Abstract
Cells secrete a large number of proteins to communicate with their surroundings. Furthermore, plasma membrane proteins and intracellular proteins can be released into the extracellular space by regulated or non-regulated processes. Here, we profiled the supernatant of 11 cell lines that are representative of different stages of breast cancer development by specifically capturing N-glycosylated peptides using the N-glyco FASP technology. For accurate quantification we developed a super-SILAC mix from several labeled breast cancer cell lines and used it as an internal standard for all samples. In total, 1398 unique N-glycosylation sites were identified and quantified. Enriching for N-glycosylated peptides focused the analysis on classically secreted and membrane proteins. N-glycosylated secretome profiles correctly clustered the different cell lines to their respective cancer stage, suggesting that biologically relevant differences were detected. Five different profiles of glycoprotein dynamics during cancer development were detected, and they contained several proteins with known roles in breast cancer. We then used the super-SILAC mix in plasma, which led to the quantification of a large number of the previously identified N-glycopeptides in this important body fluid. The combination of quantifying the secretome of cancer cell lines and of human plasma with a super-SILAC approach appears to be a promising new approach for finding markers of disease.
There has been a long-standing interest in applying proteomics to the cancer field (1). Technological advances in liquid chromatography-mass spectrometry (LC-MS) have made it feasible to profile the proteome of cancer cells to great depth (2, 3) and these developments now allow studying protein expression on a systems wide level (4). Analyses of intracellular proteins provide data on what is occurring at the intracellular level in terms of biochemical processes, signaling pathways and cellular structure. However, from a clinical perspective, focusing on proteins that are secreted by these cells is very appealing for diagnostic purposes, as they may filtrate into the peripheral blood (5). This is advantageous because peripheral blood is an easily accessible source whereas tissue biopsies are invasive and they are generally only taken when a medical condition is already suspected. Blood itself is a very complex fluid whose proteome is extremely challenging to analyze because of its very high dynamic range (6–8). Furthermore, a tumor in the initial stages would not be expected to secrete large amounts of proteins and these proteins would be severely diluted in the total blood volume (9). Therefore, discovery of biomarkers by direct analysis of blood plasma has been very difficult so far (10). A more straightforward approach would be the analysis of proteins secreted from homogeneous cell populations (11–14). Consequently, the conditioned medium of cell lines has extensively been used for the analysis of secreted cancer proteins (15). The secretome contains proteins that are actively secreted through classical and nonclassical routes but also proteins that are shed from the plasma membrane by various sheddases (12). Secretome studies are generally performed using serum-free media to reduce the initial protein contents. Further precautions are taken to minimize the contamination of intracellular proteins arising from dead cells that release their contents. Despite these caveats, the totality of proteins that are found in the conditioned medium has been referred to as the “secretome” (13).
During cancer development, the invasive capacity of the cells increases progressively. Cancer cells lose cell-cell adhesion which allows eventual release of the cell from the surrounding tissue and may facilitate metastasis to other organs. The extracellular matrix is an important factor in this process as it plays a significant role in regulating numerous cellular functions like adhesion, cell shape, migration, proliferation, polarity, differentiation and apoptosis (16, 17). Many components of the extracellular matrix change in expression during cancer development. Therefore, these changes would likely be reflected in the protein contents of the secretome.
Here, we set out to profile the proteins that are secreted by breast cancer cell lines from different stages by MS-based proteomics methods. For several reasons, we focused on N-glycosylated proteins as an appropriate handle to probe proteins that could be of clinical interest. First, proteins that use the classical secretion pathway or are shed from the membrane are typically N-glycosylated because they have passed through the endoplasmic reticulum (ER)1 and Golgi system (18). Second, glycosylation may enhance the stability of the protein and protect it from proteolytic degradation (19), which would increase the likelihood of detection away from the place where the protein was produced or secreted. Third, glycosylation has a direct relationship to cancer development (20, 21). Fourth, almost all of the currently used protein biomarkers are in fact glycoproteins, such as carcinoembryonic antigen (CEA), cancer antigen 125 (CA125), and prostate-specific antigen (PSA) (22). Finally, glycoproteins have themselves been used as therapeutic targets in cancer. These include ErbB2, targeted by trastuzumab and VEGF-A, targeted by bevacizumab (23).
Experimentally, a prime advantage of targeting glycosylation is the fact that glycopeptides or glycoproteins can be efficiently enriched over nonglycosylated molecules. In proteomics, enrichment targeted to N-glycosylation has typically been performed using hydrazide chemistry (24–26) or lectin based enrichment (27, 28). Our group has previously used the ‘filter aided sample preparation’ (FASP) as a basis of N-glycopeptide enrichment (29). The filter membrane in FASP can be employed to physically retain mixtures of lectins, which do not need to be coupled to beads. N-glycopeptides are first bound to the lectins and in a subsequent step simultaneously deglycosylated and released from the lectins. The complexity of the sample is thereby reduced to a level where extensive fractionation is dispensable and the highly enriched fraction of previously N-glycosylated peptides can readily be analyzed in a single high-resolution LC-MS run. We have used N-glyco-FASP to determine N-glycosylation sites in several mouse tissues (29) and in evolutionary distant model organisms (30). Here we adapted the method to supernatants of cell lines and we used the latest generation of Orbitrap analyzers for MS detection. Furthermore, to allow accurate quantification of differences in abundance levels between different secretomes, we spiked an internal standard of a super-SILAC mix (31) containing the conditioned medium of three heavy stable isotope labeled cell lines into all the conditioned medium samples. We collected the conditioned medium from a panel of eleven breast cell lines that were representative of five different cancer stages, from healthy to metastatic cells. The method was further applied to the analysis of blood plasma to verify its applicability in a body fluid context.
EXPERIMENTAL PROCEDURES
Cell Culturing and SILAC Labeling
Primary human mammary epithelial cells (HMEC) were obtained from the European Collection of Cell Cultures (ECACC, Salisbury, UK; HMEpC1) and from Lonza (HMEpC2; Basel, Switzerland); HMT-3522-S1 and MFM223 cells were obtained from ECACC. HCC202, and HCC2218 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA); HCC1143, HCC1937, and HCC1599 cells were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ); MCF-10a and MDA-MB-453 cells were kindly provided by Axel Ulrich (Max-Planck Institute of Biochemistry, Martinsried, Germany). HMEC cells were grown in mammary epithelial cell growth medium (ECACC); MCF-10a cells were cultured in DMEM/F12 supplemented with 5% horse serum, 20 ng/ml EGF, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone and 0.1 μg/ml choleratoxin; HMT-3522-S1 cells were cultured in DMEM/F12 supplemented with 250 ng/ml insulin, 10 μg/ml transferrin, 0.1 μm sodium selenite, 0.1 nm 17β-estradiol, 5 μg/ml ovine prolactin, 0.5 μg/ml hydrocortisone and 10 ng/ml EGF; HCC1143, HCC1937, HCC202, HCC2218, and HCC1599 were grown in RPMI supplemented with 10% FBS; MFM223 cells were grown in MEM supplemented with 10% FBS; MDA-MB-453 cells were cultured in l-15 supplemented with 10% FBS. All cells were cultured with Pen/Strep at 37 °C and under 5% CO2, except for MDA-MB-453 which were culture under 0% CO2.
For the super-SILAC mix, HCC1143, HCC1937, and HCC2218 were SILAC labeled by culturing in RPMI with the natural lysine and arginine replaced by heavy isotope labeled amino acids, l-13C615N4-arginine (R10; 13C6 98%, 15N4, 98%) and l-13C615N2-lysine (K8; 13C6 98%, 15N2, 98%). Labeled amino acids were purchased from Cambridge Isotope Laboratories (Andover, MA). The media were supplemented with 10% dialyzed serum. Cells were cultured for approximately five passages in the SILAC medium for complete incorporation of the heavy isotopes. Incorporation was checked by separate LC MS/MS analysis of supernatant. For each of the proteins discussed in the results, incorporation of the glycopeptide standard was better than 95%.
Collecting Conditioned Medium
The secretomes of the cell lines were collected as conditioned media. In short, 1.5 million cells were seeded in 100 mm culture dishes and left overnight for cell attachment in normal growth medium. After 24 h each medium was replaced by growth medium in which serum and other stimulatory proteins were omitted. After another 24 h the medium was collected and 4 ml of this collected conditioned medium was mixed 1:1 with the super-SILAC mix which consisted of the pooled conditioned medium of SILAC labeled HCC1143, HCC1937, and HCC2218. HCC2218 and HCC1599 grow in suspension and their conditioned medium was collected by taking the supernatant after spinning down at 1000 rpm for 5 min. Viability of all cell lines before, during and after serum starvation was monitored using the trypan blue exclusion technique. The conditioned medium was filtered through a 0.22 μm Durapore polyvinylidene difluoride Membrane (Millipore, Billerica, MA) and concentrated to 500 μl on Amicon Ultra 4, 30,000 molecular weight cutoff centrifugal filter units. This procedure was performed in quintuplicate for all the eleven cell lines.
Blood Plasma
Approximately 300 μg of plasma from four different female donors (obtained from the Biobank der Blutspender, Munich, Germany) was processed in duplicate. Two of the donors were later diagnosed with breast cancer. The super-SILAC internal standard was spiked-in before the N-glyco FASP procedure.
N-glyco FASP
To 500 μl of concentrated conditioned medium, 60 mg of urea and 16 μl of 1 m dithiotreitol were added, mixed and incubated at 56 °C for 15 min. This mixture was applied to an Ultracel YM-10 10,000 molecular weight cut-off centrifugal filter (Millipore, Billerica, MA) spun down and washed two times with 200 μl of 2 m urea in 0.1 m Tris/HCl pH 8.5. 100 μl of 0.05 m iodoacetamide was added and left for 30 min at RT in the dark. Two washes with 2 m urea in 0.1 m Tris/HCl pH 8.0 were performed and finally 10 μl of 0.5 μg/μl sequencing grade modified trypsin (Promega, Mannheim, Germany) and 100 μl of 2 m urea in 0.1 m Tris/HCl pH 8.0 were added and left for digestion overnight at 37 °C.
