Background: orfX is a gene of unknown function and is conserved in all staphylococci.
Results: OrfX methylates 70 S ribosomes, and the crystallographic dimer binds two molecules of the substrate S-adenosyl-l-methionine, one in each active site.
Conclusion: OrfX is a staphylococcal ribosomal methyltransferase of the RlmH type.
Significance: This is the first time that an RlmH-type methyltransferase has been co-crystallized with its substrate.
Keywords: Bacteria, Enzyme Structure, RNA Methylation, RNA Methyltransferase, Staphylococcus aureus, RlmH, OrfX
Abstract
The gene orfX is conserved among all staphylococci, and its complete sequence is maintained upon insertion of the staphylococcal chromosome cassette mec (SCCmec) genomic island, containing the gene encoding resistance to β-lactam antibiotics (mecA), into its C terminus. The function of OrfX has not been determined. We show that OrfX was constitutively produced during growth, that orfX could be inactivated without altering bacterial growth, and that insertion of SCCmec did not alter gene expression. We solved the crystal structure of OrfX at 1.7 Å and found that it belongs to the S-adenosyl-l-methionine (AdoMet)-dependent α/β-knot superfamily of SPOUT methyltransferases (MTases), with a high structural homology to YbeA, the gene product of the Escherichia coli 70 S ribosomal MTase RlmH. MTase activity was confirmed by demonstrating the OrfX-dependent methylation of the Staphylococcus aureus 70 S ribosome. When OrfX was crystallized in the presence of its AdoMet substrate, we found that each monomer of the homodimeric structure bound AdoMet in its active site. Solution studies using isothermal titration calorimetry confirmed that each monomer bound AdoMet but with different binding affinities (Kd = 52 ± 0.4 and 606 ± 2 μm). In addition, the structure shows that the AdoMet-binding pocket, formed by a deep trefoil knot, contains a bound phosphate molecule, which is the likely nucleotide methylation site. This study represents the first characterization of a staphylococcal ribosomal MTase and provides the first crystal structure of a member of the α/β-knot superfamily of SPOUT MTases in the RlmH or COG1576 family with bound AdoMet.
Introduction
orfX has previously been of interest primarily because the insertion site of the staphylococcal chromosome cassette mec (SCCmec)2 mobile genomic island is within its C terminus (Fig. 1) (1). SCCmec contains mecA, the gene mediating resistance to all β-lactam antibiotics in staphylococci (methicillin resistance). The orfX gene product has been suspected to play an important role in bacterial growth and survival because it, or its homolog, is present in every sequenced coagulase-positive or coagulase-negative staphylococcal genome. Because the SCCmec insertion or att site comprises the terminal nucleotides of the gene, the gene is always left intact when SCCmec inserts. However, as the name given the gene implies, no function had previously been assigned to the gene product until recent annotation of sequenced genomes began designating the gene as a methyltransferase based on homology searches (2, 3).
FIGURE 1.
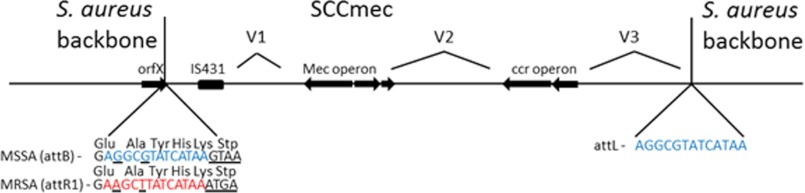
Schematic of the general arrangement of SCCmec. The insertion of SCCmec into the C terminus of orfX at the attachment site attB changes it to attR1. The terminal five amino acids and stop codon are unchanged even though the DNA sequence is altered. Also shown are the regions that define a SCCmec: the mec operon containing mecA, the gene responsible for β-lactam resistance; the cassette chromosome recombinase (ccr) operon that facilitates the insertion and excision of SCCmec; and three hypervariable regions between them. The sequences of the mec and ccr operons define the SCCmec type. A more detailed description of the different SCCmec types can be found at the International Working Group on the Staphylococcal Cassette Chromosome Elements. This figure is not to scale and does not show all of the genes in the region.
The crystal structure of OrfX has been solved as part of a project to determine the structure of random bacterial proteins, but the structure was deposited in the structural database as a “hypothetical protein,” with no function ascribed and no structural analysis performed (4). On the basis of a possible role for OrfX in SCCmec insertion and excision and the lack of biologic confirmation of its function as a methyltransferase, we sought to identify its activity and perform an analysis of the relationship between OrfX structure and function.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Media, and Culture Conditions
Four Staphylococcus aureus strains were used in this study: RN450, derived from strain NCTC8325 (5); AW2, an RN4220 (6)-derived strain with the first 380 bp of orfX deleted (7); RN450M, an RN450 derivative in which SCCmec type I from COL was inserted at the attB site (8); and AW18, a derivative of RN450M in which orfX was deleted in the same manner as described for AW2 (6). The strains were grown in brain heart infusion (BHI) broth (BD Biosciences) at 37 °C unless stated otherwise. The strains Escherichia coli XL1-Blue (Novagen, Madison, WI) and E. coli BL21 Rosetta (Novagen) were used for the cloning and expression of OrfX, respectively, and were grown in LB broth or agar (USB Corp., Cleveland, OH) supplemented with kanamycin (Sigma-Aldrich).
Antimicrobial Susceptibility Testing
The susceptibility of strains AW18 and RN450M to oxacillin was determined using oxacillin Etest strips (bioMérieux, Durham, NC) following the manufacturer's instructions.
Cloning, Expression, and Purification of OrfX
The orfX gene from strain RN450M was PCR-amplified using primers OrfX_F3_EcoRI (5′-ccg gaa ttc atg aaa atc acc att tta gct-3′) and OrfX_R1_NotI (5′-ata aga atg cgg ccg ctt act tat gat acg cct ctc ctc g-3′) with Platinum High Fidelity Taq polymerase (Invitrogen) and cloned into pET30a+ (Novagen), cut with EcoRI and NotI (New England Biolabs, Ipswich, MA), generating an N-terminal His6 tag, and introduced into E. coli XL1-Blue cells. This construct was then introduced into the expression strain E. coli BL21 Rosetta. For protein expression, E. coli BL21 Rosetta cells containing pET30a+-OrfX were inoculated into 4 liters of LB broth and grown with no antibiotic selection at 37 °C to an absorbance of 0.5 and then induced with 100 μm isopropyl β-d-thiogalactopyranoside (Bioline, Taunton, MA) and grown for a further 5 h. The cells were harvested by centrifugation at 6000 rpm for 10 min. The cell pellet was resuspended in 100 ml of 50 mm sodium phosphate (pH 7.5) and lysed by three passages through an EmulsiFlex-C3 French press operating (Avestin Inc., Ottawa, Canada) at >20,000 p.s.i. The lysate was centrifuged at 16,000 rpm for 30 min, and 657 g/liter ammonium sulfate was slowly added to the supernatant and stirred overnight at 4 °C. The saturated supernatant was then centrifuged at 16,000 rpm for 30 min, and the pellet was resuspended in 50 ml of 50 mm sodium phosphate (pH 7.5) and dialyzed three times with 2 liters of 50 mm sodium phosphate (pH 7.5) and 10 mm imidazole over 24 h. The precipitate was then removed from the sample by centrifugation at 16,000 rpm for 30 min, and the supernatant was run over a nickel-nitrilotriacetic acid column (Qiagen, Valencia, CA) and washed with 50 mm sodium phosphate (pH 7.5) and 300 mm NaCl plus increasing concentrations of imidazole (Sigma-Aldrich) as follows: 10 mm (100 ml), 30 mm (100 ml), 75 mm (50 ml), 150 mm (50 ml), 300 mm (50 ml), and 500 mm (50 ml). Flow-through fractions from the 10 and 30 mm washes were discarded, the remaining buffers were collected, and fractions exceeding an A280 of 1.0 were pooled and dialyzed in 2 liters of 50 mm sodium phosphate (pH 7.5) and 300 mm NaCl for 24 h. The N-terminal His6 tag was excised using the enzyme EKmax (Invitrogen) according to the manufacturer's directions, dialyzed in 2 liters of 50 mm sodium phosphate (pH 7.5) and 300 mm NaCl for 24 h, and centrifuged at 16,000 rpm for 30 min at 4 °C. The cleaved N-terminal His6 tag was separated from OrfX by passing the supernatant over a nickel-nitrilotriacetic acid column and collecting OrfX in the flow-through fraction. The pure protein was then concentrated to 15 mg/ml and stored at 4 °C, as freezing caused precipitation.
