Abstract
Trace metals such as copper, iron, zinc, and manganese play important roles in several biochemical processes, including respiration and photosynthesis. Using a label-free, quantitative proteomics strategy (MSE), we examined the effect of deficiencies in these micronutrients on the soluble proteome of Chlamydomonas reinhardtii. We quantified >103 proteins with abundances within a dynamic range of 3 to 4 orders of magnitude and demonstrated statistically significant changes in ∼200 proteins in each metal-deficient growth condition relative to nutrient-replete media. Through analysis of Pearson's coefficient, we also examined the correlation between protein abundance and transcript abundance (as determined via RNA-Seq analysis) and found moderate correlations under all nutritional states. Interestingly, in a subset of transcripts known to significantly change in abundance in metal-replete and metal-deficient conditions, the correlation to protein abundance is much stronger. Examples of new discoveries highlighted in this work include the accumulation of O2 labile, anaerobiosis-related enzymes (Hyd1, Pfr1, and Hcp2) in copper-deficient cells; co-variation of Cgl78/Ycf54 and coprogen oxidase; the loss of various stromal and lumenal photosynthesis-related proteins, including plastocyanin, in iron-limited cells; a large accumulation (from undetectable amounts to over 1,000 zmol/cell) of two COG0523 domain-containing proteins in zinc-deficient cells; and the preservation of photosynthesis proteins in manganese-deficient cells despite known losses in photosynthetic function in this condition.
The investigation of cellular responses to micronutrient deficiency has provided important insights into the utilization of metals in biochemistry. Copper, iron, manganese, and zinc (denoted by their elemental symbols Cu, Fe, Mn, and Zn, without reference to their ionic state) are among the most widely studied metals, as they are integral to many metabolic pathways and processes. A systematic examination of proteins with known three-dimensional structures belonging to one of 1,371 Enzyme Commission groups (defined as groups of proteins known to catalyze the same reaction) showed that 47% of these proteins require metals, often as part of the catalytic center (1). Aside from magnesium, which is thought to bind to enzymes only transiently, zinc, iron, and manganese are the three most highly utilized metals by enzymes, and copper is the seventh most utilized (1). These four metals are of particular importance to plant biochemistry, as they each play a role in photosynthesis. Copper-containing plastocyanin transfers electrons from Photosystem (PS)1 II to PS I; iron is found in all photosynthetic electron transport complexes, in ferredoxin, and as heme and iron-sulfur centers in many enzymes; manganese is required for the water splitting reaction of PS II; and zinc is utilized by carbonic anhydrases, which help to concentrate inorganic carbon at the site of carbon fixation (2–4).
We have used Chlamydomonas reinhardtii, a single-celled alga, as a photosynthetic reference organism for understanding trace metal metabolism (3). Among other advantages, Chlamydomonas grows in culture in defined medium from which it is straightforward to withhold the nutrient under study. In contrast to other plant models, Chlamydomonas can grow (photo)heterotrophically on acetate as a source of reduced carbon when photosynthetic activity is compromised by mutation or the absence of an essential metal co-factor (5). Chlamydomonas has also emerged as a reference organism for the study of the genetics and genomics of bio-fuel production (6). Most studies of triacylglyceride (TAG) synthesis in Chlamydomonas have focused primarily on nitrogen-deficiency-induced TAG overaccumulation (7), but more recent studies indicate that other stress conditions such as iron and zinc deficiency also result in TAG overaccumulation (8).
Traditional biochemical and classical genetic approaches have revealed considerable information about responses to micronutrient deficiencies in Chlamydomonas (3). The copper deficiency response in Chlamydomonas is controlled via the activity of a transcriptional regulator, CRR1 (9, 10). Examples of CRR1-controlled responses include the replacement of plastocyanin with heme-containing cytochrome c6 and increases in the abundance and activity of coporphyrinogen oxidase III (a product of the CPX1 gene, often abbreviated to coprogen oxidase, that is involved in tetrapyrrole biosynthesis) and copper uptake proteins Ctr1, Ctr2, and a related soluble protein Ctr3 (11). Recently, several CRR1-independent changes in transcript abundance in copper-deficient cells were also noted, and many of these overlap with changes that occur in anaerobic Chlamydomonas cells (12). Specifically, HYD1, HYDEF, HYDG, HCP2, HCP3, and PFR1 transcripts, which are involved in hydrogenase assembly, are increased under both copper deficiency and O2 deficiency conditions (12, 13). The proteins that correspond to these transcripts are O2 labile, and thus it is unclear whether the increase in HYD1, HYDEF, HYDG, HCP2, HCP3, and PFR1 transcripts will actually result in an increase in the corresponding proteins.
As is the case for the copper nutrition regulon, responses to iron deficiency include up-regulation of iron assimilation proteins such as Ftr1 (a ferric ion transporter), ferroxidase (Fox1), Fea1, and Fea2 and elevation of the abundance of ferritins, which are believed to buffer iron released from iron-deficiency-induced degradation in heterotrophic cells of abundant iron proteins, including ferredoxin, an abundant iron–sulfur protein in chloroplasts, and PS I with three Fe4S4 centers (14–16). Interestingly, although many PS I proteins are known to decrease under iron limitation (17), changes in the transcripts of these proteins are not observed (18). Also, the magnitude of changes in the abundance of iron-containing proteins from iron-deficient/limited cells differs greatly among proteins. For example, ferredoxin is known to decrease substantially in iron-starved cells (15), whereas the iron-containing superoxide dismutase is only mildly affected, suggesting that some iron-proteins are more dispensable than others (19). This illustrates the need for proteomics surveys to identify changes at the protein level that are not apparent at the transcript level and to help determine the dispensability of iron-proteins relative to one another.
Manganese-deficient cells have reduced manganese-containing superoxide dismutase (MnSOD1) activity, and in severely deficient conditions PS II activity is reduced as well, so that such cultures require acetate for growth. The oxygen evolving enhancer proteins (Oee1, Oee2, Oee3) appear to be less tightly associated with the thylakoid membrane in this situation and are found in soluble fractions of cell extracts (20).
Zinc deficiency has not been studied extensively in Chlamydomonas to date. However, recent work has shown that mRNAs encoding COG0523 family proteins, whose functions are unknown but hypothesized to involve metal trafficking, are increased in zinc-deficient growth conditions (21). Zinc-binding carbonic anhydrases, which interconvert CO2 and bicarbonate as part of the carbon-concentrating mechanism, are important for photosynthetic function at atmospheric levels of CO2. They are decreased in abundance in zinc-deficient cells (22).
Availability of the Chlamydomonas genome has made possible the initiation of large-scale examinations of changes to the transcriptome and proteome under nutritional deficiency conditions. Prior examinations of the metal-deficient transcriptomes in Chlamydomonas provide excellent examples of the application of the genome data. From these studies, a great deal of information was obtained on the effect of metal deficiency on the abundance of thousands of transcripts (13). Although transcript levels provide important information, they may describe only a subset of all responses to metal deficiency. For instance, although plastocyanin abundance is decreased dramatically in copper deficiency, the corresponding mRNAs are not (23). The need for proteome data to close the knowledge gap between responses at the transcript level and responses at the protein level is clear.
Recent advances in proteomics of Chlamydomonas have led to the analysis of various subproteomes, including those from the mitochondria (17, 24), chloroplast (17), centriole (25), eyespot apparatus (26–28), anaerobic responsive proteins (29), high-light responsive proteins (30), thioredoxin interacting proteins (31), and others (17, 24–32). To date, more than 2,000 Chlamydomonas proteins have been identified via mass spectrometry (MS)-based proteomics techniques. Of special interest are two comparative proteomics studies that examined the iron-responsive proteome of the chloroplast and mitochondria in Chlamydomonas (17) and a study of the heat-shock response in the total soluble proteome (33). In both studies, stable isotope labeling was used to examine changes in the proteome via the relative quantification of proteins. The iron deficiency study found that proteins involved in photosynthesis were reduced in abundance, whereas major respiratory proteins of the mitochondria either did not change or were increased (17), but proteins outside the chloroplast and mitochondria were not examined. In the heat-shock study, over 1,100 proteins were quantified from the total soluble lysate, allowing for a more global look at the Chlamydomonas proteome (33).
In the present contribution, we employed a label-free, absolute quantitative proteomics technique termed MSE on the Chlamydomonas soluble proteome (post-ribosomal supernatant) to further advance our understanding of the effects of micronutrient deficiency. The MSE data-independent scanning mode allows for parallel collection of precursor and product ion data measured by oscillating between high and low energies in the collision cell of a tandem mass spectrometer (34–37). The absolute quantification of proteins, as described by Silva and coworkers, is accomplished with MSE data by utilizing a universal response factor (34). To our knowledge, this work represents the first absolute quantitative examination of the soluble subproteome of Chlamydomonas. This study allows a quantitative comparison of a large proteomic dataset to a recently published transcriptome (13), thus providing a means to assess the degree of correlation between RNA and proteins in Chlamydomonas. This study also allows the examination of multiple metal-dependent proteomes, which provides a better understanding of the inter-relationship between various metal metabolisms.
EXPERIMENTAL PROCEDURES
Strains and Cultures
Chlamydomonas reinhardtii strain 2137 was grown in Tris acetate-phosphate (TAP) medium at 24 °C with shaking (180 rpm) and under continuous light (∼50 to 100 μmol/m2/s, 2:1, cool white:warm white light). For copper deficiency studies, a trace element solution was prepared as described by Quinn and Merchant (38). Trace element solutions for the manganese nutrition experiments were made as described by Allen et al., except that we also examined an intermediate nutritional condition with 0.05 μm MnCl2 present (20). Iron-limited (0.25 μm FeSO4), -deficient (1 μm FeSO4), and -replete (20 μm FeSO4) media were prepared as described by Moseley et al. (18). For zinc deficiency studies, a revised trace element solution was used containing 25 mm EDTA-Na2, 28.5 μm (NH4)6Mo7O24, 0.1 mm Na2SeO3, 2.5 mm ZnSO4 in 2.75 mm EDTA, 6 mm MnCl2 in 6 mm EDTA, 20 mm FeCl3 in 22 mm EDTA, and 2 mm CuCl2 with zinc in EDTA withheld in zinc-deficient medium (8). For each experiment, cells were collected at a density of ∼3 × 106 to 6 × 106 cell ml−1. For each experimental condition, biological triplicates were grown in separate flasks to account for biological variation among cultures.
Sample Preparation and MSE Analysis
MSE quantitative proteomics measurements were performed as previously described, with slight modifications (13). In brief, cells were broken via two freeze-thaw cycles at −80 °C. Insoluble material was removed after centrifugation at 16,000 × g for 10 min at 4 °C followed by a second centrifugation at 253,000 × g for 20 min at 4 °C. For each sample, ∼30 μg of protein was separated via denaturing gel electrophoresis (4% to 12% NuPage Bis-Tris gels; Invitrogen) and visualized via Coomassie blue staining (Bio-Rad). Each gel lane was divided into ∼3 mm bands, and individual bands were subjected to in-gel trypsin digestion (sequencing grade modified trypsin; Promega, Madison, WI). Tryptic peptides were extracted into 50/50 water/acetonitrile solution containing 2.5% (v/v) formic acid and lyophilized. Peptides were then resuspended in a 1% formic acid solution containing 25 fmol/μl bovine serum albumin (BSA) trypsin digestion standard (MassPREP BSA Digestion Standard; Waters Corporation, Milford, MA).
The peptides were analyzed as described elsewhere (13) using an ultra-performance liquid chromatography (UPLC) system (Waters nanoAcquity UltraPerformance UPLC) coupled to a Waters Xevo quadrupole time-of-flight mass spectrometer. Peptides were separated via UPLC with a 5 μm Symmetry C18 180 μm × 20 mm reversed-phase trap column in-line with a 1.7 μm BEH130, 75 μm × 100 mm reversed-phase C18 analytical column. Peptides were eluted using a 60 min 3% acetonitrile/0.1% formic acid to 40% acetonitrile/0.1% formic acid gradient at a flow rate of 0.3 μl/min to the electrospray ionization (ESI) mass spectrometer. [Glu1]-Fibrinopeptide B was used as a mass calibration standard (100 fmol/μl) and was infused via a separate ESI sprayer, and the standard peptide was measured at 1 min intervals during the LC-MS experiment. The LC-mass spectrometer was operated in the MSE data independent acquisition mode (34, 35, 37). The collision energy was continually switched between low (6 eV) and elevated energy (ramped from 15 to 40 eV) during alternating scans (m/z 50–2,000). The correlation of product ions to precursor ions was achieved by using reconstructed retention times (i.e. alignment of LC retention times of the precursors and products) and chromatographic peak shapes. To ensure adequate coverage of the proteome from each band, at least two 1 μl replicate injections from each gel band were analyzed via LC-MS.
Database Searches
Protein Lynx Global Server (PLGS version 2.4; Waters) was used to process the LC-MS raw data and determine protein identification and quantification. The quantification of protein levels was achieved via the addition of an internal protein standard (BSA trypsin digest standard) to which the data set was normalized (34, 35, 39).
The MS data were searched using the PLGS algorithm described by Li and coworkers (40). Briefly, the calibrated mass spectra are centroided, de-isotoped, and charge-state-reduced to yield a monoisotopic mass for each peptide and its tentatively associated product ions. The direct correlation of a precursor ion and a potential product ion is initially achieved through the alignment of drift times, followed by a further correlation process during the database searching that is based on the physicochemical characteristics of peptides when they undergo collisionally activated dissociation. Searches were limited to trypsin proteolysis fragments, and peptide precursor and product ion mass tolerances were set to 20 ppm and 40 ppm, respectively. Other search parameters included the minimum number of peptide matches (1), minimum number of product ions per peptide (2), minimum number of product ions per protein (2), maximum number of missed tryptic cleavage sites (2), and maximum false positive rate for identification (4%) (40). Carbamidomethylation of cysteine residues was set as a fixed modification, and methionine oxidation, asparagine and glutamine deamidation, and N-terminal acetylation were set as variable modifications. Additional stringency was applied to the data by requiring that a protein be detected in at least two different conditions in order to be considered identified and be observed in two of three biological replicates in each condition in order to be considered quantifiable. The average abundances of proteins are thus based on two or three different measurements.
Three different Chlamydomonas gene model sets are presently in use and are based on two different genome assemblies. For each of the two assemblies, there are multiple gene model predictions. The catalogue track contains the best gene model at each locus as determined by users or by automated annotation. Catalogue track gene models from the version 3 genome assembly are called FM3.1 models (14,723 entries), and catalogue track gene models from the version 4 assembly are called the FM4 (16,837 entries) and Augustus 10.2 (Au10.2) model sets (17,430 entries). The Au10.2 set represents a new set of gene models predicted by an algorithm separate from that used for deriving FM3.1 and FM4. MS data were searched against each of the three Chlamydomonas protein databases modified by the replacement of some catalogue models with individual user-curated models and supplemented with sequences for keratin, trypsin, BSA, and chloroplast and mitochondrial proteins from the National Center for Biotechnology Information. Quantification was determined using up to the three most abundant shared peptides as described by Silva et al. (34–36).
