Background: Cardiolipin is required for in vivo respiratory supercomplex formation in Saccharomyces cerevisiae.
Results: Supercomplex III2IV2 reconstitution from purified complexes III and IV was dependent on addition of cardiolipin over their tightly bound amounts. Electron microscopy confirmed supercomplex organization.
Conclusion: Supercomplex III2IV2 formation is absolutely contingent on cardiolipin presence in the membrane.
Significance: This minimal system provides understanding of lipid-dependent supercomplex dynamics in vivo.
Keywords: Cardiolipin, Electron Microscopy (EM), Mitochondria, Respiratory Chain, Yeast, Saccharomyces cerevisiae, Complex III, Complex IV, In Vitro Reconstitution, Supercomplex
Abstract
Here, we report for the first time in vitro reconstitution of the respiratory supercomplexes from individual complexes III and IV. Complexes III and IV were purified from Saccharomyces cerevisiae mitochondria. Complex III contained eight molecules of cardiolipin, and complex IV contained two molecules of cardiolipin, as determined by electrospray ionization-mass spectrometry. Complex IV also contained Rcf1p. No supercomplexes were formed upon mixing of the purified complexes, and low amounts of the supercomplex trimer III2IV1 were formed after reconstitution into proteoliposomes containing only phosphatidylcholine and phosphatidylethanolamine. Further addition of cardiolipin to the proteoliposome reconstitution mixture resulted in distinct formation of both the III2IV1 supercomplex trimer and III2IV2 supercomplex tetramer. No other anionic phospholipid was as effective as cardiolipin in supporting tetramer formation. Phospholipase treatment of complex IV prevented trimer formation in the absence of cardiolipin. Both trimer and tetramer formations were restored by cardiolipin. Analysis of the reconstituted tetramer by single particle electron microscopy confirmed native organization of individual complexes within the supercomplex. In conclusion, although some trimer formation occurred dependent only on tightly bound cardiolipin, tetramer formation required additional cardiolipin. This is consistent with the high cardiolipin content in the native tetramer. The dependence on cardiolipin for supercomplex formation suggests that changes in cardiolipin levels resulting from changes in physiological conditions may control the equilibrium between individual respiratory complexes and supercomplexes in vivo.
Introduction
It is accepted that the mitochondrial respiratory chain in mammalian cells is organized as a supramolecular structure (“respirasome” (1)) composed of individual redox complexes. Complexes I, III,3 and IV (NADH:ubiquinone oxidoreductase, ubiquinol:cytochrome c oxidoreductase (CIII),4 and cytochrome c oxidase (CIV), respectively) in different stoichiometric ratios form active supercomplexes, which after extraction from the membrane by mild detergents can be separated and detected by blue native-PAGE (BN-PAGE) (2–6). This is consistent with the suggestion that the respirasome is a dynamic assembly, the aggregation states of which can respond to variations in the demand for energy under different physiological conditions (7, 8). Electron transfer in the respiratory chain occurs either by direct substrate channeling of CoQ and cytochrome c between the respective individual complexes within a supercomplex or less efficiently via diffusion and random collision of the two low molecular weight electron carriers with the individual dissociated complexes (for review see Refs. 2, 3).
The Saccharomyces cerevisiae respiratory chain lacks the energy-transducing transmembrane complex I (9). Instead S. cerevisiae has several peripheral NADH dehydrogenases, which funnel electrons into the respiratory chain. Solubilization of the S. cerevisiae mitochondrial membrane with mild detergents such as digitonin produces supercomplexes III2IV1 and III2IV2 lacking the dehydrogenases as observed with BN-PAGE (4).