Western Blotting
A flask containing 50 ml of BHI broth was seeded with 1 ml of overnight culture of RN450M and grown with shaking at 37 °C for 8 h. 1 ml of culture was removed every 30 min, the A600 was read, the bacteria were pelleted at 6000 rpm for 10 min, and the pellet was stored at −20 °C. The pellets were resuspended in 100 μl of lysis buffer (100 mm sodium phosphate, 10 mm Tris-HCl, and 8 m urea (pH 8.0) (Sigma-Aldrich)) using a QIAexpressionist system (Qiagen) and lysed in a bead beater (FastPrep FP120, Qbiogene, Carlsbad, CA) using a 2-ml Lysing Matrix B tube (MP Biomedicals, Solon, OH) for 20 s at 4.0 speed. The A280 of each sample was then adjusted using lysis buffer to 0.064 (the lowest reading from the growth curve) so that all samples had the same absorbance. The samples were separated on a 10–20% Tricine gel (Invitrogen) using 1× Novex Tricine/SDS running buffer (Invitrogen), transferred to a PVDF membrane, and incubated with polyclonal rabbit antiserum against OrfX (Invitrogen). Bands were visualized by binding with secondary antibody (anti-rabbit IgG (whole molecule)-peroxidase, produced in goat, Sigma-Aldrich) and a 3,3′-diaminobenzidine substrate system with nickel enhancement (Vector Labs, Burlingame, CA) following the manufacturer's instructions.
Growth Curve
Two strains, the parent (RN450) and the orfX mutant (AW2), were grown at both 18 and 37 °C with constant shaking. Three separate tubes of each organism were incubated at the two temperatures. Samples were taken every hour, and the A600 was read until the reading stopped increasing, indicating that the culture had reached stationary phase. For those cultures growing at 18 °C, the incubations had to be staggered because of the prolonged growth phase at the lower temperature. This was accomplished by timing each incubation so that the next portion of the growth phase could be sampled between 8 a.m. and 6 p.m. over the 3 days it took for the culture to reach stationary phase. Readings at each time point were averaged, and the mean ± S.D. was determined.
Purification of rRNA
4 liters of BHI broth was seeded with 200 ml of an overnight culture of RN450 or AW2. The culture was grown to an absorbance of 0.6, and the bacteria were pelleted by centrifugation at 6000 rpm for 15 min. The pellets were then resuspended in 30 ml of buffer containing 16% (w/v) sucrose in 6 mm MgCl2, 60 mm NH4Cl, 60 mm KCl, 50 mm Tris-HCl (pH 8.0), and 6 mm β-mercaptoethanol, and 70 S ribosomes were purified as described previously (9). Briefly, cells were lysed by five passages through an EmulsiFlex-C3 French press. The S-30 lysate was centrifuged at 12,000 × g, and the supernatant was layered onto a 15–30% (w/w) sucrose gradient and centrifuged at 39,000 × g for 22 h. Ribosomal particles were precipitated from gradient fractions, analyzed at 254 nm absorbance, precipitated with 2.5 volumes of ice-cold ethanol, and collected by centrifugation at 5000 rpm. 70 S ribosomal pellets were resuspended in buffer containing 12 mm MgCl2, 60 mm NH4Cl, 60 mm KCl, 20 mm Tris-HCl (pH 8.0), and 6 mm β-mercaptoethanol and stored at −80 °C.
OrfX Methylation Assay
A 50-μl reaction mixture containing 10 pmol of 70 S ribosomes from RN450 or AW2 in buffer containing 40 mm Tris-HCl (pH 7.2), 40 mm NH4Cl, 8 mm MgOAc, and 1 mm dithiothreitol was incubated at 42 °C for 5 min and then chilled on ice. Subsequently, 10 pmol of OrfX, 0.2 mm cold S-adenosyl-l-methionine (AdoMet; Sigma-Aldrich), and 10 nm [methyl-3H]AdoMet (Sigma-Aldrich) were added to the chilled 70 S ribosomes, and the reaction was allowed to proceed for 1 h at 37 °C. The 50-μl reaction was transferred to Whatman paper, the reaction was quenched, and excess AdoMet was washed from the filter paper with 5% ice-cold TCA (Fisher). The filter paper was then washed twice with 5% TCA for 5 min and twice with 100% ethanol. The filter papers were left to dry at room temperature for 2 h, and the dried filter paper was placed in a scintillation vial with 3 ml of scintillation liquid (Fisher). The scintillation vial was read in a TriCarb 2810TR liquid scintillation analyzer (PerkinElmer Life Sciences). Each experimental reaction and the following controls were carried out in triplicate: OrfX and radiolabeled AdoMet with no 70 S ribosomes, RN450 70 S ribosomes and radiolabeled AdoMet with no OrfX, and AW2 70 S ribosomes and radiolabeled AdoMet with no OrfX.
Isothermal Titration Calorimetry
Isothermal titration calorimetry was performed on purified OrfX protein with AdoMet in a MicroCal iTC200 system with the following settings: experimental mode = highest quality, n = 1, cell = nucleic acid, syringe = protein, Kd = 0.0000001, ΔH = 5, and temperature = 37 °C. OrfX and AdoMet were dissolved in 50 mm sodium phosphate (pH 7.5). Preliminary experiments demonstrated no interaction between the buffer and OrfX or AdoMet. Further experimentation showed that an AdoMet concentration of 1.182 mm and an OrfX concentration of 0.0726 mm gave the best curves. AdoMet concentration was calculated using a molar extinction coefficient of 15,400 at A254 (10), and OrfX concentration was calculated using an extinction coefficient of 14,440 at A280, with the assumption from the crystal structure that OrfX was in its dimeric form in solution. The experiment was then performed using one injection of 1.0 μl followed by 21 injections of 1.5 μl of OrfX into 200 μl of AdoMet with constant stirring of the reaction vessel. Data were analyzed using Origin 7 SR4 (11, 12).
Real-time Quantitative PCR
RNA was prepared from strains AW2 and RN450 by seeding 0.5 ml of an overnight culture in 25 ml of BHI broth, growing the culture to an absorbance of 0.6, and pelleting at 6000 rpm for 10 min. The pellet was then suspended in 500 μl of lysis buffer, and 500 μl of acid phenol/chloroform was added (Sigma-Aldrich). The bacteria were lysed in a bead beater using a 2-ml of Lysing Matrix B tube for 40 s at 6.0 speed, and the lysate was centrifuged at 13,800 rpm for 10 min. 400 μl of the aqueous phase was removed to a new tube containing 250 μl of 100% ethanol, mixed, and transferred to an RNeasy column (Qiagen) for RNA purification following the instruction manual. The total RNA was then treated with Ambion DNase (Invitrogen). Quantitative RT-PCR was performed on the AW2 and RN450 samples at the Virginia Commonwealth University Nucleic Acid Research Facilities using primers and probes for 16 S rRNA and OrfX.
Crystallization and Data Collection of OrfX
OrfX (16 mg/ml in 20 mm sodium phosphate at pH 7.3) and AdoMet (100 mm in distilled water) solutions were mixed with a final substrate concentration of 2 mm and incubated on ice for 2 h. Crystallization of the OrfX-AdoMet complex was achieved using the hanging-drop vapor-diffusion technique. Initial crystallization conditions were Crystal Screen and Crystal Screen 2 (Hampton Research, Aliso Viejo, CA) at 290 K. Crystals grew within 1 week in the solvents PEG-550MME and PEG-2000MME. X-ray quality crystals with a thin plate-like morphology grew in 20025% PEG-550MME and 5 mm ZnSO4 solutions buffered with 0.1 m MES (pH 6.0).
