Abstract
While large-scale efforts have rapidly advanced the understanding and practical impact of human genomic variation, the latter is largely unexplored in the human microbiome. We therefore developed a framework for metagenomic variation analysis and applied it to 252 fecal metagenomes of 207 individuals from Europe and North America. Using 7.4 billion reads aligned to 101 reference species, we detected 10.3 million single nucleotide polymorphisms (SNPs), 107,991 short indels, and 1,051 structural variants. The average ratio of non-synonymous to synonymous polymorphism rates of 0.11 was more variable between gut microbial species than across human hosts. Subjects sampled at varying time intervals exhibited individuality and temporal stability of SNP variation patterns, despite considerable composition changes of their gut microbiota. This implies that individual-specific strains are not easily replaced and that an individual might have a unique metagenomic genotype, which may be exploitable for personalized diet or drug intake.
Introduction
With the increasing availability of individual human genomes, various theoretical and practical aspects of genomic variation can be deduced for individuals and the human population as a whole1,2. Like sequenced human genomes, the number of human gut metagenomes (currently mostly derived from Illumina shotgun sequencing of stool samples) is increasing exponentially. Given the importance of the gut microbiota in human health3,4 and a growing number of studies reporting associations between gut microbiota and diseases5-8, an understanding of genomic variation in gut microbial populations will likely trigger applications towards human well-being and disease.
For example, in the common gut commensal Escherichia coli, just three point mutations in two genes can confer clinically relevant antibiotic resistance5, and natural variation in a single gene can lead to pathogenic adaptation8. Even within pathogenic species in the gut, closely related coexisting strains can exhibit different pathogenic potentials due to minor genomic variation7. These examples illustrate how genomic variation within gut microbes could confer phenotypes that require personalized care or treatment of the host.
Studies based on 16S ribosomal RNA gene surveys or whole metagenome shotgun sequencing characterized taxonomic and functional compositions of healthy individuals’ and patients’ gut microbiota at the genus or species level6,9-12. Variation in taxonomic abundance as well as functions encoded by these gut microbiota have been described between individuals6,11 and used to stratify individuals according to their gut community compositions into enterotypes13. However, genomic variation within species, which leads to their phenotypic diversity and adaptations to different environments, has only been studied in a few taxa, such as Citrobacter spp7.
An early landmark study on a small dataset described metagenomic variation in an acidic biofilm microbiome of low complexity14. The population structure for one species in that habitat was studied and positive selection was observed in some genes15. Another recent study resolved multiple clinical isolates of methicillin-resistant Staphylococcus aureus and delineated its epidemiology and microevolution based on genomic variation16. With the availability of hundreds of deeply sequenced human gut metagenomes9,11,17, sufficient data are becoming available for quantitative analyses of the genetic structure of complex microbial communities, allowing the study of many species at the same time.
Here, we analyzed 1.56 terabases of sequence data from 252 stool samples from 207 individuals (Supplementary Table 1; Supplementary Notes) obtained from the MetaHIT project (71 Danish, 39 Spanish; all sampled once11), the NIH Human Microbiome Project (94 US-American; 51 individuals sampled once, 41 sampled twice, and two sampled three times9), and Washington University (three US-American samples; all sampled once12). Our goals were to (i) develop a framework for genomic variation analysis using metagenomic shotgun data, (ii) gather basic knowledge on the genomic variation landscape in gut metagenomes, and (iii) gain insights into the individuality, temporal stability, and biogeography of metagenomic variation.
Results
Framework for metagenomic variation analysis
We used 1,497 prokaryotic genomes to generate a set of reference genomes (Supplementary Table 2) for the analysis of genomic variation in gut microbial species in 252 samples (on average 6.2 ± 4.1 Gbp were analyzed). Pairwise comparisons of 40 universal marker genes18,19 identified in these genomes were performed to create a set of 929 clusters based on a 95% DNA identity threshold recommended for identifying species20. The genome recruiting the highest number of reads in a cluster was selected as reference for that species (see Methods and Supplementary Information).
Using the same 95% identity threshold, we mapped 7.4 billion metagenomic reads (42% of the total, 91% thereof uniquely) with an average length of 80 bp to the 929 reference genomes (Supplementary Tables 1 and 3). To avoid mapping artifacts (for example caused by high coverage of prophages), we required ≥40% of each reference genome to be covered by reads (corresponding to the gene content similarity between two strains of E. coli21). The resulting 101 prevalent species with base pair coverage from 12x to 32,400x (Fig. 1 and Supplementary Fig. 1) were subjected to genomic variation analysis.
