Background: Cell surface nucleolin (NCL) is a promising target for development of anticancer agents.
Results: A novel pathway that includes ανβ3 integrin and leads to cell surface NCL localization and cell migration has been identified.
Conclusion: ανβ3 can be used as a biomarker for the use of NCL antagonists.
Significance: This pathway is active in endothelial and glioma cells, as well as in human glioblastomas.
Keywords: Angiogenesis, Cancer Biology, Cell Migration, Endothelial Cell, Integrins, Signal Transduction, Tyrosine-protein Kinase (Tyrosine Kinase), Tyrosine-protein Phosphatase (Tyrosine Phosphatase), Pleiotrophin
Abstract
The multifunctional protein nucleolin (NCL) is overexpressed on the surface of activated endothelial and tumor cells and mediates the stimulatory actions of several angiogenic growth factors, such as pleiotrophin (PTN). Because αvβ3 integrin is also required for PTN-induced cell migration, the aim of the present work was to study the interplay between NCL and αvβ3 by using biochemical, immunofluorescence, and proximity ligation assays in cells with genetically altered expression of the studied molecules. Interestingly, cell surface NCL localization was detected only in cells expressing αvβ3 and depended on the phosphorylation of β3 at Tyr773 through receptor protein-tyrosine phosphatase β/ζ (RPTPβ/ζ) and c-Src activation. Downstream of αvβ3, PI3K activity mediated this phenomenon and cell surface NCL was found to interact with both αvβ3 and RPTPβ/ζ. Positive correlation of cell surface NCL and αvβ3 expression was also observed in human glioblastoma tissue arrays, and inhibition of cell migration by cell surface NCL antagonists was observed only in cells expressing αvβ3. Collectively, these data suggest that both expression and β3 integrin phosphorylation at Tyr773 determine the cell surface localization of NCL downstream of the RPTPβ/ζ/c-Src signaling cascade and can be used as a biomarker for the use of cell surface NCL antagonists as anticancer agents.
Introduction
Nucleolin (NCL)5 is a multifunctional protein with well characterized roles in the organization of nucleolar chromatin, packaging of pre-rRNA, rDNA transcription, and ribosome assembly (1). It also acts as a shuttling protein between the cytoplasm and nucleus (2) and is overexpressed on the plasma membrane of cancer (3–5), as well as activated endothelial (6–8) cells. The importance of cell surface NCL has been suggested by studies showing that functional blockade or down-regulation of cell surface NCL in endothelial cells inhibits migration and capillary-tubule formation (3, 8) and causes endothelial cell apoptosis (7), whereas targeting NCL on the plasma membrane of cancer cells seems to be an effective way to inhibit cancer cell growth and angiogenesis in various in vitro and in vivo experimental models (3, 9, 10). The potential significance of targeting cell surface NCL is being proved by the fact that the guanosine-rich quadruplex-forming oligodeoxynucleotide AS1411 (11) and the NUCANT peptide (3, 12) that interact with surface NCL, are currently being tested in Phase I/II clinical trials. The signals, however, that mobilize NCL from the nucleus to the cell surface still remain unclear and not all tumor cells express cell surface NCL.
Cell surface NCL interacts with receptors associated with malignancies, such as ErbB1, facilitating their activation and leading to enhanced cell growth (4). Moreover, it binds a variety of ligands that play critical roles in tumorigenesis and angiogenesis, such as hepatocyte growth factor (13), endostatin (14), tumor homing peptide F3 (6), laminin (15), P-selectin (16), and midkine (17). We have recently shown that NCL interacts with the heparin-binding growth factor pleiotrophin (PTN) in both chorioallantoic membrane vessels and human endothelial cells. This interaction is taking place on the cell surface and down-regulation or blockade of cell surface NCL abolishes PTN-induced endothelial cell migration (18).
Integrins are cell surface heterodimeric receptors that mediate the physical and functional cell-cell or cell-matrix interactions. Among several integrins, αvβ3 is the most abundant and influential receptor regulating angiogenesis, and its tumor cell expression is correlated with disease progression in various tumor types (19). Growing evidence supports a central role for cooperative signaling between αvβ3 integrin and growth factor receptors, such as ErbB-2 (20), platelet-derived growth factor receptor β (21, 22), and vascular endothelial growth factor receptor 2 (21, 23), mediating tumor cell adhesion, migration, invasion, and survival, as well as endothelial cell activation (19). In the same line, we have previously shown interaction of αvβ3 with the PTN receptor protein-tyrosine phosphatase β/ζ (RPTPβ/ζ), which is required for PTN-induced endothelial cell migration (24).
The secreted heparin-binding growth factor PTN has established roles in cancer development, either directly by acting on cancer cells, or indirectly by affecting tumor angiogenesis (25). PTN binding to its receptor RPTPβ/ζ in endothelial cells leads to dephosphorylation and thus activation of c-Src (26), leading to Tyr773 phosphorylation of β3 integrin and PTN-induced endothelial cell migration (24). Because both αvβ3 (24) and NCL (18) are required for PTN-induced cell migration, the aim of the present study was to investigate interplay between NCL and αvβ3 that regulates human endothelial and glioma cell migration. Our data show that αvβ3 expression and β3 phosphorylation at Tyr773, as a result of the RPTPβ/ζ/c-Src signaling cascade, induces cell surface localization of NCL through phosphoinositide 3-kinase (PI3K), shedding light into the mechanisms that underlie NCL overexpression on the surface of activated endothelial and cancer cells. Moreover, evidence is presented that αvβ3 integrin may be a useful biomarker to be used for identification of patients that could benefit from strategies targeting cell surface NCL.
EXPERIMENTAL PROCEDURES
Materials
Human recombinant PTN was from PeproTech, Inc. (Rocky Hill, NJ) or was prepared as previously described (27). Human recombinant vascular endothelial growth factor (VEGF) was expressed in Sf9 insect cells and purified by cation exchange and heparin-affinity chromatography, as previously reported (28). HB-19 and Nucant 6L pseudopeptides were from Polypeptide Laboratories (Strasbourg, France). Plasmids encoding human wild-type β3, β3Y773F, β3Y785F, β3Y773F/Y785F, and αν have been previously described (29).
Cell Culture
Human umbilical vein endothelial cells (HUVEC), human glioma M059K and U87MG cells, rat glioma C6 cells, and Chinese hamster ovary (CHO) cells (ATTC, CRL 9096 deficient in endogenous integrin β3) were cultured as previously described (24). Stable CHO cell clones expressing wild-type β3, β3Y773F, β3Y785F, or β3Y773F/Y785F were generated as previously described (29). Cell culture reagents were from BiochromKG (Seromed, Germany). All cultures were maintained at 37 °C, 5% CO2, and 100% humidity. When cells reached 70–80% confluence, they were serum starved for 16 h before performing migration assays, lysed for immunoprecipitation experiments or fixed for immunofluorescence studies.
RNA Interference
Cells were grown to 50% confluence in medium without antibiotics. Transfection was performed in serum-free medium for 4 h using annealed RNA for RPTPβ/ζ (26), NCL (18), αv and β3 (24). JetSI-ENDO (Polyplus Transfection, Illkirch, France) was used as transfection reagent. Cells were incubated for another 48 h in serum-containing medium and lysed, serum starved, or fixed before further experiments. Double-stranded negative control siRNA (Ambion, Austin, TX) was also used in all experiments.
Transient Transfection
M059K cells were transfected with pCDNA3.1 vector, wild-type β3, or wild-type αv constructs using jetPEI HUVEC (Polyplus Transfection). At 50% confluence, cells were incubated with DNA and jetPEI-HUVEC in an N/p = 5 ratio for 4 h at 37 °C, as previously described (24). The transfection medium was replaced by fresh serum-containing medium, and 24 h later, the cells were examined for NCL expression in different cell compartments by Western blot or immunofluorescence analyses.
Immunofluorescence
Cells were fixed with 3.7% paraformaldehyde in phosphate-buffered saline (PBS), pH 7.4, for 10 min and permeabilized with PBS containing 0.1% Triton. After being washed 3 times with PBS, the cells were blocked with PBS containing 3% bovine serum albumin (BSA) and 10% fetal bovine serum (FBS) for 1 h at room temperature. The cells were stained with primary antibodies against RPTPβ/ζ (1:250, BD Biosciences, San Diego, CA), αvβ3 (1:500, Merck Millipore, Billerica, MA), or/and NCL (1:1,000, Sigma). Nuclei were stained with Draq5 (Biostatus Limited, Leicestershire, UK). Fluorescent Alexa secondary antibodies (Molecular Probes, Carlsbad, CA) were used at the concentration of 1:500, and the cells were mounted with Mowiol 4-88 (Merck Millipore) and visualized at 21 °C with Leica SP5 (×63 objective with a numerical aperture of 1.4) confocal microscope.
