Abstract
In response to genotoxic stress, a transient arrest in cell cycle progression enforced by the DNA damage checkpoint (DDC) signaling pathway positively contributes to genome maintenance1. Because hyperactivated DDC can lead to a persistent and detrimental cell cycle arrest2,3, cells must tightly regulate the activity of DDC kinases. Despite their importance, the mechanisms for monitoring and modulating DDC signaling are not fully understood. Here we show that DNA repair scaffolding proteins Slx4 and Rtt107 prevent lesions generated during DNA replication from aberrantly hyperactivating DDC signaling in Saccharomyces cerevisiae. Upon replication stress, cells lacking Slx4 or Rtt107 exhibit hyperactivation of the downstream DDC kinase Rad53 while activation of the upstream DDC kinase Mec1 remains normal. An Slx4-Rtt107 complex counteracts the checkpoint adaptor Rad9 by physically interacting with Dpb11 and phospho-H2A, two positive regulators of Rad9-dependent Rad53 activation. Reduction of DDC signaling by hypomorphic mutations in RAD53 and H2A rescue the hyper-sensitivity of slx4Δ or rtt107Δ cells to replication stress. We propose that the Slx4-Rtt107 complex modulates Rad53 activation via a competition-based mechanism that balances the engagement of Rad9 at replication-induced lesions. Our findings reveal that DDC signaling is monitored and modulated through the direct action of DNA repair factors.
Slx4 is an evolutionarily conserved DNA repair scaffolding protein important for the cellular response to exogenous DNA damaging agents4,5,6,7, and mutations in human SLX4 were recently linked to Fanconi anemia8,9. In budding yeast, slx4Δ cells are highly sensitive to methyl methanesulfonate (MMS)7, a DNA alkylating agent that induces replication blocks and DDC activation. While investigating the activation status of the budding yeast DDC kinase Rad53 in slx4Δ cells, we noticed that MMS treatment leads to hyperphosphorylation, thus hyperactivation, of Rad53 when compared to wild-type cells (Fig. 1a and Supplementary Fig. 1a), consistent with a previous report10. Phosphorylation of histone H2a (Hta1/Hta2), a substrate of the upstream DDC kinase Mec1, at serine 129 (here referred as H2apS129) was not increased in slx4Δ cells, however (Fig. 1a, lower panel). These results suggest that the hyperactivation of Rad53 in slx4Δ cells is not caused by increased damage-induced Mec1 signaling but results from improper downstream regulation of Rad53 activation. To test this possibility, we compared the phosphoproteome of wild-type and slx4Δ cells following MMS treatment using quantitative mass-spectrometry. While most of the detected Mec1 targets are phosphorylated to the same extent in both cell types, Rad53-dependent phosphorylation is significantly increased in slx4Δ cells (Supplementary Fig. 1b), further supporting that Slx4 plays a role in specifically blocking Rad53 hyperactivation. Because activation of Rad53 in response to MMS is mostly dependent on the checkpoint adaptor Rad9 (Fig. 1b and Supplementary Fig. 2), Slx4 likely counteracts Rad9-dependent Rad53 activation.
Figure 1. Slx4 counteracts Rad9-dependent Rad53 activation.
a, Western blot showing phosphorylation of Rad53-HA and histone H2aS129. b, Western blot showing phospho-status of Rad53-HA in indicated strains. c, Schematic representation of the Rad53 protein. d, MMS sensitivity assay of strains containing the indicated Flag-tagged RAD53 alleles. Similar results were obtained with untagged Rad53 strains (Supplementary Fig. 3). e, Western blot showing phospho-status of Rad53-Flag. f, Model for the role of Slx4 in uncoupling Rad53 activation from Mec1 signaling by counteracting Rad9.
