Abstract
Trypanosoma brucei developed a sophisticated life cycle to adapt to different host environments. Although developmental differentiation of T. brucei has been the topic of intensive research for decades, the mechanisms responsible for adaptation to different host environments are not well understood. We developed stable isotope labeling by amino acids in cell culture in trypanosomes to compare the proteomes of two different life cycle stages. Quantitative comparison of 4364 protein groups identified many proteins previously not known to be stage-specifically expressed. The identification of stage-specific proteins helps to understand how parasites adapt to different hosts and provides new insights into differences in metabolism, gene regulation, and cell architecture. A DEAD-box RNA helicase, which is highly up-regulated in the bloodstream form of this parasite and which is essential for viability and proper cell cycle progression in this stage is described as an example.
Trypanosoma brucei is a unicellular eukaryotic parasite that causes African sleeping sickness in humans and “nagana” in livestock in sub-Saharan Africa. Trypanosomes have developed a complex life cycle, which includes two very different host environments: the vascular system and tissue fluids of mammals and the intestinal tract and salivary glands of the vector, the tsetse fly. To shuttle between these two hosts, trypanosomes differentiate from the so-called bloodstream form (BSF)1 in the mammalian host to the procyclic form (PCF), which is adapted to life in the insect vector (1). The differentiation process of BSF can be divided into two steps. First, proliferating cells, called “long slender,” differentiate into cell cycle-arrested cells, called “short stumpy” (SS) in the mammalian host, which are pre-adapted to transmission to the insect vector. On ingestion by the tsetse fly during a bloodmeal, SS differentiate into procyclic forms and resume proliferation in the insect's midgut. Eventually, the parasites develop into epimastigote forms and migrate to the salivary glands where they attach to the epithelia. Epimastigotes differentiate into nonproliferative metacyclic forms, which are pre-adapted to transmission to another mammal during blood feeding of the fly. The life cycle is completed after metacyclic forms differentiate to long slender BSF and re-enter cell cycle progression. This sophisticated differentiation process involves coordinated changes of the parasite's gene expression, in order to provide host specific surface proteins and to adapt metabolism, morphology and organelle activity (1, 2). Thus, many stage-specifically expressed proteins are important for infectivity of the mammalian host and are of medical interest as putative drug targets against the parasite. Although this has previously been extensively analyzed by transcriptomics (3, 4), the genomic organization of trypanosomes suggests that transcriptome studies will not reveal a complete picture of adaption of the two life stages. Most genes of trypanosomes are organized in polycistronic units and gene expression is controlled almost exclusively post-transcriptionally on the level of RNA stability, translation efficiency and/or protein turnover (5). We therefore performed a large-scale study to compare mRNA abundance and protein levels. Stable isotope labeling by amino acids in cell culture (SILAC) allows exact quantitation of proteome differences and has recently been extended to the labeling of complete model organisms including yeast (6, 7), Caenorhabditis elegans (8, 9), Drosophila melanogaster (10) and mice (11). We established protocols for incorporation of stable isotopes for Trypanosoma brucei in different life cycle stages and report the identification of stage-specifically expressed proteins by comparing the proteomes of the PCF and BSF life cycle stages in SILAC-labeled T. brucei.
EXPERIMENTAL PROCEDURES
Insect Form Growth
Procyclic form trypanosomes (MiTat 1.4) were cultivated in SILAC SDM-79 composed of SDM-79 basic medium (12) without lysine and arginine and complemented with 7.5 mg/l hemin, 100,000 U/l penicillin, 100 mg/l streptomycin, 10 mm glycerin, 10% (v/v) dialyzed FCS (PAA) and different concentrations of lysine and arginine (Supplemental Fig. S1). For the proteome experiment, a final concentration of 40 mg/l Arg and Lys was used. Cells were kept in logarithmic growth throughout the experiment.
Bloodstream Form Growth
Bloodstream form trypanosomes (MiTat 1.4) were cultivated in modified HMI-9 (13) SILAC medium: IMDM medium without lysine, arginine, and glutamine (PAA) complemented with 136 mg/l hypoxanthine, 82.2 mg/l bathucoproine sulfonate, 0.2 mm beta-mercaptoethanol, 39 mg/l thymidine, 100,000 U/l penicillin, 100 mg/l streptomycin, 182 mg/l cysteine, 584 mg/l l-glutamine, 10% v/v dialyzed FCS (GIBCO, Carlsbad, CA) and different concentrations of lysine and arginine (Supplemental Fig. S1). For the proteome experiment, a final concentration of 40 mg/l Arg and Lys was used. Cells were kept in logarithmic growth throughout the experiment.
