A., Romaschin A. D., Honey R. J., Stewart R., Pace K., Youssef Y., Chow T. F., Yousef G. M. (2009) Differential protein expressions in renal cell carcinoma: new biomarker discovery by mass spectrometry. J. Proteome. Res. 8, 3797–3807 [DOI] [PubMed] [Google Scholar]
- 19. Khella H. W., White N. M., Faragalla H., Gabril M., Boazak M., Dorian D., Khalil B., Antonios H., Bao T. T., Pasic M. D., Honey R. J., Stewart R., Pace K. T., Bjarnason G. A., Jewett M. A., Yousef G. M. (2012) Exploring the role of miRNAs in renal cell carcinoma progression and metastasis through bioinformatic and experimental analyses. Tumour Biol. 33, 131–140 [DOI] [PubMed] [Google Scholar]
- 20. White N. M., Khella H. W., Grigull J., Adzovic S., Youssef Y. M., Honey R. J., Stewart R., Pace K. T., Bjarnason G. A., Jewett M. A., Evans A. J., Gabril M., Yousef G. M. (2011) miRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. Br. J. Cancer 105, 1741–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. White N. M., Yousef G. M. (2011) Translating molecular signatures of renal cell carcinoma into clinical practice. J. Urol. 186, 9–11 [DOI] [PubMed] [Google Scholar]
- 22. Pavlovich C. P., Schmidt L. S. (2004) Searching for the hereditary causes of renal-cell carcinoma. Nat. Rev. Cancer 4, 381–393 [DOI] [PubMed] [Google Scholar]
- 23. Sarto C., Marocchi A., Sanchez J. C., Giannone D., Frutiger S., Golaz O., Wilkins M. R., Doro G., Cappellano F., Hughes G., Hochstrasser D. F., Mocarelli P. (1997) Renal cell carcinoma and normal kidney protein expression. Electrophoresis 18, 599–604 [DOI] [PubMed] [Google Scholar]
- 24. DeSouza L. V., Grigull J., Ghanny S., Dube V., Romaschin A. D., Colgan T. J., Siu K. W. (2007) Endometrial carcinoma biomarker discovery and verification using differentially tagged clinical samples with multidimensional liquid chromatography and tandem mass spectrometry. Mol. Cell. Proteomics. 6, 1170–1182 [DOI] [PubMed] [Google Scholar]
- 25. DeSouza L., Diehl G., Rodrigues M. J., Guo J., Romaschin A. D., Colgan T. J., Siu K. W. (2005) Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res. 4, 377–386 [DOI] [PubMed] [Google Scholar]
- 26. Ralhan R., DeSouza L. V., Matta A., Chandra T. S., Ghanny S., Datta G. S., Bahadur S., Siu K. W. (2008) Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol. Cell. Proteomics. 7, 1162–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ralhan R., Masui O., DeSouza L. V., Matta A., Macha M., Siu K. W. (2011) Identification of proteins secreted by head and neck cancer cell lines using LC-MS/MS: strategy for discovery of candidate serological biomarkers. Proteomics 11, 2363–2376 [DOI] [PubMed] [Google Scholar]
- 28. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 29. Tang W. H., Shilov I. V., Seymour S. L. (2008) Nonlinear fitting method for determining local false discovery rates from decoy database searches. J. Proteome Res. 7, 3661–3667 [DOI] [PubMed] [Google Scholar]
- 30. Voisin S. N., Krakovska O., Matta A., DeSouza L. V., Romaschin A. D., Colgan T. J., Siu K. W. (2011) Identification of novel molecular targets for endometrial cancer using a drill-down LC-MS/MS approach with iTRAQ. PLoS One 6, e16352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang N., Li L. (2008) Exploring the precursor ion exclusion feature of liquid chromatography-electrospray ionization quadrupole time-of-flight mass spectrometry for improving protein identification in shotgun proteome analysis. Anal. Chem. 80, 4696–4710 [DOI] [PubMed] [Google Scholar]
- 32. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ito K., Ralph S. J. (2012) Inhibiting galectin-1 reduces murine lung metastasis with increased CD4(+) and CD8 (+) T cells and reduced cancer cell adherence. Clin. Exp. Metastasis 29, 561–572 [DOI] [PubMed] [Google Scholar]
