Abstract
The study of metabolically labeled or probe-modified proteins is an important area in chemical proteomics. Isolation and purification of the protein targets is a necessary step before MS identification. The biotin-streptavidin system is widely used in this process, but the harsh denaturing conditions also release natively biotinylated proteins and non-selectively bound proteins. A cleavable linker strategy is a promising approach for solving this problem. Though several cleavable linkers have been developed and tested, an efficient, easily synthesized, and inexpensive cleavable linker is a desirable addition to the proteomics toolbox. Here, we describe the chemical proteomics application of a vicinal diol cleavable linker. Through easy-to-handle chemistry we incorporate this linker into an activity-based probe and a biotin alkyne tag amenable for bioorthogonal ligation. With these reagents, background protein identifications are significantly reduced relative to standard on-bead digestion.
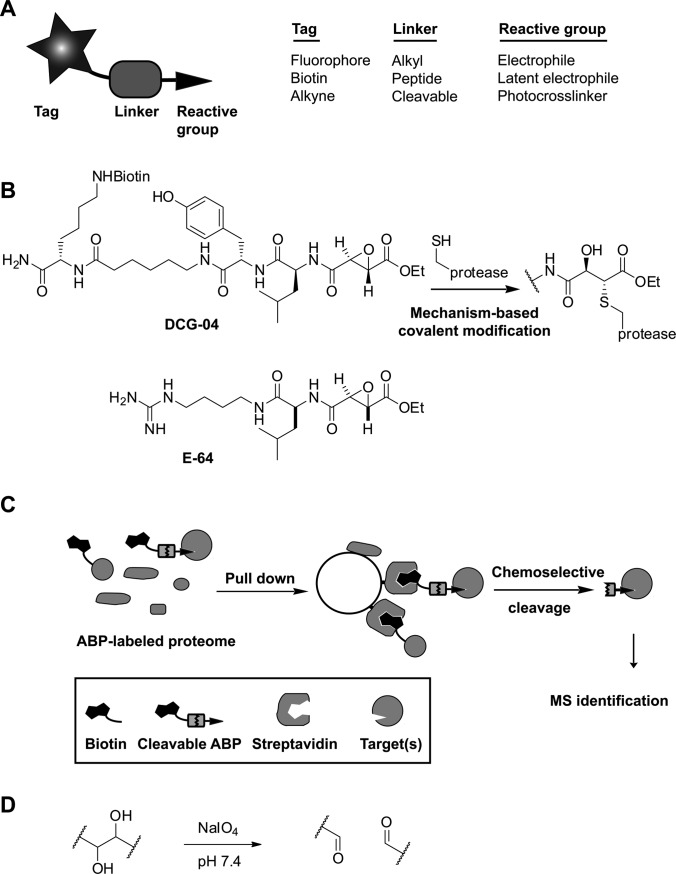
The covalent modification of proteins by small molecules within a complex proteome is a major theme in chemical biology and proteomics. An effective method for the detection of posttranslational modifications of proteins is the metabolic incorporation of modified biomolecules such as tagged carbohydrates or lipids (1). Reversible interactions of enzyme inhibitors, natural products, or drugs can be detected when one appends photocrosslinking agents, thereby facilitating target discovery (2, 3). A particularly interesting example of protein labeling is activity-based protein profiling (ABPP)1 (4, 5), which utilizes the intrinsic catalytic activity of a target enzyme for the covalent attachment of an affinity or visualization tag. ABPP makes use of small molecules (activity-based probes (ABPs)) that react with the active form of a specific enzyme or enzyme class by means of a “warhead,” which is often derived from a mechanism-based enzyme inhibitor (Fig. 1A). DCG-04, for example, is based on the naturally occurring inhibitor E-64 and targets the papain family of cysteine proteases via covalent attachment of the epoxysuccinate group to the active site cysteine (Fig. 1B) (6).
Fig. 1.
The cleavable linker strategy in ABPP. A, the elements of an ABP. B, the example ABP DCG-04, an epoxysuccinate-containing probe for clan CA cysteine proteases. DCG-04 is based on the naturally occurring protease inhibitor E-64. C, schematic strategy of cleavable linker-mediated target identification. D, the cleavage mechanism of a vicinal diol.
Bulky fluorophore or biotin tags on chemical probes might interfere with efficient protein binding. Moreover, they can negatively influence the cell permeability of probes, which therefore limits their applicability in in vitro experiments. Bioorthogonal chemistries, such as the Bertozzi-Staudinger ligation (7) and the 1,3-bipolar cycloaddition of an azide and an alkyne (click chemistry) (8), allow tandem labeling strategies in which a biotin or a fluorophore is attached to an enzyme probe complex in a separate step. Consequently, the probes themselves only carry azide or alkyne groups as “mini-tags.” Tandem labeling using bioorthogonal chemistry has now become a widely used strategy to label biomolecules in lysates and in live cells (9–11).
An essential step in ABPP, as well as in other chemical proteomics approaches, is the elucidation of the tagged proteins. This usually involves a biotin-mediated enrichment step followed by mass-spectrometry-based identification. Although the streptavidin-biotin interaction allows efficient enrichment as a result of the strong binding affinity (Kd ∼ 10−15 m), it also has limitations. The quantitative elution of biotinylated proteins requires harsh conditions (12), which lead to contamination of the sample by endogenous biotinylated and non-specifically bound proteins. These other proteins will be identified together with the real protein targets. Given that subsequent target validation with secondary assays can be a costly and time-consuming process, a reduction in false positive identifications is highly desirable. For cleaner protein identification, cleavable linker strategies (13) that allow the selective release of target proteins have been developed (Fig. 1C). The commercially available disulfide linker can be cleaved under mild conditions, but it suffers from premature cleavage in reducing media such as the intracellular environment and reducing buffers used for click chemistry and in vitro reactions of cysteine proteases. Therefore, a variety of alternative linkers for proteomics applications have been reported, including a sterically hindered disulfide (14), diazobenzenes (15–19), hydrazones (20, 21), silanes (22), light sensitive linkers (23–25), tobacco etch virus protease sensitive linkers (26, 27), and a levulinoyl-based linker (28). The synthesis of some of these linkers is lengthy or difficult to scale up, which limits their general application in chemical proteomics.