Cell Lysis
Equal amounts (5 × 107 cells) of BSF and PCF trypanosomes were separately harvested by centrifugation (1400 × g, 10 min) and resuspended in 0.5 ml PBS. Cell suspensions of “heavy” isotope labeled BSF were mixed with “light” isotope labeled PCF and vice versa. After centrifugation (1400 × g, 10 min) cell pellets were resuspended in lysis buffer (50 mm Tris, pH 7.3; 4% SDS), boiled at 95 °C for 10 min and stored at −80 °C.
MS Sample Preparation
To assess incorporation efficiency, 107 cells were lysed in 200 μl lysis buffer (modified RIPA) and centrifuged for 1 min at 13,000 × g to remove cell debris. Two microliter of the lysis supernatant was added to 20 μl of 8 m urea, before reduction with 0.5 mm dithiothreitol and alkylation with 3 mm iodoacetamide for 30 min each at RT. After an initial 3 h Lys-C (Wako) digest, the sample was diluted to 200 μl with 50 mm ammonium bicarbonate buffer and incubated with 0.5 μg sequencing grade modified trypsin (Promega, Charbonnières, France) overnight at RT. The digested samples were desalted and stored on stage tips (14). For obtaining a comparative trypanosome proteome, 15 μg of either stage in the respective labeled version was combined and boiled in 2 × LDS sample buffer (Invitrogen, Carlsbad, CA) before loading on a 4–12% denaturing SDS Novex Gel (Invitrogen). Lanes were cut in 12 slices and processed as described previously (15).
MS Measurement and Analysis
Peptides were analyzed by nanoflow liquid chromatography on an EASY-nLC system coupled to an LTQ-Orbitrap XL (Thermo Fisher). Peptides were separated on a C18-reversed phase column, packed with Reprosil (Dr. Maisch), which was directly mounted on the electrospray ion source. We used a 160 min gradient from 2% to 60% acetonitrile in 0.5% acetic acid at a flow rate of 200 ml/min. For the pilot experiment the LTQ-Orbitrap XL was operated in a data dependent mode with top10 MS/MS spectra acquisition method in the linear ion trap per MS full scan in the orbitrap. For the final experiment the same samples were measured on a Q-Exactive (Thermo Fisher) benchtop orbitrap operated with a top10 MS/MS data dependent HCD fragmentation. All raw files (pilot and final experiments) were processed with MaxQuant 1.2.2.0 (16) using standard settings and searched against the annotated protein database of Trypanosoma brucei TREU927 (version 3.3, 9826 entries) downloaded from tritrypDB (http://tritrypdb.org) with the Andromeda search engine (17) integrated into the MaxQuant software suite. Enzyme search specificity was Trypsin/P for tryptic digest (Promega) and LysC for digest with lysyl endopeptidase (Wako Chemicals). Up to two miscleavages for each peptide were allowed. Carbamidomethylation on cysteines was set as fixed modification whereas methionine oxidation and protein N-acetylation was considered as variable modifications. The search was performed with an initial mass tolerance of 6 ppm mass accuracy for the precursor ion and 0.5 Da for CID MS/MS spectra and 20 ppm for HCD MS/MS spectra, respectively. False discovery rate was fixed at 1% on peptide and protein level. Identifications were matched between runs with a 2 min window. Before statistical analysis, peptides mapped to known contaminants, reverse hits and protein groups only identified by side were removed. Only protein groups identified with at least two, one of them unique, peptides and two quantitation events were considered for data analysis.
Data Analysis
RNA-seq data (18): The provided gene identifiers were matched to the Trypanosoma brucei TREU927 version 3.3 gene entries (http://tritrypdb.org). Nineteen of 7578 entries could not be transferred. For values labeled with BSF only an arbitrary average expression ratio of 50 was assumed, whereas for values of PCF only the inversed value of 0.02 was used. For comparison of the proteome and the transcriptome, the normalized transcript fold change was plotted against the SILAC ratios of the first experiment using Perseus (version 1.2.2.0).
GO Annotation
Gene ontology (GO) annotations for Trypanosoma brucei (version 1.83) were parsed from tritryp DB and matched to the proteome data using Perseus (version 1.2.2.0). Comparison of GO annotation was performed in R (version 2.0.8) using a custom made script.