The crystals belong to the C2221 space group with unit cell dimensions a = 77.71, b = 84.20, and c = 55.68 Å. The X-ray data set was obtained at 100 K on an R-axis IV++ image plate detector using CuKα x-rays (λ = 1.54 Å) from a Rigaku Micro-MaxTM-007 x-ray source equipped with VariMax confocal optics operating at 40 kV and 20 mA. Prior to data collection, the crystals were transferred with a cryoloop into the mother liquid solutions containing additional 5% glycerol and 1 mm AdoMet for ∼10 s and subsequently flash-cooled in a cold nitrogen gas stream from a Rigaku X-stream CryoSystem. Crystals diffracted to 1.7 Å resolution, and the data set was processed and scaled with Rigaku D*TREK software.
Structure Determination
Structure determination was carried out by molecular replacement with the program Phaser v1.2 (13) using the unliganded crystal structure of a hypothetical protein deposited in the Protein Data Bank with code 1VH0. Several cycles of refinement using CNS (with each cycle consisting of positional and/or annealing and/or composite omit map and/or individual B-factor refinement with intermittent manual model correction) led to final R-factor/Rfree of 20.7/24.7% at 1.7 Å. A total of 247 water molecules were also added to the model during the refinements. The model also contains one molecule each of AdoMet, phosphate, and PEG-550MME, all refined with full occupancies. Refinement statistics are summarized in Table 1. The graphic program COOT (14) was used for model building and/or correction.
TABLE 1.
Refinement parameters for the OrfX structure with bound AdoMet
r.m.s.d., root mean square deviation.
| Data collection statistics | |
| Space group | C2221 |
| Cell dimensions (Å) | 77.79, 84.29, 55.73 |
| Resolution (Å) | 28.58–1.70 (1.76–1.70) |
| No. of measurements/unique reflections | 165,135/19,664 (1862) |
| I/σI | 19.5 (5.4) |
| Completeness (%) | 95.8 (93.8) |
| Rmerge (%)a | 5.9 (34.7) |
| Structure refinement | |
| Resolution limit (Å) | 26.43–1.70 (1.76–1.70) |
| No. of reflections | 19,662 (1858) |
| Rfactor (%) | 20.7 (36.6) |
| Rfree (%)b | 24.7 (37.6) |
| r.m.s.d. standard geometry | |
| Bond lengths (Å) | 0.018 |
| Bond angles | 1.7° |
| Dihedral angles | |
| Most favored regions | 95.2 |
| Allowed regions | 4.8 |
| Average B-factors | |
| All atoms | 30.5 |
| Protein alone | 29.3 |
| PEG-550MME | 49.7 |
| Phosphate | 38.7 |
| Water | 38.3 |
a Rmerge = ΣhklΣi|Ihkli − 〈Ihkli〉|/ΣhklΣi〈Ihkli〉.
b Rfree was calculated with 5% of excluded reflection from the refinement.
Construction and Purification of OrfX Mutants
The three most likely candidate catalytic bases were identified by analysis of the crystal structure, and primers were designed to introduce the following mutations in the pET30a+-OrfX plasmid: D49A, E77A, and E86A. Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) following the manufacturer's protocol. The mutations were confirmed by DNA sequencing. The plasmids were then purified and transferred to E. coli BL21 Rosetta cells as described above. Protein expression and purification were performed following procedures similar to those described above for the wild type. Purified OrfX and three OrfX mutants were subjected to the methylation assay described above.
Statistics
Student's t test and standard deviation using Microsoft Excel were used. χ2/degrees of freedom and modeling the substrate binding activity were performed using Origin 7 software.
RESULTS
OrfX Expression and Its Role in Growth and Antibiotic Susceptibility
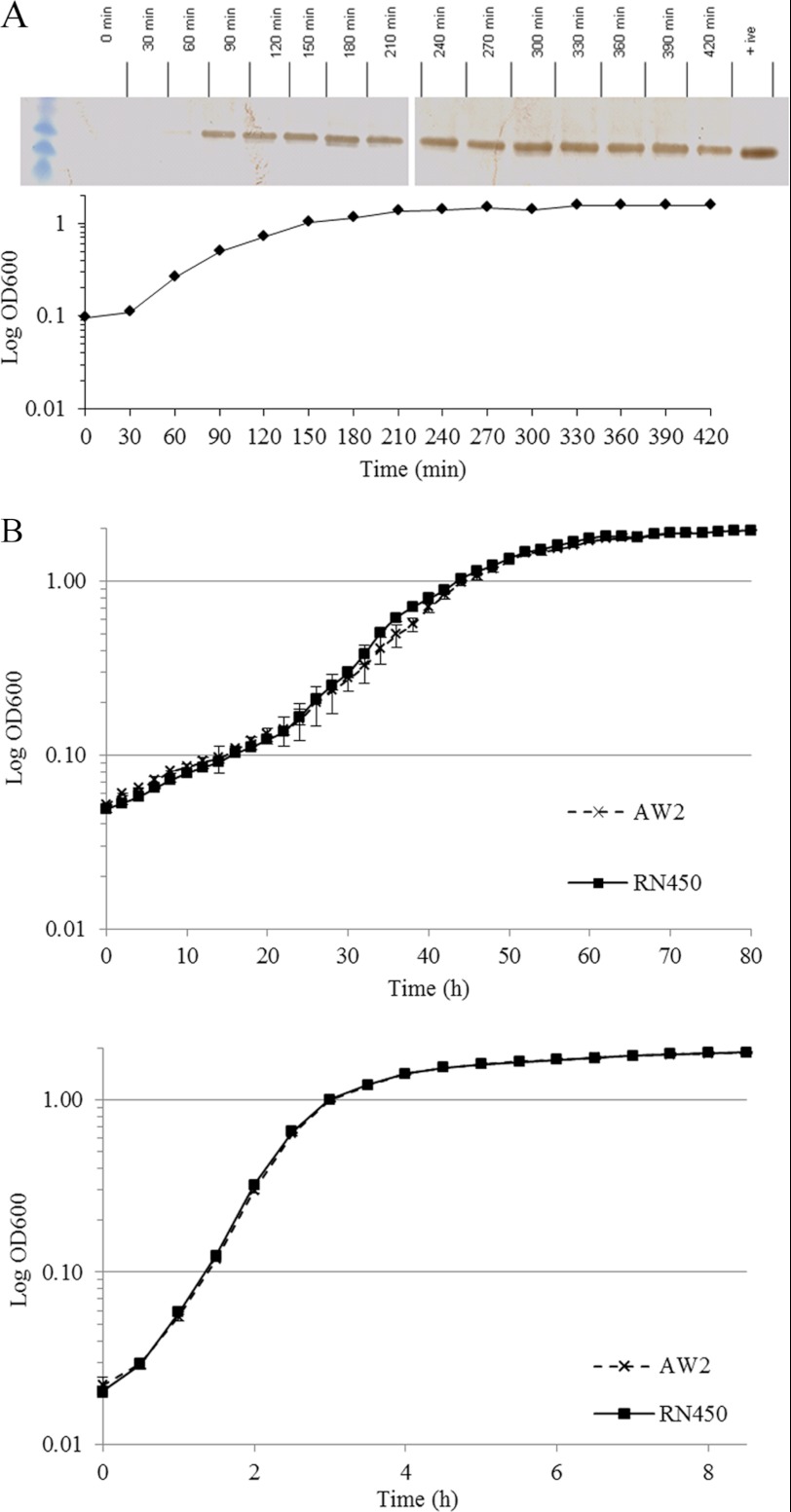
OrfX expression was monitored by Western blotting every 30 min over 7 h through one growth cycle at 37 °C. Expression was shown to be constitutive with no change in protein quantity through the log and stationary phases (Fig. 2A). There was also no difference in orfX transcription, as monitored by quantitative RT-PCR, during growth or between RN450 and RN450M. The latter experiment showed that the insertion of SCCmec into the att site, which is located in the orfX C terminus, had no effect on gene expression or on protein production (data not shown). There was also no obvious effect of the loss of OrfX on growth. The growth of RN450 was compared with that of AW2, the isogenic orfX deletion strain, over 80 h at 18 °C and over 8 h at 37 °C. Both RT-PCR and Western blotting confirmed that OrfX was not produced in AW2. Growth was the same for both strains at both temperatures (Fig. 2B). Finally, inactivation of orfX in RN450M had no effect on the susceptibility of this strain to oxacillin, mediated by the mecA gene in SCCmec.
