Abstract
Skp2 SCF complex displays E3 ligase activity and oncogenic activity by regulating protein ubiquitination and degradation, in turn regulating cell cycle entry, senescence and tumorigenesis. The maintenance of the integrity of Skp2 SCF complex is critical for its E3 ligase activity. The Skp2 F-box protein is a rate-limiting step and key factor in this complex, which binds to its protein substrates and triggers ubiquitination and degradation of its substrates. Skp2 is found to be overexpressed in numerous human cancers, which plays an important role in tumorigenesis. The molecular mechanism by which the function of Skp2 and Skp2 SCF complex is regulated remains largely unknown. Here we show that Foxo3a transcription factor is a novel and negative regulator of Skp2 SCF complex. Foxo3a is found to be a transcriptional repressor of Skp2 gene expression by directly binding to the Skp2 promoter, thereby inhibiting Skp2 protein expression. Surprisingly, we found for the first time that Foxo3a also displays a transcription-independent activity by directly interacting with Skp2 and disrupting Skp2 SCF complex formation, in turn inhibiting Skp2 SCF E3 ligase activity and promoting p27 stability. Finally, we show that the oncogenic activity of Skp2 is repressed by Foxo3a overexpression. Our results not only reveal novel insights into how Skp2 SCF complex is regulated, but also establish a new role for Foxo3a in tumor suppression through a transcription-dependent and independent manner.
Introduction
Skp2 (S-phase kinase-associated protein-2) belongs to the F-Box protein family that forms a complex with Skp1, Cullin1 (Cul-1), and Rbx1 to constitute the Skp2 SCF complex (1, 2). The Skp2 SCF complex displays ubiquitin ligase (E3) activity, which binds to protein substrates and triggers their ubiquitination and proteosome-dependent degradation. Skp2 recognizes protein substrates and brings them to the Skp2 SCF complex for subsequent ubiquitination and degradation (1, 2). Although several proteins have been reported to be Skp2 substrates, p27, a cell cycle inhibitor, is most well characterized and represents a physiological target for Skp2. This notion is supported by the genetic evidence showing that Skp2-/- MEFs display elevated p27 protein levels and a marked defect in proliferation, and concomitant loss of p27 rescues proliferative defects and reduction in body and organ size observed in Skp2-/- mice (3, 4).
Importantly, Skp2 is found to overexpress in a variety of human cancer and associate with cancer metastasis (1, 2, 5), suggesting that Skp2 overexpression may contribute to cancer development and metastasis. In support of this notion, we found that Skp2 overexpression promotes cancer development and metastasis by regulating p27 expression and RhoA expression (6, 7). Moreover, using the genetic mouse tumor models, Skp2 deficiency profoundly restricts cancer development by regulating either cellular senescence or apoptosis depending on distinct cell and tissue contexts (8, 9). Thus, Skp2 displays an oncogenic activity which targeting may be a promising therapeutic strategy for cancer treatment.
Skp2 level is tightly orchestrated during the cell cycle. It is low in early G1 phase, but increases at the G1 to S phase transition, which inversely correlates with the level of Cdh1, a coactivator for the ubiquitin ligase anaphase promoting complex/cyclosome (APC/C). The degradation of Skp2 is regulated by Cdh1 through Cdh1-mediated ubiquitination and degradation (10, 11). Although Skp2 plays a critical role in cell cycle transition and oncogenic activity, how Skp2 gene expression and its protein activity are regulated remains largely unclear. Understanding the molecular mechanism by which Skp2 gene expression and Skp2 SCF E3 ligase activity are orchestrated may result in an important therapeutic application.
Foxo3a is a Forkhead box (Fox) transcription factor that contains a conserved DNA binding domain capable of binding to DNA to induce or repress gene expression. Overexpression of Foxo3a causes cell cycle arrest and apoptosis by inducing or repressing expression of several genes such as p27, Bim, FasL, and Gadd45 (12, 13). Interestingly, loss of Foxo3a expression or cytosolic relocalization is found in various human cancer types (14). Overexpression of Foxo3a inhibits breast cancer development, whereas silencing its expression promotes breast cancer development in xenograft model (14, 15). These results suggest that loss of foxo3a functions may contribute to cancer development. Interestingly, Foxo3a activity is negatively regulated by oncogenic kinase, such as Akt, IKK, and ERK (12, 13), which are known to be activated in human cancers, raising the possibility that these oncogenic kinases may trigger Foxo3a inactivation during cancer development.
In this study, we aim to decipher the mechanism by which Skp2 gene expression and the formation of Skp2 SCF complex are regulated. We find here that Foxo3a is not only a novel transcriptional repressor for Skp2 gene expression, but also a repressor for the formation of Skp2 SCF complex, offering a new mechanistic insight how Foxo3a may serve as a tumor suppressive activity.
