Background: Fascin is the main actin-bundling protein in filopodia.
Results: Biochemical, cryo-electron tomographic, and x-ray crystal structural data reveal the unique actin-binding characteristics of fascin.
Conclusion: There are two major actin-binding sites on fascin and there is a concerted conformational change between the actin-binding sites.
Significance: These data will advance our understanding of the function of fascin in filopodial formation.
Keywords: Cell Invasion, Cell Migration, Cell Motility, Signal Transduction, Tumor Metastases
Abstract
Filopodia are cell surface protrusions that are essential for cell migration. This finger-like structure is supported by rigid tightly bundled actin filaments. The protein responsible for actin bundling in filopodia is fascin. However, the mechanism by which fascin functions in filopodial formation is not clear. Here we provide biochemical, cryo-electron tomographic, and x-ray crystal structural data demonstrating the unique structural characteristics of fascin. Systematic mutagenesis studies on 100 mutants of fascin indicate that there are two major actin-binding sites on fascin. Crystal structures of four fascin mutants reveal concerted conformational changes in fascin from inactive to active states in the process of actin bundling. Mutations in any one of the actin-binding sites impair the cellular function of fascin in filopodial formation. Altogether, our data reveal the molecular mechanism of fascin function in filopodial formation.
Introduction
Cell motility plays a central role in both normal and disease processes, including development, wound healing, inflammation, and tumor metastasis (1, 2). Cell movement requires highly dynamic rearrangements of the actin filament network (3). Among the morphological structures supported by actin filaments, the two most prominent and best characterized protrusive organelles are lamellipodia and filopodia, which are fundamental to cell shape and motility events (4). Filopodia are cylindrical cell surface protrusions that extend beyond the leading edge of migrating cells (4, 5). They contain long actin filaments cross-linked into parallel bundles by actin-binding proteins. Filopodia enable migrating cells to navigate their surroundings and determine the direction of migration and destination. Growth factors stimulate the formation of filopodia in fibroblasts to direct fibroblast migration and wound healing. In developing neurons, filopodia extend from the growth cone at the leading edge and is required for proper navigation. Filopodia can act as phagocytic tentacles and pull bound objects toward the cell for phagocytosis in macrophages. Filopodia can also enhance the invasive ability of tumor cells (6).
Fascin is the primary actin cross-linker in filopodia and shows no amino acid sequence homology with other actin-binding proteins (7–12). It has a molecular mass of ∼55 kDa and functions as a monomer (9). It is required to maximally cross-link the actin filaments into straight, compact, and rigid bundles, and it imparts distinct mechanical stiffness to actin bundles (13, 14). Fascin holds 10–30 parallel actin filaments together to form the 60–200-nm diameter filopodia as observed by electron microscope (4). Metazoans have three fascin forms (fascin-1, -2, and -3) encoded by three different genes. Although fascin-1 (referred as fascin here) is expressed in a few types of cells, fascin-2 is specifically expressed in retina and hair-cell stereocilia, and fascin-3 is only found in the testis (15). Elevated levels of fascin have been found in metastatic tumors and are correlated with clinically aggressive phenotypes, poor prognosis, and shorter survival (16–21). Previously we and others have solved the x-ray crystal structure of the wild-type fascin (22–24). Although there are no internal homologous amino acid sequence repeats, the crystal structure of fascin consists of four structurally homologous tandem β-trefoil folds that form a two-lobed structure with pseudo 2-fold symmetry. The molecular mechanism by which fascin bundles F-actin filaments is not clear although both the N- and C-terminal residues are required for its actin-bundling function (25).
Previously, based on the x-ray crystal structure of fascin, we used point mutagenesis to identify residues His392, Lys471, and Ala488 to be critical for the actin-bundling activity of fascin (23). Jansen et al. (24) employed structure-guided mutagenesis studies to reveal that fascin mutants K22E/K43E/R398E and R271E/K353E/K358E were defective in bundling actin filaments. In this report, we have used systematic mutagenesis studies and examined 100 fascin mutants. Two major actin-binding sites were identified in fascin. Cryo-electron tomography of in vitro reconstituted unconstrained three-dimensional bundles formed by fascin and actin filaments showed that the bundles display a hexagonal packing as observed for the bundles from native filopodia. Furthermore, taking advantage of the identified inactive fascin mutants, we have solved the x-ray crystal structures of four inactive fascin mutants and have defined the inactive configuration of fascin. Moreover, we have performed cell biological studies to demonstrate the requirement of all of the identified actin-binding sites for fascin function in filopodial formation in cells.
EXPERIMENTAL PROCEDURES
Human Fascin-1 Expression and Purification
The recombinant human fascin-1 was expressed as GST fusion protein in Escherichia coli. Typically, 1 liter of 2YT medium with antibiotic was inoculated with 3 ml overnight in BL21/DE3 culture transformed with pGEX4T-Fascin1 plasmid and grown at 37 °C until A600 reached ∼0.8. The culture was then transferred to 22 °C and 0.1 mm isopropyl 1-thio-β-d-galactopyranoside was added for induction. After overnight induction, the bacteria were harvested by centrifugation at 5,000 × g for 10 min. The bacteria pellet was snap frozen with liquid nitrogen and suspended in 30 ml of 1× PBS supplemented with 0.2 mm PMSF, 1 mm DTT, 1% Triton X-100, and 1 mm EDTA. After sonication, the suspension was centrifuged at 15,000 × g for 60 min to remove the cell debris. The supernatant was then incubated with 4 ml of glutathione beads (Sigma) at 4 °C for 2 h. After an extensive wash with PBS, the beads were resuspended in 10 ml of thrombin cleavage buffer (20 mm Tris, pH 8.0, 150 mm NaCl, 2 mm CaCl2, 1 mm DTT). Human Fascin-1 was released from the beads by incubation with 40–100 units of thrombin overnight at 4 °C to remove the GST tag. After centrifugation, 0.2 mm PMSF was added to the supernatant to inactivate the remnant thrombin activity. The fascin protein was further purified with a Superdex 200 gel filtration column and concentrated with Centricon to about 80 mg/ml. The typical yield from a 1-liter culture is about 40 mg.
F-Actin-bundling Assay
Actin-bundling activity was measured by a low-speed centrifugation assay and fluorescence microscopy. In the low-speed centrifugation assay, monomeric rabbit G-actin was induced to polymerize at room temperature in F-actin buffer (20 mm Tris-HCl, pH 8, 1 mm ATP, 1 mm DTT, 2 mm MgCl2, and 100 mm KCl). Recombinant fascin proteins (wild-type or mutant) were subsequently incubated with F-actin for 60 min at room temperature and centrifuged for 30 min at 10,000 × g in an Eppendorf 5415D table top centrifuge. Both supernatants and pellets were dissolved in an equivalent volume of SDS sample buffer, and the amount of actin was determined by SDS-PAGE. We measured the intensities of fascin proteins in Coomassie-stained gels and then calculated the relative actin-bundling activity, P, by the following formula: P = 100 × Mp/Mpc, where P is the relative actin-bundling activity, Mp is the percentage of actin present in pellet when mixed with different concentrations of fascin protein, calculated by (intensity in pellet)/(intensity in pellet + intensity in supernatant), and Mpc is the percentage of actin that is present in pellet when mixed with 0.25 μm fascin, which is used at a saturated concentration determined in our control experiment.