Western Blot Analysis
Proteins were analyzed by SDS-PAGE and transferred to Immobilon P membranes. Blocking was performed by incubating the membranes with Tris-buffered saline (TBS), pH 7.4, with 0.05% Tween (TBS-T), containing 5% nonfat dry milk in all cases. Membranes were incubated with primary antibodies for 16 h at 4 °C under continuous agitation, washed 3 times with TBS-T, and incubated with secondary antibodies for 1 h at room temperature. Detection of immunoreactive bands was performed using the enhanced chemiluminescence (ECL) detection kit (Pierce Biotechnology, Rockford, IL). The protein levels that corresponded to the immunoreactive bands were quantified using the ImagePC image analysis software (Scion Corp., Frederick, MD).
Immunoprecipitation Assay
Cells were lysed with RIPA buffer, as previously described (24). Three mg of total protein were incubated with primary antibody for 16 h at 4 °C under continuous agitation. Protein A- and protein G-agarose beads (Merck Millipore) were added, samples were further incubated for 2 h at 4 °C, and beads with bound proteins were collected by centrifugation and washed twice with ice-cold PBS. Immunoprecipitated proteins were resuspended in 50 μl of SDS loading buffer and analyzed by Western blot.
Immunohistochemistry
Human Brain Cancer Tissue Array (USBiomax Inc., Rockville, MD) sections were deparaffinized with xylene (3 × 5-min incubations) followed by treatment with serial dilutions of ethanol (100, 100, 95, and 95%, 10 min each) and by two changes of ddH2O. Antigen unmasking was achieved by boiling the slides (95–99 °C) for 10 min in 10 mm sodium citrate, pH 6.0. Sections were rinsed three times with ddH2O, immersed in 3% H2O2 for 20 min, washed twice with ddH2O and once with TBST (TBS, 0.1% Tween 20), and blocked for 30 min with Image-iT FX signal enhancer (InVitrogen, Carlsbad, CA). Mouse anti-CD51/CD61 and rabbit anti-NCL antibodies (Abcam, Cambridge, MA) were diluted 1:50 and 1:100, respectively, in blocking solution (TBST, 5% normal goat serum, Cell Signaling Technology, Inc., Beverly, MA) and incubated with the sections overnight at 4 °C. Following incubation with the antibodies, sections were washed three times with TBST and incubated for 1 h at room temperature with the corresponding Alexa fluorescent secondary antibodies diluted in blocking solution (1 μg/ml). Sections were finally washed three times, 5 min each, with TBST and mounted with DAPI-containing Vectashield mounting medium (Vector Labs, Burlingame, CA). Images were captured with a Nikon 80i Upright Microscope equipped with a Nikon Digital Sight DS-Fi1 color camera, using the Metamorph image acquisition software. All images were captured and processed using identical settings. Co-localization analysis was performed using Intensity Correlation Analysis plug-in within WCIF ImageJ software (30). Results are expressed with the Mander's overlap coefficient (ranges between 1 and 0, with 1 being high and 0 being low co-localization) and Pearson's correlation coefficient (value close to 1 indicates reliable co-localization).
Biotinylation of Cell Membrane Proteins
HUVEC were washed twice with PBS and incubated with 0.5 mg/ml of NHS-d-biotin for 40 min at 4 °C, as previously described (24). Three mg of total proteins from the cell lysate were used for immunoprecipitation with primary antibody or 0.5 mg of streptavidin magnetic particles (Roche Applied Science) for 24 h at 4 °C. The immune complexes or the particles were subjected to SDS-PAGE and Western blot analysis for NCL.
Migration Assays
Migration assays were performed as described previously (24) in 24-well microchemotaxis chambers (Corning, Inc., Lowell, MA) using uncoated polycarbonate membranes with 8-μm pores. Serum-starved CHO cells were harvested and resuspended at a concentration of 105 cells/0.1 ml in serum-free medium containing 0.25% BSA. HUVEC, U87MG, M059K, and C6 cells grown in serum-containing medium were harvested and resuspended at a concentration of 5 × 104 cells/0.1 ml (except from HUVEC that were resuspended at a concentration of 105 cells/0.1 ml) in serum-containing medium. In all cases, the bottom chamber was filled with 0.6 ml of the corresponding medium and the tested substances. The upper chamber was loaded with 0.1 ml of medium containing the cells and incubated for 4 h at 37 °C. After completion of the incubation, the filters were fixed and stained with 0.33% toluidine blue solution. The cells that migrated through the filter were quantified by counting the entire area of each filter, using a grid and an Optech microscope at ×20 objective (Optech Microscope Services Ltd., Thame, UK).
PI3K p85 ELISA
The levels of total and phosphorylated PI3K p85 were quantified using Fast Activated Cell-based ELISA (Active Motif, Carlsbad, CA). Briefly, cells were cultured in 96-well plates 1 day prior to manipulation. Serum-starved CHO stably transfected cells were treated with 100 ng/ml of PTN for 10 min, fixed, and incubated with anti-phospho and anti-total p85 antibodies, according to the manufacturer's instructions.
Subcellular Fractionation
Subcellular fractions of cells comprising cytosolic, nuclear, and cell membrane extracts were prepared as previously described (18). Cell monolayers in 100-mm plates were washed extensively with PBS before being scraped and pelleted. Washed cells (30 × 109) were then disrupted in a hypotonic solution (10 mm Hepes, pH 6.9, 10 mm KCl, 2 mm MgCl2, 0.1 mm PMSF) on ice. Nuclei were pelleted at 400 × g for 5 min and washed twice in PBS before extraction in the lysis buffer (10 mm Tris-HCl, pH 7.6, 400 mm NaCl, 1 mm EDTA, 0.1 mm PMSF, and 1% Triton X-100). This suspension was centrifuged at 12,000 × g for 10 min and the pellet was referred to as the nuclear fraction. The supernatant obtained after pelleting intact nuclei was further centrifuged at 14,000 × g for 30 min and the supernatant corresponding to the cytoplasmic fraction was recovered, whereas the pellet was resuspended in lysis buffer containing 150 instead of 400 mm NaCl. This latter suspension was recentrifuged at 14,000 × g for 30 min to separate the cytoskeletal (the pellet) and membrane (supernatant) fractions. Equivalent amounts of total protein from all fractions were immunoprecipitated for RPTPβ/ζ or αvβ3 and then analyzed by Western blot analysis for NCL.
Mass Spectrometry
Three mg of total protein from cell lysates were immunoprecipitated for αvβ3 and subjected to reduction by pre-treatment with dithiothreitol. After SDS-PAGE, proteins were visualized by silver staining. Relevant bands were excised and treated for in-gel digestion as described (31). Briefly, the silver was destained using Farmer's reagent and trypsin (porcine, modified, sequence grade, Promega, Madison, WI) was introduced to the dried gel pieces. After overnight tryptic digestion, the peptides were bound to a C18 μZipTip and after washing, they were eluted with acetonitrile containing the matrix (alfa-cyano 4-hydroxycinnamic acid) directly onto the target plate. The mass list was generated by MALDI-TOF mass spectrometry on an Ultraflex TOF/TOF from Bruker Daltonics. The search for identity was performed using the search engine ProFound by scanning the current version of the NCBI nr sequence database. The spectrum was internally calibrated using autolytic tryptic peptides, and the error was set at ±0.02 Da. One missed cleavage was allowed, and methionine could be oxidized. The significance of the identity was judged from the search engines scoring system. The occurrence of the few missed cuts was either on a terminal basic residue or surrounded by acidic amino acid residues.
In Situ Proximity Ligation Assay
For detection of protein-protein interactions, in situ proximity ligation assay (PLA) was performed. The components used (Olink Bioscience Uppsala, Sweden) were as follows: anti-mouse PLA plus probe, anti-rabbit PLA minus probe, and Detection Reagents Orange. HUVEC, M059K, and U87MG cells were grown on chamber slides (Ibidi® μ-Chamber 12 well on glass slides, Martinsried, Germany). After reaching 80% confluence, the assay was performed according to the manufacturer's instructions. Briefly, after fixation and blocking, the cells were incubated with the primary antibodies: mouse anti-PTN (1:500, Abnova, Heidelberg, Germany), rabbit anti-RPTPβ/ζ (1:250, Santa Cruz Biotechnology Inc., Santa Cruz, CA), rabbit anti-αv (1:500, Merck Millipore), goat anti-β3 (1:50, Merck Millipore), and mouse anti-NCL (1:50, Santa Cruz Biotechnology Inc.). Subsequently, the cells were incubated with secondary antibodies conjugated with oligonucleotides. After hybridization and ligation of the oligonucleotides, the DNA was amplified. A detection mixture detected the amplicons, resulting in red fluorescence signals. Nuclei were counterstained with Draq5, cells were mounted with Mowiol 4-88 and visualized at 21 °C with Leica SP5 confocal microscope.
