Background: The BH3-only protein Bim is conventionally considered a proapoptotic protein because it induces Bax/Bak oligomerization on mitochondria.
Results: Bim is constitutively up-regulated in cancer cells via an E2F1-dependent mechanism. Silencing of Bim induces cancer cell apoptosis.
Conclusion: Bim phosphorylation and its sequestration by prosurvival proteins Bcl-xL/Mcl-1 may suppress proapoptotic function of Bim.
Significance: Bim may possess prosurvival functions in epithelial cancer cells.
Keywords: Apoptosis, Bcl-2 Family Proteins, Cancer, Cell Signaling, Mitochondria, Bax/Bak, Bcl-xL/Mcl-1, Bim, E2F1, Microtubules
Abstract
Proapoptotic Bcl-2 homology 3-only protein Bim plays an important role in Bax/Bak-mediated cytochrome c release and apoptosis. Here, we provide evidence for a novel prosurvival function of Bim in cancer cells. Bim was constitutively overexpressed in multiple prostate and breast cancer cells as well as in primary tumor cells. Quantitative real time PCR analysis showed that Bim was transcriptionally up-regulated. We have identified eight endogenous E2F1-binding sites on the Bim promoter using in silico analysis. Luciferase assay demonstrated that Bim expression was E2F1-dependent as mutation of the E2F1-binding sites on the Bim promoter inhibited luciferase activities. In support, E2F1 silencing led to the loss of Bim expression in cancer cells. Bim primarily localized to mitochondrial and cytoskeleton-associated fractions. Bim silencing or microinjection of anti-Bim antibodies into the cell cytoplasm resulted in cell rounding, detachment, and subsequent apoptosis. We observed up-regulation of prosurvival proteins Bcl-xL and Mcl-1, which sequester Bim in cancer cells. In addition, a phosphorylated form of Bim was also elevated in cancer cells. These findings suggest that the constitutively overexpressed Bim may function as a prosurvival molecule in epithelial cancer cells, and phosphorylation and association with Bcl-xL/Mcl-1 block its proapoptotic functions.
Introduction
Bcl-2 family proteins play a critical role in apoptosis by regulating permeabilization of the mitochondrial membrane (1). Proapoptotic BH33-only proteins such as Bim bind to antiapoptotic proteins and thus allow the proapoptotic multidomain proteins, Bax and Bak, to form channels on the mitochondrial membrane leading to cytochrome c release and apoptosis (2). Bim interacts with Bax and Bak, suggesting that Bim can directly activate Bax/Bak to form pores on the mitochondrial membrane (3–5). Two groups independently identified Bim (6, 7), and since then multiple isoforms of Bim have been reported (8–10). All Bim isoforms that possess a BH3 domain are proapoptotic (6, 8–11). It has a canonical BH3 domain (LEDIGD) and also retains the dynein-binding domain, DKSTQT (10). Normally, Bim is sequestered to the microtubule-associated dynein motor complex by binding to 8-kDa dynein light chain 8 (LC8 or DLC1), also called PIN (12, 13). BimEL has also been shown to bind directly to the microtubule in addition to LC8 binding (14). These findings raise the possibility that proapoptotic Bcl-2 family proteins, including Bim, may possess nonapoptotic or prosurvival function.
Indeed, multiple proapoptotic proteins such as Bax/Bak (15–17), Bid (18), Bad (19), Noxa (20), and Puma (18) have been reported to possess nonapoptotic/prosurvival functions. Bim is expressed in multiple lineages of cells, including hematopoietic, epithelial, and neuronal cells (21). A significant number of Bim-null mice, and even some of the Bim+/− animals, die in utero prior to E9.5, suggesting that Bim plays a critical role in development (11). Among the BH3 domain-containing proteins whose genes have been knocked out, including Bad, Bid, Bim, Blk, Bmf, Puma, and Noxa, Bim is the only BH3 protein whose deletion leads to embryonic lethality. It remains unknown, however, what is the physiological role of Bim during development and how do Bim-deficient animals die? Interestingly, Bim deficiency can rescue defective kidney development caused by Bcl-2 deficiency (22). Bim plays an essential role in leukocyte homeostasis as well as in preventing autoimmunity by eliminating autoreactive lymphocytes (11, 23–26). These findings suggest that Bim plays a physiological role in promoting cell survival in addition to its well known function in apoptosis induction.
Here, we show that Bim is highly up-regulated in prostate and breast cancer cells, and its up-regulation is mediated by E2F1 transcription factor. Bim was mostly associated with membranous structures such as mitochondrial membrane and performed prosurvival functions. Silencing of Bim induced cancer cell detachment and caused subsequent cell apoptosis. Bim was sequestered by prosurvival proteins Bcl-xL and Mcl-1, and phosphorylated forms of Bim protein were also detected/elevated in cancer cells, potentially explaining why constitutively overexpressed Bim did not kill cancer cells. Together, our findings identify a novel prosurvival function for Bim in addition to its well known apoptosis-inducing function.
EXPERIMENTAL PROCEDURES
Cells and Reagents
NHP1–NHP4 represent four NHP epithelial strains. PPC1, LNCaP, C4-2, C5, PC3, DU145, and MDA2b are prostate cancer cell lines (10). HME87 (human mammary epithelial) are primary strains, and B26, B27, B28, and B42 are primary organoid isolates. MB231, MB435, and MB453 refer to MDA-MB-231, MDA-MB-435, and MDA-MB-453 cells, respectively. Human mammary epithelial cells, NHP8, NHP9, and MCF-7 cells were obtained commercially or from collaborators.
Rat monoclonal anti-Bim antibody was bought from Alexis Biochemicals (San Diego). Rabbit polyclonal anti-Bim antibody was purchased from Calbiochem. Rabbit polyclonal anti-Bax was from Pharmingen. Rabbit polyclonal anti-Bcl-XL and Bak and mixed goat polyclonal anti-lamin A and -lamin B antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal anti-Bcl-2 was bought from Dako (Carpinteria, CA). Rabbit polyclonal anti-LC8 was kindly provided by Dr. S. King. The mouse monoclonal anti-IC-74 and goat polyclonal anti-LDH antibodies were purchased form Chemicon (Temecula, CA). Mouse monoclonal anti-XIAP antibody was bought from BD Transduction Laboratories. Rabbit polyclonal anti-Bap31 and mouse monoclonal anti-Cox II antibodies were obtained from Molecular Probes. Anti-Bim, phosphorylated-Bim (Ser-69), Mcl-1, and E2F1 rabbit polyclonal antibodies were purchased from Cell Signaling. Anti-phosphorylated Bim (Ser-87) polyclonal antibody was obtained from Bioss Inc. (Woburn, MA). Anti-dynein (DLC2–1G7) antibody was procured from Novus Biologicals. Monoclonal antibody to DLC1 (10D6)-LC8 was obtained from A. G. Scientific, Inc. (San Diego). Goat anti-rat IgM-HRP conjugate was obtained from Enzo Life Sciences. Anti-Bcl-xL rabbit antibody was purchased from BD Biosciences. Anti-calnexin rabbit antibody was bought from Abcam Inc. Mouse monoclonal anti-β-tubulin antibody was bought from Sigma. The secondary antibodies, goat anti-mouse or -rabbit and sheep anti-rat IgG conjugated to horseradish peroxidase, were purchased from GE Healthcare. Liposome FuGENE 6 was bought from Roche Applied Science.
Semiquantitative RT-PCR Analysis
Total RNA was isolated using TRIzol (Invitrogen). RT was performed using 2 μg of total RNA at 42 °C for 2 h in a volume of 20 μl containing random hexamers and Superscript II reverse transcriptase (Invitrogen). One μl of cDNA was used for PCR. The details were described previously (27).
Analysis of Bim mRNA Expression by Real Time RT-PCR
Cancer and normal cells were harvested in TRI Reagent (Molecular Research Center, Inc.). Total RNA was isolated according to the manufacturer's instructions. cDNA was prepared from 3 μg of total RNA using a SuperScriptIII first-strand synthesis system (Invitrogen) following the manufacturer's instructions. Real time reverse transcription-PCR (RT-PCR) was done using SYBR Green Supermix (Bio-Rad) on a Prism 7300 sequence detection system (Applied Biosystems) following the manufacturers' instructions. The fold-change in expression levels (using GAPDH and actin as control) was determined by comparative CT method. The primers for Bim, GAPDH, and actin were used as described previously (28, 29). The primers Bim forward, GGCCCCTACCTCCCTACA, and Bim reverse, GGGGTTTGTGTTGATTTGTCA; GAPDH forward, AGCCACATCGCTCAGACAC, and GAPDH reverse, GCCCAATACGACCAAATCC; and actin forward, CTTCGTCGCACATTGTGTCT, and actin reverse, GACAGCGCCAAGTGAAGC were used to amplify the mRNA expression levels. Both forward and reverse primers were used at a final concentration of 400 nmol/liter. PCR products were electrophoresed in 1.2% agarose gels.
Whole Cell Lysate Preparation and Western Blotting
Protein extraction and Western blotting were carried out as described previously (10, 30). The blots were stripped and reprobed for the indicated proteins in the respective figures.
Quantification of Apoptosis and Caspase Activity Measurement
Apoptotic cells were counted based on live cell staining with DAPI to label apoptotic nuclei (31). In addition, both live and dead cells were counted using trypan blue dye (30). DEVDase and LEHDase activities were measured as described previously (31).
Immunohistochemistry
For tissue section staining, following de-paraffinization and dehydration, slides were incubated in 3% hydrogen peroxide to block endogenous peroxidase activity. For antigen retrieval, slides were incubated in 10 mm citrate buffer, pH 6.0, for 15 min in a microwave oven. Then slides were sequentially incubated in blocking solution (10% goat serum in PBS, 30 min), primary antibody (rabbit polyclonal anti-Bim; 1:1,000 for 1 h), secondary antibody (goat anti-rabbit IgG conjugated to HRP; 1:4,000), and substrate (3,3′-diaminobenzidine).
Subcellular Fractionation
Heavy mitochondria (HM), light mitochondria (LM), cytosol, and microsome preparation were described previously (31). Briefly, cells were harvested, washed, and homogenized in buffer A as described previously (32) with protease inhibitors (Sigma) in a glass Pyrex homogenizer followed by clearing unbroken cells and nuclear fractions. HM, LM, cytosol, and microsome fractions were obtained through differential centrifugation at 1,500 × g, 10,000 × g, and 100,000 × g, respectively. In some experiments, mitochondria, cytosol, and ER were purified as described previously (33). HM, LM, and microsome pellets were then solubilized in TNC buffer (10 mm Tris acetate, pH 8.0, 0.5% Nonidet P-40, and 5 mm CaCl2) containing protease inhibitor. For cytoskeleton and cytoskeleton-associated protein (CAP) preparation, the cells were harvested in TNC buffer and centrifuged at 12,000 rpm for 10 min. The pellet was dissolved in 200 μl of cytoskeleton extraction buffer (CEB: 600 mm KCl, 150 mm MgCl2, 50 mm MES, 10 μg/ml DNase, 10 μg/ml RNase, 20 mm PMSF, and 1% Triton X-100) containing protease inhibitors, followed by the centrifugation at 12,000 rpm for 10 min. Pellets obtained after three rounds of CEB extraction were saved as cytoskeleton, whereas pellets obtained after addition of 1 volume of acetone in the supernatant of CEB extraction was considered as CAP. Both of these pellets were dissolved in SDS loading buffer for SDS-PAGE and Western blotting.
Nuclei were isolated as described in Sigma technical bulletin nuclei isolation kit. Briefly, the cells were harvested in Nuclei EZ lysis buffer. After vortexing, cells were centrifuged at 500 × g for 5 min. The pellet was then resuspended in Nuclei EZ lysis buffer by vortexing. After centrifugation, the resulting pellet was dissolved in 200 μl of Nuclei EZ storage buffer for Western blot analysis.