In fluorescence microscopy, monomeric G-actin was polymerized as described earlier. F-actin was mixed with recombinant fascin protein in F-buffer (20 mm Tris-HCl, pH 8, 1 mm ATP, 1 mm DTT, 2 mm MgCl2, and 100 mm KCl) and incubated at room temperature for 60 min. Actin was then labeled by adding 2% Alexa 488-phalloidin to the mixture. The samples were applied to a coverslip that was freshly coated with 1 mg/ml of poly-d-lysine. After a 60-min incubation, the coverslip was mounted on a slide and the image was taken on fluorescence microscopy. Three randomly selected fields (×63 objectives) were photographed, and the bundles were counted.
Actin-binding Assay
Actin-binding activity was measured by a high-speed centrifugation assay. In a high-speed centrifugation assay, monomeric rabbit G-actin (10 μm) was induced to polymerize at room temperature in F-actin buffer (20 mm Tris-HCl, pH 8, 1 mm ATP, 1 mm DTT, 2 mm MgCl2, and 100 mm KCl). Recombinant fascin proteins (wild-type or mutant) were subsequently incubated with F-actin for 60 min at room temperature and centrifuged for 30 min at 150,000 × g. Both supernatants and pellets were dissolved in an equivalent volume of SDS sample buffer, and the amount of actin was determined by SDS-PAGE.
Cryo-electron Tomography
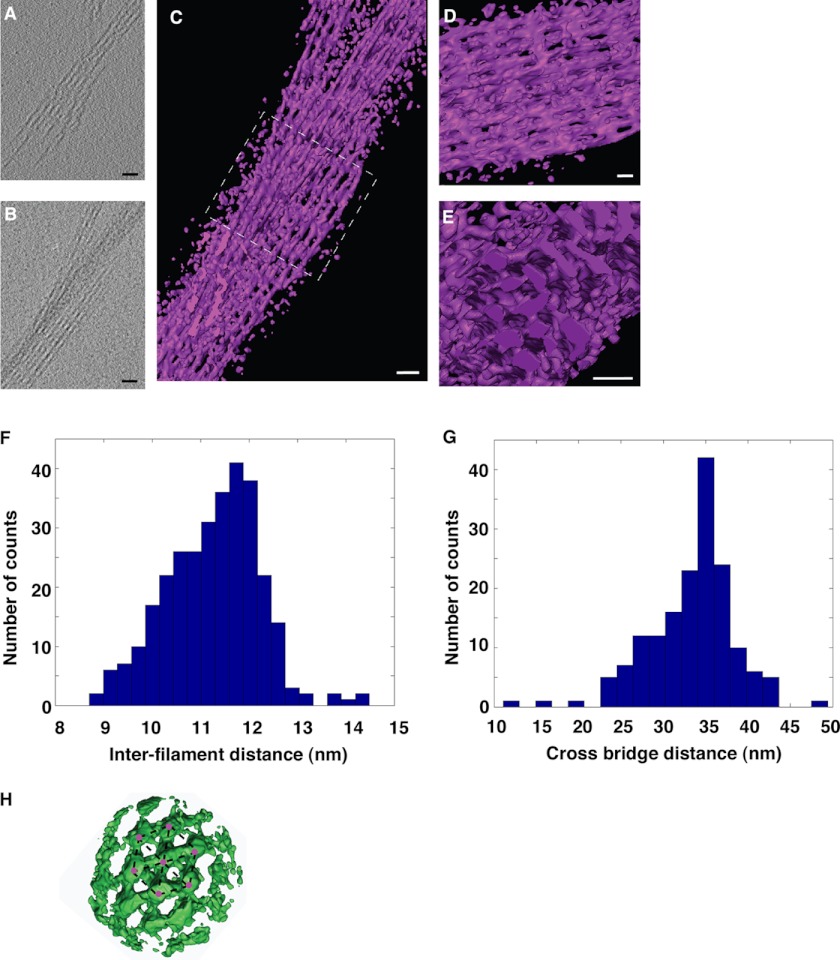
One mg/ml of G-actin was incubated for 1 h with stoichiometric amounts of fascin (in actin polymerization buffer), which led to the assembly of actin/fascin bundles suitable for frozen hydrated specimens. Typically, a 3-ml aliquot of the bundles was placed on a glow discharged 2/2 Quantifoil grid, blotted, and plunge-frozen in liquid ethane. Images were recorded on a JEOL 3200FSC electron microscope operating at 300 kV with an in-column energy filter and collected at a magnification of either 50 or 25 kX, aiming for a defocus value of 4 μm, and using a 20-eV slit for the energy filter. Tilt series were recorded using the SerialEM program on a 4k× 4k CCD camera set to 2× binning, giving an effective pixel size of 4.4 Å and 8.8 Å at specimen angles between ±60 with 2 degree increments (26). The total specimen dose was limited to 60–80 electrons/Å2 for each complete tilt series. Tomogram reconstructions were performed using the eTomo software suite. Statistics regarding the number of filaments per bundle, inter-filament spacing, and cross-bridge distances were calculated. Boxes containing filaments and cross-bridges were manually picked, extracted from the tomograms, and iteratively aligned and averaged using Xmipp and MATLAB (Mathworks), equipped with the TOM and AV3 software packages (27, 28). Visualization and representations were generated using Amira (Visage Imaging).
Crystallization
The concentrated protein stock solution was diluted with the protein buffer (20 mm Tris, pH 8.0, 40 mm KBr, 0.5 mm EDTA, 1 mm DTT) to 15 mg/ml prior to crystallization. Crystallization was performed by the vapor diffusion hanging drop method at 20 °C. Fascin crystallized in 100 mm Hepes, pH 8.0, 16% PEG4000, 1% isopropyl alcohol. Crystals were briefly transferred to the cryo-solution, which consisted of the crystallization solution implemented with 15% glycerol and flash-cooled in liquid nitrogen.
Data Collection, Structure Determination, and Refinement
Diffraction data of four fascin mutants presented in this paper were collected at 100 K using synchrotron radiation (λ = 1.000 Å) at beamline X6A, National Synchrotron Light Source (Brookhaven, NY). All data reduction was performed with HKL2000 and the crystals belong to space group C2. The corresponding data collection statistics are shown in the supplemental Table S1. The structures of all fascin mutants were solved by molecular replacement with PHASER of the CCP4 suite using the wild-type fascin structure (PDB code 3LLP)6 as the search model (29). Model refinements were performed with REFMAC5 followed by employing the program COOT for iterative cycles of rebuilding based on σ-A weighted 2Fo − Fc and Fo − Fc maps (30, 31). Solvent molecules were identified based on the Fo − Fc difference map and the corresponding mutated residues were corrected as well. The final steps of refinement have also incorporated TLS restraints for all mutant structures (32). A total of two mostly complete molecules were observed per asymmetric unit except for the several highly flexible loops with poorly resolved electronic density. Refinement statistics are also presented in supplemental Table S1, and the stereochemical quality of the refined structures was validated using MolProbity (33). Global alignment of various structural models of fascin was performed using PyMOL (super_align) (DeLano Scientific LLC) and all structural model figures were created with PyMOL as well.