Statistical Analysis
All experiments were performed at least 3 independent times. Where applicable, the significance of variability between the results from each group and the corresponding control was determined by unpaired t test or analysis of variance. All results are expressed as mean ± S.E. from at least 3 independent experiments.
RESULTS
αvβ3 Expression and β3 Tyr773 Phosphorylation Are Required for Cell Surface NCL Localization
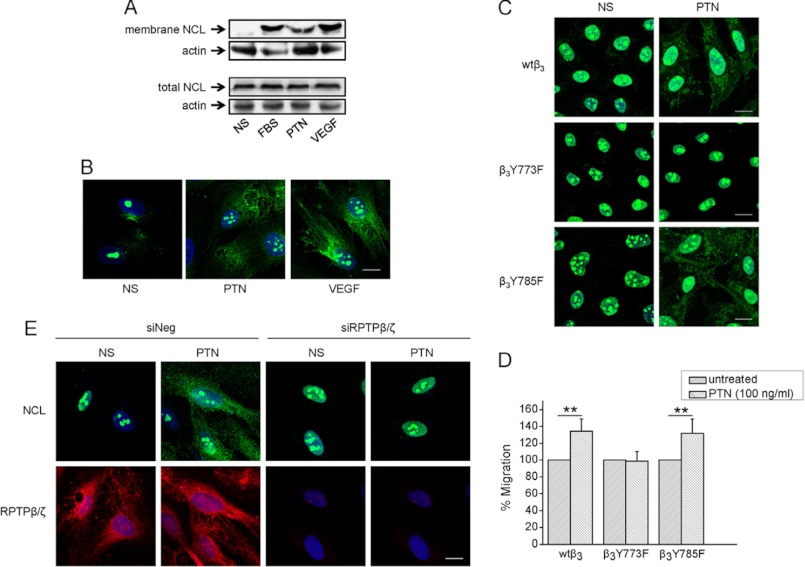
To examine the cell surface localization of NCL in different types of cells that differentially express αvβ3, fluorescence immunostaining of endothelial and glioma cells for NCL was performed. As shown in Fig. 1A, NCL was not detected in the extranuclear compartments of M059K and C6 cells that do not express ανβ3 (24), whereas it was localized on the plasma membrane of HUVEC (also supplemental Fig. S1) and U87MG cells that express αvβ3 (24). To investigate whether expression of αvβ3 is playing a role in cell surface localization of NCL, we down-regulated β3 expression in U87MG cells by using siRNA, or overexpressed β3 in M059K, as well as β3 in CHO cells that do not normally express it (Fig. 1, B and C), as previously described (24). Down-regulation of β3 but not αv in U87MG cells resulted in decreased extranuclear NCL, as evidenced by immunofluorescence microscopy (Fig. 1B) and biotin labeling of cell membrane proteins (Fig. 1D). In the same line, overexpression of wild-type β3 in M059K and CHO cells resulted in extranuclear localization of NCL, as also evidenced by immunofluorescence microscopy (Fig. 1B) and biotin labeling of cell membrane proteins (Fig. 1D).
FIGURE 1.
Expression of αvβ3 correlates with cell surface NCL localization. A, representative immunofluorescence images of M059K, C6, HUVEC, and U87MG cells cultured in serum-containing medium and stained for NCL (green) and nucleus (blue). B, representative immunofluorescence images for NCL (green) and αvβ3 (red) of U87MG cells treated with a negative control siRNA or siRNAs for β3 or αv; mock-transfected M059K glioma cells, M059K cells transiently transfected to overexpress wild-type β3 or αv; mock-transfected CHO cells, CHO cells stably transfected to overexpress wild-type β3 or mutant β3Y773F, β3Y785F, or β3Y773F/Y785F. C, cell lysates after treatment of U87MG cells with a negative control siRNA or siRNAs for β3 or αv, or after overexpression of wild-type β3 or αv in M059K cells, or different forms of β3 in CHO cells, were analyzed by Western blot for αv, β3, or actin (used as a loading control). D, for detection of cell surface NCL, intact cells were incubated with biotin, cell lysates were immunoprecipitated for magnetic streptavidin beads, and immunoprecipitates were analyzed by Western blot for NCL (membrane NCL). Whole cell protein extracts were analyzed by Western blot for NCL (total NCL) or actin, used as a loading control. siNeg, cells transfected with a negative control siRNA; siβ3, cells transfected with siRNA for β3; siαv, cells transfected with siRNA for αv; vector, cells negative for β3, transfected with the plasmid vector; wtβ3, cells overexpressing wild-type β3; β3Y773F/Y785F, cells overexpressing double mutant β3Y773F/Y785F; β3Y773F, cells overexpressing single mutant β3Y773F; β3Y785F, cells overexpressing single mutant β3Y785F; wtαv, cells overexpressing wild-type αv. Scale bars in all cases correspond to 10 μm.
To determine whether β3 Tyr773 or/and Tyr785 is responsible for the cell surface localization of NCL, we used CHO cells overexpressing β3Y773F, β3Y785F, or β3Y773F/Y785F instead of wild-type β3 and performed immunofluorescence assays. As shown in Fig. 1, B and D, NCL was localized in the extranuclear fraction of CHO cells overexpressing β3Y785F, similarly to CHO cells overexpressing wild-type β3, whereas in cells overexpressing β3Y773F or β3Y773F/Y785F it was restricted in the nucleus. In all cases, the total cellular amounts of NCL were not affected by the presence or absence of αvβ3 integrin (Fig. 1D).
PTN Induces Cell Surface NCL Localization through RPTPβ/ζ-mediated αvβ3 Activation
Because cell surface localization of NCL is induced by angiogenic growth factors, such as VEGF (8), we questioned whether PTN alters the intracellular trafficking of NCL. HUVEC displayed increased levels of cell surface NCL upon PTN stimulation, similarly to the known effect of serum (2) or VEGF (Fig. 2A). This increase was not due to increased protein levels of NCL (Fig. 2A) and was confirmed by immunofluorescence studies (Fig. 2B). Notably, PTN-induced cell surface NCL localization was not observed in CHO cells overexpressing β3Y773F (Fig. 2C). Accordingly, PTN induced migration of CHO cells overexpressing wild-type β3 or β3Y785F, but had no effect on CHO cells overexpressing β3Y773F (Fig. 2D), suggesting that cell surface localization of NCL through β3 Tyr773 phosphorylation relates to PTN-induced cell migration.
FIGURE 2.
PTN induces cell surface NCL localization through RPTPβ/ζ-mediated αvβ3 activation. A, serum-starved HUVEC were stimulated with PTN (100 ng/ml) or VEGF (10 ng/ml) or FBS (15%) for 5 h at 37 °C and then incubated with biotin, to label cell surface proteins, as described under “Experimental Procedures.” In the upper panel, cell lysates were immunoprecipitated with magnetic streptavidin beads and immunoprecipitates were analyzed by Western blot for NCL. In the lower panel, total cell lysates were analyzed by Western blot for NCL. Actin was used in both cases as a loading control. B, representative immunofluorescence images stained for NCL (green) and nucleus (blue) in serum-starved HUVEC treated with PTN or VEGF for 5 h at 37 °C. C, representative immunofluorescence images stained for NCL (green) and nucleus (blue) in serum-starved CHO cells expressing wild-type β3 (wtβ3), β3Y773F, and β3Y759F, treated with PTN for 5 h at 37 °C. D, serum-starved CHO cells were treated with human recombinant PTN (100 ng/ml) and cell migration assays were performed as described under “Experimental Procedures.” Data are the mean ± S.E. percentage change in number of migrating cells versus the corresponding untreated cells (default = 100). **, p < 0.01. E, down-regulation of RPTPβ/ζ expression by siRNA, significantly decreased PTN-induced cell surface localization of NCL. NS, nonstimulated cells; siNeg, cells transfected with a negative control siRNA; siRPTPβ/ζ, cells transfected with siRNA for RPTPβ/ζ. Scale bars in all cases correspond to 10 μm.