FIGURE 2.
A, OrfX expression monitored throughout the growth cycle of S. aureus. Shown is a Western blot of samples taken at each of the time points indicted and by diamonds in the growth curve below. B, growth curves comparing the growth of the parent (RN450) and orfX mutant (AW2) at either 18 °C (upper panel) or 37 °C (lower panel) in enriched BHI broth.
OrfX-mediated Ribosomal Methylation
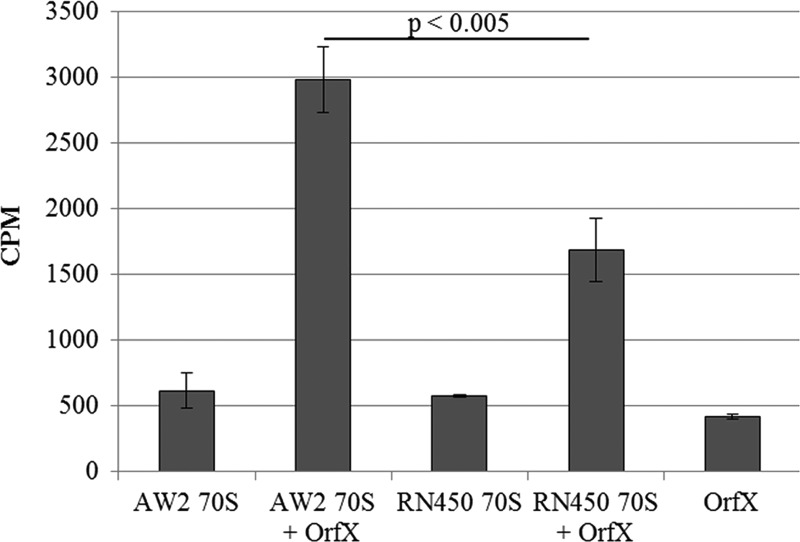
Comparison of a published crystal structure of OrfX (9) with structural databases showed that it has high structural homology to an E. coli methyltransferase, YbeA, the rlmH gene product. To confirm that, like YbeA, OrfX can methylate ribosomes, we incubated purified OrfX with 70 S ribosomes from both a strain with intact orfX (RN450) and an isogenic mutant with orfX inactivated (AW2). Both reactions contained the AdoMet substrate with a 3H-radiolabeled donor methyl group. The radioactive counts from AW2 ribosomes, which would have had more unmethylated target nucleotides available for OrfX methylation, were significantly higher than the cpm from RN450 ribosomes, which were methylated by OrfX in vivo prior to extraction (Fig. 3).
FIGURE 3.
Methylation of ribosomes from either the orfX mutant (AW2) or the parent (RN450) with a fully functional orfX (see Fig. 1). The bars are radioactive counts (vertical axis) from ribosomes exposed to OrfX incubated with substrate (AdoMet) containing a 3H-labeled methyl group. Error bars indicate the S.D. of the results from three repeated experiments. The difference between the results obtained with the two strains was statistically significant (p < 0.005; Students t test). Also shown are the controls with 70 S ribosome, with radiolabeled AdoMet and no OrfX, and with OrfX with radiolabeled AdoMet and no 70 S ribosomes.
Structure Determination
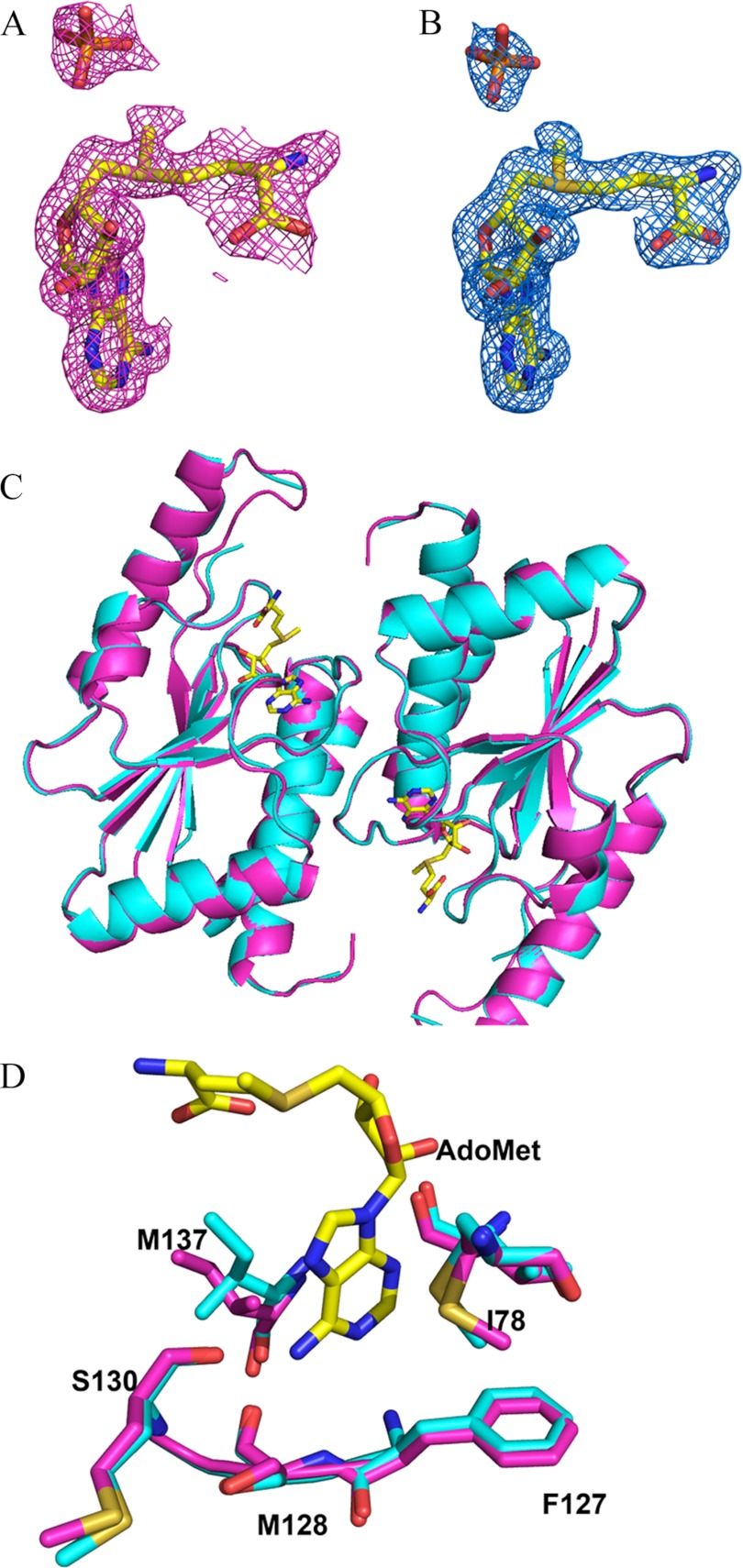
To define the interaction of OrfX with substrate and to be able to predict OrfX-AdoMet-ribosome interactions we solved the crystal structure of the 159-amino acid protein with bound AdoMet (Fig. 4, A and B) at 1.7 Å resolution. A search of the structural database with the OrfX sequence using the program FUGUE identified a recently deposited hypothetical protein structure (Protein Data Bank code 1VH0 (4)) as having a similar structure. It became obvious that the unliganded 1VH0 and OrfX were the same. Because OrfX crystallized in a different space group, molecular replacement was performed with 1VH0 as the search molecule. The starting solved model was then refined to a final crystallographic R-factor/Rfree of 20.7/24.7%. The asymmetric unit contains one monomer, and the functional dimer can be obtained by application of a crystallographic 2-fold symmetry. In addition to the bound AdoMet at the active site, the model also contains a phosphate ion (Fig. 4, A and B) at the active site and a PEG-550MME molecule (used for crystallization) at the surface of the protein. Refinement and other crystallographic parameters are listed in Table 1. OrfX (AdoMet) and 1VH0 (native) structures remain essentially the same, with a root mean square deviation of 0.3 Å for the backbone atoms (Fig. 4, C and D).