Immunofluorescence Microscopy
Cells were seeded onto lamini-coated glass coverslips in starvation condition (no serum) for 2 h. They were fixed with 3.7% formaldehyde in PBS for 10 min at room temperature, permeabilized with 0.1% Triton X-100 for 5 min, and then washed 3 times with PBS. Actin filaments were labeled with 2% Alexa 488-phalloidin. The coverslips were then mounted onto slides and imaged using Zeiss LSM510 confocal microscopy.
Statistical Analysis
Data are expressed as mean ± S.D. and analyzed by Student's t test with significance defined as p < 0.05.
RESULTS
Identification of Two Major Actin-binding Sites on Fascin
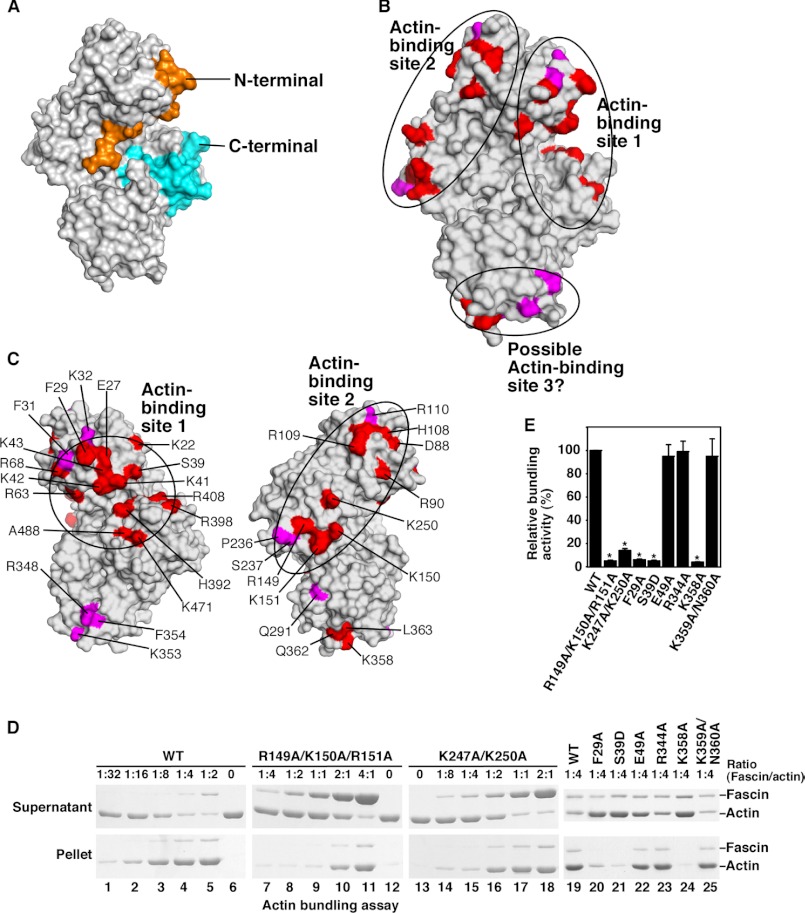
Previous studies have shown that purified fascin proteins could initiate the formation of filopodial-like actin filamental structures in vitro (10, 34). To understand why fascin (among all actin bundling or cross-linking proteins) is uniquely selected for filopodial formation, we have investigated the biochemical and biophysical properties of fascin. We and others have recently solved the x-ray crystal structure of fascin (22–24) (Fig. 1A). Fascin functions as a monomer to bundle actin filaments, and it has been proposed that fascin has two actin-binding sites for this bundling activity (25). Previous amino acid sequence deletion studies implied that both the N and C termini of fascin contribute to actin binding (25). However, from the fascin crystal structure, these N and C termini do not represent two different actin-binding sites. Rather, both the N and C termini are located in the same actin-binding site (Fig. 1A).
FIGURE 1.
Identification of two major actin-binding sites on fascin. A, surface presentation of the structure of fascin. The N and C termini were labeled in orange and blue, respectively. B and C, surface presentation of the fascin structure showing the two major actin-binding sites identified from systematic mutagenesis studies. Critical residues involved in actin bundling are indicated. D, low-speed centrifugation assays of the actin-bundling activity of fascin and its representative mutants. G-actin (1 μm) and different concentrations of fascin or fascin mutant proteins were used. E, quantification of the data from low-speed centrifugation assays (ratio of fascin/actin as 1:4). Results are mean ± S.D. (n = 3, *, p < 0.001). Residues showing strong effects (>80% decrease in the actin-bundling activity) are labeled in red. Residues showing moderate effects (>50 but <80% decreases) are labeled in magenta.
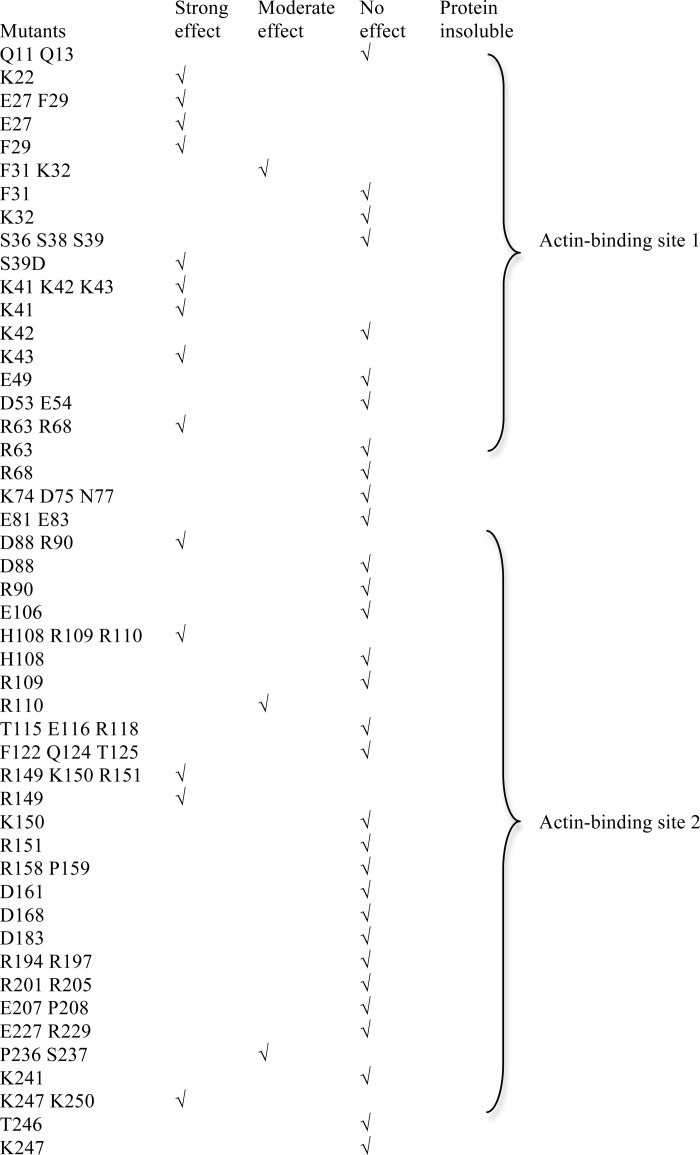
To understand how fascin interacts with actin filaments to form filopodia, we decided to first unbiasedly and systematically identify all the actin-binding sites on fascin. Systematic alanine scanning mutagenesis were done on all solvent accessible residues that are not involved in maintaining the tertiary structure of fascin based on the crystal structure of fascin (Figs. 1, B–E, and 2, A–F, and Table 1). We used two different approaches to examine the actin-bundling activity of 100 fascin mutant proteins. In the low-speed co-sedimentation assay, G-actins and individual F-actin filaments are in the supernatant after low-speed centrifugation, whereas the actin bundles formed in the presence of fascin are in the pellet (9, 23). From these actin-bundling activity assays, we observed that some mutations such as K22A, E27A, F29A, R149A/K150A/R151A, K247A/K250A, and K358A significantly (>80%) decreased the actin-bundling activity of fascin (Fig. 1, D and E, and Table 1). The defective actin-bundling activity was not due to overall structural changes because these mutant fascin proteins and wild-type fascin displayed similar intrinsic fluorescence emission spectrums, acrylamide fluorescence quenching (supplemental Fig. S1, A and B), and x-ray crystal structures (see below). Some fascin mutants such as R110A and Q291A moderately (>50% but <80%) reduced the