PCR-mediated C-terminal in situ Tagging
For recombination-based tagging, 3–5 μg of purified PCR product was transfected into 1 × 107 BSF cells (MiTat 1.4) as described elsewhere (19). Stable transformants were selected in a batch with 0.1 μg/ml puromycin for 1 week prior to further analysis.
Western Blotting
Protein extracts of 2 × 106 cells were separated by SDS-PAGE and blotted onto a PVDF membrane. Blocking was performed in 5% milk powder in PBS/0.1% TWEEN-20. Primary antibodies (α-Ty BB2 and α-TbH3) were diluted in PBS/0.1% TWEEN-20. Blots were analyzed using the LI-COR Odyssey Imager after incubation with secondary antibodies (IRDye 800 α-mouse and IRDye 680 α-rabbit, Biomol).
Differentiation of BSF to PCF
Differentiation of BSF was performed as described elsewhere (20). Briefly, BSF were grown to a density of 2.5 –3 × 106/ml and kept in stationary phase for 24 h. Cells were harvested (1400 × g, 10 min, 37 °C) and resuspended in DTM complemented with 6 mm cis-aconiate at a cell density of 5 × 106/ml. Two days post-induction of differentiation, cells were stepwise diluted with SDM-79. Western blot samples were taken after 4 days of differentiation.
RNAi-mediated Depletion of DEAD-box Helicase
The helicase RNAi cell line used in this work was generated using the pTrypRNAi Gate vector as described in details elsewhere (21). Briefly, a fragment of the helicase ORF (Tb09.211.4430, nucleotide position 415–994) was cloned head-to-tail downstream of a tetracycline-inducible PARP promoter. The construct was digested with NotI prior to transfection. Transfection and drug selection were described previously (22). RNAi was induced by adding 1 μg/ml tetracycline to the cell culture. For microscopic analysis, cells were stained with 10 mm sulfo-NHS-ATTO488 (Atto-Tec) in PBS for 10 min at 4 °C. Pictures were taken using the IMIC microscope (TILL Photonics, Gräfelfing, Germany) and processed with Huygens Essential software 4.1.
RESULTS AND DISCUSSION
Stable Isotope Labeling of Trypanosomes
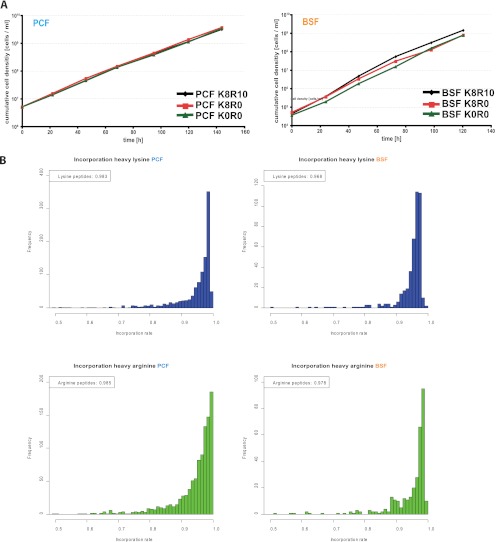
Trypanosomes have two major life cycle stages: the blood stream form (BSF) and the procyclic insect form (PCF, Fig. 1 A). Both stages can be cultured in vitro, although each form requires different cell culture conditions. We thus established individual SILAC protocols for BSF and PCF. First, the concentrations of lysine and arginine were titrated to guarantee optimal growth conditions (Supplemental Fig. S1) as well as complete incorporation of the isotope enriched amino acids (Fig. 1B). Incorporation rates for isotope-enriched arginine and lysine reached more than 97%. Although arginine to proline conversion has been reported to be a significant issue in yeast, C. elegans and selected vertebrate cell lines, we did not observe any conversion in either PCF or BSF (Supplemental Fig. S2). Having established the SILAC-trypanosomes, we labeled BSF with heavy lysine and heavy arginine and combined them with PCF grown in light condition. In a label-switch experiment, heavy labeled PCF trypanosomes were mixed with light labeled BSF. To increase proteome coverage by using a different protease, we performed an identical experiment with heavy lysine. We combined an equal number of cells (107), rather than equal protein amounts prior to lysis in SDS, to allow direct comparison of individual protein concentrations in the two life cycle stages (Supplemental Fig. S3).
Fig. 1.