Immunostaining and Double Immunostaining Microscopy
PPC1 cells (5,000) were plated on circle glass coverslips. An indirect immunofluorescence protocol using tyramide amplification for Bim was applied. Briefly, cells cultured on glass coverslips were fixed (4% paraformaldehyde), permeabilized (1% Triton), and then blocked in 10% goat whole serum. Cells were then incubated with polyclonal anti-Bim antibody (1:4,000) followed by biotinylated goat anti-rabbit IgG (1:1,000). Then cells were reacted with streptavidin-conjugated horseradish peroxidase (1:1,000) followed by incubation with tyramide/fluorescein (1:1,000).
For double immunofluorescence, cells on the coverslips were fixed, permeabilized, and blocked with 10% goat whole serum. Then cells were incubated with the first primary antibodies (1:1,000) and secondary antibody (goat anti-rabbit or goat anti-mouse IgG-Alexa Fluor 594) (34). Cells were re-blocked with 10% of goat serum, followed by the indirect immunofluorescence to Bim described above. Finally, cells were incubated with 1 μg/ml DAPI for nuclei labeling. MitoTracker was incubated with cells on coverslips for 1 h at 37 °C before Bim immunostaining.
RNAi Down-regulation of Bim
Bim siRNA were purchased from Dharmacon (catalog no. MU-004383-00) or chemically synthesized by Ambion (Invitrogen). The siRNAs at final concentration of 40 nm were transfected with FuGENE 6 (Roche Applied Science). The cells were counted for detachment and were harvested at different time points (0, 24, 48, and 72 h) for whole cell lysate preparation.
Microinjection of Bim Antibody and Bim siRNA
Cells were microinjected in a 6-cm tissue culture dish using an Eppendorf pressure injector (model 5246) and micromanipulator model (model 5171). Microinjection needles (inner diameter, ∼0.1 μm) were purchased from Eppendorf and loaded using Eppendorf microloaders. Dextran-Texas Red (Mr 10,000 lysine-fixable, Molecular Probes; 0.2% final concentration) was mixed with anti-Bim antibody or Bim siRNAs and injected into the cytoplasm of PC3 cells (pressure, 40–80 hectopascals; time, 0.3 s). Cells were replenished with fresh medium immediately after microinjection. A total of 100–150 cells were microinjected for each condition and were counted for detachment. Apoptosis was determined based on morphology and DAPI staining (35).
Generation of Bim or E2F1 or Bcl-xL-silenced Stable Cells Using shRNA Lentiviral Vectors
Bim (shRNA sequence, 5′-CAGGCTGAACCTGCAGATA-3′), E2F1 (shRNA sequence, 5′-GTGATTTATTTATTGGGAA-3′), and Bcl-xL (shRNA sequence, 5′-CTCACTCTTCAGTCGGAAA-3′ or 5′ AAATTCTAGAAAACTAGCT-3′)-specific shRNA and control shRNAs expressing lentiviral particles were obtained from the Roswell Park Cancer Institute shRNA core resources and were directly utilized to infect cells at a multiplicity of infection of 3. After 48 h, puromycin (1 μg/ml) was added to the medium to select Bim or E2F1 or Bcl-xL-silenced cells (30).
Chromatin Immunoprecipitation (ChIP) Assays
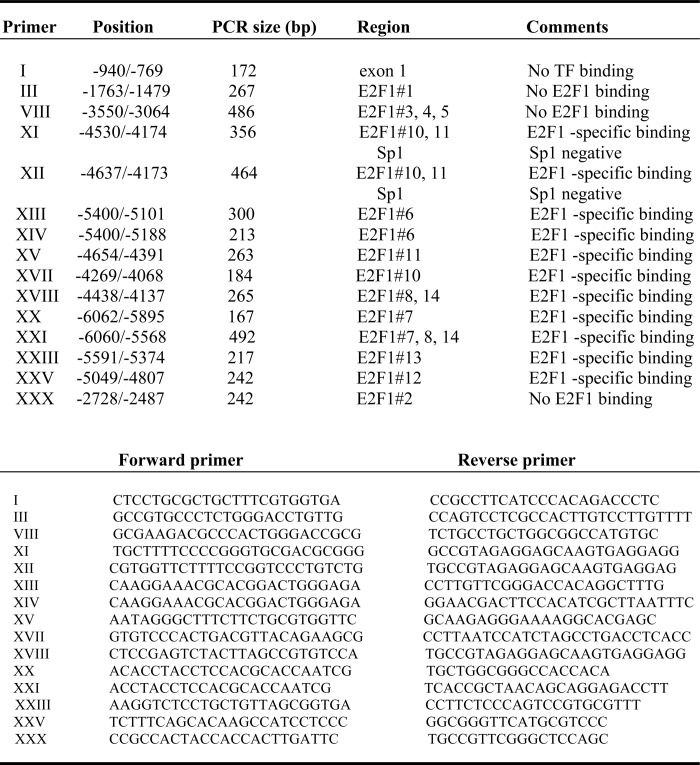
ChIP assays were performed as described previously (36) according to the manufacturer's instructions (Upstate, Charlottesville, VA). PC3 or PPC1 cells were fixed, washed, and harvested followed by shearing of genomic DNA using sonication. 20 μl of sonicated DNA was purified and used as input DNA control. Sheared DNA was cleared, and chromatin-bound DNAs were immunoprecipitated using E2F1 or FOXO3a or Sp1 or RbIgG and protein A beads. The eluted DNA from the beads was precipitated and analyzed by PCR using multiple primer sets as described in Fig. 3A and Table 1.
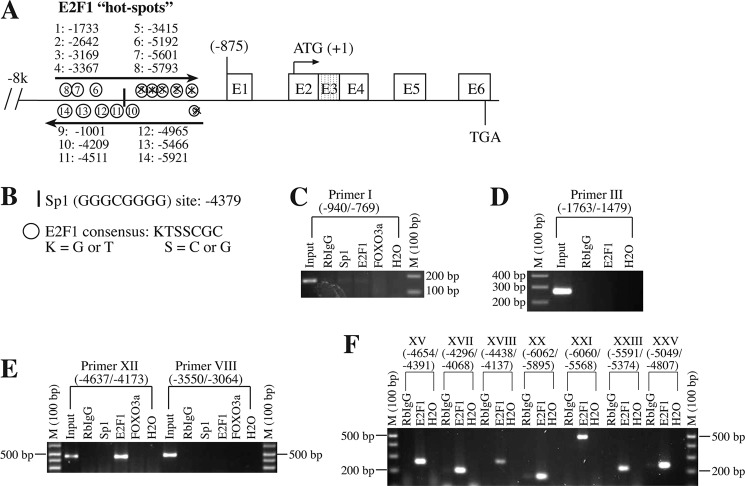
FIGURE 3.
E2F1 binds to the Bim promoter at multiple sites. A and B, putative E2F1-binding sites on the Bim promoter were determined by in silico analysis using KTSSCGC consensus sequence. C–F, ChIP analysis determines multiple E2F1-binding sites on the Bim promoter. Briefly, cancer cells were subjected to chromatin cross-linking followed by immunoprecipitation with anti-E2F1, anti-Sp1, anti-FOXO3a, and RbIgG. Bim promoter fragments were amplified by using PCR primers as described in Table 1. Positive controls (input DNA) or negative controls (H2O) were used in the amplification of the Bim promoter. Crossed sites in A (i.e. 1–5 and 9 predicted sites) indicate that these sites do not show binding with E2F1 based on ChIP analysis.
TABLE 1.
ChIP assay primer summary
Analysis of Bim Promoter Activity by Luciferase Assays
Bim promoter fragments of 1.6, 2.6, and 5.1 kb upstream of the translation start site (TSS) were cloned in pGL3 vector. To determine the E2F1-dependent Bim expression, dual-luciferase assays were performed with Renilla as internal control. In brief, cells in triplicate were transfected with firefly luciferase plasmids (control and indicated Bim promoter constructs) and 10 ng of Renilla luciferase (internal control) using Lipofectamine 2000 (Invitrogen) per the manufacturer's instructions. After 24 h, luciferase assay was performed using the Dual-Glo luciferase assay system (Promega) on a Veritas Luminometer (Molecular Devices) as recommended by the manufacturer. Promoter activity was further validated by mutation of two E2F1-binding sites on the promoter at −4965/−4963 or −4782/−4780 by replacing CGC with TTT or CCC with TTT, respectively.
Immunoprecipitation (IP)
Whole cell lysates were precleared with mouse or rabbit (depending on the primary antibodies used) IgG-conjugated agarose beads, followed by immunoprecipitation using primary antibodies against Bim or Bcl-xL or Mcl-1 or β-tubulin or Ms-IgG (MOPC) or Rb-IgG. Finally, the beads were washed and analyzed by Western blotting system (30, 37).
Clonogenic Survival Assays
Cells (0.5 × 103/well) were plated in 6-well tissue culture dishes, and medium was changed every 72 h. When colonies became visible (∼2 weeks), cells were fixed and stained with Giemsa (1:10 in distilled water at room temperature) and counted. The stained plates were pictured using the GS800 densitometer (Bio-Rad).
Statistical Analysis
Data represent means ± S.D. of at least three independent experiments. Statistical analysis was performed by analysis of variance using Sigma Stat. Significant changes (p < 0.05 or 0.01) are represented by an asterisk.
RESULTS
Bim Is Constitutively Overexpressed in Multiple Cancer Cells
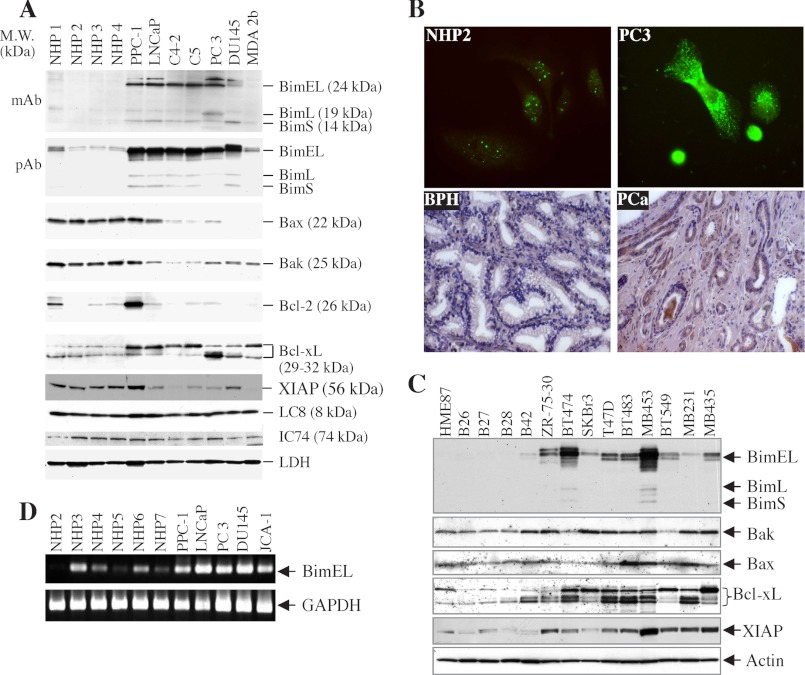
We previously observed that multiple prostate cancer cells express higher levels of Bim (10, 34), indicating the possibility that overexpression of Bim may be required for the survival of prostate cancer cells. Therefore, to further explore this interesting phenomenon, we utilized a panel of NHP epithelial cells and prostate cancer cells to examine the expression levels of Bim. As shown in Fig. 1A, Western blot analysis using either a monoclonal or a polyclonal Bim antibody revealed an increased level of Bim protein in prostate cancer cells, whereas Bim was undetectable in NHP cells. All three major Bim isoforms (i.e. BimEL, BimL, and BimS) were up-regulated in prostate cancer cells, although the up-regulation of BimEL was the most prominent (Fig. 1A). NHP1–NHP4 cells were derived from normal donors and all prostate cancer cells from metastases (10, 34, 38, 39). Comparison with several other NHP (including NHP5 and NHP6) and prostate cancer cell lines (including MDA 2a, JCA-1, and Tsu-Pr) similarly revealed increased expression of Bim proteins in prostate cancer cells (data not shown) (10, 34).