The digested peptides were eluted from the filters using two times 50 μl 1 × Binding Buffer (BB, 20 mm Tris/HCl pH 7.6, 1 mm MnCl2, 2 mm CaCl2, 1 m NaCl). Forty-five microliters of 3 mg/ml of wheat germ agglutinin (WGA; Sigma-Aldrich, Taufkirchen, Germany) and concanavalin A (Con A; Sigma-Aldrich, Taufkirchen, Germany) in 2 × BB and 6 μl of 50 mm of PMSF was added and left for 1h at RT. Afterwards the mixture was filtered through a Vivacon 500 30,000 molecular weight cutoff centrifugal filter (Sartorius Stedim, Göttingen, Germany). The flow through was collected as non-glycosylated secretome (and analyzed in quadruplicate), while the filtrate was washed with first 1 × BB followed by 0.05 m ammonium bicarbonate in H218O. Two microliters N-glycosidase F (Roche Diagnostics, Mannheim, Germany) was added and incubated at 37 °C for 3 h. The released deglycosylated peptides were then eluted. Before mass spectrometry analysis, both the non-glycosylated and deglycosylated fractions were desalted using C18 StageTips (32).
LC-MS
The samples were analyzed using LC-MS instrumentation consisting of an Easy nano-flow HPLC system (Thermo Fisher Scientific, Odense, Denmark) coupled via a nanoelectrospray ion source (Thermo Fisher Scientific, Bremen, Germany) to either an LTQ-Orbitrap Elite (33) or Q Exactive (34) mass spectrometer (both Thermo Fisher Scientific, Bremen, Germany). Peptide separation was performed on a 20 cm column with 75 μm inner diameter packed in-house with ReproSil-Pur C18-AQ 1.8 μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). Peptides were loaded in buffer A (0.5% acetic acid (v/v)) and eluted with a 140 min linear gradient of buffer B (80% acetonitrile, 0.5% acetic acid (v/v)) at 200 nL/min (5–30% buffer B in 90 min; 30–60% buffer B in 10 min; 60–95% B in 5 min; 5 min 95% buffer B). Mass spectra were acquired in a data-dependent manner, with an automatic switch between MS and MS/MS using a top 10 method. MS spectra were acquired in the Orbitrap analyzer with a mass range of 300–1650 m/z and 120,000 resolution at m/z 400 (Orbitrap Elite) or 300–1750 m/z and 70,000 resolution at m/z 200 (Q Exactive). HCD peptide fragments, acquired at 30 (Orbitrap Elite) or 25 normalized collision energy (Q Exactive), were analyzed at high resolution in the Orbitrap.
Data Analysis
The raw files were processed using the MaxQuant computational proteomics platform (35) version 1.2.2.9. The fragmentation spectra were searched against the Human IPI database v3.68 (87083 entries, common contaminants were added to this database, including Con A and WGA) using the Andromeda search engine (36) with the precursor and fragment mass tolerances set to 6 and 20 ppm, respectively, tryptic cleavage specificity with up two missed cleavages, minimal peptide length of six amino acids, carbamidomethyl (C) as fixed modification and oxidation (M) only as variable modification for the nonglycosylated fractions and oxidation (M) and deamidation 18O (N, +2.99826 Da) for the deglycosylated fractions. Leucines were replaced by isoleucines. False discovery rate, determined by using a reversed database, was set to 1% for peptide, modification site and protein identifications. Specifying the FDR independently for peptides and proteins ensures that we obtain the desired proportion of false positive proteins, independent of peptide statistics. Peptides that belong to proteins that did not make it above the independently specified protein FDR threshold were removed from the dataset. The actual, final, FDR of the peptide data set is therefore lower (3–5 times lower) than 1%. Peptides are assigned to protein groups, rather than proteins. Matching between runs from the same mass spectrometer and the same sample (i.e. nonglycosylated or deglycosylated) was performed with a 2 min. retention time window. Quantification was performed using the heavy super-SILAC mix as internal standard and ratios were normalized to this mix and expressed here as L/H (i.e. sample/super-SILAC internal standard). For the blood plasma analysis, the option “re-quantify” was disabled. For cases where no ratio could be determined, an arbitrary Log2 value of 7 or 9 was given, depending on whether a signal was seen in the light or heavy SILAC channel.
All the statistical analyses of the MaxQuant output tables were performed with the Perseus program (versions 1.2.3.3 and 1.2.7.4), which is a component of the MaxQuant distribution. The tables were filtered to remove contaminants and reversed sequences. Furthermore, only modified asparagines within the canonical sequence motif N!PS/T/C were accepted as true glycosylation sites. This extra restriction, together with the data set being enriched for N-glycosylated peptides, results in a FDR at N-glycosylation site identification and localization below 1% (29, 37). Hierarchical clustering on Z-scored values was based on Euclidean distance and average linkage clustering.
RESULTS AND DISCUSSION
Enrichment of N-glycosylated Peptides from Conditioned Medium of Breast Cancer Cell Lines
We used a panel of cell lines that were isolated from human breast tumors representative of various TNM stages, to profile differences in the secretome during breast cancer development. To investigate generic breast cancer related changes, rather than cell line specific differences, we selected per cancer stage two to three cell lines derived from different patients (4). Primary human mammary epithelial cells from two different sources were selected as control cell lines. MCF-10a and HMT-3522-S1 cells represent pre-malignant cells, HCC1143 and HCC1937 cells stage II tumors, HCC202, HCC2218, and HCC1599 cells stage III tumors and, finally, MFM223 and MDA-MB-453 are metastatic cells from pleural-effusions (Supplemental Table S1). This panel of cell lines covers stages in the progression to tumor phenotypes and includes triple negative and ErbB2 over-expressing cells.
The secretome of the different cell lines was collected as conditioned medium after 24 h of incubation. To reduce the amount of background proteins in the conditioned medium, supplementation of serum proteins and other growth factors to the culture medium was omitted. An initial test showed that none of the cell lines suffered from a significant reduction in viability on 24 h of serum and growth factor starvation (EXPERIMENTAL PROCEDURES).
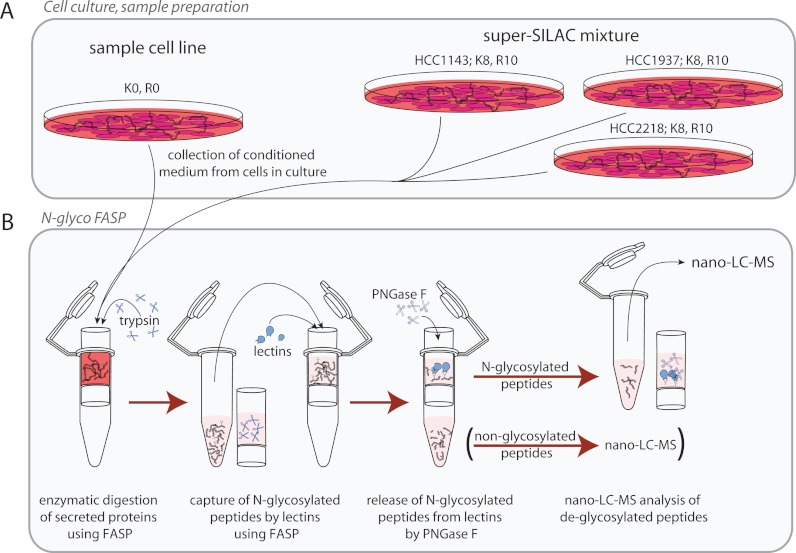
To enable SILAC-based quantification of differences in abundance levels of secreted proteins and peptides between the different cell lines, we collected conditioned medium from three representative different heavy stable isotope labeled cell lines. This so-called super-SILAC mix (31) was then mixed with the conditioned medium of the tested cell lines as an internal spike-in standard (38) (Fig. 1A).
Fig. 1.
Overview of the experimental workflow. A, The secretome was collected as conditioned medium from cell lines in culture. HCC1143, HCC1937 and HCC2218 were also cultured under SILAC conditions to generate a super-SILAC mix as an internal standard. B, N-glycosylated peptides were enriched using N-glyco FASP. Proteins were first digested to peptides and N-glycosylated peptides were then captured using lectins. Finally, N-glycosylated peptides were released by deglycosylation using PNGase F and the deglycosylated peptides were analyzed by LC-MS.
The conditioned medium was collected and processed in quintuplicate replicates. After collection and mixing with the super-SILAC mix, the conditioned medium was filtered and concentrated. Proteins in the concentrate were then digested with trypsin using the FASP method. After the digestion, N-glycosylated peptides were captured on a 30 kDa filter by two broad spectrum lectins—concanavalin A and wheat germ agglutinin. The N-glycosylated peptides were thereby separated from non-glycosylated peptides (N-glyco FASP, Fig. 1B)(29). Next, N-glycosylated peptides were released from the lectins by deglycosylation and analyzed by highly sensitive LC-MS using the latest generation linear ion trap Orbitrap or quadrupole Orbitrap instrumentation (33, 34). As the deglycosylation reaction was performed in H218O, the former site of glycosylation could be recognized by the 2.98826 Da increase of the mass of asparagine, representing the deamidation of this amino acid with incorporation of one heavy oxygen (39, 40).