Ideally, a cleavable linker is stable under a wide variety of conditions, is efficiently and selectively cleaved, and can be synthesized in a low number of easy chemical transformations. We aimed to meet these requirements by using a vicinal diol as a cleavable linker system. When vicinal diols are treated with sodium periodate (NaIO4), the carbon–carbon bond is cleaved (Fig. 1D). Periodate treatment of proteins can result in side-reactions, such as the cleavage of linked carbohydrates or the oxidation of N-terminal serine and threonine residues. However, these N-termini rarely occur in proteins and are therefore of minor concern. In general, the mild, neutral conditions of periodate cleavage are compatible with proteins. This has been illustrated in the past, for example, by its application in the detection of protein–protein interactions (29) and the creation of unliganded MHC class I molecules (30). In this article, we report the chemical proteomics application of diol cleavable linker probes. We show that the synthesis of the linker and its probe derivatives is straightforward, that the linker is compatible with tandem click labeling, that enrichment and release of probe targets is efficient, and that the identification of targets takes place with significantly lower background than in on-bead digestion protocols.
EXPERIMENTAL PROCEDURES
Synthesis of Cleavable Linker Building Block
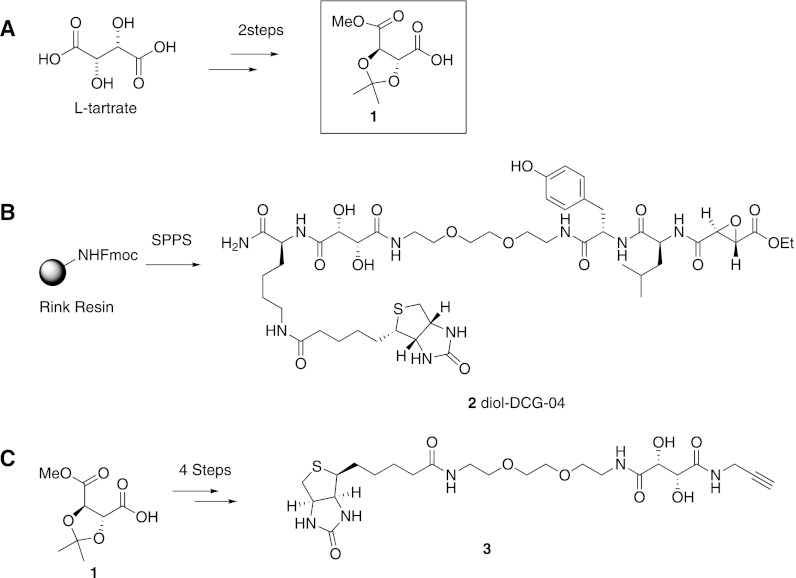
Building block 1 was synthesized from l-tartaric acid. First, methyl and isopropylidene protecting groups were introduced via reaction with methanol, 2,2-dimethoxypropane, and a catalytic amount of p-toluenesulfonic acid at elevated temperature. The fully protected tartrate was purified by means of vacuum distillation and obtained as a light yellow oil in 74% yield. Next, one methyl ester was saponified through reaction with one equivalent of potassium hydroxide. Building block 1 was isolated via extraction as a colorless oil in 68% yield.
Synthesis of Diol-DCG-04
Elongation of Rink resin with Fmoc-ε-biotinyl-lysine and cleavable building block 1 under the influence of diisopropylcarbodiimide/hydroxybenzotriazole was followed by treatment with neat 1,8-diamino-3,6-dioxaoctane, which replaced the methyl ester of the tartrate under formation of an amide bond. Further elongation was achieved with Fmoc-tyrosine, Fmoc-leucine, and ethyl (2S, 3S)-epoxysuccinate (6). Diol-DCG-04 (2) was cleaved from the resin and purified via HPLC.
Synthesis of Alkyne-biotin Reagent 3
Building block 1 was coupled to propargylamine with 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU)/N,N-diisopropyl-N-ethylamine (DIEA). After extraction and silica column purification, the product was reacted with an excess of 1,8-diamino-3,6-dioxaoctane in toluene at 80 °C. The resulting compound was purified (silica column chromatography) and coupled to biotin under the influence of HBTU and DIEA in dimethylformamide. The solvent was evaporated and the residue heated in 90% acetic acid for deprotection of the isopropylidene group. The solvent was removed, and the final product was purified via HPLC.
Synthetic Details and Compound Characterization
The detailed synthetic procedures of the linker and probes and their characterization are provided in the supplementary material.
Lysates and Cultures
For the rat liver homogenate, a piece of rat liver was cut into small chunks and combined with lysis buffer (50 mm acetate pH 5.5, 5 mm MgCl2, 250 mm sucrose, and 2 mm DTT). The liver/buffer mixture was dounced 15× on ice. After douncing, the mixture was centrifuged (2500 rcf, 4 °C for 10 min) to remove unlysed tissue and then centrifuged (19,000 rcf, 4 °C, for 20 min) to remove cell debris. The supernatant was collected, snap-frozen in liquid nitrogen, and stored at −80 °C. The protein concentration was determined via the Bradford or DC protein assay (Bio-Rad).
RAW 264.7 cells were cultured in DMEM containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin and maintained in a humidified 37 °C incubator with 5% CO2. For in situ labeling, cells were treated with probes with a final DMSO concentration of 0.1%. To generate lysates, cells were washed twice with cold phosphate-buffered saline (PBS), harvested with trypsin or via the use of a cell scraper, and collected by means of centrifugation. Cell pellets were then washed with PBS and lysed with 0.1% triton-X 100 in sodium phosphate buffer (100 mm sodium phosphate, pH 7.4) or 0.1% Triton-X 100 in sodium acetate buffer (pH 5.5, 2 mm DTT, 50 mm NaOAc, and 5 mm MgCl2) at 4 °C for 0.5 h. The mixture was centrifuged (19,000 rcf, 4 °C, for 20 min) to remove unlysed cells and cell debris. The supernatant was snap-frozen in liquid nitrogen and stored at −80 °C. Protein concentration was determined via Bradford or DC protein assay (Bio-Rad).