actin-bundling activity, whereas others had a minimal (<20%) effect on the actin-bundling activity (Table 1). The mutations that reduced the actin-bundling activity are clustered on three distinct surface areas (Fig. 1, B and C). One (here, termed actin-binding site 1) is formed by residues from the N and C termini of fascin, and includes the cleft formed by β-trefoil folds 1 and 4 (Fig. 1, B and C). Within this actin-binding site 1, Ser39 is the phosphorylation site of protein kinase C and this phosphorylation decreases the actin-bundling activity of fascin (25, 35). Indeed, whereas the fascin(S39A) mutant had no effect on the actin-bundling activity, the phosphomimetic fascin(S39D) mutant significantly decreased the actin-bundling activity of fascin (Table 1). The second site (here, termed actin-binding site 2) is opposite to actin-binding site 1, and includes residues from β-trefoil folds 1 and 2 (Fig. 1, B and C). A smaller surface area, at the bottom of the fascin molecule depicted in Fig. 1, B and C, involves residues from β-trefoil fold 3 (Fig. 1B). This region may represent a potential third actin-binding site or is part of the other two major actin-binding sites. Hence, we uncovered two major spatially distinct actin-binding sites on fascin, and all four β-trefoil folds are involved in actin bundling.
FIGURE 2.
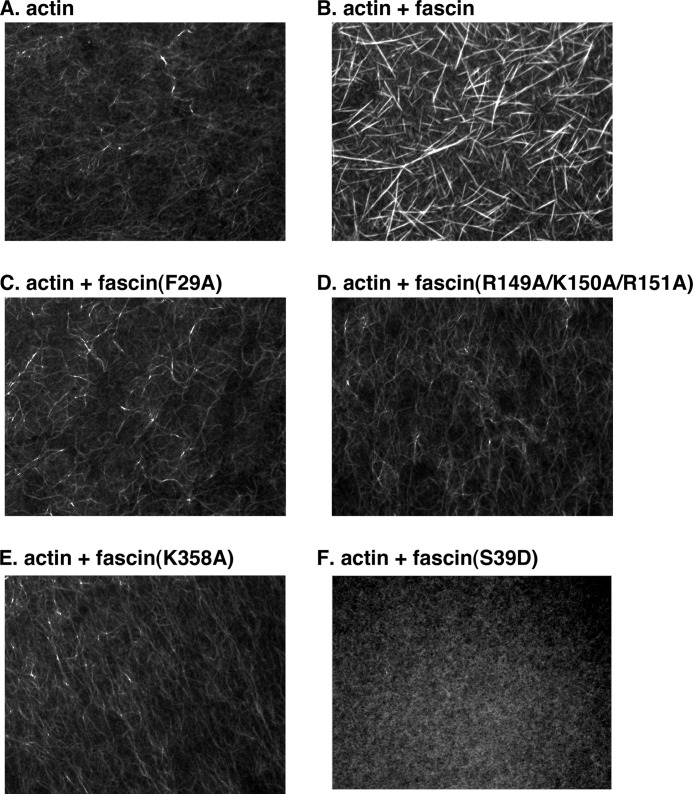
Actin-bundling activity assay by fluorescence microscopy. A, actin alone. B, actin together with wild-type fascin proteins. C, actin with mutant fascin(F29A) proteins. D, actin with mutant fascin(R149A/K150A/R151A) proteins. E, actin with mutant fascin(K358A) proteins. F, actin with mutant fascin(S39D). Images are representative of three independent experiments.
TABLE 1.
Actin-bundling activity of fascin mutants
The actin-bundling activity of each fascin mutant was studied with the low-speed co-sedimentation assay. Strong effect (>80% decrease of the bundling activity compared with the wild-type fascin), moderate effect (>50%, but <80% decrease of the bundling activity), and no effect (<20% decrease of the bundling activity) were used to group the mutants. Experiments for each mutant were repeated at least three times.
To confirm the above results using a different assay, we directly visualized actin filaments under fluorescence microscopy (Fig. 2). In the absence of fascin, actin alone displayed overall weak fluorescent intensity and thin filaments (Fig. 2A). Addition of fascin generated longer and thicker filaments with much stronger fluorescent intensity (Fig. 2B). Fascin mutants that are defective in actin bundling such as fascin(F29A), fascin(R149A/K150A/R151A), fascin(K358A), and fascin(S39D), showed weaker fluorescent intensities and thinner filaments (Fig. 2, C–F). Together, these two complementary approaches identified amino acid residues important for the actin-bundling activity of fascin and uncovered two major actin-binding sites on fascin.
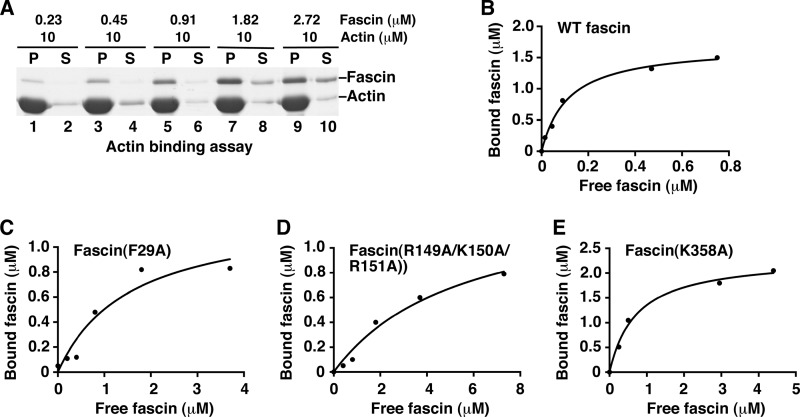
To directly measure the actin-binding activity of some representative fascin mutants, we used the high-speed co-sedimentation assay. In this high-speed co-sedimentation assay, fascin proteins bound to F-actin filaments (bundled and unbundled) are in the pellet after high-speed centrifugation (9, 23). From these actin-binding activity assays, we observed that wild-type fascin has a Kd of 0.15 μm for actin filaments (Fig. 3, A and B). This value is the same as previously reported (35). Under similar experimental conditions, fascin(F29A), fascin(R149A/K150A/R151A), and fascin(K358A) have Kd values of 1.5, 5.5, and 0.72 μm, respectively (Fig. 3, C–E). These data reflect a decrease in the actin binding constant by 10-, 37-, or 4.8-fold by fascin(F29A), fascin(R149A/K150A/R151A), and fascin(K358A) mutants, respectively. Therefore, in addition to their defects in actin bundling, these fascin mutants have impaired ability in actin binding.
