Abstract
Changes in the actin cytoskeleton, especially the formation of cross-linked actin networks (CLANs) are thought to contribute to the increased intraocular pressure observed in primary open-angle and steroid-induced glaucoma. To better understand the effects of glucocorticoids, we employed a shotgun method to analyze global changes in the cytoskeleton and integrin signaling pathways following dexamethasone (DEX) treatment of human trabecular meshwork (HTM) cells. RNA and cell lysates were obtained from HTM cells incubated with or without DEX. Changes in protein expression were determined by mass spectrometry (MS) following differential centrifugation of cell lysates to enrich for low-abundance cytoskeletal and signaling proteins, proteolytic digestion, and a titanium dioxide column to enrich for phosphopeptides. Results were validated by Western blots. Changes in RNA levels were determined with gene arrays and RT-PCR. Overall, MS identified 318 cytoskeleton associated proteins. Five of these proteins (PDLIM1, FGFR1OP, leiomodin-1, ZO-2 and LRP16A) were only detected in DEX-treated cells by MS. However, only PDLIM1 showed a statistically significant increase at the RNA level. Other proteins with differences at both the RNA and protein levels included β3 integrin, caveolin-1, Borg2, raftlin1, PI-3 kinase regulatory subunit α, transgelin, and filamin B. By immunofluorescence microscopy filamin B and PDLIM1 showed enhanced expression in human trabecular meshwork cells, but only PDLIM1 demonstrated significant localization within CLANs. Finally, MS showed that some of the cytoskeleton proteins (Borg2, leiomodin-1, LRP16A, raftlin1 and CKAP4) contained phosphorylated residues. This study suggests that DEX affects the expression of cytoskeleton proteins at the transcriptional and translational level and shows that a combined genomic and proteomic approach can be used for rapid analysis of proteins in the TM. It also shows that DEX altered the expression of components (PDLIM1 and β3 integrins) involved in CLAN formation and provides new findings into the effects of glucocorticoids on the cytoskeleton.
Steroid-induced glaucoma is an iatrogenic condition resulting from the use of glucocorticoids. Glucocorticoids such as dexamethasone (DEX)1 raise intraocular pressure (IOP) in ∼40% of patients in the general population, and ∼6% of these patients will go on to develop glaucoma (1, 2). This condition is similar to primary open angle glaucoma (1–3), and is caused by a restriction in fluid outflow through the trabecular meshwork (TM), resulting in an imbalance between the amount of aqueous humor produced and the amount drained. This imbalance results in a higher IOP.
It is thought that an alteration in the cytoskeletal structure or contractile properties of TM cells may result in the disruption of normal fluid flow. In support of this idea, cross-linked actin networks, referred to as CLANs, have been observed with increased frequency in the TM of glaucomatous patients and in glucocorticoid treated anterior segments as well as in TM cells in culture. CLANs are thought to alter the contractility of the TM by holding the cells in a rigid conformation, making the cells unresponsive to the change in pressure and blocking the aqueous humor outflow pathway (1, 4, 5). Thus, agents such as H7 and the latrunculins A and B, which disrupt the organization of the cytoskeleton, decrease IOP in porcine and monkey cultured anterior segments (6–9).
Control of the actin cytoskeleton is mediated by the Rho family of small GTPases. The Rho effector ROCK has been shown to play a part in TM contractility and modulation of IOP. Inhibition of ROCK using a dominant negative mutant or the inhibitor Y-27632 causes TM cells to “relax” by decreasing actin stress fiber formation and phosphorylation of myosin light chain (MLC) (10, 11). ROCK inhibition also decreases IOP in cultured human and porcine anterior segments (10, 11). In contrast, constitutively active RhoA (RhoA V14) increases stress fiber formation and MLC phosphorylation, and increases IOP in cultured porcine anterior segments (12).
Previous studies have suggested that DEX up-regulates and activates a β3 integrin signaling pathway that induces CLAN formation (13). This signaling cascade includes Src, the Rho family GTPase Rac1, and the Rac1 guanine nucleotide exchange factor (GEF) Trio (4). Other components of this signaling pathway activated by αvβ3 integrin signaling or DEX-treatment are unknown, but may include the atypical G-protein-coupled receptor CD47 and a PI-3 kinase-mediated β1 integrin signaling pathway.
Genomic and proteomic analyses are powerful new tools to rapidly study changes associated with glaucoma. Microarray analyses of TM cells identified many genes that are up-regulated by DEX in multiple studies, including myocilin (MYOC), angiopoietin-like 7 (ANGPTL7), insulin-like growth factor binding protein 2 (IGFBP2), monoamine oxidase A (MAOA), and α1-antichymotrypsin (SERPINA3) (14–18). Other studies have used proteomic techniques such as 2-D gel electrophoresis and mass spectrometry (MS) to evaluate the proteome of ocular tissues including vitreous humor from normal patients (19), immortalized glaucomatous TM cells (20), immortalized normal TM cells (21), and normal human and porcine TM tissues (22) and cells (21). This latter study found that DEX down-regulated RhoGDI, an important regulator of the cytoskeleton and integrin signaling.
In this study, we examined the possibility of using combined genomic and proteomic studies to analyze changes associated with the cytoskeleton and integrin signaling following DEX treatment. Identification of these proteins is essential because they may be useful targets for inhibiting CLAN formation and possibly steroid-induced glaucoma. However, analysis of the integrin/cytoskeleton signaling network in the TM has been hampered by the low abundance of some of these proteins. Using gene microarrays and MS of cellular fractions, we have identified an approach suitable for the rapid analysis of proteins low in abundance in the TM that are involved in the integrin/cytoskeleton signaling network. Proteins identified to be up-regulated by DEX include filamin B and PI-3 kinase, which are involved in β3 integrin signaling. Another protein identified is caveolin-1, a mediator of integrin signaling that may be associated with primary open-angle glaucoma (POAG) (23). Two novel proteins previously unidentified to be up-regulated by DEX are PDLIM1 and Borg2, both of which regulate stress fiber formation. Both filamin B and PDLIM1 were found within CLANs and PDLIM1 demonstrated a significant DEX-dependent increase in CLAN localization. Finally, this study confirmed that the β3 integrin subunit which is involved in CLAN formation was up-regulated at both the gene and protein level following DEX treatment.
EXPERIMENTAL PROCEDURES
Cell Culture
Two cell strains (HTM-1 and HTM-2) from the same donor were used. Both strains were isolated from a 27-year old donor as previously described (24–26) and grown to confluency in low glucose DMEM (Sigma-Aldrich, Co.), 15% fetal bovine serum (Atlanta Biologicals), 2 mm l-glutamine (Sigma-Aldrich, Co.), 1% amphoteracin B (Mediatech, Herndon, VA), 0.05% gentamicin (Mediatech) and 1 ng/ml FGF-2 (Peprotech). After reaching confluency, HTM monolayers were cultured in the presence of 500 nm DEX or vehicle (0.1% ethanol) in media lacking FGF and containing 10% FBS.
Protein Isolation
Cells grown for 4–5 days in the presence of DEX or ethanol were lifted using Cell Dissociation Solution (Sigma-Aldrich, Co.) and replated onto dishes precoated with 10 μg/ml fibronectin for 1 h. Cells were then washed and lysed with ice-cold lysis buffer (50 mm Tris pH 7.9, 7.5 mm MgCl2, 50 mm NaCl, 0.5% Nonidet P-40, 0.5 mm dithiothreitol (DTT), 1 mm Na3VO4, 6.4 mm Na4P2O7). All buffers contained 1 × Complete Mini EDTA-Free protease inhibitor tablets and phosSTOP phosphatase inhibitor tablets (Roche Diagnostics Corp). Lysates were fractionated using the centrifugation procedure outlined in Supplemental Fig. S1 (27). Insoluble nuclear material was removed by centrifugation at 1000 × g for 10 min at 4 °C. The soluble cytoplasmic and membrane-associated fractions were separated by centrifugation at 100,000 × g for 3 h. The pellet containing the membrane fraction was resuspended in 50 mm Tris pH 8.3, 6 m Urea, 0.1% Nonidet P-40, 0.5 mm DTT, 1 mm Na3VO4, 6.4 mm Na4P2O7. The protein concentration in each fraction was measured using a BCA or Bradford assay.
For MS analysis, the cytoplasmic and membrane-associated fractions were split into two aliquots. One aliquot from each fraction was used to analyze the total proteins in that sample whereas the other was used to enrich for phosphorylated proteins (see below). Proteins from the membrane-associated and soluble cytoplasmic fractions were then precipitated with ice cold 20% trichloroacetic acid. Precipitated proteins from each fraction were resuspended in 50 mm Tris, pH 7.9, 8 m urea, and 50 mm Tris, pH 8.3, 6 m urea, 0.1% Nonidet P-40, 0.5 mm DTT, respectively. All samples were stored at −80 °C.
Sample Preparation
Cleavable surfactant (ProteaseMax, Promega Corp.) was added to cytoplasmic and membrane-associated fractions to a final concentration of 0.02%. Samples <300 μl were diluted with 50 mm NH4HCO3, pH 7.8 to dilute the urea and sonicated on ice at 30 s intervals. After maximal solubilization, samples were further diluted to 1 m urea with 50 mm NH4HCO3, pH 7.8 and proteins were reduced and alkylated with DTT and iodoacetamide as described (28). A second aliquot of DTT was added to quench alkylation.