FIGURE 1.
Bim is constitutively up-regulated in cancer cells. A, equal amounts of protein using whole cell lysates prepared from NHPs and prostate cancer cells were subjected to Western blotting for the indicated proteins. B, NHPs or PC3 cells plated on coverslips were subjected to immunolabeling using tyramide amplification as described under “Experimental Procedures.” Thin sections of BPH or primary prostate tumor (PCa) specimens were subjected to immunohistochemistry using rabbit polyclonal antibody (pAb). C, whole cell lysates prepared from normal breast epithelial and breast cancer cells were subjected to Western blotting for the indicated proteins. HME87 (human mammary epithelial) is a primary strain, whereas B26, B27, B28, and B42 are primary organoid isolates. MB231, MB435, and MB453 refer to MDA-MB-231, MDA-MB-435, and MDA-MB-453 cells, respectively. D, semiquantitative RT-PCR for BimEL (upper panel) and GAPDH (lower panel) was performed in several NHP and prostate cancer cell lines.
To further demonstrate increased Bim expression in prostate cancer cells, we carried out immunofluorescence and immunohistochemical staining. As shown in Fig. 1B, increased Bim expression was detected in PC3 cells compared with NHP2 cells. Similar staining revealed increased Bim expression in other prostate cancer cells (i.e. PPC-1 and Du145) compared with NHP (i.e. NHP4 and NHP6) cells (data not shown). Increased Bim staining was also observed in clusters of high grade (Gleason score 8) primary prostate tumor cells in tissue sections compared with either BPH or normal prostatic glands (Fig. 1B, lower panels; and data not shown). To quantify the global Bim protein expression level in normal, benign, and cancerous prostate tissues, we employed ChromaVision (Advanced Cellular Imaging System, San Juan Capistrano, CA) to quantify Bim expression. Briefly, we collected 7 normal prostate, 15 BPH samples, and 20 prostate cancer tissues (Gleason scores 7–9). Five serial sections from each sample were stained for Bim using the polyclonal antibody and then lightly counter-stained for nuclei using hematoxylin. The stained slides were loaded onto the imaging station, and all glandular structures on the whole section were digitally captured. The total brown-colored chromogen (i.e. 3,3′-diaminobenzidine) level was first obtained by scanning the whole slide, which generally contained 15–36 glandular structures. The expression levels were assigned a number between 0 and 250 by the Advanced Cellular Imaging System Quantification software. The relative Bim expression levels from these experiments were 254 ± 87 (means ± S.D.) for the normal prostate group (n = 35), 295 ± 61 for the BPH group (n = 75), and 16,774 ± 419 for the prostate cancer group (n = 100) (p < 0.001; F test). These observations together suggest that Bim protein is constitutively overexpressed in prostate cancer cells and tumor samples.
To determine whether up-regulation of Bim is unique to prostate cancer cells, we carried out Western blotting on a panel of normal breast epithelial cells (i.e. HME87, B26, B27, B28, and B42) and breast cancer cells (i.e. ZR75-30, BT474, SKBr3, T47D, BT483, MDA-MB-453, BT549, MDA-MB-231, and MDA-MB-435) (Fig. 1C). Similar to prostate cancer cells, breast cancer cells expressed much higher levels of BimEL than normal mammary epithelial cells (Fig. 1C). In addition, some breast cancer cells also up-regulated another faster migrating protein (just beneath the BimEL band) that most likely represents a Bim isoform. Three breast cancer cell lines, i.e. ZR75–30, BT474, and MDA-MB-453, also expressed a band above the BimEL (Fig. 1C), which most likely represents the phosphorylated BimEL (40, 41). Several prostate cancer cell lines also expressed the phosphorylated Bim (Fig. 1A). Collectively, these results suggest that Bim, especially BimEL (referred to throughout as “Bim”), is up-regulated in both prostate and breast cancer cells.
Transcriptional Up-regulation of Bim in Cancer Cells
To determine whether constitutively up-regulated Bim is due to transcriptional activation of the Bim gene, we first performed semi-quantitative RT-PCR and observed that Bim mRNA was up-regulated in prostate cancer cells compared with NHP cells (Fig. 1D). These findings suggest transcriptional up-regulation of Bim in prostate cancer cells. To conclusively demonstrate that Bim is transcriptionally up-regulated, we performed real time RT-PCR analysis, and the results confirmed the up-regulation of Bim mRNA levels in both prostate cancer (Fig. 2A) and breast cancer (Fig. 2B) cells compared with the respective normal cells.
FIGURE 2.
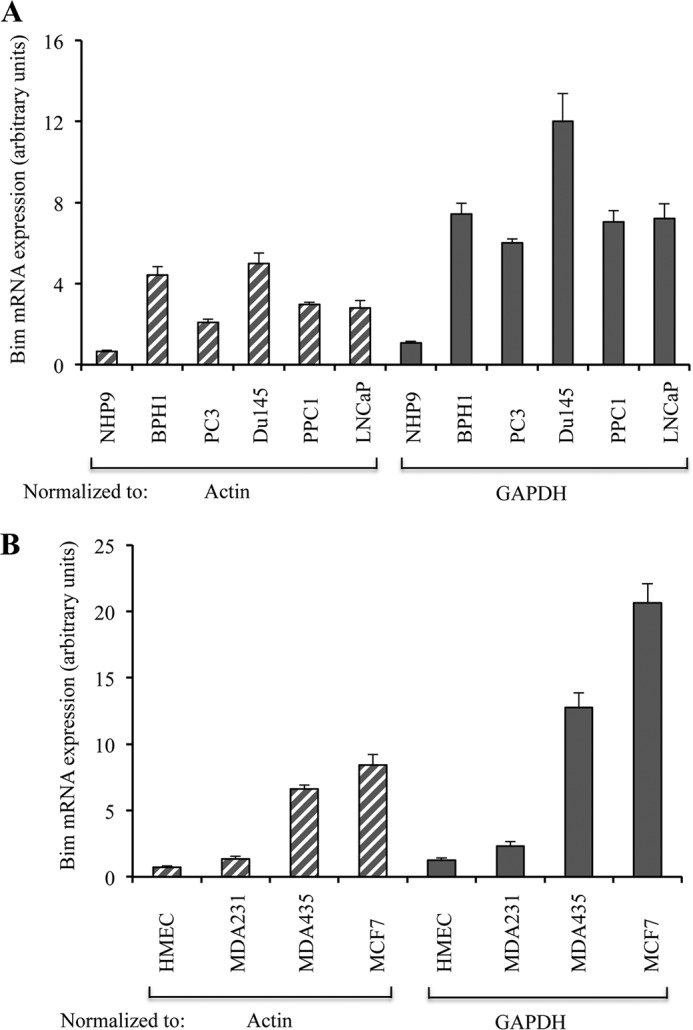
Bim is transcriptionally up-regulated in cancer cells. A, equal amounts RNA from normal NHP9 cells and BPH1, PC3, Du145, PPC1, and LNCaP cells were converted to cDNA and subjected to real time PCR analysis to measure the expression of Bim using actin or GAPDH as internal controls. B, equal amounts of RNA from normal human mammary epithelial cells (HMEC) and MDA-MB231 (MDA231), MDA-MB435 (MDA-435), and MCF-7 cells were converted to cDNA and subjected to real time PCR analysis to measure the expression of Bim using actin or GAPDH as internal controls. Data are average of triplicates ± S.D.
Cancer Cells Also Show Abnormal Expression of Other Apoptotic Proteins
How do cancer cells avoid apoptotic cell death in the presence of significantly up-regulated levels of Bim proteins? One possibility is that cancer cells may also possess abnormal expression and/or function of other apoptosis-related molecules such as multidomain Bcl-2 proteins or inhibitors of apoptosis. To explore this possibility, we reprobed the membrane blots for some of the key apoptosis-related proteins. As shown in Fig. 1A, prostate cancer cells expressed lower levels of Bak and Bax as compared with normal counterparts. In fact, Du145 and MDA 2b cells lacked Bax expression due to mutations in the Bax gene (10, 42). Furthermore, prostate cancer cells overexpressed Bcl-xL, which was detected as two major bands and one minor band migrating at 29–32 kDa (Fig. 1A). These results suggest that prostate cancer cells not only up-regulate Bim but also up-regulate Bcl-xL and down-regulate Bax and Bak, thus nullifying the prodeath effects of Bim up-regulation in cancer cells.
Similarly, Western blot analysis revealed a significant increase in the levels of Bcl-xL isoforms in breast cancer cells (Fig. 1C). In contrast to prostate cancer cells, some breast cancer cells also expressed higher levels of Bax and/or Bak than the normal mammary epithelial cells (Fig. 1C). However, all breast cancer cells expressed much higher levels of XIAP (Fig. 1C), a potent inhibitor of activated caspase-9 and caspase-3 (43–46), suggesting that breast cancer cells have evolved antiapoptotic mechanisms by increasing not only Bcl-xL levels but also XIAP to inhibit the activated caspases. Taken together, the results from both prostate and breast cancer cells suggest that abnormal expression of other antiapoptotic proteins may represent one of the mechanisms for cancer cell survival in the presence of increased expression of Bim.
E2F1 Binds at Multiple Sites on the Bim Promoter
How do cancer cells constitutively activate the Bim gene? It is well established that cancer cells, in association with their rapid proliferation, possess increased activity of E2F1 transcription factor (47, 48), which is known to promote expression of proapoptotic BH3-only proteins, including Bim during stress or apoptotic stimulation (49, 50). To determine whether cancer cells adopt E2F1-mediated overexpression of Bim, we first performed in silico analysis of the Bim promoter region for potential E2F1-binding site(s). Sequence analysis of genomic sequences spanning 8 kb upstream of the TSS of the human Bim promoter identified a total of 14 “E2F1 hot spots” as mentioned in Fig. 3A. For these E2F1 hot spots, we used KTSSCGC as a consensus sequence of E2F1 binding, where K represents G or T and S represents C or G (Fig. 3B). In contrast to E2F1, we observed only one potential binding site for Sp1 at −4379 with a consensus sequence GGGCGGGGG for Sp1 binding (Fig. 3B).
To demonstrate directly whether E2F1 binds to the Bim promoter in cells, we performed ChIP assays using E2F1, RbIgG, and Sp1 antibodies. Multiple PCR primers representing different binding sites as described in Table 1 were used to amplify the bound DNA sequence. We observed multiple E2F1-specific binding sites on the Bim promoter as presented in Table 1 and Fig. 3, E and F. Out of 14 E2F1 hot spots identified by in silico analysis, hot spots 1–5 and 9 were not confirmed in vivo (Fig. 3, A and C–F; and data not shown; Table 1), suggesting that these putative E2F1 sites were not occupied by the endogenous E2F1. In contrast, ChIP assays indicated that the putative E2F1 sites 6–8 and 10–14 were bound by endogenous E2F1 (Fig. 3, E and F; Table 1). Sp1 binding at −4379 on the Bim promoter was not detected by ChIP analysis (Fig. 3, C–F).
It has been reported that FOXO3a also binds to the Bim promoter (51) and regulates Bim expression. However, our ChIP analysis in prostate cancer cells (Fig. 3, C–F) failed to demonstrate FOXO3a binding to the Bim promoter, suggesting that the FOXO3a regulation of Bim expression may likely be cell type-specific. Taken together, our findings suggest that E2F1 may represent a primary transcription factor responsible for increased Bim expression in unstimulated prostate (and probably breast) cancer cells.