- 34. Kim H. J., Jeon H. K., Cho Y. J., Park Y. A., Choi J. J., Do I. G., Song S. Y., Lee Y. Y., Choi C. H., Kim T. J., Bae D. S., Lee J. W., Kim B. G. (2012) High galectin-1 expression correlates with poor prognosis and is involved in epithelial ovarian cancer proliferation and invasion. Eur. J. Cancer 48, 1914–1921 [DOI] [PubMed] [Google Scholar]
- 35. Camby I., Belot N., Lefranc F., Sadeghi N., de L. Y., Kaltner H., Musette S., Darro F., Danguy A., Salmon I., Gabius H. J., Kiss R. (2002) Galectin-1 modulates human glioblastoma cell migration into the brain through modifications to the actin cytoskeleton and levels of expression of small GTPases. J. Neuropathol. Exp. Neurol. 61, 585–596 [DOI] [PubMed] [Google Scholar]
- 36. Lu J., Guo H., Treekitkarnmongkol W., Li P., Zhang J., Shi B., Ling C., Zhou X., Chen T., Chiao P. J., Feng X., Seewaldt V. L., Muller W. J., Sahin A., Hung M. C., Yu D. (2009) 14–3-3zeta cooperates with ErbB2 to promote ductal carcinoma in situ progression to invasive breast cancer by inducing epithelial-mesenchymal transition. Cancer Cell 16, 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moiseeva E. P., Spring E. L., Baron J. H., de Bono D. P. (1999) Galectin 1 modulates attachment, spreading and migration of cultured vascular smooth muscle cells via interactions with cellular receptors and components of extracellular matrix. J. Vasc. Res. 36, 47–58 [DOI] [PubMed] [Google Scholar]
- 38. Pollard T. D., Borisy G. G. (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 39. Witke W. (2004) The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 14, 461–469 [DOI] [PubMed] [Google Scholar]
- 40. Wu M. H., Hong T. M., Cheng H. W., Pan S. H., Liang Y. R., Hong H. C., Chiang W. F., Wong T. Y., Shieh D. B., Shiau A. L., Jin Y. T., Chen Y. L. (2009) Galectin-1-mediated tumor invasion and metastasis, up-regulated matrix metalloproteinase expression, and reorganized actin cytoskeletons. Mol. Cancer Res. 7, 311–318 [DOI] [PubMed] [Google Scholar]
- 41. Zou L., Hazan R., Roy P. (2009) Profilin-1 overexpression restores adherens junctions in MDA-MB-231 breast cancer cells in R-cadherin-dependent manner. Cell Motil. Cytoskeleton 66, 1048–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Noma N., Simizu S., Kambayashi Y., Kabe Y., Suematsu M., Umezawa K. (2012) Involvement of NF-kappaB-mediated expression of galectin-3-binding protein in TNF-alpha-induced breast cancer cell adhesion. Oncol. Rep. 27, 2080–2084 [DOI] [PubMed] [Google Scholar]
- 43. Fan C., Fu Z., Su Q., Angelini D. J., Van E. J., Johns R. A. (2011) S100A11 mediates hypoxia-induced mitogenic factor (HIMF)-induced smooth muscle cell migration, vesicular exocytosis, and nuclear activation. Mol. Cell. Proteomics. 10, M110.000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Keshamouni V. G., Michailidis G., Grasso C. S., Anthwal S., Strahler J. R., Walker A., Arenberg D. A., Reddy R. C., Akulapalli S., Thannickal V. J., Standiford T. J., Andrews P. C., Omenn G. S. (2006) Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J. Proteome Res. 5, 1143–1154 [DOI] [PubMed] [Google Scholar]
- 45. Cheng C. W., Wang H. W., Chang C. W., Chu H. W., Chen C. Y., Yu J. C., Chao J. I., Liu H. F., Ding S. L., Shen C. Y. (2012) MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res. Treat. 134, 1081–1093 [DOI] [PubMed] [Google Scholar]
- 46. Diamandis M., White N. M., Yousef G. M. (2010) Personalized medicine: marking a new epoch in cancer patient management. Mol. Cancer Res. 8, 1175–1187 [DOI] [PubMed] [Google Scholar]
- 47. Yousef G. M. (2012) Personalized cancer genomics: the road map to clinical implementation. Clin. Chem. 58, 661–663 [DOI] [PubMed] [Google Scholar]