Samples were digested with trypsin (sequencing grade, Promega) at an enzyme-to-substrate ratio of 1:50 or 1:75 overnight at 37 °C and the digestion was quenched by addition of trifluoroacetic acid (TFA) to a pH of 2. Digests were desalted by solid-phase extraction (Sep-Pak tC18, Waters Corp.) and dried. Samples for total protein analysis were reconstituted in 0.5% acetic acid at a final concentration of 2 μg/μl, whereas samples for phosphopeptide analysis were reconstituted in 100 μl of a solution containing 58% acetonitrile, 0.07% TFA, and 300 mg/ml lactic acid.
Phosphopeptide Enrichment
Phosphopeptides were isolated as previously described (28–30) using custom made titanium dioxide (TiO2) enrichment tips. Phosphopeptides were eluted sequentially; first with 1% NH4OH and then with 30% acetonitrile, 0.038% TFA into a single tube. Eluates were acidified with TFA and dried by a vacuum concentrator. Enriched phosphopeptides were reconstituted with 0.25% TFA.
LC/MS/MS and Database Searching
HPLC was performed using an Agilent 1100 series system featuring an isocratic loading pump and nano-flow gradient pump. The entire phospho-enriched sample or 16 μg of the total protein digest was separated on a C18 column as described (28, 31). Because peptide phosphorylation is often present at substoichiometric levels and the amount of material recovered from phosphopeptide enrichment can be low, the entire quantity of phosphopeptides was loaded in each biological replicate for phosphopeptide detection. Conversely, from total protein analysis, there was additional material remaining for one biological replicate and this was used to perform a technical replicate analysis. Samples were washed from the autosampler onto a 0.3 mm x 5 mm Stablebond C18 trapping cartridge (Agilent Technologies, Santa Clara, CA) at a flow rate of 15 μl/min. For elution, peptides were gradient eluted onto an in-house fabricated 15-cm resolving column packed with 3 μm Magic-C18 beads (Michrom Bioresources, Auburn, CA) and incorporating a laser pulled tip (P-2000, Sutter Instrument, Novato, CA). Peptide elution used solvents comprised of 0.1 m acetic acid in water (solvent A) and 0.1 m acetic acid, 95% acetonitrile in water (solvent B). The gradient consisted of a 20 min loading and desalting period with column equilibration at 1% solvent B, an increase to 40% B over 195 min, ramp to 60% B over 20 min, increase to 100% B in 5 min and hold for 3 min. The column was then re-equilibrated at 1% B for 30 min. The flow rate for peptide elution and re-equilibration was 200 nl/min.
Mass spectrometry was performed using an LTQ-Orbitrap XL hybrid instrument (ThermoFisher, San Jose, CA) operated in data-dependent mode. Survey scans were acquired in the Orbitrap region of the instrument at a resolving power of 100,000 over the m/z range 300–2000. Following acquisition, raw data files were converted to the open source mzXML format (32) using the Trans-Proteomic Pipeline software suite version 4.4 (33) and then converted to Mascot generic format. Mascot version 2.2.07 was used for database searching (Matrix Science, www.matrixscience.com) and peptide sequence assignments. Mascot search parameters required a precursor ion mass tolerance of 30 ppm and a fragment ion mass tolerance of 0.5 daltons. Additionally, precursors arising from first or second 13C isotope were considered. Peptides were required to be fully tryptic on both ends (cleavage after lysine or arginine residues). Fixed modifications consisted of carbamidomethylation of cysteine. Variable modifications considered were oxidation of methionine residues and deamidation of asparagine or glutamine residues as well as phosphorylation of serine, threonine and tyrosine residues. The database used was the UniProt human protein database (downloaded on Sept. 6, 2010, http://www.uniprot.org/) containing 35026 protein sequences. This database was then reversed to yield a nonsense “decoy” database and appended to the forward sequences. A list of known common contaminant proteins such as trypsin and human keratin were also appended for a total of 70,130 sequences in the searched database. The appended “decoy” portion of the database was used to calculate false discovery rates for peptide and protein identification. Results were imported into the Scaffold software (Version 3, Proteome Software) for curation, quantitative analysis, and visualization. Scaffold's spectral counting was employed to compare relative protein abundances between treated and untreated samples in each experiment as the basis for normalization of the spectral counts for all the other LC/MS/MS data in that experiment. The functions and cellular localizations of identified proteins were based on the NCBI GO Annotations and Ontology database. Phosphorylation sites and consensus motifs were validated using PhosphoMotif Finder database (http://www.hprd.org) and PhosphoSitePlus® (www.phosphosite.org) (34). The data associated with this manuscript may be downloaded from ProteomeCommons.org Tranche using the following hash: DzOJRE0nJj+l7HokcNP3oq3iBcaPkLnMRCkjukv87GRvIYnN0nCCnZHpNuyvYjGEi9iOO.
Immunoblot Analysis
Western blot analysis of whole cell lysates or fractions from HTM cells was done as previously described (35) using the ECL Plus Western blotting detection kit (GE Healthcare) according to the manufacturer's instructions. All primary antibodies were from Abcam unless otherwise noted. Primary antibodies were: β3 integrin (mAb EP2417Y, 1:1000); FKBP5 pAb (Sigma-Aldrich, 1:1000); SDHA (2E3 mAb, 1:5000); PDLIM1 mAb (1:500); caveolin-1 pAb (1:2000); CDC42EP3-N-terminal pAb (1:500); filamin B pAb (EMD Millipore Corp., 1:500); SM22α (mAb 1B8, 1:500); CKAP4 pAb (1:1000); PI3 Kinase p85 alpha mAb (1:1000); PI3 kinase p110β (mAb Y384, 1:500). The myocilin mAb 7.1 (1:4200) (36) was a kind gift from Dr. Michael Fautsch, (Mayo Clinic, Rochester, MN).
RNA Isolation, Reverse Transcription and Real-Time PCR
Total RNA was isolated from confluent cultures of HTM cells treated with or without 500 nm DEX for 6 days using the QIAshredder and RNeasy Plus Mini Kits (Qiagen Inc.). Total RNA (2 μg) was reverse transcribed with the RETROscript reverse transcription kit using random primers (Invitrogen) according to the manufacturer's instructions. The RNA integrity was checked using the Agilent 2100 BioAnalyzer (Agilent Technologies).
Real-time PCR experiments using the synthesized cDNA were performed using an Applied Biosystems 7300 real-time PCR system (Invitrogen., Inc.) with SYBR Green PCR Master Mix (Invitrogen). The PCR profile used was 2 min at 50 °C and 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Data were normalized to SDHA or GAPDH and the fold change compared with ethanol treatment was determined.
Microarrays
Poly-A RNA controls were prepared using the Affymetrix® GeneChip® Eukaryotic Poly-A RNA Control Kit (Affymetrix). The poly-A RNA controls and T7-(N)6 Primers were then added to 300 ng of total RNA and cDNA synthesis was initiated using the GeneChip® WT cDNA Synthesis Kit. First-cycle, cRNA synthesis and cleanup was performed using the GeneChip® WT cDNA Amplification Kit and the GeneChip® Sample Cleanup Module. Second-cycle, first-strand cDNA synthesis and cRNA hydrolysis used the GeneChip® WT cDNA Synthesis Kit. The single-stranded cDNA was then fragmented and labeled using the GeneChip® WT Terminal Labeling Kit. Hybridization of labeled, single-stranded cDNA samples to Affymetrix's GeneChip® Human Gene 1.0 ST Arrays was done by the University of Wisconsin Biotechnology Center. Statistical analysis of the raw data used the open source software Bioconductor in R to generate a list of genes that showed a p value ≤ 0.05 and a fold change of more than 2 or less than −2. DAVID Bioinformatics Resources was used to identify gene names from the probe IDs composing the array (37, 38).
Spreading Assays and Immunofluorescence Microscopy
Confluent HTM monolayers pretreated with either DEX or ethanol for 5–7 days were serum-starved for 24 h and then replated in the continued presence of DEX or ethanol onto coverslips precoated with 20 nm fibronectin as previously described (4, 5, 13). Cells were allowed to spread for 3 h before fixation/permeablization in 4% p-formaldehyde plus 0.18% TX-100 for 15 min. Fixed cells were incubated with Alexa 488-conjugated phalloidin (Invitrogen) together with either rabbit polyclonal antibodies (pAbs) against human PDLIM1 (2 μg/ml; Abcam) or filamin B (10 μg/ml; FLN B, Abcam) or rabbit IgG (10 μg/ml) as a control. Rabbit pAbs were detected with Alexa 546-conjugated goat anti-rabbit IgG (Invitrogen). Nuclei were labeled with Hoechst 33342.
CLAN-positive cells (CPCs) were identified as previously described (4). Fifty CPCs were counted from each coverslip and checked for colocalization of either PDLIM1 or filamin B. Data were pooled from four experiments and a total of 6 coverslips for each condition were analyzed. Fluorescence was observed with a Zeiss Axioplan 2 epifluorescence microscope equipped with an Axiocam HRm digital camera. Images were acquired using Axiovision version 4.8.1 image acquisition software. Statistical analysis was performed using a paired t test.