Detection of Capture and Release
Rat-liver homogenates (1 mg/ml total protein) were incubated with diol-DCG-04 or DCG-04 (1 μm) for 0.5 h. As an input sample, 50 μl labeled lysate was taken out and treated with 16 μl 4 X SB. The free probes in the remaining lysate were removed by Zeba spin desalting columns (Thermo Scientific). The eluate (in PBS buffer) was shaken with 10 μl Ultralink streptavidin (Thermo Scientific) for 2h and centrifuged to collect the supernatant. The beads were washed with PBS (five times) and divided into two aliquots. One was treated with 1 X SB (5 min at 90 °C). The other was treated with 1 mm NaIO4 (50 μl) in sodium phosphate buffer (100 mm sodium phosphate buffer, pH 7.4) in the dark for 1 h, and the supernatant was collected. Then the beads were washed with 0.1% SDS (three times) and PBS (three times). 1X SB buffer (66 μl) was added and heated at 90 °C for 5 min. 30 μl of each sample was loaded onto a 15% SDS gel, and proteins were visualized via streptavidin Western blot.
For the detection of cleavage by silver stain, rat liver homogenate (13.5 mg of total protein) in sodium acetate buffer (2.5 ml; pH 5.5) was incubated with diol-DCG-04 (10 μm) for 2 h. Unreacted probe was removed via filtration over a PD10 column (GE Healthcare), and the eluate (in PBS buffer) was shaken with Ultralink streptavidin slurry (100 μl) for 1 h. The streptavidin beads were subsequently washed with PBS buffer containing 1 m NaCl, 0.1% SDS, and 10% EtOH (three times each). Finally the beads were washed with PBS buffer three times. Proteases labeled by the probe were released from the beads either by direct boiling with 1 X SB (105 μl) or by 10 mm NaIO4 in sodium phosphate buffer (40 μl; 2 × 40 min) in the dark. To the cleavage supernatant was added 4 X SB (25 μl). After the chemical release, the beads were washed with 1 m NaCl, 0.1% SDS, 10% EtOH, and PBS (three times). After washing, the beads were treated with 1 X SB (105 μl) and boiled at 90 °C to determine the cleavage efficiency. 40 μl of each sample was loaded on a 15% SDS gel, and all the proteins were visualized by means of silver staining. In order to check the nature of the cleavage (i.e. specific chemical cleavage or unspecific release by denaturation), the samples were diluted 100× with 1 X SB, and 30 μl was loaded onto gel for streptavidin Western blot.
Click Chemistry Mediated Labeling
To an azido-E-64 (4) labeled proteome in sodium phosphate buffer, pH 7.4, biotin-diol-alkyne (20–50 μm), tris-(benzyltriazolylmethyl)amine (50 μm), tris-(2-carboxyethyl)phosphine (1 mm), and CuSO4 (1 mm) were added, and the reaction mixture was incubated at room temperature for 30 to 60 min. The reaction was stopped either by adding 4 X SB or by running a PD-10 column (GE Healthcare).
On-bead Digestion or Chemical Release for Diol-DCG-04
Rat liver homogenate (3 mg) was incubated with diol-DCG-04 (10 μm) for 2 h. Unreacted probe was removed via filtration over a PD10 column (GE Healthcare), and the eluate (in PBS buffer) was shaken with Ultralink streptavidin slurry (30 μl) for 4 h. Beads were separated from the unbound fraction via centrifugation. The beads were sequentially washed with a series of buffers containing 1% SDS in PBS, 4 m urea in H2O, 1 m NaCl in PBS, and 10% EtOH in PBS, and finally the beads were washed with PBS buffer three times. After washing, beads were used for either on-bead digestion or chemical elution. For on-bead digestion, streptavidin beads with bound proteins were resuspended in 100 μl of denaturating buffer (50 mm ammonium hydrogen carbonate, 6 m urea). Bound proteins were reduced in the presence of 10 mm DTT for 1 h. Samples were alkylated by the addition of 200 mm iodoacetamide (20 μl) and incubated for 1 h in the dark. Unreacted iodoacetamide was neutralized by the addition of 200 mm DTT (20 μl). The urea concentration was reduced by the addition of distilled H2O (800 μl). Samples were incubated with trypsin overnight at 37 °C and purified on a Vivapure C18 spin column (Sartorius, Goettingen, Germany). Chemical elution was performed via the addition of 50 μl elution buffer (10 mm sodium periodate in 100 mm sodium phosphate buffer, pH 7.4) for 30 min in the dark. The cleavage buffer was removed and replaced with fresh solution. The two eluted supernatants were combined and desalted with a Zeba spin desalting column. To the eluate (100 μl; 100 mm ammonium hydrogen carbonate) was added 12 m urea (100 μl), and the sample was prepared for LC-MS/MS analysis as described for the on-bead digestion.
On-bead Digestion or Chemical Release after Tandem Labeling
For in vitro proteome labeling, the probe azide-E64 (4, final concentration of 10 μm) was added to RAW 264.7 cell lysate (3.2 mg in 2.5 ml buffer; pH 5.5, 50 mm NaOAc, 2 mm DTT, and 5 mm MgCl2) for 1 h. Unreacted probe was removed via filtration over a PD10 column (GE Healthcare), and the eluate (in sodium phosphate buffer, pH 7.4) was adjusted to 5 ml total volume with sodium phosphate buffer. After click chemistry, unreacted probe was removed via filtration over a PD10 column (GE Healthcare), and the eluate (in PBS buffer) was shaken with Ultralink streptavidin slurry (50 uL) for 4 h. On-bead digestion and chemical release followed by digestion were carried out according to the protocol described above. For in situ proteome labeling, RAW 264.7 cells were labeled in DMEM (six-well plate, 1 ml for each well) with azide-E64 8 (5 μm) for 1 h, harvested, and washed with PBS, and a lysate was made using a 0.1% triton-X100-containing sodium phosphate buffer (pH 7.4). 4.1 mg labeled protein was used for the comparison of on-bead digestion and chemical release following the protocol for in vitro proteome labeling.