- 48. Minamida S., Iwamura M., Kodera Y., Kawashima Y., Ikeda M., Okusa H., Fujita T., Maeda T., Baba S. (2011) Profilin 1 overexpression in renal cell carcinoma. Int. J. Urol. 18, 63–71 [DOI] [PubMed] [Google Scholar]
- 49. Dallmann K., Junker H., Balabanov S., Zimmermann U., Giebel J., Walther R. (2004) Human agmatinase is diminished in the clear cell type of renal cell carcinoma. Int. J. Cancer 108, 342–347 [DOI] [PubMed] [Google Scholar]
- 50. Hwa J. S., Park H. J., Jung J. H., Kam S. C., Park H. C., Kim C. W., Kang K. R., Hyun J. S., Chung K. H. (2005) Identification of proteins differentially expressed in the conventional renal cell carcinoma by proteomic analysis. J. Korean Med. Sci. 20, 450–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xinzhou H., Ning Y., Ou W., Xiaodan L., Fumin Y., Huitu L., Wei Z. (2011) RKIp inhibits the migration and invasion of human prostate cancer PC-3M cells through regulation of extracellular matrix. Mol. Biol. (Mosk.) 45, 1004–1011 [PubMed] [Google Scholar]
- 52. Yang L. P., Jiang S., Liu J. Q., Miao X. Y., Yang Z. L. (2012) Association of immunostaining of galectin-3 and Sambucus nigra agglutinin with invasion, metastasis and poor progression of gallbladder adenocarcinoma. Hepatogastroenterology 59 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 53. Wang Y. G., Kim S. J., Baek J. H., Lee H. W., Jeong S. Y., Chun K. H. (2012) Galectin-3 increases the motility of mouse melanoma cells by regulating MMP-1 expression. Exp. Mol. Med. 44, 387–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gillies R. J., Robey I., Gatenby R. A. (2008) Causes and consequences of increased glucose metabolism of cancers. J. Nucl. Med. 49 Suppl 2, 24S–42S [DOI] [PubMed] [Google Scholar]
- 55. Huang S. K., Darfler M. M., Nicholl M. B., You J., Bemis K. G., Tegeler T. J., Wang M., Wery J. P., Chong K. K., Nguyen L., Scolyer R. A., Hoon D. S. (2009) LC/MS-based quantitative proteomic analysis of paraffin-embedded archival melanomas reveals potential proteomic biomarkers associated with metastasis. PLoS One 4, e4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ramaswamy S., Ross K. N., Lander E. S., Golub T. R. (2003) A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33, 49–54 [DOI] [PubMed] [Google Scholar]
- 57. Moiseeva E. P., Williams B., Goodall A. H., Samani N. J. (2003) Galectin-1 interacts with beta-1 subunit of integrin. Biochem. Biophys. Res. Commun. 310, 1010–1016 [DOI] [PubMed] [Google Scholar]
- 58. Camby I., Decaestecker C., Lefranc F., Kaltner H., Gabius H. J., Kiss R. (2005) Galectin-1 knocking down in human U87 glioblastoma cells alters their gene expression pattern. Biochem. Biophys. Res. Commun. 335, 27–35 [DOI] [PubMed] [Google Scholar]
- 59. Aitken A. (2006) 14–3-3 proteins: a historic overview. Semin. Cancer Biol. 16, 162–172 [DOI] [PubMed] [Google Scholar]
- 60. Gronborg M., Kristiansen T. Z., Iwahori A., Chang R., Reddy R., Sato N., Molina H., Jensen O. N., Hruban R. H., Goggins M. G., Maitra A., Pandey A. (2006) Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol. Cell. Proteomics. 5, 157–171 [DOI] [PubMed] [Google Scholar]
- 61. Wu N., Zhang W., Yang Y., Liang Y. L., Wang L. Y., Jin J. W., Cai X. M., Zha X. L. (2006) Profilin 1 obtained by proteomic analysis in all-trans retinoic acid-treated hepatocarcinoma cell lines is involved in inhibition of cell proliferation and migration. Proteomics 6, 6095–6106 [DOI] [PubMed] [Google Scholar]
- 62. Ma C. Y., Zhang C. P., Zhong L. P., Pan H. Y., Chen W. T., Wang L. Z., Andrew O. W., Ji T., Han W. (2011) Decreased expression of profilin 2 in oral squamous cell carcinoma and its clinicopathological implications. Oncol. Rep. 26, 813–823 [DOI] [PubMed] [Google Scholar]