FIGURE 4.
A, initial difference electron density map (with coefficients Fo − Fc shown at the 2. 8σ level) of the OrfX structure before the bound AdoMet molecule was added to the model. B, final electron density map (with coefficients 2Fo − Fc shown at the 1.0 σ level) of the OrfX structure. All maps are superimposed with the final refined models. C, least-squares superposition of the OrfX structure (blue) with that of the Protein Data Bank 1VH0 structure (magenta). D, least-squares superposition of the active site structures of OrfX (blue) and 1VH0 (magenta).
Overall Structure of OrfX
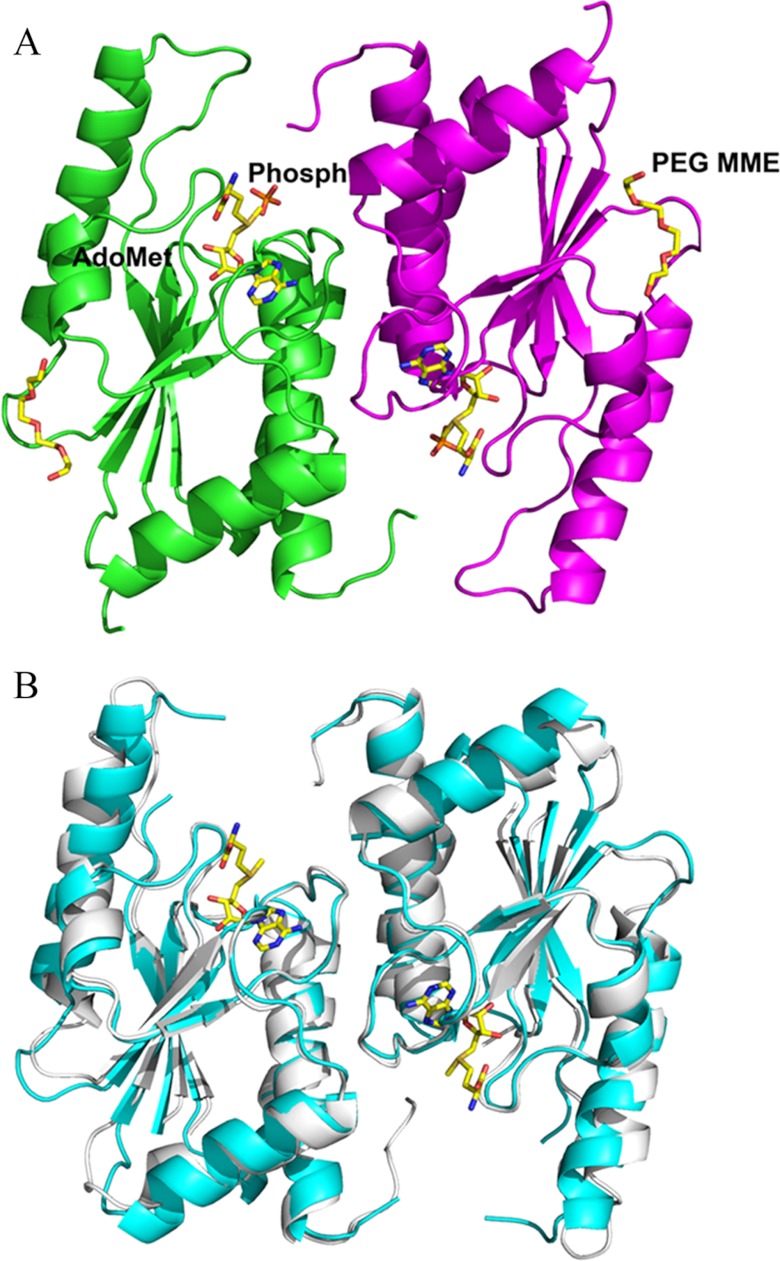
OrfX is a single-domain protein with a buried surface area of 1290 Å2 of the total area of the dimer (Fig. 5A). Each monomer displays the structural elements characteristic of α/β-knot methyltransferases consisting of an all parallel five-stranded β-sheet (β2-β1-β4-β3-β5) and flanked on one side by helices α2 and α4 and on another side by helices α1, α3, and α5. Within the β-sheet is the deep trefoil knot that binds a well defined AdoMet and a phosphate molecule (Fig. 5A). The knot is formed by the threading of the last 34 residues (positions 121–157) through a 37-residue knotting loop structure (residues 74–105).
FIGURE 5.
Overall structure of OrfX (Protein Data Bank code 4FAK). A, ribbon diagram of the dimeric OrfX structure, also with bound AdoMet (yellow stick), a phosphate molecule (brown stick), and PEG-550MME (yellow stick). Monomers A and B are colored green and magenta, respectively. B, least-squares superposition of the OrfX structure (blue) with the YbeA structure (gray).
OrfX dimerizes in an antiparallel fashion with close packing of α1 (residues 13–22), α5, the loop between β5 and α5 (residues 127–156), and the loop between β2 and α3 from each subunit. Although the AdoMet is located mostly in the cavity formed by the deep trefoil knot from one monomer, the other monomer provides residues from the N-terminal α3 and the C terminus of the protein to partially cover the active site.
As expected, a database search using Dali (15) identified three other structurally related members of the α/β-knotted methyltransferase COG1576 family, all with z-scores above 22 and root mean square deviations ranging between 0.7 and 1.7 Å. These include YydA (a 159-residue protein structure from Bacillus subtilis, Protein Data Bank code 1TO0), YbeA (a 155-residue protein structure from E. coli, code 1NS5), and Tm0844 (a 151-residue protein structure from E. coli, code 1O6D). Like OrfX, the other COG1576 family members are also homodimers both in solution and in crystals (16). The dimer interface residues of all members are invariant. They are also single-domain proteins, displaying similar structural elements as well as AdoMet-binding sites (Fig. 5B) (see below). Sequence analysis resulted in identities of OrfX with YbeA (code 1NS5) of 30.8%, with YydA (code 1TO0) of 60.4%, and with Tm0844 (code 1O6D) of 32.9%.