Identifications from FM3.1 and FM4 databases were converted to Au10.2 identifiers by mutual best blast hits using the Algal Functional Annotation Tool (41). All quantifications reported were derived from Au10.2 database searches, with one exception. Because Au10.2 and FM3 were based on different genome assemblies, it is possible that proteins detected in FM3 searches do not exist in the Au10.2 database. Thus, identifications unique to FM3 (vide infra) were maintained in the dataset. All other proteins not identified in Au10.2 searches were not examined further.
Functional Analysis
Proteins were separated into subgroups based on whether they showed a statistically significant (p < 0.05 by Student's t test) increase or decrease of at least 2-fold in magnitude of a protein in the metal deficiency condition. Proteins that were detected in only one of the conditions could not be assigned statistical significance and were included in the analysis only if their abundances were over 20 zmol/cell. In the case of manganese and iron deficiency studies, we focused on differences observed between the metal-replete condition and the metal-limited (0 μm Mn and 0.25 μm Fe) conditions. Protein lists were analyzed using the “Enriched Ontology Terms” function of the Algal Functional Annotation Tool (41). Proteins that could not be clustered using this function were manually investigated and grouped based on their Au10.2 annotations.
Functional categories were based on those utilized by Karpowicz et al., with slight modifications (42). The “pigments” category was expanded to include other co-factor metabolism components such as folate, quinone, tocopherol, iron–sulfur centers, thiamin, and molybdenum co-factor. This new grouping was renamed “secondary metabolites and co-factors.” It should be noted, however, that the “secondary metabolites and co-factors” do not include proteins that bind or utilize co-factors; rather, they are proteins involved in the degradation, synthesis, or insertion of co-factors. All other categories remained with similar classification rationale applied. “Protein metabolism” included any enzyme involved in protein or amino acid synthesis, degradation, or modification. “Nucleic acid metabolism” referred to any protein involved in nucleotide synthesis/degradation, DNA or RNA binding, and transcriptional regulators. “Photosynthesis” was related to proteins involved in the photosynthetic apparatus and the carbon-concentrating mechanism. “Transport” included any transport proteins, channels, or assimilation factors that interact directly with transporters. “Redox” proteins are involved in electron transfer (not including photosynthetic proteins) and other reduction/oxidation reactions. “Signaling” included kinases, sensory proteins, and factors involved in signal transduction. The “lipid metabolism” category included proteins involved in fatty acid, membrane, and storage lipid synthesis, degradation, and membrane trafficking. “Carbohydrate metabolism” included proteins related to sugar or starch synthesis and/or degradation including glycolysis/gluconeogenesis. “Cell cycle” referred to proteins that are involved in cell development and cell division. “Other” proteins were those with functions not related to the categories described here. “Unknown” included proteins with no known function.
Protein Expression and Purification and Antibody Production
The CGL78/YCF54 expression construct was generated using nested PCR and the Gateway recombinational cloning system (Invitrogen, Carlsbad, CA) as described elsewhere (43). The segment of A. thaliana CGL78 (At5g58250) encoding amino acids S73–V211 was amplified from the cDNA clone U63260 (ABRC) using Phusion polymerase (New England Biolabs, Ipswich, MA). The forward primer (CGL78.S73.For.) added a 5′ extension encoding a tobacco etch virus (TEV) protease cleavage site, and the reverse primer (CGL78.V211.Rev.) added a C-terminal hexahistidine tag followed by a stop codon. This initial PCR product was then amplified using a second set of primers (PE-277 and PE-278) to introduce AttB1 and AttB2 recombination sites. The final PCR product was gel purified and recombined into the donor vector pDONR201 and subsequently into the expression vector pKM596 (43) to produce a His-tagged maltose binding protein fusion. The expression construct was checked via DNA sequencing (Genewiz, South Plainfield, NJ).
The expression plasmid was transformed into E. coli BL21-Gold (DE3) cells (Novagen, EMD Chemicals, Billerica, MA) harboring pRK603, an expression vector for the TEV protease. Cells were grown at 37 °C in LB supplemented with ampicillin (100 μg/ml) and kanamycin (30 μg/ml) to an OD600 of 1.0, at which point the temperature was shifted to 18 °C and protein expression was induced by the addition of isopropyl 1-thio-β-D-galactopyranoside (IPTG) to a concentration of 1 mm. Cells were left to grow overnight and were harvested via centrifugation the following day. The cell pellet was resuspended in lysis buffer (50 mm Tris/Cl pH 8.0, 300 mm NaCl, 10% glycerol, 20 mm imidazole) supplemented with protease inhibitor mixture (Sigma), 100 μm PMSF, DNase, lysozyme, and 10 mm β-mercaptoethanol and broken using a French press. The lysate was clarified via centrifugation (35,000 × g, 30 min, 4 °C), and the supernatant was incubated with nickel-nitrilotriacetic acid agarose beads (Qiagen, Valencia, CA) for 60 min at 4 °C. The beads were washed extensively with lysis buffer, and bound protein was eluted with 300 mm imidazole in lysis buffer. CGL78 was further purified via size exclusion chromatography using a HiLoad Superdex S-200 column (GE Healthcare) equilibrated with 50 mm Tris/Cl pH 8.0 containing 300 mm NaCl and 10% glycerol. Peak fractions were analyzed via SDS-PAGE, and those containing pure CGL78 were pooled and concentrated. The purified protein was used directly for antiserum production in rabbits (at Agrisera AB, Vännäs, Sweden, and the antibodies are available from the vendor).
A synthetic codon-optimized ZCP2 gene (Genscript, Piscataway, NJ) was cloned NdeI-XhoI in pET28b (Novagen) to allow expression of ZCP2 as a fusion protein with an N-terminal hexahistidine tag. The expression construct was checked via DNA sequencing (Genewiz).
The ZCP2 expression plasmid was transformed into E. coli BL21-Gold (DE3) cells (Novagen). Cells were grown at 37 °C in Terrific Broth supplemented with kanamycin (30 μg/ml) and glucose (20 mm) to an OD600 of 1.0, at which point the temperature was shifted to 18 °C and protein expression was induced by addition of IPTG to a concentration of 1 mm. Cells were left to grow overnight and were harvested via centrifugation the following day. The cell pellet was resuspended in supplemented lysis buffer (see above) and broken by means of sonication. The crude lysate was clarified via centrifugation (35,000 × g, 30 min, 4 °C), and the supernatant was loaded onto a HisTrap Ni2+ chelation affinity column (GE Healthcare) equilibrated with Buffer A (20 mm Tris/Cl pH 8.0, 1 m NaCl, 10% glycerol, 10 mm imidazole). The column was washed extensively with Buffer A, and the target protein was eluted with a linear gradient of imidazole (10–300 mm) in this buffer. The eluted protein was concentrated and purified further via size exclusion chromatography using a HiLoad Superdex S-200 column (GE Healthcare) equilibrated with 20 mm Tris/Cl pH 8.0 containing 1 m NaCl and 10% glycerol. The fractions containing ZCP2 still contained numerous impurities as determined via SDS-PAGE, so selected fractions were pooled, dialyzed against 20 mm Tris/Cl pH 8.0 containing 100 mm NaCl and 10% glycerol (Buffer C), and loaded on a Q-Sepharose column (GE Healthcare) equilibrated with this buffer. Bound proteins were eluted with a linear gradient of NaCl (0.1–0.5 M) in Buffer C. Fractions were analyzed via SDS-PAGE, and those containing pure ZCP2 were pooled and concentrated. The purified protein was used directly for antiserum production in rabbits (service provided by Covance Inc., Denver, PA).
Immunoblot Analysis
Immunoblot analysis for Cgl78/Ycf54, coprogen oxidase, ferredoxin, and plastocyanin was performed as described elsewhere (11); for Cgl78/Ycf54, SDS was omitted from the transfer buffer, and the transfer time was reduced to 45 min. Primary antibodies were used at the following dilutions: anti-Cgl78/Ycf54 (1:1,000), anti-coprogen oxidase (1:1,000) (44), anti-ferredoxin (1:1,000), and anti-plastocyanin (1:1,000). Plastocyanin was also immunodetected via chemiluminescence as described elsewhere (15) using primary antibody at a 1:1,000 dilution in blocking buffer.
Transmembrane Domain Prediction
A hidden Markov model based algorithm, TMHMM, was used to predict the presence of transmembrane domains in the Chlamydomonas proteome (45). The algorithm was accessed via the Web site of the Center for Biological Sequence Analysis.
RESULTS
Chlamydomonas Cultures Showed Clear Phenotypes for Metal Deficiency
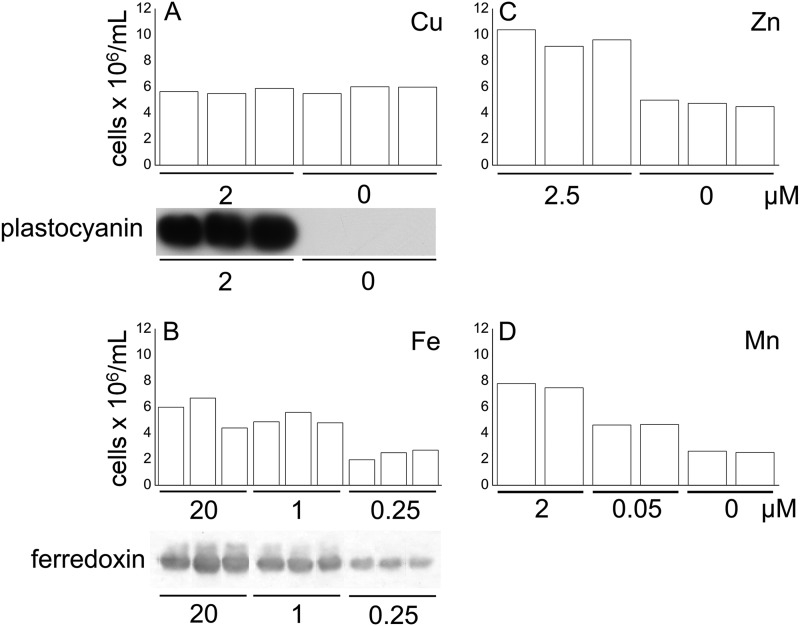
For each deficiency situation, we validated the establishment of deficiency and the physiology of the culture based on phenotype and the expression of sentinel genes as noted in previous studies (18, 20, 46). In each case, we noted a pattern of growth essentially identical to those described previously for each condition (18, 20, 46) (Fig. 1). The accumulation of plastocyanin in the copper-replete cells combined with its absence in the copper-depleted cells and the presence of ferredoxin in iron-replete cells versus its reduced abundance in the iron-depleted cells (Figs. 1A and 1B, respectively) are consistent with previous reports (15, 23). Proteins were then isolated from each condition, separated via gel electrophoresis, and identified and quantified as described in the “Experimental Procedures” section.
Fig. 1.
Verification of the trace metal nutritional status. Cell density was measured in (A) copper-, (B) iron-, (C) zinc-, and (D) manganese-replete and -deficient cells, as described elsewhere (18, 20, 46). Immunoblots showing the abundance of plastocyanin and ferredoxin were also used to indicate nutritional status (A, B).
Au10.2 Gene Model–based Protein Database Outperforms FM 3.1 and FM 4 Databases
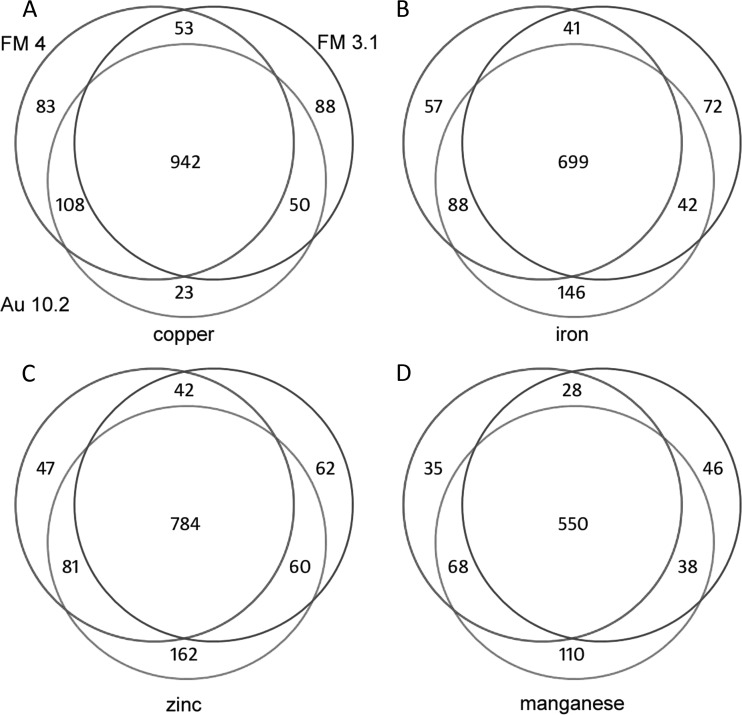
Three different gene models (FM 3.1, FM4, and Au10.2), based on two genome assemblies, are available. Because protein sequence data were not considered in the generation of these gene model sets, we reasoned that proteomics data could provide an independent means to assess the accuracy of each gene model set. To determine which gene model set was best, we systematically compared results from each database qualitatively and quantitatively.
In general, protein quantifications were not significantly affected by our sequence database selection (Table I). Fewer than 6% of all proteins measured showed a significant difference in their abundances when analyzed using Au10.2, FM3.1, or FM4 gene model set databases. The average change in abundance arising from database selection was found to be less than the biological variation among replicates (Table I). Thus, database selection does not affect whether a protein shows a statistically significant difference between growth conditions. Also, the qualitative examination of protein identifications determined in the three database searches showed a large overlap. Fig. 2 indicates that there is ∼75% to 80% overlap in the identified proteins from each of the three databases across the four metal deficiency experiments. Despite the similarities, in three out of four cases, Au10.2 provided ∼10% more identifications on average than FM4 and 16% more than FM3.1. Considering this, it seems that the Au10.2 gene model sets outperform both FM3.1 and FM4 databases, and this suggests that Au10.2 provides the most accurate gene models of those considered. As a result, we chose to use the Au10.2 model for the full analysis of the proteomics data. In addition, because the FM3.1 model arises from a different assembly, we acknowledge that it contains gene models that do not exist in any version 4 assembly model set. Therefore, if a protein was uniquely identified using the FM3.1 database, it was retained in our study.