SILAC-labeling of T. brucei. A, Cumulative growth of trypanosomes labeled with heavy amino acids compared with normal cell culture conditions. B, Histograms of the incorporation rate distribution for the selected 40 mg/L arginine and 40 mg/L lysine concentration.
Comparative Proteomics of Bloodstream- and Insect-form Trypanosomes
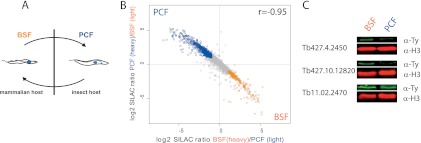
Analysis of the lysine and arginine labeling, digested with trypsin, identified 3889 and quantified 3458 protein groups, whereas labeling with lysine followed by digestion with LysC resulted in quantification of 3047 protein groups of 3370 identified ones (Supplemental Fig. S3B and S2C). Reproducibility was very high (Pearson coefficient 0.88 and 0.94, respectively) for the label switch (Supplemental Fig. S3B). Excellent correlation between the double and single label experiment (Pearson coefficient 0.92, Supplemental Fig. S3C) allowed combining both experiments to increase the proteome coverage (Fig. 2B). In total, we quantified 4081 of 4364 identified protein groups (Supplemental Tables S1 to S4). A large number of genes in T. brucei originate from gene duplications and have nearly identical sequences. As a consequence, around 10% of our protein groups contain more than one database entry resulting in a final number of 5036 identified and 4718 quantified proteins with an FDR of 0.01 from the trypanosome protein database. As we combined an equal amount of cells for each experiment and performed complete lysis in SDS, our SILAC ratios directly reflect the protein amount of the different stages. With a twofold cutoff, 970 protein groups (23.8% of the quantified proteome) are stage-specifically enriched for PCF and 386 protein groups (9.5% of the quantified proteome) for BSF. The total of 33.2% are significantly higher than the 14% of regulated transcripts in a similar experimental setup using the same cut-off (18) and also higher than previous transcript-based estimates (23, 24). Structural proteins such as histone H3 and α-tubulin, which are expected to be equally abundant in both stages given equal numbers of cells, indeed had SILAC ratios of around 1 (Supplemental Fig. S4). Next, we used in situ tagging to follow expression levels of three proteins down-regulated in PCF by Western blot analysis during early differentiation (Fig. 2C). In agreement with the SILAC results, two candidates were rapidly down-regulated 3 days after differentiation from BSF to PCF. A third protein (Tb11.02.2470) was still detectable at day 3, but was not expressed in an established PCF cell line. This suggests the protein is indeed down regulated as measured by SILAC but that it might be necessary for differentiation and therefore persists for longer than the 3 days time point.
Fig. 2.
Comparative proteomics of bloodstream- and insect-form trypanosomes. A, Trypanosoma brucei differentiates from an insect procyclic form (PCF) to bloodstream forms (BSF) in mammals during its life cycle. B, Label-switch experiment with stable isotope-labeled trypanosomes shows high correlation (pearson coefficient 0.92) with 970 protein groups up-regulated (SILAC ratio >2) in PCF and 386 more than twofold up-regulated BSF protein groups. C, A random selection of stage specific proteins were tagged and investigated during the differentiation process for validation by Western blot analysis.
Regulation of Protein Expression in Trypanosomes
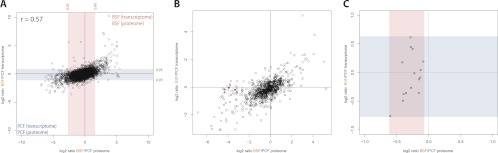
We used a recently reported RNA-Seq study comparing transcript abundance between PCF and BSF (18) to correlate the transcriptome differences with our observed protein expression differences. We found only moderate correlation (Pearson coefficient 0.57) between the transcriptome and the proteome. This suggests a strong regulatory effect of translation efficiency and protein stability on the protein levels between PCF and BSF trypanosomes. Underscoring this observation, we found that the differences between PCF and BSF are five times larger (0.9 quantile) at the proteome level compared with the transcriptome. To investigate whether differences between the transcriptome and proteome are apparent for certain biological groups or metabolic functions or if some proteins are exclusively regulated by translation or protein stability, we conducted a comparison of gene ontology annotation (Fig. 3B). Some proteins of the oxidative stress response and citrate acid cycle are up-regulated in PCF without corresponding changes in RNA abundance. Additionally, the proteasome subunits have an equal SILAC ratio between PCF and BSF, whereas the mRNA transcript abundance showed stronger variation between both forms arguing that the proteasome complex is regulated by protein stability.