Protein Identification
Nanoflow LC-MS/MS of tryptic peptides was performed by coupling an Eksigent nanoLC-Ultra 1D+ (Eksigent, Dublin, CA) to an LTQ Orbitrap Velos (Thermo Scientific, Bremen, Germany). Tryptic peptides were eluted from the Vivapure spin columns with 50/50 water/acetonitrile (ACN) (20 μl 0.1% formic acid (FA)), and 10 μl were injected for each analysis. Peptides were delivered to a trap column (100 μm inner diameter × 2 cm, packed with 5 μm C18 resin, Reprosil PUR AQ, Dr. Maisch, Ammerbuch, Germany) at a flow rate of 5 μl/min in 100% buffer A (0.1% (FA) in HPLC grade water). After 10 min of loading and washing, peptides were transferred to an analytical column (75 μm× 40 cm C18 column, Reprosil PUR AQ, 3 μm, Dr. Maisch, Ammerbuch, Germany) and separated using a 110 min gradient from 7% to 35% of buffer B (0.1% FA in ACN) at a flow rate of 300 nl/min. The mass spectrometer was operated in data dependent mode, automatically switching between MS and MS2. Full scan MS spectra were acquired in the Orbitrap at 30,000 resolution, and tandem MS spectra were acquired at 7500 resolution. Internal calibration was performed using the ion signal (Si(CH3)2O)6H+ at m/z 445.120025 present in ambient laboratory air. Tandem mass spectra were generated for up to ten peptide precursors using higher energy collision dissociation (31). Precursors were dynamically excluded from fragmentation for 20 s, and unassigned charge states and singly charged ions were rejected.
Data Processing and Analysis
Peak picking and processing of raw MS data were performed using the Mascot Distiller software (version 2.3, Matrix Science, London, UK), and peaklist files were submitted to Mascot (version 2.3.01, Matrix Science, London, UK) for peptide and protein identification. The Mascot database search was performed against either the SwissProt database (version 57) with taxonomy restricted to Rattus for the rat liver lysate samples (7479 proteins searched) or the UniProtKB mouse complete proteome set (download date of October 26, 2010; 73,688 proteins searched) for the RAW cell samples including protein sequences of common laboratory contaminants and with the Mascot built-in target-decoy database search option enabled. Search parameters included a precursor tolerance of 10 ppm and a fragment tolerance of 0.02 Da; allowed a maximum of two missed cleavages; and accounted for the oxidation of methionine (+15.9949 Da), the carbamidomethylation of cysteine residues (+57.0215 Da), the oxidation of cysteine (+15.9949 Da), and the modification by cleaved diol-DCG-04 with and without hydration (+623.2928 and +641.3034 Da) or clicked and cleaved azido-E64 with and without hydration (+467.2254 and +485.2360 Da), all as variable modifications. The Mascot search result files were analyzed using Scaffold version 3.6.2 (Proteome Software Inc., Portland, OR). Threshold parameters (protein probability, 99%; minimum number of peptides, 2; peptide probability, 95%) were highly conservative and resulted in a peptide and protein false discovery rate of 0%.
RESULTS
Synthesis of a Diol Cleavable Linker and Incorporation into Chemical Probes
We decided to explore tartrate as a vicinal diol-containing cleavable linker. Tartrate is small relative to most other reported cleavable linkers and is therefore likely to have little influence on probe reactivity. Its hydrophilic nature might increase the solubility of probes in aqueous buffers and reduce nonspecific interactions with hydrophobic surfaces of proteins (32). More important, it is stable under acidic, basic, and reducing conditions, and it is therefore compatible with many buffer systems used in biochemistry.
Methyl-2,3-O-isopropylidene-l-tartrate (1) was selected as a synthetic building block for incorporation into chemical probes, because it can be obtained on large scale and in good yield from inexpensive l-tartaric acid in two straightforward protecting group manipulation steps (Fig. 2A). Building block 1 can be used in both solution and solid phase synthesis. To illustrate this, we performed a solid support synthesis of diol-DCG-04 (2; Fig. 2B), which is a cleavable version of DCG-04. For tandem labeling of azide probes, we designed the alkyne containing cleavable biotin reagent 3, which was synthesized in solution in four steps (Fig. 2C).
Fig. 2.
Synthesis of the cleavable linker building block and its probe derivatives. A, the synthesis of cleavable building block 1 from l-tartaric acid can be achieved in two easy protecting group manipulation steps: protection of the functional groups with an isopropylidene and methyl esters, followed by selective saponification of one methyl ester. B, diol-DCG-04 (2) was made via Fmoc-based solid phase peptide synthesis. C, cleavable alkyne biotin reagent 3, which is amenable for click chemistry mediated labeling of azido-tagged proteins, can be made in four steps from building block 1.
Evaluation of Diol-DCG-04 Labeling and Cleavage
We tested whether the diol cleavable linker within the context of a probe could be cleaved by periodate. To this end, we incubated ABP 2 with 10 mm sodium periodate and identified the nature of the cleavage products. We observed formation of the aldehyde, which is the expected cleavage product, as well as the hydrate of the aldehyde (supplemental Fig. S1). In order to check for possible side reactions with proteins and peptides, we also performed cleavage in the presence of a model peptide, which gratifyingly did not result in any side reactions (supplemental Fig. S1).
At this point, we evaluated the capture and release of proteases from a whole proteome. We first made a direct comparison of the diol-containing cathepsin ABP 2 with the parent compound DCG-04. In a rat liver proteome, which contains a variety of previously identified cathepsins (33), both probes yielded a similar, activity-based labeling pattern, indicating that the cleavable linker does not influence the specificity and potency of the ABP (supplemental Fig. S2). The labeled cathepsin proteases were also efficiently depleted from the proteome via incubation with immobilized streptavidin. Treatment with sodium periodate led to release of the diol-DCG-04-labeled cathepsins but had no effect on DCG-04-labeled ones (Fig. 3A).
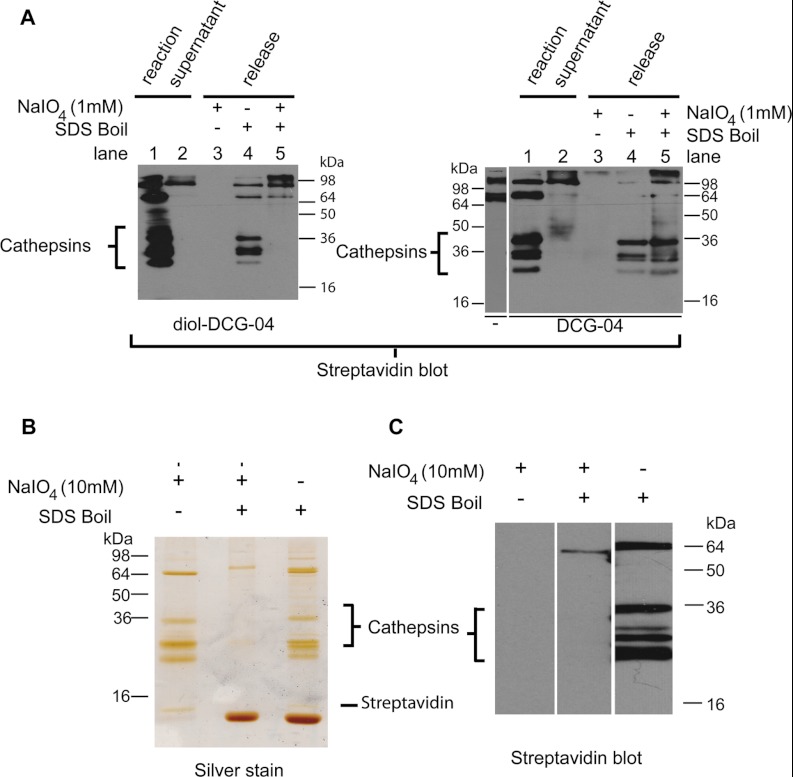
Fig. 3.