To test whether the sensitivity of slx4Δ cells to MMS is caused mostly by aberrant Rad53 hyperactivation, we used hypomorphic alleles of rad53 that reduce Rad53 activation, reasoning that they would rescue the MMS sensitivity of slx4Δ cells. Rad53 has two FHA domains that redundantly bind to phosphorylated Rad9 to mediate Rad53 activation11 (Fig. 1c), and mutations in the FHA2 domain promote a stronger reduction in MMS-induced Rad53 activation than mutations in the FHA1 domain12. Whereas a mutation (R70A) in the FHA1 domain of Rad53 had no effect on the MMS sensitivity of slx4Δ cells, a mutation (R605A) in the FHA2 domain reduced the sensitivity of slx4Δ cells (Fig. 1d). Consistent with our hypothesis that Rad53 hyperactivation is the cause of the MMS sensitivity of slx4Δ cells, mutation of the FHA2 domain reduced Rad53 activation in slx4Δ cells to a level similar to wild-type (Fig. 1e). Collectively, these results suggest that Slx4 has a crucial role in preventing excessive Rad9-dependent activation of Rad53 (Fig. 1f). Because the levels of MMS used here require that cells pass through S phase for Rad53 to become active13, our results suggest that Slx4 counteracts the Rad9-Rad53 pathway in response to replication-induced lesions. The fact that combined deletion of the SLX1 and RAD1 genes, which encode nucleases known to associate with Slx4, leads to lower MMS sensitivity and Rad53 activation compared to slx4Δ cells (Supplementary Fig. 4) supports a nuclease-independent function for Slx4 during the cellular response to MMS-induced replication stress.
We have recently reported that upon replication stress Slx4 binds Dpb1114, a replication factor involved in DDC activation15,16. Because Dpb11 binds Rad9 to positively regulate Rad9-dependent Rad53 activation17,18, we assessed whether the Slx4-Dpb11 interaction plays a role in counteracting Rad53 activation during MMS treatment. We previously showed that phosphorylation of Slx4 by Mec1 mediates the interaction with Dpb11, and that an slx4 mutant lacking seven Mec1 consensus phosphorylation sites (slx4-7MUT) is unable to stably interact with Dpb1114. As shown in Fig. 2a, Rad53 is hyperactivated in slx4-7MUT cells, supporting a model in which the Slx4-Dpb11 interaction is important to prevent Rad53 hyperactivation. Next, we tested whether the Dpb11-Slx4 interaction inhibits the ability of Rad9 to bind Dpb11 in wild-type and slx4Δ cells. Deletion of SLX4 leads to a significant increase in the MMS-induced interaction between Dpb11 and Rad9 (Fig. 2b and Supplementary Fig. 5), suggesting that Slx4 and Rad9 compete for Dpb11 binding. Dpb11 contains two pairs of BRCT domains that bind phosphorylated motifs. We found that recombinant BRCT domains 1 and 2 (BRCT1/2) of Dpb11 are able to bind both phosphorylated Slx4 and Rad9 from MMS-treated yeast cell lysates (Fig. 2c), consistent with a model where Slx4 and Rad9 compete for BRCT1/2 binding. BRCT1/2 of Dpb11 was previously shown to directly interact with CDK-dependent phosphorylation sites in Sld3 to initiate DNA replication19,20. To better understand the mechanism of the Slx4-Dpb11 interaction, we looked for a CDK targeting motif in Slx4 that resembles the Dpb11 binding region in Sld3. Notably, Slx4 contains proline-directed phospho-sites that align well with serines 600 and 622 of Sld3 important for Dpb11-Sld3 interaction19,20 (Supplementary Fig. 6). We tested the importance of the proline-directed sites within the Sld3-like region in Slx4 and found that serine 486 of Slx4 is crucial for the MMS-induced interaction with Dpb11 (Fig. 2d). In addition, three canonical Mec1 phosphorylation sites (S/T-Q sites) important to mediate robust Slx4-Dpb11 interaction14 are located within or nearby the