Table I. For each metal condition, protein quantification was database independent. Only proteins identified in all three databases were compared under each condition.
| Micronutrient status | Number of significant changesa | Total number of proteins comparedb | Percentage of significant changes | Average % changec | |
|---|---|---|---|---|---|
| Metal | μm in medium | Au10.2 vs. FM3.1 | |||
| Cu | 0 | 24 | 839 | 2.9 | 16 |
| 2 | 20 | 782 | 2.6 | 17 | |
| Zn | 0 | 31 | 692 | 4.5 | 16 |
| 2.5 | 15 | 625 | 2.4 | 15 | |
| Fe | 0.25 | 14 | 434 | 3.2 | 14 |
| 1 | 17 | 534 | 3.2 | 14 | |
| 20 | 21 | 553 | 3.8 | 14 | |
| Mn | 0 | 2 | 418 | 0.5 | 14 |
| 0.05 | 8 | 404 | 2.0 | 14 | |
| 2 | 6 | 408 | 1.5 | 16 | |
| Au10.2 vs. FM4 | |||||
| Cu | 0 | 12 | 833 | 1.4 | 12 |
| 2 | 14 | 791 | 1.8 | 13 | |
| Zn | 0 | 21 | 692 | 3.0 | 15 |
| 2.5 | 7 | 625 | 1.1 | 14 | |
| Fe | 0.25 | 8 | 434 | 1.8 | 14 |
| 1 | 11 | 534 | 2.1 | 13 | |
| 20 | 31 | 553 | 5.6 | 18 | |
| Mn | 0 | 4 | 418 | 0.96 | 16 |
| 0.05 | 11 | 404 | 2.72 | 15 | |
| 2 | 4 | 408 | 0.98 | 15 | |
| FM3.1 vs. FM4 | |||||
| Cu | 0 | 14 | 839 | 1.7 | 15 |
| 2 | 14 | 796 | 1.8 | 16 | |
| Zn | 0 | 24 | 692 | 3.5 | 17 |
| 2.5 | 15 | 625 | 2.4 | 16 | |
| Fe | 0.25 | 10 | 434 | 2.3 | 16 |
| 1 | 22 | 534 | 4.1 | 16 | |
| 20 | 29 | 553 | 5.2 | 19 | |
| Mn | 0 | 5 | 418 | 1.2 | 18 |
| 0.05 | 10 | 404 | 2.5 | 17 | |
| 2 | 7 | 408 | 1.7 | 19 | |
a Significance determined by Student's t-test with a 95% confidence interval.
b Proteins were selected for comparison only if identified using all three databases.
c Calculated as (Average abundanceDatabase1 − Average abundanceDatabase2)/(Average abundanceall values).
Fig. 2.
FM3.1, FM4, and Au10.2 database searches identify many of the same proteins. Venn diagrams showing the overlap among identifications for proteins that were detected in two of three biological replicates from each database search for each metal deficiency: A, copper; B, iron; C, zinc; and D, manganese. Please see panel A for labeling of the different databases.
Overview of Proteomics Data
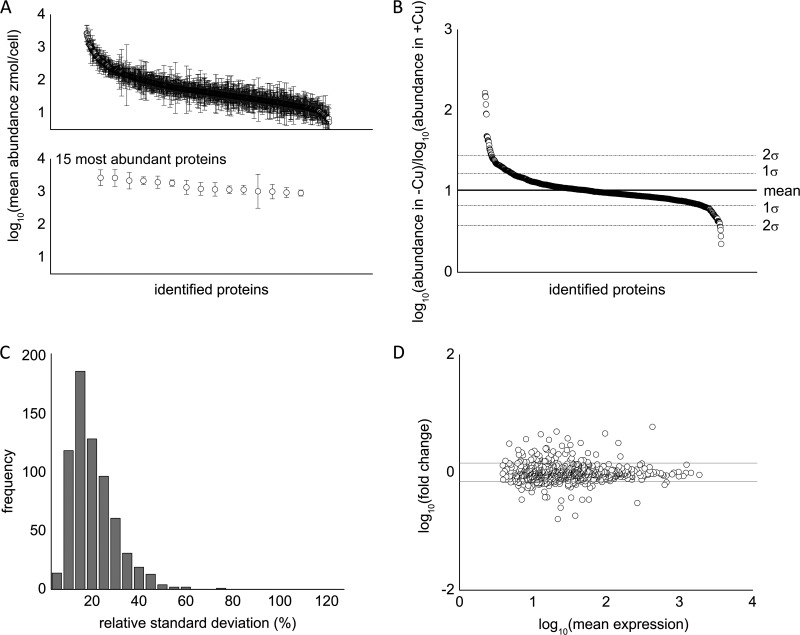
Over 2,000 proteins were identified among all of the metal deficiency studies (Table II, supplemental Table S6). Because we required that a protein be detected in at least two of three biological replicates in each study, the number of high-confidence proteins identified was reduced to ∼1,000 on average, ranging from over 1,200 in the copper study to slightly less than 900 identified in our manganese study, which is on a scale similar to other large proteomes published on Chlamydomonas. We compiled a list of all proteins detected in at least three out of four of our control growth conditions (replete for all metals) and determined the average abundance for each protein in this list. The measured abundances of proteins showed a dynamic range of 3 to 4 orders of magnitude (Fig. 3A), which is consistent with other dynamic range estimates using the label-free MSE proteomics strategy (39). A representative plot showing the mean abundance of all proteins in a dataset, along with a cutoff of 1 standard deviation, shows that ∼80% of the proteins did not show a statistically significant change in abundance in response to nutritional deficiency (Fig. 3B). Furthermore, 80% of the proteins showed a relative standard deviation in abundance of less than 25%, with an average relative standard deviation of 18% throughout the whole dataset (Fig. 3C). We also examined reproducibility by plotting the fold change against the average abundance (Fig. 3D). In general, data from all four experiments show good reproducibility, with less variation observed for highly abundant proteins. The average coefficient of variance of each dataset ranged between 0.28 and 0.30 (vide infra). This compares favorably to other label-free techniques, such as the Exponentially Modified Protein Abundance Index, which has an error of ∼60% (47).
Table II. A survey of proteomics studies in Chlamydomonas and the number of proteins identified.
| Source | Number of proteins identified |
|---|---|
| Mühlhaus et al., 2011 (33) | 3,433 |
| Terashima et al., 2010 (29) | 2,315 |
| Hsieh et al., 2012 (91) | 2,250 |
| Atteia et al., 2009 (24) | 496 |
| Förster et al., 2006 (30) | 444 |
| Wagner et al., 2006 (27) | 328 |
| Naumann et al., 2007 (17) | 233 |
| Schmidt et al., 2006 (92) | 202 |
| Keller et al., 2005 (25) | 61 |
| Lemaire et al., 2004 (31) | 55 |
| Wagner et al., 2008 (28) | 39 |
| Yamaguchi et al., 2003 (93) | 30 |
| Michelet et al., 2008 (94) | 25 |
| Yamaguchi et al., 2002 (95) | 21 |
Fig. 3.
Statistical overview of reproducibility of the proteomics dataset. A, top: representative log10 ratio of protein abundance of metal-deficient cells versus metal-replete cells. Approximately 80% of identified proteins are within 1 standard deviation of the mean change in abundance. Bottom: the top 15 most abundant proteins were identified and are, from left to right, eukaryotic translation elongation factor 1 α 3 (Cre06.g263450), eukaryotic translation elongation factor 1 α 2 (132905), ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (41179049), isocitrate lyase (Cre06.g282800), fructose-1,6-bisphosphate aldolase (Cre05.g234550), S-adenosyl homocysteine hydrolase (Cre03.g204250), cobalamin-independent methionine synthase (Cre03.g180750), 2-cys peroxiredoxin (Cre06.g257601), glyceraldehyde-3-phosphate dehydrogenase (Cre01.g010900), enolase (Cre12.g513200), pre-apoplastocyanin (Cre03.g182551), ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (108283), oxygen-evolving enhancer protein 2 (Cre12.g550850), phosphoenolpyruvate carboxykinase (Cre02.g141400), and unnamed protein (Cre12.g528000). B, representative plot of the ratio of protein abundance between copper-deficient and copper-replete cells. Roughly 20% of the data lie outside 1 standard deviation, and only 5% of data lie outside 2 standard deviations. C, histogram of the relative standard deviation (RSD). The average %RSD was 18%. D, representative mean difference scatterplots after pooling biological replicates from metal-deficient and metal-replete conditions.
Our criteria were that the difference in protein abundance between growth conditions must be statistically significant (p < 0.05 by Student's t test) and at least 2-fold or greater in magnitude in order to define a change in protein abundance. It should be noted that we maintained some proteins in our analysis that did not show a 2-fold change. This was done only if the change in protein abundance was statistically significant and if changes to the abundance of the protein in question helped to provide additional insight into the effect of the metal deficiency on a pathway. Instances in which this was done are clearly indicated in supplemental Tables S1–S4. Additionally, proteins that were detected in only one of the conditions could not be assigned statistical significance and therefore were included in the analysis only if their abundances were over 20 zmol/cell. We also included some proteins previously examined as part of a copper deficiency transcriptome study (13) in our analysis for completeness.
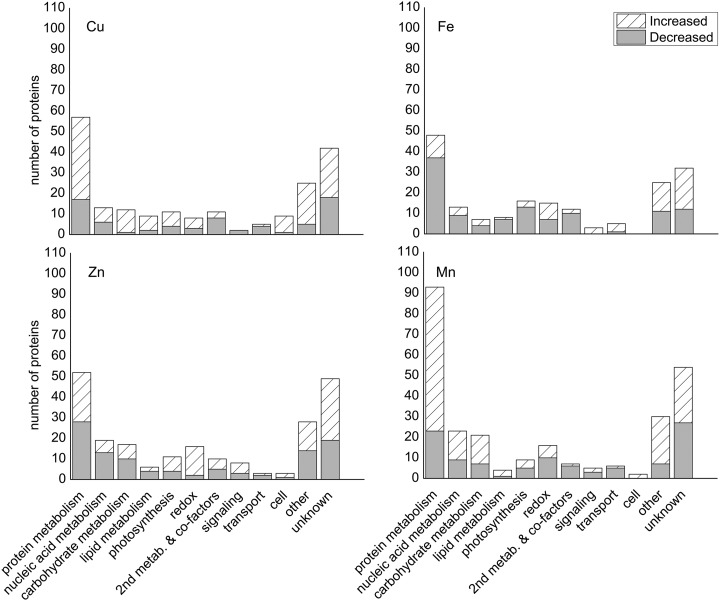
Based on these criteria, we identified 204 proteins that changed in abundance under copper deficiency, 184 in iron limitation, 222 in zinc deficiency, and 270 in manganese limitation, suggesting that only 20% of proteins analyzed changed in our deficiency studies. The proteins that changed under each condition were distributed among several functional categories, with the “protein metabolism” and “unknown” categories being the most well represented (Fig. 4). We estimated our false discovery rate by randomly permuting the data 1,000 times and comparing the number of changes observed in the permutated runs to the number of changes in the real experiment. Using this method, we estimated a 20% false discovery rate in determining when a change in abundance occurred for our data.
Fig. 4.
Distribution of proteins among functional groups for each metal deficiency. Functional distribution of proteins changed under (A) copper, (B) iron, (C) zinc, and (D) manganese deficiency. Functional group definitions are described in the “Experimental Procedures” section.
Using TMHMM, we examined the prevalence of transmembrane domains in our proteomics dataset. The TMHMM method, when tested on a set of 645 proteins with known three-dimensional structures, was able to accurately discriminate between soluble and membrane proteins with an accuracy of 99% (45). In our dataset, between 5% and 6% of all proteins in each study were found to contain at least a single transmembrane domain; this is in comparison to the 18% of all proteins in the genome with a single transmembrane domain. This suggests a good degree of enrichment of soluble proteins in our sample, as expected from the experimental design. However, some membrane-associated proteins that are important to metal homeostasis in Chlamydomonas are still detectable in our study. For example, ferroxidase is a copper-containing protein involved in high-affinity iron uptake that has a single transmembrane domain between amino acids 47 and 68 (48). We detected several peptides from this protein in our study, suggesting that some degradation occurs during processing that allows us to estimate the abundance of the protein despite it being membrane bound.
As an additional method to assess the accuracy of our quantification measurements, we examined the stoichiometry of proteins from the same complex. We examined the Rubisco large and small subunits, the 20S proteasome α subunit complex, and the CF1 ATP synthase α and β subunits; these represent a highly abundant soluble complex, a low-abundance soluble complex, and a membrane-associated complex, respectively. Each of the subunits in these complexes should be present in a 1:1 ratio with the other proteins from the same complex mentioned in this assessment (49–51). In general, we observed good agreement between the expected and experimentally determined ratios (Table III).
Table III. Stoichiometry for Rubisco, CF1, and 20S proteasome α complexes.
| Protein | Average abundance (zmol/cell) | Ratio |
|---|---|---|
| RbcL | 3,253 | – |
| RbcS (1 + 2) | 2,185 | 0.7 |
| CF1 α | 395 | – |
| CF1 β | 303 | 0.8 |
| 20S Proteasome α subunit A | 21 | 1.0 |
| 20S Proteasome α subunit B | 23 | 1.0 |
| 20S Proteasome α subunit C | 22 | – |
| 20S Proteasome α subunit D | 20 | 0.9 |
| 20S Proteasome α subunit E | 38 | 1.8 |
| 20S Proteasome α subunit F | 15 | 0.7 |
| 20S Proteasome α subunit G | 34 | 1.5 |
Rubisco large subunit, CF1 α, and 20S proteasome α subunit C were used as the reference for each protein complex, respectively. Subunit ratios between all proteins within each complex are expected to be 1:1.
Validation of Protein Abundance Changes
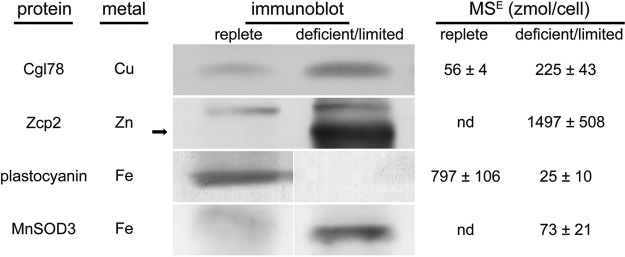
We compared our quantitative proteomics data to previously published work describing several known responses to each micronutrient deficiency condition. We focused on proteins that are either primarily localized to the soluble fraction or, in the case of the oxygen evolving enhancer proteins (Oee), easily solubilized from the membrane-associated fraction. Of the proteins selected, we observed agreement in the direction of the change in 19 of 20 instances across the four different metal deficiency conditions examined (Table IV). Four metal nutrition-dependent changes in protein abundance that had not been previously demonstrated in Chlamydomonas were corroborated by means of immunoblot analysis (Fig. 5). The proteins selected for this validation included Zcp2 (in the zinc study), Cgl78 (in the copper study), MnSOD3 (in the iron study), and plastocyanin (in the iron study) and are described in more detail in the sections below. It should be noted that the immunoblot results were from samples that are independent of the samples used for proteomics, thus validating the reproducibility of the biology.