To test the hypothesis that PTN-induced cell surface NCL localization depends on β3 Tyr773 phosphorylation through RPTPβ/ζ, down-regulation of RPTPβ/ζ expression in HUVEC by siRNA was performed as previously described (26), and inhibited PTN-induced cell surface NCL localization (Fig. 2E), suggesting that RPTPβ/ζ-mediated β3 Tyr773 phosphorylation is involved.
c-Src and PI3K That Lie Up- and Downstream of αvβ3, Respectively, Are Required for PTN-induced Cell Surface NCL Localization
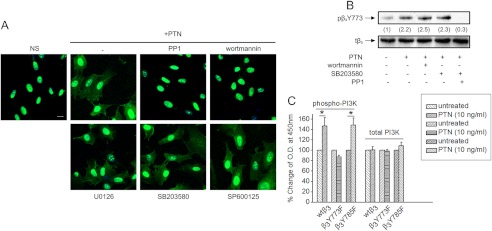
To further elucidate signaling molecules that affect PTN-induced cell surface NCL localization, we used inhibitors of several signaling molecules known to affect cell migration. As shown in Fig. 3A, inhibition of c-Src (also supplemental Fig. S2) and PI3K abolished PTN-induced cell surface localization of NCL, whereas inhibition of p38, JNK, and ERK½ had no effect. PTN is known to activate c-Src, PI3K, and ERK½ (26), but did not induce p38 or JNK activation in HUVEC (supplemental Fig. S3).
FIGURE 3.
Signaling molecules required for PTN-induced cell surface localization of NCL. A, representative immunofluorescence images stained for NCL (green) and nucleus (blue) in serum-starved HUVEC stimulated with human recombinant PTN (100 ng/ml) for 5 h at 37 °C, in the presence or absence of inhibitors for c-Src (PP1 10 μm), PI3K (wortmannin 100 nm), p38 (SB203580, 10 μm), JNK (SP600125 20 nm), and ERK½ (U0126 20 nm), respectively. Scale bar corresponds to 10 μm. B, serum-starved HUVEC pretreated for 15 min with PP1, wortmannin, and SB203580 were stimulated with PTN for 10 min, lysed, and analyzed by Western blot for phospho-β3 Tyr773 (pβ3Y773) and total β3 (tβ3) integrin. Numbers in parentheses denote the average-fold change of the ratio pβ3(Tyr773):tβ3 compared with nonstimulated cells (set as default 1) of at least three independent experiments. C, levels of phosphorylated PI3K were assayed by the Fast Activated Cell-based ELISA PI3 Kinase p85 kit. Data are the mean ± S.E. percentage change in phosphorylated PI3K versus the corresponding untreated cells (default = 100). *, p < 0.05.
To investigate whether PI3K is up- or downstream of αvβ3, the effect of PI3K inhibition in PTN-induced β3 Tyr773 phosphorylation was studied. The p38 inhibitor was used as a negative control and the c-Src inhibitor as a positive control, because c-Src is known to lay upstream of β3 Tyr773 phosphorylation (24). As shown in Fig. 3B, PI3K inhibition did not affect PTN-induced β3 Tyr773 phosphorylation, suggesting that PI3K lies downstream of αvβ3. This was confirmed by ELISA for activated PI3K, using CHO cells overexpressing wild-type β3, β3Y773F, or β3Y785F subunit. PTN significantly induced PI3K activation in CHO cells overexpressing wild-type β3 or β3Y785F, whereas it had no effect in cells overexpressing β3Y773F (Fig. 3C).
Cell Surface NCL Directly Interacts with αvβ3
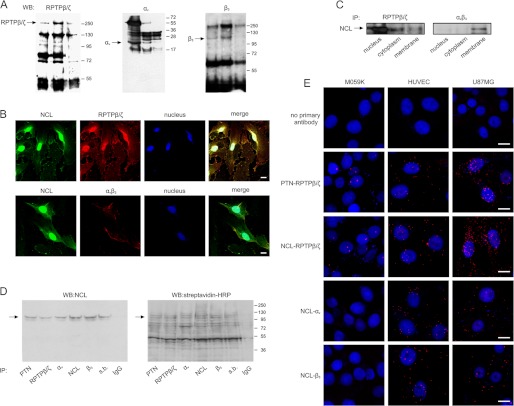
To study whether NCL directly associates with αvβ3, immunoprecipitation assays were performed in HUVEC lysates. NCL was found to interact with αvβ3 by mass spectrometry (Table 1) and Western blot (Fig. 4A) analysis. In the former, among the proteins co-immunoprecipitated with αvβ3, NCL (accession number AAA59954) was found to be a binding partner with a minimum sequence coverage of 19%. This relatively low sequence coverage can be explained by the particular structure of NCL, which is very rich in acidic (24.9%) and basic (16.3%) residues in highly basic regions, e.g. the C terminus (32), producing tryptic fragments that are too short for analysis by MALDI-TOF. Co-localization of αvβ3 with NCL on the cell surface was observed by immunofluorescence studies (Fig. 4B), whereas co-immunoprecipitation experiments using different cellular fractions (Fig. 4C) or cell lysates of biotinylated cells (Fig. 4D), as well as PLA assays (Fig. 4E), demonstrated direct formation of NCL-αvβ3 complexes in cells expressing αvβ3.
TABLE 1.
Identification of NCL (accession number AAA59954) by peptide mass fingerprint analysis (see text for details)
Monoisotopic uncharged masses are shown.
| Measured mass | Computed mass | Error | Residues start | Residues to | Missed cut | Peptide sequence |
|---|---|---|---|---|---|---|
| Da | ||||||
| 811.448 | 811.455 | −0.007 | 555 | 561 | 0 | LELQGPR |
| 939.502 | 939.506 | −0.004 | 430 | 437 | 0 | GIAYIEFK |
| 999.526 | 999.535 | −0.008 | 334 | 342 | 0 | NDLAVVDVR |
| 1159.572 | 1159.575 | −0.003 | 458 | 467 | 0 | SISLYYTGEK |
| 1560.671 | 1560.673 | −0.002 | 611 | 624 | 0 | GFGFVDFNSEEDAK |
| 1593.741 | 1593.734 | 0.007 | 524 | 537 | 0 | GYAFIEFASFEDAK |
| 1775.822 | 1775.825 | −0.003 | 348 | 362 | 1 | KFGYVDFESAEDLEK |
| 1990.981 | 1990.984 | −0.003 | 404 | 420 | 1 | VTQDELKEVFEDAAEIR |
| 2199.017 | 2199.017 | 0.000 | 578 | 597 | 1 | GLSEDTTEETLKESFDGSVR |
| 2345.120 | 2345.120 | 0.000 | 625 | 645 | 1 | EAMEDGEIDGNKVTLDWAKPK |
FIGURE 4.
Cell surface NCL directly interacts with αvβ3 and RPTPβ/ζ. A, HUVEC were lysed with RIPA buffer and immunoprecipitated for NCL, RPTPβ/ζ, αv, or β3 integrin subunit. Immunoprecipitates (IP) were analyzed by Western blot (WB) for the presence of RPTPβ/ζ, αv, or β3 integrin subunit as shown. Immunoprecipitation for IgG was used as a negative control in every case. B, representative immunofluorescence images showing colocalization (yellow) of NCL (green) and RPTPβ/ζ (red) or αvβ3 (red) on the surface of HUVEC. Nuclei are shown blue. C, total proteins from nuclear, cytosolic, and membrane fractions of U87MG cells were immunoprecipitated for RPTPβ/ζ or αvβ3 and analyzed by Western blot for NCL. D, intact U87MG cells were incubated with biotin and cell lysates were immunoprecipitated for PTN, RPTPβ/ζ, αv, β3, NCL, magnetic streptavidin beads (s.b.) or IgG. Immunoprecipitates were analyzed by Western blot for NCL (left). The same blot was stripped and biotinylated proteins were detected by incubation with peroxidase-conjugated streptavidin (right). The arrow shows the band that corresponds to NCL. E, in situ PLA signals were detected as red dots, indicating the direct formation of NCL-RPTPβ/ζ, NCL-αv, and NCL-β3 complexes in different cell types. The interaction of PTN with its receptor RPTPβ/ζ was used as a positive control. Omission of the primary antibodies was used as negative control. Scale bars in all cases correspond to 10 μm.
Interestingly, by using the same experimental approaches, NCL was also found to co-localize and co-immunoprecipitate with RPTPβ/ζ, not only on the cell membrane, but also in the cell cytoplasm and the nucleus in HUVEC and U87MG cells that express αvβ3 (Fig. 4, A–E). In M059K cells, NCL-RPTPβ/ζ complexes were limited and restricted in the cell nucleus (Fig. 4E).