Results
Foxo3a is a transcriptional repressor for Skp2 gene expression
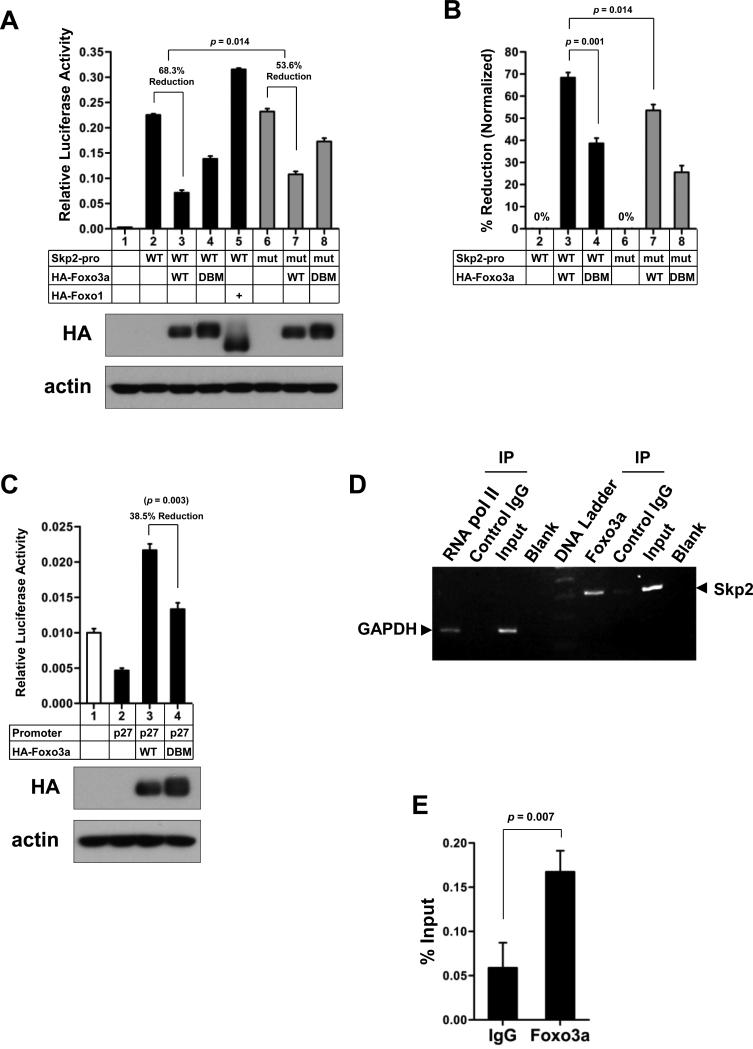
To understand how Skp2 gene expression is regulated, we analyzed the Skp2 promoter region and found that the Skp2 promoter consists of two potential consensus binding sites [744TAGAGTAAATT754; 997TTATGTAAACA1007; Accession number DQ021501; foxo3a consensus binding site: A/G TAAA T/C A/T] (16) for Foxo3a, a tumor suppressor involved in cell-growth arrest and apoptosis (12, 13). We therefore hypothesized that Foxo3a may be a transcription factor that regulates Skp2 gene expression. To test this hypothesis, we performed the luciferase reporter assay to examine whether Foxo3a regulates Skp2 promoter activity and found that overexpression of Foxo3a significantly reduced the Skp2 promoter activity (Fig. 1A and B). To further test the specificity of the Foxo3a-mediated transcriptional repression of the Skp2 promoter, we applied both the DNA binding mutant (DBM) form of Foxo3a, Foxo3a-H212R, which cannot bind to the consensus Forkhead responsive element (17), and the mutant Skp2 promoter construct, which harbors mutations in both two potential Foxo3a consensus binding sites, to the luciferase reporter assay. As shown in Fig. 1A and B, while Foxo3a suppressed the Skp2 promoter activity (68.3% reduction), Foxo3a-DBM compromised this activity (38.6% reduction), indicating that Foxo3a is a transcriptional repressor of Skp2. Surprisingly, Foxo1, another member of the Foxo family transcription factors, failed to suppress Skp2 promoter activity (Fig. 1A, lane 5). As for the mutant Skp2 promoter, the suppression effect of Foxo3a-WT on the Skp2 promoter activity was reduced by the mutations of the identified Foxo3a binding sites, and the reduction rate was dropped from 68.3% to 53.6% (Fig. 1A and B). Both moderately compromised effects highly suggest that there are likely other unrecognized non-consensus Foxo3a binding sites present within the ~4 kb Skp2 promoter. The defective function of Foxo3a-DBM was confirmed using the p27 promoter luciferase construct in Fig. 1C. To determine whether Foxo3a is a transcription repressor that directly binds to Skp2 promoter, we next performed the ChIP assay and found that Foxo3a directly bound to the Skp2 promoter region. Accordingly, our results suggest that Foxo3a is a novel transcriptional repressor for Skp2.
Figure 1. Foxo3a is a transcription repressor of Skp2.
(A) HEK293T cells were transfected with the indicated plasmids for 48 h and harvested for the Skp2 reporter assay. The relative luciferase activity was presented as a ratio of the firefly luciferase activity to the Renilla luciferase activity and expressed as mean ± s.d. (n=3). (B) The data from (A) were recalculated and presented as the percentage of reduction normalized to each mock control. WT or mutant Skp2 promoter only is set as 0% respectively. The sample number is correlated to the number in (A). The results are shown as mean ± s.d. (n=3). (C) Equal aliquots of chromatin extracts from PC-3 cells were subjected to immunoprecipitation with anti-Foxo3a antibody, rabbit control IgG or blank with PBS. The DNA associated with immunoprecipitates was isolated and used as templates for PCR-amplifying the Skp2 promoter region. RNA Polymerase II antibody was provided by the kit as a positive control. Upon immunoprecipitation of chromatin with an antibody against RNA Polymerase II, the promoter region of a known target gene, GAPDH, was detected by PCR. (D) The same DNA purified from (B) was used as templates for real-time quantitative PCR in triplicate. The results are presented as a percentage relative to the input (mean ± s.d.).