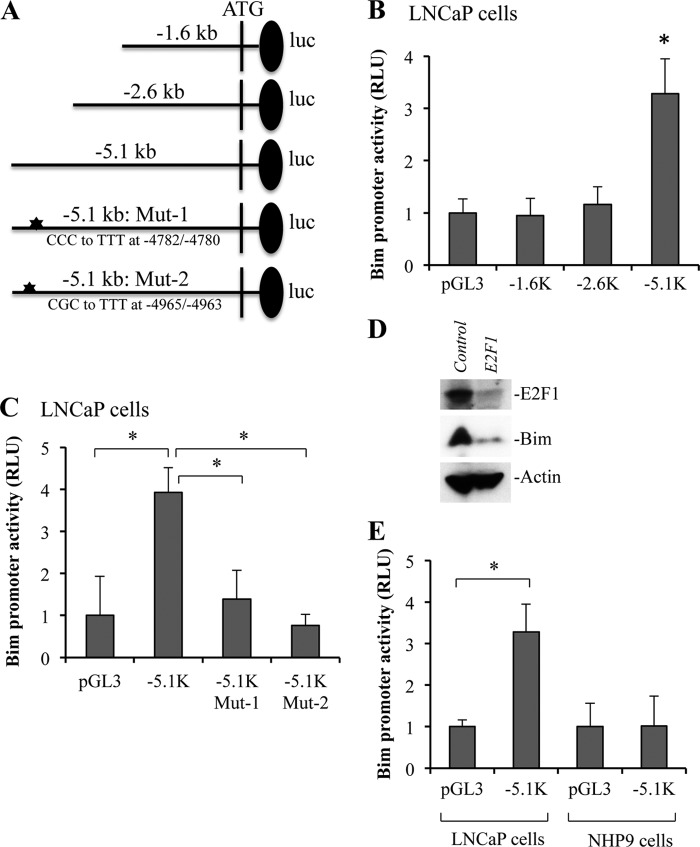
Genomic Sequence within −5.1 to −2.6 kb Confers Bim Promoter Activity
To characterize whether all or only some E2F1-binding sites were necessary for activation of the Bim, we cloned three genomic DNA fragments (i.e. −5.1, −2.6, and −1.6 kb) spanning the Bim promoter region into pGL3-luciferase reporter vector for the analysis of Bim promoter activity (Fig. 4A). After transfection into LNCaP cells, a strong promoter activity was observed with the −5.1-kb fragment (Fig. 4B), which encompassed potential E2F1 sites 1–5 and 9–12 (Fig. 3A). In contrast, very little or no promoter activity was observed with the −2.6 and −1.6-kb fragments (Fig. 4B). Similar results were obtained in PPC1 and MDA-MB231 cells (data not shown). Because the −2.6-kb promoter fragment contained the putative E2F1-binding site 1 (Fig. 3A), lack of promoter activity in the −2.6-kb fragment (Fig. 4B) thus confirmed our ChIP data showing lack of E2F1 binding to site 1. Combining the ChIP and luciferase data, the strong promoter activity associated with the −5.1-kb fragment most likely results from E2F1 sites 10–12. To further demonstrate that E2F1-binding sites on the Bim promoter are indeed required for Bim expression, we mutated CGC to TTT at hot spot 12 (−4965/−4963). Because of the GC-rich regions, we could not design primers for other hot spots. In addition, we also mutated CCC to TTT between hot spots 11 and 12 at −4782/−4780, which was part of sequence (GCCCCAAA) similar to E2F1 consensus (Fig. 4A). We observed that mutation of either E2F1-binding site significantly inhibited Bim promoter activity in LNCaP and other cancer cells (Fig. 4C and data not shown).
FIGURE 4.
E2F1-dependent expression of Bim in cancer cells. A, Bim promoter constructs of indicated fragment size were cloned in pGL3-basic vector and E2F1-binding sites were mutated as shown. B and C, LNCaP cells were transfected with −1.6, −2.6, or −5.1-kb Bim promoter luciferase (luc) constructs (B) or −5.1-kb fragment and its mutants (C) as described under “Experimental Procedures” or empty vector together with Renilla luciferase construct. D, MDA-MB231 cells were infected with control or E2F1 shRNA lentiviral particles at a multiplicity of infection of 5. Stable cells were subjected to Western blotting to detect indicated proteins. Actin serves as loading control. E, NHP9 normal cells and LNCaP cancer cells were transfected with −5.1-kb Bim promoter construct. Luciferase activities in B, C, and E were determined at 24 h after transfection. RLU, relative luciferase units. Data are mean ± S.D., n = 3; *, p < 0.01.
Finally, to demonstrate that Bim expression is regulated by E2F1 at protein levels, we silenced E2F1 and observed reduced levels of Bim protein in E2F1-silenced cells (Fig. 4D). Also, consistent with the increased expression of Bim only in cancer cells (Fig. 1), we detected very little or no −5.1-kb Bim promoter activity in normal human prostate epithelial cells, although robust promoter activity was observed in PPC-1, LNCaP, PC3, and Du145 prostate cancer cells (Fig. 4E; data not shown).
Bim Associates with Cytoskeleton (Microtubule), LC8-based Motor Molecules, and Mitochondrial Membranes
Once we established that multiple cancer cell types express high levels of Bim and its expression is regulated by E2F1, we began investigating the functions of Bim up-regulation in cancer cells. In unstressed cells, Bim is sequestered by the microtubule-based dynein motor complex through LC8 and by direct binding to the microtubules (12–14), suggesting that up-regulated Bim in cancer cells may function as a nonapoptotic or a prosurvival protein. To explore whether Bim overexpression in cancer cells is also associated with higher expression of dynein motor complex or microtubules, we examined the expression levels of two integral components of the dynein motor complex, LC8 and IC74 (52–54). As shown in Fig. 1A, both LC8 and IC74 proteins were expressed at similar levels in NHP and prostate cancer cells.
To determine the subcellular localization of up-regulated Bim in cancer cells, we isolated cytoskeleton (i.e. Triton-insoluble fraction), CAP (i.e. Nonidet P-40-insoluble but Triton-soluble proteins extracted from the cytoskeleton preparations), nucleus, cytosol (i.e. the 100,000 × g supernatant), and three membrane fractions, i.e. HM (1,500 × g pellet, enriched in heavy mitochondria and some ER), LM (10,000 × g pellet, enriched mainly with mitochondria), and microsome (100,000 × g pellet, which contains mainly ER). The subcellular fractions were characterized based on specific marker proteins (10, 31, 39). For example, Bap31 for ER, COX II (cytochrome c oxidase subunit II) for mitochondria, nuclear lamins (A/B) for nuclei, and LDH (lactate dehydrogenase) for the cytosol (Fig. 5A). Unfractionated whole cell lysate was used as input control. We observed that in unstimulated cancer cells, Bim was localized mainly in the cytoskeleton and membrane fractions (Fig. 5A). Because the cytoskeleton was prepared by extracting Nonidet P-40-insoluble components with Triton X-100 and high salt (39), our results suggest that Bim associates tightly with the filamentous cytoskeleton, whereas some amounts of Bim could be extracted as CAP. Bim was not detected in cytosol, nucleus, or microsome (Fig. 5A).
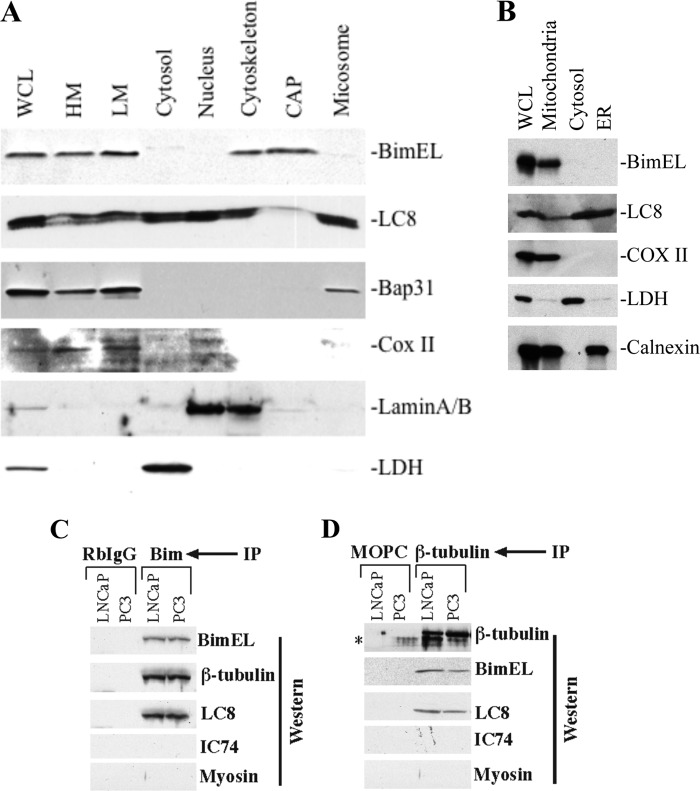
FIGURE 5.
Constitutively overexpressed Bim was localized to the cytoskeleton and mitochondrial fractions. Bim interacts with microtubule and microtubule-associated dynein motor complex protein LC8. A, PPC-1 cells (5 × 107) were subjected to biochemical fractionation, and equal amounts of proteins were separated on SDS-PAGE for Western analysis. WCL, whole cell lysate. B, cytosol, mitochondria, ER, and whole cell lysates were prepared from LNCaP cells followed by detection of the indicated protein by Western analysis. C, whole cell lysates from PC3 and LNCaP cells were subjected to RbIgG or Bim (C) and MsIgG (MOPC) or β-tubulin (D) immunoprecipitation (IP) followed by Western blotting for the indicated proteins. *, nonspecific band.
In contrast to Bim, LC8 was detected as a doublet in every fraction with the highest amounts in the cytosol, nucleus, and microsome (Fig. 5A). These findings are consistent with the idea that LC8 binds many other proteins in addition to Bim (55–57). As expected, the marker proteins were detected in the whole cell lysate as well as in the expected fractions, i.e. LDH in the cytosol, Bap31 in the membrane fractions, COX II in HM and LM, and lamin A/B in the nucleus and cytoskeleton (Fig. 5A; note that cell nuclei are known to interact with cytoskeleton so that these two often co-fractionate). Immunolabeling experiments also demonstrated Bim co-localization with LC8 (data not shown). We performed another independent subcellular fractionation experiment to verify that constitutively expressed Bim in cancer cells was not localized in the cytosol and ER but primarily to mitochondria (Fig. 5B).
Because the cytoskeleton is enriched in microtubules and the membrane fractions (both HM and LM) are enriched in dynein motor complexes, constitutively overexpressed Bim in prostate cancer cells might directly associate with microtubules and LC8-containing dynein motors, as reported in other cells (12, 14). To test this possibility, we carried out co-IP experiments using PC3 and LNCaP cells. As shown in Fig. 5C, IP using the polyclonal antibody to Bim pulled down β-tubulin and LC8 but not IC74 or myosin. Reciprocal IP using a mAb against β-tubulin co-precipitated BimEL and LC8 but not IC74 or myosin (Fig. 5D). In contrast, both RbIgG and MOPC21 control did not immunoprecipitate any specific products (Fig. 5, C and D). These results suggest that in prostate cancer cells the constitutively overexpressed Bim protein is associated with the microtubules and the microtubule/vesicle-based, LC8-containing dynein motor complexes.
Together, the above results indicate that the overexpressed Bim in cancer cells is localized to various subcellular compartments and sequestered by microtubules and dynein motor complexes. These observations also raise the possibility that the constitutively up-regulated Bim may play a prosurvival role in cancer cells.