Correlation between Cell Surface NCL Localization and αvβ3 Expression in Glioblastomas
To further confirm that αvβ3 expression is a prerequisite for cell surface NCL localization, we performed immunohistochemistry in tissue microarrays, containing samples from normal brains, grade 2 astrocytomas and grade 4 glioblastomas. As shown in Fig. 5, the localization pattern of both NCL and αvβ3 seems to correlate in all cases, although obvious diversity between samples can be observed. Intensity correlation analysis revealed increased co-localization in astrocytomas and the highest co-localization in glioblastomas compared with normal brains.
FIGURE 5.
Correlation between cell surface NCL localization and αvβ3 expression in glioblastomas. A, representative immunofluorescence images showing colocalization (yellow) of NCL (red) and αvβ3 (green) in different samples from a tissue microarray containing human normal brains, grade 2 astrocytomas, and grade 4 glioblastomas. Each line corresponds to tissue from a different patient of the corresponding grade. Nuclei are shown blue. Magnification in all images is ×20. Scale bars in all cases correspond to 100 μm. B, co-localization of ανβ3 and NCL analysis was performed using Intensity Correlation Analysis-WCIF ImageJ software. Results are expressed with the Mander's overlap coefficient (R) and Pearson's correlation coefficient (Rr), as described under “Experimental Procedures.”
Expression of αvβ3 Determines the Biological Effect of Cell Surface NCL Targeting Agents on Endothelial and Cancer Cell Migration
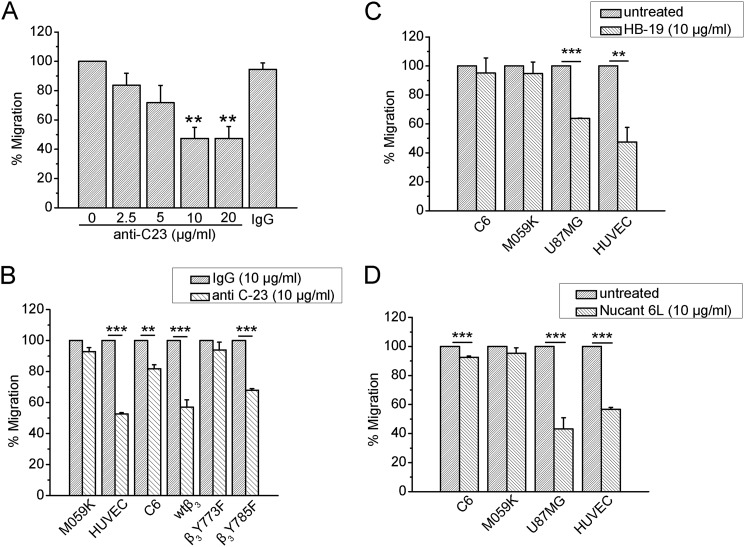
Based on the observation that NCL is preferentially localized on the surface of cells expressing αvβ3, we wanted to evaluate the efficacy of anti-cell surface NCL strategies in relationship to αvβ3 expression. Anti-C23 monoclonal antibody raised against NCL, which has been shown to dramatically inhibit angiogenic functions of VEGF stimulated HUVEC (3), as well as HB-19 and Nucant 6L pseudopeptides, which are known to target and down-regulate only surface NCL (3, 10), were tested for their effect on migration of cells differentially expressing αvβ3. As shown in Fig. 6A, the anti-C23 antibody caused a concentration-dependent inhibition of U87MG cell migration, which was maximal at 10 μg/ml. Anti-C23 antibody significantly inhibited migration of all tested cells expressing activated αvβ3, such as U87MG, HUVEC, CHO wtβ3, and CHO β3Y785F cells, whereas it did not affect or affected to a limited degree migration of M059K, C6, and CHO β3Y773F cells (Fig. 6B). Similarly, HB-19 and Nucant 6L pseudopeptides significantly inhibited migration of U87MG cells and HUVEC, whereas they had a minor effect on the migration of M059K and C6 cells (Fig. 6, C and D).
FIGURE 6.
Targeting of cell surface NCL is effective only in cells expressing αvβ3. A, U87MG cells were treated with various concentrations of the monoclonal anti-C23 antibody against NCL and with a high concentration (20 μg/ml) of human IgG. Different cell types that expressed or not αvβ3 were treated with anti-C23 antibody (10 μg/ml) (B), HB-19 pseudopeptide (10 μg/ml) (C), and Nucant 6L pseudopeptide (10 μg/ml) (D). Cell migration assays were performed in FBS-containing medium using microchemotaxis chambers as described under “Experimental Procedures.” Data are the mean ± S.E. percentage change in number of migrating cells versus the corresponding untreated cells (default = 100). Asterisks in A denote statistical significant differences from untreated cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
Extranuclear distribution of NCL has been observed in both activated endothelial (6) and cancer (3–5) cells. However, not all tumor cells express cell surface NCL and the mechanisms involved are still unclear. Molecules known to affect tumor growth or/and angiogenesis that have been reported to increase cell surface NCL localization are serum (2), VEGF (8), laminin-1 (33), and low density lipoprotein receptor-related protein 1 (10). In the present study it is shown that PTN also increases cell surface NCL localization, and that expression of αvβ3 and β3 Tyr773 phosphorylation are required for the redistribution of NCL from the nucleus to the cell membrane. VEGF has been shown to induce expression (34) and c-Src-mediated phosphorylation of β3 at Tyr773 (35), which based on our data, provide an explanation for the VEGF-stimulated NCL cell surface localization. Laminin-1-induced increase of NCL cell surface localization relies on an integrin-dependent cascade (33), supporting our notion that integrins regulate NCL cell surface localization. In the same line are also data showing that many of the molecules that have been described to bind and act through cell surface NCL are also described to act through integrins, and especially αvβ3. This is true for VEGF (34, 35), laminins (15, 36, 37), hepatocyte growth factor (13, 38), endostatin (14, 39), and PTN (18, 24).
Interestingly, β3 but not αv, appears to be the subunit responsible for NCL cell surface localization. Cell surface NCL is only observed in β3-overexpressing CHO and M059K cells, but not αv overexpressing cells. In the same line, down-regulation of β3 abolishes cell surface localization of NCL in U87MG cells. However, down-regulation of αv does not cause such an effect. The latter could be attributed to formation of a functional complex of β3 with its other partner subunit αIIb, which can be ectopically expressed in several tumor cell lines (40).
Cell surface NCL localization depends on the Tyr773 phosphorylation of β3 integrin subunit. PTN induces β3 Tyr773 phosphorylation through RPTPβ/ζ and c-Src activation (24). Down-regulation of RPTPβ/ζ or c-Src inhibition completely abolishes PTN-induced β3 Tyr773 phosphorylation (24) and cell surface localization of NCL (present study), suggesting that both RPTPβ/ζ and c-Src lie upstream of ανβ3. c-Src is also known to lie upstream of and be responsible for VEGF-induced αvβ3 phosphorylation (23), although possible involvement of RPTPβ/ζ is currently being investigated. Downstream of αvβ3 phosphorylation, NCL cell surface localization is mediated by PI3K (Fig. 7). The latter has important roles in cell migration (41) and the remodeling of actin filaments induced by growth factors (42) and integrins (43), and is involved in PTN-induced endothelial cell migration (26). Both the regulation of nucleocytoplasmic shuttling of NCL by phosphorylation (44) and the reported physical interaction between NCL and PI3K (16, 45), indicates a possible direct regulation of NCL by PI3K upon β3 Tyr773 phosphorylation. Alternatively, PI3K may drive NCL membrane shuttling through regulation of other targets that interact with NCL. For example, it has been shown that proteins containing pleckstrin homology domains are recruited in the membrane following PI3K activation in response to different growth factors (46). Although NCL does not possess related sequences, it has been proposed as a potential ligand for such proteins through its acidic motifs (47).