Global Results for Detected N-Glycosylation Sites
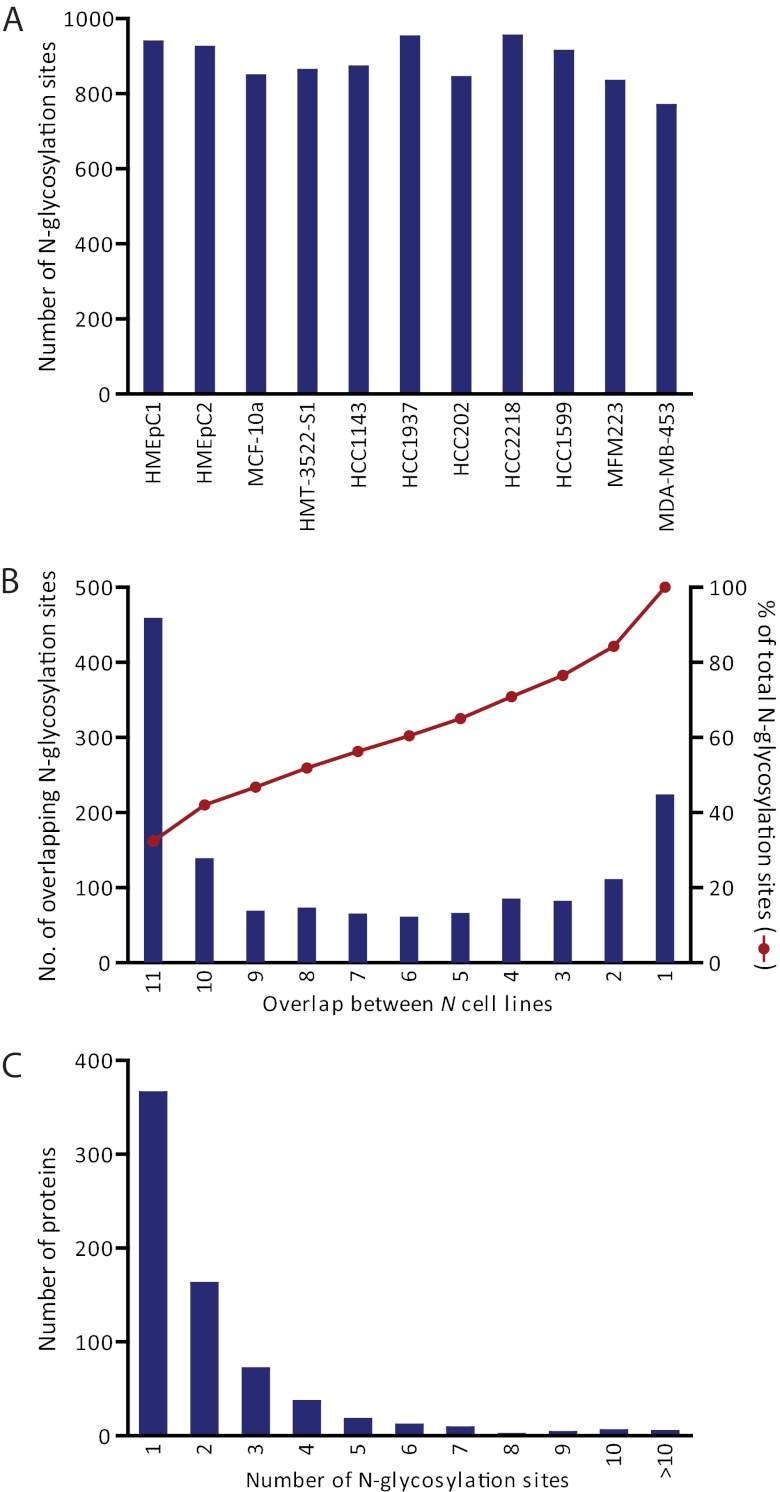
Combining LC-MS data from the analysis in quintuplicate of the secretome of the 11 cell lines, we obtained 45,824 peptide spectrum matches with a 1% FDR, which incorporated the characteristic asparagine mass increment and whose N-glycosylation site agreed with the canonical N!PS/T/C motif. This amounted to a data set of 1398 unique N-glycosylation sites (Supplemental Table S2). The use of replicates combined with a consistent spiked-in super-SILAC mix resulted in a fairly large overlap of identifications between the different cell lines: on average, 881 N-glycosylation sites were identified and quantified per cell line (Fig. 2A) with almost one third appearing in all cell lines and more than half of the sites in at least eight of the analyzed cell lines (Fig. 2B). When combining the cell lines for each cancer stage, more than half of the sites were found in each of them.
Fig. 2.
Summary statistics of identified N-glycosylation sites. A, Number of unique N-glycosylation sites identified in each cell line. B, Overlap of N-glycosylation sites between different cell lines. C, Number of N-glycosylation sites identified per protein.
In total, 1253 of the N-glycosylation sites that we identified were annotated in Uniprot (release 2012_01). However, for almost 60% of these annotated sites, knowledge is based on prediction or similarity. Therefore, 868 (62%) N-glycosylation sites from our data set can be considered as new experimentally confirmed N-glycosylation sites, a substantial increase compared with the little more than 2300 sites that were annotated based on empirical data before. These N-glycosylation sites mapped to 701 different protein groups. Of these, approximately half were identified with one N-glycosylation site and one fifth with two N-glycosylation sites (Fig. 2C). The maximum number of N-glycosylation sites per protein was twenty for Alpha-2-macroglobulin receptor. Other proteins with ten or more identified N-glycosylation sites include laminin subunit alpha-5, cadherin family member 8, attractin, cytotactin, sortilin-related receptor, cadherin family member 7, gastric mucin, CD109, and laminin B2 chain.
Many identified glycoproteins are known secreted or membrane proteins, including components of the extracellular matrix such as proteoglycans, fibronectin, and laminins. Among the identified glycoproteins we find some of the membrane proteins that are used to classify the cell lines. For example, ErbB2 expression levels are high in HCC202 and HCC2218, whereas other growth factor receptors are found with relatively higher expression in the metastatic cell lines (MFM223, MDA-MB-453).
Using ion trap instrumentation, Yen et al. recently performed an N-glycosylation centered study on 14 different breast cancer cell lines to detect differences in glycosylation between normal breast cells (HMEC) and breast cancer cells based on spectral counting (41). For some 63% of the glycoproteins identified in that dataset, we found at least one N-glycosylation site. More recently, Drake et al. compared the N-glycosylated secretome of luminal and triple negative breast cancer cell lines using the lectins SNA and AAL and based their quantification on spectral counting (42). Even though different cell lines were used, approximately half of the identified sites were also present in our data set. Of the 83 putative triple negative-specific glycoproteins, twenty also had a significantly higher expression in our comparison of triple negative versus ErbB2 expressing cell lines (see below). Although this reasonable overlap with these previous studies is encouraging, we here went one step further by performing quantification based on a super-SILAC internal standard, which we used to quantitatively determine secretion profiles as a function of cancer stage.
Increased Precision by Super-SILAC Internal Standard
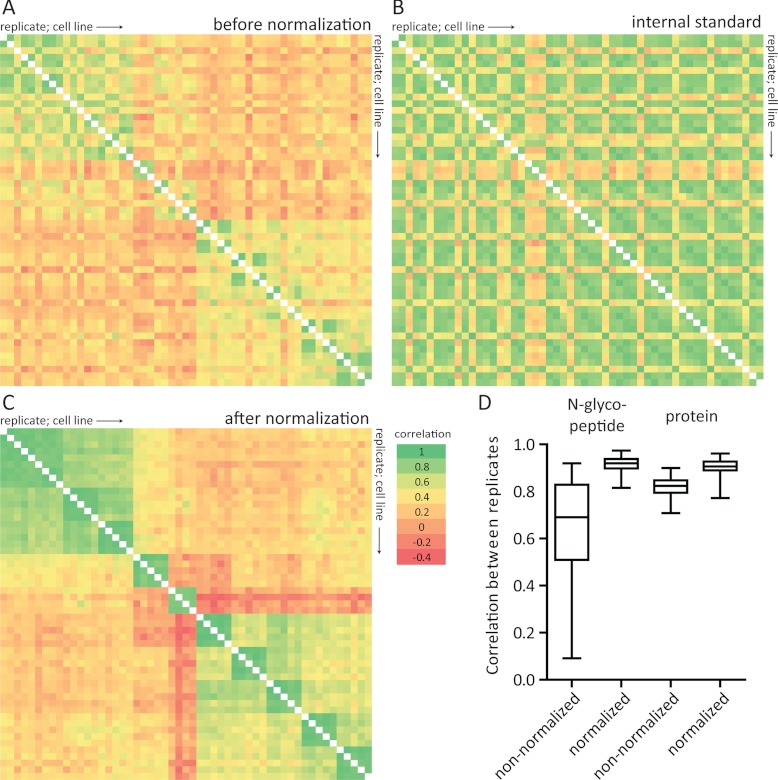
In this study, ample replicates were used to minimize the effects of biological and technical variation. The inclusion of an internal super-SILAC standard further allowed for normalization of technical variance. Variations that were expected include plate-to-plate variability of the same cell line and technical variation from N-glyco FASP being performed in different batches and different LC-MS instrumentation (nanoLC-Orbitrap Elite and Q Exactive). When comparing the correlation of intensities between replicates of the light SILAC channel, i.e. the samples before normalization by the internal super-SILAC standard, a certain degree of variation is apparent. However, similar variability can be seen in the super-SILAC channel. When performing normalization using the super-SILAC internal standard, correlation between the replicates clearly increased, and differences between the cell lines were augmented (Fig. 3). This demonstrates the power of using an internal standard, such as the super-SILAC mix, for increased quantification precision. The effect of the internal standard is lower at the proteome level, which was based on quantification of both enriched and non-enriched supernatant (see below). This is because in protein quantification, variation may be balanced out by different peptides. In contrast, quantification of N-glycosylation sites usually involves only one peptide, which, moreover, may be more prone to variation compared with nonglycosylated peptides because of the additional steps of lectin-capture and release with PNGase F. Remarkably, normalization with the internal standard elevated the correlation between replicates to the level of protein quantification.
Fig. 3.