AdoMet Interactions
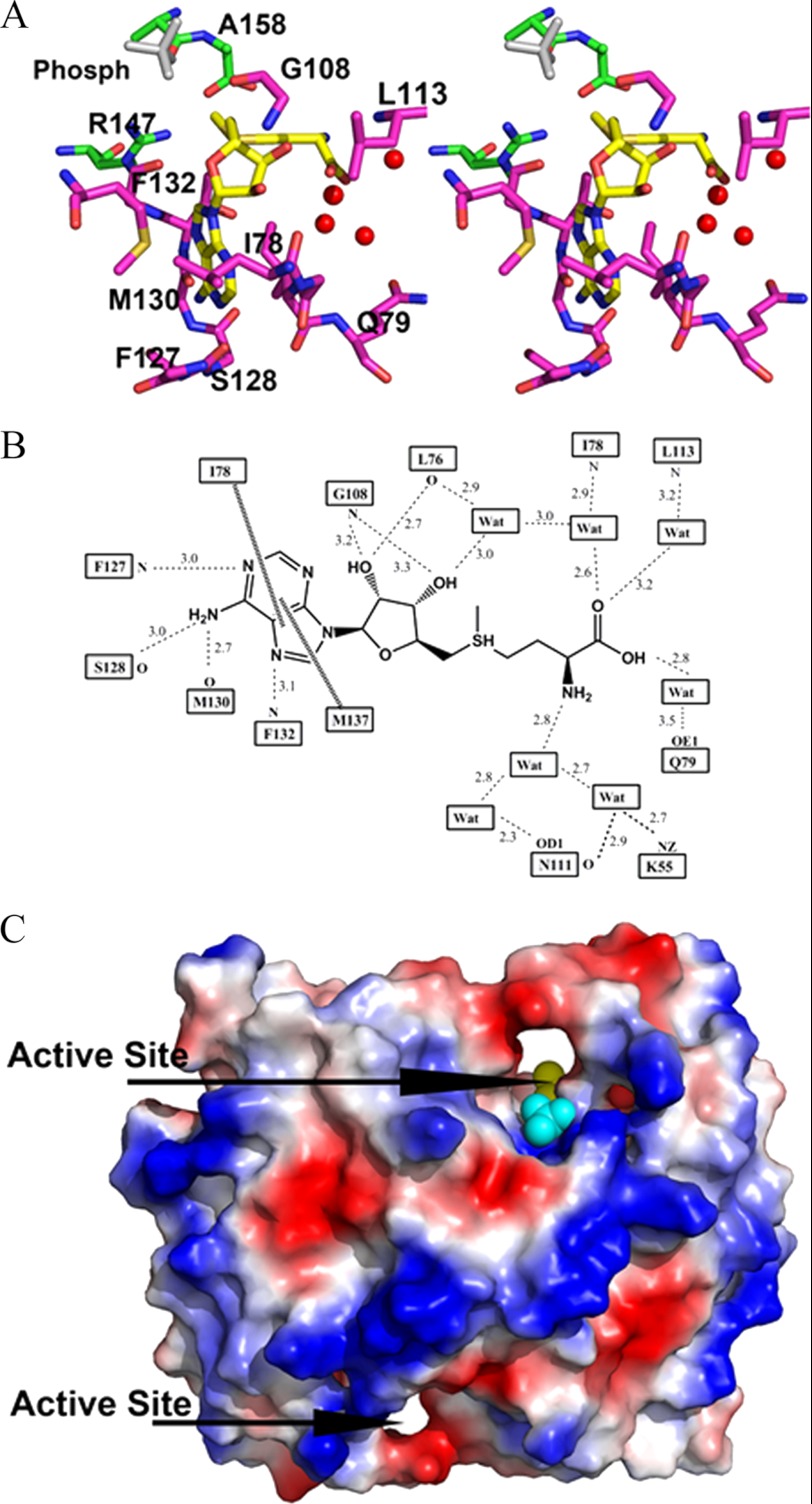
OrfX is the first RlmH or COG1576 family member to have been co-crystallized with AdoMet, although a putative model for the interaction between the protein and AdoMet has been proposed for YbeA (17). The active site is formed by the two subunits, with the adenine portion deeply buried and the positively charged methyl group and carboxylate moiety partly exposed (Fig. 6, A and B). Only the adenine and ribose portions of AdoMet make direct interaction with the protein (Fig. 6, A and B). This includes hydrogen bond interactions exclusively with the protein backbone residues of Met-130, Ser-128, Phe-127, Phe-132, Leu-76, and Gly-108. The side chains of Met-137 and Ile-78 form hydrophobic interactions with AdoMet by clamping on each side of the adenine ring. The amine and carboxylate portions of the methionine moiety make only water-mediated hydrogen bond interactions with the backbone or side chains of Leu-113, Ile-78, and Gln-79. Compared with the unliganded OrfX structure (Protein Data Bank code 1VHO), only Met-137 and Ile-78 show any significant movement toward the bound AdoMet (Fig. 4, C and D). The active site geometry and residues are conserved among the COG1576 family members, with the exception of Met-137 and Ile-78, which are Lys and Thr, respectively, in 1O6D.
FIGURE 6.
Active site and AdoMet cofactor-binding mode of OrfX. A, stereo view of the AdoMet-binding site and the bound AdoMet. Monomers A and B of the protein are as yellow and green sticks, respectively. AdoMet is shown as a yellow stick, and the water molecules are red spheres. B, two-dimensional schematic diagram showing interactions between AdoMet and the protein residues or water molecules. Dotted lines indicate hydrogen bond interactions, and dashed lines indicate hydrophobic contacts. C, surface representations of the OrfX-AdoMet complex. The two active sites are shown, with one of them showing the bound AdoMet and phosphate molecules as yellow and cyan spheres, respectively. The protein structure is colored according to the distribution of electrostatic potential from red (−8 kiloteslas) to blue (+8 kiloteslas.
OrfX-AdoMet Association
Predictions of OrfX-AdoMet interactions made from the crystal structure of OrfX and from models of other SPOUT family methyltransferases (MTases) were assessed using isothermal titration calorimetry. We sought to demonstrate that OrfX interacts with the substrate AdoMet. Fig. 7 shows that OrfX does indeed bind its substrate AdoMet and that it is statistically more likely that both sites of the homodimer are filled at the same time. Isothermal titration calorimetry data modeled with Origin 7 software gave a value of n = 1.04 ± 0.03 for a single-site model with χ2/degrees of freedom of 959. However, further modeling of the isothermal titration calorimetry data allowing for sequential binding of AdoMet to the active site of one monomer and then the subsequent filling of a second active site gave a lower, more statistically likely χ2/degrees of freedom of 726. This sequential binding model also fits the crystal data more closely, where we see both active sites of the homodimer filled with AdoMet molecules.
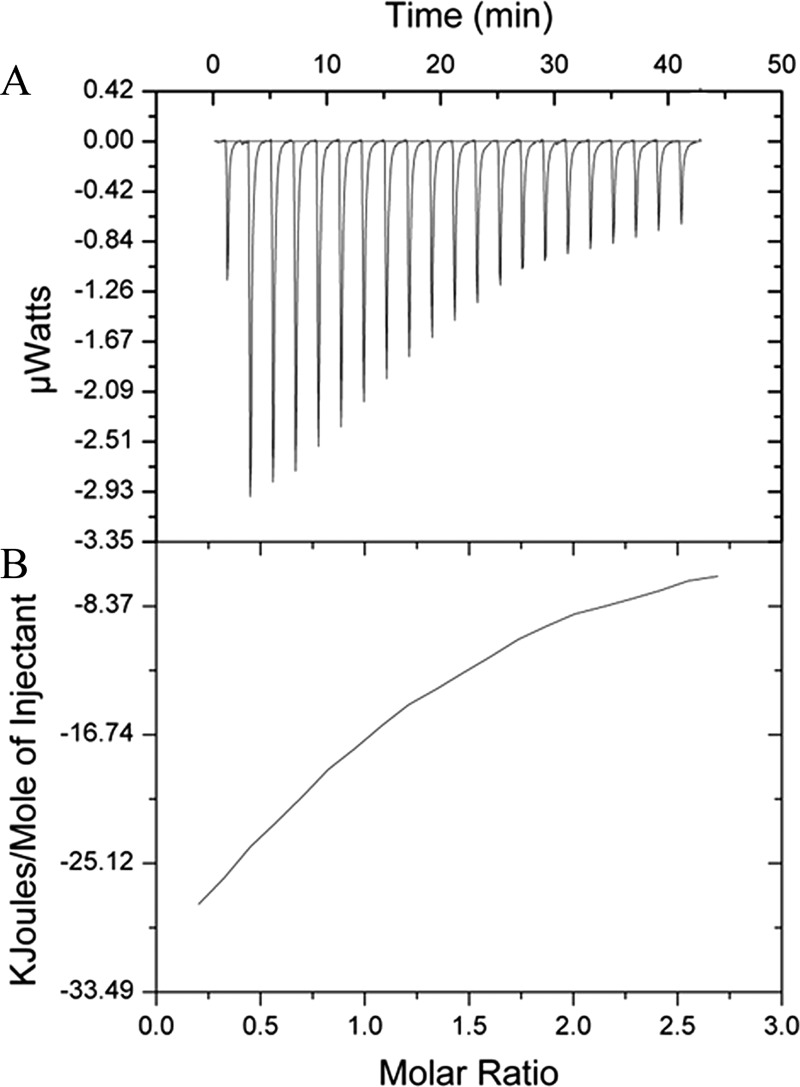
FIGURE 7.