Fig. 3.
Regulation of protein expression in trypanosomes. A, Comparison of the expression differences in the transcriptome and the proteome between PCF and BSF trypanosomes shows moderate correlation (Pearson coefficient 0. 57) for individual proteins and their transcript. B, Comparison of the ratios between PCF and BSF expression in transcriptome and proteome for gene ontology terms shows that the transcriptome is a good proxy for the regulation of many proteins like members of the glycolytic pathways in BSF and mitochondrial proteins in PCF. However, for some individual GO terms like oxidative stress response and citrate acid cycle differences between transcriptome and proteome are observable. C, Although the SILAC ratio for the protein subunits of the proteasome show an identical ratio between PCF and BSF, their transcripts show different ratios (blue: regulatory subcomplex, red: core subcomplex).
New Insights Into the Parasite's Host Adaptation Machinery
Trypanosomes have to master a difficult task when they shuttle from the intestinal tract of the insect vector to the vascular system of the mammalian host. They face large environment differences and host-specific defense mechanisms during the life cycle, which force them to adapt many basic biological processes like metabolism, organelle activity, cell cycle regulation and endocytic activity. Many of these processes have been intensively investigated but the mechanisms involved in the parasite's adaptation machinery remain unclear. To better understand how the parasite adjusts to the challenges in different hosts, we searched for novel stage-specifically regulated proteins. We found that several proteins are strongly stage-specifically expressed without significant changes of their corresponding mRNA (Supplemental Table S4). For example, a GPI inositol deacylase precursor (Tb927.10.4780), which is most likely involved in GPI anchor processing, is only moderately regulated on at the mRNA level (3.5-fold up-regulated) although protein levels are more than 30-fold higher in BSF compared with PCF (Supplemental Table S4). Interestingly, a putative ATP-dependent DEAD-box RNA helicase (Tb09.211.4430) is strongly up-regulated in BSF independent of mRNA levels. Recently, a homolog of the DEAD-box RNA helicase DHH1 was shown to be essential for the correct expression of many developmentally regulated mRNAs in insect-stage trypanosomes (25). Database search suggests that the BSF-specific RNA helicase is also an ATP-dependent DEAD-box RNA helicase. Hence, it might also be involved in regulation of trypanosome mRNAs in the mammal host. Another protein identified as BSF-specific is a cysteine peptidase (Tb927.10.2440). Although this protein was already found in the BSF phosphoproteome (26) and in a set of mitochondrial proteins in PCF (27), it was not described to be stage-specific. However, at the protein level it is 25-fold up-regulated in BSF suggesting that this peptidase has different functions in different hosts. We also found an oxidoreductase that is highly up-regulated in parasites in the mammal host (Tb927.10.9360). Several oxidoreductases have been described in trypanosomes already (27). However, most of them are associated with the mitochondrial respiration system and are up-regulated in PCF. Oxidoreductases represent a large and functionally heterogeneous group of proteins. This BSF-specific oxidoreductase displays weak homology with 2-hydroxyglutarate dehydrogenases and might be involved in stage-specific energy metabolism. Furthermore, several other hypothetical proteins with unknown functions are less than twofold regulated at the mRNA level but more than 10-fold at the protein level in BSF suggesting a specific role in the adaptation process to the mammal host.
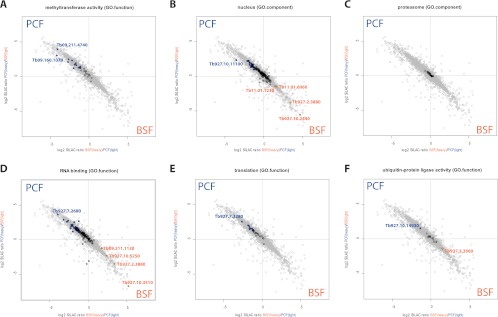
To gain functional insights into the differences between the life cycle stages more systematically, we performed GO analysis. Our data not only confirm reported differences of previous organellar proteomic studies (28, 29) but interestingly uncovered additional classes of proteins that were not expected to be stage specific or associated with the differentiation process. For example, several methyltransferases and ribosomal proteins are differentially abundant (Fig. 4A). The observed stage-specifically expressed nuclear proteins might shed light on known but yet unexplained differences in nuclear architecture and chromatin structure (Fig. 4B). Finally, proteasome composition and abundance does not seem to be different in the two life cycle stages (Fig. 4C), but differentially expressed proteins that are involved in RNA binding, translation and protein modification like ubiquitination are, providing hints to the mechanisms by which the proteomes change during the differentiation process (Fig. 4D and 4E).