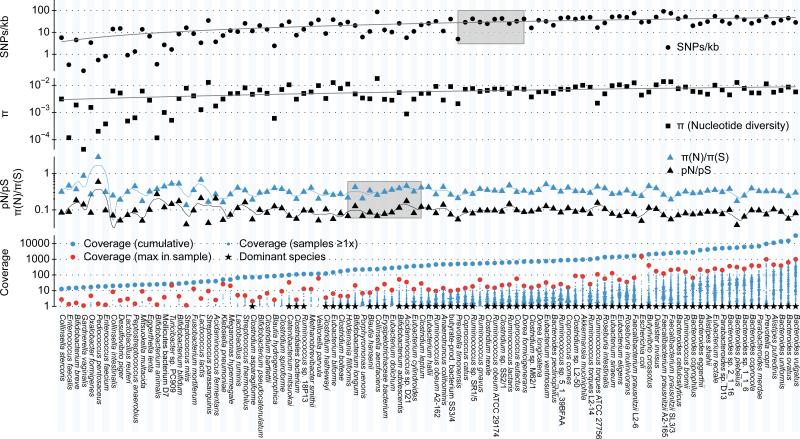
Figure 1. Genomic variation statistics for 101 gut microbial species prevalent in 252 samples from 207 individuals.
Genomic variation statistics were calculated for 101 prevalent gut microbial species, operationally defined as having ≥10x cumulative (over all samples) base pair coverage with at least one sample exhibiting a genome coverage of ≥40%. The 66 dominant species (indicated by *), which account for 99% of the mapped reads, were used for analyses that required high base pair coverage. Species names are given without strain specifications unless this would result in duplicate entries. The blue point cloud plots show the coverages (≥1x) in all samples, with the blue dot above indicating the cumulative coverage and the red dot the maximum coverage across all samples. Gray shaded areas indicate the level of base pair coverage at which abundance effects have only minor effects on SNPs/kb and pN/pS ratios of the pooled samples (Supplementary Information). SNP counts appear to saturate at approximately 500x, with minor increases at higher coverages likely due to the sampling of rare variants at low rates. In individual samples pN/pS is largely stable from a coverage of 10x onward (Supplementary Fig. 7), corresponding to approximately 200x cumulative coverage in our sample set. Nucleotide diversity π follows SNPs/kb closely, as does the derived measure of π(N)/π(S) with respect to pN/pS.
To enable comparative analyses in multiple metagenomes and to identify low frequency variants not detected when analyzing them individually, we used multi-sample calling22 to identify single nucleotide polymorphisms (SNPs), short indels (1 - 50 bp) and structural variations (SVs, >50 bp) in each genome, although SVs were largely underestimated due to small insert sizes (Supplementary Information). We only called variants with allele frequency ≥1% (the conventional definition of polymorphism2) and supported by ≥4 reads. False positive rates were estimated at 0.71% for SNPs and 1.04% for SVs (Supplementary Information, Supplementary Tables 4 and 5, Supplementary Fig. 2).
Genomic variation in prevalent gut species
We identified 10.3 million SNPs in 101 genomes (3.1% of the total 329 Mb positions) across 252 samples from 207 subjects, almost as many as the 14.4 million SNPs recently identified in 179 human genomes2. Within an individual the rate was lower (on average 1.21%, see Supplementary Table 6), yet SNPs/kb increased with base pair coverage when samples were pooled (Supplementary Fig. 3). We also identified 107,991 indels and 1,051 SVs in these 101 species (Supplementary Information). Their relative ratios to SNPs (10,485 short indels and 102 SVs per million SNPs) were robust across species and individuals (Supplementary Fig. 4). Subsequent analyses were restricted to SNPs due to their orders of magnitude higher count over other variation types.