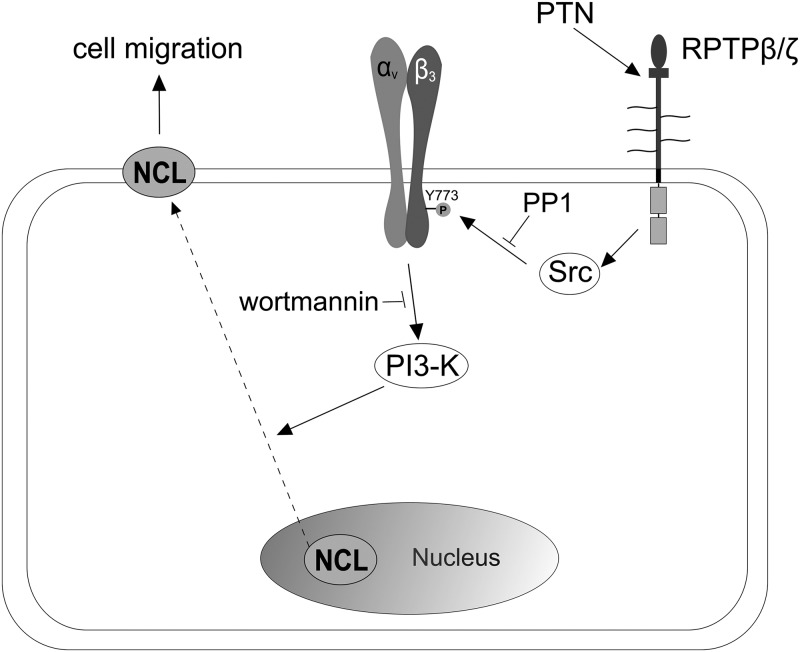
FIGURE 7.
Schematic representation of the proposed mechanism leading to αvβ3-mediated NCL cell surface localization upon PTN stimulation. Binding of exogenous PTN to its receptor RPTPβ/ζ on the surface of endothelial or cancer cells, leads to c-Src activation, Tyr773 phosphorylation of β3 integrin subunit, and PI3K phosphorylation. Activated PI3K induces, either directly or indirectly, the translocation of NCL from the nucleus to the cell membrane. Cell surface NCL interacts with both RPTPβ/ζ and αvβ3 and as previously shown (18), is required for PTN-induced cell migration.
It is well known that ERK½ is activated by αvβ3 and participates in integrin-mediated cell migration (48). The present study shows that ERK½ is not involved in PTN-induced cell surface NCL localization, although it is required for PTN-induced endothelial cell migration (26). One possible explanation is that ERK½ may lay downstream of αvβ3-mediated cell surface NCL localization. Although few data exist on the participation of cell surface NCL to activation of cell signaling pathway(s), it has been shown that cell surface NCL enhanced signaling through the mitogen-activated protein kinase (MAPK) pathway (49), in agreement with our data.
Interestingly, cell surface NCL directly interacts with αvβ3 and RPTPβ/ζ on the cell surface. This is in line with the emerging notion that cell surface NCL may participate in functional complexes with other receptors, such as urokinase plasminogen activator receptor (50) and ErbB receptors (4). Interaction of NCL with αvβ3 has been previously observed in MDA-MB-231 and HeLa cancer cells by bioconjugated semiconductor quantum dots; however, it was then discussed as a methodology artifact (51). In the present study, by using several different techniques, this interaction has been clearly demonstrated. One possibility for the functional significance of these interactions is activation of cell signaling pathways. Besides activation of MAPKs (49), plasma membrane NCL has been shown to increase cytoplasmic Ca2+ levels (52), or activate PI3K (53). On the other hand, accumulating evidence suggests that signaling events related to cell migration require ligand/receptor endocytosis (54). VEGF and VEGF receptor 2 internalization are required for endothelial recovery in a wound healing assay (55), whereas low platelet-derived growth factor (PDGF) concentrations induce cell migration by promoting clathrin-mediated endocytosis of PDGF receptor (56). Moreover, functions of integrins and their signaling leading to cell migration are strictly controlled by their endocytic trafficking (57). The well demonstrated role of NCL in the internalization of specific ligands and transport to the cell nucleus (6, 14, 17, 18, 58) prompt the speculation that NCL mediates the translocation of PTN or/and RPTPβ/ζ inside the cell, thus triggering or amplifying signaling events. This hypothesis is strengthened by the fact that both PTN (18) and RPTPβ/ζ (present study) co-localize with NCL in the cell nucleus, whereas down-regulation of NCL expression caused significant reduction in nuclear localization of PTN (18) and RPTPβ/ζ (supplemental Fig. S4). The functional significance of this internalization is currently under investigation.
The relevance of αvβ3-regulated cell surface NCL localization to human cancer was addressed by analyzing brain cancer tissue microarrays. Increased αvβ3 expression was observed in grade 4 glioblastomas compared with grade 2 astrocytomas and normal brain, in line with previous data showing that αvβ3 expression was significantly increased in high-grade glioblastomas compared with low-grade gliomas (59). Extranuclear NCL expression was also increased in grade 4 glioblastomas, and in all cases correlated with αvβ3, further strengthening our notion that αvβ3 is playing a role for NCL cell surface targeting.
It was recently shown that the phosphorylation of NCL by protein kinase C-ξ and casein kinase 2 mediates the surface translocation of NCL via regulating its interaction with heat sock cognate 70 (hsc70) (60). Although NCL phosphorylation (1, 53, 61) and glycosylation (52) have been mentioned to play a role in NCL cell surface localization, the mechanisms remain mostly unexplored. In the present study we did not look for NCL phosphorylation; however, we showed that β3 Tyr773 phosphorylation is required for NCL cell surface localization. αvβ3 integrin is found to form a complex with hsc70 in lipid raft microdomains of the cytoplasmatic membrane of susceptible intestinal epithelial cells, and both act as receptors for rotavirus entry (62). Therefore, it is tempting to speculate that αvβ3 and hsc70 may cooperate for cell surface NCL localization and in that case direct or indirect NCL phosphorylation by protein kinase C-ξ and casein kinase 2 may have a role.
Based on the observation that NCL is preferentially localized on the surface of cells that express αvβ3, we evaluated the efficacy of an anti-NCL antibody, as well as HB-19 and Nucant 6L pseudopeptides on cell migration, in relationship to αvβ3 expression, in the presence of serum that is known to increase levels of cell surface NCL (Ref. 2 and Fig. 2A). Several cell lines were tested and significant reduction of cell migration was observed only in cells that express αvβ3. This is in line with data showing that a pseudopeptide that is known to bind the C-terminal RGG domain of the cell surface-expressed NCL and block its function, similarly to HB-19 and Nucant 6L pseudopeptides, completely abolished PTN-induced HUVEC migration (18) and data showing that the surface NCL antagonists, HB-19 and related NUCANT pseudopeptides, exert different inhibitory effects in different tumor cell types. Although the reason for the latter observation has not been studied, this has been attributed to different levels of surface NCL or/and different NCL partners in tumor cells (10). Our data clearly show that levels of cell surface NCL are different among different types of cells and correlate with αvβ3 expression, suggesting that αvβ3 could be a biomarker for prediction of the biological outcome of cell surface NCL antagonists.
In summary, the current study unravels at least one of the pathways that participate in cell surface localization of NCL in endothelial and glioma cells, and provides evidence that αvβ3 expression could be a useful biomarker for targeting cell surface NCL as a therapeutic strategy in treatment of gliomas or/and other types of angiogenesis-dependent cancers.
Acknowledgments
We thank the Advanced Light Microscopy facility of the Medical School, University of Patras (especially Dr. Zoi Lygerou) for use of the Leica SP5 confocal microscope, and the Nikon Imaging Center, Harvard Medical School, Boston, MA, for use of the Nikon Eclipse Microscope 80i.
This work was supported by the European Union (European Social Fund) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF)-Research Funding Program: “Heracleitus II, and Investing in Knowledge Society through the European Social Fund.”

This article contains supplemental Figs. S1–S4.
- NCL
- nucleolin
- HUVEC
- human umbilical vein endothelial cells
- MAPK
- mitogen-activated protein kinase
- PTN
- pleiotrophin
- RPTPβ/ζ
- receptor protein-tyrosine phosphatase β/ζ
- PLA
- proximity ligation assay.