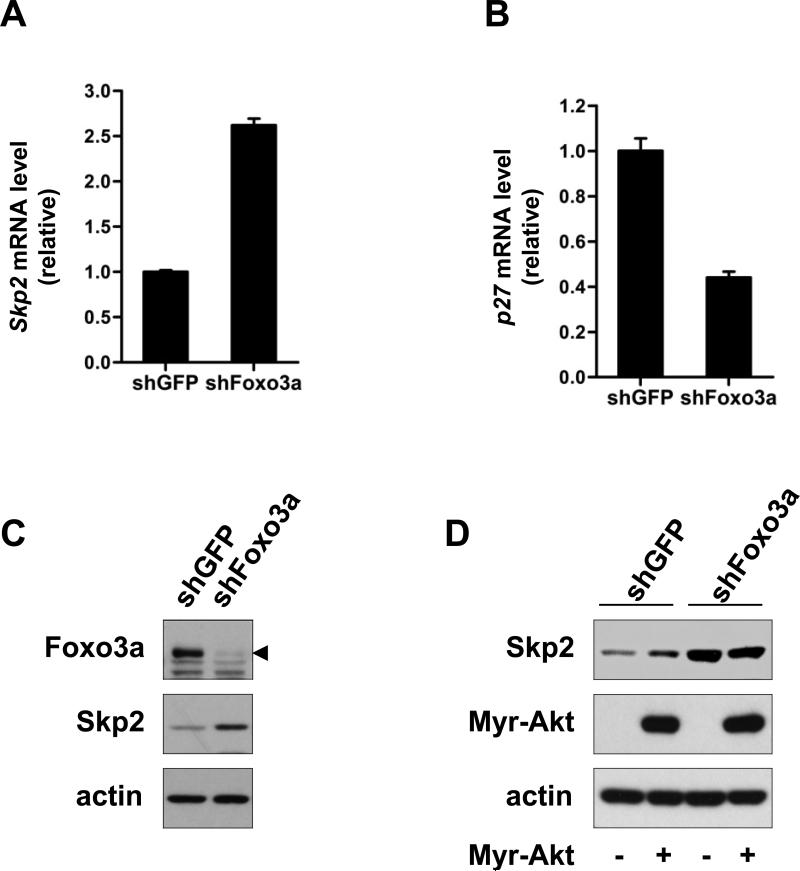
To rule out the possibility of Foxo3a-mediated repression of Skp2 gene expression may be due to the overexpression artifact, we therefore utilized the lentiviral infection system to knock down Foxo3a expression. In support of the notion that Foxo3a is a negative repressor for Skp2 gene expression, the real-time PCR result revealed that Foxo3a knockdown profoundly enhanced the level of Skp2 mRNA (Fig. 2A). As a positive control, we showed that Foxo3a knockdown reduced p27 mRNA level, consistent with the fact that Foxo3a is a transcription factor for p27 (Fig. 2B). Similarly, Foxo3a knockdown also markedly promoted Skp2 protein expression (Fig. 2C). Taken together, these results suggest that Foxo3a is a transcription repressor for Skp2 gene expression. To further examine whether Akt signaling, which has been shown to positively regulate Skp2 expression (18-20), has an impact on Foxo3a transcription-dependent activity on Skp2. As shown in Figure 2D, while constitutively activated Akt (Myr-Akt) upregulated Skp2 protein level (comparing lane 1 and 2) in GFP control knockdown cells, it failed to induce Skp2 expression in Foxo3a knockdown cells (comparing lane 3 and 4). Since Akt is known to downregulate Foxo3a activity, this result suggests that the effect of Akt on Skp2 expression is likely though its effect on Foxo3a inhibition.
Figure 2. Foxo3a regulates Skp2 gene expression and protein level.
(A, B) Total RNA from GFP control and Foxo3a knockdown DU145 cells was isolated, and Skp2 (A) and p27 (B) mRNA levels were measured by real-time quantitative PCR. The results are normalized to the internal control (GAPDH) and presented as mean ± s.d. (n=3). (C) GFP control and Foxo3a knockdown PC-3 cells were harvested for IB analysis. The arrowhead indicates the Foxo3a bands. (D) GFP control and Foxo3a knockdown PC-3 cells as in (C) were transfected with or without HA-Myr-Akt plasmid for 48 h and harvested for IB analysis.
Foxo3a interacts with Skp2 and inhibits Skp2 SCF E3 ligase activity
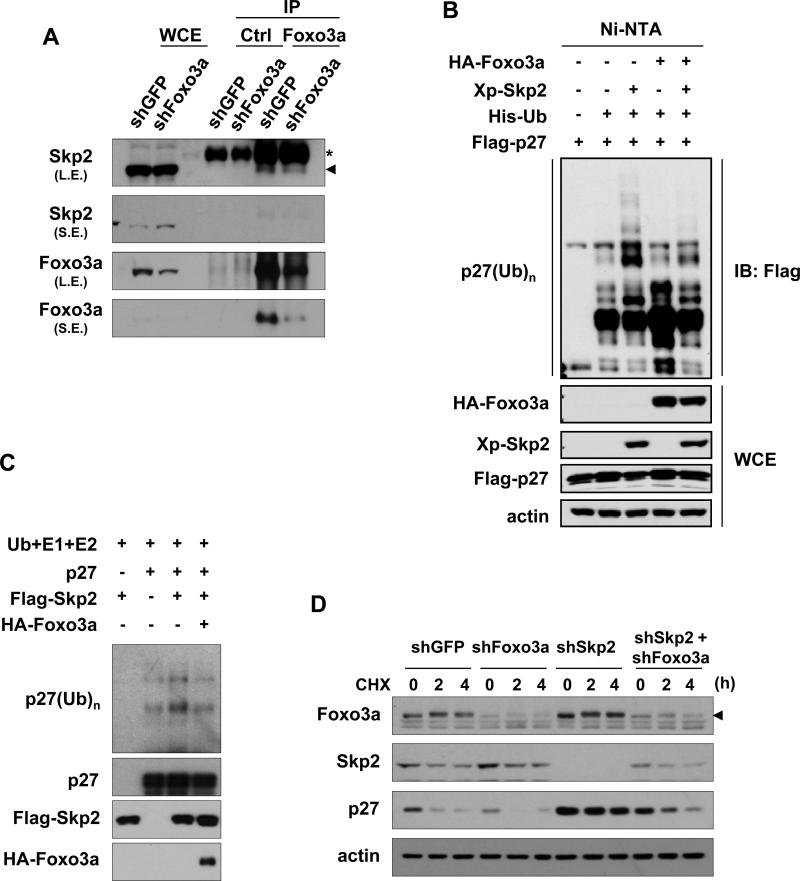
As aforementioned, Skp2 is an F-box protein that forms the Skp2 SCF complex with Skp1, Cul-1 and Rbx1 to constitute an E3 ligase activity. The Skp2 SCF E3 ligase activity is required for Skp2's oncogenic activity. The factor controlling Skp2 SCF complex formation presumably affects Skp2 SCF E3 ligase activity. Skp2 is shown to interact with Foxo1 and target it for ubiquitination and degradation (21). We therefore determined whether Skp2 may interact with Foxo3a as well. In vivo co-immunoprecipitation experiments revealed that endogenous Foxo3a could form a complex with endogenous Skp2, and their interaction was profoundly attenuated by Foxo3a knockdown (Fig. 3A).
Figure 3. Foxo3a interacts with Skp2 and inhibits Skp2 SCF E3 ligase activity.