We annotated the genes of the prevalent genomes using orthologous groups (OGs) from eggNOG23 (Supplementary Information) and found that the OGs with the highest SNP density were enriched in functions related to conjugal transfer of antibiotic resistance (Supplementary Table 7). For example, the OG with the highest average SNP density across samples was the Clindamycin resistance transfer factor BtgA (NOG119724), required for conjugative transmission of plasmids. Mutations commonly accrued from the process of conjugation may account for increased diversity among conjugation-associated functions24. Additionally, CRISPR-associated proteins, responsible for conferring resistance in bacteria, were also found among the OGs with high SNP densities (Supplementary Information and Supplementary Table 7).
The large number of SNPs provided the opportunity to compare, for the first time at such scale, the evolution of different coexisting species across a large cohort of individuals. To evaluate selective constraints in these species in their natural habitat, we estimated the ratio of non-synonymous to synonymous polymorphism rates25,26 (pN/pS) within each species in every sample (Fig. 1; Supplementary Information). pN/pS characterizes selective constraint at the level of a population contrary to the more commonly used dN/dS that characterizes it between individual species26. To validate pN/pS ratios, we estimated genetic variation using the sample size-independent nucleotide diversity π, and found that π is highly correlated with SNPs/kb (Fig. 1 and Supplementary Fig. 5). The derived measures of π(N)/π(S) and π(non-degenerate sites)/π(four-fold degenerate sites), the latter of which is less dependent on specific properties of mutation spectra such as transition and transversion ratios, were coherent with pN/pS (Supplementary Fig. 6).
The pN/pS ratio of a genome within a sample remained stable at coverages higher than 10x (Supplementary Fig. 7) – yet another indication of few false SNP calls – and was on average 0.11, but varied considerably for different species (0.04 to 0.58) in accordance with dN/dS ratios estimated independently in a number of interspecies comparisons between closely related bacteria and archaea27,28.
pN/pS across gut species and individuals
Since meaningful comparison of genomic variation requires both breadth (across samples) and depth (in number of base pairs) of sequencing, we focused on the 66 most dominant species that attracted >99% of the reads (Fig. 1). Their relatively low pN/pS ratios were constant across different hosts (Fig. 2a and Supplementary Table 8), which may indicate similar selective constraints across individuals. Thus, the evolution of gut species is most likely dominated by long-term purifying selection and drift rather than rapid adaptations to specific host environments. The wide range of these ratios across species may suggest that different gut species face different evolutionary constraints.
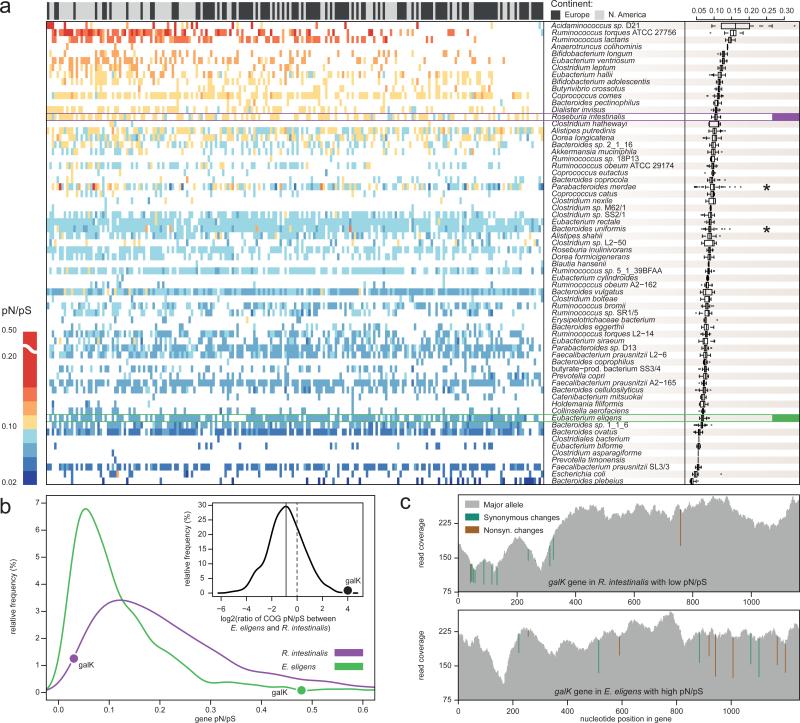
Figure 2. pN/pS ratios of 66 dominant species reveal more variation between species than between individuals.