Effect of normalization using the super-SILAC internal standard. A, Correlation of intensities of light N-glycosylated peptides intensities between different replicates and cell lines (light channel of SILAC measurements only). Samples are ordered horizontally and vertically according to replicates and cell line (from HMEpC1 until MDA-MB-453). B, Correlation of intensities of heavy isotope labeled N-glycosylated peptides (super-SILAC internal standard) intensities between different replicates and cell lines. C, Correlation of N-glycosylated peptide ratios between different replicates and cell lines after normalization using the super-SILAC internal standard. Correlations between replicates (“green blocks” near the diagonal) and cell lines from the same cancer stages are increased, whereas differences with other cell lines are augmented. D, Correlation between replicates from each cell line at the N-glycosylated peptide level (from panel A and C) and protein level (based on nonglycosylated and glycosylated peptides) before and after normalization using the super-SILAC internal standard. Pearson's r was calculated between all replicates from each cell line. Two aberrant measurements (one from HMEpC2 and one from HCC1937) were omitted.
Differences Between Proteins from N-glycosylation Enriched and Nonenriched Samples
In addition to the N-glycosylated peptides captured by the lectins, we also analyzed the noncaptured, and thus nonglycosylated, peptides (Fig. 1B, Supplemental Tables S3 and S4). This allows us to compare differences in features between glycosylated and non-glycosylated proteins and to assess the beneficial effect of enriching for N-glycosylated peptides. Of the 701 glycoproteins, 366 were only identified by their N-glycosylated peptides, whereas the remaining 335 were also identified by nonglycosylated peptides. The latter represent less than 10% of the 3482 proteins that we identified in the supernatant in total and less than half of the glycoproteins detectable when using enrichment. This suggests that the enrichment of N-glycosylation peptides efficiently focuses the analysis on a particular subset of proteins that may otherwise remain undetected.
To determine what subset of proteins was enriched using N-glyco FASP we looked at the predicted cellular localization of the proteins using Gene Ontology (GO) annotation (43). In total, 75% of the identified N-glycosylated proteins had a GO cellular compartment (GOCC) term that included the keywords “extracellular” and/or “plasma membrane” versus 31% in the non-enriched dataset (Table I). Moreover, the GOCC terms “intrinsic to membrane” and “extracellular region” were enriched 3.1 and 2.6 times in the N-glycosylated dataset compared with the whole dataset, respectively (Benjamini-Hochberg corrected p values 6.3 × 10−152 and 3.2 × 10−37).
Table I. Percentage of proteins with GOCC terms or predicted signal peptide and pathway of secretion.
| % of proteins | N-glyco dataset | Non-enriched dataset |
|---|---|---|
| GOCC “extracellular”, “plasma membrane” | 75 | 31 |
| Signal peptide predicted | 84 | 21 |
| Non-classical secretion predicted | 9 | 31 |
| Secretion predicted | 93 | 53 |
Proteins are N-glycosylated in the ER and Golgi apparatus. The route through ER and Golgi system delivers proteins to the plasma membrane and extracellular space and is part of the classical secretion pathway (18). As most of the proteins that enter the ER have an N-terminal signal sequence used to target them for translocation across the ER membrane, we expected that N-glycosylated proteins would therefore be more likely than nonglycosylated proteins to contain a signal peptide. SignalP 4.0 can be used to predict whether the protein amino acid sequence contains a signal peptide (44), whereas SecretomeP 2.0 delivers ab initio predictions of nonclassical secretion (45). Confirming our expectation, the N-glycosylation enriched data set contained more proteins with a predicted signal peptide (84% versus 21% for the nonglycosylation enriched dataset), whereas the percentage of proteins predicted to be nonclassically secreted was lower (9% versus 31%, Table I). Finally, the percentage of proteins that was predicted to be secreted was 93% for the N-glycosylation enriched data set compared with 53% of the non-enriched dataset. This demonstrates that enriching N-glycosylated peptides is advantageous as it focuses the analysis on actively (classically) secreted and plasma membrane proteins.
Determining Dynamics of N-glycosylation
The abundance changes that we detect at the N-glycosylation site level can have various underlying causes. In principle, the used lectins may have different affinity toward the glycan-chain after modification of the glycan-structure. In our method, we may neglect these changes because the used lectins for capturing N-glycosylated peptides select for glycan characteristics that are rather common. If they are not because of glyco-structure, the detected differences may relate to actual differences in occupancy of N-glycosylation sites whereas the overall protein expression remains the same. Or, the differing abundance may be related to the changed expression, secretion or shedding of the whole protein. We can differentiate between these two scenarios by comparing abundance changes of N-glycosylated peptides with the total protein changes. When we plot the ratios at the N-glycosylation site level against the protein ratios (that were calculated excluding the N-glycosylated peptides), we observe a high correlation (Supplemental Fig. S1; average Pearson's correlation coefficient 0.84). This suggests that most of the changes that we detected at N-glycosylation level are in fact caused by changes in protein abundance. This is in agreement with a previous study in which a significant part of the apparent N-glycosylation dynamics correlated well with protein dynamics as measured by RNAseq (42). It is also in agreement with the general character of N-glycosylation as a rather stable, co-translational modification. In the remainder of this manuscript, we will therefore discuss expression levels of proteins, synonymous with the N-glycosylation sites.
Hierarchical Clustering to Identify Cell Line and Cancer Stage Specific Features
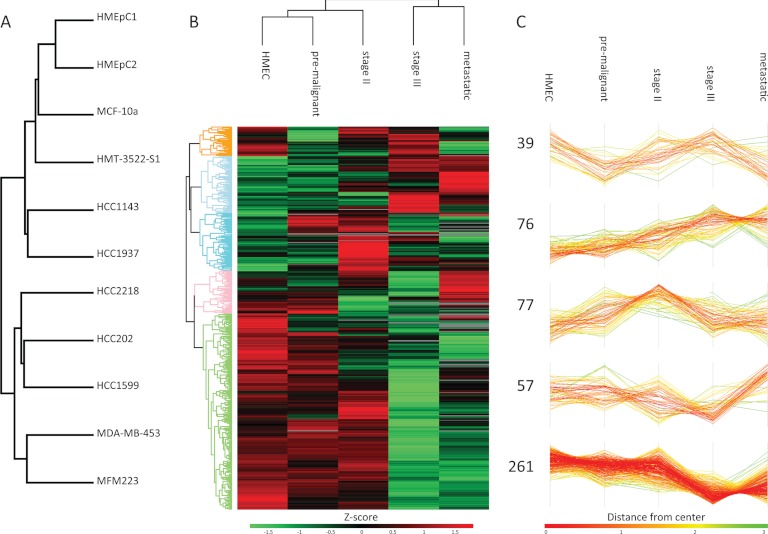
The “one shot” analysis of N-glycopeptides allowed us a quintuplicate analysis which in turn provides an excellent basis for statistical analysis of differences between cell lines. By one-way ANOVA, 722 N-glycosylation sites showed a significant difference (Benjamini Hochberg FDR < 0.05) in at least one of them. We then performed hierarchical clustering based on Euclidean distance on the Z-scored mean values of the N-glycosylation sites per cell line. Even though this clustering was unsupervised, cell lines that belong to the same cancer stage were grouped together and the secretomes of the Stage III and metastatic cells were separated from the lower grade and non-tumor cells (Fig. 4A). This is encouraging because the selected cell lines differ in the required growth medium, the supplier of the cell line, the adherence to the culture plate and expression of estrogen receptor, progesterone receptor (PR) and ErbB2, which all may affect the conditioned medium that was collected. In contrast, in our recent study of the proteome of these cells the clustering was not always by cancer stage (4). Our results demonstrate that in the N-glycosylated secretome, cancer stage specific factors are dominating the secretome rather than cell culture-related aspects.
Fig. 4.
Hierarchical clustering of N-glycosylation abundance profiles. A, Hierarchical clustering based on Euclidean distance at the cell line level. B, Hierarchical clustering based on Euclidean distance at the cancer stage level of N-glycosylation sites that showed a significant difference between healthy mammary epithelial cells (HMEC) and at least one other cancer stage. C, Five main clusters extracted from B).
From the previous clustering analysis it became clear that the variation between the five cancer stages was larger than the variation between cell lines within the same cancer stage. Therefore, we decided to combine the replicates within the cancer stages to elucidate N-glycosylation differences that may be indicative of breast cancer development. The 510 N-glycosylation sites that showed a significant difference (Benjamini Hochberg corrected students t test < 0.05) between at least one cancer stage and healthy epithelial cells were filtered followed by hierarchical clustering of the Z-scored values to identify patterns in regulation (Fig. 4B). Five main clusters can be observed (Fig. 4C). Cluster I contains N-glycosylation sites with a mixed regulation, but often low in pre-malignant and metastatic cell lines. In cluster II the expression of N-glycosylation sites is continuously increasing with the aggressiveness of the cancer stages. Cluster III shows the highest expression in Stage II cancer cells, whereas cluster IV has the highest expression in the metastatic cells. Finally, cluster V, encompassing slightly more than 50% of the data set, shows a trend of expression that is the lowest in Stage III and metastatic cancer cells. GO-term enrichment analysis showed that the GOCC term “integral to membrane” is enriched in cluster II (1.7 times, 3.1 × 10−6 Benjamin-Hochberg FDR), whereas cluster V had significantly more proteins with the “extracellular space” tag (1.5 times, 5.6 × 10−11 Benjamin-Hochberg FDR). Many of the sites in cluster V map to proteins involved in cell-cell interaction and adhesion. The fact that their abundance is lower in the later stage cancerous cell lines suggests negative involvement of these proteins in the release of the cell from the surrounding tissue.