(A) GFP control and Foxo3a knockdown HEK293 cells were collected and lysed for immunoprecipitation with Foxo3a or IgG control antibodies, followed by IB analysis. The asterisk indicates the heavy chain of the antibodies, and the arrowhead indicates the bands of the immunoprecipitated Skp2. L.E.: longer exposure; S.E.: shorter exposure. (B) HEK293T cells were transfected with the indicated plasmids for 48 h, treated with MG132 for 6 h, and harvested for the in vivo ubiquitination assay. (C) Immunoprecipitated p27 was incubated with ubiquitin, E1, E2 enzymes and purified Flag-Skp2 in the presence or absence of purified HA-Foxo3a in the in vitro system. Ubiquitinated p27 was detected by IB with anti-ubiquitin antibody. (D) GFP control, Foxo3a, Skp2 or Foxo3a/Skp2 knockdown DU145 cells were treated with 20 μg/ml cycloheximide (CHX) for the indicated time and harvested for IB analysis. The arrowhead indicates the Foxo3a bands.
As Foxo3a interacts with Skp2 under a physiological condition, we next sought to determine whether Foxo3a regulates Skp2 E3 ligase activity. To this goal, we performed both the in vivo and in vitro p27 ubiquitination assays to determine whether Foxo3a regulates Skp2-mediated p27 ubiquitination. Fig. 3B showed that p27 underwent the basal ubiquitination in the mock-transfected cells, and Skp2 overexpression readily promoted p27 ubiquitination in vivo. Notably, Foxo3a overexpression suppressed Skp2-mediated p27 ubiquitination (Fig. 3B). Also, the presence of Foxo3a inhibited Skp2-mediated ubiquitination of p27 in the in vitro system (Fig. 3C). Therefore, Foxo3a interacts with Skp2 and negatively regulates Skp2 SCF E3 ligase activity.
Foxo3a negatively regulates the formation Skp2 SCF complex
In support of this notion that Foxo3a regulates Skp2 SCF E3 ligase activity, we found that Foxo3a knockdown markedly promoted endogenous p27 degradation compared to control knockdown (Fig. 3D), revealing that higher Skp2 SCF E3 ligase activity is present in Foxo3a knockdown cells. Strikingly, Skp2 silencing completely rescued p27 stability in Foxo3a knockdown cells (Fig. 3D). Accordingly, these results underscore the important role of Foxo3a in suppressing Skp2 SCF E3 ligase activity, in turn promoting p27 stability.
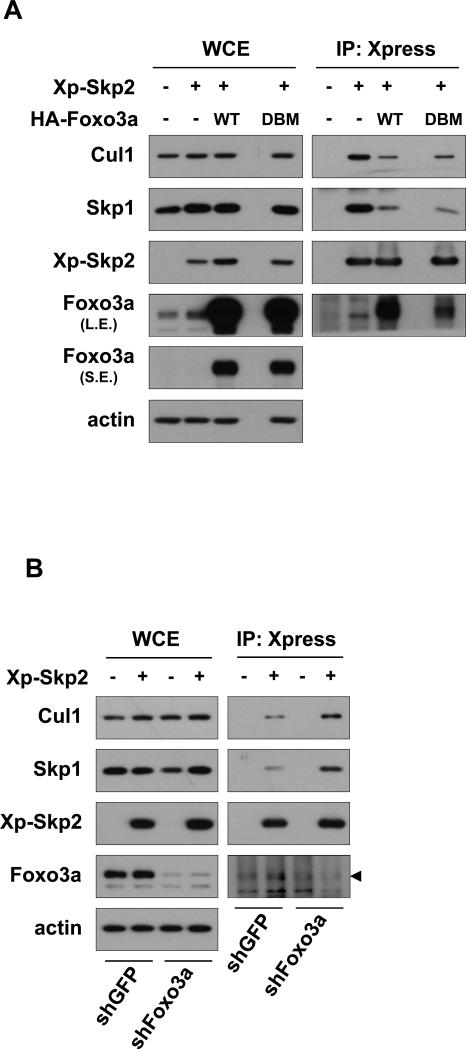
Skp2 SCF E3 ligase activity is tightly regulated by the integrity of Skp2 SCF complex. As our result show that Foxo3a represses Skp2 SCF E3 ligase activity, it is conceivable that Foxo3a may regulate the formation of Skp2 SCF complex. Indeed, Foxo3a overexpression inhibited the ability of Skp2 to interact with Skp1 and Cul-1 (Fig. 4A). Conversely, Foxo3a knockdown promoted the interaction of Skp2 with Skp1 and Cul-1 (Fig. 4B). Taken together, our results suggest that Foxo3a interacts with Skp2 and prevents the formation of Skp2 SCF complex, in turn suppressing Skp2 SCF E3 ligase activity.
Figure 4. Foxo3a negatively regulates the formation Skp2 SCF complex.
(A) HEK293T cells transfected with the indicated plasmids for 48 h were harvested for immunoprecipitation assay using anti-Xpress antibody, followed by IB analysis. L.E.: longer exposure; S.E.: shorter exposure. (B) GFP control and Foxo3a knockdown HEK293 cells transfected with or without Xp-Skp2 plasmid for 48 h were harvested for immunoprecipitation assay with anti-Xpress antibody, followed by IB analysis. The arrowhead indicates the Foxo3a bands.