Supercomplex formation strongly depends on membrane phospholipid composition being most dependent on cardiolipin (CL), which is uniquely found in mitochondrial and other energy-transducing membranes (for review see Ref. 10). Reduced CL levels or alterations in the landscape of CL species with respect to fatty acid composition (generally loss of CL species with a high content of unsaturated fatty acids) results in reduced formation of both individual complexes and supercomplexes in a number of pathological states as follows: aging (11), neurodegenerative diseases (12), ischemia followed by reperfusion, induction of apoptosis (13–15), heart failure (16–18), cancer (19), and Barth syndrome (male-inherited defect in TAZ1 necessary for remodeling newly synthesized CL to its highly unsaturated forms) (10, 12, 20–23). Oxidative stress accompanied by lipid peroxidation particularly of CL occurs in the above diseases resulting in reduced supercomplex formation (3, 12). The failure to couple respiratory complexes further increases oxidative stress setting up a vicious cycle leading to reduced energy production and cell death. Thus, the study of the exact molecular mechanism by which CL supports supercomplex formation and stability will provide fundamental information mandatory to understanding of pathological alterations in mitochondrial metabolism.
We previously demonstrated that yeast mutants (Δcrd1) lacking CL failed to display the native III2IV2 supercomplex on BN-PAGE after extraction of mitochondria by digitonin (24); trace amounts of the III2IV1 trimer form of the supercomplex were present. Kinetic analysis of electron transfer from reduced CoQ to O2 is consistent with channeling of cytochrome c between CIII and CIV in intact wild type yeast mitochondria (25, 26). However, in mitochondria lacking CL, the kinetics of electron transfer supports random collision between respiratory complexes and the low molecular weight carriers consistent with a lack of supercomplex formation (26). Mutations in the TAZ gene in yeast and somatic cells (Barth syndrome in humans) result in a dramatic broadening in the spectrum of CL species with respect to fatty acid composition accompanied by reduction in supercomplex formation (21, 22). In Barth syndrome patients, the disruption in the interaction of CIII with CIV is more pronounced than complex I with CIII (20). In S. cerevisiae, site-directed mutagenesis demonstrated that CL molecules tightly bound to a specific site on CIII were important for stabilization of the interactions between CIII and CIV in the supercomplex (27). Therefore, CL appears to play a central role in supercomplex formation.
Recent structural studies of the purified mammalian and yeast respiratory chain supercomplexes by single particle cryo-electron microscopy (cryo-EM) (28, 29) and cryo-electron tomography (30) resulted in three-dimensional density maps and construction of pseudoatomic models for these structures showing specific arrangements of the individual complexes in the supercomplexes. One of the interesting features in the arrangement of the individual complexes in the respirasomes revealed in these studies is the presence of spaces between transmembrane domains of individual complexes, which may be filled with lipids. Consistent with these observations, about 200 CL molecules were estimated to be present in the purified bovine respirasome (I1III2IV1) (28), and about 50 CL molecules were determined in the III2IV2 supercomplex from S. cerevisiae (29). In each case, this level of CL is in great excess over the amount of CL tightly associated and integrated into the structure of the individual purified complexes.
In this study, we employed a minimal system composed of purified CIII and CIV from S. cerevisiae mitochondria and liposomes of different phospholipid composition to study the dependence on CL for supercomplex formation. We demonstrate for the first time the reconstitution of the supercomplexes III2IV1 and III2IV2 from individual CIII and CIV in proteoliposomes and specific dependence of III2IV2 formation on liposomes containing CL in strong preference over other anionic phospholipids. Using EM and single particle analysis, we demonstrated the proper structural arrangement of CIII and CIV within the III2IV2 supercomplex reconstituted in and purified from the proteoliposomes. Thus, CL is essential for supercomplex formation in addition to its occurrence as an integral part of individual CIII and CIV.