From a biomarker perspective cluster II-IV would be more valuable as these are positive (increasing) markers for cancer development. Cluster II and IV have highest expression in the metastatic cell lines, whereas proteins in cluster III could be considered early indicators. Within these clusters are known cancer makers such as CEA and CEACAM1, as well as proteins suggested to be biomarkers in other studies. This is a powerful positive control for our approach and indicates that the N-glycosylated secretome of the cell lines may be a potential source to identify disease markers. In the following, we discuss proteins that were found in clusters II-IV and have a known or potential role in cancer development and may therefore mark carcinogenic changes in breast cancer.
Growth Factor Pathways
The growth factor receptors ErbB2, ErbB3, and FGFR1 were found in the clusters with higher abundance in metastatic cells. They have clearly been associated with breast cancer development and the success of cancer treatment whereby trastuzumab (Herceptin) targets ErbB2, whereas amplification of FGFR1 causes tamoxifen resistance (46). There is crosstalk between EGFR and NTRK2 (or Trkβ), which in the context of ovarian cancer enhances cell migration and proliferation (47). Silencing the expression of neurotrophin tyrosine kinase receptor related protein (NTRKR1) was shown to impair the growth and survival of human breast cancer cells (48).
Other proteins that are involved in growth factor signaling and that appeared in cluster II are PRSS14, LIV-1, and LTBP1. PRSS14 is a protease that was suggested to function as an epithelial membrane activator for other proteases and latent growth factors and its expression was associated with various types of tumors (49). LIV-1 (or estrogen-regulated protein 1) is an effector molecule downstream of soluble growth factors and has previously been associated with breast cancer and its metastatic spread to regional lymph nodes (50). Furthermore, it promotes the epithelial-mesenchymal transition in human pancreatic, breast, and prostate cancer cells (51). Finally, LTBPs are required for the proper folding and secretion of TGF-β (52). The expression of both proteins was reported to be synchronized in ovarian and breast cancer cells (53).
TSPAN-enriched Microdomain
Fourteen of the identified glycoproteins belong to the family of tetraspanin (TSPAN) proteins or are recruited into the organized membrane structures known as TSPAN-enriched microdomains (54). In these domains, TSPAN proteins assemble with similar or different TSPANs, integrins, cytokine receptors, Ig superfamily members, cytosolic signaling molecules, gangliosides, and proteases such as ADAMs and MMPs. Their roles include signal transduction, cell-cell adhesion, cytoskeletal anchoring and protein trafficking. Therefore it is not surprising that TSPANs have been associated with cancer development. In this study, TSPAN1, 8, 13, 15, 24, and 27 were identified, but not with a sufficient number of quantitative values to determine a significant change compared with the healthy control stage. However, TSPAN3, 6, 29, and 30 did show a significant increase in abundance on cancer development. Among the direct TSPAN interactors that we identified were EWI-2 (in cluster II), EWI-F and EpCAM (not significantly quantified), CD44 (cluster V), and various integrin subunits with varying dynamics. In general, TSPAN8 (CO-029) and 24 (CD151) are considered to promote motility of tumor cells whereas TSPAN27 (CD82), 29 (CD9), and 30 (CD63) appear to limit the dissemination of tumor cells (54). It is therefore striking to see TSPAN29 and 30 and EWI-2 (which is a direct partner of TSPAN27 and TSPAN29) appear in cluster II, with a higher expression in the later cancer development stages. This is, however, in accordance with previous proteomics data of the same cell lines (4), suggesting that different TSPAN members may not only have different roles in cancer development but also cell type specific roles. In support of this notion, we observed some cell line specific abundance differences for these proteins. TSPAN29 and EWI-2, for example, are significantly higher in abundance in MFM223 and HCC202 than their respective cancer stage members (Supplemental Fig. S2).
Semaphorins
Another class of proteins of which many members were identified are the semaphorins, proteins that are secreted or membrane anchored and that can be shed from the membrane by ADAMs and MMPs. The semaphorins were first described as axon guidance factors in the nervous systems, but semaphorin receptors were found to be expressed in various cell types including endothelial cells and many types of cancer (55). In this study, we identified semaphorins 3C, 4B, 4C, 4D, 5A, 7A, and their receptors NRP1 and plexin A1, A2, B1, B2, and D1. With a significant quantitative difference compared with the HMECs, semaphorins 3C, 4B, and plexin B1, B2, and NRP1 were all members of cluster II-IV whereas Semaphorin 7A was found in cluster V. Activation of plexins by semaphorins modulates cell adhesion and induces changes in the organization of the cytoskeleton of the target cells (55). Some semaphorins and their receptors have been considered tumor suppressors whereas others are known to activate tumor formation. For example, overexpression of NRP1, which is a receptor for the secreted semaphorins type 3 and pro-angiogenic factors such as VEGF and HGF, was shown to induce rapid tumor growth and progression in the context of prostate cancer and colon carcinoma (56). NRP1 appeared in cluster III with the highest abundance in the early cancer stages. VEGF-C and VEGF-E appeared in cluster V with a decrease in the abundance over the cancer stages, whereas semaphorin 3C showed the opposite profile. Regulation by semaphorins can be context dependent; for example, Semaphorin 4D can be pro- or antimigratory, depending on the presence of ErbB2 or MET (57).
Additional Proteins With Potential Breast Cancer Links
Prolactin induced protein (PIP or GCDFP15) was first isolated from human breast gross cystic fluid (58) and it has a mitogenic effect on breast cancer cells (59). This secreted glycoprotein has been suggested as a marker for apocrine breast carcinoma and prognosis predictor (60–62). Zinc α2-glycoprotein (AZGP1) forms a complex with PIP (63) and increased protein expression was found in well-differentiated tumors in which PIP levels were higher (64). In our data set, both AZGP1 and PIP appear in cluster II with higher abundance in the later stages of cancer, although AZGP1 does not seem to fully correlate with PIP levels in different cell lines (Supplemental Fig. S2).
The expression of phosphatase leukocyte common antigen related (LAR), a transmembrane tyrosine-protein, has been shown to increase in breast cancer (65). In accordance with these observations, we also see an increase in LAR expression in the later stages of cancer development. We identified 11 N-glycosylation sites on the extracellular matrix protein cytotactin (or tenascin, TNC). Their expression was increased mostly in the metastatic cell lines and HCC1143. TNC is a protein that binds to fibronectin, periostin, integrin cell adhesion receptors, and syndecan membrane proteoglycans. Breast cancer cells have been shown to produce TNC to support the fitness of initiating cells during the establishment of metastasis in lungs (66). That may be the reason that their expression is high in the later, metastatic stages of breast cancer cells (Supplemental Fig. S2).
Breast Cancer Specificity
A clinically useful positive marker for disease should be up-regulated in expression specifically in that disease but not in related diseases. Additionally, it would be of interest if the proposed marker was specific to the tissue in which the disease originated. However, in general many of the proteins that are detected with a differential regulation during cancer development are proteins that are rather generically involved in cancer progression. In our data set, for example, we observe an increase in expression of ephrin-A3, ICOS ligand, and inositol monophosphatase 3. However, according to immunohistochemistry data from the Human Protein Atlas (67) all of the 65 different cell types and all cancer tissues that were tested stained positive for these proteins. ErbB2 is more specific with expression detected by antibodies in 40% of cancer tissues other than breast cancer and 81% of breast cancer tissue. PIP had the highest specificity in breast cancer; only four out 66 test cell types and only 4% of the cancer tissues showed some expression of the protein by antibody staining. Interestingly, most of these were breast cancer tissues. Note that not all proteins that we identified have been evaluated in the Human Protein Atlas yet.
Differences Between Breast Cancer Cell Types (triple negative and ErbB2 expressing cell lines)
So far, we based analysis of the secreted glycoproteins on breast cancer stage, however, in a more detailed analysis we noticed several proteins that seemed to differ significantly between cell lines within the same cancer stage group. These different cell lines represent different breast cancer types, and therefore as an example, we decided to investigate possible difference between ErbB2 expressing or triple negative. In total, 132 N-glycosylation sites showed a significant differences between these types of which 96 also differed significantly compared with the healthy breast epithelial cells. A total of 98 of the N-glycosylation sites were higher in abundance in the triple negative cell lines. Cluster III contains N-glycosylation sites for which the expression was highest in the pre-malignant cell lines, HCC1143 and HCC1937. Almost 40% of these sites also appear in the analysis of the difference between ErbB2 positive and triple negative and may therefore be explained by their expression of hormonal receptors. Of the above discussed proteins only SEMA4B, NTRKR1, and CD44 had a significantly higher expression in triple negative cell lines, whereas PIP has a significantly higher expression in the ErbB2 expressing cells.
N-glycosylated Secretome Versus Cellular Proteome
The abundance change of proteins in the secretome can be related to altered protein expression, but also to differential secretion or shedding. To distinguish these scenarios, one can compare the secretome data with intracellular data. Previously, our group analyzed the intracellular proteome of the panel of cell lines that was used in this study to identify cancer stage specific differences (4). Of the protein groups identified in the current study, 461 (representing 1031 N-glycosylation sites) overlapped with the Geiger et al. dataset. Unsupervised hierarchical clustering with the averaged ratios of proteins of the different cell lines grouped the proteome and the N-glycosylated secretome next to each other in almost all cases (Supplemental Fig. S3). This demonstrates that proteomic quantification of the cellular and secreted proteomes both capture essential aspects of cancer stage related changes and that the differences between the different cell lines are larger than the differences between the same cell line in the two proteomic approaches. The secretome of the cancer cell lines therefore appears, at least partly, to be a reflection of the intercellular protein state and vice versa. In the previous study we strove for a deep and comprehensive analysis of the cellular proteome and identified more than 8000 proteins. The data set reported here is significantly smaller as no fractionation was performed and the analysis was focused on capturing N-glycosylated peptides, but interestingly still more than 30% of the proteins were not found in the large proteome dataset. This is likely the result of focusing on the secretome versus cellular proteome in combination with enriching for N-glycosylated peptides. These features of N-glyco FASP of cancer cell secretomes prompted us to investigate the method for detecting and quantifying the potential breast cancer biomarkers directly in human blood as described next.