a) A heatmap of pN/pS ratios for the 66 dominant species (rows) and 207 individuals (columns; only the first time-point per individual) is shown and summarized by species (boxplots on the right). Rows and columns are ordered by their mean pN/pS ratios, which vary considerably between species, but have a tighter bandwidth across samples. Two genomes that are exceptions to this trend (indicated by *) might indicate higher strain diversity. The panel above the heatmap indicates the continent of residence for each individual. A significant difference was found in the mean pN/pS ratios between the two continents, although this is likely an effect of lower sequencing depths of European samples (Supplementary Table 8) that leads to missing data points in some samples (see for example top right corner). b) The distributions of average pN/pS ratios of individual genes from Roseburia intestinalis and Eubacterium eligens (both highlighted in (a)) illustrate that, while base pair coverages are similar, the pN/pS ratio of R. intestinalis is higher in general. The relative pN/pS ratios of orthologous groups in the two species are shown in the inset, the average log2 ratio indicated by the solid line and the random expectation by the dashed line. Outliers can be revealed this way, like the galactokinase gene (galK) whose pN/pS is among the lowest in R. intestinalis and the highest in E. eligens. c) Illustration of low and high pN/pS ratios in galK genes from R. intestinalis (top panel) and E. eligens (bottom panel). The cumulative read coverage is shown in grey with synonymous (green) and non-synonymous (brown) changes marked at the nucleotide positions they occur.
To investigate how different gut species respond to the pressure from the gut environment, we compared the pN/pS ratios of individual genes in Roseburia intestinalis and Eubacterium eligens, which differed considerably in their overall mean pN/pS ratios (0.236 vs. 0.131 from 106 and 147 samples, respectively) despite having comparable average base pair coverages (Supplementary Information). While 75% of the genes in R. intestinalis had systematically higher pN/pS ratios compared to their orthologs in E. eligens, few others showed considerable deviations (Fig. 2c; Supplementary Table 9) suggesting differing evolutionary constraints for these genes. For example, galK, the gene encoding galactokinase, an essential enzyme in the Leloir pathway for galactose metabolism in most organisms29, was among the lowest in terms of pN/pS ratio in R. intestinalis, but among the highest in E. eligens (0.03 and 0.48, respectively; Fig 2b,c). Although present in E. eligens, this gene may not exert its main function (see also30), since E. eligens cannot ferment galactose, nor the galactose-containing disaccharides lactose and melibiose31. On the other hand, R. intestinalis is known to ferment melibiose32 implying that its galK gene is functional (Supplementary Information). Thus, the same gene can be under tight negative selection in one species, but under more relaxed negative selection in another.
Our framework allowed us additionally to obtain information on all genes in each sample (Supplementary Information). As expected, we found that housekeeping genes had usually lower pN/pS ratios. For example, the DNA-dependent RNA polymerase beta subunit gene was consistently among the genes exhibiting the lowest pN/pS ratios across samples and species (Supplementary Table 10). Less obvious examples included genes related to type IV secretion systems used to transfer DNA between microbes33 and involved in host interactions of both pathogenic33 and commensal bacteria34, specifically in anti-inflammatory responses and immune modulation35. Their low pN/pS ratio suggests that maintaining genome plasticity and antibiotic resistance through conjugative transposition is essential in the constantly changing environment of the gut and that the interaction with the host immune system is under purifying selection (Supplementary Table 10). We also found a few conserved unknown, but apparently gut microbe-specific proteins, which exhibited low pN/pS ratios, suggesting that they perform important, yet hitherto unexplored functions (Supplementary Table 11).
Among the genes or OGs with consistently the highest pN/pS ratios were many transposases and antimicrobial resistance genes including the gut-specific gene bile salt hydrolase (BSH)36 (Supplementary Table 10). Conjugated bile acids (CBA) secreted by the hosts repress microbial growth and up-regulate the host mucosal defense system. BSHs are involved in the initial reaction in the metabolism of CBAs by gut microbes36. Their high pN/pS ratio may be indicative for the genomic plasticity necessary to metabolize and survive the variety of different bile acids present in the gut37.