Down-regulation of Endogenous Bim by siRNA Induces Cell Detachment and Anoikis
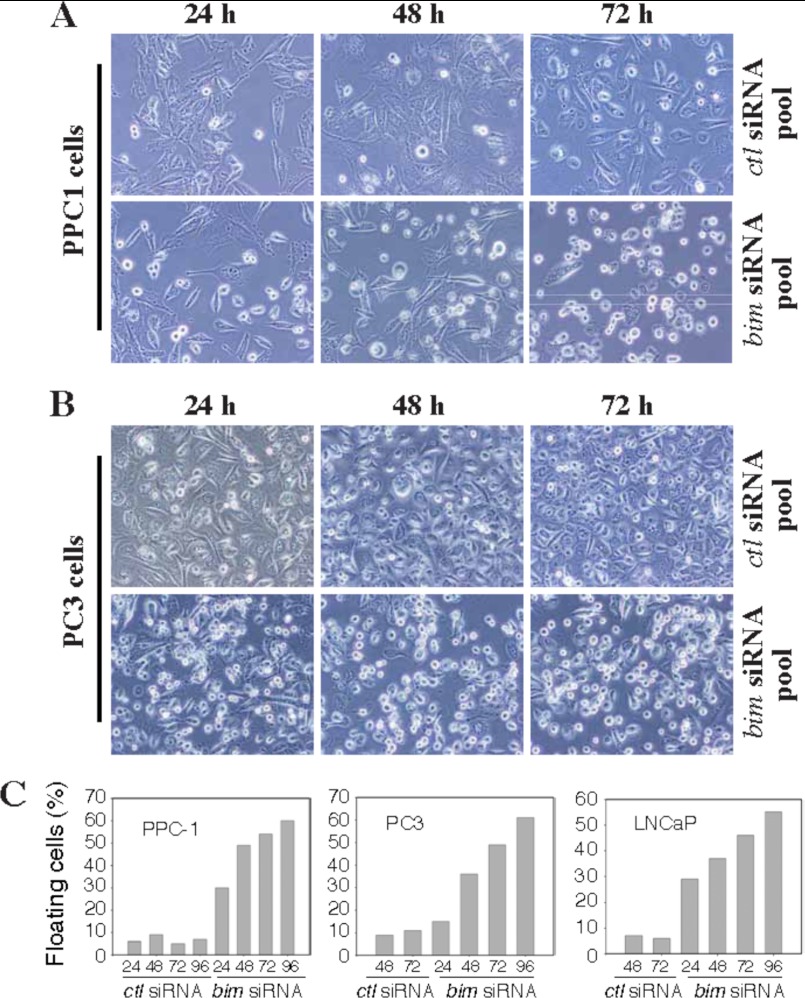
Having established that cancer cells constitutively up-regulate Bim in an E2F1-dependent manner, we began investigating the physiological function of up-regulated Bim in cancer cells by carrying out loss of functions experiments. PPC1, PC3, and LNCaP cells were transfected with a pool of four Bim siRNAs for different time intervals, which down-regulated Bim protein levels by ∼90% at 24 h and onward (data not shown). A nonspecific siRNA was used as control. Surprisingly, Bim silencing caused cells to detach and undergo apoptosis (Fig. 6). Although control siRNA did not affect cancer cells, in contrast Bim siRNA treatment led to detachment of a significant number of cells from the culture dishes (Fig. 6, A and B). Quantification of ∼500 PPC1, PC3, and LNCaP cells demonstrated that about 30–60% of cells were detached upon transfection with Bim siRNAs (Fig. 6C). Similarly, detachment of PPC1 cells was observed using an additional four siRNAs targeted to different regions of the Bim mRNA but not with their respective scrambled siRNA controls (data not shown).
FIGURE 6.
Down-regulation of Bim causes cell rounding and detachment. PPC1 (A) and PC3 (B) cells were transfected with control siRNA or Bim siRNA, and images were captured at multiple time periods. Original magnification, ×100. C, PPC1, PC3, and LNCaP cells were transfected with control or Bim siRNA, and floating cells (500 cells for each conditions) were counted at various time periods. Data are presented as percentage floating cells.
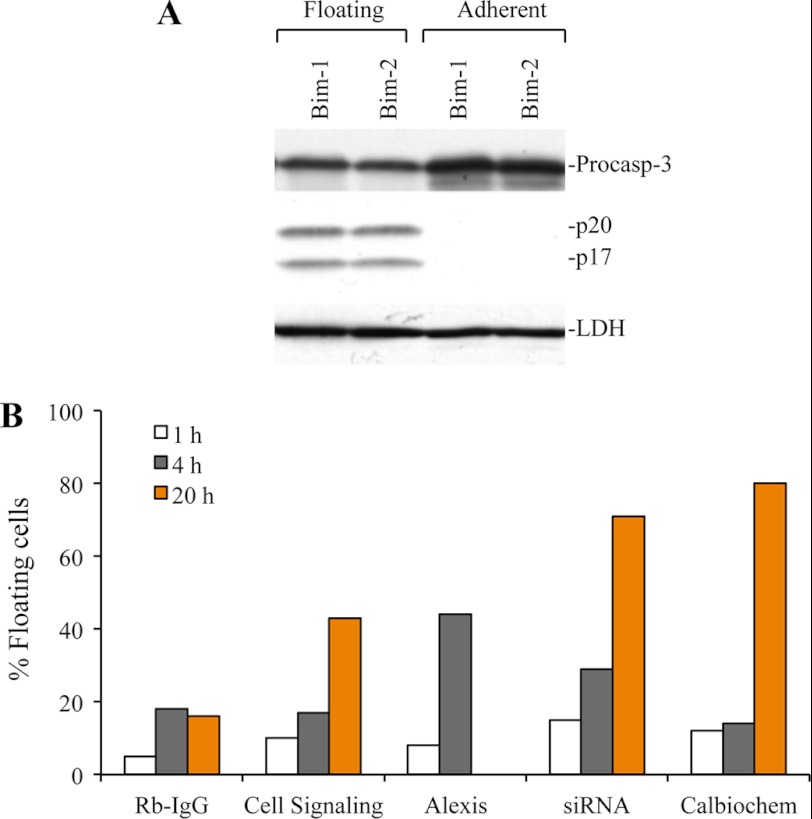
Because detachment of cells is known to induce apoptotic cell death, a phenomenon called anoikis (58, 59), we determined whether floating cells indeed underwent apoptosis. PPC1 and LNCaP cells were transfected with Bim siRNAs, and both floating and adherent cells were harvested and subjected to Western blotting for caspase-3. We observed caspase-3 processing only in floating cells but not in adherent cells (Fig. 7A). To confirm that inhibition of Bim expression and/or functions promote detachment and apoptosis, we microinjected cancer cells with three different anti-Bim antibodies with Rb-IgG as control. We used live cell labeling with DAPI to determine apoptotic cell death during microinjection (35). We observed that the microinjected anti-Bim antibodies, in a time-dependent manner, induced detachment of cells to varying degrees (Fig. 7B). We also microinjected Bim siRNAs, which also caused cell detachment and apoptosis (Fig. 7B).
FIGURE 7.
Bim silencing causes cell detachment and subsequent apoptosis. A, PPC1 cells were transfected with control or Bim siRNA, and after 48 h of transfection, floating and attached cells were collected. Whole cell lysates were subjected to Western blotting for caspase-3. LDH was used as loading control. B, PC3 cells were microinjected with multiple Bim antibodies or RbIgG antibody or Bim siRNA in the cytoplasm of cells. Cells were labeled live with DAPI, and floating cells were counted to monitor apoptosis after microinjection.
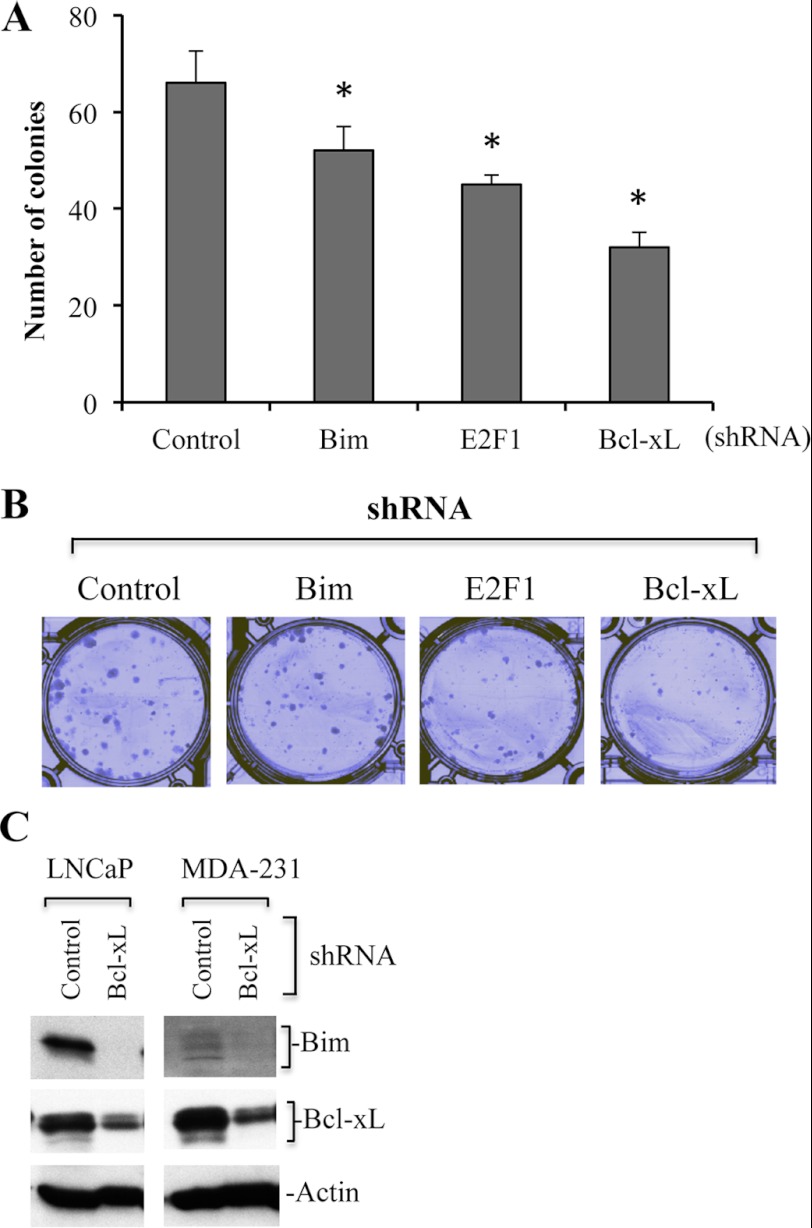
To further demonstrate that Bim is required for cancer cell survival, we performed clonogenic assays using Bim silenced MDA-MB231 cells. We observed that Bim knockdown significantly reduced the number of colonies (Fig. 8, A and B). Because Bim expression is regulated by E2F1, we also used E2F1-silenced MDA-MB231 cells for clonogenic assays. The results showed that E2F1 silencing reduced the number of colonies compared with control shRNA-infected cells (Fig. 8, A and B).
FIGURE 8.
Bim or E2F1 or Bcl-xL silencing inhibits colony formation of cancer cells. A and B, Bim or Bcl-xL or E2F1-silenced MDA-MB231 cancer cells were plated at a density of 500 cells/well in a 6-well plate. Medium was replaced with fresh culture medium every 72 h. When colonies became visible (∼2 weeks), cells were fixed and stained with Giemsa (1:10 in distilled water at room temperature) and counted. Representative images are shown in B. C, control or Bcl-xL shRNA LNCaP and MDA-MB231 cells were subjected to Western blotting to detect levels of Bim expression. Actin serves as loading control. Data are mean ± S.D., n = 3; *, p < 0.05.