RESULTS
Prolonged treatment with glucocorticoids such as DEX have been reported to induce the formation of CLANs in TM cells in culture (39) and in vivo (40) leading to the idea that alterations in the organization of the cytoskeleton could be responsible for some glaucomas. To analyze how prolonged treatment with DEX could trigger the formation of CLANs, TM cells were dosed for 4–6 days with DEX, which is generally the minimum amount of time needed to see the formation of CLANs in vitro (39).
Before using MS to screen for changes in the expression of cytoskeleton-associated proteins following DEX treatment, cell lysates were fractionated using a differential centrifugation procedure (Supplemental Fig. S1A) to separate nuclear proteins from membrane-associated proteins and soluble cytoplasmic proteins (27). This was done to enrich for proteins that might be in low abundance and/or associated with the plasma membrane. The purity of the cytoplasmic and membrane-associated fractions is shown in Supplemental Fig. S2. Both the soluble cytoplasmic and membrane-associated fractions showed very little of the nuclear marker HDAC2. PI-3 kinase was found predominantly in fractions containing soluble cytoplasmic proteins whereas caveolin-1 was found in the membrane-associated fraction and not the cytoplasmic fraction. As expected, RhoDGI was found in both the cytoplasmic and membrane-associated fractions. Caveolin-1 and, to a lesser degree, Rho GDI were enriched in the membrane-associated fraction compared to levels seen in whole cell lysates suggesting that the fractionation procedure aided in the detection of proteins lower in abundance. Proteins in both the soluble cytoplasmic and membrane-associated fractions were then analyzed by MS. The nuclear fraction was not analyzed by MS.
Overall, MS analysis identified 1644 proteins across all experiments with a protein false discovery rate of 0.6% and assigned peptide sequences to 116078 spectra with a peptide false discovery rate of <0.1%. These false discovery rates were obtained with Scaffold filters set at minimum protein probability of 90%, a minimum number of two peptides, and a minimum peptide probability of 50%. A comparison of all three biological replicates and one technical replicate indicated that 95% of the proteins in the cytoplasmic fraction and 90% of the proteins in the membrane fraction were detected in both control and DEX treated cells in > two runs. A Scaffold file showing all the proteins detected with a high scoring confidence is included as a supplement.
Based on GO NCBI Ontology and Genecard databases, 318 of these proteins were determined to be associated with the cytoskeleton either as a structural element or involved in the regulation of it (Supplemental Tables S1 and S2). The majority of these proteins were found associated with the total membrane or soluble cytoplasmic fractions. However, 41 of these proteins (Supplemental Table S2) were only detected in samples that had been passed through the TiO2 column to enrich for phosphorylated peptides. This indicates that the phospho-enrichment protocol aided in the identification of cytoskeleton associated proteins presumably because they were lower in abundance.
Because the contractile properties of the actomyosin network have been implicated as a potential cause of glaucoma, especially in steroid-induced glaucoma, we specifically looked for proteins involved in the assembly of the actomyosin network. As shown in Fig. 1, the MS analyses identified many proteins that are specifically involved in the regulation of the actomyosin network including many components of focal adhesions. All of these proteins were identified with >95% confidence. Among the proteins of particular interest were components in the β3 integrin signaling pathway such as the β3 integrin subunit, Rac1, and the catalytic subunit of PI-3 kinase. These proteins have previously been shown to be involved in the formation of the CLAN-like structure observed in both glaucomatous and steroid treated tissues (1, 4, 5).
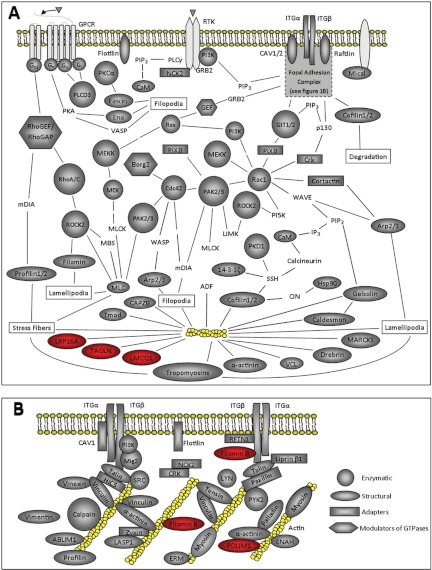
Fig. 1.
Interactome of cytoskeleton proteins associated with the actomyosin network. A, Schematic of the signaling pathways involved in the regulation of actomyosin network. The actomyosin network is indicated in yellow. Proteins detected by LC/MS/MS are shaded in gray. The gray shape of the protein indicates the function of that protein whereas the color red indicates that this protein was up-regulated by DEX at the protein level. The key shown in 1B indicates the function of all the proteins shown in both Figs. 1A and 1B. Proteins not associated with a gray shape were not found by MS. For some of these proteins several members of the family were identified. Subunits of the Arp2/3 complex found were ARPC2, ARPC4, ARPC5, ARC1B. Filamin A, B, and C were found. Myosin Va, 6, 9, 9B, 10, and 18A were found. Guanine exchange factors (GEFs) that were found are ARHGEF7, 11, 12, and 17. Tropomyosins (TPM) found were TPM1, TPM3, TPM4. Integrin subunits (ITGs) found were α1, α3, α4, β1, β3, β5. ARHGAP5 was the only GAP found. B, Schematic of the focal adhesion proteins identified by MS. ERM indicates that all three members of the family - ezrin, radixin and moexin were found. Both α-actinin-1 and −4 were found. Except for ENAH all proteins were identified at the 95% confidence level by MS.
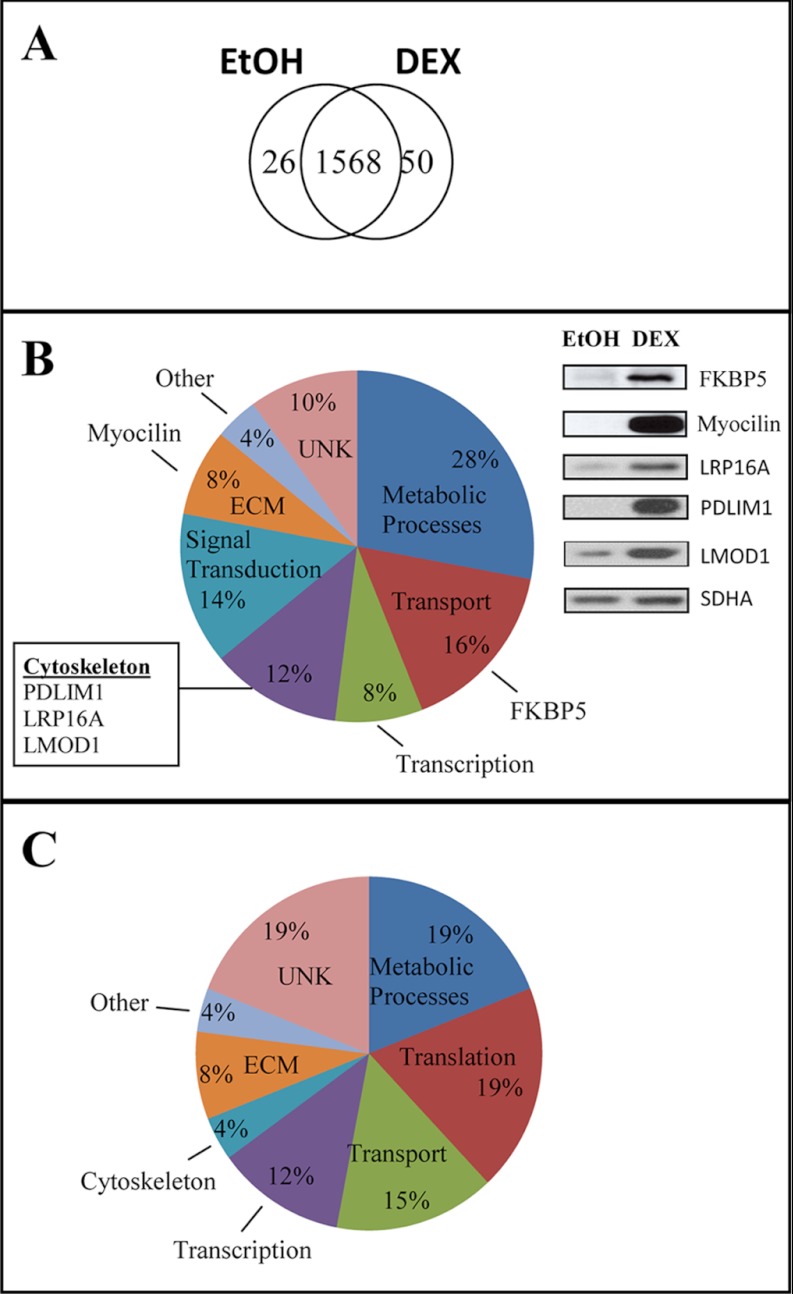
Overall, the MS analysis showed that 50 proteins were found only in the DEX-treated samples (Fig. 2A). Supplemental Table S4 lists all the proteins found. Two proteins consistently found only in the DEX treated samples were myocilin, which has previously been shown to be up-regulated by DEX (41), and FKBP5, which was predicted to be up-regulated based on previous genomic data (15). As expected, Western blot analysis verified that both these proteins were up-regulated by DEX (Fig. 2B).
Fig. 2.