Temporal stability of individual SNP patterns
Several studies on adult human gut microbial samples from a few individuals have suggested that within-individual differences over time are smaller compared to between-individual differences in microbial species composition and abundance38-40. Within a larger cohort, individuality of host-associated microbiota has been reported based on 16S rRNA gene profiling of fingertip-associated communities41, while other studies on a few samples have investigated the persistence of specific strains over time42,43. However, intra-species variation at nucleotide resolution at whole genome level and accompanying changes in species abundances within the human gut over long time periods (>1 year) have not been studied yet in large cohorts. It is unclear if the concept of resident strains is common to other prevalent species, if host-specific strains are retained over time, and how fast they evolve inside the gut environment.
To explore these questions, we used 88 gut metagenomes from 43 healthy US-American subjects (a subset of our cohort) from whom at least two samples were obtained at different time-points with no antibiotics treatment in between (Supplementary Table 12). To measure how similar the subpopulations (strains) of the dominant species were between two samples, we estimated the fixation indices (FST) between the populations (Supplementary Information). Since this measure depends on allele frequencies, which cannot be determined accurately at low base-pair coverage, we also estimated the fraction of alleles shared between the samples out of all polymorphic sites (only 49 genomes that accrued 40% genome coverage in at least two samples were used and genomes with >10x base pair coverage were downsampled to 10x; Supplementary Information and Supplementary Fig. 3). Since the fraction of shared SNPs depends on the number of variable sites, we developed a heuristic allele sharing similarity score that takes into account both (Supplementary Information).
When we compared all 252 samples, FST was significantly lower and allele sharing significantly stronger between different samples from the same individuals than between samples from different individuals (Mann-Whitney: P<0.001 for both; see Figs. 3a and 3b and Supplementary Information). The same trend was observed, albeit much weaker, based on species compositions (Fig. 3c), in line with previous observations from microbial composition-based results38-40. Intra-individual variation being smaller than inter-individual variation does, however, not require that samples from the same individual are more similar to each other than to any other sample in the tested cohort. Our results showed for both measures of variation similarity that all but one of the 88 multi-time-point samples had the highest similarity to another sample from the same individual, which was not true when comparing species abundance over time (Fig. 3c and Supplementary Information). This implies that species abundance in gut microbiota cannot serve as a fingerprint of an individual whereas variation patterns might.
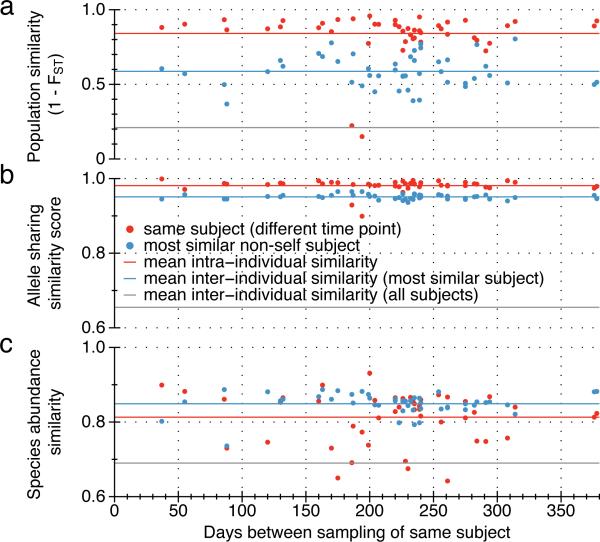
Figure 3. Individuality and temporal stability of genomic variation patterns.
Samples from 43 individuals that were sampled at different time intervals (red dots) were compared with the most similar sample from a different individual (blue dots) in terms of (a) population similarity that takes allele frequencies into account, (b) allele sharing similarity score that takes SNP counts and the ratio of shared SNPs into account (Supplementary Information) and (c) species abundance similarity measured using the Jensen-Shannon Distance13 (JSD). Most similar sample is the one with the lowest FST value in (a), the highest allele sharing similarity score in (b) and the lowest JSD in (c). The three similarity measures are plotted against the number of days between the sampling time-points. The mean across all intra-individual, best inter-individual, and all inter-individual similarities are shown as red, blue, and green dashed lines. For both population similarity and allele sharing similarity between samples from the same individual, all but one sample (resulting in two outliers due to comparisons with two other time-points, see Supplementary Table 12) shared the highest similarity with another sample of the same individual providing strong evidence for individuality of SNP sharing patterns. No decline of similarity over time could be observed.