Elution of enriched rat liver lysate cathepsins. A, cathepsins labeled by diol-DCG-04 (left gel) or DCG-04 (right gel) can be efficiently pulled down by streptavidin beads (compare lanes 1 and 2). They are released by boiling (lane 4). Treatment with NaIO4 (lane 3) does not show any cathepsin bands, because they have lost their biotin (diol-DCG-04; left gel) or are not eluted (DCG-04; right gel). Subsequent boiling of these samples releases the remaining biotinylated proteins. For DCG-04, both endogenously biotinylated proteins and probe-labeled cathepsins are eluted. For diol-DCG-04, only endogenously biotinylated proteins are eluted, with none of the cathepsins, indicating efficient release by NaIO4. B, selective (10 mm NaIO4) or non-selective (SDS sample buffer boil) release of cathepsins from streptavidin beads detected via silver staining. Boiling of the beads after selective release does not elute additional cathepsin proteins. C, streptavidin Western blot of the same samples as in the silver staining. Note that the NaIO4 cleaved proteins are invisible because of the chemoselective removal of the biotin part.
Because cleavage of the diol linker results in loss of the biotin used for detection, cleaved proteins are not visible by means of streptavidin blot. We therefore also applied silver staining in order to detect the released cathepsins upon selective release (NaIO4 treatment) or non-selective release (boiling in SDS sample buffer). In an effort to optimize the cleavage conditions, we released target proteins from the immobilized streptavidin using different periodate concentrations and incubation times. Although 1 mm periodate was reported to release diol-containing ligands from MHC class I molecules (30), this concentration (for 2 h or overnight) did not result in the efficient cleavage of proteins from streptavidin beads (supplemental Fig. S3). Incubation with 10 mm periodate (2 × 40 min) eluted the cathepsin targets, as well as three proteins of ∼10, 65, and 90 kDa (Fig. 3B). These proteins were also visible in the SDS-treated sample. However, a large amount of free streptavidin, along with a faint smear of non-selectively bound high molecular weight proteins, was detected in addition, indicating that undesired proteins were bound to the streptavidin beads. Subsequent boiling of the periodate-treated sample did not release any additional cathepsins, showing the efficiency of the elution. Generally, cathepsins are N-glycosylated on the pro-part and the mature chain (34), and their glycans will also be oxidized at the diol-functionalities. One concern we had was Schiff base formation between the oxidized diols and the streptavidin beads. However, the similar intensity of the cleaved cathepsins resulting from chemical cleavage and non-selective release (SDS-boil) shows that this process has no influence on the efficiency of the elution (Fig. 3B). To prove that the cleavage conditions were chemoselective and not due to disruption of the streptavidin–biotin interaction, the same samples were also analyzed via streptavidin Western blotting (Fig. 3C). No biotinylated proteins were detected in the periodate treated sample, confirming that the cleavage took place between the reactive part of the ABP and the biotin moiety.
Labeling of Cathepsins with Azido-E64 in Vitro and in Situ
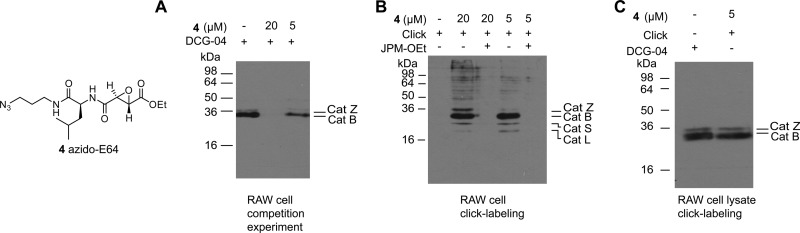
Although biotinylated ABPs are valuable tools for the study of enzymes in cell and tissue lysates, they have limited applicability in situ and in vivo because of their poor cell permeability. Tandem labeling was introduced in ABPP in 2003 by the groups of Cravatt, who used copper-catalyzed click chemistry for the detection of serine hydrolases (35, 36), and Overkleeft, who utilized a Bertozzi-Staudinger ligation for labeling of proteasomes in live cells (37). We here used the cathepsin ABP azido-E64 (38) (4, Fig. 4) to illustrate that the diol cleavable linker is compatible with tandem labeling and can be used to identify targets in live cells. When cells of the macrophage cell line RAW 264.7 were incubated with 20 μm azido-E64 for 30 min, no residual active cathepsins were detected as shown by streptavidin Western blotting after cell lysis and treatment with the cathepsin probe DCG-04 (Fig. 4A). This confirms that azido-E64 is cell permeable and reacts quantitatively with the expressed cathepsins. Following lysis of treated cells, click chemistry with alkyne tag 3 indeed showed cathepsin bands between 22 and 36 kDa (Fig. 4B). All of these bands can be competed away by the pan-cathepsin inhibitor JPM-OEt, indicating that the labeling takes place in an activity-dependent manner. When desired, click chemistry detection can also be performed on cell lysates (Fig. 4C). Interestingly, we observed that azido-E64 labels more protein targets in situ (whole cells) than in vitro (lysates; see Figs. 4B and 4C).
Fig. 4.
Streptavidin Western blots of cathepsin labeling in RAW 264.7 cells. A, cells were treated with azido-E64 (20 μm, 0.5 h or 5 μm, 1 h). Lysates were prepared and labeled with DCG-04 to detect residual active cathepsins. B, reaction of cathepsins in situ; then lysates were made and detection took place using diol-reagent 3. C, click labeling of cathepsins in vitro using diol reagent 3.