Fig. 4.
Gene ontology analysis of differentially regulated proteins. Two dimensional expression blot shows some proteins of different GO categories to be stage-specifically expressed. Individual proteins of these selected groups are annotated: A, methyltransferase activity; B, nuclear proteins; C, proteasome; D, RNA binding proteins; E, proteins involved in translation; F, ubiquitin-ligases.
Identification of New Potential Drug Targets
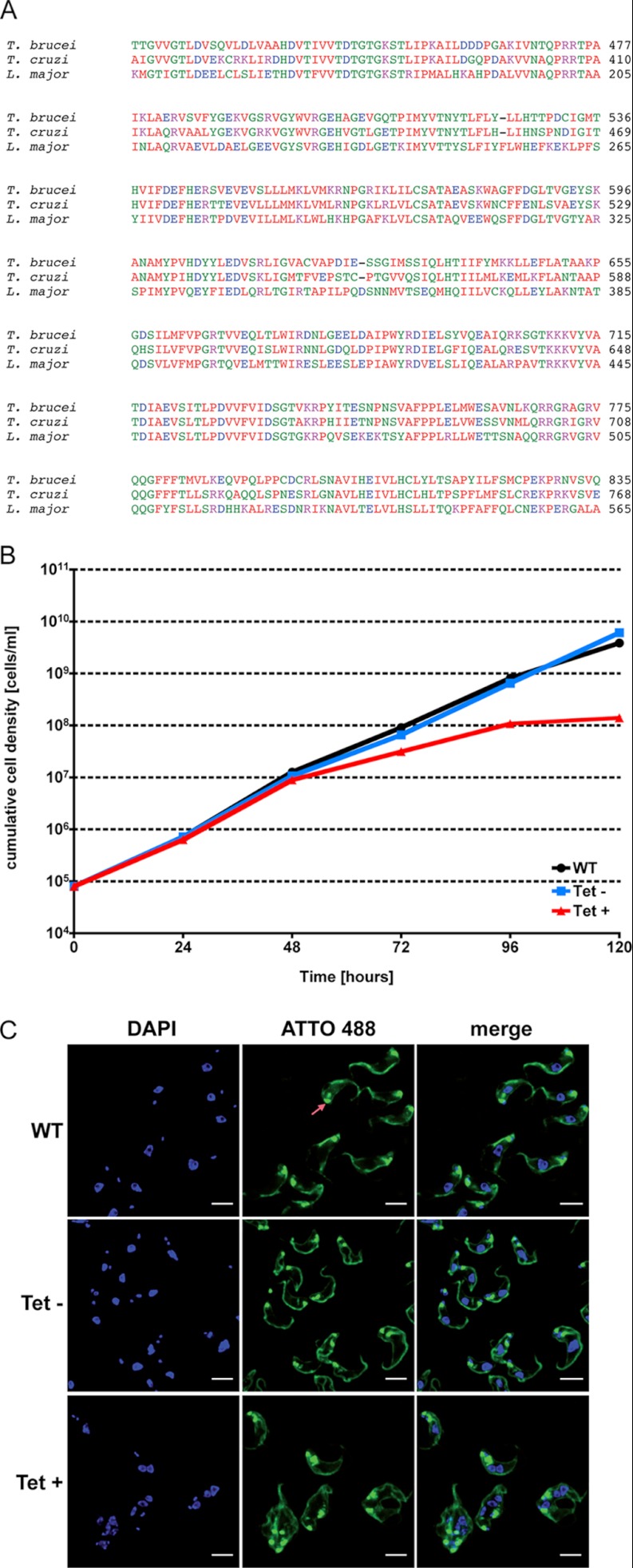
This study was initiated to gain new insights into the adaptation machinery of trypanosomes to better understand the sophisticated life cycle of this parasite. Additionally, this data can be exploited to find new potential drug targets in trypanosomes and related parasites. As a proof of principle, we wanted to test whether the DEAD-box RNA helicase (Tb09.211.4430) might qualify as a putative drug target. The helicase is highly up-regulated in BSF and potential homologues can also be found in the related kinetoplastid parasites Leishmania major and Trypanosoma cruzi (Fig. 5A). Homologs in humans could not be identified using NCBI BLAST (data not shown). We used an inducible RNAi system to deplete the helicase in the human infective stage of T. brucei. The parasites showed a clear growth phenotype and cells with an aberrant morphology 72 h post RNAi induction confirming that the helicase is essential for viability in BSF (Fig. 5B). To further investigate the morphological changes, we stained the surface of the cells with Atto488 NHS-ester and the DNA with DAPI. Interestingly, multiple nuclei and kinetoplasts could be detected suggesting repeated replication and segregation of these organelles without cytokinesis (Fig. 5B). Deep sequencing of these cells after RNAi induction is expected to shed more light on this phenotype in the future and might help to unravel the mechanisms of mRNA regulation in trypanosomes in their different host environments.