We also tested if differences in FST and allele sharing decreased over time, which may suggest a divergence of the strains or a horizontal transmission of strains from the environment, but the individual-specific variation patterns remained stable over all the time intervals monitored (Fig. 3). Although this stability should be verified for longer periods as well as when antibiotic treatment or other gut microbiota-challenging events have taken place, our observation suggests that healthy human individuals retain specific strains (see also Supplementary Table 12 and 13) for at least one year.
In contrast to the strong evidence for individuality and temporal stability of SNP patterns, we did not observe a significant geographic separation between European and US samples (Supplementary Fig. 8; Supplementary Table 14). This implies that long-term horizontal transmission of at least some dominant gut microbial strains cause geographic mixing over time. The strongest continental separation, based on FST, was seen in Bacteriodes coprocola (Fig. 4), which was also the only genome with sufficient amounts of data that showed continental separation based on the allele sharing score (Supplementary Fig. 8; Supplementary Tables 14 and 15).
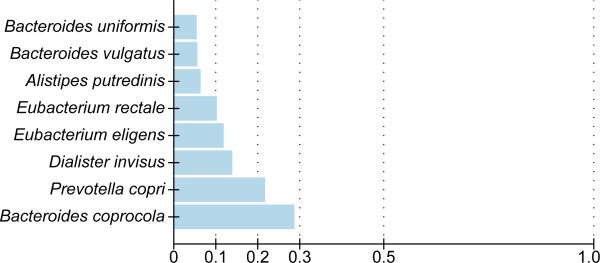
Figure 4. Inter-continental comparison of gut microbial species.
Between continent FST values for eight genomes with ≥10 samples representing each continent are shown. Bacteroides coprocola was the species with the highest FST value, implying a separation between the B. coprocola populations in Europe and North America (see also Supplemental Material; all data available in Supplementary Table 14).
Discussion
We have established a framework for gut microbial genomic variation analysis using metagenomic data and identified in a single analysis, involving 252 stool samples from 207 human individuals, almost as many SNPs in the human gut microbiome as the 1000 Genomes Project recorded in 179 human individuals over several years2. The stable pN/pS ratios of gut microbial species across individuals suggest that host conditions (such as diet, genetic differences, and immune tolerance) have a minor influence on the evolution of species compared to constraints common to the human population (such as gut physiology, anaerobic conditions, and pH). In the 66 dominant species, the analysis of more than 229,000 genes comprising about 8,000 OGs pinpointed consistently fast or slow evolving genes across individuals (Supplementary Table 10). However, further studies are needed in order to interpret different selection types at the gene level.
The availability of time-point data revealed that individual-specific variation patterns were remarkably stable over time (Fig. 3a,b), which was much less the case for similarities in species abundance – for almost 60% of the samples, a sample from a different individual was the most similar (Fig. 3c). Thus, the metagenomic variation patterns observed here support the hypothesis that a healthy individual retains specific strains for extended periods of time. This suggests that each individual has a metagenomic variation profile that could be unique even in very large cohorts. It should be noted that the maximal sampling period was only one year, and 43 individuals might not be sufficient to trace horizontal transmissions of strains. The likelihood of the latter is supported by the apparent absence of clear continental stratification (despite different sampling and sequencing protocols of the European and the US samples), although for one out of eight species analysed, Bacteroides coprocola, we provide preliminary evidence (Fig. 4; Supplementary Figure 8). Geographical stratification has been described for Helicobacter pylori44,45, and weak, but detectable signals have also been observed in some bacterial pathogens46,47. Thus, we expect more gut microbial stratification patterns to emerge when larger datasets under standardized sampling and sequencing protocols become available, although it remains to be tested which factors (such as geographic separation, diseases, host-genetic and life-style/diet factors) shape the distribution of gut microbial strains and segregating SNPs within the population. The absence of clear geographic stratification implies that stable differences in variation patterns of gut species are not explained by large-scale structures of local microbial populations. They may rather be a result of genetic drift due to population bottlenecks that could occur not only during the colonization of the infant gut but also by processes causing community shifts during adult life stages, followed by a rapid population growth accompanied by purifying selection. This model suggests that the source of genetic variation in human gut microbial populations is less likely to be new mutations within the host than the variation in the initial colonizing populations or transmissions from the environment. This would imply that most allelic variants analyzed in this study segregate at time scales greatly exceeding human generation time.