Distribution of proteins in HTM cultures. A, Venn diagram of the distribution of proteins based on the combined MS analysis. To detect proteins found only in one treatment group with greater sensitivity, lower scoring peptide matches were included to enhance the discrimination between the two data sets. B, Distribution of proteins found only in DEX-treated cultures. Insert shown is a Western blot showing the expression of myocilin, FKBP5, PDLIM1, and LRP16A in DEX and ethanol treated samples. SDHA was used as a loading control. C, Distribution of proteins found only in ethanol-treated cultures.
In addition to myocilin and FKBP5, the MS analysis suggested that several other proteins were up-regulated by DEX. Fig. 2B shows the distribution of proteins found only in the DEX-treated cells from all three biological replicates and one technical replicate. Although the largest percentage of them was involved in metabolic processes, DEX also affected proteins associated with transcription, transport, signal transduction and the ECM. In addition to myocilin and FKBP5, three proteins (PDLIM1, leiomodin-1 and LRP16A) that regulate actin polymerization (Fig. 1) were only found in the DEX-treated samples. Western blot analysis confirmed that both PDLIM1 and LRP16A expression was up-regulated by DEX (Fig. 2). The leucine-rich repeat containing protein 16A (LRP16A) is also known as CARMIL or capping Arp2/3 and myosin-I-linker (www.genecards.org). Although MS data suggested that leiomodin-1 was only found in DEX-treated cells, Western blot analysis did not support this finding and showed that it was also detected in the control cells albeit at a much lower level. This illustrates the complexity and dynamic range of protein expression detected by MS. Two other cytoskeleton-associated proteins that the MS data suggested may be up-regulated in DEX-treated cells included the microtubule-anchoring protein FGFR1 oncogene partner (FGFR1OP) and tight junction protein ZO-2 (Supplemental Table S4).
Out of the 26 proteins found only in ethanol-treated controls by MS, ENAH was the only cytoskeleton-associated protein found (Supplemental Table S5) suggesting that it was down-regulated by DEX. However, Western blot analysis did not support this MS finding because ENAH was detected in DEX-treated samples at a similar level (data not shown). The distribution of the proteins in the control cells is shown in Fig. 2C. Supplemental Table S5 lists all the proteins detected only in the ETOH-treated controls.
To aid in the identification of proteins affected by DEX, we compared MS data with gene microarray data. The gene microarray studies indicated that relatively few genes for cytoskeleton proteins or proteins involved in the regulation of the cytoskeleton were affected by DEX treatment (Table I). Overall, only 21 genes involved in the regulation of the cytoskeleton were found to be altered in DEX-treated cultures at a statistically significant level. Seventeen of these genes were up-regulated by DEX and 4 genes were down-regulated (Table I). The majority of these genes showed small changes in RNA expression of around two- to three-fold. However, some genes were up-regulated by DEX greater than four-fold in both cell strains. They were: NEDD9, PDLIM1, β3 integrin, and the PI-3 Kinase regulatory subunit α. This finding supports a previous study (15), which also showed that RNA levels for NEDD9 as well as filamin B, nebulette, and transgelin were up-regulated by DEX more than twofold.
Table I. Changes in genes involved in assembly of the cytoskeleton. HTM1 and HTM2 are separate cell strains. The fold change (FC) for HTM1 is the average of 5 biological replicates. The FC for HTM2 is the average of 3 biological replicates. Abbreviations: ND, not detected; MS, mass spectrometry. * p < 0.05, † p < 0.01, ‡ p < 0.001.
| GenBank | Name | Function | FC HTM1 | FC HTM2 | MS Protein Detected |
|---|---|---|---|---|---|
| NM_006403.3 | NEDD9 | Docking protein involved in tyrosine-kinase-based signaling related to cell adhesion, β1 integrin signaling | 6.4‡ | 7.4‡ | ND |
| NM_020992.2 | PDLIM1 | Binds α-actinin; recruits Clik1 kinase to F-actin | 5.4‡ | 5.5‡ | Yes |
| NM_000212.2 | β3 Integrin | Binds ECM, signals to regulate cytoskeleton | 4.8‡ | 6.7‡ | Yes |
| NM_181523.2 | PI-3 Kinase (regulatory subunit α) | Phosphorylates phosphatidylinositol; involved in integrin, GPCR, RTK and MAPK signaling | 4.1‡ | 4.3‡ | Catalytic subunit |
| NM_001753.4 | Caveolin-1 | Scaffolding protein involved in adhesion; regulates MAPK signaling | 3.8† | 3.8‡ | Yes |
| NM_003603.5 | SORBS2 | Adaptor protein that promotes signaling complexes which regulates stress fibers | 3.6† | 4.2‡ | SORBS3 |
| NM_002652.2 | Prolactin-induced protein | Belongs to the PIP family; interacts with AZGP1 | 3.3* | 4.6‡ | ND |
| NM_006449.3 | CDC42 effector protein 3; Borg2 | Regulation of F-actin & pseudopodia formation | 3.1* | 3.5‡ | Yes |
| NM_001098424.1 | Discs, large homolog 1; DLG1 | Scaffolding protein for membrane recruitment of receptors, channels, signaling proteins | 2.6* | 3.3‡ | Isoform 2 |
| NM_001164317.1 | Filamin B | Binds membrane proteins to actin cytoskeleton; may promote branching of actin filaments | 2.5* | 3.1‡ | Yes |
| NM_001635.3 | Amphiphysin | Control the association of membrane with cytoskeleton | 2.5† | 3.2‡ | ND |
| NM_014945.2 | ABLIM3 | Mediates interaction between F-Actin & ABRA | 2.5* | 2.6‡ | ABLIM1 |
| NM_003633.2 | ENC (with BTB-like domain) | Actin-binding protein involved in differentiation of neural crest cells | 2.4‡ | 4.3‡ | ND |
| NM_015150.1 | Raftlin, lipid raft linker 1 | May stabilize lipid rafts | 2.3‡ | 2.4‡ | Yes |
| NM_006393.2 | Nebulette | Component of focal adhesion complexes | 2.3‡ | 2.2* | LASP1 |
| NM_001001522.1 | Transgelin | Actin cross-linking/gelling protein | 2.1* | 1.9† | Yes |
| NM_001233.4 | Caveolin-2 | Scaffolding protein involved in adhesion; regulates MAPK signaling | 2.1* | 1.7† | ND |
| NM_006825.3 | Cytoskeleton associated protein, CKAP | Stabilizes microtubules | −1.4* | −1.5† | Yes |
| NM_000638.3 | Vitronectin | Extracellular matrix protein | −2.1† | −1.9† | ND |
| NM_014981.1 | Myosin, heavy chain 15 | Creates contractile force with F-actin | −2.7† | −2.1‡ | ND |
| NM_001001548.2 | CD36 | Thrombospondin receptor | −10.2‡ | −5.3† | ND |
The majority of the cytoskeleton genes found to be affected by DEX in this microarray study encoded for actin binding proteins or regulated the assembly of actin filaments. Some of the proteins, like CKAP4, are associated with microtubules, whereas others like caveolin-1, caveolin-2 and raftlin1 are involved in forming lipid domains. We also found several signaling molecules associated with the assembly of the actin cytoskeleton and CLAN formation, including the β3 integrin subunit, PI-3 kinase regulatory subunit α and the ECM protein vitronectin, which is a ligand for β3 integrins.
One of the genes found to be up-regulated by DEX was PDLIM1, which was also found to be up-regulated by DEX based on the MS data and Western blot analysis (Fig. 2). Other cytoskeleton-associated proteins identified in the MS analyses (Table II) that were affected by DEX according to the microarray data (Table I) are Borg2, caveolin-1, CKAP4, filamin B, β3 integrin, raftlin1 and transgelin. The regulatory subunit α of PI-3 Kinase which was up-regulated at the RNA level was not detected by MS. However, MS did detect the catalytic subunit β for PI-3 Kinase (Supplemental Table S1). The other twelve proteins predicted to be up-regulated by the gene arrays (Table I) were not detected by MS. For some of these proteins, however, the MS analysis did detect a related family member such as SORBS3 (vinexin), ABLIM1, and LASP1 (Table I).