Isothermal titration calorimetry results from the binding of purified dimeric 0.0726 mm OrfX with 1.182 mm AdoMet buffered with 50 mm sodium phosphate (pH 7.5). A shows the exothermic reaction of AdoMet being injected into the cell and binding OrfX. Analysis using Origin 7 produced B. The most favorable χ2/degrees of freedom (726) was in a model predicting sequential binding with the first active site of the dimer being filled, with Kd1 = 52.1 μm, ΔH1 = −5,135 J/mol, and ΔS1 = −83.5 J/mol/degree. The second active site of the dimer was then filled, with Kd2 = 606 μm, ΔH2 = −2.41 × 104 J/mol, and ΔS2 = −15.9 J/mol/degree. This demonstrates that both active sites in the OrfX dimer are filled with an AdoMet molecule, but that first one active site is filled and then the second one.
From this, we conclude that AdoMet binds both active sites of OrfX, with the first site being filled with a Kd1 of 52.1 ± 0.4 μm and the second site being filled with a Kd2 of 606 ± 2 μm. The binding of both AdoMet molecules is exothermic, with a ΔH1 of (−5.135 × 104) ± 447 J/mol and a ΔS1 of −85.5 J/mol/degree for the binding of the first AdoMet molecule and a ΔH2 of (−2.405 × 104) ± 1.56 × 103 J/mol) and a ΔS2 of −15.9 J/mol/degree for the second AdoMet molecule. This shows that in the first active site of the homodimer, AdoMet is bound more tightly than the molecule in the second active site.
Identification of the Catalytic Amino Acid
Unlike TrmD, in which Asp-169, located at the active site of the enzyme, has been identified as the catalytic base (19), in OrfX and the rest of the RlmH family, there is no such obvious catalytic base located at the active site. Nonetheless, there are three conserved amino acids, Asp-49, Glu-77, and Glu-86, ∼5 Å removed from the active site, that could possibly move to the active site as a result of a conformational rearrangement. To test whether any of these three amino acids could be the catalytic base, constructs were made changing each to alanine in pET30a+. Each construct was then expressed, purified, and subjected to the methylation assay. None of the three mutations abolished OrfX activity. The E77A mutation significantly lowered OrfX activity but only by 30%, so whereas this base might be important, it is not the catalytic base because mutating it does not abolish activity.
DISCUSSION
Structures of Other Related Methyltransferases
In addition to the COG1576 family members, the Dali database search also identified other SPOUT superfamily MTases, with the closest being the tRNA-binding TrmD-type m1G MTases (COG0336), with a z-score of ∼11 and a root mean square deviation of ∼3 Å. The COG1576 and TrmD-type structures (using Protein Data Bank code 1UAM as an example of a TrmD structure) show a common core five-stranded parallel sheet sandwiched between two layers of α-helices. However, TrmD shows an additional α/β-fold C-terminal subdomain that is absent in the COG1576 family. The single-domain structure of the COG1576 family members corresponds to the N-terminal domain of TrmD. The TrmD C-terminal domain has been proposed to be the tRNA substrate-binding site, and its absence in the COG1576 family members has led to the suggestion that the COG1576 family is a minimalist member of the SPOUT MTase superfamily (17). The AdoMet-binding pockets in OrfX and TrmD (code 1UAM) are similar but are nevertheless part of the C-terminal subdomain, and the loop that connects the N-terminal domain and C-terminal subdomain (which are absent in OrfX) forms part of the active site in TrmD. In the COG1576 family, the C terminus mimics the loop of TrmD to form part of the active site. TrmD (code 1UAM) dimerizes in a similar fashion as OrfX, with the two monomers arranged in parallel fashion. Nevertheless, TrmD (code 1UAM) has a significantly larger buried surface area due to the additional intersubunit contacts involving the C-terminal domain.
The active site residues Leu-76 and Gly-108 in OrfX are conserved in TrmD (Protein Data Bank code 1UAM). However, Met-137 and Ile-78, which clamp on the AdoMet adenine ring in OrfX, are replaced with Pro in TrmD (code 1UAM). Also, the active site residues Ser-128, Phe-127, and Phe-132 in OrfX are replaced with Gly, Ile, and Leu, respectively. Other active site residues, including Gly-80, Gly-108, Gly-109, and Gly-112 (not necessarily making interactions with AdoMet), are highly conserved among the COG1576 family members and TrmD as well as other SPOUT MTases (17). These glycine residues have been suggested to be critical for active site structure formation (18, 19). In OrfX, the amine and carboxylate portions of the methionine moiety make water-mediated hydrogen bond interactions with the backbone or side chains of Leu-113, Ile-78, and Gln-79. In TrmD, the methionine moiety makes direct hydrogen bond interactions with the negative residues Asp-169, Asp-177, Glu-116, and Gln-90. Only the latter residue, Gln-90, is conserved in OrfX (Gln-79).
Putative Binding between OrfX and rRNA
Prior to this work, only YbeA in the COG1576 family has been functionally characterized and shown to methylate the 70 S ribosomal subunit, specifically catalyzing N3 methylation at the 1915 pseudouridine site (ψ1915) to form m3ψ1915 (17, 20). The COG1576 orthologs, all from bacteria, have also been suggested to methylate ψ1915 (17), and our functional study showed that OrfX does methylate S. aureus 70 S ribosomal subunits. There is no structural study of any of the COG1576 family members in complex with rRNA, and with the exception of the structure reported here, none of them has also been co-crystallized with AdoMet. On the basis of the ternary complex of OrfX with AdoMet and phosphate, we offer some insight as to how OrfX and the rest of the COG1576 family members may bind to rRNA for subsequent catalytic nucleotide methylation. The unique C-terminal domain in TrmD and other methyltransferases, which is absent in the COG1576 family, has been proposed to facilitate tRNA binding and specificity (21). Nonetheless, there are several conserved positive and negative patches at the AdoMet active site in the COG1576 family members that could interact with the rRNA substrate (Fig. 4C). The bound phosphate at the active site of OrfX is ∼3.5 Å from the AdoMet methyl group. Interestingly, a similar bound phosphate at the active site of TrmD (Protein Data Bank code 1UAM) has been proposed to be the putative position of the phosphate on the 5′-side of Gly-37, corresponding to the N1 methylation target. The bound phosphate in OrfX may also correspond to the phosphate of the rRNA nucleotide to be methylated. Consistent with this analysis is a previous docking study between YbeA and the 70 S ribosome that showed ψ1915 at the same location as the phosphate in OrfX (17, 20). In TrmD, Glu-116, Arg-154, and Asp-169 have been suggested to recognize the guanine ring of Gly-37, the latter residue identified as the catalytic base that deprotonates the guanine N1 prior to methylation (19). Arg-154 is strictly conserved in COG1576 family members (residue 147 in OrfX). Glu-116 is conserved in YbeA but is changed to Asn, Leu, and Tyr in Orfx, 1TO0, and 1O6D, respectively. Asp-169, the catalytic general base residue in TrmD, has no counterpart in COG15 family members due to the absence of the C-terminal domain. It is therefore unclear what residue in the COG1576 family would act as the base. As noted above, there are several highly conserved residues close to the active site that could move to the active site to act as a catalytic base as a result of a conformational rearrangement, perhaps triggered by rRNA binding. Consistently, the active sites of the COG1576 family members are relatively open compared with TrmD (code 1UAM). Similar observations have been noted for several other SPOUT class RNA MTases that do not show any obvious catalytic base at the active site (22). Nonetheless, site mutation studies of several putative catalytic bases did not lead to abolition of enzymatic activity. At this stage, it is not obvious which other amino acid could be acting as the base, and further detailed studies are clearly warranted.