The introduction of large-scale variation analysis in metagenomic data of complex communities and the discovery of individual metagenomic variation profiles open up several applications. It is now possible to screen in silico for many pathogenic or antibiotics resistance variants in the population. Once a sample has been analyzed, the data can also be used in the future given the temporal stability of SNP profiles. As it took years to identify marker genes and variations for diseases or phenotypes in the human genome, the variation landscape uncovered here can only be seen as the beginning to find molecular biomarkers including particular variants that reveal useful information for human health and well-being.
Methods Summary
Mapping to non-redundant genomes
A reference genome set representing 929 species was derived from a total of 1,511 published prokaryotic genomes, based on a median sequence identity of 95% in 40 universal, single copy marker genes18,19. Metagenomic reads from 252 samples were aligned to these 929 genomes using the same 95% sequence identity cutoff.
Coverage
For each genome, we calculated the sample-specific base pair coverage and the number of bases of the genome covered by at least one read. For a genome to be considered we required a cumulative depth of coverage of ≥10x across all samples. In order to remove species that are not present in our cohort, yet attract reads due to highly conserved regions, we required at least 40% breadth of the genome coverage (the criterion for the species to be considered present) from at least one sample.
Variation detection
We performed SNP calling on the pooled samples and only considered bases with a quality score ≥15. We required SNPs to be supported by ≥4 reads and to occur with a frequency of ≥1%. Structural variations were detected using Pindel48. False positive rates in SNP and structural variation detection were estimated using nonsense and frameshift mutations in 40 essential single copy marker genes.
π and FST
We estimated nucleotide diversity (π) and fixation indices (FST) based on allele frequencies.
pN/pS ratio
SNPs occurring in coding regions were classified as synonymous or non-synonymous. Genes from the non-redundant genomes were annotated using eggNOG orthologous groups allowing calculation of pN/pS ratios at the level of genomes, OGs and genes.
Pairwise sample comparisons
Similarity in strain populations between two samples was estimated using (i) a similarity score based on shared SNPs and (ii) FST.
Supplementary Material
Acknowledgments
The authors are grateful to Jan Korbel and the members of the Bork group at EMBL for discussions and assistance, especially Sean Powell for performing some of the computations. We thank the EMBL IT core facility and Y. Yuan for managing the high-performance computing resources. We would like to thank Jeffrey I. Gordon for providing three of the samples used. We are also grateful to the European METAHIT consortium and the NIH Common Fund Human Microbiome Project Consortium for generating and making available the data sets used in this study. The research leading to these results has received funding from EMBL, the European Community's Seventh Framework Programme via the MetaHIT (HEALTH-F4-2007-201052) and IHMS (HEALTH-F4-2010-261376) grants as well as from the National Institutes of Health grants U54HG003079 and U54HG004968.
Footnotes
Author contributions
P.B. and G.M.W. conceived the study. P.B., M.A., G.M.W., and Sha.S. designed the analyses. Si.S., Shi.S., M.A., M.M., J.T., A.Z., A.W., D.M., J.M., and K.K. performed the analyses. M.A., Shi.S., Si. S., and P.B. wrote the manuscript. All authors read and approved the manuscript.