Table II. Unweighted spectra counts. Values are the total number of spectra assigned to the protein by the Scaffold. Values represent data from three biological replicates and one technical replicate. Unless otherwise indicated all peptides were identified >95% confidence level. All the others were identified at lower confidence levels >80–94%1; >50–79%2; 20–49%3. Alternative names for the proteins are indicated in parenthesis.
| Name | Total Cytoplasmic |
Total Membrane |
Phospho-enriched Cytoplasmic |
Phospho-enriched Membrane |
||||
|---|---|---|---|---|---|---|---|---|
| DEX | EtOH | DEX | EtOH | DEX | EtOH | DEX | EtOH | |
| CDC42 effector protein 3; (Borg2) | 0 | 0 | 0 | 0 | 6 | 21 | 0 | 0 |
| Caveolin-1 | 0 | 0 | 22 | 12 | 0 | 0 | 3 | 0 |
| Cytoskeleton-associated protein 4; CKAP4 | 0 | 5 | 51 | 84 | 0 | 0 | 21 | 7 |
| Filamin-B | 117 | 19 | 27 | 0 | 24 | 0 | 0 | 0 |
| β3 Integrin | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Leiomodin-1 | 0 | 0 | 0 | 0 | 11 | 0 | 4 | 0 |
| Leucine rich protein 16A, LRP 16A; (CARMIL) | 4 | 0 | 0 | 0 | 21 | 0 | 13 | 0 |
| PDZ and LIM domain protein 1; PDLIM1 | 18 | 0 | 0 | 0 | 12 | 0 | 0 | 0 |
| Raftlin, lipid raft linker 1, (MIG2) | 8 | 13 | 0 | 0 | 4 | 13 | 12 | 0 |
| Transgelin | 21 | 7 | 22 | 0 | 3 | 0 | 0 | 0 |
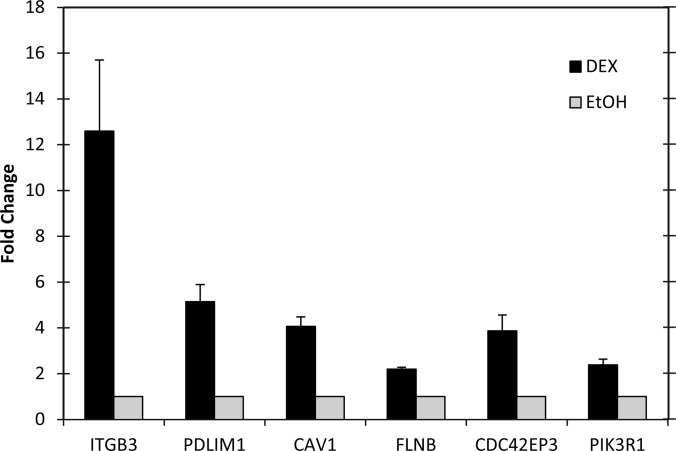
Because we are interested in how integrins may trigger CLAN formation in steroid-induced glaucoma, changes in gene expression likely to affect the organization of the actin cytoskeleton or integrin signaling were then validated using quantitative RT-PCR. The primers used for this study are shown in Table III. As shown in Fig. 3, RNA levels for the β3 integrin subunit were up-regulated 12-fold compared with the ethanol-treated controls (p < 0.05). RNA levels for caveolin-1, cdc42 effector protein 3 (CDC42EP3, Borg2), filamin B, PDLIM1, and the PI-3 kinase regulatory subunit α were also up-regulated in DEX-treated cells compared with controls (p < 0.05, Fig. 3) supporting the microarray results.
Table III. Intron-spanning primers used for RT-qPCR.
| Gene | Forward Primer (5′→3′) | Reverse Primer (5′→ 3′) |
|---|---|---|
| CAV1 | ACCCACTCTTTGAAGCTGTTG | GAACTTGAAATTGGCACCAGG |
| FLNB | GCCTGTGGATAATGCACGAGA | GGC TCGATTCCTCTGCCATA |
| PDLIM1 | CCCAGCAGATAGACCTCCAG | TCTGAGCTTCCAAGTGTGTCATA |
| CDC42EP3 | TTTCTTCAAGGGAACTACGAGC | GAGGGCGTTTCTGTGAACAC |
| PIK3R1 | GCTTCTCTGACCCATTAACCTTC | CCTTTGGTATTTGGATACTGGA |
| GAPDH | ATGGGGAAGGTGAAGGTCG | GGGGTCATTGATGGCAACAATA |
| ITGB3 | GTGACCTGAAGGAGAATCTGC | TTCTTCGAATCATCTGGCC |
| SDHA | TGGGAACAAGAGGGCATCTG | CCACCACTGCATCAAATTCATG |
Fig. 3.
PCR Validation of microarray data. Real-time PCR of β3 integrin (ITGB3), PDLIM1, caveolin-1 (Cav1), filamin B, CDC42EP3 (Borg2), and PI-3 kinase regulatory domain (PIK3R1) in the presence or absence of DEX. Data was normalized to the housekeeping gene GAPDH or SDHA. The fold change shown is the average of 3–5 runs; bars indicate standard error.
These microarray analyses also revealed many of the major genes previously reported to be up- or down-regulated by DEX (Supplemental Table S3). Thus the gene array results obtained in this study appear to mirror those observed in other TM cell strains indicating that our results are not specific for these HTM cell strains. Some of the major genes found in this study that were commonly seen in other studies were angiopoietin-like 7 (ANGPTL7), α-1-antichymotrypsin (SERPINA3), myocilin (MYOC), mono-amine oxidase A (MAOA), apolipoprotein D (APOD), and chitinase-3-like (CH13L1). However, it should be noted that not all cytoskeleton genes previously determined to be up-regulated by DEX were detected in this study. This probably reflects the heterogeneity of the cellular response to DEX. Two cytoskeleton genes that we did not detect a significant change in but which had previously been shown (15) to be affected by DEX were γ2-actin (ACTG2) and α2-actin (ACTA2).
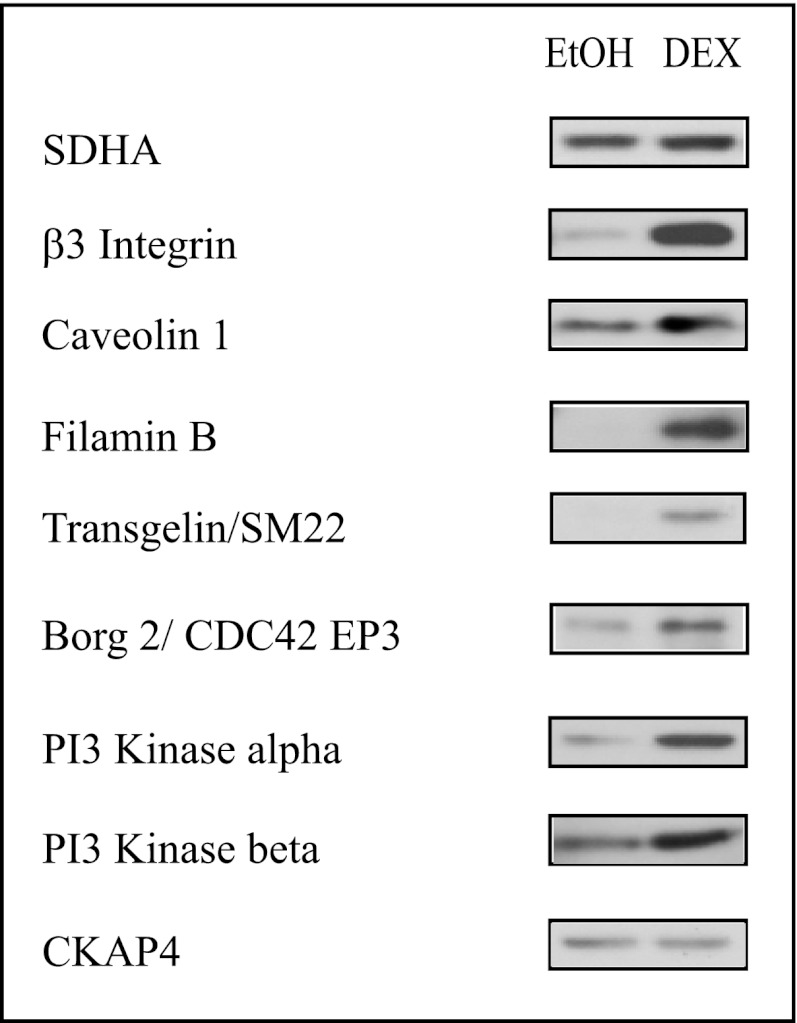
Western blots were then used to validate expression of the proteins detected by MS and predicted to be affected based on the gene arrays (Table II). As shown in Fig. 4, there is a corresponding increase in the levels of β3 integrin, filamin B, transgelin, and the catalytic subunit β of PI-3 kinase in the DEX-treated samples compared with the ethanol samples. We also found an increase in the level of expression of the PI-3 kinase regulatory subunit α, even though this subunit was not detected by MS. On the other hand, CKAP4 did not show any significant difference by Western blot analysis. Interestingly, the genes for leiomodin-1 and LRP16A did not show a >2 fold significant change in gene expression (data not shown). However, leiomodin-1, a member of the tropomodulin family, was up-regulated in the DEX-treated cells and LRP16A was only found in the DEX-treated samples by MS (Fig. 2). This suggests that DEX might be regulating expression of some proteins at the translational level and not at the transcriptional level.
Fig. 4.
Western blot analyses of proteins predicted to be up-regulated by DEX. Whole cell lysates were obtained from cultures treated with or without DEX for 6 days. All the proteins except CKAP4 showed increased expression in DEX-treated cultures. CKAP4 which did not show a statistically significant change (>twofold) at genomic level did not show a change by Western blot analysis. SDHA was used as a loading control.
MS analysis of the phospho-enriched fractions identified a total of 1253 unique phosphopeptides. Some of the phosphorylated peptides were found in the cytoskeletal proteins listed in Figs. 2B and 4. Among the proteins found to contain phosphorylated residues were CKAP4, Borg2, leiomodin-1, FGFR10P, LRP16A, and raftlin 1. Interestingly, some of the sites in raftlin 1 were only seen in DEX-treated samples suggesting that DEX was affecting the phosphorylation of this protein. Because leiomodin-1, LRP16A, and FGFR10P were not found in the control, it is not possible to say whether DEX was affecting their phosphorylation patterns. Surprisingly, caveolin-1 which was also found in the phospho-enriched fractions did not contain any phosphorylated peptides (data not shown). Similarly, none of the PDLIM1 peptides found in the phospho-enriched fractions contained any phosphorylated residues. It is not known why the peptides for PDLIM1 and caveolin-1 were found in that fraction, though the peptide identified from caveolin-1 may have bound to the titanium dioxide particles because of the presence of a di-aspartate in its sequence.