Sld3-like Dpb11 binding region in Slx4. We conclude that the Slx4-Dpb11 interaction is mediated by the coordinated action of Mec1 and of a proline-directed kinase at a motif likely targeted by BRCT1/2 of Dpb11. While we could still detect residual binding between Dpb11 and slx4-7MUT, we could not detect interaction between Dpb11 and slx4S486A suggesting that phosphorylation of serine 486 plays a more important role in mediating the Slx4-Dpb11 interaction. This is also supported by the fact that slx4S486A leads to Rad53 hyperactivation (Fig. 2e) and a higher MMS sensitivity than slx4-7MUT (Fig. 2f). Interestingly, previous reports showed that proline-directed sites in Rad9 are also important for the Dpb11-Rad9 interaction17,18 (see Supplementary Fig. 7). To further test the model that Slx4 antagonizes Rad53 activation by binding Dpb11 and outcompeting Rad9, we overexpressed Slx4 using an ADH1 or a TDH3 promoter and monitored different steps of checkpoint activation. In an early step in checkpoint activation, Rad9 assembles into a ternary complex with Dpb11 and Mec1 and is hyperphosphorylated by Mec118. We first monitored Rad9 phospho-status in cells expressing Slx4 from its endogenous promoter and in cells overexpressing Slx4. Overexpression of wt, but not S486A mutated, Slx4 significantly inhibited the MMS-induced hyperphosphorylation of Rad9 (Fig. 2g), as revealed by the strong reduction of the slower migrating band at 45 and 60 min after release from α-factor arrest. We then monitored the effect of Slx4 overexpression in Rad53 activation in response to MMS treatment (Supplementary Fig. 8a). While overexpression of Slx4 leads to a small but consistent reduction in Rad53 activation early in the response, we could not observe the same effect at later time points. We speculated that after 40 min, the Rad53 activation observed in Slx4 overexpressing cells was being mediated by a parallel mechanism based on Dot1-mediated histone H3-K79 methylation, which can promote the recruitment of Rad9 to sites of lesions independently of Dpb1121. We therefore monitored Rad53 activation in cells lacking DOT1 and found that Slx4 overexpression significantly reduces Rad53 activation also at later time points (Fig. 2h); this effect is dependent on serine 486 of Slx4 (Supplementary Fig. 8b,c). Next, we determined if Slx4 overexpression specifically disrupts the Rad9-Dpb11 interaction. We could detect hyperphosphorylated Rad9 in a Dpb11 pull-down from cells expressing Slx4 from its endogenous promoter, but we could not detect Rad9 in a Dpb11 pull-down using cells overexpressing Slx4 (Fig. 2i). This effect was specific for serine 486 in Slx4. These findings support our model that Slx4 counteracts DDC signaling by binding Dpb11 and preventing its stable interaction with Rad9 (Fig. 2j).
Figure 2. Slx4 binding to Dpb11 counteracts the Dpb11-Rad9 interaction and Rad53 activation.
a, Western blot showing the phospho-status of Rad53-HA. b, Co-immunoprecipitation (co-IP) between Dpb11 and Rad9 (see also Supplemental Figure 5). c, Pull-down of Slx4-Flag or Rad9-HA from yeast lysates using recombinant BRCT1/2 and BRCT3/4 from Dpb11. d, Interaction of the indicated Flag-tagged slx4 mutants with Dpb11-HA. e, Rad53 phospho-status in the indicated slx4 mutants. Experiment was performed as described in Fig. 1a. f, MMS sensitivity assay. g, Rad9 phospho-status in cells expressing Slx4 from the endogenous SLX4 promoter or ADH1 promoter. h, Rad53 phospho-status in dot1Δ cells expressing Slx4 from the endogenous SLX4 promoter or TDH3 promoter. Cells were arrested in α-factor and released for 45 min in media containing MMS. i, Co-IP between Dpb11 and Rad9. j, Model for mechanism by which Slx4 counteracts Rad53 activation.