REFERENCES
- 1. Srivastava M., Pollard H. B. (1999) Molecular dissection of nucleolin's role in growth and cell proliferation. New insights. FASEB J. 13, 1911–1922 [PubMed] [Google Scholar]
- 2. Hovanessian A. G., Puvion-Dutilleul F., Nisole S., Svab J., Perret E., Deng J. S., Krust B. (2000) The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp. Cell Res. 261, 312–328 [DOI] [PubMed] [Google Scholar]
- 3. Destouches D., El Khoury D., Hamma-Kourbali Y., Krust B., Albanese P., Katsoris P., Guichard G., Briand J. P., Courty J., Hovanessian A. G. (2008) Suppression of tumor growth and angiogenesis by a specific antagonist of the cell-surface expressed nucleolin. PLoS One 3, e2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Di Segni A., Farin K., Pinkas-Kramarski R. (2008) Identification of nucleolin as new ErbB receptors-interacting protein. PLoS One 3, e2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoja-Łukowicz D., Przybyło M., Pocheć E., Drabik A., Silberring J., Kremser M., Schadendorf D., Laidler P., Lityńska A. (2009) The new face of nucleolin in human melanoma. Cancer Immunol. Immunother. 58, 1471–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christian S., Pilch J., Akerman M. E., Porkka K., Laakkonen P., Ruoslahti E. (2003) Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vessels. J. Cell Biol. 163, 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fogal V., Sugahara K. N., Ruoslahti E., Christian S. (2009) Cell surface nucleolin antagonist causes endothelial cell apoptosis and normalization of tumor vasculature. Angiogenesis 12, 91–100 [DOI] [PubMed] [Google Scholar]
- 8. Huang Y., Shi H., Zhou H., Song X., Yuan S., Luo Y. (2006) The angiogenic function of nucleolin is mediated by vascular endothelial growth factor and nonmuscle myosin. Blood 107, 3564–3571 [DOI] [PubMed] [Google Scholar]
- 9. El Khoury D., Destouches D., Lengagne R., Krust B., Hamma-Kourbali Y., Garcette M., Niro S., Kato M., Briand J. P., Courty J., Hovanessian A. G., Prévost-Blondel A. (2010) Targeting surface nucleolin with a multivalent pseudopeptide delays development of spontaneous melanoma in RET transgenic mice. BMC Cancer 10, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krust B., El Khoury D., Nondier I., Soundaramourty C., Hovanessian A. G. (2011) Targeting surface nucleolin with multivalent HB-19 and related Nucant pseudopeptides results in distinct inhibitory mechanisms depending on the malignant tumor cell type. BMC Cancer 11, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Soundararajan S., Wang L., Sridharan V., Chen W., Courtenay-Luck N., Jones D., Spicer E. K., Fernandes D. J. (2009) Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells. Mol. Pharmacol. 76, 984–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Destouches D., Page N., Hamma-Kourbali Y., Machi V., Chaloin O., Frechault S., Birmpas C., Katsoris P., Beyrath J., Albanese P., Maurer M., Carpentier G., Strub J. M., Van Dorsselaer A., Muller S., Bagnard D., Briand J. P., Courty J. (2011) A simple approach to cancer therapy afforded by multivalent pseudopeptides that target cell-surface nucleoproteins. Cancer Res. 71, 3296–3305 [DOI] [PubMed] [Google Scholar]
- 13. Tate A., Isotani S., Bradley M. J., Sikes R. A., Davis R., Chung L. W., Edlund M. (2006) Met-independent hepatocyte growth factor-mediated regulation of cell adhesion in human prostate cancer cells. BMC Cancer 6, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi H., Huang Y., Zhou H., Song X., Yuan S., Fu Y., Luo Y. (2007) Nucleolin is a receptor that mediates antiangiogenic and antitumor activity of endostatin. Blood 110, 2899–2906 [DOI] [PubMed] [Google Scholar]
- 15. Kibbey M. C., Johnson B., Petryshyn R., Jucker M., Kleinman H. K. (1995) A 110-kDa nuclear shuttling protein, nucleolin, binds to the neurite-promoting IKVAV site of laminin-1. J. Neurosci. Res. 42, 314–322 [DOI] [PubMed] [Google Scholar]
- 16. Reyes-Reyes E. M., Akiyama S. K. (2008) Cell-surface nucleolin is a signal transducing P-selectin binding protein for human colon carcinoma cells. Exp. Cell Res. 314, 2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Said E. A., Krust B., Nisole S., Svab J., Briand J. P., Hovanessian A. G. (2002) The anti-HIV cytokine midkine binds the cell surface-expressed nucleolin as a low affinity receptor. J. Biol. Chem. 277, 37492–37502 [DOI] [PubMed] [Google Scholar]
- 18. Koutsioumpa M., Drosou G., Mikelis C., Theochari K., Vourtsis D., Katsoris P., Giannopoulou E., Courty J., Petrou C., Magafa V., Cordopatis P., Papadimitriou E. (2012) Pleiotrophin expression and role in physiological angiogenesis in vivo. Potential involvement of nucleolin. Vascular Cell 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Desgrosellier J. S., Cheresh D. A. (2010) Integrins in cancer. Biological implications and therapeutic opportunities. Nat. Rev. Cancer 10, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Falcioni R., Antonini A., Nisticò P., Di Stefano S., Crescenzi M., Natali P. G., Sacchi A. (1997) α6β4 and α6β1 integrins associate with ErbB-2 in human carcinoma cell lines. Exp. Cell Res. 236, 76–85 [DOI] [PubMed] [Google Scholar]
- 21. Borges E., Jan Y., Ruoslahti E. (2000) Platelet-derived growth factor receptor β and vascular endothelial growth factor receptor 2 bind to the β3 integrin through its extracellular domain. J. Biol. Chem. 275, 39867–39873 [DOI] [PubMed] [Google Scholar]
- 22. Patsenker E., Popov Y., Wiesner M., Goodman S. L., Schuppan D. (2007) Pharmacological inhibition of the vitronectin receptor abrogates PDGF-BB-induced hepatic stellate cell migration and activation in vitro. J. Hepatol. 46, 878–887 [DOI] [PubMed] [Google Scholar]
- 23. Mahabeleshwar G. H., Feng W., Reddy K., Plow E. F., Byzova T. V. (2007) Mechanisms of integrin-vascular endothelial growth factor receptor cross-activation in angiogenesis. Circ. Res. 101, 570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mikelis C., Sfaelou E., Koutsioumpa M., Kieffer N., Papadimitriou E. (2009) Integrin αvβ3 is a pleiotrophin receptor required for pleiotrophin-induced endothelial cell migration through receptor protein tyrosine phosphatase β/ζ. FASEB J. 23, 1459–1469 [DOI] [PubMed] [Google Scholar]
- 25. Papadimitriou E., Mikelis C., Lampropoulou E., Koutsioumpa M., Theochari K., Tsirmoula S., Theodoropoulou C., Lamprou M., Sfaelou E., Vourtsis D., Boudouris P. (2009) Roles of pleiotrophin in tumor growth and angiogenesis. Eur. Cytokine Netw. 20, 180–190 [DOI] [PubMed] [Google Scholar]
- 26. Polykratis A., Katsoris P., Courty J., Papadimitriou E. (2005) Characterization of heparin affin regulatory peptide signaling in human endothelial cells. J. Biol. Chem. 280, 22454–22461 [DOI] [PubMed] [Google Scholar]
- 27. Papadimitriou E., Polykratis A., Courty J., Koolwijk P., Heroult M., Katsoris P. (2001) HARP induces angiogenesis in vivo and in vitro. Implication of N or C terminal peptides. Biochem. Biophys. Res. Commun. 282, 306–313 [DOI] [PubMed] [Google Scholar]
- 28. Héroult M., Bernard-Pierrot I., Delbé J., Hamma-Kourbali Y., Katsoris P., Barritault D., Papadimitriou E., Plouet J., Courty J. (2004) Heparin affin regulatory peptide binds to vascular endothelial growth factor (VEGF) and inhibits VEGF-induced angiogenesis. Oncogene 23, 1745–1753 [DOI] [PubMed] [Google Scholar]
- 29. Schaffner-Reckinger E., Gouon V., Melchior C., Plançon S., Kieffer N. (1998) Distinct involvement of β3 integrin cytoplasmic domain tyrosine residues 747 and 759 in integrin mediated cytoskeletal assembly and phosphotyrosine signaling. J. Biol. Chem. 273, 12623–12632 [DOI] [PubMed] [Google Scholar]