- 63. Chan C. M., Wong S. C., Lam M. Y., Hui E. P., Chan J. K., Lo E. S., Cheuk W., Wong M. C., Tsao S. W., Chan A. T. (2008) Proteomic comparison of nasopharyngeal cancer cell lines C666–1 and NP69 identifies down-regulation of annexin II and beta2-tubulin for nasopharyngeal carcinoma. Arch. Pathol. Lab. Med. 132, 675–683 [DOI] [PubMed] [Google Scholar]
- 64. Zoidakis J., Makridakis M., Zerefos P. G., Bitsika V., Esteban S., Frantzi M., Stravodimos K., Anagnou N. P., Roubelakis M. G., Sanchez-Carbayo M., Vlahou A. (2012) Profilin 1 is a potential biomarker for bladder cancer aggressiveness. Mol. Cell. Proteomics. 11, M111.009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kuo P. L., Huang M. S., Cheng D. E., Hung J. Y., Yang C. J., Chou S. H. (2012) Lung cancer-derived galectin-1 enhances tumorigenic potentiation of tumor-associated dendritic cells by expressing heparin-binding EGF-like growth factor. J. Biol. Chem. 287, 9753–9764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huang L., Zheng M., Zhou Q. M., Zhang M. Y., Jia W. H., Yun J. P., Wang H. Y. (2011) Identification of a gene-expression signature for predicting lymph node metastasis in patients with early stage cervical carcinoma. Cancer 117, 3363–3373 [DOI] [PubMed] [Google Scholar]
- 67. Li Y., Zou L., Li Q., Haibe-Kains B., Tian R., Li Y., Desmedt C., Sotiriou C., Szallasi Z., Iglehart J. D., Richardson A. L., Wang Z. C. (2010) Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat. Med. 16, 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lin C. C., Chen L. C., Tseng V. S., Yan J. J., Lai W. W., Su W. P., Lin C. H., Huang C. Y., Su W. C. (2011) Malignant pleural effusion cells show aberrant glucose metabolism gene expression. Eur. Respir. J. 37, 1453–1465 [DOI] [PubMed] [Google Scholar]
- 69. Hamaguchi T., Iizuka N., Tsunedomi R., Hamamoto Y., Miyamoto T., Iida M., Tokuhisa Y., Sakamoto K., Takashima M., Tamesa T., Oka M. (2008) Glycolysis module activated by hypoxia-inducible factor 1alpha is related to the aggressive phenotype of hepatocellular carcinoma. Int. J. Oncol. 33, 725–731 [PubMed] [Google Scholar]
- 70. Tang Z., Yuan S., Hu Y., Zhang H., Wu W., Zeng Z., Yang J., Yun J., Xu R., Huang P. (2012) Over-expression of GAPDH in human colorectal carcinoma as a preferred target of 3-bromopyruvate propyl ester. J. Bioenerg. Biomembr. 44, 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tsutsumi S., Fukasawa T., Yamauchi H., Kato T., Kigure W., Morita H., Asao T., Kuwano H. (2009) Phosphoglucose isomerase enhances colorectal cancer metastasis. Int. J. Oncol. 35, 1117–1121 [DOI] [PubMed] [Google Scholar]
- 72. Funasaka T., Hogan V., Raz A. (2009) Phosphoglucose isomerase/autocrine motility factor mediates epithelial and mesenchymal phenotype conversions in breast cancer. Cancer Res. 69, 5349–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Colas E., Perez C., Cabrera S., Pedrola N., Monge M., Castellvi J., Eyzaguirre F., Gregorio J., Ruiz A., Llaurado M., Rigau M., Garcia M., Ertekin T., Montes M., Lopez-Lopez R., Carreras R., Xercavins J., Ortega A., Maes T., Rosell E., Doll A., Abal M., Reventos J., Gil-Moreno A. (2011) Molecular markers of endometrial carcinoma detected in uterine aspirates. Int. J. Cancer 129, 2435–2444 [DOI] [PubMed] [Google Scholar]
- 74. Lovat P. E., Corazzari M., Armstrong J. L., Martin S., Pagliarini V., Hill D., Brown A. M., Piacentini M., Birch-Machin M. A., Redfern C. P. (2008) Increasing melanoma cell death using inhibitors of protein disulfide isomerases to abrogate survival responses to endoplasmic reticulum stress. Cancer Res. 68, 5363–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ashrafian H., O'Flaherty L., Adam J., Steeples V., Chung Y. L., East P., Vanharanta S., Lehtonen H., Nye E., Hatipoglu E., Miranda M., Howarth K., Shukla D., Troy H., Griffiths J., Spencer-Dene B., Yusuf M., Volpi E., Maxwell P. H., Stamp G., Poulsom R., Pugh C. W., Costa B., Bardella C., Di Renzo M. F., Kotlikoff M. I., Launonen V., Aaltonen L., El-Bahrawy M., Tomlinson I., Pollard P. J. (2010) Expression profiling in progressive stages of fumarate-hydratase deficiency: the contribution of metabolic changes to tumorigenesis. Cancer Res. 70, 9153–9165 [DOI] [PubMed] [Google Scholar]