FIGURE 3.
Actin-binding activity assay for fascin and some fascin mutants. A and B, actin-binding assay for wild-type fascin. G-actin (10 μm) and different concentrations of fascin proteins were used. After ultracentrifugation, fascin and actin proteins in the supernatant (S) and pellet (P) were separated by SDS-PAGE (A) and analyzed with the plot of bound fascin versus free fascin. C, actin with mutant fascin(F29A) proteins. D, actin with mutant fascin(R149A/K150A/R151A) proteins. E, actin with mutant fascin(K358A) proteins. One representative experiment of three is shown.
Cryo-electron Tomography of Reconstituted Bundles of Fascin and F-actin
Next we used cryo-electron tomography to analyze the in vitro reconstituted unconstrained three-dimensional bundles formed by fascin and actin filaments. Fascin/actin bundles consisted of a hexagonal lattice of parallel actin filaments, where fascin formed the cross-bridges (Fig. 4, A–E). The distance between actin filaments within the bundle was 11–12 nm (Fig. 4F). The interval between the cross-bridges was ∼35 nm (Fig. 4G). These values are similar to the ones observed for fascin/actin bundles isolated from native tissues (36, 37). Within the hexagonal packing, fascin interacts with two actin filaments (Fig. 4H). It is not clear whether fascin uses the same two actin-binding sites or different combinations from the potential three actin-binding sites to form these fascin/actin bundles. A higher-resolution structure is needed to answer these questions.
FIGURE 4.
Cryo-electron tomography of in vitro reconstituted F-actin and fascin bundles. A, a 4.4-Å thick slice (XY plane) from a tomogram containing a bundle of filaments. B, projection along the Z axis of the same tomogram. The total thickness of this tomogram was 110 nm. The entire tomogram was used for the Z-projection. C, three-dimensional rendering of the bundle where filaments and periodic connections between them are visible. D, zoom in and slightly rotated view of the area inside the dashed square in C. E, orthogonal view with respect to the one in D, showing actin filaments arranged in a hexagonal packing. Scale bar in all the images is 30 nm. F, histogram of inter-filament distances. The average is 11.2 ± 0.9 nm (mean ± S.D.; number of measurements = 308). G, histogram of distances between cross-bridges. The average is 33.3 ± 5.1 nm (mean ± S.D.; number of measurements = 166). H, surface rendering front view of the average electronic density obtained by subtomogram averaging. 147 boxes containing filaments and cross-bridges were manually picked, extracted from the tomograms, and iteratively aligned and averaged. Up to 10 actin filaments with several cross-bridges between them can be distinguished clearly. The hexagonal packing of the filaments (red dots) is also visible (indicated by dashed lines). Scale bar is 30 nm.
X-ray Crystal Structures of the Inactive State of Fascin
To understand the structural basis for the function of fascin in actin bundling, we have solved the x-ray crystal structures of four fascin mutants that are defective in actin-bundling activity. Phosphorylation of Ser39 inhibits the actin-bundling activity (25, 35). The phosphomimetic mutant fascin(S39D) has abolished the actin-bundling activity (Fig. 2 and Table 1). Therefore, the structure of fascin(S39D) should represent an “inactive” or “low active” conformation. We have solved the fascin(S39D) crystal structure at a resolution of 2.2 Å (Fig. 5 and supplemental Table S1 and Fig. S2A). There are two fascin(S39D) molecules in one asymmetric crystallization unit and these two molecules have similar conformations. When compared with the structure of wild-type fascin that we previously solved at 1.8-Å resolution (PDB code 3LLP) under the same experimental condition (23), fascin(S39D) has a similar conformation as one (defined as Chain B) of the two wild-type fascin molecules in the asymmetric crystallization unit, and shows some structural changes from the other molecule (defined as Chain A), especially in the β-trefoil fold 2 (supplemental Fig. S2, B and C). The crystal structures of wild-type fascin have also been solved by two other laboratories under different conditions (PDB codes 1DFC and 3P53) (22, 24). Comparisons of these structures show that the two molecules in an asymmetric unit of PDB structure code 1DFC are similar to each other and to Chain A of our PDB structure 3LLP (supplemental Fig. S3A). The two molecules in an asymmetric unit of PDB structure 3P53 are similar to each other and to Chain B of our PDB 3LLP, and to the structure of fascin(S39D) (supplemental Fig. S3B). These structural studies suggest that the structure of fascin is dynamic, and different conformations exist and are influenced by residue mutations or the surrounding environment. For clear and understandable descriptions of these structural conformations, here we refer to the conformation of fascin(S39D) (and Chain B from 3LLP and the two molecules from 3P53) as the inactive or low active conformation, the conformation of Chain A of wild-type fascin in 3LLP and the two molecules in 1DFC as the “active“ or “high active” conformation.
FIGURE 5.
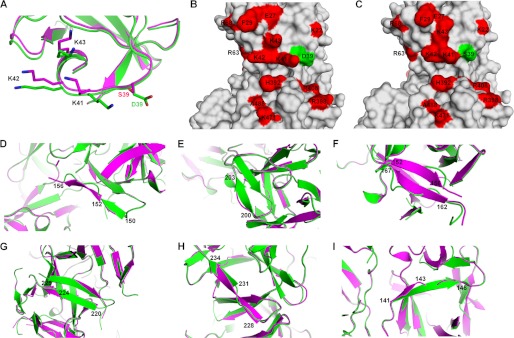
Conformational changes in four mutant fascin proteins. A–I, comparison of the structures of wild-type fascin (Chain A of PDB code 3LLP, in magenta) and fascin(S39D) (Chain A, in green). A, ribbon diagram of the region around Ser39. B and C, surface representations of fascin(S39D) (B) and wild-type fascin (C) in the area of Ser39. Critical residues involved in actin bundling are labeled in red. Ser39 and Asp39 are labeled in green. D, ribbon diagram of the area of residues 150–156 showing the conformational changes in this area between fascin(S39D) and the wild-type fascin. E, conformational changes in the area of residues 200–203. F, comparison of the regions of residues 152 to 162 of fascin(S39D) and wild-type fascin. G, conformational changes of residues 220–225. H, structures of regions of residues 228–234. I, structural differences in the region of residues 141–146 between fascin(S39D) and wild-type fascin.