Three other residues, Arg-39, Asp-50, and Tyr-115, have also been proposed to be important in the binding of Gly-36 in TrmD. These residues correspond to Lys-10 (or Thr in 1TO0), Glu-41 (or Lys or Arg in YbeA and 1O6D), and Ser-110 (or Pro in YbeA and 1O6D) in the COG1576 family, respectively. Close to the phosphate molecule in OrfX, we also observed a totally conserved His-134 in the COG1576 family, which is replaced with Leu in TrmD (code 1UAM). This residue may play a role in RNA binding and/or base recognition.
The above analysis suggests some similarities in the binding of RNA between TrmD and COG1576 family members and highlights the close evolutionary pathway between the two SPOUT MTase families. Nevertheless, the differences in the active site residue and/or conformation between TrmD and OrfX and, for that matter, among the COG1576 family members could determine whether the protein may bind to tRNA or rRNA, as well as affect RNA substrate specificity.
Conservation of orfX
The conservation of orfX among staphylococci suggests that it is an essential gene, but we could not identify any growth defect in vitro at either 18 or 37 °C, nor could we clearly show that there was a fitness defect in vitro in an orfX mutant in competition with the parent (data not shown). However, an effect of the loss of ribosomal methylation may be seen only in vivo. With the ribosomal MTase KsgA, there is no clear growth defect in vitro of a ksgA mutant in E. coli, but when the gene is inactivated in either Erwinia amylovora, a pear fruit pathogen, or in Yersinia pseudotuberculosis, there is a loss of virulence. One E. amylovora ksgA mutant, recovered by assessing resistance to the drug targeting KsgA (kasugamycin), did not differ in growth rate from its parent but had markedly reduced pear necrotic lesions (23). Y. pseudotuberculosis ksgA mutants both failed to kill mice and were outcompeted in vivo when mixed with the wild-type strain (24). There may be a similar reduction in either virulence or fitness in orfX mutants when these strains are evaluated in experimental S. aureus infection models. Experiments are under way to define the effects of orfX on virulence and fitness in animal models.
Acknowledgment
The structural biology resources were supported in part by National Institutes of Health Grant CA 16059-28 from NCI to the Virginia Commonwealth University Massey Cancer Center.
This work was supported, in whole or in part, by National Institutes of Health Grant 2R01 AI035705 from NIAID (to G. L. A.).
The atomic coordinates and structure factors (code 4FAK) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SCCmec
- staphylococcal chromosome cassette mec
- BHI
- brain heart infusion
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- AdoMet
- S-adenosyl-l-methionine
- MTase
- methyltransferase.
REFERENCES
- 1. Noto M. J., Kreiswirth B. N., Monk A. B., Archer G. L. (2008) Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus. J. Bacteriol. 190, 1276–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tkaczuk K. L., Dunin-Horkawicz S., Purta E., Bujnicki J. M. (2007) Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics 8, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sass P., Berscheid A., Jansen A., Oedenkoven M., Szekat C., Strittmatter A., Gottschalk G., Bierbaum G. (2012) Genome sequence of Staphylococcus aureus VC40, a vancomycin- and daptomycin-resistant strain, to study the genetics of development of resistance to currently applied last-resort antibiotics. J. Bacteriol. 194, 2107–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Badger J., Sauder J. M., Adams J. M., Antonysamy S., Bain K., Bergseid M. G., Buchanan S. G., Buchanan M. D., Batiyenko Y., Christopher J. A., Emtage S., Eroshkina A., Feil I., Furlong E. B., Gajiwala K. S., Gao X., He D., Hendle J., Huber A., Hoda K., Kearins P., Kissinger C., Laubert B., Lewis H. A., Lin J., Loomis K., Lorimer D., Louie G., Maletic M., Marsh C. D., Miller I., Molinari J., Muller-Dieckmann H. J., Newman J. M., Noland B. W., Pagarigan B., Park F., Peat T. S., Post K. W., Radojicic S., Ramos A., Romero R., Rutter M. E., Sanderson W. E., Schwinn K. D., Tresser J., Winhoven J., Wright T. A., Wu L., Xu J., Harris T. J. (2005) Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins 60, 787–796 [DOI] [PubMed] [Google Scholar]
- 5. Novick R. (1967) Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33, 155–166 [DOI] [PubMed] [Google Scholar]
- 6. Kreiswirth B. N., Löfdahl S., Betley M. J., O'Reilly M., Schlievert P. M., Bergdoll M. S., Novick R. P. (1983) The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712 [DOI] [PubMed] [Google Scholar]
- 7. Wang L., Safo M., Archer G. L. (2012) Characterization of DNA sequences required for the CcrAB-mediated integration of staphylococcal cassette chromosome mec, a Staphylococcus aureus genomic island. J. Bacteriol. 194, 486–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Niemeyer D. M., Pucci M. J., Thanassi J. A., Sharma V. K., Archer G. L. (1996) Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J. Bacteriol. 178, 5464–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ero R., Peil L., Liiv A., Remme J. (2008) Identification of pseudouridine methyltransferase in Escherichia coli. RNA 14, 2223–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farrar C. E., Clarke S. (2005) Diet-dependent survival of protein repair-deficient mice. J. Nutr. Biochem. 16, 554–561 [DOI] [PubMed] [Google Scholar]
- 11. Todd M. J., Gomez J. (2001) Enzyme kinetics determined using calorimetry: a general assay for enzyme activity? Anal. Biochem. 296, 179–187 [DOI] [PubMed] [Google Scholar]
- 12. Freyer M. W., Lewis E. A. (2008) Isothermal titration calorimetry: experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol. 84, 79–113 [DOI] [PubMed] [Google Scholar]
- 13. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 14. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 15. Holm L., Sander C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 16. Mallam A. L., Jackson S. E. (2007) A comparison of the folding of two knotted proteins: YbeA and YibK. J. Mol. Biol. 366, 650–665 [DOI] [PubMed] [Google Scholar]
- 17. Purta E., Kaminska K. H., Kasprzak J. M., Bujnicki J. M., Douthwaite S. (2008) YbeA is the m3ψ methyltransferase RlmH that targets nucleotide 1915 in 23S rRNA. RNA 14, 2234–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J. N., Björk G. R. (1999) Structural alterations of the tRNA(m1G37)methyltransferase from Salmonella typhimurium affect tRNA substrate specificity. RNA 5, 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahn H. J., Kim H. W., Yoon H. J., Lee B. I., Suh S. W., Yang J. K. (2003) Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition. EMBO J. 22, 2593–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schuwirth B. S., Borovinskaya M. A., Hau C. W., Zhang W., Vila-Sanjurjo A., Holton J. M., Cate J. H. (2005) Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834 [DOI] [PubMed] [Google Scholar]
- 21. Persson B. C., Jäger G., Gustafsson C. (1997) The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2′-O-methyltransferase activity. Nucleic Acids Res. 25, 4093–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen H. Y., Yuan Y. A. (2010) Crystal structure of Mj1640/DUF358 protein reveals a putative SPOUT-class RNA methyltransferase. J. Mol. Cell. Biol. 2, 366–374 [DOI] [PubMed] [Google Scholar]
- 23. McGhee G. C., Sundin G. W. (2011) Evaluation of kasugamycin for fire blight management, effect on nontarget bacteria, and assessment of kasugamycin resistance potential in Erwinia amylovora. Phytopathology 101, 192–204 [DOI] [PubMed] [Google Scholar]
- 24. Bergman M. A., Loomis W. P., Mecsas J., Starnbach M. N., Isberg R. R. (2009) CD8+ T cells restrict Yersinia pseudotuberculosis infection: bypass of anti-phagocytosis by targeting antigen-presenting cells. PLoS Pathog. 5, e1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]