Cancer Cells Express Phosphorylated Bim Protein and Mcl-1/Bcl-xL Sequesters Bim in Cancer Cells
We next performed several experiments to further explore the interrelationship between Bim with several antiapoptotic Bcl-2 family proteins in the context of nonapoptotic functions of Bim (Figs. 8 and 9). Immunoblotting studies in Fig. 1 (A and C) provided some clues as to why up-regulated Bim in cancer cells does not elicit proapoptotic function; perhaps a portion of Bim proteins is phosphorylated and/or sequestered by antiapoptotic Bcl-2 family proteins. It has been shown that phosphorylation of Bim at Ser-69 or Ser-87 promotes cancer cell survival (60–62). In support of the first conjecture, both phosphorylated forms of Bim (i.e. Ser-69 and Ser-87) were detected in prostate and breast cancer cells at higher levels than in the corresponding normal cells (Fig. 9A). Of interest, breast tumors from patients also expressed higher levels of Bim and phosphorylated Bim than the normal breast tissues (Breast-N; Fig. 9A).
FIGURE 9.
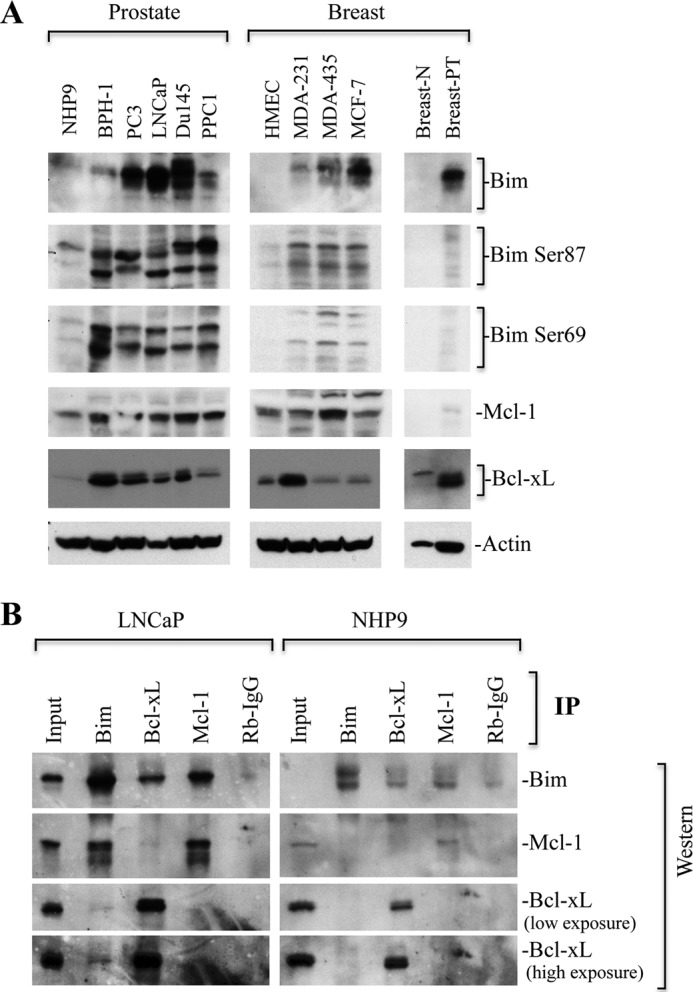
Bim phosphorylation is accompanied by overexpression of Bcl-xL/Mcl-1, and Bim is sequestered by Mcl-1 and Bcl-xL. A, whole cell lysates prepared from normal prostate NHP9 cells, BPH1, PC3, LNCaP, PPC1 cells; breast normal human mammary epithelial cells (HMEC), MDA-MB231 (MDA-231), MDA-MB435 (MDA-435), MCF-7 cells; breast normal tissue (breast-N), and breast primary tumor (breast-PT) were subjected to Western blotting for indicated proteins. Actin serves as loading control. B, equal amounts of protein from LNCaP prostate cancer and NHP-9 normal prostate epithelial cells were immunoprecipitated with anti-Bim or Bcl-xL or Mcl-1 antibodies along with Rb- or Ms-IgG controls. Western blot analysis for indicated proteins was performed.
To test the second conjecture, we first determined the levels of three prosurvival Bcl-2 family proteins Bcl-xL, Bcl-2, and Mcl-1. We observed increased levels of Bcl-xL and/or Mcl-1 in cancer cells as compared with noncancerous cells (Fig. 9A). However, we were unable to detect Bcl-2 in the cancer cells examined (data not shown). Because we observed high levels of Bcl-xL and Mcl-1 in cancer cells, we investigated whether up-regulated Bcl-xL and Mcl-1 might sequester Bim and neutralize its proapoptotic functions. Indeed, IP analysis revealed that a higher amount of Bim was precipitated in LNCaP cells by antibodies to Bcl-xL and Mcl-1 compared with NHP9 cells (Fig. 9B). Similarly, Bim IP also pulled down high levels of Mcl-1 as well as Bcl-xL, although to a lesser extent as compared with Mcl-1 (Fig. 9B).
To further correlate prosurvival function, we silenced Bcl-xL and observed that the number of colonies was highly reduced upon Bcl-xL silencing (Fig. 8, A and B). To demonstrate whether prosurvival function of Bim is associated with its interaction with Bcl-xL, we determined the level of Bim in Bcl-xL silenced cells and observed reduced/undetectable expression of Bim in Bcl-xL-silenced cells (Fig. 8C).
DISCUSSION
Pro-apoptotic BH3-only protein Bim is frequently induced upon treatment with anticancer therapeutics. However, Bim induction-based anticancer therapy has many limitations such as tumor cell selectivity or diversity of Bim controlling systems in cancer cells (63, 64). In this study, we first report that Bim is constitutively overexpressed both at protein and mRNA levels in various prostate and breast cancer cells. We further show multiple E2F1-binding sites on the Bim promoter, which are involved in Bim up-regulation in cancer cells. The overexpressed Bim localizes to the mitochondria and cytoskeleton. Importantly, down-regulation of Bim either by siRNA-mediated Bim silencing or by injecting anti-Bim antibodies results in cell rounding, substrate detachment, and subsequent apoptosis. Together, our findings demonstrate a surprising prosurvival function of Bim, which needs to be taken into consideration for future Bim-based cancer therapy.
How is Bim up-regulated in cancer cells? We observed that E2F1 is primarily responsible for higher expression of Bim in cancer cells. This conclusion is based on identification of 14 E2F1 “hot spots” by in silico analysis using E2F1 consensus sequence (KTSSCGC, where K = G or T and S = C or G). Previous reports identified four key E2F1-binding sites by genomic sequence analysis spanning −2.5 kb upstream of the TSS (49, 50). Our findings suggest the existence of multiple E2F1-binding sites up to −6 kb upstream of TSS. Promoter analysis using several cancer cell types demonstrates that these new E2F1-binding sites upstream of −2.6 kb are primarily responsible for Bim promoter activity, suggesting that the higher expression of Bim in cancer cells, but not in normal epithelial cells, is regulated by E2F1. This conclusion is further supported by the lack of Sp1 and FOXO3a binding to the −5.1-kb Bim promoter in cancer cells.
Although it has previously been shown that Bim protein expression could be regulated by FOXO3a (51) or Sp1 (65), our findings suggest that, in prostate cancer cells, E2F1 is primarily responsible for Bim overexpression. In support, E2F1 silencing causes reduced expression of Bim causing similar biological effects as observed upon Bim-silencing in cancer cells. For example, Bim or E2F1 silencing showed increased cell detachment and apoptosis (data not shown).
Bim has been traditionally considered to be one of the key proteins required for Bax activation, cytochrome c release, and apoptosis. The higher expression of Bim in cancer cells raises two key questions. First, why is the up-regulated Bim unable to induce apoptosis under normal (i.e. unstimulated) conditions? Second, what is the potential physiological function(s) of the up-regulated Bim in cancer cells? Multiple pieces of evidence suggest that the effect of Bim up-regulation could be nullified by higher expression of prosurvival proteins such as Bcl-xL and Mcl-1. For example, up-regulation of multiple isoforms of prosurvival protein Bcl-xL (66) and up-regulation of Mcl-1 suggest that Bim may be sequestered on the mitochondrial membrane by Bcl-xL and/or Mcl-1 and unable to perform proapoptotic function. Indeed, IP analysis clearly demonstrated that higher levels of Bim were sequestered by Bcl-xL and Mcl-1 in cancer cells as compared with normal noncancerous cells. Because Bim directly binds and activates proapoptotic multidomain proteins Bax/Bak (4, 5), a low level of Bax/Bak may be a limiting factor for the proapoptotic function of Bim. Sequestration of Bim by the microtubule-associated dynein motor complex or direct association with microtubule further supports that Bim may not be available in sufficient amounts to induce apoptotic cell death under unstimulated conditions. In addition, up-regulation of XIAP, which inhibits active caspase-9 and -3, suggests that Bim-mediated cytochrome c release and caspase activation may not be sufficient to induce apoptosis.
Our findings establish a prosurvival function of Bim in unstimulated cancer cells. How up-regulated Bim in cancer cells possesses prosurvival functions is not known, but various possibilities exist; for example, Bim overexpression in cancer cells may be associated with its polymorphism causing accumulation of the Bim isoform that lacks proapoptotic functions (67, 68). Although Bim associates with LC8, a component of the microtubule-associated dynein motor complex (12), Bim overexpression does not correlate with the expression of LC8 in cancer cells, suggesting that prosurvival function of Bim is not solely related to the association with LC8. The majority of Bim was observed in the mitochondrial fraction, which is consistent with the previous findings (66). Cell detachment from the tissue culture dishes and their subsequent apoptosis suggest that Bim may be playing prosurvival function on the mitochondrial membrane. Early up-regulation of Bim during stress or apoptotic stimulation represents proapoptotic function, but it is noteworthy that Bim up-regulation happens early during stress or apoptotic stimulation, whereas cytochrome c release and apoptosis occur at a later stage of apoptosis stimulation (27, 31, 69). Thus, early up-regulation of Bim could be a prosurvival mechanism, although it contributes to apoptotic cell death at later stages when prosurvival mechanisms have been compromised. These findings are consistent with the concept that various proapoptotic proteins may also possess nonapoptotic or prosurvival functions. For example, caspase-8, Fas, and FADD have been associated with increased proliferation and survival (70–72). Apaf-1, a key adaptor protein for caspase activation, might be involved in DNA damage checkpoint response (73). Proapoptotic multidomain proteins, Bax, Bak, and Bad, a BH3-only pro-apoptotic protein, also play an important role in cell survival (15, 17, 19).
How does Bim silencing in unstimulated cancer cells cause cell death? Although the exact underlying mechanisms remain to be elucidated, our findings suggest that the primary response to Bim knockdown in unstimulated cancer cells is cell rounding and detachment rather cell death. In other words, our data suggest that cell death is anoikis consequent to cell detachment. A plausible explanation here is that the constitutively overexpressed Bim protein in cancer cells, by associating with microtubules, mitochondria, and perhaps other cytoskeleton elements and by participating in LC8-containing dynein motor functions, plays a critical role in regulating the overall integrity and cellular shape and adhesion. Interaction of mitochondria with microtubules or cytoskeleton networks regulates mitochondrial respiration and apoptosis, and microtubule-based motor proteins such as dynein and kinesin drive mitochondrial movement on the microtubules (74). Therefore, lack of Bim in unstressed cancer cells would destabilize the cytostructure and dynein motor complexes causing cell detachment (i.e. anoikis) and mitochondria dysfunction causing ultimate apoptosis.
Phosphorylation of Bim at Ser-69 by ERK1/2 (60, 61) and at Ser-87 by Akt promotes cancer cell survival (62). Therefore, Bim silencing will lead to the removal of prosurvival phosphorylated Bim causing apoptosis induction in cancer cells. Both phosphorylated and unphosphorylated forms of Bim are constitutively overexpressed in cancer cells. The unphosphorylated form of Bim has high affinity to the dynein LC8, thus remains sequestered by microtubules (14). In contrast, phosphorylated forms of Bim are freely available, which possess prosurvival functions. Based on these, we suggest that Bim silencing may also reduce the levels of prosurvival phosphorylated Bim, contributing to cell death.