Foxo3a inhibited Skp2-mediated cell proliferation
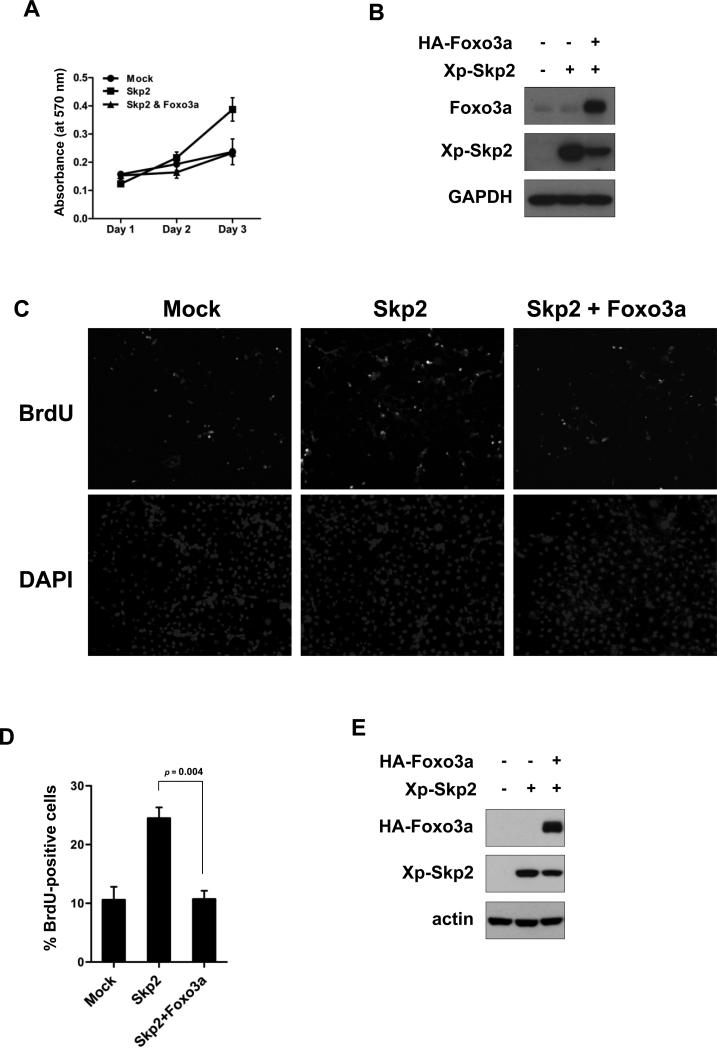
Skp2 displays an oncogenic activity by targeting p27 ubiquitination and degradation. As Foxo3a inhibits Skp2 SCF E3 ligase towards p27 ubiquitination and degradation, we next determined whether Foxo3a has an impact on Skp2-mediated cell proliferation. To this end, we applied the growth curve assay and the BrdU incorporation assay to examine cell proliferation. We found that, as expected, Skp2 overexpression profoundly promoted cell proliferation (Fig. 5). Strikingly, introduction of Foxo3a markedly suppressed Skp2-mediated cell proliferation (Fig. 5). Therefore, out results suggest that Foxo3a negatively regulates Skp2-mediated cell proliferation.
Figure 5. Foxo3a inhibits Skp2-mediated cell proliferation.
(A, B) HeLa cells transfected with the indicated plasmids were cultured for 24 h and plated for the MTT assay (A) and IB analysis (B). (C-E) COS-1 cells were transfected with the indicated plasmids. At 24 hours post-transfection, the cells were serum-starved for 24 h, refreshed with 10% FBS for 18 h, and then subjected to the BrdU incorporation assay (C, D) and IB analysis (E). The percentage of BrdU-positive cells was scored. The representative images were shown in (C), and the quantitative results were expressed as mean ± s.d. (n=3).
Discussion
Although there are redundant roles among Foxo family transcription factors, the mice deficient for all Foxo transcription including Foxo1, Foxo3, and Foxo4 develop thymic lymphomas and hemangiomas (22), suggesting that Foxo family transcription factors are bona fide tumor suppressors. In line with this notion, Foxo3a is shown to display tumor suppressive activity which overexpression induces cell cycle arrest and apoptosis, while its silencing promotes cell growth and survival. As a transcription factor, Foxo3a is generally thought to act as a tumor suppressor in a manner dependent on its transcriptional activity. Foxo3a triggers cell cycle arrest by inducing p27 and p21 gene expression, while it turns on Bim, FasL and GADD45 to elicit apoptosis (12, 13). Whether Foxo3a-mediated tumor suppressive functions can also act through its transcription-independent activity remains largely unknown. Our finding showing that Foxo3a interacts with Skp2 and disrupts the formation of Skp2 SCF complex and Skp2 SCF E3 ligase activity reveals a novel transcription-independent activity of Foxo3a in tumor suppression.
Although several transcription factors, such as E2F1 (23), NF-kB (24, 25), SP1(26), CBF1 (27), GABP (GA-binding protein) (28), and FoxM1 (29), can serve as transcription factors for Skp2, little is known about the transcription repressors for Skp2. In this study, we present the evidence that Foxo3a is a transcription repressor for oncogenic Skp2. Our study therefore provides a clue to this puzzle and reveals that Foxo3a is a novel transcription repressor for Skp2. As Skp2 level is known to be regulated by growth factors or serum stimulation, which also repress Foxo3a expression, it is formally possible that these stimuli may inhibit Foxo3a expression, in turn promoting Skp2 gene expression.
Interestingly, Foxp3, a Fox transcription factor, is also shown to binds to Skp2 promoter and represses Skp2 gene expression (30); therefore, Skp2 gene expression may be generally repressed by Forkhead box (Fox) transcription factors. However, this may not be a general phenomenon since we show that Foxo1 fails to suppress Skp2 gene expression (Fig. 1A). Nevertheless, it requires more thorough analysis to examine whether other Foxo family transcription factors can also serve as Skp2 transcription repressors.
Notably, PTEN/Akt signaling is known to regulate Skp2 gene expression, although the molecular mechanism responsible for it remains to be determined. As Akt kinase is known to phosphorylate Foxo3a and repress its transcription activity, it is highly possible that Akt induces Skp2 gene expression through phosphorylating and inhibiting Foxo3a activity. Consistent with this notion, we demonstrate that Akt may regulate Skp2 gene expression through its effect on Foxo3a inhibition (Fig. 2D).
The integrity of Skp2 SCF complex formation is important for Skp2 SCF E3 ligase activity; however, how this complex is regulated remains largely unclear. Earlier studies showed that neddylation of Cul-1 positively regulates Skp2 SCF complex formation and its E3 ligase activity by preventing the binding of Cul-1 to Cand1, a negative regulator for the Skp2 SCF complex. Cand1 preferentially interacts with unneddylated Cul-1, thus preventing Cul-1 from binding to Skp1 and Skp2 (31-33). In addition, the formation of the Skp2 SCF complex can be induced by the PI3K/Akt signal (34), and recent studies reveal that Akt interacts with Skp2 and induces S72 phosphorylation of Skp2, which is critical for Skp2 stability, Skp2 SCF complex formation and Skp2 SCF E3 ligase activity (7, 35). How Skp2 phosphorylation at Ser 72 regulates Skp2 SCF complex and its E3 ligase activity is not yet clear.