N-glyco FASP Applied to Blood Plasma
One large stumbling block in the proteomics analysis of plasma is the enormous quantitative dynamic abundance range of proteins. Albumin, for example, takes up about 50% of the total mass of plasma proteins and is present at 35–50 mg/ml levels, whereas other clinically interesting proteins are present at only pg/ml levels (6). For this reason, plasma is often depleted of the highest abundant proteins, such as albumin, immunoglobulins, and complement components, before proteomics analysis. However, this risks losing proteins that may bind to the proteins that are depleted (68). We reasoned that using the N-glyco FASP strategy to reduce the complexity of the sample, may also reduce the dynamic range challenge in plasma, as has already been shown with other glyco-capture approaches (26). Although many of the high abundance plasma proteins (but not albumin) are N-glycosylated (22), the removal of their non-N-glycosylated peptides should result in a significant reduction of their share in the final analyzed sample.
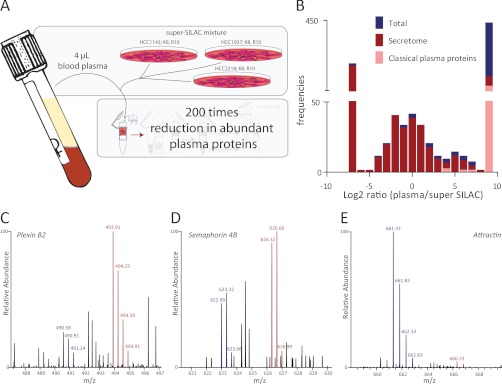
We first set out to compare the identified proteins prior and after enrichment of N-glycosylated peptides and for this purpose we digested plasma of female blood donors. An aliquot was used for determination of the plasma proteome, whereas the remainder was enriched for N-glycosylated peptides using N-glyco FASP (Fig. 5A). More than 800 proteins were identified from both the non-enriched and N-glycosylation enriched fractions. Among them we found as the most abundant ones almost all proteins that were listed by Anderson et al. as “classical plasma proteins” (6). These include the proteins that are depleted by, for example the Hu-6, Hu-14, and Proteoprep 20, multiaffinity removal systems (68). To test whether these classical plasma proteins are relatively reduced after enriching for only their N-glycopeptides, we summed the MS intensities of their peptides and normalized for the total peptide intensity present in the samples. Interestingly, this indeed revealed a twofold decrease in relative intensity after enrichment of N-glycosylated peptides. Considering that the input for the N-glyco FASP for a single analysis can be about a hundred times more, this leads to an about 200 times reduced intensity of the high abundance proteins compared with a total proteome measurement. This then allowed the identification in single, 2h LC MS/MS runs of many lower abundant glycoproteins such as CD63, ALCAM, Plnxb2, and CD49 and 180 additional proteins that may be classified as “tissue leakage” proteins.
Fig. 5.
N-glyco FASP applied to blood plasma. A, Blood plasma was mixed with the breast cancer cell secretome super-SILAC mix. The enrichment of N-glycosylated peptides, and thereby removal of nonglycosylated peptides, reduced the intensity of abundant plasma proteins by about 200 times. B, Distribution of ratios of N-glycosylation sites between the super-SILAC internal standard and plasma. Sites without a signal in the light or heavy SILAC channel were assigned a Log2 ratio of –7 and 9, respectively. Overlapping sites with the secretome data set are labeled in red, whereas classical plasma proteins (as taken from Anderson et al. (6)) are labeled in pink. C, D, E, example MS spectra of N-glycosylated peptides with differing ratios. C, IHVTIYN(de)CSFGR (3+), plexin B2, average ratio −1.75; D, FEAEHISN(de)YTAIIISR (3+), semaphorin 4B, average ratio 0.0; E, IDSTGN(de)VTNEIR (2+), attractin, average ratio 4.3.
Quantification of N-glycoproteins in Blood by Super-SILAC
Apart from the difficulties imposed by the large dynamic range of the plasma proteome, accurate quantification in body fluid is likewise challenging. The super-SILAC strategy has been employed with tissues and post-translational modifications (38, 69), but it may seem to be impossible to apply to the quantification of body fluids. To evaluate the use of the super-SILAC mix in blood plasma we analyzed in duplicate the plasma of four female donors to which we added an approximately equal amount of the super-SILAC mix composed from the secretome of SILAC labeled cell lines. From this analysis we identified 925 unique N-glycosylation sites (Supplemental Tables S5, S6, and S7). Three classes of quantification results would be expected: proteins that occur in both the secretome of breast cancer cells and in human plasma should occur as SILAC pairs in the analysis. Proteins that are largely unique to plasma, such as the classical plasma proteins discussed above can be recognized by not having an equivalent heavy labeled counter-peptide. Finally, proteins that are unique to breast cancer cells and not present in the donor plasma would only have the heavy form of the SILAC peptides. On inspecting the results, we indeed found these three classes of glycopeptides (Fig. 5B).
Because the apparent SILAC-ratios may be very high or very low for classical plasma proteins and secretome proteins that do not appear in the plasma, some precautions need to be taken not to introduce artifacts. For example, during quantification in MaxQuant, the “re-quantify” option shifts the isotope pattern of the detected SILAC partner to the location of the inferred missing SILAC partner to integrate the signal at that position and thereby determine a minimum ratio. Because there may be no partner in this case, the re-quantify option should be turned off. Furthermore, although in general the incorporation of isotopically labeled lysine and arginine in the super-SILAC mix is high, there may be proteins that are not fully labeled. A small, but detectable, fraction of the light version of the peptide may therefore remain and this can be determined by analyzing the super-SILAC mix on its own. Finally, typically used normalization methods are not applicable, because no normal distributions are expected between super-SILAC N-glycopeptides and plasma N-glycopeptides. For practical analysis purposes, the N-glycosylation sites for which no ratio could be determined because of missing light or heavy labeled counter-peptides, received an value of –7 or 9 (the minimum and maximum ratios that were quantified) depending on whether only the light or the heavy version was found (Fig. 5B). With only one or two exceptions, for the classical plasma proteins no counter-peptide could be detected in the super-SILAC mix, validating our analysis strategy. Given the above described challenges of plasma proteome analysis, we were encouraged to see that more than half of the N-glycosylation sites overlapped with the 1398 sites found in in-depth measurements of the cell line secretomes. Taking a Log2 ratio of –2 as a very conservative lower threshold for positive proof of presence of the light peptide, 180 N-glycosylation sites were found with both a signal from the plasma and the super-SILAC internal standard. Of these, 164 sites (91%) were also quantified in the secretome study. When mapped on the N-glyco secretome, these plasma sites have a small bias toward the more abundant part of the distribution but cover most of the dynamic range (Supplemental Fig. S4A). For these overlapping sites we also have information about their dynamics in breast cancer development. In total, 101 of the sites overlapping with the secretome and with a ratio between –2 and 9 were present in one of the five clusters discussed above, with no apparent preference.
Overall a surprisingly high share of the N-glycosylation sites identified in the secretome of breast cancer cell lines was also identifiable and quantifiable in the blood plasma. For example, from the proteins that we discussed above with an abundance increasing in breast cancer cell lines we identify here in the plasma, NTRK2, PRSS14, LTBP1, TSPAN30, CD44, semaphorins 4B and 7A, plexin B1, B2, and D1, NRP, AZGP1, and cytotactin. This indicates that numerous potentially clinically relevant proteins that are secreted by cell lines are also present in blood plasma at detectable levels. Often, antibody based techniques, such as ELISA, have been used to validate findings from secretome studies in body fluids (11, 13, 23). Here, however, by spiking the super-SILAC mix from the secretome of cell lines into the plasma, we can simultaneously determine whether proteins are tissue-derived and quantify the abundance of many N-glycosylation sites at once.
CONCLUSION
Here we have shown that the enrichment of N-glycosylated peptides using N-glyco FASP allowed for a focus on a specific subset of classically secreted or shed proteins. In this manner a reasonably deep analysis could be obtained in a “single-shot” LC-MS fashion in a very short analysis time. While this method requires additional sample handling compared with direct shotgun analysis, this may often be outweighed by the above advantages, especially when analyzing blood. In general, N-glycosylation proved to not be a transient modification as barely any difference was observed between N-glycosylation occupancy and changes of the protein abundance itself. Quantification at the N-glycosylation site level therefore in most cases acts as a proxy for protein quantification, but in a much more feasible way given the reduced complexity of the sample after enrichment of N-glycosylated peptides. Including an internal standard, such as the heavy isotope labeled super-SILAC mix in the N-glyco FASP analysis further allows normalizing the data for technical variation between replicates, thus increasing quantification precision.
The total N-glycosylated secretome profiles that we measured were cell line and cancer stage specific and clustered as such. This implies that there are indeed cancer stage specific differences in glycoproteins that are secreted or shed into the supernatant. Further proof of the usability of the conditioned medium for finding differences between breast cancer cell lines came from the identification and quantification of several known markers for breast-cancer such as CEA and CEACAM.
Of the detected N-glycosylation sites, 510 showed a significant difference in abundance in one of the cancer stages compared with the normal breast epithelial cells. Many of the sites that had an abundance that was lower in the advanced cancer stages were mapped to proteins that are involved in cell adhesion, which suggests a (negative) role in the release of cancerous cells from the surrounding tissue. Other N-glycosylation sites were found to have a higher abundance in the later cancer stages, including proteins from the TSPAN and semaphorin family, proteins involved in growth factor signaling and other proteins that have been associated with breast cancer development. In general, cell lines within a cancer stage group showed similar N-glycosylation profiles, but some glycoproteins proved to be cell line specific, partly correlated to the expression of estrogen receptor, PR, and ErbB2. The fact that our quantification of the N-glycosylated secretome found a class of proteins with a higher expression in the later breast cancer stages suggests that this workflow could be used for biomarker discovery, provided that the sensitivity and specificity of these candidates was independently verified in large patient cohorts.