Although many of the phosphorylated residues identified in the proteins in Table IV have already been identified in other MS studies (www.phosphosite.org), some of the phosphorylated residues found for raftlin1 had not previously been identified. To determine if these phosphorylated residues could represent valid phosphorylation sites, we analyzed the sequences surrounding these novel phosphorylation sites using the PhosphoMotif database. Based on this database, all the phosphorylated sites in raftlin 1 contained sequence motifs that could be recognized by known serine/threonine kinases.
Table IV. Phosphorylation pattern of proteins.
Phosphorylated residues are underlined. Sequences in bold italics indicate that these phosphorylated residues were reported in other MS studies (database from www.phosphosite.org). Plain text indicates that these sites were not previous identified. For all the proteins, the phosphorylated peptides were found in at least two of the three biological replicates. The number in parenthesis indicates the number of peptides found.
| Name | EtOH | DEX |
|---|---|---|
| CKAP4 | 455LEGLGSSEADQDGLASTVR473 (n = 3) | 455LEGLGSSEADQDGLASTVR473 |
| 455LEGLGSSEADQDGLASTVR473 | ||
| CDC42EP3 | 87ANSTSDSVFTEPSPVLK104 | 87ANSTSDSVFTEPSPVLK104 (n = 3) |
| (Borg 2) | 105NAISLPTIGGsQALMLPPLLSPVTFNASK131 | 105NAISLPTIGGsQALMLPPLLSPVTFNASK131 |
| 105NAISLPTIGGsQALMLPPLLSPVTFNASK131 | ||
| 105NAISLPTIGGsQALMLPPLLSPVTFNASK131 | ||
| Leiomodin-1(LMOD1) | 10QVSEDPDIDSLLETL24 (n = 4) | |
| 10QVSEDPDIDSLLETL24 | ||
| FGFR1OP | 139EKGPTTGEGALDLSDVHSPPKSPEGK165 (n = 3) | |
| LRP 16A | 1080SERPPTILMTEEPSSPK1096 | |
| 1278TASRPDDIPDSPSSPK1293 (n = 2) | ||
| Raftlin, 1 | 195GDHASLENEKPGTGDVCSAPAGR217 | 195GDHASLENEKPGTGDVCSAPAGR217 |
| 218NQSPEPSSGPR228 | ||
| 236QPSSPSGEGDGGELSPQGVSK256 | ||
| 236QPSSPSGEGDGGELSPQGVSK256 |
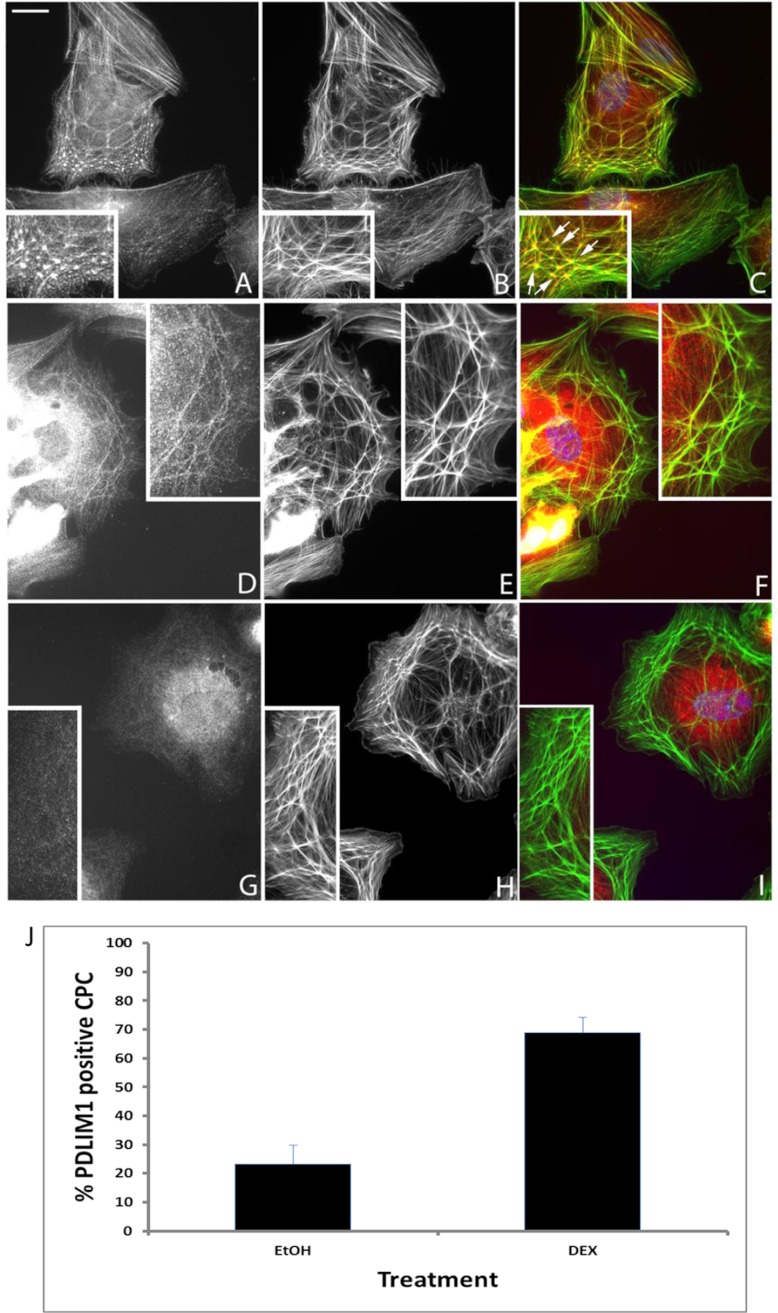
Finally, because both filamin B and PDLIM1 are involved in actin polymerization, we looked to see if DEX affected their expression or localization within CLANs. In both ethanol (data not shown) and DEX-treated cells, PDLIM1 was localized prominently within CLAN vertices as well as along the actin filaments making up the CLAN spokes (Fig. 5A–5C). Treatment with DEX not only significantly increased the percentage of cells expressing PDLIM1 from 25 ± 11% in the ethanol-treated controls to 70 ± 12% (p < 0.0001) in DEX-treated cells, but it also increased localization within CLANs. In the ethanol treatment group only 23% of CPCs contained PDLIM1 within CLANs whereas 69% (p < 0.0001) of CPCs in the DEX treatment group contained PDLIM1 within CLANs (Fig. 5J). Almost 75% of the ethanol-treated cells and 30% of the DEX-treated cells with CLANs, however, lacked detectable PDLIM1, suggesting that this protein is not necessary for CLAN formation.
Fig. 5.
Localization of PDLIM1 and filamin B in DEX-treated HTM cells. HTM cells treated with DEX for 6 days before being plated on fibronectin for 3 h were double-labeled with anti-PDLIM1 (A), anti-filamin B (D), rabbit IgG (G) antibodies and phalloidin (B, E, H). Merged images (C, F, I) are also shown. PDLIM1 colocalized with F-actin within CLANs and along stress fibers in both ethanol-treated (not shown) and DEX-treated cells, however, not all cells were positive for PDLIM1 (J). Although some CLANs did not contain filamin B, most CLANs ethanol contain filamin B regardless of the treatment. Insets show regions of cells with CLANs. Arrows point to five hubs or “vertisomes” of a CLAN that are interconnected by actin filaments; bar = 20 μm.
In contrast, localization of filamin B which was only occasionally found within CLANs (Fig. 5D) remained at 12–15% of CLAN-positive cells (not shown) in both ethanol- and DEX-treated cells. This lack of colocalization contrasted with filamin A whose localization completely mirrored F-actin labeling in earlier studies (5). Overall, DEX increased CLAN formation in TM cells as previously reported [13] from 3 ± 3% in ethanol-treated cells to 10 ± 5% in DEX-treated cells (p < 0.0001).
DISCUSSION
In this study, we showed that a combined genomic and proteomic approach can be used to rapidly identify the effects of DEX on the expression and activity of the actin cytoskeleton at the transcriptional, translational, and post-translational levels. In particular, this study has shown that at least 21 genes, nine proteins and four phosphoproteins known to affect the organization of the cytoskeleton were affected by DEX in HTM cells. The changes occurred at different stages along the pathways that regulate the formation of the actomyosin network and involve both cytoskeleton-binding proteins as well as signaling molecules in these regulatory pathways. The changes were not restricted to actomyosin filaments as the DEX treatment also affected other cytoskeleton networks such as microtubules and intermediate filaments. This indicates that the entire cytoskeleton architecture of the TM is likely to be affected by DEX.
Surprisingly, only a relatively small number of cytoskeleton proteins, less than 5% of the total cytoskeleton proteins found by MS, are affected at the transcriptional level by DEX and most of them only showed small changes in expression. Of the proteins observed in the DEX-treated cells, 17 of these proteins had been shown to have a greater than twofold increase at the gene level in this and other studies (14, 15, 17, 42). Two of these proteins, Borg2 and PDLIM1 have not previously been reported to be up-regulated at either the gene or protein level in TM cells. The significance of Borg2 in CLAN formation and glaucoma remains to be determined. Borg2 (also called cdc42 effector protein 3) is involved in Rho family GTPase signaling pathways, which regulate the actin cytoskeleton.