Table IV. Proteomics discoveries are validated in the literature. Quantification via MSE-based proteomics shows agreement with several previously reported responses of Chlamydomonas to various metal-deficiency growth conditions.
| Model IDa | Gene name | Description | Experimental conditionb | Δc | Evidenced | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Cu | R | D | – | |||||
| Cre03.g182551 | PCY1 | Plastocyanin | 1271 ± 200 | n.d. | – | ↓ | IB | 23 |
| Cre16.g651050 | CYC6 | Cytochrome c6 | n.d. | 283 ± 41 | – | ↑ | IB, RNA-Seq | 13, 23 |
| Cre02.g085450 | CPX1 | Coprogen III oxidase | 94 ± 43 | 940 ± 136 | – | ↑ | IB, RNA-Seq | 13, 44 |
| Cre09.g393150 | FOX1 | Ferroxidase | 13 ± 5 | n.d. | – | ↓ | IB | 44, 48 |
| Cre17.g700950 | FDX5 | Ferredoxin 5 | n.d. | 7 ± 2 | – | ↑ | IB, RNA-Seq | 13, 15 |
| Zn | R | D | L | |||||
| Cre04.g223100 | CAH1 | Carbonic anhydrase | 171 ± 81 | n.d. | n.m. | ↓ | qPCR | Unpublished data |
| 117458 | ZCP1 | COG0523 domain | n.d. | 1588 ± 226 | n.m. | ↑ | qPCR | 21 |
| Fe | R | D | L | |||||
| Cre09.g387800 | FER1 | Pre-apoferritin | 22 ± 2 | 68 ± 15 | 53 ± 11 | ↑ | IB, qPCR | 16 |
| Cre12.g546550 | FEA1 | Fe-assimilating protein | 124 ± 21 | 735 ± 34 | 1356 ± 115 | ↑ | IB, qPCR | 14 |
| Cre12.g546600 | FEA2 | Fe-assimilating protein | n.d. | 97 ± 16 | 512 ± 77 | ↑ | IB, qPCR | 14 |
| Cre14.g626700 | PETF | Ferredoxin | 87 ± 35 | 30 ± 27 | n.d. | ↓ | IB, qPCR | 15 |
| Cre06.g306350 | FDX3 | Ferredoxin 3 | n.d. | n.d. | 45 ± 7 | ↑ | IB | 15 |
| Cre09.g393150 | FOX1 | Ferroxidase | n.d. | 11 ± 1 | 25 ± 21 | ↑ | IB | 48 |
| Cre16.g676150 | MSD3 | Mn superoxide dismutase | n.d. | 10 ± 3 | 73 ± 21 | ↑ | A, IB, qPCR | 19, 20 |
| Cre06.g257601 | PRX1 | 2-Cys peroxiredoxin | 839 ± 77 | 834 ± 70 | 1042 ± 54 | ↑ | SILAC, IB | 17 |
| Mn | R | D | L | |||||
| Cre02.g096150 | MSD1 | Mn superoxide dismutase | 48 ± 48 | n.d. | n.d. | ↓ | A, qPCR | 20 |
| Cre16.g676150 | MSD3 | Mn superoxide dismutase | n.d. | n.d. | 27 ± 3 | ↑ | qPCR | 20 |
| Cre02.g132800 | PSBO | Oxygen-evolving enhancer protein 1 | 1056 ± 276 | 1235 ± 298 | 1730 ± 447 | ↑ | IB | 20 |
| Cre12.g550850 | PSBP1 | Oxygen-evolving enhancer protein 2 | 1589 ± 176 | 2296 ± 259 | 2013 ± 817 | ↑ | IB | 20 |
| Cre08.g372450 | PSBQ | Oxygen evolving enhancer protein 3 | 655 ± 120 | 807 ± 281 | 788 ± 360 | ↑ | IB | 20 |
Protein abundance is reported in the “Experimental Condition” column in zmol/cell.
n.d., not detected; n.m., not measured; ↓, decrease; ↑, increase; IB, immunoblot; A, activity; SILAC, stable isotope labeling of amino acids in cell culture.
a Reported by Augustus 10.2 identifiers or, if an Augustus 10.2 model does not exist, by FM3.1 identifiers.
b R, metal replete; D, metal deficient; L, metal limited.
c Response reported as the change observed in the micronutrient-deficient condition relative to the replete status.
d Type of evidence available.
Fig. 5.
Changes in protein abundance measured by MSE-based quantification are verified by immunoblot analysis. Protein abundance from metal-replete (left columns) cells and from metal-deficient/limited (right columns) cells compared by means of both immunoblot analysis and MSE-based quantification.
RNA and Protein Abundance Correlations Are Similar to Those Noted for Other Organisms
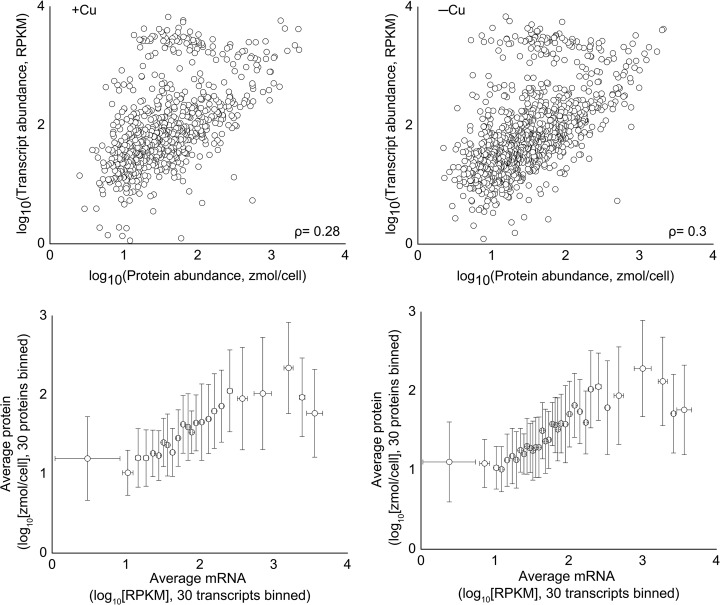
Taking advantage of the availability of both a large proteomics dataset and a recently published transcriptomics dataset (13), we sought to determine the level of correlation between RNA and protein abundance under standard growth conditions (for the dataset acquired from the copper nutrition study). We assessed the correlation between protein and RNA abundance by calculating the Pearson correlation coefficient (ρ) of the data, where a ρ of +1 represents a perfect positive linear relationship; ρ = +0.30 was calculated for our protein-RNA abundance data for the copper study (Fig. 6, top). To minimize the effects of noise and the dynamic range differences between the transcriptomics and proteomic datasets (5 to 6 orders of magnitude and 3 to 4 orders of magnitude, respectively) and the differences in vivo, the data were binned as described by Lu et al. (52) and as shown in Fig. 6 (bottom). We also determined the Pearson coefficient between the proteomic and transcriptomics datasets for our copper-deficient samples and found a similar correlation (ρ = +0.28), suggesting that this overall correlation is not dependent on the nutritional state. However, transcripts that changed significantly under copper deficiency showed a much stronger correlation to their corresponding proteins (ρ = +0.73). This behavior is similar to that observed in an earlier study with Drosophila in which the correlation between the change in protein and transcript abundance improved when limiting the dataset to include only transcripts that changed significantly (53). These results suggest that although RNA abundance is only a moderate predictor of protein abundance, changes in RNA abundance are often good indicators of changes in protein abundance.
Fig. 6.
RNA and protein levels are moderately correlated under steady state conditions. (Top row) RNA and protein abundance were log transformed then compared via scatter plot. The Pearson correlation was also calculated for each dataset. (Bottom row) Data were sorted by RNA abundance and binned in groups of 30. For the +Cu data, R2 = 0.78, and for −Cu, R2 = 0.77.
Copper Responsive Proteome
In Chlamydomonas, there are three abundant copper enzymes: plastocyanin, which is involved in photosynthetic electron transfer between the two photosystems and is the most abundant copper-protein in Chlamydomonas; cytochrome oxidase, which is utilized in respiration; and ferroxidase, which is involved in high-affinity iron assimilation. In addition, copper is also found in matrix metalloproteases, in amine oxidases, in blue copper proteins, as a transitory place-holder for molybdenum in MoCo biosynthesis enzymes, and in a variety of other proteins (3, 13, 54). Chlamydomonas can survive copper deficiency (Fig. 1A) because of its ability to replace plastocyanin with cytochrome c6, a copper-independent substitute (23). In addition, a copper-containing amine oxidase may be replaced by a flavin-containing amine oxidase (13). These and other responses to copper deficiency are controlled by the transcription factor CRR1 (9).
Previously, we examined responses to copper deficiency at the transcript level (13) and used proteomics data to validate some of the copper nutrition-dependent changes. We found that 17 of 18 changes observed at the transcript level were recapitulated in the proteome (supplemental Table S1). Here we summarize some of those previous findings and describe additional changes at the protein level. These additional changes either complement changes observed at the transcript level or demonstrate responses that were not observed based on transcriptome analysis alone.
Recent transcriptome studies have revealed that there is a copper-deficiency induced increase in the abundance of HYD1, HYDEF, HYDG, HCP2, HCP3, and PFR1 transcripts, which are related to anaerobic responses (12, 13). Proteins that correspond to these transcripts, however, cannot function in aerobic conditions because their active site clusters are O2 labile. We hypothesized in prior work that these transcripts are mis-expressed because copper deficiency results in faulty O2 sensing (13). Therefore, we asked the question of whether the increase in transcript levels also resulted in the accumulation of these proteins. We were able to detect three of these gene products—Hyd1, Pfr1, and Hcp2—and found that they were all increased under copper-deficiency conditions. This shows that the expression of these proteins is controlled at the level of their transcript abundance, regardless of whether the protein can actually function in these conditions. Another subset of anaerobically controlled genes that are also copper regulated encodes prolyl 4-hydroxylases (55). Transcriptome evidence also indicated that five genes for prolyl 4-hydroxylases showed increased expression in copper-deficient conditions, of which four are CRR1 targets. We examined the proteome to uncover any evidence of copper responsiveness of prolyl 4-hydroxylases at the protein level. Although we could not detect any of the gene products from the five copper-responsive genes listed in the previous study, we identified three other prolyl 4-hydroxylases (Phx7, Phx13, and Phx18) whose abundances are increased under copper-deficient growth conditions (Table V). However, an accurate magnitude of the change could not be assessed because the proteins were detected only in copper-deficient conditions, and only one of them, Phx18, accumulated to a level that met our cutoff criterion of 20 zmol/cell (see “Experimental Procedures”). Nevertheless, this study indicates the operation of post-transcriptional mechanisms that impact prolyl hydroxylases in Chlamydomonas reinhardtii in addition to the previously described transcriptional response.
Table V. Proteins with altered accumulation under copper deficiency and implicated in anaerobic acclimation.
| Protein ID (Au10.2) | Gene name | Description | Protein amount (zmol/cell) |
Fold changea | |
|---|---|---|---|---|---|
| +Cu | −Cu | ||||
| Fermentation pathway enzymes | |||||
| Cre20.g758200 | ADH1 | Alcohol/acetaldehyde dehydrogenase | 114 ± 13 | 217 ± 38 | 2 |
| Cre02.g129550 | HCP2 | Hybrid-cluster protein | n.d. | 15 ± 8 | n/a |
| Cre01.g044800 | PFL1 | Pyruvate-formate lyase | 280 ± 95 | 475 ± 75 | 2 |
| Cre03.g199800 | HYDA1 | Iron hydrogenase | n.d. | 22 ± 24 | n/a |
| Cre09.g396650 | PAT2 | Phosphate acetyltransferase | n.d. | 34 ± 5 | n/a |
| Cre11.g473950 | PFR1 | Pyruvate-ferredoxin oxidoreductase | n.d. | 88 ± 43 | n/a |
| Prolyl 4-hydroxylases | |||||
| Cre03.g160200 | PHX7 | Prolyl 4-hydroxylase | n.d. | 2 ± 1 | n/a |
| Cre10.g424900 | PHX13 | Prolyl 4-hydroxylase | n.d. | 13 ± 9 | n/a |
| Cre14.g626200 | PHX18 | Prolyl 4-hydroxylase | n.d. | 23 ± 13 | n/a |
Fermentative pathway enzymes have been described elsewhere (29).
n.d., not detected; n/a, not applicable.
a Calculated as the ratio of protein abundance in −Cu versus the protein abudance in +Cu.
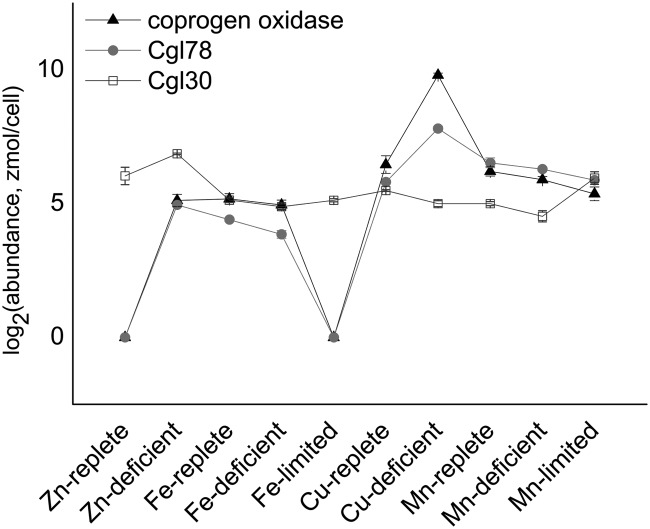
The expression of genes CPX1, CRD1, and CAO for three enzymes in the tetrapyrrole pathway is increased with copper deficiency (13, 56, 57). In the present study, only the CPX1 gene product was detected, as the other two proteins are membrane associated (57). The abundance of coprogen oxidase increased, in accordance with a prior study that demonstrated increased enzyme activity and immunoreactive material (Table II) (56). In addition to coprogen oxidase, we also observed changes to three other enzymes of the tetrapyrrole biosynthetic pathway: urogen decarboxylase isoforms 1 and 2, and PBG deaminase 1 (Cre11.g467700, Cre02.g076300, and Cre16.g663900, respectively) (supplemental Table S1). In copper deficiency, we observed a decrease in isoform 1 of urogen decarboxylase but an increase in isoform 2. A total of three well-expressed genes encode urogen decarboxylase isozymes, but the pattern of expression of each gene is not known, nor is it known whether there are functional differences in the corresponding gene products. It is possible that this represents a compensatory change and thus is the first indication of how the urogen decarboxylase isozymes might be regulated relative to each other. Again, these findings suggest that in addition to CRR1-dependent transcriptional mechanisms that impact tetrapyrrole metabolism, there are also mechanisms that operate post-transcriptionally.