MS Identification of Protein Targets
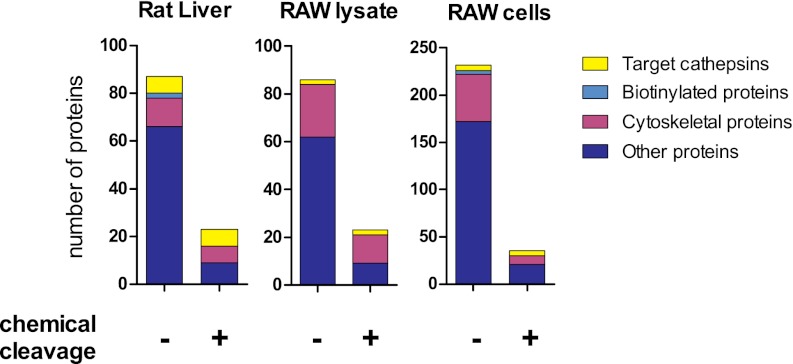
In order to show that the cleavable linker can reduce protein contaminations in protein identification by MS, we compared chemoselective release with on-bead digestion, a commonly used SDS-PAGE-free method for the identification of affinity-purified proteins. A diol-DCG-04 treated rat liver proteome was incubated with strepavidin beads and extensively washed to remove unbound proteins. The streptavidin beads were then divided into two parts and subjected to either an on-bead trypsin digestion or a chemoselective release followed by a trypsin digestion. LC-MS/MS analysis revealed the presence of seven cathepsins in both samples (Table I), which is two more than in previous reports (33). The periodate cleavage conditions are likely to oxidize the identified proteins, primarily at methionine residues. In the target cathepsins, we indeed found more oxidized methionines in the sample resulting from chemical cleavage than in that obtained via on-bead digestion (supplemental Table S1). However, this did not have an influence on the identification of the tryptic peptides (supplemental Table S2). Cytoskeletal proteins such as actin, keratins, and tubulins are abundant in any eukaryotic cell and were found as background in all samples (Fig. 5). Endogenously biotinylated proteins, in contrast, were detected only in the on-bead digestion, and not in the chemoselective release. A significant part of the other background proteins found in the on-bead digestion comprised housekeeping enzymes (for instance, dehydrogenases). These apparently remained stuck to the streptavidin beads despite stringent washes and were detected by the sensitive MS equipment. Overall, samples obtained via chemical cleavage showed a nearly 80% reduction in background proteins relative to on-bead digestion samples (Fig. 5).
Table I. Cathepsins identified in rat liver lysate. Listed are the number of unique peptides and the sequence coverage (in brackets) of all cathepsin targets of diol-DCG-04 identified from a rat liver lysate. A full list of identified proteins is given in supplemental Table S3.
| Protein name | Accession number | On-bead digestion | Chemical cleavage |
|---|---|---|---|
| Cathepsin Z | Q9R1T3 | 11 (30%) | 10 (32%) |
| Cathepsin B | P00787 | 11 (28%) | 9 (24%) |
| Cathepsin C | P80067 | 8 (23%) | 7 (22%) |
| Cathepsin H | P00786 | 8 (26%) | 9 (26%) |
| Cathepsin S | Q02765 | 5 (17%) | 3 (15%) |
| Cathepsin J | Q63088 | 5 (14%) | 2 (9.3%) |
| Cathepsin L1 | P07154 | 10 (32%) | 2 (5.4%) |
Fig. 5.
Overview of the background reduction in the proteomic analysis of ABP targets in rat liver, RAW cell lysate, and RAW cells. After labeling and removal of free probe, proteomes were incubated with streptavidin beads. Beads were collected, washed, and subjected to either on-bead digestion or chemical cleavage followed by digestion. Tryptic peptides were analyzed via LC-MS/MS. Contaminating non-rat or non-mouse proteins are not depicted. Proteins are classified as target cathepsins and background originating from endogenously biotinylated proteins, cytoskeletal proteins, and other proteins.
Next, we turned out attention to the identification of probe targets labeled via bioorthogonal chemistry. Lysates or live cells of RAW 264.7 macrophages were treated with azido-E-64 and subjected to click chemistry with alkyne biotin tag 3. As shown in Fig. 4, labeling in whole cells resulted in the detection of more protein target bands than in cell lysates. Indeed, tandem MS identified cathepsin Z and B both in vitro and in situ (Tables II and III, respectively). Cathepsins H, F, S, and L1 were identified only in situ. Cathepsins H, S, and L1 correspond to the lower running gelbands (Fig. 4B), which match the molecular weights of their mature forms (24, 24, and 19 kDa, respectively). Although cathepsin F was identified only in the on-bead digestion, overall comparable sequence coverages of the targets in the RAW 264.7 samples were obtained for chemical release and on-bead digestion experiments (Tables II and III). The important benefit of the cleavable linker is again illustrated by the reduction of background proteins by up to 87% (Fig. 5).
Table II. Cathepsins identified in RAW 264.7 lysate. Listed are the number of unique peptides and the sequence coverage (in brackets) of all cathepsin targets of azido-E-64 identified from a RAW 264.7 lysate. A full list of identified proteins is given in supplemental Table S4.
Table III. Cathepsins identified in RAW 264.7 cells. Listed are the number of unique peptides and the sequence coverage (in brackets) of all cathepsin targets of azido-E-64 identified from experiments with live RAW 264.7 cells. A full list of identified proteins is given in supplemental Table S5.
DISCUSSION
Developments in bioconjugation technologies have given enormous impetus to the study of small molecule modifications of proteins including post-translational modifications and ABPs. For the enrichment of such modified proteins, the streptavidin–biotin system is widely used. However, elution conditions are generally harsh and suffer from background. A variety of cleavable linkers have been developed to aid the release of protein targets. Most of these studies aimed at achieving mild cleavage conditions. Although this has resulted in interesting chemistries for target capture and release, the construction of most linker building blocks involved multi-step organic syntheses ranging from 3 (15) to more than 10 consecutive chemical transformations (28, 39). Here, we showed the application of tartrate derivative 1, which can be made on large scale in two uncomplicated steps from l-tartrate, as a cleavable linker building block. Synthetic intermediate 5 (see supplemental Fig. S4), which gives access to building block 1 in one saponification step, is also commercially available, although not as inexpensive as l-tartrate. Cleavable building block 1 is compatible with Fmoc-based solid phase synthesis. Thus, the availability of the linker building block is not a limiting factor in the probe synthesis, and the probe synthesis itself can be performed through easy-to-handle peptide chemistry that may be handled in laboratories without an extensive background in organic chemistry.