In a study of blood plasma from female donors, we for the first time extended the super-SILAC approach to body fluids. We showed that the super-SILAC approach can distinguish classical plasma proteins from tissue leakage proteins by their SILAC ratios. We confirmed that similar proteins as found in the condition medium of cell lines can be detected in plasma underlining the clinical usefulness of the secretome of cell lines. Even in this initial study, about a hundred N-glycosites associated with proteins that had significant changes with breast cancer stage in the cell line secretome, were also quantifiable in plasma and this number can surely be extended with further method development. For instance, some 120 sites were quantified in the N-glycosylated secretome of the cell lines but these heavy peptides were nevertheless not detected after mixing with plasma. We suspect that their abundance in the super-SILAC mix may have been too low to be detected and in the secretome study their abundance was indeed in the lower half of the abundance distribution (Supplemental Fig. S4B). For these, MRM-based targeted approaches could be employed to improve S/N (70, 71) or for the quadrupole-Orbitrap instrumentation employed here, selected ion monitoring (SIM) scans (34) could be used. Further studies could incorporate analyses of plasma comparing healthy donors and breast cancer patients to trace the eventual altered abundance of these markers in the peripheral blood.
Supplementary Material
Acknowledgments
We thank the members of the Department of Proteomics and Signal Transduction for support and fruitful discussions. Especially, we thank Dorota Zielinska for technical support and Herbert Schiller for critically reading the manuscript.
Footnotes
* This work was supported by the Max Planck Society of the Advancement of Science, by the European Commission's 7th Framework Program (grant agreement HEALTH-F4–2008-201648/PROSPECTS) and the Munich Center for Integrated Protein Science (CIPSM). P.J.B was supported by the Netherlands Organisation for Scientific Research (NWO).
 This article contains supplemental Figs. S1 to S4 and Tables S1 to S7.
This article contains supplemental Figs. S1 to S4 and Tables S1 to S7.
Data availability - Supplementary data is available with this publication at the MCP web site. Raw MS files are uploaded to Tranche (www.proteomecommons.org; hash: RM8Oo51NEQ3U/XQZwiIItSL5AGMmzHI1+oaqeR5XAaFRkGVffkVzfKlXOeI1XU1m4CH8jFSMFY6hscxvgn+V3dGzsiIAAAAAAABYNw==).
1 The abbreviations used are:
- ER
- endoplasmic reticulum
- CA125
- cancer antigen 125
- CEA
- carcinoembryonic antigen
- Con A
- concanavalin A
- FASP
- filter aided sample preparation
- FDR
- false discovery rate
- GO
- gene ontology
- GOCC
- gene ontology cellular compartment
- HMEC
- human mammary epithelial cells
- PIP
- prolactin-inducible protein
- PR
- progesterone receptor
- SILAC
- stable isotope labeling by amino acids in cell culture
- SIM
- selected ion monitoring
- WGA
- wheat germ agglutinin.
REFERENCES
- 1. Hanash S. M., Baik C. S., Kallioniemi O. (2011) Emerging molecular biomarkers[mdash]blood-based strategies to detect and monitor cancer. Nat. Rev. Clin. Oncol. 8, 142–150 [DOI] [PubMed] [Google Scholar]
- 2. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Paabo S., Mann M. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., Aebersold R. (2011) The quantitative proteome of a human cell line. Mol. Syst. Biol. 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Geiger T., Madden S. F., Gallagher W. M., Cox J., Mann M. (2012) Proteomic portrait of human breast cancer progression identifies novel prognostic markers. Cancer Res. 72, 2428–2439 [DOI] [PubMed] [Google Scholar]
- 5. Clark H. F., Gurney A. L., Abaya E., Baker K., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Currell B., Deuel B., Dowd P., Eaton D., Foster J., Grimaldi C., Gu Q. M., Hass P. E., Heldens S., Huang A., Kim H. S., Klimowski L., Jin Y. S., Johnson S., Lee J., Lewis L., Liao D. Z., Mark M., Robbie E., Sanchez C., Schoenfeld J., Seshagiri S., Simmons L., Singh J., Smith V., Stinson J., Vagts A., Vandlen R., Watanabe C., Wieand D., Woods K., Xie M. H., Yansura D., Yi S., Yu G. Y., Yuan J., Zhang M., Zhang Z. M., Goddard A., Wood W. I., Godowski P. (2003) The Secreted Protein Discovery Initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: A bioinformatics assessment. Genome Res. 13, 2265–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson N. L., Anderson N. G. (2002) The human plasma proteome. Mol. Cell. Proteomics 1, 845–867 [DOI] [PubMed] [Google Scholar]
- 7. Anderson N. L., Polanski M., Pieper R., Gatlin T., Tirumalai R. S., Conrads T. P., Veenstra T. D., Adkins J. N., Pounds J. G., Fagan R., Lobley A. (2004) The human plasma proteome. Mol. Cell. Proteomics 3, 311–326 [DOI] [PubMed] [Google Scholar]
- 8. Farrah T., Deutsch E. W., Omenn G. S., Campbell D. S., Sun Z., Bletz J. A., Mallick P., Katz J. E., Malmström J., Ossola R., Watts J. D., Lin B., Zhang H., Moritz R. L., Aebersold R. (2011) A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol. Cell. Proteomics 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berven F. S., Ahmad R., Clauser K. R., Carr S. A. (2010) Optimizing performance of glycopeptide capture for plasma proteomics. J. Proteome Res. 9, 1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Q., Faca V., Hanash S. (2011) Mining the plasma proteome for disease applications across seven logs of protein abundance. J. Proteome Res. 10, 46–50 [DOI] [PubMed] [Google Scholar]
- 11. Wu C. C., Hsu C. W., Chen C. D., Yu C. J., Chang K. P., Tai D. I., Liu H. P., Su W. H., Chang Y. S., Yu J. S. (2010) Candidate serological biomarkers for cancer identified from the secretomes of 23 cancer cell lines and the human protein atlas. Mol. Cell. Proteomics 9, 1100–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dowling P., Clynes M. (2011) Conditioned media from cell lines: A complementary model to clinical specimens for the discovery of disease-specific biomarkers. Proteomics 11, 794–804 [DOI] [PubMed] [Google Scholar]
- 13. Karagiannis G. S., Pavlou M. P., Diamandis E. P. (2010) Cancer secretomics reveal pathophysiological pathways in cancer molecular oncology. Mol. Oncol. 4, 496–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Makridakis M., Vlahou A. (2010) Secretome proteomics for discovery of cancer biomarkers. J. Proteomics 73, 2291–2305 [DOI] [PubMed] [Google Scholar]
- 15. Pavlou M. P., Diamandis E. P. (2010) The cancer cell secretome: A good source for discovering biomarkers? J. Proteomics 73, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 16. Radisky D. C. (2005) Epithelial-mesenchymal transition. J. Cell Sci. 118, 4325–4326 [DOI] [PubMed] [Google Scholar]
- 17. Zent R., Pozzi A. (2009) Cell-extracellular matrix interactions in cancer, Springer [Google Scholar]
- 18. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (2008) Essentials of glycobiology, 2nd edition [PubMed] [Google Scholar]
- 19. Lisowska E. (2002) The role of glycosylation in protein antigenic properties. Cell. Mol. Life Sci. 59, 445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lau K. S., Dennis J. W. (2008) N-Glycans in cancer progression. Glycobiology 18, 750–760 [DOI] [PubMed] [Google Scholar]
- 21. Ohtsubo K., Marth J. D. (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 22. Drake P. M., Cho W., Li B. S., Prakobphol A., Johansen E., Anderson N. L., Regnier F. E., Gibson B. W., Fisher S. J. (2010) Sweetening the pot: Adding glycosylation to the biomarker discovery equation. Clin. Chem. 56, 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whitmore T. E., Peterson A., Holzman T., Eastham A., Amon L., McIntosh M., Ozinsky A., Nelson P. S., Martin D. B. (2012) Integrative analysis of N-linked human glycoproteomic data sets reveals PTPRF ectodomain as a novel plasma biomarker candidate for prostate cancer. J. Proteome Res. 11, 2653–2665 [DOI] [PubMed] [Google Scholar]
- 24. Zhang H., Li X. J., Martin D. B., Aebersold R. (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 [DOI] [PubMed] [Google Scholar]
- 25. Zhang H., Yi E. C., Li X.-j., Mallick P., Kelly-Spratt K. S., Masselon C. D., Camp D. G., Smith R. D., Kemp C. J., Aebersold R. (2005) High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol. Cell. Proteomics 4, 144–155 [DOI] [PubMed] [Google Scholar]
- 26. Zhang H., Liu A. Y., Loriaux P., Wollscheid B., Zhou Y., Watts J. D., Aebersold R. (2007) Mass spectrometric detection of tissue proteins in plasma. Mol. Cell. Proteomics 6, 64–71 [DOI] [PubMed] [Google Scholar]
- 27. Hirabayashi J., Kasai K.-i. (2002) Separation technologies for glycomics. J. Chromatography B Analyt. Technol. Biomed. Life Sci., 771, 67–87 [DOI] [PubMed] [Google Scholar]
- 28. Bunkenborg J., Pilch B. J., Podtelejnikov A. V., Wisniewski J. R. (2004) Screening for N-glycosylated proteins by liquid chromatography mass spectrometry. Proteomics 4, 454–465 [DOI] [PubMed] [Google Scholar]
- 29. Zielinska D. F., Gnad F., Wiśniewski J. R., Mann M. (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 [DOI] [PubMed] [Google Scholar]
- 30. Zielinska D. F., Gnad F., Schropp K., Wiśniewski J. R., Mann M. (2012) Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol. Cell 46, 542–548 [DOI] [PubMed] [Google Scholar]