Comparisons of the inactive and active conformations of fascin reveal the following structural changes. Most of the major conformational changes are in β-trefoil fold 2 around the actin-binding site 2 (Fig. 5). Residues Lys150–Tyr152 are forming a loop followed by a β-strand from Ala153 to Ser156 in the active conformation, whereas in the inactive conformation, it is switched to a β-strand from Lys150 to Tyr152 followed by a loop from Ala153 to Ser156 (Fig. 5D). The region of Gly200–Val203 is a β-strand in the inactive conformation, rather than a loop in the active conformation (Fig. 5E). The region of Glu162–Arg167 is a loop in the inactive conformation, rather than a β-strand in the active conformation (Fig. 5F). The inactive conformation has shorter β-strands in the Lys220–Arg224 and Leu231–Ser234 (Fig. 5, G and H) regions, and a longer β-strand in the Gln141–Ser146 (Fig. 5I) region, than the active conformation. The β-strand Gln141–Ser146 links actin-binding site 1 to actin-binding site 2. Although it is not clear which of these conformational changes are responsible for the decreased activity of fascin, the localization of these changes in β-trefoil fold 2 around actin-binding site 2 indicates an important role for this region in modulating the activity of fascin.
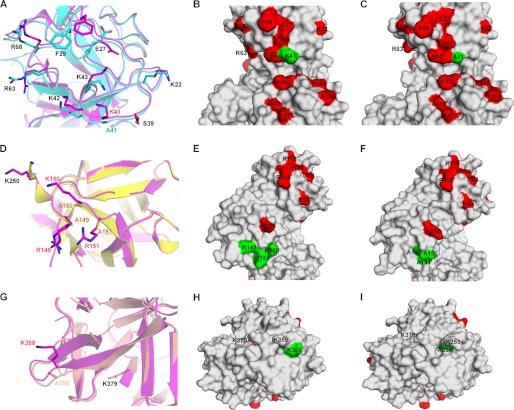
We selected fascin(K41A), fascin(R149A/K150A/R151A), and fascin(K358A) as representatives from actin-binding sites 1, 2, and the potential site 3, respectively. Strikingly, the crystal structures of all these three fascin mutants show the inactive conformation (supplemental Fig. S4). As in fascin(S39D), the major structural changes were in β-trefoil fold 2 around actin-binding site 2, even though the mutations were in different β-trefoil folds (supplemental Fig. S4). These data suggest that the mutations induce allosteric effects to stabilize the inactive conformation. In addition to the above conformational changes in β-trefoil fold 2, these individual mutations had effects on their local environment. In fascin(S39D), the introduction of a negative charged residue changed the orientation of the side chain of the positively charged residue Lys41, which is critical for actin bundling (Fig. 5, A–C, and Table 1). In fascin(K41A), the side chain positions of several nearby critical residues including Glu27, Phe29, Lys43, and Arg63 were changed (Fig. 6, A–C). In fascin(R149A/K150A/R151A), the β-sheet strand from 150 to 156 was longer and the side chain of the critical Lys250 was rotated away from actin-binding pocket 2, compared to wild-type fascin (Chain A in 3LLP) (Fig. 6, D–F). In fascin(K358A), mutation of Lys358 to Ala caused this region to become more disordered and the loop from Ser357 to Leu363 was longer than that in the active form of fascin (Fig. 6, G–I). These data provide a structural basis for the defects in actin bundling by these fascin mutations.
FIGURE 6.
Structural comparisons of mutant and wild-type fascin proteins. A–C, ribbon diagrams (A) or surface representations (B and C) of the structures of the region around Lys41 for fascin(K41A) (Chain A) (A in cyan, and C) and wild-type fascin (Chain A from PDB code 3LLP, A in magenta, and B). D–F, ribbon diagrams (D) or surface representations (E and F) of the structures of the region around Arg149–Arg151 for fascin(R149A/K150A/R151A) (Chain A) (D in yellow, and F) and wild-type fascin (D in magenta, and E). G–I, ribbon diagrams (G) or surface representations (H and I) of the structures of the region around Lys358 for fascin(K358A) (Chain A) (G in wheat, and I) and wild-type fascin (Chain A from PDB code 3LLP, G in magenta, and H).
Functional Effect of Fascin Mutations in Actin-binding Sites on Filopodial Formation
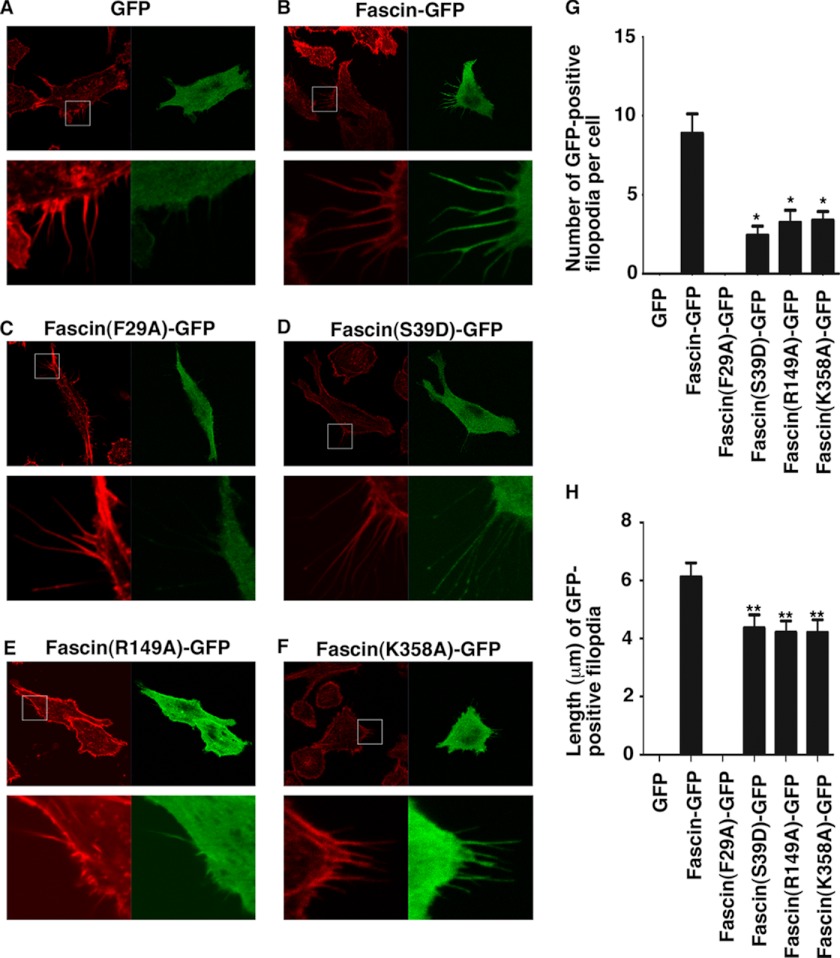
Finally we investigated the biological effect of fascin mutations on filopodial formation. GFP-tagged fascin mutants were transfected into 4T1 breast tumor cells. Filopodia containing GFP-fascin were quantified in terms of numbers of filopodia and the length of filopodia (Fig. 7). Although GFP alone did not produce any GFP-containing filopodia, GFP-fascin induced the formation of GFP-fascin-containing filopodia (Fig. 7, A, B, G, and H). Fascin mutants F29A (representing the actin-binding site 1), R149A (representing the actin-binding site 2), and K358A (representing the potential actin-binding site 3) induced fewer and shorter filopodia compared with GFP-fascin (Fig. 7, C and E–H). These data demonstrate the requirement of the identified actin-binding sites for fascin function in filopodial formation in cells. Furthermore, the enhanced filopodial tip localization of fascin(S39D) has been previously observed with the fascin(S39E) mutant (38) (Fig. 7D), indicating that, although PKC phosphorylation of Ser39 decreases its actin-bundling activity, this phosphorylation may increase the association of fascin with a protein(s) in the tip complex. Moreover, fascin(F29A) has an additional defect of localization in filopodia and no fascin(F29A)-GFP signal was observed in any filopodia (Fig. 7C). This may imply that Phe29 is involved in the translocation of fascin to filopodia. Previously Rab35 has been reported to directly interact with fascin and this interaction might be involved in the translocation/trafficking of fascin (39). Together these cellular studies confirm the biological function in filopodial formation of actin-binding sites identified from our biochemical and structural studies.