Fig. 5.
The putative DEAD-box RNA helicase is essential for viability in BSF. A, Alignment of core protein domains of homologs of the T. brucei RNA helicase (Tb09.211.4430) in L. major (LmjF.35.2270) and T. cruzi (Tc00.1047053510655.30). Alignments were performed with ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). B, Cumulative growth of parasites after induction of RNAi (red line), without induction (blue line) and of the parental strain (WT, black line). C, Images of WT parasites (top panel), un-induced RNAi cells (middle panel) and 96 h after induction (bottom panel). The surface of the cells was stained with sulfo-NHS-ATTO488 (green) and the DNA with DAPI (blue). The bright signal of the flagella pocket is marked with an arrow. Scale bars represent 6 μm.
In conclusion, we here demonstrate the feasibility to perform metabolic labeling of trypanosomes with lysine and arginine and obtain highly accurate quantitation of more than 4000 proteins in two life cycle stages of T. brucei. Our study confirms that the transcriptome only serves as a moderate proxy for the changes in actual protein levels and that further quantitative proteomics studies might provide new insights into the biology of this parasite. The importance of proteomics analysis in trypanosomes is emphasized by the publication of a similar study while this work was under revision (30). This data set can serve as a resource to the community to gain insight into the development of the parasite and the molecular bases of different morphology, motility and metabolism of life cycle stages. In fact, we already identified several proteins that might be associated with important biological processes such as GPI-anchoring, translation regulation and energy metabolism, which are specifically adapted in T. brucei to survive in the mammalian host.
Supplementary Material
Acknowledgments
We thank Markus Engstler, Michael Boshart, and Nicolai Siegel for carefully reading the manuscript and Thanatip Viturawong for help with bioinformatics analysis.
Footnotes
* This work was supported by funds of the Max Planck Society and the European Union (PROSPECTS, HEALTH-F4–2008-201648) to the Mann Laboratory. The Janzen Laboratory was supported by the DFG grant JA1013/2 and the SFB TR5.
 This article contains supplemental Figs. S1 to S4 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S4 and Tables S1 to S4.
1 The abbreviations used are:
- BSF
- Bloodstream forms
- GO
- Gene Ontology
- PCF
- Procyclic forms
- SDS
- sodium dodecyl sulfate
- SILAC
- stable isotope labeling by amino acids in cell culture
- T. brucei
- Trypanosoma brucei.