Slx4 forms a tight complex with the BRCT domain-containing protein Rtt107, a DNA repair scaffold that stabilizes the Dpb11-Slx4 interaction14. Analysis of the phospho-status of Rad53 in rtt107Δ cells revealed that DDC signaling is hyperactivated (Fig. 3a), suggesting that Rtt107 contributes to a similar function as Slx4 in counteracting Rad53 activation. Rtt107 recognizes H2apS129 via its C-terminal pair of BRCT domains (BRCT5/6) and this region is essential for Rtt107’s role in the MMS response (Supplementary Figs 9 and 10)22. Furthermore, mutation of serine 129 to alanine (h2a-S129A) abrogated the phosphorylation of Slx4 upon MMS treatment (Fig. 3b), suggesting that the recruitment of Rtt107 to H2apS129 occurs upstream of Slx4-Dpb11 complex formation. Because H2apS129 can function as a positive regulator of Rad9-dependent Rad53 activation23, we reasoned that the hyperactivation of Rad53 in rtt107Δ cells could be suppressed by h2a-S129A. rtt107Δ cells expressing the h2aS129A mutant exhibit reduced Rad53 activation and MMS sensitivity than rtt107Δ cells expressing wild-type H2a (Fig. 3c,d). Disruption of the Rad9-H2a interaction by mutation of the C-terminal pair of BRCT domains of Rad9 (K1088M) partially rescues MMS-sensitivity of rtt107Δ cells (Supplementary Fig. 11a). A similar rescue is also observed when overexpressing BRCT5/6 of Rtt107 (Supplementary Fig. 11b). Taken together, these results reveal that Rtt107 also counteracts DDC signaling. The anti-checkpoint function of Rtt107 depends on its recognition of H2apS129, a step that is further required for the assembly of the Slx4-Dpb11 complex. Thus, the Slx4-Rtt107 complex functions as a negative regulator of Rad53 kinase activation by physically interacting with two positive regulators of the Rad9 adaptor, Dpb11 and H2apS129.
Figure 3. Rtt107 counteracts Rad9-dependent Rad53 activation by binding phosphorylated histone H2a.
a, Western blot showing the phospho-status of Rad53-HA. b, Western blot showing Slx4-Flag phospho-status. c, Western blot showing phospho-status of Rad53 and H2aS129. Indicated strains expressing HA-tagged RAD53 were arrested in nocodazole and released for 60 min in media containing MMS. d, MMS sensitivity assay.
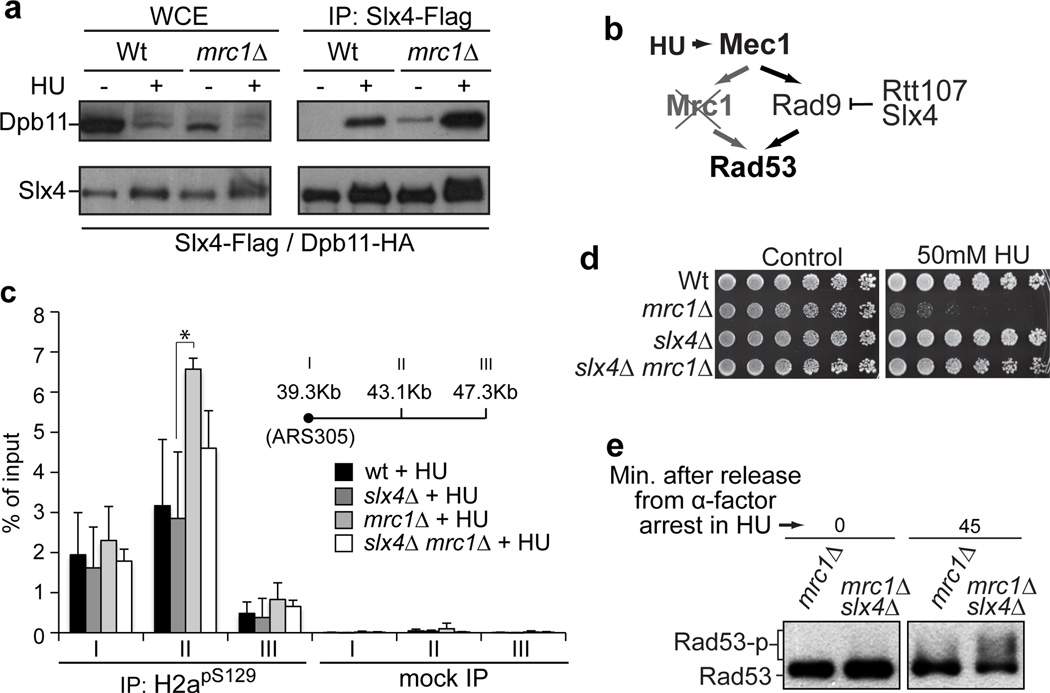
To further test the anti-checkpoint function of the Slx4-Rtt107 complex, we developed an alternative experimental setup in which the presence of Slx4 would sensitize cells to replication stress by reducing DDC signaling below a level required for proper cellular response. For that, we used cells lacking Mrc1, a checkpoint adaptor that works in parallel with Rad9. Mrc1 mediates Rad53 activation at stalled replication forks and therefore plays a more central role