FIGURE 7.
Effect of fascin mutations on filopodial formation. 4T1 breast tumor cells were transfected with GFP (A), fascin-GFP (B), fascin(F29A)-GFP (C), fascin(S39D)-GFP (D), fascin(R149A)-GFP (E), or fascin(K358A)-GFP (F) plasmids. After staining with phalloidin (red color), the number of GFP-positive filopodia in each cell (G) and the length of GFP-positive filopodia (H) were tabulated for 20–49 cells for each transfection. In A–F, the bottom panels were the enlarged images of the boxed area in the top panels. Data in G and H were shown in mean ± S.D. *, p < 0.0001. **, p < 0.01.
DISCUSSION
Our mutational studies, cryo-electron tomography-based reconstructions of the fascin/actin bundles, and x-ray crystal structural studies of fascin mutants provide the molecular basis for the unique structural and mechanical properties of fascin. We have identified two major actin-binding sites on fascin. All four β-trefoil folds of fascin contribute to these actin-binding sites. A point mutation in Drosophila fascin (the singed mutant) in Gly409 (to Glu) (equivalent to Gly393 in human fascin 1, next to the critical His392 in actin-binding site 1) reduced fascin activity in vivo, implying that this actin-binding site 1 is functionally critical (40). Furthermore, a point mutation of Arg109 to His in fascin-2 (corresponding to Arg110 in fascin-1 in the actin-binding site 2) led to early-onset hearing loss in mice (fascin-2 is the main actin-bundling protein in hair-cell stereocilia) (41). Also, fascin(K22E/K43E/R398E) and fascin(R271E/K353E/K358E) mutants had been shown to be defective in actin bundling (24). These mutated residues are within our identified actin-binding site 1 and the potential site 3. Moreover, all identified actin-binding sites are positively charged, consistent with the surface of the actin filaments as negatively charged. Structures of the fascin-actin complex would be helpful in determining the conformation of fascin within the actin/fascin bundles.
Our data also suggest that fascin has dynamic conformation and exists in equilibrium among different configurations including inactive and active configurations. Because wild-type fascin has been crystallized in inactive and active conformations under different experimental conditions, it is likely that individual fascin proteins are in constant motion, sampling active, inactive, and possibly other conformations. Protein kinase C phosphorylation of Ser39 and mutations of residues in the actin-binding sites stabilize the inactive conformation. Furthermore, our structural data provide a mechanistic explanation for the cooperativity among the actin-binding sites. It has been shown that the actin-binding activity of fascin is cooperative (35). Our data reveal that fascin mutants in all actin-binding sites display the same inactive configuration. This indicates that any impairment in one actin-binding site leads to a concerted conformational change, thus disabling the other actin-binding sites. The actin-binding sites are conformationally interconnected. Hence, it is likely that binding of one F-actin filament at one actin-binding site stabilizes fascin at the active configuration and enhances the binding of actin filaments at the other actin-binding sites. The relative populations of the inactive and active states of fascin may be modulated by fascin interaction with actin polymers.
Human fascin expression is low or absent in normal adult epithelial cells, but is elevated in all forms of human carcinoma studied to date during the epithelial-mesenchymal transitions (19, 42). Overexpression of fascin confers invasive properties on tumor cells, contributes to a more aggressive clinical course of cancer, and is a significant independent prognostic indicator of poor outcome (6, 19). Hence, fascin could be a new molecular target for therapeutic intervention. Although there are several different actin cross-linker proteins, fascin is specifically overexpressed in many types of metastatic tumor cells including breast, prostate, ovarian, lung, liver, gastric, pancreatic, and brain cancers (17, 18, 43, 44). Fascin is also one of the gene signatures that correlate with breast cancer metastasis to the lung (45). Given the essential role of fascin in tumor metastasis, inhibitors of the interaction between fascin and actin filaments, and small molecules that stabilize the inactive conformation of fascin could be explored to prevent tumor metastasis.
Acknowledgments
We thank members of our laboratory for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA136837.
The atomic coordinates and structure factors (code 4GOV, 4GOY, 4GP0) have been deposited in the Protein Data Bank (http://wwpdb.org/).

This article contains supplemental Table S1 and Figs. S1–S4.
- PDB
- Protein Data Bank.
REFERENCES
- 1. Hall A. (2009) The cytoskeleton and cancer. Cancer Metastasis Rev. 28, 5–14 [DOI] [PubMed] [Google Scholar]
- 2. Swaney K. F., Huang C. H., Devreotes P. N. (2010) Eukaryotic chemotaxis. A network of signaling pathways controls motility, directional sensing, and polarity. Annu. Rev. Biophys. 39, 265–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pollard T. D., Cooper J. A. (2009) Actin, a central player in cell shape and movement. Science 326, 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mattila P. K., Lappalainen P. (2008) Filopodia. Molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9, 446–454 [DOI] [PubMed] [Google Scholar]
- 5. Mogilner A., Rubinstein B. (2005) The physics of filopodial protrusion. Biophys. J. 89, 782–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Machesky L. M., Li A. (2010) Fascin. Invasive filopodia promoting metastasis. Commun. Integr Biol. 3, 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Otto J. J., Kane R. E., Bryan J. (1979) Formation of filopodia in coelomocytes. Localization of fascin, a 58,000 dalton actin cross-linking protein. Cell 17, 285–293 [DOI] [PubMed] [Google Scholar]
- 8. Bryan J., Kane R. E. (1978) Separation and interaction of the major components of sea urchin actin gel. J. Mol. Biol. 125, 207–224 [DOI] [PubMed] [Google Scholar]
- 9. Yamashiro-Matsumura S., Matsumura F. (1985) Purification and characterization of an F-actin-bundling 55-kilodalton protein from HeLa cells. J. Biol. Chem. 260, 5087–5097 [PubMed] [Google Scholar]
- 10. Vignjevic D., Yarar D., Welch M. D., Peloquin J., Svitkina T., Borisy G. G. (2003) Formation of filopodia-like bundles in vitro from a dendritic network. J. Cell Biol. 160, 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vignjevic D., Kojima S., Aratyn Y., Danciu O., Svitkina T., Borisy G. G. (2006) Role of fascin in filopodial protrusion. J. Cell Biol. 174, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adams J. C. (2004) Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 16, 590–596 [DOI] [PubMed] [Google Scholar]