Besides our results relating copper nutrition and anaerobiosis and the effect of copper on tetrapyrrole biosynthesis, we observed three additional effects of copper deficiency in Chlamydomonas. One such effect was a decrease in copper deficiency of three isoprenoid pathway enzymes (supplemental Table S1)—Pps1, Pps2, and Pps4—that are responsible for the synthesis of farnesyl pyrophosphate, geranylgeranyl pyrophosphate, and solanesyl pyrophosphate, respectively. Pps1 was decreased 2-fold, Pps2 was decreased 3-fold, and Pps4 was not detected (less than 1.7 zmol/cell) in copper-deficient cells. These enzymes are involved in pathways leading to the synthesis of sterols, quinones, phytol, and prenylated proteins. It should be noted that these proteins are believed to be membrane associated, and therefore it is difficult to determine whether a decrease in these proteins represents an actual decrease in their total abundance or results from altered association with the membrane, perhaps due to increased isoprenoid pathway activity. A second observation in copper deficiency was the decrease of two enzymes in the S-adenosylmethionine (SAM) biosynthesis pathway, 5,10-methylenetetrahydrofolate reductase (from 293 zmol/cell to 180 zmol/cell) and cobalamin-independent methionine synthase (from 999 zmol/cell to 525 zmol/cell) (Cre10.g433600 and Cre03.g180750, respectively). At the same time, a third member of the pathway, serine hydroxymethyltransferase isoform 1, was increased 3-fold (supplemental Table S1). These results are interesting because the accumulation of another enzyme in the pathway, S-adenosylhomocysteine hydrolase, had been previously shown to be copper dependent, and copper is a non-competitive inhibitor of the enzyme in mouse liver (58, 59). Although S-adenosylhomocysteine hydrolase was detected in our proteomics study, we did not observe a change in its abundance, which contrasts with results from the mouse model. Lastly, our proteomics study also showed that two selenocysteine tRNA synthases (encoded by TSS1, Cre02.g082850 and TSS2, Cre03.g189400) and a selenium binding-protein (SBD1, Cre03.g185550) are decreased in copper deficiency by ∼2-fold (supplemental Table S1). Previous studies have shown that selenium-containing glutathione peroxidase activity levels are reduced in copper-deficient rats and chicken relative to animals given a copper-replete diet (60, 61). In the rat study, it was further shown that this might have resulted from a reduced ability to retain selenium in the cell (61). Although the connections between copper and selenium metabolism have not been established firmly in algae, these data suggest that there might well be one in Chlamydomonas.
Iron Responsive Proteome
A prior study of iron deficiency in Chlamydomonas detailed three distinct nutritional states (18). With 20 μm Fe in the growth medium, Chlamydomonas cells show normal growth and are considered as iron replete. Iron-deficient (1–3 μm) cells do not show visible phenotypes but are identified by increases in the expression of several genes encoding components of high-affinity iron uptake (48). In addition to these biochemical markers, iron-limited Chlamydomonas cells are chlorotic and growth inhibited (18). Fluorescence rise and decay kinetics indicate that the iron-deficient cultures are mildly affected with respect to PS I function, whereas iron-limited cells have lost PS II and PS I function (18). To assess the changes to the proteome of cells in these nutritional states, we compared the proteome of wild-type cells grown photoheterotrophically in the presence of 20 μm, 1 μm, and 0.25 μm supplemental iron and collected from batch cultures at a density of ∼5 × 106 cells/ml. Growth of the cells was found to be similar to what was previously reported (18), and the accumulation of ferredoxin also closely followed values in the published literature (15) (Fig. 1B).
One of the most apparent effects of iron deficiency/limitation in Chlamydomonas is that on the photosynthetic apparatus (17, 18, 62). Other studies describe a remodeling of PS I, in which light harvesting complexes are disconnected from photosystem components (18), and previous proteomics study showed that there was also a decrease in several photosystem I and light harvesting complex proteins (62), but little change was observed for proteins involved in respiration or related to PS II function (17). These earlier studies, however, were limited to the examination of enriched chloroplast fractions with no evaluation of iron-deficiency/limitation-induced effects on metabolism and/or the entire system. Our study demonstrates additional effects of iron deficiency/limitation that have not been described previously.
Interestingly, functional analysis showed that a large subset of proteins classified under the “photosynthesis” category is decreased in abundance (Fig. 4, supplemental Table S2). This included LciB (Cre10.g452800) and LciC (Cre06.g307500), representing components of inorganic carbon assimilation; plastocyanin (Cre03.g182551), Oee1 (Cre02.g132800), and Oee3 (Cre08.g372450), proteins associated with light reactions; and Calvin cycle enzymes Pgk1 (Cre22.g763250) and Prk1 (Cre12.g554800). Many of the changes to these proteins are ∼2-fold (Oee1, Pgk1, and Prk1) or 3-fold (LciB and Oee3). LciC was detected at 41 zmol/cell in iron-replete cells, but it was not detected in iron-limited cells. Plastocyanin was reduced significantly from 797 zmol/cell in iron-replete conditions to 25 zmol/cell in iron-limited conditions, a change that was confirmed by immunoblot analysis of independent samples (Fig. 5). These results indicate that the previously noted changes in thylakoid membrane protein abundance extend also to stromal and lumenal proteins of the photosynthetic apparatus (17, 63). It is worth noting that only Oee3 and Prk1 decrease under iron deficiency (1 μm Fe), and even then less than a 2-fold change was observed relative to iron-replete cells (20 μm Fe). All other proteins described above were decreased only under iron-limitation conditions (0.25 μm Fe). Therefore, it is likely that the Calvin cycle, carbon-concentrating mechanism (CCM), and light reactions are affected only when the cells are growth-limited by the absence of Fe.
Previously, we demonstrated that changes in the abundance of thylakoid membrane proteins resulted from induced proteolysis (18). Consistent with this earlier result, four proteases were found whose abundance increased in iron deficiency/limitation, including two matrix metalloproteases (isoforms 1 and 13, Cre17.g718500 and Cre60.g792000, respectively), a pepsin-type aspartyl protease (Cre04.g226850), and an unnamed protease (Cre17.g728100) (supplemental Table S2). Reduced protein synthesis is likely to be a contributing factor, as we noted a decrease in 11 amino acid metabolism proteins and seven tRNA synthetases (supplemental Table S2). There also was a concomitant decrease in the abundance of urogen decarboxylase isozyme 1, PBG deaminase 1, and coprogen oxidase; all three are enzymes in the tetrapyrrole pathway, and this is consistent with the reduction in chlorophyll and heme proteins in this condition. This might occur as a flux control step so that iron can be preserved for other purposes such as oxidative stress defense in iron-containing superoxide dismutase.
Because PS I is a prime target of iron deficiency because of the presence of three Fe4S4 clusters, its compromised function in this situation is likely to result in elevated superoxide production via the Mehler reaction (18, 62). Superoxide generated by iron-less PS I may be detoxified via a plastid-localized MnSOD, with the resulting hydrogen peroxide further detoxified by peroxidases, which rely on either glutathione or ascorbate. Monodehydroascorbate reductase functions to regenerate ascorbate from monodehydroascorbate. In fact, the ascorbate-glutathione pathway has been proposed as a major detoxification pathway of superoxide (64). An iron-deficiency/limitation-inducible MnSOD activity has been previously observed and attributed to MnSOD3, based on the accumulation of the MSD3 transcript under the same conditions and immunoblotting with an MnSOD3-specific antibody (19, 20). Not surprisingly, chloroplast anti-oxidant mechanisms are up-regulated in the iron-deficient/limited stress situation. MnSOD3 is undetectable in iron-replete cells, but its levels increase to 10 zmol/cell in iron-deficient cells and 73 zmol/cell in iron-limited cells (Fig. 5). Our protein data therefore support the association of inducible superoxide dismutase activity to MnSOD3 levels (Fig. 5). Similarly, monodehydroascorbate reductase 1 increases from 70 zmol/cell to 224 zmol/cell to 1,234 zmol/cell, and glutathione synthetase 1 increases from 34 zmol/cell to 45 zmol/cell to 95 zmol/cell (from iron replete to iron deficient to iron limited, respectively) (supplemental Table S2).
In addition, Hsp 70B (supplemental Table S2), a protein that has been previously shown to act as a photoprotectant and which is involved in PS II repair in Chlamydomonas, increases in iron-limited cells to 1,216 zmol/cell from 466 zmol/cell in iron-replete cells (30, 65). Schroda et al. showed that strains over-expressing Hsp 70B had reduced rates of PS II photoinhibition and that Hsp 70B under-expressing strains showed slower rates of PS II repair relative to wild-type (65). Although iron-limited cells prepare for photooxidative damage via PS I remodeling, it is noteworthy that additional mechanisms may be employed. Interestingly, although two components of the ascorbate-glutathione pathway (monodehydroascorbate reductase and MnSOD3) are increased already in iron deficiency, changes in the abundance of Hsp 70B do not occur until iron-limitation conditions are sensed. This might further suggest that the ascorbate-glutathione pathway is the primary anti-oxidant defense mechanism in response to decreased iron availability.
We also observed an apparently non-photosynthesis-related effect of iron deficiency/limitation. Four components of the SAM synthesis pathway—5,10-methylenetetrahydrofolate reductase, cobalamin-independent methionine synthase, serine hydroxymethyl transferase isoform 2, and S-adenosylhomocysteine hydrolase (Cre03.g204250)—were decreased under iron-limited conditions by small but statistically significant amounts. The effect of iron nutrition on SAM metabolism might be based on the activity of adenosylmethionine-dependent Fe4S4 cluster proteins. One such Fe4S4 cluster protein is biotin synthase, the transcripts of which are known to be reduced under iron deficiency in Saccharomyces cerevisiae. The decrease in biotin synthase transcripts is accompanied by an increase in VHT1, encoding for a high-affinity biotin transporter, presumably as a mechanism of iron sparing (66). Additionally, other adenosylmethionine-dependent Fe4S4 cluster proteins such as pyruvate formate lyase activase, fumarate-nitrate reductase, and others described by Cheek and Broderick (67) may be affected in a similar way. Thus, the possibility exists that iron deficiency might result in a loss of many such iron–sulfur proteins and consequently a reduction in the need for SAM. Unfortunately, we did not detect any radical SAM proteins in our dataset to validate this theory.
Zinc Responsive Proteome
Cells were grown (photo)heterotrophically in TAP medium in triplicate cultures, and their zinc content was reduced by two transfers into growth medium with no supplemental zinc (referred to as first and second “rounds,” respectively). This process reduces the internal zinc content to 33% relative to zinc-replete cultures.2 By analogy to the studies on iron and manganese deficiency, cells from the first round are more mildly deficient than are cells from the second round, with the more depleted Zn cultures showing evidence of a stress response. Therefore, we used protein samples from the first round of growth in zinc-poor medium to distinguish the primary targets. The reduced growth rate of such cultures shows that even a single transfer into zinc-depleted medium is sufficient to disrupt normal cell function and thus trigger a zinc deficiency response (Fig. 1C).
Carbonic anhydrases constitute a major group of proteins that bind zinc. The classical function of the carbonic anhydrases is to interconvert CO2 and bicarbonate to facilitate CO2 assimilation for carbon fixation, and a subclass of these proteins is involved in the CCM, which enables photosynthetic growth under limiting CO2 conditions (68, 69). Of the 12 predicted carbonic anhydrases, only 3 were detected in our dataset: Cah1 (Cre04.g223100), Cah3 (Cre09.g415700), and Cah8 (Cre09.g405750). Cah1 was detected at 171 ± 81 zmol/cell in zinc-replete growth conditions but was not recovered under zinc deficiency (Table IV and supplemental Table S3). Previous work showed that cah1 null mutants in Chlamydomonas do not exhibit severe growth phenotypes, even under low CO2 growth conditions (70). Therefore, carbonic anhydrase 1 is a dispensable enzyme and, as an abundant enzyme, would make a good target for degradation if the cell were to scavenge intracellular zinc for recycling and redistribution (22). In contrast to Cah1, Cah8, which was recently characterized (71), was detected in zinc-deficient cells but not in zinc-replete cells (supplemental Table S3). Cah3 was detected, but its abundance did not change significantly in response to zinc nutrition.
We also observed three proteins that were detected in zinc-deficient proteomes but were not detected under zinc-replete conditions (supplemental Table S3). Each protein was detected at levels near the upper end of the dynamic range of our study (over 1,000 zmol/cell). These three proteins do not have a known function; one is unannotated (Cre07.g352000), and the other two are COG0523 domain-containing proteins (named Zcp1 and Zcp2, for zinc-responsive COG0523 domain-containing proteins). Although ZCP1 (117458) and ZCP2 (Cre02.g118400) transcript abundances were previously shown to be increased with zinc deficiency, this is the first report of a corresponding change in protein abundance dependent on zinc levels. The increase in Zcp2 abundance was confirmed in independent experiments via immunoblot analysis (Fig. 5). The function of COG0523 domain-containing proteins is not well understood, although members of this family are involved in cobalamin biosynthesis, activation of an iron-containing nitrile hydratase, and the bacterial zinc deficiency response (21, 72, 73). Despite the apparent diversity of functions carried out by COG0523 proteins, all known functionalities are related to metal metabolism (21).
Interestingly, 2-fold abundance increases (on average) for 14 redox proteins with zinc deficiency were detected (supplemental Table S3). Of special interest is thioredoxin 5 (Cre09.g391900), which is undetectable in zinc-replete cells but accumulated to 22 zmol/cell in zinc-deficient cells. Several targets of thioredoxin 5, identified by Lemaire et al., are also affected by zinc deficiency (31), including Icl1, Acs3, Gln2, ferredoxin, and Rpn12 (Table VII). Although some are increased and others are decreased, precluding a more precise explanation, it is interesting that so many thioredoxin 5 interacting proteins are affected. It was suggested that thioredoxin 5 might play a role in the insertion of the iron–sulfur cluster into ferredoxin (31). Indeed, this is supported by 3-fold increases in ferredoxin levels and increases in several ferredoxin interacting proteins, ferredoxin-NADP+ reductase, ferredoxin thioredoxin reductase, and ferredoxin-sulfite reductase (Cre11.g476750, Cre03.g193950, Cre16.g693150; supplemental Table S3).
Table VII. Several thioredoxin targets show altered accumulation under zinc deficiency. Proteins identified by Lemaire et al. (31) were also found to change under zinc deficiency.
| Protein ID (Au10.2) | Gene name | Description | Protein amount (zmol/cell) |
Fold changea | |
|---|---|---|---|---|---|
| +Zn | −Zn | ||||
| Cre12.g530650 | GLN2 | Glutamine synthetase | 117 ± 18 | 39 ± 28 | 0.3 |
| Cre06.g282800 | ICL1 | Isocitrate lyase | 2,658 ± 729 | 1,381 ± 243 | 0.5 |
| Cre14.g626700 | PETF | Ferredoxin | 84 ± 58 | 212 ± 53 | 2.5 |
| Cre07.g353450 | ACS3 | Acetyl-CoA synthetase/ligase | 384 ± 95 | 164 ± 70 | 0.4 |
| Cre17.g708300 | RPN12 | 26S proteasome regulatory subunit | 9 ± 4 | 20 ± 5 | 2.2 |
a Calculated as the ratio of protein abundance in −Zn versus the protein abundance in +Zn.