than Rad9 in Rad53 activation in response to hydroxyurea (HU)24. Upon HU treatment, mrc1Δ cells rely solely on Rad9 to activate Rad53. As shown in Fig. 4a, the interaction of Slx4 with Dpb11 was enhanced in mrc1Δ cells, especially in response to HU, suggesting that the Slx4-Rtt107 complex is actively counteracting Rad9 in mrc1Δ cells (Fig. 4b). Consistent with our finding that the Slx4-Dpb11 interaction requires H2apS129, high levels of H2apS129 were found to accumulate in the proximity of origins of replication in mrc1Δ cells after HU treatment (Fig. 4c). Because we expect that Rad53 kinase activity is limiting in mrc1Δ cells, we reasoned that deletion of SLX4 would be beneficial in HU-treated mrc1Δ cells by allowing more activation of Rad53 via Rad9. Consistent with this, mrc1Δslx4Δ cells display significantly higher HU resistance compared to mrc1Δ cells (Fig. 4d), and this correlates with higher Rad53 activation (Fig. 4e). Taken together, these results reveal that Slx4 sensitizes mrc1Δ cells to replication stress and provide strong additional evidence that the Slx4-Rtt107 complex counteracts Rad9 in response to replication-induced lesions (see detailed model in Supplementary Fig. 12). Because this function of the Slx4-Rtt107 complex is dependent on Mec1-dependent phosphorylation, we propose that this complex is involved in a mechanism we have named DAMP, for Dampens checkpoint Adaptor Mediated Phospho-signaling, by which DDC signaling self-monitors its activation state. We speculate that by uncoupling upstream Mec1 signaling from downstream Rad53 activation, DAMP could allow Mec1 to maintain control over specific effectors, such as repair enzymes, without an aberrant arrest in cell cycle progression.
Figure 4. Slx4 sensitizes mrc1Δ cells to HU-induced replication stress.
a, Co-IP of Dpb11 and Slx4 after 2 hr treatment with 0.1M HU. b, Model for the action of Slx4 and Rtt107 in mrc1Δ cells. c, HU treatment of mrc1Δ cells leads to H2apS129 accumulation near an origin of replication. ChIP analysis of H2apS129 at neighboring ARS305 regions I, II and III on the indicated strains exposed to 0.2 M HU. Data are presented as mean + s.e.m. (n=3). Asterisk indicates a statistically significant difference, as analyzed by unpaired, two-tailed student’s t test (P=0.025). d, HU sensitivity assay. e, Western blot for the phospho-status of Rad53-HA upon treatment with 10 mM HU.
METHODS SUMMARY
Yeast strains and plasmids used are described in Supplementary Tables 3 and 4. Mass spectrometry-based phosphorylation analyses of whole cell lysates or purified Rad53, as well as other procedures used for cell growth and synchronization, genotoxin treatment, and ChIP analysis are detailed in the Methods section.