PDLIM1, on the other hand, appears to be a component of CLANs. These studies are the first to show that PDLIM1 can be found localized within the vertices of CLANs. Previous studies have suggested that the vertices consist of a molecular complex (the “vertisome”) of α-actinin, syndecan-4 and phosphatidylinositol 4,5-bisphosphate (PIP2) that may regulate CLAN formation (5). Hence, as a component of CLAN vertices, PDLIM1 could participate in regulating CLAN formation. PDLIM1 is a known cytoskeletal adaptor protein that interacts with actin filaments (43) and can recruit the kinase CLIK1 to actin stress fibers as well as bind α-actinin (44–46). It also contains a PDZ domain which would enable it to interact with syndecan-4 (47, 48) another component found within vertices of CLANs (5). Although the role of PDLIM1 in CLAN formation in DEX-treated cells can only be speculated on, it would appear to be a gain of function as CLAN formation can occur in the absence of PDLIM1. One possible role is that it serves to inhibit disassembly of the CLAN and thus stabilize the structure.
Not all of the proteins that were up- or down-regulated by DEX at the genomic level were detected by the MS analysis, despite the enrichment procedure. Presumably, this is because the proteins were either too low in abundance to be detected or were not digestible by trypsin and hence could not be detected by MS. Those that were detected appeared to show modest changes in the translational and post-translational level with the exception of the proteins observed only in either DEX or control cell lysates. This suggests that changes in the cytoskeleton and contractile properties of the TM following DEX treatment are just as likely to be caused by changes in the activity of the actin cytoskeleton as expression levels.
Out of all of the proteins observed only in the DEX-treated cells, two proteins (LRP16A and leiomodin-1) could have a significant impact on the contractile properties of the TM. Both LRP16A and leiomodin-1 promote actin polymerization, which has been shown to play a role in regulating IOP (49). Leiomodin-1 is a member of the tropomodulin family of proteins which modulate the binding of tropomyosin to actin filaments and promotes the capping of the slow growing end of actin filaments (50). It is known to be highly expressed in eye muscle as well as skeletal muscle. One of its functions is to bind the capping protein CAPZA2 thereby preventing the uncapping of actin barbed ends (51). Neither of these proteins showed any significant changes at the gene level in this study or in any other reported microarray studies. Thus, DEX appears to be affecting the translation of these proteins and significant changes in their activity on the actin cytoskeleton following DEX would not be detected by microarray analysis alone.
The prevalence of CLANs in both glaucomatous and steroid-treated anterior segments and HTM cell cultures suggests a role for this actin structure in increased IOP. Interestingly, several of the proteins affected by DEX in this study are either found within CLANs or could participate in the formation of them. One of the major proteins affected at both the transcriptional and translational level is filamin B. In addition to being up-regulated at the total protein level, filamin B also showed a phosphorylation pattern following DEX treatment (Table II). Filamins are a family of actin-binding proteins that can cross-link actin filaments. This study showed that filamin B, like filamin A (5), can also colocalize with actin in CLANs. In addition to binding actin filaments, filamins can also bind to the cytoplasmic tail of β3 integrin and modulate signals from them (52). These properties potentially make filamin B an important modulator of CLAN formation.
Two other components found to be up-regulated were the β3 integrin subunit and PI-3 kinase. It has been previously shown that activation of β3 integrin can trigger CLAN formation in nonglaucomatous cells (4, 5), and that this protein is up-regulated and activated on the cell surface following DEX treatment (13). Previous work has also shown that treatment with the PI-3 kinase inhibitor LY294002 significantly decreased CLAN formation (4). Thus, up-regulation of these proteins could increase CLAN formation.
The use of a TiO2 column to enrich for phosphoproteins not only helped us screen and identify proteins that are lower abundance in TM cells, but these phosphoproteomic studies also showed that DEX can affect the phosphorylation state of proteins. All the phosphorylation sites identified in these proteins match known consensus sequences for either serine or threonine phosphorylation by a number of kinases including PKA, PKC, calmodulin-dependent protein kinase II, MAPKs, casein kinase I, and casein kinase II (PhosphoMotif database). Although the effects of these phosphorylation patterns on the proteins remains to be determined, they could have far-reaching effects on signaling pathways and assembly of the actomyosin network as phosphorylation events are known to regulate the activity of proteins.
In summary, the MS study allowed us to rapidly screen the TM proteome and showed that the effects of DEX treatment on the actomyosin cytoskeleton are most likely the result of a complex assortment of changes at the transcriptional, translational and post-translational levels. Analyzing which types of families of proteins are affected should provide new insights into the effect of DEX on the TM.
Supplementary Material
Footnotes
* This work was funded by the National Center for Research Resources, the American Recovery Reinvestment Act and NEI-NIH grants EY017006, EY0020490 (D.M.P.), and a Core grant to the Department of Ophthalmology and Visual Sciences (P30 EY016665).
 This article contains supplemental Figs. S1 and S2 and Tables S1 to S5.
This article contains supplemental Figs. S1 and S2 and Tables S1 to S5.
1 The abbreviations used are:
- DEX
- dexamethasone
- TM
- trabecular meshwork
- LC/MS/MS
- liquid chromatography-tandem MS
- ECM
- extracellular matrix
- CLANs
- cross-linked actin networks.
REFERENCES
- 1. Clark A. F., Wordinger R. J. (2009) The role of steroids in outflow resistance. Exp. Eye Res. 88, 752–759 [DOI] [PubMed] [Google Scholar]
- 2. Zhang X., Clark A. F., Yorio T. (2005) Regulation of glucocorticoid responsiveness in glaucomatous trabecular meshwork cells by glucocorticoid receptor-β Invest. Ophthal. Vis. Sci. 46, 4607–4616 [DOI] [PubMed] [Google Scholar]
- 3. Dickerson J. E., Jr., Steely H. T., Jr., English-Wright S. L., Clark A. F. (1998) The effect of dexamethasone on integrin and laminin expression in cultured human trabecular meshwork cells. Exp Eye Res. 66, 731–738 [DOI] [PubMed] [Google Scholar]
- 4. Filla M. S., Schwinn M. K., Sheibani N., Kaufman P. L., Peters D. M. (2009) Regulation of cross-linked actin network (CLAN) formation in human trabecular meshwork (HTM) cells by convergence of distinct β1 and β3 integrin pathways. Invest. Ophthal. Vis. Sci. 50, 5723–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Filla M. S., Woods A., Kaufman P. L., Peters D. M. (2006) β1 and β3 integrins cooperate to induce syndecan-4 containing cross-linked actin networks (CLANs) in human trabecular meshwork (HTM) cells. Invest. Ophthal. Vis. Sci. 47, 1956–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sabanay I., Tian B., Gabelt B. T., Geiger B., Kaufman P. L. (2006) Latrunculin B effects on trabecular meshwork and corneal endothelium structure in the monkey eye. Exp Eye Res. 82, 236–246 [DOI] [PubMed] [Google Scholar]
- 7. Sabanay I., Tian B., Gabelt B. T., Geiger B., Kaufman P. L. (2004) Functional and structural reversibility of H-7 effects on the conventional aqueous outflow pathway in monkeys. Exp. Eye Res. 78, 137–150 [DOI] [PubMed] [Google Scholar]
- 8. Peterson J. A., Tian B., Geiger B., Kaufman P. L. (2000) Effect of latrunculin-B on outflow facility in monkeys. Exp. Eye Res. 70, 307–313 [DOI] [PubMed] [Google Scholar]
- 9. Borras T., Rowlette L. L., Erzurum S. C., Epstein D. L. (1999) Adenoviral reporter gene transfer to the human trabecular meshwork does not alter aqueous humor outflow. Relevance for potential gene therapy of glaucoma. Gene Ther. 6, 515–524 [DOI] [PubMed] [Google Scholar]
- 10. Rao P. V., Deng P., Maddala R., Epstein D. L., Li C. Y. (2005) Expression of dominant negative Rho-binding domain of rho-Kinase in organ cultured human eye anterior segments increase aqueous humor outflow. Mol. Vis. 11, 288–297 [PubMed] [Google Scholar]
- 11. Rao P. V., Deng P. F., Kumar J., Epstein D. L. (2001) Modulation of aqueous humor outflow facility by the Rho kinase-specific inhibitor Y-27632. Invest. Ophthal. Vis. Sci. 42, 1029–1037 [PubMed] [Google Scholar]