Several CRR1-controlled responses were observed to be affected by zinc deficiency, albeit to a lesser extent than what was observed with copper-deficient cells (Table VI). These responses include a decrease in plastocyanin, an increase in coprogen oxidase, and an increase in Cgl78 (Table IV, Figs. 1 and 5). We also detected Ctr3, a soluble member of the Chlamydomonas CTR3 family, in the zinc-deficient samples, but at low abundance (16 zmol/cell) (11). In addition to the CRR1-controlled responses, additional signs of copper deficiency were observed. Pyruvate phosphate dikinase 1, for example, is increased in zinc deficiency, as it is in copper-deficient cells (supplemental Tables S1 and S3). Two of the components of the anaerobically controlled fermentative pathway discussed previously, Adh1 and Pfl1, were increased in this dataset as well. Other proteins in this pathway (Pat2 and Ack2) did not show any change under zinc deficiency. Lastly, two out of three prolyl 4-hydroxylases (Phx18 and Phx7) that increased with copper deficiency (supplemental Table S1) were also increased with zinc deficiency (from an undetectable amount in zinc-replete cells to 30 zmol/cell and 11 zmol/cell, respectively, in zinc-deficient cells).
Table VI. The zinc-deficient proteome recapitulates in part the copper-deficient proteome.
| Protein IDa | Gene name | Description | +Cu (zmol/cell) | −Cu (zmol/cell) | Fold changeb | +Zn (zmol/cell) | −Zn (zmol/cell) | Fold changeb |
|---|---|---|---|---|---|---|---|---|
| Cre02.g085450 | CPX1 | Coprogen III oxidase | 88 ± 54 | 899 ± 164 | 10 | n.d. | 35 ± 18 | n/a |
| Cre17.g701700 | FAB2 | Plastid acyl-ACP desaturase | 127 ± 17 | 631 ± 62 | 5 | 62 ± 19 | 87 ± 31 | 1.4 |
| Cre12.g490500 | CGL78 | Ycf54, aerobic cyclase subunit | 56 ± 4 | 225 ± 43 | 4 | n.d. | 31 ± 0.7 | n/a |
| Cre12.g546550 | FEA1 | Fe-assimilating protein 1 | 520 ± 64 | 239 ± 172 | 0.5 | 634 ± 144 | 331 ± 167 | 0.5 |
| 536235 | DJ-1/PfpI family | 19 ± 11 | 50 ± 6 | 3 | 21 ± 20 | n.d. | n/a | |
| Cre10.g424750 | PPD1 | Pyruvate phosphate dikinase | 39 ± 17 | 168 ± 30 | 4 | 37 ± 28 | 134 ± 23 | 3.6 |
| Cre03.g182551 | PCY1 | Plastocyanin | 1350 ± 261 | n.d. | n/a | 1458 ± 508 | 725 ± 86 | 0.5 |
| Cre20.g758200 | ADH1 | Alcohol/acetaldehyde dehydrogenase | 114 ± 13 | 217 ± 38 | 2 | 27 ± 16 | 113 ± 29 | 4.1 |
| Cre01.g044800 | PFL1 | Pyruvate-formate lyase | 280 ± 95 | 475 ± 75 | 2 | 172 ± 24 | 239 ± 32 | 1.4 |
| Cre14.g626200 | PHX7 | Prolyl 4-hydroxylase | n.d. | 2 ± 1 | n/a | n.d. | 11 ± 14 | n/a |
| Cre14.g626200 | PHX18 | Prolyl 4-hydroxylase | n.d. | 23 ± 13 | n/a | n.d. | 30 ± 5 | n/a |
n.d., not detected; n/a, not applicable.
a Protein IDs are based on Augustus 10.2 nomenclature, except in cases in which proteins were only identified with FM3.1 database searches.
b The ratio of protein abundance under deficiency versus protein abundance under replete conditions.
Manganese Responsive Proteome
Three growth conditions were examined in the manganese study. In addition to the replete (2 μm supplemental Mn) and deficient (0 μm supplemental Mn) conditions described previously (20), we examined the proteome of cells grown in 0.05 μm supplemental Mn (Fig. 1D). Based on preliminary fluorescence induction and decay kinetics measurements, it is suggested that PS II is still functional with 0.05 μm Mn, although to a decreased extent relative to the Mn-replete conditions (data not shown). This phenotype is reminiscent of the iron deficiency situation (18), and the examination of this intermediate Mn level might help reveal early primary responses.
In Chlamydomonas, iron deficiency results in various biochemical changes such as the accumulation of assimilation factors like Fea1 and ferroxidase. Only after cells become iron limited are the dramatic effects on photosynthesis (described in brief above) observed. Similar to this, manganese deficiency induces changes to the proteome before the complete loss of photosynthetic function. Several examples of proteins significantly increasing or decreasing in the 0.05 μm Mn condition relative to the replete situation were observed (supplemental Table S4); subunits of the 20S proteasome increase (vide infra), whereas MnSOD isoform 1 is decreased, as noted previously (20).
Manganese is abundant in the photosynthetic apparatus and in MnSODs; accordingly, these are the primary targets of nutritional manganese deficiency and limitation (20). Manganese-limited cells cannot grow photoautotrophically and are growth inhibited in photoheterotrophic conditions. This is likely to be a consequence of an overall decrease in photosynthesis quantum efficiency. At the biochemical level, components of the oxygen evolving enhancer complex were found to disassociate from chloroplast membranes, most likely due to the absence of the manganese cluster (20). MnSOD activity also decreases as manganese levels in the growth medium are decreased. Each of these results is supported by the proteomics data (Table IV).
It is also important to note that photosynthetic activity, lost in manganese limitation as measured by room temperature chlorophyll fluorescence induction kinetics, is quickly recovered (within ∼1 h) upon the addition of supplemental manganese to the growth medium. The addition of chloroplast protein biosynthesis inhibitor chloramphenicol also did not prevent PS II recovery (20). These lines of evidence suggest that most of the photosynthetic apparatus remains intact under conditions of manganese deficiency. Accordingly, we observed very little change in the levels of proteins involved in photosynthetic function with manganese deficiency (supplemental Table S4). The components of multiple protein complexes related to metabolism were increased in manganese-deficient and manganese-limited cells relative to the replete situation (supplemental Table S4). These include the 20S and 26S proteasomes and the T-complex chaperonin. Specifically, seven subunits of the 20S proteasome (Poa1, Poa2, Poa3, Pob3, Poa4, Poa5, and Poa7, which correspond to Cre17.g705400, Cre08.g373250, Cre10.g418100, Cre01.g030850, Cre17.g724350, Cre14.g619550, and Cre10.g424400, respectively), three subunits of the 26S proteasome (Rpn7, Rpn9, and Rpn12, corresponding to Cre13.g581450, Cre15.g644800, and Cre17.g708300, respectively), and four protein subunits of the T-complex chaperonin (Cct1, Cct2, Cct5, and Cct8, corresponding to Cre10.g439100, Cre09.g416750, Cre03.g156750, and Cre03.g168450, respectively) were increased. Increasing proteasome levels suggest an overall increase in the amount of proteolysis occurring in manganese-deficient cells, possibly due to increased levels of oxidative damage, as the proteasome is known to act on oxidized proteins (74). Previous studies have shown that manganese-deficient cells are more sensitive to H2O2 treatment than are cells grown in manganese-replete conditions, and that several oxidative stress response genes were up-regulated (20). Interestingly, whereas several of the 20S proteasome subunits can be detected in 0.05 μm Mn, none of the 26S proteasome subunits was detected until the 0 μm Mn condition, suggesting that the accumulation of the 20S proteasome precedes the accumulation of the 26S proteasome complex. The T-complex, which acts as a protein chaperone for a variety of cytosolic proteins, may be up-regulated either as a response to this increasingly proteolytic environment or to protect proteins from the proposed increase in oxidative damage (75).
Under manganese-deficient growth conditions, we also observed a reduction in importin-α isoform 1 and importin-β isoforms 1 and 7 (supplemental Table S4). These proteins were detected at 46, 23, and 21 zmol/cell, respectively, under replete conditions but were undetectable under manganese limitation. The importin complex comprises two soluble subunits that help deliver large molecules to the nuclear pore complex and is involved in signaling events. Thus, it is possible that manganese-limited cells might have some signaling defects. For example, studies of Arabidopsis mutants of MOS6 (a plant importin-α protein) and SAD2 (an importin β family member) show enhanced susceptibility to pathogens and abscisic acid stress, respectively (76).
COP I Coat Proteins Are Increased in All Four Metal Deficiency Conditions
Responses common to the four different trace metal deficiencies examined in this study included an increase of several subunits of the non-clathrin coat (COP I) protein complexes (Table VIII). These coat proteins are involved in retrograde trafficking between the Golgi apparatus and the endoplasmic reticulum. Across the various micronutrient experiments, we observed that all the COP I proteins showed increased abundance in two or more deficiency conditions, with the only decrease in abundance observed being for CopD1 in the zinc deficiency condition The increase of COP I subunit proteins may be attributed either to increased synthesis of the proteins or to increased partitioning of the proteins to the soluble fraction.
Table VIII. Abundance (zmol/cell) of coatamer protein subunits across all four studies of metal nutrition deficiency.
| Accession | Gene name | μm Cu |
μm Zn |
μm Fe |
μm Mn |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 0 | 2.5 | 0 | 20 | 1 | 0.25 | 2 | 0.05 | 0 | ||
| Cre13.g565850 | COPA1 | 7 ± 1 | 57 ± 56 | n.d. | n.d. | n.d. | n.d. | 18 ± 10 | n.d. | n.d. | 24 ± 12 |
| Cre03.g210600 | COPB1 | n.d. | n.d. | n.d. | 33 ± 6 | n.d. | n.d. | n.d. | n.d. | n.d. | 63 ± 47 |
| Cre02.g118500 | COPB2 | 8 ± 1 | 32 ± 17 | n.d. | 18 ± 3 | n.d. | n.d. | n.d. | n.d. | n.d. | 27 ± 15 |
| Cre02.g089100 | COPD1 | 13 ± 5 | 37 ± 11 | 18 ± 18 | n.d. | 6 ± 1 | 15 ± 8 | 16 ± 1 | n.d. | n.d. | n.d. |
| Cre13.g592450 | COPE1 | 16 ± 2 | 34 ± 9 | 15 ± 3 | 16 ± 6 | 7 ± 1 | 7 ± 1 | 21 ± 3 | n.d. | n.d. | n.d. |
| Cre06.g310750 | COPG1 | n.d. | n.d. | n.d. | 18 ± 3 | n.d. | n.d. | 12 ± 5 | n.d. | n.d. | 38 ± 32 |
| Cre08.g371450 | COPZ1 | n.d. | 32 ± 6 | 31 ± 17 | 25 ± 3 | n.d. | n.d. | 6 ± 1 | n.d. | n.d. | n.d. |
n.d., not detected.
DISCUSSION
Changes at the RNA Level Can Serve as Reliable Predictors of Changes at the Protein Level but Do Not Reveal All Protein Level Changes
The Pearson coefficient between transcript and protein data showed a moderate correlation (ρ = 0.28 for copper-replete and ρ = 0.30 for copper-deficient cells). Given that the dynamic range of the proteomics study (3 to 4 orders of magnitude) is significantly less than that of the transcriptomics study (roughly 5 to 6 orders of magnitude), the low-to-moderate correlation is likely due to the limited dynamic range of the proteomics study. With over 15,000 genes in the Chlamydomonas genome and ∼12,300 soluble proteins estimated (using TMHMM), only 23% of all gene products were detected and quantified. Our transcript–protein correlation estimates agree with other studies of other organisms ranging from E. coli and yeast to mouse and human (52, 53, 77–80). Our analysis also suggests that this overall correlation does not change depending on the nutritional state. Therefore, RNA abundance is a moderate predictor of protein abundance as it is in other organisms.
However, changes in RNA abundance are good predictors of changes in protein levels. In cases in which the transcript showed a significant change (identified by Castruita et al. (13)), the correlation between the transcript and protein abundance was determined to be fairly strong (ρ = 0.73). Thus, for Chlamydomonas, significant changes in RNA abundance can denote a complementary change at the protein level. In other words, changes in RNA abundance can serve as a reliable predictor of changes at the protein level. Studies on yeast, corn, and Drosophila further support this notion (53, 81, 82).
Despite this correlation, changes in RNA abundance do not always result in corresponding changes in protein levels, especially in cases in which RNA abundance is decreased. In a recent study on yeast, the decrease in RNA abundance showed poor correlation with changes in the protein abundance (81). This is because protein half-life, a key factor that determines the magnitude of a response with respect to protein abundance, is independent of RNA abundance (83). A decrease in RNA abundance can prevent the synthesis of new protein, but it does not impact preexisting protein. Changes in protein levels then become evident only if proteolysis is induced in parallel.
Some changes in protein amount cannot be explained by changes in transcriptome levels, such as those of proteins whose half-lives are changed (e.g. by ubiquitination or induced proteolysis) or when rates of synthesis are changed by mechanisms operating at the translational level. Some excellent examples include plastocyanin in copper-deficient Chlamydomonas and carbonic anhydrase in zinc-deficient cells (84). Additionally, only 60% of protein changes in yeast are explained by changes in the transcript (81), studies in Drosophila showed only a 30% overlap in significant changes at both protein and transcript levels (53), and work in mice estimates that only 43% of changes to the protein are directly based on changes to the transcriptome (77). This is also true for Chlamydomonas, as many of the protein changes observed were not accompanied by significant changes in the corresponding transcripts (13). Despite this, transcriptomics can still serve as a strong predictor of changes at the protein level, as the correlation between changes in transcript and protein abundance is high.
Putative Oxidative Stress Responses with Metal Deficiency
Among the four metal deficiency/limitation conditions tested, only iron and zinc deficiency appeared to result in an oxidative stress response. Copper-deficient cells did not show clear trends with regard to oxidative stress responses and did not display visible stress phenotypes such as reduced growth or changes in pigmentation. In manganese deficiency, reduced MnSOD activity is apparent (20), but there is no compensatory response known.
In both iron and zinc deficiency, there are clear indications of a global oxidative stress response. With iron deficiency, this is observed via up-regulation of the ascorbate-glutathione pathway, whereas the response appears to be much less focused on a single pathway in zinc deficiency. Much of the iron-deficiency-induced stress response is focused on the prevention of superoxide damage, likely generated from PS I activity. This observation supports previous reports that the major detoxification pathway for PS I is the ascorbate-glutathione pathway (64). For zinc-deficient cells, the abundance of 14 different redox proteins increased, and many of these proteins are implicated in oxidative stress defense but show no other common metabolic associations. One of the redox proteins that increases in zinc-deficient cells is thioredoxin 5, which has been shown to be induced by metal stress (Hg and Cd) but not in response to oxidative stress generators, suggesting that metal-deficiency-induced stress might be occurring in the cell. Indeed, zinc-deficient cells tend to hyper-accumulate several metals including copper, iron, and manganese.2
The Effect of Metal Deficiency on Photosynthetic Function
Each of the metals selected for this study is linked to photosynthetic function (2–4). Some of the metal-deficiency effects found in the proteomics data were expected and represent known responses. For example, copper deficiency resulted in the loss of plastocyanin and accumulation of cytochrome c6, and zinc-containing carbonic anhydrase isoform 1 was decreased under zinc deficiency (Table IV). The proteomics results for iron- and manganese-related growth conditions, however, reveal differences in how photosynthesis is affected in metal-deficient Chlamydomonas.