Methods
Yeast strains and plasmids
Strains generated in this study are derived either from MBS164 or MBS191 (both congenic to S288C) or W303 (where indicated). Unless indicated, all tags were inserted at the C-terminus of the corresponding genes by homologous recombination at the genomic loci and verified by western blot. Tagged strains were assayed for sensitivity to MMS to ensure they behaved similar to wild-type. Sensitivity assays were independently confirmed in strains derived from freshly sporulated diploids. Standard cloning methods were used to generate the plasmids for this study. Plasmids containing domains of Dpb11 or Rtt107 tagged at the N-terminus with a PATH tag (2 × protein A + TEV cleavage site + 6×His)25 were based on pET21a (Novagen) vector. HA-tagged full length Rtt107, along with its native promoter, was cloned into pRS416 (Stratagene) to generate pMBS163. Wild-type alleles cloned into a pFA6a vector (Addgene) were linearized before integration into the respective endogenous loci. A pYES2/NT C vector (Invitrogen) containing full length or C-terminal BRCTs of Rtt107 was used for over-expression. SLX4 and DPB11 constructs containing an ADH1 or a TDH3 promoter were generated by fusing the respective promoters (800 bp upstream the start codon) to the corresponding ORF. The resulting PCR products were subsequently cloned into the pRS416 or pFA6a vectors. All point mutations were generated via site directed mutagenesis using either QuikChange Multi Site-Directed Mutagenesis (Stratagene) or the PFU Ultra II (Agilent) Kits. All yeast strains and plasmids used in this study are described in Supplementary Tables 3 and 4 and are available upon request.
Cell synchronization and genotoxin treatment
Yeast cells were grown in yeast peptone dextrose (YPD) or drop-out media at 30°C. Log phase cultures (O.D.600 ≈ 0.3) were subjected to alpha factor (0.5 ug/ml) or nocodazole (1.5 ug/ml) treatment for G1 or G2/M arrest, respectively. Cells were then washed and resuspended in warm media containing the indicated genotoxins.
Western blotting and immunoprecipitation
For western blotting, about 50 mg of frozen cell pellets were lysed by bead beating at 4°C in lysis buffer (50 mM Tris-HCl pH 7.5, 0.2% Tergitol, 150 mM NaCl, 5 mM EDTA, 1 mM PMSF, Complete EDTA free protease inhibitor cocktail (Roche), PhosSTOP (Roche)). SDS loading buffer with 60 mM DTT was added. Samples were separated by standard SDS-polyacrylimide gel electrophoresis (SDS-PAGE). Proteins were detected using the following antibodies: anti-Rad53 (yc-19, 1:10000, Santa Cruz), anti-H2a-pS129 (07-0745, 1:10000, Millipore), anti-HA (12CA5, 1:10000, Roche), anti-FLAG (M2, 1:5000, Sigma). For immunoprecipitation (IP), approximately 100 mg of frozen cell pellets were lysed by bead beating at 4°C in lysis buffer (50 mM Tris-HCl pH 7.5, 0.2% Tergitol, 150 mM NaCl, 5 mM EDTA, 1 mM PMSF, Complete EDTA free protease inhibitor cocktail (Roche), 5 mM sodium fluoride, 10 mM B-glycerol-phosphate). After adjusting protein concentrations to around 6 mg/ml, inputs were aliquoted and lysates were incubated with either anti-HA or anti-FLAG agarose resin (Sigma) for 2 to 3 h at 4°C. After 3 washes in lysis buffer, bound proteins were eluted with 3 resin volumes of SDS elution buffer (100 mM Tris-HCl pH 8.0, 1% SDS) for HA-IP or FLAG peptide (Sigma) solution (0.5ug/mL in 100 mM Tris, 0.2% Tergitol) for FLAG-IP. SDS loading buffer with DTT was added and samples were analysed by western blotting with indicated antibodies.
Pull-down with recombinant BRCT domain
Protein domains (for Dpb11, BRCT1/2: amino acids 1–270 and BRCT3/4: amino acids 271–582) containing an N-terminal PATH tag (see Plasmids) were expressed in E. coli, bound to human IgG-agarose resin (GE healthcare) and then used as bait for pull-downs from yeast lysates as previously described25.
SILAC labeling of yeast
For mass spectrometry experiments, cells were grown in (−) Arg (−) Lys dropout media (‘‘light’’ version complemented with normal arginine and lysine; ‘‘heavy’’ version complemented with lysine 13C6, 15N2 and arginine 13C6, 15N4) for at least 5 generations.