- 30. Li Q., Lau A., Morris T. J., Guo L., Fordyce C. B., Stanley E. F. (2004) A syntaxin 1, Gαo, and N-type calcium channel complex at a presynaptic nerve terminal. Analysis by quantitative immunocolocalization. J. Neurosci. 24, 4070–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hellman U. (2002) in Mass Spectrometry and Hyphenated Techniques in Neuropeptide Research (Silberring J., Ekman R., eds) pp. 259–275, John Wiley & Sons, Inc., New York [Google Scholar]
- 32. Lapeyre B., Amalric F., Ghaffari S. H., Rao S. V., Dumbar T. S., Olson M. O. (1986) Protein and cDNA sequence of a glycine-rich, dimethylarginine-containing region located near the carboxyl-terminal end of nucleolin (C23 and 100 kDa). J. Biol. Chem. 261, 9167–9173 [PubMed] [Google Scholar]
- 33. Turck N., Lefebvre O., Gross I., Gendry P., Kedinger M., Simon-Assmann P., Launay J. F. (2006) Effect of laminin-1 on intestinal cell differentiation involves inhibition of nuclear nucleolin. J. Cell Physiol. 206, 545–555 [DOI] [PubMed] [Google Scholar]
- 34. Senger D. R., Ledbetter S. R., Claffey K. P., Papadopoulos-Sergiou A., Peruzzi C. A., Detmar M. (1996) Stimulation of endothelial cell migration by vascular permeability factor/vascular endothelial growth factor through cooperative mechanisms involving the αvβ3 integrin, osteopontin, and thrombin. Am. J. Pathol. 149, 293–305 [PMC free article] [PubMed] [Google Scholar]
- 35. Somanath P. R., Malinin N. L., Byzova T. V. (2009) Cooperation between integrin αvβ3 and VEGFR2 in angiogenesis. Angiogenesis 12, 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lian J., Dai X., Li X., He F. (2006) Identification of an active site on the laminin α4 chain globular domain that binds to αvβ3 integrin and promotes angiogenesis. Biochem. Biophys. Res. Commun. 347, 248–253 [DOI] [PubMed] [Google Scholar]
- 37. Oikawa Y., Hansson J., Sasaki T., Rousselle P., Domogatskaya A., Rodin S., Tryggvason K., Patarroyo M. (2011) Melanoma cells produce multiple laminin isoforms and strongly migrate on α5 laminin (s) via several integrin receptors. Exp. Cell Res. 317, 1119–1133 [DOI] [PubMed] [Google Scholar]
- 38. Rahman S., Patel Y., Murray J., Patel K. V., Sumathipala R., Sobel M., Wijelath E. S. (2005) Novel hepatocyte growth factor (HGF) binding domains on fibronectin and vitronectin coordinate a distinct and amplified Met-integrin induced signalling pathway in endothelial cells. BMC Cell Biol. 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rehn M., Veikkola T., Kukk-Valdre E., Nakamura H., Ilmonen M., Lombardo C., Pihlajaniemi T., Alitalo K., Vuori K. (2001) Interaction of endostatin with integrins implicated in angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 98, 1024–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen Y. Q., Trikha M., Gao X., Bazaz R., Porter A. T., Timar J., Honn K. V. (1997) Ectopic expression of platelet integrin αIIb β3 in tumor cells from various species and histological origin. Int. J. Cancer 72, 642–648 [DOI] [PubMed] [Google Scholar]
- 41. Cain R. J., Ridley A. J. (2009) Phosphoinositide 3-kinases in cell migration. Biol. Cell 101, 13–29 [DOI] [PubMed] [Google Scholar]
- 42. Wennström S., Hawkins P., Cooke F., Hara K., Yonezawa K., Kasuga M., Jackson T., Claesson-Welsh L., Stephens L. (1994) Activation of phosphoinositide 3-kinase is required for PDGF-stimulated membrane ruffling. Curr. Biol. 4, 385–393 [DOI] [PubMed] [Google Scholar]
- 43. Mercurio A. M., Rabinovitz I. (2001) Towards a mechanistic understanding of tumor invasion. Lessons from the α6β4 integrin. Semin. Cancer Biol. 11, 129–141 [DOI] [PubMed] [Google Scholar]
- 44. Ginisty H., Sicard H., Roger B., Bouvet P. (1999) Structure and functions of nucleolin. J. Cell Sci. 112, 761–772 [DOI] [PubMed] [Google Scholar]
- 45. Huddleson J. P., Ahmad N., Lingrel J. B. (2006) Up-regulation of the KLF2 transcription factor by fluid shear stress requires nucleolin. J. Biol. Chem. 281, 15121–15128 [DOI] [PubMed] [Google Scholar]
- 46. Lemmon M. A., Ferguson K. M. (2000) Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 350, 1–18 [PMC free article] [PubMed] [Google Scholar]
- 47. Burks D. J., Wang J., Towery H., Ishibashi O., Lowe D., Riedel H., White M. F. (1998) IRS pleckstrin homology domains bind to acidic motifs in proteins. J. Biol. Chem. 273, 31061–31067 [DOI] [PubMed] [Google Scholar]
- 48. Eliceiri B. P., Klemke R., Strömblad S., Cheresh D. A. (1998) Integrin αvβ3 requirement for sustained mitogen-activated protein kinase activity during angiogenesis. J. Cell Biol. 140, 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Inder K. L., Hill M. M., Hancock J. F. (2010) Nucleophosmin and nucleolin regulate K-Ras signaling. Commun. Integr. Biol. 3, 188–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dumler I., Stepanova V., Jerke U., Mayboroda O. A., Vogel F., Bouvet P., Tkachuk V., Haller H., Gulba D. C. (1999) Urokinase-induced mitogenesis is mediated by casein kinase 2 and nucleolin. Curr. Biol. 9, 1468–1476 [DOI] [PubMed] [Google Scholar]
- 51. Ko M. H., Kim S., Kang W. J., Lee J. H., Kang H., Moon S. H., Hwang do W., Ko H. Y., Lee D. S. (2009) In vitro derby imaging of cancer biomarkers using quantum dots. Small 5, 1207–1212 [DOI] [PubMed] [Google Scholar]
- 52. Losfeld M. E., Khoury D. E., Mariot P., Carpentier M., Krust B., Briand J. P., Mazurier J., Hovanessian A. G., Legrand D. (2009) The cell surface expressed nucleolin is a glycoprotein that triggers calcium entry into mammalian cells. Exp. Cell Res. 315, 357–369 [DOI] [PubMed] [Google Scholar]
- 53. Barel M., Balbo M., Le Romancer M., Frade R. (2003) Activation of Epstein-Barr virus/C3d receptor (gp140, CR2, CD21) on human cell surface triggers pp60src and Akt-GSK3 activities upstream and downstream to PI 3-kinase, respectively. Eur. J. Immunol. 33, 2557–2566 [DOI] [PubMed] [Google Scholar]
- 54. Sadowski L., Pilecka I., Miaczynska M. (2009) Signaling from endosomes. Location makes a difference. Exp. Cell Res. 315, 1601–1609 [DOI] [PubMed] [Google Scholar]
- 55. Santos S. C., Miguel C., Domingues I., Calado A., Zhu Z., Wu Y., Dias S. (2007) VEGF and VEGFR-2 (KDR) internalization is required for endothelial recovery during wound healing. Exp. Cell Res. 313, 1561–1574 [DOI] [PubMed] [Google Scholar]
- 56. De Donatis A., Comito G., Buricchi F., Vinci M. C., Parenti A., Caselli A., Camici G., Manao G., Ramponi G., Cirri P. (2008) Proliferation versus migration in platelet-derived growth factor signaling. The key role of endocytosis. J. Biol. Chem. 283, 19948–19956 [DOI] [PubMed] [Google Scholar]
- 57. Caswell P., Norman J. (2008) Endocytic transport of integrins during cell migration and invasion. Trends Cell Biol. 18, 257–263 [DOI] [PubMed] [Google Scholar]
- 58. Legrand D., Vigié K., Said E. A., Elass E., Masson M., Slomianny M. C., Carpentier M., Briand J. P., Mazurier J., Hovanessian A. G. (2004) Surface nucleolin participates in both the binding and endocytosis of lactoferrin in target cells. Eur. J. Biochem. 271, 303–317 [DOI] [PubMed] [Google Scholar]
- 59. Schnell O., Krebs B., Wagner E., Romagna A., Beer A.J., Grau S. J., Thon N., Goetz C., Kretzschmar H. A., Tonn J. C., Goldbrunner R. H. (2008) Expression of integrin αvβ3 in gliomas correlates with tumor grade and is not restricted to tumor vasculature. Brain Pathol. 18, 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ding Y., Song N., Liu C., He T., Zhuo W., He X., Chen Y., Song X., Fu Y., Luo Y. (2012) Heat shock cognate 70 regulates the translocation and angiogenic function of nucleolin. Arterioscler. Thromb. Vasc. Biol. 32, e126–134 [DOI] [PubMed] [Google Scholar]
- 61. Garcia M. C., Williams J., Johnson K., Olden K., Roberts J. D. (2011) Arachidonic acid stimulates formation of a novel complex containing nucleolin and RhoA. FEBS Lett. 585, 618–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Guerrero C. A., Moreno L. P. (2012) Rotavirus receptor proteins Hsc70 and integrin αvβ3 are located in the lipid microdomains of animal intestinal cells. Acta Virol. 56, 63–70 [DOI] [PubMed] [Google Scholar]