- 13. Tilney L. G., Connelly P. S., Vranich K. A., Shaw M. K., Guild G. M. (1998) Why are two different cross-linkers necessary for actin bundle formation in vivo and what does each cross-link contribute? J. Cell Biol. 143, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Claessens M. M., Bathe M., Frey E., Bausch A. R. (2006) Actin-binding proteins sensitively mediate F-actin bundle stiffness. Nat. Mater 5, 748–753 [DOI] [PubMed] [Google Scholar]
- 15. Kureishy N., Sapountzi V., Prag S., Anilkumar N., Adams J. C. (2002) Fascins, and their roles in cell structure and function. Bioessays. 24, 350–361 [DOI] [PubMed] [Google Scholar]
- 16. Maitra A., Iacobuzio-Donahue C., Rahman A., Sohn T. A., Argani P., Meyer R., Yeo C. J., Cameron J. L., Goggins M., Kern S. E., Ashfaq R., Hruban R. H., Wilentz R. E. (2002) Immunohistochemical validation of a novel epithelial and a novel stromal marker of pancreatic ductal adenocarcinoma identified by global expression microarrays. Sea urchin fascin homolog and heat shock protein 47. Am. J. Clin. Pathol. 118, 52–59 [DOI] [PubMed] [Google Scholar]
- 17. Pelosi G., Pasini F., Fraggetta F., Pastorino U., Iannucci A., Maisonneuve P., Arrigoni G., De Manzoni G., Bresaola E., Viale G. (2003) Independent value of fascin immunoreactivity for predicting lymph node metastases in typical and atypical pulmonary carcinoids. Lung Cancer 42, 203–213 [DOI] [PubMed] [Google Scholar]
- 18. Hashimoto Y., Shimada Y., Kawamura J., Yamasaki S., Imamura M. (2004) The prognostic relevance of fascin expression in human gastric carcinoma. Oncology 67, 262–270 [DOI] [PubMed] [Google Scholar]
- 19. Hashimoto Y., Skacel M., Adams J. C. (2005) Roles of fascin in human carcinoma motility and signaling. Prospects for a novel biomarker? Int. J. Biochem. Cell Biol. 37, 1787–1804 [DOI] [PubMed] [Google Scholar]
- 20. Yoder B. J., Tso E., Skacel M., Pettay J., Tarr S., Budd T., Tubbs R. R., Adams J. C., Hicks D. G. (2005) The expression of fascin, an actin-bundling motility protein, correlates with hormone receptor-negative breast cancer and a more aggressive clinical course. Clin. Cancer Res. 11, 186–192 [PubMed] [Google Scholar]
- 21. Zigeuner R., Droschl N., Tauber V., Rehak P., Langner C. (2006) Biologic significance of fascin expression in clear cell renal cell carcinoma. Systematic analysis of primary and metastatic tumor tissues using a tissue microarray technique. Urology 68, 518–522 [DOI] [PubMed] [Google Scholar]
- 22. Sedeh R. S., Fedorov A. A., Fedorov E. V., Ono S., Matsumura F., Almo S. C., Bathe M. (2010) Structure, evolutionary conservation, and conformational dynamics of Homo sapiens fascin-1, an F-actin cross-linking protein. J. Mol. Biol. 400, 589–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen L., Yang S., Jakoncic J., Zhang J. J., Huang X. Y. (2010) Migrastatin analogues target fascin to block tumour metastasis. Nature 464, 1062–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jansen S., Collins A., Yang C., Rebowski G., Svitkina T., Dominguez R. (2011) Mechanism of actin filament bundling by fascin. J. Biol. Chem. 286, 30087–30096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ono S., Yamakita Y., Yamashiro S., Matsudaira P. T., Gnarra J. R., Obinata T., Matsumura F. (1997) Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J. Biol. Chem. 272, 2527–2533 [DOI] [PubMed] [Google Scholar]
- 26. Mastronarde D. N. (2005) Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 [DOI] [PubMed] [Google Scholar]
- 27. Nickell S., Förster F., Linaroudis A., Net W. D., Beck F., Hegerl R., Baumeister W., Plitzko J. M. (2005) TOM software toolbox. Acquisition and analysis for electron tomography. J. Struct. Biol. 149, 227–234 [DOI] [PubMed] [Google Scholar]
- 28. Förster F., Medalia O., Zauberman N., Baumeister W., Fass D. (2005) Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl. Acad. Sci. U.S.A. 102, 4729–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Painter J., Merritt E. A. (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 33. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) MOLPROBITY. Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee K., Gallop J. L., Rambani K., Kirschner M. W. (2010) Self-assembly of filopodia-like structures on supported lipid bilayers. Science 329, 1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamakita Y., Ono S., Matsumura F., Yamashiro S. (1996) Phosphorylation of human fascin inhibits its actin binding and bundling activities. J. Biol. Chem. 271, 12632–12638 [DOI] [PubMed] [Google Scholar]
- 36. DeRosier D., Mandelkow E., Silliman A. (1977) Structure of actin-containing filaments from two types of non-muscle cells. J. Mol. Biol. 113, 679–695 [DOI] [PubMed] [Google Scholar]
- 37. Spudich J. A., Amos L. A. (1979) Structure of actin filament bundles from microvilli of sea urchin eggs. J. Mol. Biol. 129, 319–331 [DOI] [PubMed] [Google Scholar]
- 38. Aratyn Y. S., Schaus T. E., Taylor E. W., Borisy G. G. (2007) Intrinsic dynamic behavior of fascin in filopodia. Mol. Biol. Cell 18, 3928–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang J., Fonovic M., Suyama K., Bogyo M., Scott M. P. (2009) Rab35 controls actin bundling by recruiting fascin as an effector protein. Science 325, 1250–1254 [DOI] [PubMed] [Google Scholar]
- 40. Cant K., Cooley L. (1996) Single amino acid mutations in Drosophila fascin disrupt actin bundling function in vivo. Genetics 143, 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shin J. B., Longo-Guess C. M., Gagnon L. H., Saylor K. W., Dumont R. A., Spinelli K. J., Pagana J. M., Wilmarth P. A., David L. L., Gillespie P. G., Johnson K. R. (2010) The R109H variant of fascin-2, a developmentally regulated actin crosslinker in hair-cell stereocilia, underlies early-onset hearing loss of DBA/2J mice. J. Neurosci. 30, 9683–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grothey A., Hashizume R., Sahin A. A., McCrea P. D. (2000) Fascin, an actin-bundling protein associated with cell motility, is upregulated in hormone receptor negative breast cancer. Br. J. Cancer 83, 870–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cao D., Ji H., Ronnett B. M. (2005) Expression of mesothelin, fascin, and prostate stem cell antigen in primary ovarian mucinous tumors and their utility in differentiating primary ovarian mucinous tumors from metastatic pancreatic mucinous carcinomas in the ovary. Int. J. Gynecol. Pathol. 24, 67–72 [PubMed] [Google Scholar]
- 44. Rodríguez-Pinilla S. M., Sarrió D., Honrado E., Hardisson D., Calero F., Benitez J., Palacios J. (2006) Prognostic significance of basal-like phenotype and fascin expression in node-negative invasive breast carcinomas. Clin. Cancer Res. 12, 1533–1539 [DOI] [PubMed] [Google Scholar]
- 45. Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., Massagué J. (2005) Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]