Purification of phosphopeptides by IMAC
For the purification of Rad53 phosphopeptides, approximately 0.6 g of cell pellet of wild-type (grown in "heavy" media) or slx4Δ (grown in "light" media) strains carrying Rad53-HA was lysed by bead beating at 4°C in 4 mL of lysis buffer (50 mM Tris-HCl pH 7.5, 0.2% Tergitol, 150 mM NaCl, 5 mM EDTA, Complete EDTA-free protease inhibitor cocktail (Roche), 5 mM sodium fluoride, 10 mM B-glycerol-phosphate). Lysate was incubated with anti-HA agarose resin (Sigma) for 4 h at 4°C. After 3 washes with lysis buffer, bound proteins were eluted with 3 resin volumes of elution buffer (100 mM Tris-HCl pH 8.0, 1% SDS). Eluted proteins from light or heavy media were mixed together, reduced, alkylated and precipitated. Proteins were resuspended in 2M urea, 12.5 mM Tris-HCl pH 8.0, and digested with trypsin for 16 h at 37°C. Phosphopeptides were enriched using an "in-house" IMAC column, then eluted with 10% Ammonia, 10% acetonitrile and dried in a speed-vac.
Phosphoproteome analysis
Approximately 0.6 g of cell pellet of wild-type (grown in "light" media) and slx4Δ (grown in "heavy" media) strains was lysed by bead beating at 4°C in 4 mL of lysis buffer (50 mM Tris-HCl pH 7.5, 0.2% Tergitol, 150 mM NaCl, 5 mM EDTA, Complete EDTA-free protease inhibitor cocktail (Roche), 5 mM sodium fluoride, 10 mM B-glycerol-phosphate). Protein lysate was denatured with 1% SDS, reduced, alkylated and then precipitated with 3 volumes 50% acetone, 50% ethanol. Proteins were solubilized with 2M urea, 50mM Tris-HCl, pH 8.0, 150mM NaCl, followed by the addition of TPCK treated trypsin. Digestion was performed overnight at 37°C and then trifluoroacetic acid and formic acid were added to a final concentration of 0.2%. Peptides were de-salted with Sep-Pak C18 column (Waters), dried in a speed-vac and ressuspended in 1% acetic acid. Phosphopeptides were enriched by IMAC as previously described26,27,28 and then reconstituted in 85 uL of a solution containing 80% acetonitrile and 1% formic acid and fractionated by Hydrophilic Interaction Liquid Chromatography (HILIC) as previously described26 before analysis by LC-MS/MS. Over 3570 phosphopeptides were identified/quantified and the results for phosphopeptides containing a phosphorylation site known to be Mec1 targets or Rad53-dependent phosphorylation27,29 were selected.
Mass spectrometry analysis
IMAC elutions or HILIC fractions were dried in a speed-vac, reconstituted in 0.1% trifluoroacetic acid and analyzed by LC-MS/MS using 125 uM ID capillary C18 column and an Orbitrap XL mass spectrometer coupled with an Eksigent nano-flow system. Database search was performed using the Sorcerer system (Sagen) running Sequest program. After searching a target-decoy budding yeast database, results were either filtered based on Probability Score to achieve a 1% false positive rate or manually inspected. Quantitation of heavy / light peptide isotope ratios was performed using the Xpress program as previously described27.
Chromatin immunoprecipitation (ChIP)
Cultures were grown in YPD to an O.D.600 of ≈ 0.3, arrested in G1 with alpha-factor for 2 h and released in the presence of 200 mM of HU for 1 h. Cultures were formaldehyde (1% final) crosslinked for 20 min followed by quenching with 125 mM glycine. Protein-DNA complexes were immunoprecipitated with an yeast specific anti-phospho-histone (H2a-pS129) antibody (07-745, Millipore). qPCR was performed with the purified DNA from the immunoprecipitate as previously described30 using pair of primers designed to amplify the genome sequences shown in Fig. 4C. Primer sequences are available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (RO1-GM097272 to M.B.S.; F31-GM093588 to P.O.). F.M.B.O. was supported by a Cornell Fleming Research Fellowship; C. J. M. was supported by an HHMI Institutional Undergraduate Education Grant to Cornell. The authors would like to thank Beatriz Almeida for technical assistance. We thank R. Weiss, S. Emr, A. Bretscher, G. Balmus and P. Russell for comments on the manuscript.