Our study identifies Foxo3a as a novel, negative regulator for Skp2 SCF complex and its E3 ligase activity. We show that Foxo3a overexpression disrupts the Skp2 SCF complex formation, whereas Foxo3a silencing promotes it. The inhibitory effect of Foxo3a on Skp2 SCF complex formation is independently of its transcriptional activity, as the transcriptional deficient mutant of Foxo3a can still repress Skp2 SCF complex formation as efficiently as wild-type Foxo3a does (Fig. 4A). Our result therefore reveals a novel, previously unrecognized transcriptional-independent activity of Foxo3a in disassembling Skp2 SCF complex. It is likely that Foxo3a may interrupt the interaction between Skp2 and Skp1 by directly interacting with Skp2. Another possibility is that Foxo3a may bind to Skp2 and recruits Cand1 to the Skp2 complex, in turn preventing Skp2 SCF complex formation. Importantly, our study offers another potential explanation for how PI3K/Akt regulates Skp2 SCF complex. We speculate that PI3K/Akt may trigger Foxo3a phosphorylation and prevents Foxo3a/Skp2 interaction, thereby facilitating Skp2 SCF complex formation. Interestingly, Foxo3a, like Foxo1, is reported to be a substrate for Skp2 (36). We postulate that upregulated Skp2 SCF E3 ligase activity by PI3K/Akt can further inhibit Foxo3a function through promoting ubiquitination and degradation of Foxo3a.
p27 displays tumor suppressive activity by inducing cell cycle arrest and senescence (8, 37, 38). The expression of p27 is frequently reduced and/or lost during cancer progression, which inversely correlates with Skp2 expression (39-41). p27 gene expression is known to be orchestrated by Foxo3a (42). Foxo3a serves as a transcription factor for p27 by binding to the p27 promoter region and induce p27 gene expression. Importantly, our study suggests that Foxo3a not only regulates p27 gene expression, but also controls p27 stability. We show that Foxo3a promotes p27 stability by disrupting the formation of Skp2 SCF complex and Skp2 SCF E3 ligase activity. Thus, our study reveals a novel insight into how Foxo3a displays tumor suppressive activity. Given that Foxo3a is often inactivated in human cancers, it is likely that the loss of p27 expression seen in advanced human cancers may be, in part, due to the loss or inactivation of Foxo3a.
In summary, our findings reveal that Foxo3a is not only a novel transcription repressor for Skp2, but also a negative regulator for Skp2 SCF complex formation and its E3 ligase activity, in turn promoting p27 stability. Our study therefore provides an important mechanistic insight into how Foxo3a orchestrates cell cycle arrest and survival, thereby favoring tumor suppression.
Methods
Cell culture and reagents
HEK293T, HEK293, DU145, PC-3 HeLa and COS-1 cells were cultured in DMEM containing 10% fetal bovine serum (FBS). Xp-Skp2, HA-Myr-Akt, His-ubiquitin and pcDNA3-p27 plasmids were described previously (7). HA-Foxo3a-WT, HA-Foxo3a-DBM (Addgene plasmid #8352) and HA-Foxo1 (Addgene plasmid #10693) plasmid were gifts from Drs. M.E. Greenberg and W.R. Sellers. The Skp2 promoter plasmid, kindly provided by Dr. V.A. Boussiotis, was described elsewhere (26). The mutant Skp2 promoter construct was generated by the site-directed mutagenesis kit (Stratagene) in accordance with the manufacturer's standard procedure using the wild-type Skp2 promoter construct as a template.
Skp2 reporter assay
HEK293T cells were transfected with indicated plasmids by using Lipofectamine 2000 (Invitrogen) for 48 h, and the luciferase activity was measured by using the dual luciferase system (Promega) as described elsewhere (43, 44). Each sample was analyzed in triplicate. The relative luciferase activity was expressed as a ratio of the firefly luciferase activity to the Renilla luciferase activity.
ChIP assay
The ChIP assay was performed according to the manufacturer's instructions (Upstate) (6). A DNA-protein complex was sheared by sonication. One percent of sheared DNA-protein complex was kept as an input DNA sample. Anti-Foxo3a (Cell Signaling), anti-GFP (Cell Signaling) (as rabbit control IgG), anti-RNA polymerase II (Upstate) and normal mouse IgG (Upstate) were used for immunoprecipitation. Standard PCR was performed using primers: Skp2 forward primer (5’-CTGTACTCTACATGGGAAAAGTGG-3’) and Skp2 reverse primer (5’-TATTCTCTGTGGTGGTGGCAGCTA-3’). PCR products were run in 1% agarose gel. Enrichment of promoter binding levels was also analyzed by real-time PCR in triplicate and expressed as a percentage relative to the input. The quantified result is presented as mean ± s.d.
Real-time RT-PCR
Total RNA was isolated from GFP control and Foxo3a-knockdown DU145 cells with TRIzol (Invitrogen). cDNA was synthesized by using the SuperScript First-Strand Synthesis System (Invitrogen). Skp2 and p27 mRNA levels were measured by applying the synthesized cDNA to real-time PCR with SYBR green (Applied Biosystems). The primers used for real-time PCR are as follows: Skp2 forward primer (5’-GGTGTTTGTAAGAGGTGGTA-3’), Skp2 reverse primer (5’-GAGACAGTATGCCGTGGA-3’), p27 forward primer (5’-CCGGCTAACTCTGAGGACAC-3’), p27 reverse primer (5’-AGAAGAATCGTCGGTTGCAG-3’), GAPDH forward primer (5’-GATTCCACCCATGGCAAATTC-3’) and GAPDH reverse primer (5’-CTTCTCCATGGTGGTGAAGAC-3’). The quantified results were presented as mean ± s.d.
Viral infection
For lentiviral short hairpin RNA (shRNA) infection, HEK293T cells were co-transfected with shRNAs against GFP, luciferase (Luc), Foxo3a or Skp2 along with the packing plasmid (deltaVPR8.9) and the envelope plasmid (VSV-G) using the calcium phosphate precipitation method. Two days after transfection, virus particles were used to infect the indicated cell lines. All the infected cells were selected in the medium containing 2 μg/ml puromycin for 5-7 days. The following lentiviral shRNAs were used for transfection: Skp2-lentiviral shRNA (5’-GCCTAAGCTAAATCGAGAGAA-3’) and Foxo3a-lentiviral shRNA (5’-GTCACTGCATAGTCGATTCAT-3’) and Foxo3a-lentiviral shRNA-2 (5’-CAGACCCTCAAACTGACACAA-3’).