- 12. Zhang X., Clark A. F., Yorio T. (2008) FK506-Binding Protein 51 Regulates Nuclear Transport of the Glucocorticoid Receptor β and Glucocorticoid Responsiveness. Invest. Ophthal. Vis. Sci. 49, 1037–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Filla M. S., Schwinn M. K., Nosie A. K., Clark R. W., Peters D. M. (2011) Dexamethasone-associated cross-linked actin network (CLAN) formation in human trabecular meshwork (HTM) cells involves b3 integrin signaling. Invest. Ophthal. Vis. Sci. 52, 2952–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fan B. J., Wang D. Y., Tham C. C., Lam D. S., Pang C. P. (2008) Gene Expression profiles of human trabecular meshwork cells induced by triamcinolone and dexamethasone. Invest. Ophthal. Vis. Sci. 49, 1886–1897 [DOI] [PubMed] [Google Scholar]
- 15. Rozsa F. W., Reed D. M., Scott K. M., Pawar H., Moroi S. E., Kijek T. G., Krafchak C. M., Othman M. I., Vollrath D., Elner V. M., Richards. J. E. (2006) Gene expression profile of human trabecular meshwork cells in response to long-term dexamethasone exposure. Mol. Vis. 12, 125–141 [PubMed] [Google Scholar]
- 16. Lo W. R., Rowlette L. L., Caballero M., Yang P., Hernandez M. R., Borras T. (2003) Tissue differential microarray analysis of dexamethasone induction reveals potential mechanisms of steroid glaucoma. Invest. Ophthal. Vis. Sci. 44, 473–485 [DOI] [PubMed] [Google Scholar]
- 17. Ishibashi T., Takagi Y., Mori K., Naruse S., Nishino H., Yue B. Y., Kinoshita S. (2002) cDNA microarray analysis of gene expression changes induced by dexamethasone in cultured human trabecular meshwork cells. Invest. Ophthal. Vis. Sci. 43, 3691–3697 [PubMed] [Google Scholar]
- 18. Leung Y. F., Tam P. O., Lee W. S., Lam D. S., Yam H. F., Fan B. J., Tham C. C., Chua J. K., Pang C. P. (2003) The dual role of dexamethasone on anti-inflammation and outflow resistance demonstrated in cultured human trabecular meshwork cells. Mol. Vis. 9, 425–439 [PubMed] [Google Scholar]
- 19. Wu C. W., Sauter J. L., Johnson P. K., Chen C. D., Olsen T. W. (2004) Identification and localization of major soluble vitreous proteins in human ocular tissue. Am. J. Ophthal. 137, 655–661 [DOI] [PubMed] [Google Scholar]
- 20. Steely H. T., Dillow G. W., Bian L., Grundstad J., Braun T. A., Casavant T. L., McCartney M. D., Clark A. F. (2006) Protein expression in a transformed trabecular meshwork cell line: proteome analysis. Mol. Vis. 12, 372–383 [PubMed] [Google Scholar]
- 21. Yu M., Sun L., Peng W., Chen Z., Lin X., Liu X., Li M., Wu K. (2010) Protein expression in human trabecular meshwork: downregulation of RhoGDI by dexamethasone in vitro. Mol. Vis. 16, 213–223 [PMC free article] [PubMed] [Google Scholar]
- 22. Picciani R., Junk A. K., Bhattacharya. S. K. (2008) Technical Brief: a novel strategy for enrichment of trabecular meshwork protease proteome. Mol. Vis. 14, 871–877 [PMC free article] [PubMed] [Google Scholar]
- 23. Finger J. H. (2011) Primary open-angle glaucoma genes. Eye 25, 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Filla M. S., David G., Weinreb R. N., Kaufman P. L., Peters D. M. (2004) Distribution of syndecans 1–4 within the anterior segment of the human eye: expression of a variant syndecan-3 and matrix-associated syndecan-2. Exp Eye Res. 79, 61–74 [DOI] [PubMed] [Google Scholar]
- 25. Polansky J. R., Weinreb R., Alvarado J. A. (1981) Studies on human trabecular cells propagated in vitro. Vis. Res. 21, 155–160 [DOI] [PubMed] [Google Scholar]
- 26. Polansky J. R., Weinreb R. N., Baxter J. D., Alvarado J. (1979) Human trabecular cells. I. Establishment in tissue culture and growth characteristics. Invest. Ophthal. Vis. Sci. 18, 1043–1049 [PubMed] [Google Scholar]
- 27. Lund R., Leth-Larsen R., Jensen O. N., Ditzel H. J. (2009) Efficient isolation and quantitative proteomic analysis of cancer cell plasma membrane proteins for identification of metastasis-associated cell surface markers. J. Proteome Res. 8, 3078–3090 [DOI] [PubMed] [Google Scholar]
- 28. Kline K. G., Barrett-Wilt G. A., Sussman. M. R. (2010) In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. USA. 107, 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sugiyama N., Masuda T., Shinoda K., Nakamura A., Tomita M., Ishihama Y. (2007) Phosphopeptide enrichment by aliphatic acid modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell. Proteomics 6, 1103–1109 [DOI] [PubMed] [Google Scholar]
- 30. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, prefractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 31. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann. M., (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 32. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 33. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amanchy R., Kandasamy K., Mathivanan S., Periaswamy B., Reddy R., Yoon W. H., Joore J., Beer M. A., Cope L., Pandey A. (2011) Identification of novel phosphorylation motifs through an integrative computational and experimental analysis of the human phosphoproteome. J. Proteomics Bioinform. 4, 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peterson J. A., Sheibani N., David G., Garcia-Pardo A., Peters D. M. (2005) Heparin II domain of fibronectin uses α4β1 integrin to control focal adhesion and stress fiber formation, independent of syndecan-4. J Biol Chem. 280, 6915–6922 [DOI] [PubMed] [Google Scholar]
- 36. Ezzat M. K., Howell K. G., Bahler C. K., Beito T. G., Loewen N., Poeschla E. M., Fautsch M. P. (2008) Characterization of monoclonal antibodies against the glaucoma associated protein myocilin. Exp Eye Res. 87, 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 38. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clark A. F., Wilson K., McCartney M. D., Miggans S. T., Kunkle M., Howe W. (1994) Glucocorticoid-induced formation of cross-linked actin networks in cultured human trabecular meshwork cells. Invest. Ophthal. Vis. Sci. 35, 281–294 [PubMed] [Google Scholar]
- 40. Clark A. F., Brotchie D., Read A. T., Hellberg P., English-Wright S., Pang I. H., Ethier C. R., Grierson I. (2005) Dexamethasone alters F-actin architecture and promotes cross-linked actin network formation in human trabecular meshwork tissue. Cell Motility Cytoskeleton 60, 83–95 [DOI] [PubMed] [Google Scholar]
- 41. Nguyen T. D., Chen P., Huang W. D., Chen H., Johnson D., Polansky J. R. (1998) Gene structure and properties of TIGR, an olfactomedin-related glycoprotein cloned from glucocorticoid-induced trabecular meshwork cells. J Biol Chem. 273, 6341–6350 [DOI] [PubMed] [Google Scholar]
- 42. Lo W. R., Rowlette L. L., Caballero M., Yang P., Hernandez M. R., Borras T. (2003) Tissue differential microarray analysis of dexamethasone induction reveals potential mechanisms of steriod glaucoma. Invest. Ophthal. Vis. Sci. 44, 473–485 [DOI] [PubMed] [Google Scholar]
- 43. te Velthuis A.J.W., Bagowski C.P. (2007) PDZ and LIM Domain-Encoding Genes: Molecular Interactions and their Role in Development. Sci. World J. 7, 1470–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vallenius T., Luukko K., Mäkelä T. P. (2000) CLP-36 PDZ-LIM protein associates with nonmuscle alpha-actinin-1 and alpha-actinin-4. J. Biol. Chem. 275, 11100–11105 [DOI] [PubMed] [Google Scholar]
- 45. Vallenius T., Makela T.P. (2002) Clik1: a novel kinase targeted to actin stress fibers by the CLP-36 PDZ-LIM protein. J. Cell Sci. 115, 2067–2073 [DOI] [PubMed] [Google Scholar]
- 46. Bauer K., Kratzer M., Otte M., de Quintana K. L., Hagmann J., Arnold G. L., Eckerskorn C., Lottspeich F., Siess W. (2000) Human CLP36, a PDZ-domain and LIM-domain protein, binds to alpha-actinin-1 and associates with actin filaments and stress fibers in activated platelets and endothelial. Blood 96, 4236–4245 [PubMed] [Google Scholar]
- 47. Gao Y., Li M., Chen W., Simons M. (2000) Synectin, syndecan-4 cytoplasmic domain binding PDZ protein, inhibits cell migration. J. Cell Physiol. 184, 373–379 [DOI] [PubMed] [Google Scholar]
- 48. Zimmerman P., Tomatis D., Rosas M., Grootjans J., Leenaerts I., Degeest G., Reekmans G., Coomans C., David G. (2001) Characterization of syntenin, a syndecan-binding PDZ protein, as a component of cell adhesion sites and microfilaments. Mol. Biol. Cell. 13, 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tian B., Geiger B., Epstein D. L., Kaufman P. L. (2000) Cytoskeletal involvement in the regulation of aqueous humor outflow. Invest. Ophthal. Vis. Sci. 41, 619–623 [PubMed] [Google Scholar]
- 50. Conley C. A., Fritz-Six K. L., Almenar-Queralt A., Fowler V. M. (2001) Leiomodins:larger members of the tropomodulin (Tmod) gene family. Genomics 73, 127–139 [DOI] [PubMed] [Google Scholar]
- 51. Liang Y., Niederstrasser H., Edwards M., Jackson C. E., Cooper J. A. (2009) Distinct roles for CARMIL isoforms in cell migration. Mol. Biol. Cell 20, 5290–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim C., Ye F., Ginsberg M. H. (2011) Regulation of Integrin Activation. Annu. Rev. Cell Dev. Biol. 27, 321–345 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.