Footnotes
SUPPLEMENTARY INFORMATION is linked to the online version of the paper at www.nature.com/nature.
AUTHOR CONTRIBUTION P.O., F.M.B.O. and M.B.S. designed and performed experiments and analyzed the data. P.O. and M.B.S. performed the mass spectrometry experiments. F.M.B.O. performed the ChIP analysis and generated slx4 mutants. Y.L. and P.O. performed co-IPs between Dpb11 and Rad9. Y.L. performed pull-downs with BRCT domains of Dpb11. C.J.M. performed the Rtt107-H2a binding assay and experiments with Rtt107 BRCT domains. P.O. and M.B.S. performed experiments with overexpression of Slx4. P.O. and M.B.S. wrote the paper.
AUTHOR INFORMATION All data sets in this publication are available in the supplementary information. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 2.Clerici M, et al. Hyperactivation of the yeast DNA damage checkpoint by TEL1 and DDC2 overexpression. EMBO J. 2001;20:6485–6498. doi: 10.1093/emboj/20.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- 4.Fekairi S, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svendsen JM, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munoz IM, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 7.Fricke WM, Brill SJ. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoepker C, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 9.Kim Y, et al. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts TM, et al. Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol Biol Cell. 2006;17:539–548. doi: 10.1091/mbc.E05-08-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz MF, et al. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol Cell. 2002;9:1055–1065. doi: 10.1016/s1097-2765(02)00532-4. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz MF, Lee SJ, Duong JK, Eminaga S, Stern DF. FHA domain-mediated DNA checkpoint regulation of Rad53. Cell Cycle. 2003;2:384–396. [PubMed] [Google Scholar]
- 13.Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 14.Ohouo PY, Bastos de Oliveira FM, Almeida BS, Smolka MB. DNA damage signaling recruits the Rtt107-Slx4 scaffolds via Dpb11 to mediate replication stress response. Mol Cell. 2010;39:300–306. doi: 10.1016/j.molcel.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 15.Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem. 2008 doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mordes DA, Nam EA, Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci U S A. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granata M, et al. Dynamics of Rad9 chromatin binding and checkpoint function are mediated by its dimerization and are cell cycle-regulated by CDK1 activity. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfander B, Diffley JF. Dpb11 coordinates Mec1 kinase activation with cell cycle-regulated Rad9 recruitment. EMBO J. 2011;30:4897–4907. doi: 10.1038/emboj.2011.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka S, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 20.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 21.Puddu F, et al. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol. 2008;28:4782–4793. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, et al. Structure of C-terminal tandem BRCT repeats of Rtt107 protein reveals critical role in interaction with phosphorylated histone H2A during DNA damage repair. J Biol Chem. 2012;287:9137–9146. doi: 10.1074/jbc.M111.311860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Javaheri A, et al. Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling. Proc Natl Acad Sci U S A. 2006;103:13771–13776. doi: 10.1073/pnas.0511192103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alcasabas AA, et al. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
References in Methods
- 25.Smolka MB, et al. An FHA domain-mediated protein interaction network of Rad53 reveals its role in polarized cell growth. J Cell Biol. 2006;175:743–753. doi: 10.1083/jcb.200605081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albuquerque CP, et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smolka MB, et al. Dynamic changes in protein-protein interaction and protein phosphorylation probed with amine-reactive isotope tag. Mol Cell Proteomics. 2005;4:1358–1369. doi: 10.1074/mcp.M500115-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen SH, Albuquerque CP, Liang J, Suhandynata RT, Zhou H. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J Biol Chem. 2010;285:12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.