Previous reports showed that Chlamydomonas cells remodel their PS I complexes (17, 18, 62) and show a reduction of photosynthesis-related proteins in iron-limited conditions (Fig. 5) (48). Although there is no clear reason for reducing the amounts of the soluble photosynthesis-related proteins, it is possible that these are degraded as a side effect of increased proteolysis under iron limitation. For example, the abundance of thylakoid membrane proteins was reduced as a result of induced proteolysis (18), and we observed increases in the abundances of four proteases in iron deficiency/limitation (supplemental Table S2).
In contrast to iron-limitation responses, manganese-limited cells did not show a widespread loss of photosynthesis-related proteins. This offers an explanation for the disparity in photosynthetic recovery time after metal supplementation. Previous work showed that although both metal-limitation conditions resulted in a loss of photosynthetic activity, cells previously under manganese limitation recovered photosynthetic function in less than 1 h after the addition of manganese (20), whereas recovery from iron limitation required up to 2 days (17). Our results suggest that much of the photosynthetic apparatus is intact under manganese limitation.
Interconnection between Metal-deficiency Conditions: Copper and Zinc
In Chlamydomonas, we have previously noted a connection between copper and zinc metabolism (10). Specifically, the copper-sensing transcription factor CRR1 is involved in zinc homeostasis. Cells carrying a version of CRR1 lacking a C-terminal Cys-rich region hyper-accumulate zinc. This coincides with the mis-expression of ZRT genes that are normally expressed only in copper-deficient cells. In this work, we document another interaction between zinc and copper nutrition (Table VI). Many of the gene products that are up-regulated in copper deficiency are also up-regulated with zinc deficiency. This suggests either that zinc deficiency causes copper deficiency or that genes corresponding to these proteins are direct targets of the zinc regulon. The fact that plastocyanin levels were decreased with zinc deficiency (Table VI), despite the presence of adequate copper in the growth medium, suggests that the cells must have been internally copper deficient; previous study has shown that intracellular copper is required for holoplastocyanin accumulation (84). In addition, some of the changes in protein abundance observed in zinc-deficient cells mimic responses in copper-deficient cells that are not CRR1 controlled.
Covariance of Cgl78/Ycf54 with Cpx1 Hints at Functions Related to Tetrapyrrole Biosynthesis
CGL78, known as YCF54 in Synechocystis 6803 and as AT5G58250.1 in Arabidopsis, is a recently identified gene that is highly conserved but of unclear function. Its transcript is highly abundant in copper-deficient cells, where it accounts for about 0.1% of the total mRNA, making it one of the most highly expressed genes in the cell (13). CGL78 was also shown to be a target of the CRR1 transcription factor. Based on co-expression studies with coprogen oxidase, it was suggested that Cgl78 might play a role in chlorophyll metabolism or be involved with chlorophyll binding proteins (13). A recently published work in fact showed that Synechocystis 6803 Ycf54 is necessary for the function and stability of the aerobic oxidative cyclase in chlorophyll biosynthesis (85). Given that the cyclase requires at least three subunits including a soluble component, perhaps Cgl78 is one of the subunits.
Our quantitative proteomics data acquired from several growth conditions support the co-expression of Cgl78 with coprogen oxidase. The two proteins increase or decrease similarly with the different conditions. Immunoblot analysis of the copper samples best shows the dramatic change in Cgl78 abundance (Fig. 5). We also examined our proteomes for other CGL gene products that might also show covariance with coprogen oxidase. CGLs are those genes that are conserved in green lineages and were originally identified via comparison of the Chlamydomonas genome to the genomes of several other green photosynthetic organisms using a mutual BLAST hit strategy (86). The only other CGL protein identified in each of the four metal conditions was Cgl30, whose abundance did not change as a function of metal deficiency (Fig. 7). This suggests that Cgl78 function is tied to the tetrapyrrole pathway, but that CGL proteins in general are not.
Fig. 7.
Cgl78/Ycf54 abundance shows covariance with coprogen oxidase among multiple growth conditions. Cgl30 is shown as a control. Coprogen oxidase abundance is represented by triangles, Cgl78 is represented by circles, and Cgl30 is represented by squares.
Enzymes of the SAM Synthesis Pathway are Affected by Several Metal-deficiency Conditions
In Chlamydomonas, SAM is produced via the methionine synthesis pathway, in which a homocysteine precursor is converted to methionine via the activity of either a cobalamin-dependent methionine synthase encoded by METH or a cobalamin-independent methionine synthease encoded by METE. Because cobalamin was not supplemented into the medium, we expect that the cobalamin-independent synthase pathway will be utilized, especially given its average abundance of 1,529 zmol/cell. In the cobalamin-independent synthase pathway, 5-methyl tetrahydrofolate provides a methyl group to homocysteine to produce methionine. The 5-methyl tetrahydrofolate is regenerated from tetrahydrofolate from the stepwise activity of serine hydroxymethyltransferase and 5,10-tetrahydrofolate reductase. An adenosyl group is then added to methionine by SAM synthase to produce SAM, which in turn is utilized by various methyltransferases. The by-product of methyltransferase reactions is S-adenosylhomocysteine, which can be converted back to homocysteine by S-adenosylhomocysteine hydrolase encoded by SAHH.
The abundances of many of the enzymes of this pathway are decreased by both iron and copper deficiency/limitation. Under copper and iron deficiency/limitation conditions, cobalamin-independent methionine synthase and 5,10-tetrahydrofolate reductase were decreased. In addition, iron-limited cells showed decreases in S-adenosylhomocysteine hydrolase and the major serine hydroxymethyltransferase, isoform 2. We measured a 3-fold increase, however, in serine hydroxymethyltransferase isoform 1 under copper deficiency, but this likely has little impact on the pathway, as isoform 1 is a minor isoform that accumulates to levels roughly 10% of that of isoform 2 under metal-replete conditions. Zinc-deficient cells showed an approximate 2-fold increase in 5,10-tetrahydrofolate reductase, but no change in any other SAM synthesis proteins was observed. Manganese limitation appeared to have little or no effect on the levels of these proteins. Therefore, the decrease of multiple SAM synthesis proteins appears to be a response specific to copper and iron deficiency/limitation.
The effects of metal deficiency/limitation on the enzymes of this pathway are potentially important, in part because many of these proteins appear to be highly abundant. S-adenosylhomocysteine hydrolase, methionine synthase, 5,10-tetrahydrofolate reductase, and the major serine hydroxymethyltransferase are each present at levels of around 200 zmol/cell, which would place these proteins in the top 100 most abundant proteins in our dataset from all nutritional states. Additionally, the transcripts encoding enzymes of the pathway are also highly abundant at the transcript level. Both SAH and METM (which encodes SAM synthase) transcripts are consistently within the top 200 most abundant transcripts among multiple growth conditions (87).
Increases in the Levels of COP I Subunits Show a Common Metabolic Link between Metal Deficiency Conditions Tied to Lipolysis
For each of the metal deficiency/limitation conditions tested, we observed a general trend that the abundance of several members of the COP I complex increased. Studies in mice and Drosophila both showed that RNAi knockdowns of any of the individual COP I subunits, with the exception of COPE, caused an over-accumulation of lipids. As a result, it was proposed that an inability to form the COP I complex inhibits normal rates of lipid degradation (88). The relationship between lipid metabolism and the COP I complex is supported in Chlamydomonas by the detection of several COP I complex subunits in the major lipid droplet and the putative regulator of the complex, ARF1a (88–90).
Two explanations exist for the increased abundance of the COP I subunits observed under various metal deficiency conditions. First, increases in COP I protein subunits might be caused by additional de novo synthesis of the proteins. Alternatively, increases in COP I proteins might result from a relocation of the proteins from lipid bodies into the cytosol, similar to the effect observed with Oee dissociating from the thylakoid membrane under manganese deficiency. In the former case, the result suggests an increased production of vesicles in the cell, perhaps to allow for trafficking of metal transporters through the secretory pathway. In the latter case, the loss of COP I proteins from the vesicles might denote inhibition of vesicle-mediated processes such as lipolysis.
Supplementary Material
Acknowledgments
We thank Dr. Erin Greiner and Dr. Deborah Francoleon for their technical expertise and assistance with the mass spectrometry measurements. Also, we are grateful to Dr. Steven Karpowicz for his guidance on functional analysis, David Lopez for advice on utilization of the Algal Functional Annotation Tool, Rey Martin for assistance with sample preparation for the zinc studies, and Dr. Jennifer Hsieh for help with TMHMM analysis. We also thank Mark Arbing, Annie Shin, and Emmeline Kuo of the UCLA/DOE Institute for Genomics and Proteomics Protein Expression Technology Center for expression and purification of CGL78 and ZCP2, as well as Agrisera for the antibody to CGL78.
Footnotes
* This work was supported by the National Institutes of Health (Grant No. GM42143 to S.M. for the work on Cu and Zn) and the Department of Energy (Grant Nos. DE-FD02–04ER15529 to S.M. for the work on Fe and Mn and DE-FC03–02ER6342 to the UCLA/DOE Institute for Genomics and Proteomics). M.C. and D.M. acknowledge fellowship support from the NIH (Nos. F32 GM086006 and F32 GM083562, respectively). S.I.H. and J.E. were supported in part by a Ruth L. Kirschstein National Research Service Award (T32 GM07185) for UCLA's predoctoral Cellular and Molecular Biology Training Program.
 This article contains supplemental material.
This article contains supplemental material.
2 Malasarn, D., Kropat, J., Hsieh, S. I., Finazzi, G., Casero, D., Loo, J. A., Pellegrini, M., Wollman, F.-A., and Merchant, S. S., in preparation.
1 The abbreviations used are:
- Au10.2
- Augustus 10.2
- MnSOD
- manganese-containing superoxide dismutase
- MS
- mass spectrometry
- Oee
- oxygen evolving enhancer protein
- PS
- photosystem
- SAM
- S-adenosylmethionine.