Following the enrichment of protein targets by immobilized streptavidin, MS-based identification is an essential process in chemical proteomics studies. On-bead digestion protocols might lead to contamination not only by endogenously biotinylated proteins, but also by other abundant proteins, despite stringent washes. To show the benefit of the diol cleavable linker in target identification, we have used two ABPs for the clan CA family of proteases: one probe that is biotinylated, and one cell-permeable probe that can be utilized in combination with tandem labeling. Our experiments show that the chemoselective elution of probe targets leads to cleaner data and an up to 87% decrease in background proteins relative to on-bead digestion. For new and uncharacterized probes, a reduction in the amount of (false positive) hits will reduce the time necessary for target validation and speed up the overall process of target discovery. An additional benefit of a cleavable linker is the possibility of releasing or specifically enriching probe-modified peptides. The identification of such modified peptides will unambiguously prove that the corresponding proteins are genuine targets. Moreover, the site of modification can confirm the mechanism of probe incorporation. Tryptic peptides containing the active site of cathepsins were not found in our current experiments, as these are notoriously difficult to identify because of their large size (16).
Relative to lysates, the experiments in live RAW 264.7 macrophages showed additional cathepsin targets of the azido-E-64 ABP. It is known that cathepsin L is rapidly inactivated upon cell lysis (40). The loss of activity of cathepsins H, F, and S might be caused by sensitivity toward the lysis conditions or exposure to cytoplasmic inhibitors. The different results obtained for lysates and whole cells highlight the importance of target identification in living cells, as the specificity of small molecules (ABPs, natural products, or drugs) might otherwise be misjudged. It further underlines the benefit of bioorthogonal ligations in combination with the capture and release strategy mediated by cleavable linkers.
In conclusion, we have shown that a vicinal diol cleavable linker can be easily synthesized and readily incorporated into chemical probes. The elution of probe targets takes place in a mild, efficient, and chemoselective manner. In the target identifications, the linker leads to a high reduction in background proteins. Because of its compatibility with tandem labeling and target discovery in live cells, we believe that the strategy described herein will be of substantial benefit for other chemical biology and proteomics studies.
Supplementary Material
Acknowledgments
We thank Dr. O. Frank for measuring NMR, Dr. M. Fonovic for useful discussion, and Prof. Dr. D. Langosch for general support.
Footnotes
* We acknowledge financial support from the DFG (Emmy Noether program), the CIPS-M, and the Graduate Centre Weihenstephan of the TUM Graduate school. Yinliang Yang is a recipient of a PhD fellowship from the Chinese Scholarship Council.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ABPP
- activity-based protein profiling
- ABP
- activity-based probe
- Cat
- cathepsin
- DIEA
- N,N-diisopropyl-N-ethylamine
- DMEM
- Dulbecco's modified Eagle medium
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- HBTU
- 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- MHC
- major histocompatibility complex
- SB
- sample buffer
- SPPS
- solid phase peptide synthesis.
REFERENCES
- 1. Heal W. P., Tate E. W. (2010) Getting a chemical handle on protein post-translational modification. Org. Biomol. Chem. 8, 731–738 [DOI] [PubMed] [Google Scholar]
- 2. Dubinsky L., Krom B. P., Meijler M. M. (2012) Diazirine based photoaffinity labeling. Bioorg. Med. Chem. 20, 554–570 [DOI] [PubMed] [Google Scholar]
- 3. Geurink P. P., Prely L. M., van der Marel G. A., Bischoff R., Overkleeft H. S. (2011) Photoaffinity labeling in activity-based protein profiling. Top. Curr. Chem. 324, 85–113 [DOI] [PubMed] [Google Scholar]
- 4. Cravatt B. F., Wright A. T., Kozarich J. W. (2008) Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 77, 383–414 [DOI] [PubMed] [Google Scholar]
- 5. Heal W. P., Dang T. H. T., Tate E. W. (2011) Activity-based probes: discovering new biology and new drug targets. Chem. Soc. Rev. 40, 246–257 [DOI] [PubMed] [Google Scholar]
- 6. Greenbaum D., Medzihradszky K. F., Burlingame A., Bogyo M. (2000) Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem. Biol. 7, 569–581 [DOI] [PubMed] [Google Scholar]
- 7. Saxon E., Bertozzi C. R. (2000) Cell surface engineering by a modified Staudinger reaction. Science 287, 2007–2010 [DOI] [PubMed] [Google Scholar]
- 8. Kolb H. C., Finn M. G., Sharpless K. B. (2001) Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 40, 2004–2021 [DOI] [PubMed] [Google Scholar]
- 9. Prescher J. A., Bertozzi C. R. (2005) Chemistry in living systems. Nat. Chem. Biol. 1, 13–21 [DOI] [PubMed] [Google Scholar]
- 10. Best M. D. (2009) Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry 48, 6571–6584 [DOI] [PubMed] [Google Scholar]
- 11. Willems L. I., van der Linden W. A., Li N., Li K. Y., Liu N., Hoogendoorn S., van der Marel G. A., Florea B. I., Overkleeft H. S. (2011) Bioorthogonal chemistry: applications in activity-based protein profiling. Acc. Chem. Res. 44, 718–729 [DOI] [PubMed] [Google Scholar]
- 12. Rybak J. N., Scheurer S. B., Neri D., Elia G. (2004) Purification of biotinylated proteins on streptavidin resin: a protocol for quantitative elution. Proteomics 4, 2296–2299 [DOI] [PubMed] [Google Scholar]
- 13. Leriche G., Chisholm L., Wagner A. (2012) Cleavable linkers in chemical biology. Bioorg. Med. Chem. 20, 571–582 [DOI] [PubMed] [Google Scholar]