Immunoprecipitation (IP), immunoblotting (IB)
IP and IB were done essentially as described with mild modification (6, 45). For immunoprecipitation, cells were lysed in E1A lysis buffer [250 mM NaCl, 50 mM HEPES (pH 7.5), 0.1% NP-40, 5 mM EDTA, protease inhibitor cocktail (Roche)] with sonication. The following antibodies were used for IP and IB: anti-Skp2 antibody (IB, 1:1,500; Invitrogen), anti-Foxo3a antibody (IP, 1:500; IB, 1:1,000; Santa Cruz), anti-p27 antibody (IP, 1:200; IB, 1:1,000; BD Pharmingen), anti-Skp1 antibody (IB, 1:1,500, BD Transduction Lab), anti-Cul-1 antibody (IB, 1:1,500, Invitrogen), anti-β-actin antibody (IB, 1:20,000; Sigma), anti-ubiquitin antibody (IB, 1:1,000; Santa Cruz), anti-Xpress antibody (IP, 1:500; IB, 1:5,000; Invitrogen), anti-HA antibody (IP, 1:500; IB, 1:10,000; Covance) and anti-Flag antibody (M2) (IP, 1:200; IB, 1:3,000; Sigma).
Ubiquitination assays
The in vivo ubiquitination assay was performed as described elsewhere (6, 7). In brief, HEK293T cells were transfected with the indicated plasmids for 48 h, treated with MG132 for 6 h, and lysed with denaturing buffer (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole), followed by nickel bead purification and IB analysis. For p27 in vitro ubiquitination assay, Flag-Skp2 and HA-Foxo3a were isolated individually from HEK293T cell extracts and eluted from beads by FLAG and HA peptides (Sigma) respectively, followed by concentration with Amicon (Millipore). The p27 protein was immunoprecipitated from the cell extract of HEK293T transfected with pcDNA3-p27, and then incubated with ubiquitin, E1, E2 enzymes (Boston Biochem) and purified Flag-Skp2 in ubiquitination buffer [20 mM Hepes (pH7.4), 1.5 mM MgCl2, 5 mM KCl, 1 mM DTT, 1 mM ATP, protease inhibitor cocktail (Roche)] in the presence or absence of purified HA-Foxo3a. Ubiquitination reaction was carried out at 37°C for 2 h on a rotator and terminated by washing the pellets. Ubiquitinated p27 was analyzed by IB with anti-ubiquitin antibody.
MTT proliferation assay
HeLa cells were transfected with the indicated plasmids for 24 h, then were plated in 96 well plate (3,000 cells per well). In the following three days, cells were incubated with MTT (Thiazolyl Blue Tetrazolium Bromide) for 4 h. After incubation, culture medium was removed, and then the equal volume of DMSO was added to each well. Absorbance was measured by a spectrophotometer at 570 nm after shaking for 10 minutes.
BrdU incorporation assay
BrdU incorporation assay was performed with the in situ cell proliferation kit, FLUOS (Roche), and the manufacturer's instructions were followed. In brief, COS-1 cells seeded on chamber slides were transfected with the indicated plasmids. At 24 hours post-transfection, the cells were serum-starved for 24 h, and then refreshed with 10% FBS for 18 h. Following incubation with 10 μM bromodeoxyuridine (BrdU) for 1 h, the cells were subjected to immunostaining for detection of BrdU incorporation. The percentages of BrdU-positive cells were scored. The representative images were shown and the quantified result was presented as mean ± s.d.
Acknowledgment
We thank Drs. M.E. Greenberg, W.R. Sellers, V.A. Boussiotis and D. Bohmann for reagents. We are grateful to Dr. Lin's lab members for insightful comments and suggestions. Special thanks extend to S. Zhang and Dr. Z. Han for their technical support. This work was supported by the M. D. Anderson Trust Scholar Fund, the National Cancer Institute's Prostate Cancer Specialized Program of Research Excellence (SPORE) development grant at the M.D. Anderson Cancer Center, NIH RO1 grants, CPRIT grant, and DOD New Investigator Award to H.K.L.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Chan CH, Lee SW, Wang J, Lin HK. Regulation of Skp2 expression and activity and its role in cancer progression. ScientificWorldJournal. 2010;10:1001–15. doi: 10.1100/tsw.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol. 2005 Jun;16(3):323–33. doi: 10.1016/j.semcdb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. Embo J. 2000 May 2;19(9):2069–81. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakayama K, Nagahama H, Minamishima YA, Miyake S, Ishida N, Hatakeyama S, et al. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev Cell. 2004 May;6(5):661–72. doi: 10.1016/s1534-5807(04)00131-5. [DOI] [PubMed] [Google Scholar]
- 5.Chan CH, Gao Y, Moten A, Lin HK. Novel ARF/p53-independent senescence pathways in cancer repression. J Mol Med. 2011 May 19; doi: 10.1007/s00109-011-0766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan CH, Lee SW, Li CF, Wang J, Yang WL, Wu CY, et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat Cell Biol. 2010 Apr 11; doi: 10.1038/ncb2047. Apr 11. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, et al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat Cell Biol. 2009 Apr;11(4):420–32. doi: 10.1038/ncb1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010 Mar 18;464(7287):374–9. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Bauzon F, Ji P, Xu X, Sun D, Locker J, et al. Skp2 is required for survival of aberrantly proliferating Rb1-deficient cells and for tumorigenesis in Rb1+/- mice. Nat Genet. 2010 Jan;42(1):83–8. doi: 10.1038/ng.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature. 2004 Mar 11;428(6979):190–3. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- 11.Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004 Mar 11;428(6979):194–8. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- 12.Dansen TB, Burgering BM. Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 2008 Sep;18(9):421–9. doi: 10.1016/j.tcb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Yang JY, Hung MC. A new fork for clinical application: targeting forkhead transcription factors in cancer. Clin Cancer Res. 2009 Feb 1;15(3):752–7. doi: 10.1158/1078-0432.CCR-08-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004 Apr 16;117(2):225–37. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 15.Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008 Feb;10(2):138–48. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999 Mar 19;96(6):857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 17.Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ, Jr., DiStefano PS, et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002 Apr 19;296(5567):530–4. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 18.Mamillapalli R, Gavrilova N, Mihaylova VT, Tsvetkov LM, Wu H, Zhang H, et al. PTEN regulates the ubiquitin-dependent degradation of the CDK inhibitor p27(KIP1) through the ubiquitin E3 ligase SCF(SKP2). Curr Biol. 2001 Feb 20;11(4):263–7. doi: 10.1016/s0960-9822(01)00065-3. [DOI] [PubMed] [Google Scholar]