- 31. Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., Mann M. (2010) Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 [DOI] [PubMed] [Google Scholar]
- 32. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protocols 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 33. Michalski A., Damoc E., Lange O., Denisov E., Nolting D., Mueller M., Viner R., Schwartz J., Remes P., Belford M., Dunyach J.-J., Cox J., Horning S., Mann M., Makarov A. (2011) Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol. Cell. Proteomics 11, mcp.O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Michalski A., Damoc E., Hauschild J.-P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., Horning S. (2011) Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics 10, mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 36. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 37. Palmisano G., Melo-Braga M. N., Engholm-Keller K., Parker B. L., Larsen M. R. (2012) Chemical deamidation: A common pitfall in large-scale N-linked glycoproteomic mass spectrometry-based analyses. J. Proteome Res. 11, 1949–1957 [DOI] [PubMed] [Google Scholar]
- 38. Geiger T., Wisniewski J. R., Cox J., Zanivan S., Kruger M., Ishihama Y., Mann M. (2011) Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat. Protocols 6, 147–157 [DOI] [PubMed] [Google Scholar]
- 39. Gonzalez J., Takao T., Hori H., Besada V., Rodriguez R., Padron G., Shimonishi Y. (1992) A method for determination of N-glycosylation sites in glycoproteins by collision-induced dissociation analysis in fast atom bombardment mass spectrometry: Identification of the positions of carbohydrate-linked asparagine in recombinant α-amylase by treatment with peptide-N-glycosidase F in 18O-labeled water. Anal. Biochem. 205, 151–158 [DOI] [PubMed] [Google Scholar]
- 40. Küster B., Mann M. (1999) O-18-labeling of N-glycosylation sites to improve the identification of gel-separated glycoproteins using peptide mass mapping and database searching. Anal. Chem. 71, 1431–1440 [DOI] [PubMed] [Google Scholar]
- 41. Yen T. Y., Macher B. A., McDonald C. A., Alleyne-Chin C., Timpe L. C. (2011) Glycoprotein profiles of human breast cells demonstrate a clear clustering of normal/benign versus malignant cell lines and basal versus luminal cell lines. J. Proteome Res. 11, 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Drake P. M., Schilling B., Niles R. K., Prakobphol A., Li B., Jung K., Cho W., Braten M., Inerowicz H. D., Williams K., Albertolle M., Held J. M., Iacovides D., Sorensen D. J., Griffith O. L., Johansen E., Zawadzka A. M., Cusack M. P., Allen S., Gormley M., Hall S. C., Witkowska H. E., Gray J. W., Regnier F., Gibson B. W., Fisher S. J. (2012) Lectin chromatography/mass spectrometry discovery workflow identifies putative biomarkers of aggressive breast cancers. J. Proteome Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Meth. 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 45. Bendtsen J. D., Kiemer L., Fausboll A., Brunak S. (2005) Non-classical protein secretion in bacteria. BMC Microbiology 5, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Turner N., Pearson A., Sharpe R., Lambros M., Geyer F., Lopez-Garcia M. A., Natrajan R., Marchio C., Iorns E., Mackay A., Gillett C., Grigoriadis A., Tutt A., Reis-Filho J. S., Ashworth A. (2010) FGFR1 Amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 70, 2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qiu L., Zhou C., Sun Y., Di W., Scheffler E., Healey S., Kouttab N., Chu W., Wan Y. (2006) Crosstalk between EGFR and TrkB enhances ovarian cancer cell migration and proliferation. Int. J. Oncol. 29, 1003–1011 [PubMed] [Google Scholar]
- 48. Zhang S., Chen L., Cui B., Chuang H. Y., Yu J., Wang-Rodriguez J., Tang L., Chen G., Basak G. W., Kipps T. J. (2012) ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PLoS ONE 7, e31127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Uhland K. (2006) Matriptase and its putative role in cancer. Cell. Mol. Life Sci. 63, 2968–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lue H. W., Yang X., Wang R., Qian W., Xu R. Z. H., Lyles R., Osunkoya A. O., Zhou B. P., Vessella R. L., Zayzafoon M., Liu Z. R., Zhau H. E., Chung L. W. (2011) LIV-1 promotes prostate cancer epithelial-to-mesenchymal transition and metastasis through HB-EGF shedding and EGFR-mediated ERK signaling. PLoS ONE 6, e27720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thiery J. P., Acloque H., Huang R. Y., Nieto M. A. (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 [DOI] [PubMed] [Google Scholar]
- 52. Koli K., Saharinen J., Hyytiäinen M., Penttinen C., Keski-Oja J. (2001) Latency, activation, and binding proteins of TGF-β. Microscopy Res. Tech. 52, 354–362 [DOI] [PubMed] [Google Scholar]
- 53. Koli K., Keski-Oja J. (1995) 1,25-Dihydroxyvitamin D3 enhances the expression of transforming growth factor β1 and its latent form binding protein in cultured breast carcinoma cells. Cancer Res. 55, 1540–1546 [PubMed] [Google Scholar]
- 54. Zijlstra A. (2010) Tetraspanins in Cancer Cell-Extracellular Matrix Interactions in Cancer. In: Zent R., Pozzi A., eds., pp. 217–243, Springer; New York [Google Scholar]
- 55. Neufeld G., Kessler O. (2008) The semaphorins: versatile regulators of tumour progression and tumour angiogenesis. Nat. Rev. Cancer 8, 632–645 [DOI] [PubMed] [Google Scholar]
- 56. Parikh A. A., Fan F., Liu W. B., Ahmad S. A., Stoeltzing O., Reinmuth N., Bielenberg D., Bucana C. D., Klagsbrun M., Ellis L. M. (2004) Neuropilin-1 in human colon cancer: expression, regulation, and role in induction of angiogenesis. Am. J. Pathol. 164, 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Swiercz J. M., Worzfeld T., Offermanns S. (2008) ErbB-2 and Met reciprocally regulate cellular signaling via plexin-B1. J. Biol. Chem. 283, 1893–1901 [DOI] [PubMed] [Google Scholar]
- 58. Haagensen D. E., Silva J. S., Leight G. S., Dilley W. G., Wells S. A. (1981) fluoxymesterone stimulates plasma-concentrations of gross cystic-disease fluid protein in patients with metastatic breast-carcinoma. Surg. Forum 32, 413–414 [Google Scholar]
- 59. Cassoni P., Sapino A., Haagensen D. E., Naldoni C., Bussolati G. (1995) mitogenic effect of the 15-kda gross cystic-disease fluid protein (gcdfp-15) on breast-cancer cell-lines and on immortal mammary cells. Int. J. Cancer 60, 216–220 [DOI] [PubMed] [Google Scholar]
- 60. Dennis J. L., Hvidsten T. R., Wit E. C., Komorowski J., Bell A. K., Downie I., Mooney J., Verbeke C., Bellamy C., Keith W. N., Oien K. A. (2005) Markers of adenocarcinoma characteristic of the site of origin: development of a diagnostic algorithm. Clin. Cancer Res. 11, 3766–3772 [DOI] [PubMed] [Google Scholar]
- 61. Sapino A., Righi L., Cassoni P., Papotti M., Gugliotta P., Bussolati G. (2001) Expression of apocrine differentiation markers in neuroendocrine breast carcinomas of aged women. Mod. Pathol. 14, 768–776 [DOI] [PubMed] [Google Scholar]
- 62. Hahnel R., Hahnel E. (1996) Expression of the PIP/GCDFP-15 gene and survival in breast cancer. Virchows Arch. Int. J. Pathol. 429, 365–369 [DOI] [PubMed] [Google Scholar]
- 63. Hassan M. I., Bilgrami S., Kumar V., Singh N., Yadav S., Kaur P., Singh T. P. (2008) Crystal structure of the novel complex formed between zinc α2-glycoprotein (ZAG) and prolactin-inducible protein (PIP) from human seminal plasma. J. Mol. Biol. 384, 663–672 [DOI] [PubMed] [Google Scholar]
- 64. Díez-Itza I., Sánchez L. M., Teresa Allende M., Vizoso F., Ruibal A., López-Otín C. (1993) Zn-α2-glycoprotein levels in breast cancer cytosols and correlation with clinical, histological and biochemical parameters. Eur. J. Cancer 29, 1256–1260 [DOI] [PubMed] [Google Scholar]
- 65. Yang T., Zhang J. S., Massa S. M., Han X., Longo F. M. (1999) Leukocyte common antigen–related tyrosine phosphatase receptor: Increased expression and neuronal-type splicing in breast cancer cells and tissue. Mol. Carcinogenesis 25, 139–149 [PubMed] [Google Scholar]
- 66. Oskarsson T., Acharyya S., Zhang X. H., Vanharanta S., Tavazoie S. F., Morris P. G., Downey R. J., Manova-Todorova K., Brogi E., Massague J. (2011) Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med. 17, 867–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Bjorling L., Ponten F. (2010) Towards a knowledge-based Human Protein Atlas. Nat. Biotech. 28, 1248–1250 [DOI] [PubMed] [Google Scholar]
- 68. Yadav A. K., Bhardwaj G., Basak T., Kumar D., Ahmad S., Priyadarshini R., Singh A. K., Dash D., Sengupta S. (2011) A systematic analysis of eluted fraction of plasma post immunoaffinity depletion: implications in biomarker discovery. PLoS ONE 6, e24442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Monetti M., Nagaraj N., Sharma K., Mann M. (2011) Large-scale phosphosite quantification in tissues by a spike-in SILAC method. Nat. Meth. 8, 655–658 [DOI] [PubMed] [Google Scholar]
- 70. Picotti P., Aebersold R. (2012) Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Meth. 9, 555–566 [DOI] [PubMed] [Google Scholar]
- 71. Gallien S., Duriez E., Domon B. (2011) Selected reaction monitoring applied to proteomics. J. Mass Spectrom. 46, 298–312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.