REFERENCES
- 1. Fenn K., Matthews K. R. (2007) The cell biology of Trypanosoma brucei differentiation. Curr. Opin. Microbiol. 10, 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bringaud F., Riviere L., Coustou V. (2006) Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol. Biochem. Parasitol. 149, 1–9 [DOI] [PubMed] [Google Scholar]
- 3. Jensen B. C., Sivam D., Kifer C. T., Myler P. J., Parsons M. (2009) Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics 10, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Queiroz R., Benz C., Fellenberg K., Hoheisel J. D., Clayton C. (2009) Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics 10, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clayton C. E. (2002) Life without transcriptional control? From fly to man and back again EMBO J. 21, 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 7. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Frohlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 8. Larance M., Bailly A. P., Pourkarimi E., Hay R. T., Buchanan G., Coulthurst S., Xirodimas D. P., Gartner A., Lamond A. I. (2011) Stable-isotope labeling with amino acids in nematodes. Nat. Methods 8, 849–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fredens J., Engholm-Keller K., Giessing A., Pultz D., Larsen M. R., Hojrup P., Moller-Jensen J., Faergeman N. J. (2011) Quantitative proteomics by amino acid labeling in C. elegans. Nat. Methods 8, 845–847 [DOI] [PubMed] [Google Scholar]
- 10. Sury M. D., Chen J. X., Selbach M. (2010) The SILAC fly allows for accurate protein quantification in vivo. Mol. Cell. Proteomics 9, 2173–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kruger M., Moser M., Ussar S., Thievessen I., Luber C. A., Forner F., Schmidt S., Zanivan S., Fassler R., Mann M. (2008) SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 [DOI] [PubMed] [Google Scholar]
- 12. Brun R., Schonenberger M. (1979) Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36, 289–292 [PubMed] [Google Scholar]
- 13. Hirumi H., Hirumi K. (1989) Continuous cultivation of Trypanosoma brucei bloodstream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75, 985–989 [PubMed] [Google Scholar]
- 14. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 15. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 16. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 17. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 18. Siegel T. N., Hekstra D. R., Wang X., Dewell S., Cross G. A. (2010) Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 38, 4946–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oberholzer M., Morand S., Kunz S., Seebeck T. (2006) A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 145, 117–120 [DOI] [PubMed] [Google Scholar]
- 20. Overath P., Czichos J., Haas C. (1986) The effect of citrate/cis-aconitate on oxidative metabolism during transformation of Trypanosoma brucei Eur. J. Biochem. 160, 175–182 [DOI] [PubMed] [Google Scholar]
- 21. Kalidas S., Li Q., Phillips, M.A. (2011) A Gateway(R) compatible vector for gene silencing in bloodstream form Trypanosoma brucei. Mol. Biochem. Parasitol. 178, 51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burkard G., Fragoso C. M., Roditi I. (2007) Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 153, 220–223 [DOI] [PubMed] [Google Scholar]
- 23. Koumandou V. L., Natesan S. K., Sergeenko T., Field M. C. (2008) The trypanosome transcriptome is remodelled during differentiation but displays limited responsiveness within life stages. BMC Genomics 9, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. El-Sayed N. M., Myler P. J., Blandin G., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E. A., Hertz-Fowler C., Ghedin E., Peacock C., Bartholomeu D. C., Haas B. J., Tran A. N., Wortman J. R., Alsmark U. C., Angiuoli S., Anupama A., Badger J., Bringaud F., Cadag E., Carlton J. M., Cerqueira G. C., Creasy T., Delcher A. L., Djikeng A., Embley T. M., Hauser C., Ivens A. C., Kummerfeld S. K., Pereira-Leal J. B., Nilsson D., Peterson J., Salzberg S. L., Shallom J., Silva J. C., Sundaram J., Westenberger S., White O., Melville S. E., Donelson J. E., Andersson B., Stuart K. D., Hall N. (2005) Comparative genomics of trypanosomatid parasitic protozoa. Science 309, 404–409 [DOI] [PubMed] [Google Scholar]
- 25. Kramer S., Queiroz R., Ellis L., Hoheisel J. D., Clayton C., Carrington M. (2010) The RNA helicase DHH1 is central to the correct expression of many developmentally regulated mRNAs in trypanosomes. J. Cell Sci. 123, 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nett I. R., Martin D. M., Miranda-Saavedra D., Lamont D., Barber J. D., Mehlert A., Ferguson M. A. (2009) The phosphoproteome of bloodstream form Trypanosoma brucei, causative agent of African sleeping sickness. Mol. Cell. Proteomics 8, 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Panigrahi A. K., Ogata Y., Zikova A., Anupama A., Dalley R. A., Acestor N., Myler P. J., Stuart K. D. (2009) A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9, 434–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vertommen D., Van Roy J., Szikora J. P., Rider M. H., Michels P. A., Opperdoes F. R. (2008) Differential expression of glycosomal and mitochondrial proteins in the two major life-cycle stages of Trypanosoma brucei. Mol. Biochem. Parasitol. 158, 189–201 [DOI] [PubMed] [Google Scholar]
- 29. Colasante C., Robles A., Li C. H., Schwede A., Benz C., Voncken F., Guilbride D. L., Clayton C. (2007) Regulated expression of glycosomal phosphoglycerate kinase in Trypanosoma brucei. Mol. Biochem. Parasitol. 151, 193–204 [DOI] [PubMed] [Google Scholar]
- 30. Urbaniak M. D., Guther M. L., Ferguson M. A. (2012) Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS One 7, e36619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.