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. doi:10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. doi:10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 4.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. doi:10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 5.Bagel S, Hüllen V, Wiedemann B, Heisig P. Impact of gyrA and parCMutations on Quinolone Resistance, Doubling Time, and Supercoiling Degree of Escherichia coli. Antimicrobial Agents and Chemotherapy. 1999;43:868–875. doi: 10.1128/aac.43.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckburg PB, et al. Diversity of the Human Intestinal Microbial Flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. doi:10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morowitz MJ, et al. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1128–1133. doi: 10.1073/pnas.1010992108. doi:10.1073/pnas.1010992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sokurenko EV, et al. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Human Microbiome Project Consortium A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. doi:10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lay C, et al. Colonic Microbiota Signatures across Five Northern European Countries. Appl. Environ. Microbiol. 2005;71:4153–4155. doi: 10.1128/AEM.71.7.4153-4155.2005. doi:10.1128/aem.71.7.4153-4155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. doi:10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. doi:10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyson GW, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 15.Allen EE, et al. Genome dynamics in a natural archaeal population. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1883–1888. doi: 10.1073/pnas.0604851104. doi:10.1073/pnas.0604851104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. doi:10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson J, et al. The NIH Human Microbiome Project. Genome Research. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. doi:10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciccarelli FD, et al. Toward Automatic Reconstruction of a Highly Resolved Tree of Life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. doi:10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 19.Sorek R, et al. Genome-Wide Experimental Determination of Barriers to Horizontal Gene Transfer. Science. 2007;318:1449–1452. doi: 10.1126/science.1147112. doi:10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- 20.Konstantinidis KT, Tiedje JM. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol. 2007;10:504–509. doi: 10.1016/j.mib.2007.08.006. doi:10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS genetics. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. doi:10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. doi:10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller J, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucl. Acids Res. 2010;38:D190–195. doi: 10.1093/nar/gkp951. doi:10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunz BA, Glickman BW. The infidelity of conjugal DNA transfer in Escherichia coli. Genetics. 1983;105:489–500. doi: 10.1093/genetics/105.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simmons SL, et al. Population genomic analysis of strain variation in Leptospirillum group II bacteria involved in acid mine drainage formation. PLoS biology. 2008;6:e177. doi: 10.1371/journal.pbio.0060177. doi:10.1371/journal.pbio.0060177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. doi:10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 27.Friedman R, Drake JW, Hughes AL. Genome-wide patterns of nucleotide substitution reveal stringent functional constraints on the protein sequences of thermophiles. Genetics. 2004;167:1507–1512. doi: 10.1534/genetics.104.026344. doi:10.1534/genetics.104.026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novichkov PS, Wolf YI, Dubchak I, Koonin EV. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. Journal of bacteriology. 2009;191:65–73. doi: 10.1128/JB.01237-08. doi:10.1128/JB.01237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frey P. a. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1996;10:461–470. [PubMed] [Google Scholar]
- 30.Kuhner S, et al. Proteome Organization in a Genome-Reduced Bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. doi:10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 31.Holdeman LV, Moore WEC. New Genus, Coprococcus, Twelve New Species, and Emended Descriptions of Four Previously Described Species of Bacteria from Human Feces. International Journal of Systematic Bacteriology. 1974;24:260–277. doi:10.1099/00207713-24-2-260. [Google Scholar]
- 32.Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. International journal of systematic and evolutionary microbiology. 2002;52:1615–1620. doi: 10.1099/00207713-52-5-1615. [DOI] [PubMed] [Google Scholar]
- 33.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiology and molecular biology reviews : MMBR. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. doi:10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagai H, Roy CR. Show me the substrates: modulation of host cell function by type IV secretion systems. Cell Microbiol. 2003;5:373–383. doi: 10.1046/j.1462-5822.2003.00285.x. doi:285 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Kelly D, Conway S, Aminov R. Commensal gut bacteria: mechanisms of immune modulation. Trends Immunol. 2005;26:326–333. doi: 10.1016/j.it.2005.04.008. doi:10.1016/j.it.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 36.Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. doi:10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Begley M, Hill C, Gahan CGM. Bile salt hydrolase activity in probiotics. Applied and environmental microbiology. 2006;72:1729–1738. doi: 10.1128/AEM.72.3.1729-1738.2006. doi:10.1128/aem.72.3.1729-1738.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso JG, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. doi:10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. doi:10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fierer N, et al. Forensic identification using skin bacterial communities. Proceedings of the National Academy of Sciences. 2010;107:6477–6481. doi: 10.1073/pnas.1000162107. doi:10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nature reviews. Microbiology. 2010;8:207–217. doi: 10.1038/nrmicro2298. doi:10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 43.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. The ISME journal. 2007;1:56–66. doi: 10.1038/ismej.2007.3. doi:10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki R, Shiota S, Yamaoka Y. Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2012;12:203–213. doi: 10.1016/j.meegid.2011.12.002. doi:10.1016/j.meegid.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaoka Y. Helicobacter pylori typing as a tool for tracking human migration. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2009;15:829–834. doi: 10.1111/j.1469-0691.2009.02967.x. doi:10.1111/j.1469-0691.2009.02967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nature reviews. Microbiology. 2008;6:431–440. doi: 10.1038/nrmicro1872. doi:10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 47.Morelli G, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nature genetics. 2010;42:1140–1143. doi: 10.1038/ng.705. doi:10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. doi:10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.