- 76. Hao J., Wang K., Yue Y., Tian T., Xu A., Hao J., Xiao X., He D. (2012) Selective expression of S100A11 in lung cancer and its role in regulating proliferation of adenocarcinomas cells. Mol. Cell Biochem. 359, 323–332 [DOI] [PubMed] [Google Scholar]
- 77. Meding S., Balluff B., Elsner M., Schone C., Rauser S., Nitsche U., Maak M., Schafer A., Hauck S. M., Ueffing M., Langer R., Hofler H., Friess H., Rosenberg R., Walch A. (2012) Tissue based proteomics reveals FXYD3, S100A11 and GSTM3 as novel markers for regional lymph node metastasis in colon cancer. J. Pathol. [Epub ahead for print] [DOI] [PubMed] [Google Scholar]
- 78. Thongwatchara P., Promwikorn W., Srisomsap C., Chokchaichamnankit D., Boonyaphiphat P., Thongsuksai P. (2011) Differential protein expression in primary breast cancer and matched axillary node metastasis. Oncol. Rep. 26, 185–191 [DOI] [PubMed] [Google Scholar]
- 79. Wang J. W., Peng S. Y., Li J. T., Wang Y., Zhang Z. P., Cheng Y., Cheng D. Q., Weng W. H., Wu X. S., Fei X. Z., Quan Z. W., Li J. Y., Li S. G., Liu Y. B. (2009) Identification of metastasis-associated proteins involved in gallbladder carcinoma metastasis by proteomic analysis and functional exploration of chloride intracellular channel 1. Cancer Lett. 281, 71–81 [DOI] [PubMed] [Google Scholar]
- 80. Hu L., Lau S. H., Tzang C. H., Wen J. M., Wang W., Xie D., Huang M., Wang Y., Wu M. C., Huang J. F., Zeng W. F., Sham J. S., Yang M., Guan X. Y. (2004) Association of Vimentin overexpression and hepatocellular carcinoma metastasis. Oncogene 23, 298–302 [DOI] [PubMed] [Google Scholar]
- 81. Lang S. H., Hyde C., Reid I. N., Hitchcock I. S., Hart C. A., Bryden A. A., Villette J. M., Stower M. J., Maitland N. J. (2002) Enhanced expression of vimentin in motile prostate cell lines and in poorly differentiated and metastatic prostate carcinoma. Prostate 52, 253–263 [DOI] [PubMed] [Google Scholar]
- 82. Cao W., Liu N., Tang S., Bao L., Shen L., Yuan H., Zhao X., Lu H. (2008) Acetyl-coenzyme A acyltransferase 2 attenuates the apoptotic effects of BNIP3 in two human cell lines. Biochim. Biophys. Acta 1780, 873–880 [DOI] [PubMed] [Google Scholar]
- 83. Chen Y. R., Juan H. F., Huang H. C., Huang H. H., Lee Y. J., Liao M. Y., Tseng C. W., Lin L. L., Chen J. Y., Wang M. J., Chen J. H., Chen Y. J. (2006) Quantitative proteomic and genomic profiling reveals metastasis-related protein expression patterns in gastric cancer cells. J. Proteome. Res. 5, 2727–2742 [DOI] [PubMed] [Google Scholar]
- 84. Ma Z., Cao M., Liu Y., He Y., Wang Y., Yang C., Wang W., Du Y., Zhou M., Gao F. (2010) Mitochondrial F1Fo-ATP synthase translocates to cell surface in hepatocytes and has high activity in tumor-like acidic and hypoxic environment. Acta Biochim. Biophys. Sin. (Shanghai) 42, 530–537 [DOI] [PubMed] [Google Scholar]
- 85. Xie H. L., Li Z. Y., Gan R. L., Li X. J., Zhang Q. L., Hui M., Zhou X. T. (2010) Differential gene and protein expression in primary gastric carcinomas and their lymph node metastases as revealed by combined cDNA microarray and tissue microarray analysis. J. Dig. Dis. 11, 167–175 [DOI] [PubMed] [Google Scholar]
- 86. Hynninen P., Parkkila S., Huhtala H., Pastorekova S., Pastorek J., Waheed A., Sly W. S., Tomas E. (2012) Carbonic anhydrase isozymes II, IX, and XII in uterine tumors. APMIS 120, 117–129 [DOI] [PubMed] [Google Scholar]