- 14. Everley P. A., Gartner C. A., Haas W., Saghatelian A., Elias J. E., Cravatt B. F., Zetter B. R., Gygi S. P. (2007) Assessing enzyme activities using stable isotope labeling and mass spectrometry. Mol. Cell. Proteomics 6, 1771–1777 [DOI] [PubMed] [Google Scholar]
- 15. Verhelst S. H. L., Fonovic M., Bogyo M. (2007) A mild chemically cleavable linker system for functional proteomic applications. Angew. Chem. Int. Ed. Engl. 46, 1284–1286 [DOI] [PubMed] [Google Scholar]
- 16. Fonovic M., Verhelst S. H. L., Sorum M. T., Bogyo M. (2007) Proteomics evaluation of chemically cleavable activity-based probes. Mol. Cell. Proteomics 6, 1761–1770 [DOI] [PubMed] [Google Scholar]
- 17. Yang Y. Y., Grammel M., Raghavan A. S., Charron G., Hang H. C. (2010) Comparative analysis of cleavable azobenzene-based affinity tags for bioorthogonal chemical proteomics. Chem. Biol. 17, 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leriche G., Budin G., Brino L., Wagner A. (2010) Optimization of the azobenzene scaffold for reductive cleavage by dithionite; development of an azobenzene cleavable linker for proteomic applications. Eur. J. Org. Chem. 4360–4364 [Google Scholar]
- 19. Landi F., Johansson C. M., Campopiano D. J., Hulme A. N. (2009) Synthesis and application of a new cleavable linker for “click”-based affinity chromatography. Org. Biomol. Chem. 8, 56–59 [DOI] [PubMed] [Google Scholar]
- 20. Park K. D., Liu R., Kohn H. (2009) Useful tools for biomolecule isolation, detection, and identification: acylhydrazone-based cleavable linkers. Chem. Biol. 16, 763–772 [DOI] [PubMed] [Google Scholar]
- 21. Dirksen A., Yegneswaran S., Dawson P. E. (2010) Bisaryl hydrazones as exchangeable biocompatible linkers. Angew. Chem. Int. Ed. 49, 2023–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Szychowski J., Mahdavi A., Hodas J. J., Bagert J. D., Ngo J. T., Landgraf P., Dieterich D. C., Schuman E. M., Tirrell D. A. (2010) Cleavable biotin probes for labeling of biomolecules via azide-alkyne cycloaddition. J. Am. Chem. Soc. 132, 18351–18360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou H., Ranish J. A., Watts J. D., Aebersold R. (2002) Quantitative proteome analysis by solid-phase isotope tagging and mass spectrometry. Nat. Biotechnol. 20, 512–515 [DOI] [PubMed] [Google Scholar]
- 24. Orth R., Sieber S. A. (2009) A photolabile linker for the mild and selective cleavage of enriched biomolecules from solid support. J. Org. Chem. 74, 8476–8479 [DOI] [PubMed] [Google Scholar]
- 25. Kim H. Y., Tallman K. A., Liebler D. C., Porter N. A. (2009) An azido-biotin reagent for use in the isolation of protein adducts of lipid-derived electrophiles by streptavidin catch and photorelease. Mol. Cell. Proteomics 8, 2080–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Speers A. E., Cravatt B. F. (2005) A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J. Am. Chem. Soc. 127, 10018–10019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weerapana E., Speers A. E., Cravatt B. F. (2007) Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)—a general method for mapping sites of probe modification in proteomes. Nat. Protoc. 2, 1414–1425 [DOI] [PubMed] [Google Scholar]
- 28. Geurink P. P., Florea B. I., Li N., Witte M. D., Verasdonck J., Kuo C. L., van der Marel G. A., Overkleeft H. S. (2010) A cleavable linker based on the levulinoyl ester for activity-based protein profiling. Angew. Chem. Int. Ed. 49, 6802–6805 [DOI] [PubMed] [Google Scholar]
- 29. Smith R. J., Capaldi R. A., Muchmore D., Dahlquist F. (1978) Cross-linking of ubiquinone cytochrome c reductase (complex III) with periodate-cleavable bifunctional reagents. Biochemistry 17, 3719–3723 [DOI] [PubMed] [Google Scholar]
- 30. Rodenko B., Toebes M., Celie P. H., Perrakis A., Schumacher T. N., Ovaa H. (2009) Class I major histocompatibility complexes loaded by a periodate trigger. J. Am. Chem. Soc. 131, 12305–12313 [DOI] [PubMed] [Google Scholar]
- 31. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 32. Shiyama T., Furuya M., Yamazaki A., Terada T., Tanaka A. (2004) Design and synthesis of novel hydrophilic spacers for the reduction of nonspecific binding proteins on affinity resins. Bioorg. Med. Chem. 12, 2831–2841 [DOI] [PubMed] [Google Scholar]
- 33. Greenbaum D., Baruch A., Hayrapetian L., Darula Z., Burlingame A., Medzihradszky K. F., Bogyo M. (2002) Chemical approaches for functionally probing the proteome. Mol. Cell. Proteomics 1, 60–68 [DOI] [PubMed] [Google Scholar]
- 34. Katunuma N. (2010) Posttranslational processing and modification of cathepsins and cystatins. J. Signal Transduction 2010, 375345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Speers A. E., Cravatt B. F. (2004) Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 11, 535–546 [DOI] [PubMed] [Google Scholar]
- 36. Speers A. E., Adam G. C., Cravatt B. F. (2003) Activity-based protein profiling in vivo using a copper(I)-catalyzed azide-alkyne [3+2] cycloaddition. J. Am. Chem. Soc. 125, 4686–4687 [DOI] [PubMed] [Google Scholar]
- 37. Ovaa H., van Swieten P. F., Kessler B. M., Leeuwenburgh M. A., Fiebiger E., van den Nieuwendijk A. M. C. H., Galardy P. J., van der Marel G. A., Ploegh H. L., Overkleeft H. S. (2003) Chemistry in living cells: detection of active proteasomes by a two-step labeling strategy. Angew. Chem. Int. Ed. 42, 3626–3629 [DOI] [PubMed] [Google Scholar]
- 38. Hang H. C., Loureiro J., Spooner E., van der Velden A. W., Kim Y. M., Pollington A. M., Maehr R., Starnbach M. N., Ploegh H. L. (2006) Mechanism-based probe for the analysis of cathepsin cysteine proteases in living cells. ACS Chem. Biol. 1, 713–723 [DOI] [PubMed] [Google Scholar]
- 39. Gartner C. A., Elias J. E., Bakalarski C. E., Gygi S. P. (2007) Catch-and-release reagents for broadscale quantitative proteomics analyses. J. Proteome Res. 6, 1482–1491 [DOI] [PubMed] [Google Scholar]
- 40. Wilcox D., Mason R. W. (1992) Inhibition of cysteine proteinases in lysosomes and whole cells. Biochem. J. 285, 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.