Although Mcl-1 degradation and Bim up-regulation are associated with anoikis and apoptosis (75), Bim interacts with and stabilizes Mcl-1, which contribute to cancer cell survival (76). Therefore, another scenario is that silencing of Bim may promote Mcl-1 degradation, causing induction of apoptosis. Because Bim interacts with Mcl-1 and Bcl-xL, and Bcl-xL silencing causes reduced expression of Bim, it is possible that Bim may also perform prosurvival function in association with Mcl-1 and Bcl-xL.
The prosurvival functions associated with multiple proapoptotic proteins raise the possibility that anticancer agents that only harness the proapoptotic function of Bim or other BH3-only proteins may not be an efficient approach for the permeabilization of mitochondria. Although it is possible that down-regulation or silencing of Bim might have novel unknown targets promoting detachment and apoptosis, our findings demonstrate that to induce efficient apoptosis, inhibition of prosurvival function of Bim may provide a new approach for efficient cancer therapy.
Acknowledgment
We thank John Chun for helping on in silico analysis of the Bim promoter.
This work was supported, in whole or in part, by National Institutes of Health K01 Award CA123142 (to D. C.) and Grants R1-ES015888 and 1R21CA 150009 (to D. G. T.) and Support Grant P30 CA016056 from NCI (to Roswell Park Cancer Institute). This work was also supported by Department of Defense Grant W81XWH-11-1-0331 (to D. G. T.).
- BH3
- Bcl-2 homology domain 3
- Bcl-2
- B-cell lymphoma 2
- Mcl-1
- myeloid cell leukemia sequence 1
- XIAP
- X-linked inhibitor of apoptosis
- NHP
- normal human prostate
- BPH
- benign prostate hyperplasia
- E2F1
- E2F transcription factor 1
- FOXO3a
- forkhead box O3a
- LDH
- lactate dehydrogenase
- COX II
- cytochrome c oxidase subunit II
- ER
- endoplasmic reticulum
- LM
- light mitochondria
- HM
- heavy mitochondria
- CAP
- cytoskeleton-associated protein
- TSS
- translation start site
- IP
- immunoprecipitation.
REFERENCES
- 1. Westphal D., Dewson G., Czabotar P. E., Kluck R. M. (2011) Molecular biology of Bax and Bak activation and action. Biochim. Biophys. Acta 1813, 521–531 [DOI] [PubMed] [Google Scholar]
- 2. Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 3. Czabotar P. E., Colman P. M., Huang D. C. (2009) Bax activation by Bim? Cell Death Differ. 16, 1187–1191 [DOI] [PubMed] [Google Scholar]
- 4. Gavathiotis E., Reyna D. E., Davis M. L., Bird G. H., Walensky L. D. (2010) BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol. Cell 40, 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mérino D., Giam M., Hughes P. D., Siggs O. M., Heger K., O'Reilly L. A., Adams J. M., Strasser A., Lee E. F., Fairlie W. D., Bouillet P. (2009) The role of BH3-only protein Bim extends beyond inhibiting Bcl-2-like prosurvival proteins. J. Cell Biol. 186, 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O'Connor L., Strasser A., O'Reilly L. A., Hausmann G., Adams J. M., Cory S., Huang D. C. (1998) Bim. A novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 17, 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hsu S. Y., Lin P., Hsueh A. J. (1998) BOD (Bcl-2-related ovarian death gene) is an ovarian BH3 domain-containing proapoptotic Bcl-2 protein capable of dimerization with diverse antiapoptotic Bcl-2 members. Mol. Endocrinol. 12, 1432–1440 [DOI] [PubMed] [Google Scholar]
- 8. U M., Miyashita T., Shikama Y., Tadokoro K., Yamada M. (2001) Molecular cloning and characterization of six novel isoforms of human Bim, a member of the proapoptotic Bcl-2 family. FEBS Lett. 509, 135–141 [DOI] [PubMed] [Google Scholar]
- 9. Marani M., Tenev T., Hancock D., Downward J., Lemoine N. R. (2002) Identification of novel isoforms of the BH3 domain protein Bim, which directly activate Bax to trigger apoptosis. Mol. Cell. Biol. 22, 3577–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu J. W., Chandra D., Tang S. H., Chopra D., Tang D. G. (2002) Identification and characterization of Bimγ, a novel proapoptotic BH3-only splice variant of Bim. Cancer Res. 62, 2976–2981 [PubMed] [Google Scholar]
- 11. Bouillet P., Metcalf D., Huang D. C., Tarlinton D. M., Kay T. W., Köntgen F., Adams J. M., Strasser A. (1999) Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286, 1735–1738 [DOI] [PubMed] [Google Scholar]
- 12. Puthalakath H., Huang D. C., O'Reilly L. A., King S. M., Strasser A. (1999) The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol. Cell 3, 287–296 [DOI] [PubMed] [Google Scholar]
- 13. Jaffrey S. R., Snyder S. H. (1996) PIN. An associated protein inhibitor of neuronal nitric oxide synthase. Science 274, 774–777 [DOI] [PubMed] [Google Scholar]
- 14. Chen D., Zhou Q. (2004) Caspase cleavage of BimEL triggers a positive feedback amplification of apoptotic signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 1235–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lewis J., Oyler G. A., Ueno K., Fannjiang Y. R., Chau B. N., Vornov J., Korsmeyer S. J., Zou S., Hardwick J. M. (1999) Inhibition of virus-induced neuronal apoptosis by Bax. Nat. Med. 5, 832–835 [DOI] [PubMed] [Google Scholar]
- 16. Kerr D. A., Larsen T., Cook S. H., Fannjiang Y. R., Choi E., Griffin D. E., Hardwick J. M., Irani D. N. (2002) BCL-2 and BAX protect adult mice from lethal Sindbis virus infection but do not protect spinal cord motor neurons or prevent paralysis. J. Virol. 76, 10393–10400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fannjiang Y., Kim C. H., Huganir R. L., Zou S., Lindsten T., Thompson C. B., Mito T., Traystman R. J., Larsen T., Griffin D. E., Mandir A. S., Dawson T. M., Dike S., Sappington A. L., Kerr D. A., Jonas E. A., Kaczmarek L. K., Hardwick J. M. (2003) BAK alters neuronal excitability and can switch from anti- to pro-death function during postnatal development. Dev. Cell 4, 575–585 [DOI] [PubMed] [Google Scholar]
- 18. Yeretssian G., Correa R. G., Doiron K., Fitzgerald P., Dillon C. P., Green D. R., Reed J. C., Saleh M. (2011) Nonapoptotic role of BID in inflammation and innate immunity. Nature 474, 96–99 [DOI] [PubMed] [Google Scholar]
- 19. Seo S. Y., Chen Y. B., Ivanovska I., Ranger A. M., Hong S. J., Dawson V. L., Korsmeyer S. J., Bellows D. S., Fannjiang Y., Hardwick J. M. (2004) BAD is a pro-survival factor prior to activation of its pro-apoptotic function. J. Biol. Chem. 279, 42240–42249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu W., Swetzig W. M., Medisetty R., Das G. M. (2011) Estrogen-mediated up-regulation of Noxa is associated with cell cycle progression in estrogen receptor-positive breast cancer cells. PLoS ONE 6, e29466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Reilly L. A., Cullen L., Visvader J., Lindeman G. J., Print C., Bath M. L., Huang D. C., Strasser A. (2000) The proapoptotic BH3-only protein Bim is expressed in hematopoietic, epithelial, neuronal, and germ cells. Am. J. Pathol. 157, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bouillet P., Cory S., Zhang L. C., Strasser A., Adams J. M. (2001) Degenerative disorders caused by Bcl-2 deficiency prevented by loss of its BH3-only antagonist Bim. Dev. Cell 1, 645–653 [DOI] [PubMed] [Google Scholar]
- 23. Bouillet P., Purton J. F., Godfrey D. I., Zhang L. C., Coultas L., Puthalakath H., Pellegrini M., Cory S., Adams J. M., Strasser A. (2002) BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature 415, 922–926 [DOI] [PubMed] [Google Scholar]
- 24. Davey G. M., Kurts C., Miller J. F., Bouillet P., Strasser A., Brooks A. G., Carbone F. R., Heath W. R. (2002) Peripheral deletion of autoreactive CD8 T cells by cross-presentation of self-antigen occurs by a Bcl-2-inhibitable pathway mediated by Bim. J. Exp. Med. 196, 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Villunger A., Scott C., Bouillet P., Strasser A. (2003) Essential role for the BH3-only protein Bim but redundant roles for Bax, Bcl-2, and Bcl-w in the control of granulocyte survival. Blood 101, 2393–2400 [DOI] [PubMed] [Google Scholar]
- 26. Enders A., Bouillet P., Puthalakath H., Xu Y., Tarlinton D. M., Strasser A. (2003) Loss of the pro-apoptotic BH3-only Bcl-2 family member Bim inhibits BCR stimulation-induced apoptosis and deletion of autoreactive B cells. J. Exp. Med. 198, 1119–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu J. W., Chandra D., Rudd M. D., Butler A. P., Pallotta V., Brown D., Coffer P. J., Tang D. G. (2005) Induction of prosurvival molecules by apoptotic stimuli. Involvement of FOXO3a and ROS. Oncogene 24, 2020–2031 [DOI] [PubMed] [Google Scholar]
- 28. Madden D. T., Davila-Kruger D., Melov S., Bredesen D. E. (2011) Human embryonic stem cells express elevated levels of multiple pro-apoptotic BCL-2 family members. PLoS ONE 6, e28530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gogada R., Amadori M., Zhang H., Jones A., Verone A., Pitarresi J., Jandhyam S., Prabhu V., Black J. D., Chandra D. (2011) Curcumin induces Apaf-1-dependent, p21-mediated caspase activation and apoptosis. Cell Cycle 10, 4128–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gogada R., Prabhu V., Amadori M., Scott R., Hashmi S., Chandra D. (2011) Resveratrol induces p53-independent, X-linked inhibitor of apoptosis protein (XIAP)-mediated Bax protein oligomerization on mitochondria to initiate cytochrome c release and caspase activation. J. Biol. Chem. 286, 28749–28760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chandra D., Liu J. W., Tang D. G. (2002) Early mitochondrial activation and cytochrome c up-regulation during apoptosis. J. Biol. Chem. 277, 50842–50854 [DOI] [PubMed] [Google Scholar]
- 32. Zhang H., Gogada R., Yadav N., Lella R. K., Badeaux M., Ayres M., Gandhi V., Tang D. G., Chandra D. (2011) Defective molecular timer in the absence of nucleotides leads to inefficient caspase activation. PLoS ONE 6, e16379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chandra D., Choy G., Deng X., Bhatia B., Daniel P., Tang D. G. (2004) Association of active caspase 8 with the mitochondrial membrane during apoptosis. Potential roles in cleaving BAP31 and caspase 3 and mediating mitochondrion-endoplasmic reticulum cross-talk in etoposide-induced cell death. Mol. Cell. Biol. 24, 6592–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu J. W., Shen J. J., Tanzillo-Swarts A., Bhatia B., Maldonado C. M., Person M. D., Lau S. S., Tang D. G. (2003) Annexin II expression is reduced or lost in prostate cancer cells and its re-expression inhibits prostate cancer cell migration. Oncogene 22, 1475–1485 [DOI] [PubMed] [Google Scholar]