- 87. Cheng I. K., Ching A. K., Chan T. C., Chan A. W., Wong C. K., Choy K. W., Kwan M., Lai P. B., Wong N. (2010) Reduced CRYL1 expression in hepatocellular carcinoma confers cell growth advantages and correlates with adverse patient prognosis. J. Pathol. 220, 348–360 [DOI] [PubMed] [Google Scholar]
- 88. Cho-Vega J. H., Vega F., Schwartz M. R., Prieto V. G. (2007) Expression of dicarbonyl/L-xylulose reductase (DCXR) in human skin and melanocytic lesions: morphological studies supporting cell adhesion function of DCXR. J. Cutan. Pathol. 34, 535–542 [DOI] [PubMed] [Google Scholar]
- 89. Sakakura C., Takemura M., Hagiwara A., Shimomura K., Miyagawa K., Nakashima S., Yoshikawa T., Takagi T., Kin S., Nakase Y., Fujiyama J., Hayasizaki Y., Okazaki Y., Yamagishi H. (2004) Overexpression of dopa decarboxylase in peritoneal dissemination of gastric cancer and its potential as a novel marker for the detection of peritoneal micrometastases with real-time RT-PCR. Br. J. Cancer 90, 665–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kanamori T., Nishimaki K., Asoh S., Ishibashi Y., Takata I., Kuwabara T., Taira K., Yamaguchi H., Sugihara S., Yamazaki T., Ihara Y., Nakano K., Matuda S., Ohta S. (2003) Truncated product of the bifunctional DLST gene involved in biogenesis of the respiratory chain. EMBO J. 22, 2913–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang B., Hsu S. H., Frankel W., Ghoshal K., Jacob S. T. (2012) Stat3-mediated activation of miR-23a suppresses gluconeogenesis in hepatocellular carcinoma by downregulating G6PC and PGC-1alpha. Hepatology 56, 186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chen M., Zhang J., Li N., Qian Z., Zhu M., Li Q., Zheng J., Wang X., Shi G. (2011) Promoter hypermethylation mediated downregulation of FBP1 in human hepatocellular carcinoma and colon cancer. PLoS One 6, e25564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hawthorn L., Luce J., Stein L., Rothschild J. (2010) Integration of transcript expression, copy number and LOH analysis of infiltrating ductal carcinoma of the breast. BMC Cancer 10, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Han M. J., Wang H., Beer L. A., Tang H. Y., Herlyn M., Speicher D. W. (2010) A systems biology analysis of metastatic melanoma using in-depth three-dimensional protein profiling. Proteomics 10, 4450–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ross C. D., Gomaa M. A., Gillies E., Juengel R., Medina J. E. (2000) Tumor grade, microvessel density, and activities of malate dehydrogenase, lactate dehydrogenase, and hexokinase in squamous cell carcinoma. Otolaryngol. Head Neck Surg. 122, 195–200 [DOI] [PubMed] [Google Scholar]
- 96. Chaika N. V., Yu F., Purohit V., Mehla K., Lazenby A. J., DiMaio D., Anderson J. M., Yeh J. J., Johnson K. R., Hollingsworth M. A., Singh P. K. (2012) Differential expression of metabolic genes in tumor and stromal components of primary and metastatic loci in pancreatic adenocarcinoma. PLoS One 7, e32996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang L., Liu H. L., Li Y., Yuan P. (2011) Proteomic analysis of pancreatic intraepithelial neoplasia and pancreatic carcinoma in rat models. World J. Gastroenterol. 17, 1434–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yun J., Frankenberger C. A., Kuo W. L., Boelens M. C., Eves E. M., Cheng N., Liang H., Li W. H., Ishwaran H., Minn A. J., Rosner M. R. (2011) Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 30, 4500–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Fan H. Z., Liu H., Zhang C., Gao D. M., Xue Q., Chen J., Sun R. X., Liu Y. K., Yang P. Y. (2009) Comparative proteomics and molecular mechanical analysis in CDA-II induced therapy of LCI-D20 hepatocellular carcinoma model. J. Cancer Res. Clin. Oncol. 135, 591–602 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.