- 19.Reichert M, Saur D, Hamacher R, Schmid RM, Schneider G. Phosphoinositide-3-kinase signaling controls S-phase kinase-associated protein 2 transcription via E2F1 in pancreatic ductal adenocarcinoma cells. Cancer Res. 2007 May 1;67(9):4149–56. doi: 10.1158/0008-5472.CAN-06-4484. [DOI] [PubMed] [Google Scholar]
- 20.van Duijn PW, Trapman J. PI3K/Akt signaling regulates p27(kip1) expression via Skp2 in PC3 and DU145 prostate cancer cells, but is not a major factor in p27(kip1) regulation in LNCaP and PC346 cells. Prostate. 2006 May 15;66(7):749–60. doi: 10.1002/pros.20398. [DOI] [PubMed] [Google Scholar]
- 21.Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, et al. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005 Feb 1;102(5):1649–54. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007 Jan 26;128(2):309–23. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Wang C. F-box protein Skp2: a novel transcriptional target of E2F. Oncogene. 2006 Apr 27;25(18):2615–27. doi: 10.1038/sj.onc.1209286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barre B, Perkins ND. A cell cycle regulatory network controlling NF-kappaB subunit activity and function. Embo J. 2007 Nov 28;26(23):4841–55. doi: 10.1038/sj.emboj.7601899. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Schneider G, Saur D, Siveke JT, Fritsch R, Greten FR, Schmid RM. IKKalpha controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression. Embo J. 2006 Aug 23;25(16):3801–12. doi: 10.1038/sj.emboj.7601259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Appleman LJ, Chernova I, Li L, Boussiotis VA. CD28 costimulation mediates transcription of SKP2 and CKS1, the substrate recognition components of SCFSkp2 ubiquitin ligase that leads p27kip1 to degradation. Cell Cycle. 2006 Sep;5(18):2123–9. doi: 10.4161/cc.5.18.3139. [DOI] [PubMed] [Google Scholar]
- 27.Sarmento LM, Huang H, Limon A, Gordon W, Fernandes J, Tavares MJ, et al. Notch1 modulates timing of G1-S progression by inducing SKP2 transcription and p27 Kip1 degradation. J Exp Med. 2005 Jul 4;202(1):157–68. doi: 10.1084/jem.20050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imaki H, Nakayama K, Delehouzee S, Handa H, Kitagawa M, Kamura T, et al. Cell cycle-dependent regulation of the Skp2 promoter by GA-binding protein. Cancer Res. 2003 Aug 1;63(15):4607–13. [PubMed] [Google Scholar]
- 29.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005 Dec;25(24):10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuo T, Liu R, Zhang H, Chang X, Liu Y, Wang L, et al. FOXP3 is a novel transcriptional repressor for the breast cancer oncogene SKP2. J Clin Invest. 2007 Dec;117(12):3765–73. doi: 10.1172/JCI32538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bloom J, Pagano M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin Cancer Biol. 2003 Feb;13(1):41–7. doi: 10.1016/s1044-579x(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell. 2002 Dec;10(6):1511–8. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- 33.Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, et al. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002 Dec;10(6):1519–26. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- 34.Jonason JH, Gavrilova N, Wu M, Zhang H, Sun H. Regulation of SCF(SKP2) ubiquitin E3 ligase assembly and p27(KIP1) proteolysis by the PTEN pathway and cyclin D1. Cell Cycle. 2007 Apr 15;6(8):951–61. doi: 10.4161/cc.6.8.4104. [DOI] [PubMed] [Google Scholar]
- 35.Gao D, Inuzuka H, Tseng A, Chin RY, Toker A, Wei W. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat Cell Biol. 2009 Apr;11(4):397–408. doi: 10.1038/ncb1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F, Chan CH, Chen K, Guan X, Lin HK, Tong Q. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene. 2011 Aug 15; doi: 10.1038/onc.2011.347. [DOI] [PubMed] [Google Scholar]
- 37.Majumder PK, Grisanzio C, O'Connell F, Barry M, Brito JM, Xu Q, et al. A prostatic intraepithelial neoplasia-dependent p27 Kip1 checkpoint induces senescence and inhibits cell proliferation and cancer progression. Cancer Cell. 2008 Aug 12;14(2):146–55. doi: 10.1016/j.ccr.2008.06.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young AP, Schlisio S, Minamishima YA, Zhang Q, Li L, Grisanzio C, et al. VHL loss actuates a HIF-independent senescence programme mediated by Rb and p400. Nat Cell Biol. 2008 Mar;10(3):361–9. doi: 10.1038/ncb1699. [DOI] [PubMed] [Google Scholar]
- 39.Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008 Apr;8(4):253–67. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 40.Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer. 2008 Apr 1;112(7):1415–24. doi: 10.1002/cncr.23317. [DOI] [PubMed] [Google Scholar]
- 41.Sicinski P, Zacharek S, Kim C. Duality of p27Kip1 function in tumorigenesis. Genes Dev. 2007 Jul 15;21(14):1703–6. doi: 10.1101/gad.1583207. [DOI] [PubMed] [Google Scholar]
- 42.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000 Apr 13;404(6779):782–7. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 43.Chan CH, Lee SW, Li CF, Wang J, Yang WL, Wu CY, et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat Cell Biol. 2010 May;12(5):457–67. doi: 10.1038/ncb2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004 Sep 9;431(7005):205–11. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- 45.Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009 Aug 28;325(5944):1134–8. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]