REFERENCES
- 1. Andreini C., Bertini I., Cavallaro G., Holliday G., Thornton J. (2008) Metal ions in biological catalysis: from enzyme databases to general principles. J. Biol. Inorg. Chem. 13, 1205–1218 [DOI] [PubMed] [Google Scholar]
- 2. Hänsch R., Mendel R. R. (2009) Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 12, 259–266 [DOI] [PubMed] [Google Scholar]
- 3. Merchant S. S., Allen M. D., Kropat J., Moseley J. L., Long J. C., Tottey S., Terauchi A. M. (2006) Between a rock and a hard place: trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 1763, 578–594 [DOI] [PubMed] [Google Scholar]
- 4. Pilon M., Cohu C. M., Ravet K., Abdel-Ghany S. E., Gaymard F. (2009) Essential transition metal homeostasis in plants. Curr. Opin. Plant Biol. 12, 347–357 [DOI] [PubMed] [Google Scholar]
- 5. Harris E. H. (2009) Chlamydomonas in the laboratory. In The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use, pp. 241–308, Academic Press, San Diego, CA [Google Scholar]
- 6. Beer L. L., Boyd E. S., Peters J. W., Posewitz M. C. (2009) Engineering algae for biohydrogen and biofuel production. Curr. Opin. Biotechnol. 20, 264–271 [DOI] [PubMed] [Google Scholar]
- 7. Siaut M., Cuine S., Cagnon C., Fessler B., Nguyen M., Carrier P., Beyly A., Beisson F., Triantaphylides C., Li-Beisson Y., Peltier G. (2011) Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnology 11, 7–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S. S., Malasarn D. (2011) A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66, 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kropat J., Tottey S., Birkenbihl R. P., Depège N., Huijser P., Merchant S. (2005) A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. U.S.A. 102, 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sommer F., Kropat J., Malasarn D., Grossoehme N. E., Chen X., Giedroc D. P., Merchant S. S. (2010) The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell 22, 4098–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Page M. D., Kropat J., Hamel P. P., Merchant S. S. (2009) Two Chlamydomonas CTR copper transporters with a novel Cys-Met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell 21, 928–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mus F., Dubini A., Seibert M., Posewitz M. C., Grossman A. R. (2007) Anaerobic acclimation in Chlamydomonas reinhardtii. J. Biol. Chem. 282, 25475–25486 [DOI] [PubMed] [Google Scholar]
- 13. Castruita M., Casero D., Karpowicz S. J., Kropat J., Vieler A., Hsieh S. I., Yan W., Cokus S., Loo J. A., Benning C., Pellegrini M., Merchant S. S. (2011) Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23, 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allen M. D., del Campo J. A., Kropat J., Merchant S. S. (2007) FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii. Eukaryot. Cell 6, 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Terauchi A. M., Lu S.-F., Zaffagnini M., Tappa S., Hirasawa M., Tripathy J. N., Knaff D. B., Farmer P. J., Lemaire S. D., Hase T., Merchant S. S. (2009) Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J. Biol. Chem. 284, 25867–25878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Long J. C., Sommer F., Allen M. D., Lu S.-F., Merchant S. S. (2008) FER1 and FER2 encoding two ferritin complexes in Chlamydomonas reinhardtii chloroplasts are regulated by iron. Genetics 179, 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., Hippler M. (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7, 3964–3979 [DOI] [PubMed] [Google Scholar]
- 18. Moseley J. L., Allinger T., Herzog S., Hoerth P., Wehinger E., Merchant S., Hippler M. (2002) Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21, 6709–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Page M. D., Allen M. D., Kropat J., Urzica E. I., Karpowicz S. J., Hsieh S. I., Loo J. A., Merchant S. S. (2012) Fe sparing and Fe recycling contribute to increased superoxide dismutase capacity in iron-starved Chlamydomonas reinhardtii. Plant Cell 24, 2649–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Allen M. D., Kropat J., Tottey S., Del Campo J. A., Merchant S. S. (2007) Manganese deficiency in chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 143, 263–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haas C., Rodionov D., Kropat J., Malasarn D., Merchant S., de Crécy-Lagard V. (2009) A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Merchant S. S., Helmann J. D. (2012) Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation advances in microbial physiology. In (Poole R. K., ed.) Vol. 60, pp. 91–210, Advances in Microbial Physiology, Academic Press, San Diego, CA: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Merchant S., Bogorad L. (1986) Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardi. Mol. Cell Biol. 6, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Atteia A., Adrait A., Brugière S., Tardif M., van Lis R., Deusch O., Dagan T., Kuhn L., Gontero B., Martin W., Garin J., Joyard J., Rolland N. (2009) A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the α-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 26, 1533–1548 [DOI] [PubMed] [Google Scholar]
- 25. Keller L. C., Romijn E. P., Zamora I., Yates J. R., III, Marshall W. F. (2005) Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15, 1090–1098 [DOI] [PubMed] [Google Scholar]
- 26. Wagner V., Fiedler M., Markert C., Hippler M., Mittag M. (2004) Functional proteomics of circadian expressed proteins from Chlamydomonas reinhardtii. FEBS Lett. 559, 129–135 [DOI] [PubMed] [Google Scholar]
- 27. Wagner V., Geβner G., Heiland I., Kaminski M., Hawat S., Scheffler K., Mittag M. (2006) Analysis of the phosphoproteome of Chlamydomonas reinhardtii provides new insights into various cellular pathways. Eukaryot. Cell 5, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wagner V., Ullmann K., Mollwo A., Kaminski M., Mittag M., Kreimer G. (2008) The phosphoproteome of a Chlamydomonas reinhardtii eyespot fraction includes key proteins of the light signaling pathway. Plant Physiol. 146, 772–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Terashima M., Specht M., Naumann B., Hippler M. (2010) Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol. Cell. Proteomics 9, 1514–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Förster B., Mathesius U., Pogson B. J. (2006) Comparative proteomics of high light stress in the model alga Chlamydomonas reinhardtii. Proteomics 6, 4309–4320 [DOI] [PubMed] [Google Scholar]
- 31. Lemaire S. D., Guillon B., Le Maréchal P., Keryer E., Miginiac-Maslow M., Decottignies P. (2004) New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 101, 7475–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rolland N., Atteia A., Decottignies P., Garin J., Hippler M., Kreimer G., Lemaire S. D., Mittag M., Wagner V. (2009) Chlamydomonas proteomics. Curr. Opin. Microbiol. 12, 285–291 [DOI] [PubMed] [Google Scholar]
- 33. Mühlhaus T., Weiss J., Hemme D., Sommer F., Schroda M. (2011) Quantitative shotgun proteomics using a uniform 15N-labeled standard to monitor proteome dynamics in time course experiments reveals new insights into the heat stress response of Chlamydomonas reinhardtii. Mol. Cell. Proteomics 10, 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Silva J. C., Gorenstein M. V., Li G.-Z., Vissers J. P. C., Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 35. Silva J. C., Denny R., Dorschel C., Gorenstein M. V., Li G.-Z., Richardson K., Wall D., Geromanos S. J. (2006) Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol. Cell. Proteomics 5, 589–607 [DOI] [PubMed] [Google Scholar]
- 36. Silva J. C., Denny R., Dorschel C. A., Gorenstein M., Kass I. J., Li G.-Z., McKenna T., Nold M. J., Richardson K., Young P., Geromanos S. (2005) Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 77, 2187–2200 [DOI] [PubMed] [Google Scholar]
- 37. Geromanos S. J., Vissers J. P. C., Silva J. C., Dorschel C. A., Li G. Z., Gorenstein M. V., Bateman R. H., Langridge J. I. (2009) The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9, 1683–1695 [DOI] [PubMed] [Google Scholar]
- 38. Quinn J. M., Merchant S. (1998) Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 297, 263–279 [DOI] [PubMed] [Google Scholar]
- 39. Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 40. Li G. Z., Vissers J. P. C., Silva J. C., Golick D., Gorenstein M. V., Geromanos S. J. (2009) Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9, 1696–1719 [DOI] [PubMed] [Google Scholar]
- 41. Lopez D., Casero D., Cokus S., Merchant S., Pellegrini M. (2011) Algal Functional Annotation Tool: a Web-based analysis suite to functionally interpret large gene lists using integrated annotation and expression data. BMC Bioinformatics 12, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Karpowicz S. J., Prochnik S. E., Grossman A. R., Merchant S. S. (2011) The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J. Biol. Chem. 286, 21427–21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fox J. D., Waugh D. S. (2003) Maltose-binding protein as a solubility enhancer. Methods Mol. Biol. 205, 99–117 [DOI] [PubMed] [Google Scholar]
- 44. Quinn J. M., Nakamoto S. S., Merchant S. (1999) Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper response elements and increased translation from a copper deficiency-specific form of the transcript. J. Biol. Chem. 274, 14444–14454 [DOI] [PubMed] [Google Scholar]
- 45. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 46. Eriksson M., Moseley J. L., Tottey S., del Campo J. A., Quinn J., Kim Y., Merchant S. (2004) Genetic dissection of nutritional copper signaling in Chlamydomonas distinguishes regulatory and target genes. Genetics 168, 795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 48. La Fontaine S., Quinn J. M., Nakamoto S. S., Page M. D., Göhre V., Moseley J. L., Kropat J., Merchant S. (2002) Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot. Cell 1, 736–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Voges D., Zwickl P., Baumeister W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068 [DOI] [PubMed] [Google Scholar]
- 50. Taylor T. C., Backlund A., Bjorhall K., Spreitzer R. J., Andersson I. (2001) First crystal structure of rubisco from a green alga, Chlamydomonas reinhardtii. J. Biol. Chem. 276, 48159–48164 [DOI] [PubMed] [Google Scholar]
- 51. Groth G., Pohl E. (2001) The structure of the chloroplast F1-ATPase at 3.2 Å resolution. J. Biol. Chem. 276, 1345–1352 [DOI] [PubMed] [Google Scholar]
- 52. Lu P., Vogel C., Wang R., Yao X., Marcotte E. M. (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotech. 25, 117–124 [DOI] [PubMed] [Google Scholar]
- 53. Bonaldi T., Straub T., Cox J., Kumar C., Becker P. B., Mann M. (2008) Combined use of RNAi and quantitative proteomics to study gene function in Drosophila. Mol. Cell 31, 762–772 [DOI] [PubMed] [Google Scholar]
- 54. Schwarz G., Mendel R. R. (2006) Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu. Rev. Plant Biol. 57, 623–647 [DOI] [PubMed] [Google Scholar]
- 55. Schofield C. J., Ratcliffe P. J. (2004) Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5, 343–354 [DOI] [PubMed] [Google Scholar]
- 56. Hill K. L., Merchant S. (1995) Coordinate expression of coproporphyrinogen oxidase and cytochrome c6 in the green-alga Chlamydomonas reinhardtii in response to changes in copper availability. EMBO J. 14, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moseley J. L., Page M. D., Alder N. P., Eriksson M., Quinn J., Soto F., Theg S. M., Hippler M., Merchant S. (2002) Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14, 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bethin K. E., Cimato T. R., Ettinger M. J. (1995) Copper binding to mouse liver S-adenosylhomocysteine hydrolase and the effects of copper on its levels. J. Biol. Chem. 270, 20703–20711 [DOI] [PubMed] [Google Scholar]
- 59. Li M., Li Y., Chen J., Wei P. X., Liu J., Liu Q., WeiLeu W., Zhang L., Yang X., Lu J., Wang K. (2007) Copper ions inhibit S-adenosylhomocysteine hydrolase by causing dissociation of NAD+ cofactor. Biochemistry 46, 11451–11458 [DOI] [PubMed] [Google Scholar]
- 60. Aydemir T., Ötürk R., Bozkaya L. A., Tarhan L. (2000) Effects of antioxidant vitamins A, C, E and trace elements Cu, Se on CuZn SOD, GSH-Px, CAT and LPO levels in chicken erythrocytes. Cell Biochem. Funct. 18, 109–115 [DOI] [PubMed] [Google Scholar]
- 61. Jenkinson S. G., Lawrence R. A., Burk R. F., Williams D. M. (1982) Effects of copper deficiency on the activity of the selenoenzyme glutathione peroxidase and on excretion and tissue retention of 75SeO322−. J. Nutr. 112, 197–204 [DOI] [PubMed] [Google Scholar]
- 62. Naumann B., Stauber E. J., Busch A., Sommer F., Hippler M. (2005) N-terminal processing of Lhca3 is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J. Biol. Chem. 280, 20431–20441 [DOI] [PubMed] [Google Scholar]
- 63. Terauchi A., Peers G., Kobayashi M., Niyogi K., Merchant S. S. (2010) Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth. Res. 105, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Asada K. (1999) The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Biol. Plant Mol. Biol. 50, 601–639 [DOI] [PubMed] [Google Scholar]
- 65. Schroda M., Vallon O., Wollman F.-A., Beck C. F. (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11, 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Philpott C. C., Protchenko O. (2008) Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 7, 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cheek J., Broderick J. B. (2001) Adenosylmethionine-dependent iron-sulfur enzymes: versatile clusters in a radical new role. J. Biol. Inorg. Chem. 6, 209–226 [DOI] [PubMed] [Google Scholar]
- 68. Badger M. R., Price G. D. (1994) The role of carbonic anhydrase in photosynthesis. Annu. Rev. Plant Biol. Plant Mol. Biol. 45, 369–392 [Google Scholar]
- 69. Moroney J. V., Ynalvez R. A. (2007) Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryot. Cell 6, 1251–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Van K., Spalding M. H. (1999) Periplasmic carbonic anhydrase structural gene (Cah1) mutant in Chlamydomonas reinhardtii. Plant Physiol. 120, 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ynalvez R. A., Xiao Y., Ward A. S., Cunnusamy K., Moroney J. V. (2008) Identification and characterization of two closely related β-carbonic anhydrases from Chlamydomonas reinhardtii. Physiol. Plantarum 133, 15–26 [DOI] [PubMed] [Google Scholar]
- 72. Gaballa A., Wang T., Ye R. W., Helmann J. D. (2002) Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 184, 6508–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Smith K. F., Bibb L. A., Schmitt M. P., Oram D. M. (2009) Regulation and activity of a zinc uptake regulator, Zur, in Corynebacterium diphtheriae. J. Bacteriol. 191, 1595–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Davies K. J. A. (2001) Degradation of oxidized proteins by the 20S proteasome. Biochimie (Paris) 83, 301–310 [DOI] [PubMed] [Google Scholar]
- 75. Kubota H., Hynes G., Willison K. (1995) The chaperonin containing t-complex polypeptide 1 (TCP-1). Eur. J. Biochem. 230, 3–16 [DOI] [PubMed] [Google Scholar]
- 76. Meier I., Brkljacic J. (2009) The nuclear pore and plant development. Curr. Opin. Plant Biol. 12, 87–95 [DOI] [PubMed] [Google Scholar]
- 77. Graumann J., Hubner N. C., Kim J. B., Ko K., Moser M., Kumar C., Cox J., Schöler H., Mann M. (2008) Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 7, 672–683 [DOI] [PubMed] [Google Scholar]
- 78. de Sousa Abreu R., Penalva L. O., Marcotte E. M., Vogel C. (2009) Global signatures of protein and mRNA expression levels. Mol. Biosyst. 5, 1512–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Griffin T. J., Gygi S. P., Ideker T., Rist B., Eng J., Hood L., Aebersold R. (2002) Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol. Cell. Proteomics 1, 323–333 [DOI] [PubMed] [Google Scholar]
- 80. Vogel C., de Sousa Abreu R., Ko D., Le S.-Y., Shapiro B. A., Burns S. C., Sandhu D., Boutz D. R., Marcotte E. M., Penalva L. O. (2010) Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 6, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lee M. V., Topper S. E., Hubler S. L., Hose J., Wenger C. D., Coon J. J., Gasch A. P. (2011) A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 7, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li P., Ponnala L., Gandotra N., Wang L., Si Y., Tausta S. L., Kebrom T. H., Provart N., Patel R., Myers C. R., Reidel E. J., Turgeon R., Liu P., Sun Q., Nelson T., Brutnell T. P. (2010) The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 83. Berlin C. M., Schimke R. T. (1965) Influence of turnover rates on the responses of enzymes to cortisone. Mol. Pharmacol. 1, 149–156 [PubMed] [Google Scholar]
- 84. Li H. H., Merchant S. (1995) Degradation of plastocyanin in copper-deficient Chlamydomonas reinhardtii—evidence for a protease-susceptible conformation of the apoprotein and regulated proteolysis. J. Biol. Chem. 270, 23504–23510 [DOI] [PubMed] [Google Scholar]
- 85. Hollingshead S., Kopecná J., Jackson P. J., Canniffe D. P., Davison P. A., Dickman M. J., Sobotka R., Hunter C. N. (2012) Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in synechocystis PCC 6803. J. Biol. Chem. 287, 27823–27833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Maréchal-Drouard L., Marshall W. F., Qu L.-H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C.-L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernández E., Fukuzawa H., González-Ballester D., González-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J.-P., Riaño-Pachón D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C.-J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martínez D., Ngau W. C. A., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Karpowicz S. J. (2011) Genomics and Transcriptomics of Algae and Plants. PhD thesis, University of California, Los Angeles [Google Scholar]
- 88. Beller M., Sztalryd C., Southall N., Bell M., Jackle H., Auld D. S., Oliver B. (2008) COPI complex is a regulator of lipid homeostasis. PLoS Biol. 6, 2530–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Moellering E. R., Benning C. (2010) RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 9, 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nguyen H. M., Baudet M., Cuiné S., Adriano J.-M., Barthe D., Billon E., Bruley C., Beisson F., Peltier G., Ferro M., Li-Beisson Y. (2011) Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics 11, 4266–4273 [DOI] [PubMed] [Google Scholar]
- 91. Hsieh S. I., Castruita M., Malasarn D., Urzica E., Erde J., Page M. D., Yamasaki H., Casero D., Matteo Pellegrini M., Merchant S. S., Loo J. A. (2012) The Proteome of Copper, Iron, Zinc, and Manganese Micronutrient Deficiency in Chlamydomonas reinhardtii. Mol. Cell Proteomics 12, 65–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Schmidt M., Geßner G., Luff M., Heiland I., Wagner V., Kaminski M., Geimer S., Eitzinger N., Reißenweber T., Voytsekh O., Fiedler M., Mittag M., Kreimer G. (2006) Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 18, 1908–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yamaguchi K., Beligni M. V., Prieto S., Haynes P. A., McDonald W. H., Yates J. R., Mayfield S. P. (2003) Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of the proteins unique to the 70S ribosome. J. Biol. Chem. 278, 33774–33785 [DOI] [PubMed] [Google Scholar]
- 94. Michelet L., Zaffagnini M., Vanacker H., Le Maréchal P., Marchand C., Schroda M., Lemaire S. D., Decottignies P. (2008) In vivo targets of S-thiolation in Chlamydomonas reinhardtii. J. Biol. Chem. 283, 21571–21578 [DOI] [PubMed] [Google Scholar]
- 95. Yamaguchi K., Prieto S., Beligni M. V., Haynes P. A., McDonald W. H., Yates J. R., Mayfield S. P. (2002) Proteomic characterization of the small subunit of Chlamydomonas reinhardtii chloroplast ribosome: identification of a novel S1 domain-containing protein and unusually large orthologs of bacterial S2, S3, and S5. Plant Cell 14, 2957–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.