- 35. Chandra D., Bratton S. B., Person M. D., Tian Y., Martin A. G., Ayres M., Fearnhead H. O., Gandhi V., Tang D. G. (2006) Intracellular nucleotides act as critical prosurvival factors by binding to cytochrome c and inhibiting apoptosome. Cell 125, 1333–1346 [DOI] [PubMed] [Google Scholar]
- 36. Jeter C. R., Liu B., Liu X., Chen X., Liu C., Calhoun-Davis T., Repass J., Zaehres H., Shen J. J., Tang D. G. (2011) NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Oncogene 30, 3833–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chandra D., Choy G., Tang D. G. (2007) Cytosolic accumulation of HSP60 during apoptosis with or without apparent mitochondrial release: evidence that its pro-apoptotic or pro-survival functions involve differential interactions with caspase-3. J. Biol. Chem. 282, 31289–31301 [DOI] [PubMed] [Google Scholar]
- 38. Tang S., Bhatia B., Maldonado C. J., Yang P., Newman R. A., Liu J., Chandra D., Traag J., Klein R. D., Fischer S. M., Chopra D., Shen J., Zhau H. E., Chung L. W., Tang D. G. (2002) Evidence that arachidonate 15-lipoxygenase 2 is a negative cell cycle regulator in normal prostate epithelial cells. J. Biol. Chem. 277, 16189–16201 [DOI] [PubMed] [Google Scholar]
- 39. Bhatia B., Maldonado C. J., Tang S., Chandra D., Klein R. D., Chopra D., Shappell S. B., Yang P., Newman R. A., Tang D. G. (2003) Subcellular localization and tumor-suppressive functions of 15-lipoxygenase 2 (15-LOX2) and its splice variants. J. Biol. Chem. 278, 25091–25100 [DOI] [PubMed] [Google Scholar]
- 40. Biswas S. C., Greene L. A. (2002) Nerve growth factor (NGF) down-regulates the Bcl-2 homology 3 (BH3) domain-only protein Bim and suppresses its proapoptotic activity by phosphorylation. J. Biol. Chem. 277, 49511–49516 [DOI] [PubMed] [Google Scholar]
- 41. Seward R. J., von Haller P. D., Aebersold R., Huber B. T. (2003) Phosphorylation of the pro-apoptotic protein Bim in lymphocytes is associated with protection from apoptosis. Mol. Immunol. 39, 983–993 [DOI] [PubMed] [Google Scholar]
- 42. Tang D. G., Li L., Chopra D. P., Porter A. T. (1998) Extended survivability of prostate cancer cells in the absence of trophic factors: increased proliferation, evasion of apoptosis, and the role of apoptosis proteins. Cancer Res. 58, 3466–3479 [PubMed] [Google Scholar]
- 43. Huang Y., Park Y. C., Rich R. L., Segal D., Myszka D. G., Wu H. (2001) Structural basis of caspase inhibition by XIAP: differential roles of the linker versus the BIR domain. Cell 104, 781–790 [PubMed] [Google Scholar]
- 44. Riedl S. J., Renatus M., Schwarzenbacher R., Zhou Q., Sun C., Fesik S. W., Liddington R. C., Salvesen G. S. (2001) Structural basis for the inhibition of caspase-3 by XIAP. Cell 104, 791–800 [DOI] [PubMed] [Google Scholar]
- 45. Srinivasula S. M., Hegde R., Saleh A., Datta P., Shiozaki E., Chai J., Lee R. A., Robbins P. D., Fernandes-Alnemri T., Shi Y., Alnemri E. S. (2001) A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410, 112–116 [DOI] [PubMed] [Google Scholar]
- 46. Shiozaki E. N., Chai J., Rigotti D. J., Riedl S. J., Li P., Srinivasula S. M., Alnemri E. S., Fairman R., Shi Y. (2003) Mechanism of XIAP-mediated inhibition of caspase-9. Mol. Cell 11, 519–527 [DOI] [PubMed] [Google Scholar]
- 47. Zhang S. Y., Liu S. C., Al-Saleem L. F., Holloran D., Babb J., Guo X., Klein-Szanto A. J. (2000) E2F-1. A proliferative marker of breast neoplasia. Cancer Epidemiol. Biomarkers Prev. 9, 395–401 [PubMed] [Google Scholar]
- 48. Gorgoulis V. G., Zacharatos P., Mariatos G., Kotsinas A., Bouda M., Kletsas D., Asimacopoulos P. J., Agnantis N., Kittas C., Papavassiliou A. G. (2002) Transcription factor E2F-1 acts as a growth-promoting factor and is associated with adverse prognosis in non-small cell lung carcinomas. J. Pathol. 198, 142–156 [DOI] [PubMed] [Google Scholar]
- 49. Hershko T., Ginsberg D. (2004) Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J. Biol. Chem. 279, 8627–8634 [DOI] [PubMed] [Google Scholar]
- 50. Zhao Y., Tan J., Zhuang L., Jiang X., Liu E. T., Yu Q. (2005) Inhibitors of histone deacetylases target the Rb-E2F1 pathway for apoptosis induction through activation of proapoptotic protein Bim. Proc. Natl. Acad. Sci. U.S.A. 102, 16090–16095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gilley J., Coffer P. J., Ham J. (2003) FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J. Cell Biol. 162, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hirokawa N. (1998) Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279, 519–526 [DOI] [PubMed] [Google Scholar]
- 53. King S. M. (2000) AAA domains and organization of the dynein motor unit. J. Cell Sci. 113, 2521–2526 [DOI] [PubMed] [Google Scholar]
- 54. King S. M., Barbarese E., Dillman J. F., 3rd, Patel-King R. S., Carson J. H., Pfister K. K. (1996) Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J. Biol. Chem. 271, 19358–19366 [DOI] [PubMed] [Google Scholar]
- 55. Rapali P., Radnai L., Süveges D., Harmat V., Tölgyesi F., Wahlgren W. Y., Katona G., Nyitray L., Pál G. (2011) Directed evolution reveals the binding motif preference of the LC8/DYNLL hub protein and predicts large numbers of novel binders in the human proteome. PLoS ONE 6, e18818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Navarro-Lérida I., Martínez Moreno M., Roncal F., Gavilanes F., Albar J. P., Rodríguez-Crespo I. (2004) Proteomic identification of brain proteins that interact with dynein light chain LC8. Proteomics 4, 339–346 [DOI] [PubMed] [Google Scholar]
- 57. Rodríguez-Crespo I., Yélamos B., Roncal F., Albar J. P., Ortiz de Montellano P. R., Gavilanes F. (2001) Identification of novel cellular proteins that bind to the LC8 dynein light chain using a pepscan technique. FEBS Lett. 503, 135–141 [DOI] [PubMed] [Google Scholar]
- 58. Grossmann J. (2002) Molecular mechanisms of “detachment-induced apoptosis–Anoikis.” Apoptosis 7, 247–260 [DOI] [PubMed] [Google Scholar]
- 59. Rytömaa M., Martins L. M., Downward J. (1999) Involvement of FADD and caspase-8 signaling in detachment-induced apoptosis. Curr. Biol. 9, 1043–1046 [DOI] [PubMed] [Google Scholar]
- 60. Paterson A., Mockridge C. I., Adams J. E., Krysov S., Potter K. N., Duncombe A. S., Cook S. J., Stevenson F. K., Packham G. (2012) Mechanisms and clinical significance of BIM phosphorylation in chronic lymphocytic leukemia. Blood 119, 1726–1736 [DOI] [PubMed] [Google Scholar]
- 61. Luciano F., Jacquel A., Colosetti P., Herrant M., Cagnol S., Pages G., Auberger P. (2003) Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene 22, 6785–6793 [DOI] [PubMed] [Google Scholar]
- 62. Qi X. J., Wildey G. M., Howe P. H. (2006) Evidence that Ser87 of BimEL is phosphorylated by Akt and regulates BimEL apoptotic function. J. Biol. Chem. 281, 813–823 [DOI] [PubMed] [Google Scholar]
- 63. Deng J., Carlson N., Takeyama K., Dal Cin P., Shipp M., Letai A. (2007) BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell 12, 171–185 [DOI] [PubMed] [Google Scholar]
- 64. Akiyama T., Dass C. R., Choong P. F. (2009) Bim-targeted cancer therapy. A link between drug action and underlying molecular changes. Mol. Cancer Ther. 8, 3173–3180 [DOI] [PubMed] [Google Scholar]
- 65. Moore P. S., Barbi S., Donadelli M., Costanzo C., Bassi C., Palmieri M., Scarpa A. (2004) Gene expression profiling after treatment with the histone deacetylase inhibitor trichostatin A reveals altered expression of both pro- and anti-apoptotic genes in pancreatic adenocarcinoma cells. Biochim. Biophys. Acta 1693, 167–176 [DOI] [PubMed] [Google Scholar]
- 66. Zhu Y., Swanson B. J., Wang M., Hildeman D. A., Schaefer B. C., Liu X., Suzuki H., Mihara K., Kappler J., Marrack P. (2004) Constitutive association of the proapoptotic protein Bim with Bcl-2-related proteins on mitochondria in T cells. Proc. Natl. Acad. Sci. U.S.A. 101, 7681–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ng K. P., Hillmer A. M., Chuah C. T., Juan W. C., Ko T. K., Teo A. S., Ariyaratne P. N., Takahashi N., Sawada K., Fei Y., Soh S., Lee W. H., Huang J. W., Allen J. C., Jr., Woo X. Y., Nagarajan N., Kumar V., Thalamuthu A., Poh W. T., Ang A. L., Mya H. T., How G. F., Yang L. Y., Koh L. P., Chowbay B., Chang C. T., Nadarajan V. S., Chng W. J., Than H., Lim L. C., Goh Y. T., Zhang S., Poh D., Tan P., Seet J. E., Ang M. K., Chau N. M., Ng Q. S., Tan D. S., Soda M., Isobe K., Nöthen M. M., Wong T. Y., Shahab A., Ruan X., Cacheux-Rataboul V., Sung W. K., Tan E. H., Yatabe Y., Mano H., Soo R. A., Chin T. M., Lim W. T., Ruan Y., Ong S. T. (2012) A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat. Med. 18, 521–528 [DOI] [PubMed] [Google Scholar]
- 68. Cheng E. H., Sawyers C. L. (2012) In cancer drug resistance, germ line matters too. Nat. Med. 18, 494–496 [DOI] [PubMed] [Google Scholar]
- 69. Ramjaun A. R., Tomlinson S., Eddaoudi A., Downward J. (2007) Up-regulation of two BH3-only proteins, Bmf and Bim, during TGFβ-induced apoptosis. Oncogene 26, 970–981 [DOI] [PubMed] [Google Scholar]
- 70. Su H., Bidère N., Zheng L., Cubre A., Sakai K., Dale J., Salmena L., Hakem R., Straus S., Lenardo M. (2005) Requirement for caspase-8 in NF-κB activation by antigen receptor. Science 307, 1465–1468 [DOI] [PubMed] [Google Scholar]
- 71. Hu W. H., Johnson H., Shu H. B. (2000) Activation of NF-κB by FADD, Casper, and caspase-8. J. Biol. Chem. 275, 10838–10844 [DOI] [PubMed] [Google Scholar]
- 72. Chen L., Park S. M., Tumanov A. V., Hau A., Sawada K., Feig C., Turner J. R., Fu Y. X., Romero I. L., Lengyel E., Peter M. E. (2010) CD95 promotes tumour growth. Nature 465, 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zermati Y., Mouhamad S., Stergiou L., Besse B., Galluzzi L., Boehrer S., Pauleau A. L., Rosselli F., D'Amelio M., Amendola R., Castedo M., Hengartner M., Soria J. C., Cecconi F., Kroemer G. (2007) Nonapoptotic role for Apaf-1 in the DNA damage checkpoint. Mol. Cell 28, 624–637 [DOI] [PubMed] [Google Scholar]
- 74. Boldogh I. R., Pon L. A. (2007) Mitochondria on the move. Trends Cell Biol. 17, 502–510 [DOI] [PubMed] [Google Scholar]
- 75. Woods N. T., Yamaguchi H., Lee F. Y., Bhalla K. N., Wang H. G. (2007) Anoikis, initiated by Mcl-1 degradation and Bim induction, is deregulated during oncogenesis. Cancer Res. 67, 10744–10752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Czabotar P. E., Lee E. F., van Delft M. F., Day C. L., Smith B. J., Huang D. C., Fairlie W. D., Hinds M. G., Colman P. M. (2007) Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc. Natl. Acad. Sci. U.S.A. 104, 6217–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]