EXPERIMENTAL PROCEDURES
Reagents
The following synthetic phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL) and contained oleic acid as the sole fatty acid: phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidic acid (PA), phosphatidylglycerol (PG), phosphatidylserine (PS), and CL. 1,1′,2,2′-Tetramyristoyl-CL (CL (14:0)4) and 1,1′,2,2′-tetraolyoyl-CL (CL (18:1)4) were also from Avanti Polar Lipids. l-α-Phosphatidylinositol (PI) from soybean, horse heart cytochrome c, bee venom phospholipase A2, and diaminobenzidine were from Sigma. Polyclonal antibody specific for Rfc1p was a generous gift of Rosemary Stuart (Marquette University). Pre-made gels for BN-PAGE were purchased from Invitrogen. Bio-Safe Coomassie G-250 was from Bio-Rad. PVDF membranes were from Millipore. Monoclonal antibody toward the Qcr7p subunit of CIII was a generous gift from Bernard Trumpower, Dartmouth University. A polyclonal antibody to CIII or CIV was obtained from Cocalico Biologicals. Antibody recognizing the polyhistidine tag (THETM) was obtained from Genscript.
Yeast Strains and Growth Conditions
For protein production, an S. cerevisiae strain was engineered to express the Cox4p subunit of CIV with a 10-residue His tag at its C terminus. Genomic DNA from strain DL1 (31) was used as the polymerase chain reaction template. The following primers were used to obtain the upstream COX4 region: COX4 F1, 5′-AGCGATATCTCCCATCTTGG-3′, and COX4 His R1, 5′-TTAATGGTGATGGTGATGGTGATGGTGATGGTGGGTCATCATTTGG-3′. The downstream DNA segment was obtained using the primer COX4DS His F1 (5′-CATCACCATCACCATCACCATTAATCTTATCATTCAAGTTGCCCTTC-3′) and COX4DS R1 (5′-TCAATTCAACTCCCTGTGCC-3′). To make the complete product, the upstream and downstream DNA fragments were purified from an agarose gel and combined for a third polymerase chain reaction with primers COX4 F1 and COX4DS R1. The final polymerase chain reaction product was transformed into WD1 (MATα, his3, leu2, ura3, cox4::LEU2) (31) and selected for its ability to grow in nonfermentable medium (1% yeast extract, 2% peptone, 1% ethanol (v/v) and 3% glycerol, pH 5.6) to isolate transformants in which chromosomal cox4::LEU2 was replaced by COX4-His10.
Based on the crystal structure of bovine CIV containing the subunit 5b homolog (32) of yeast Cox4p, the whole Cox4p, including the C terminus, is exposed on the matrix surface of CIV with the C terminus far removed from the membrane surface or the interface with CIII. Therefore, the tag should have no influence on the interaction with CIII or CL. Indeed, the strain containing COX4-His10 showed normal levels of the supercomplexes with the same mobility on BN-PAGE as the native supercomplexes.
The native supercomplex was isolated from strain USY00b (MATα, ade2-1, his3-11-15, leu2-3-112, trp1-1, ura3-52, can1R-100, atp2::LEU2,trp1::ATP2-His6) (29) that was cultivated in YP/lactate medium containing 1% yeast extract, 2% peptone, and 3.7% of lactic acid, pH 5.6.
Protein Purification
Mitochondria were isolated from yeast cells expressing the Cox4p-His10 subunit of CIV as described previously (26) with slight modifications. Zymolase (3 mg per g of wet weight cells) was used for spheroplast production. Mitochondria (80 mg of protein) were resuspended in 10 ml of lysis buffer containing 2% dodecyl β-d-maltoside (DDM, Anatrace), 50 mm potassium acetate, 10% glycerol, 1:50 volume of protease inhibitor mixture for fungal and yeast cells (Sigma), 1.5 mm phenylmethylsulfonyl fluoride, and 30 mm HEPES-KOH, pH 7.4, for 90 min at 4 °C. After centrifugation (145,000 × g, 20 min), the supernatant was loaded on a DEAE CL-6B column (GE, 4 ml of resin) for CIII immobilization (33). The flow-through was collected and saved for CIV purification (see below). The column was washed with 50 mm MOPS, pH 6.9, containing 150 mm NaCl, and 0.015% DDM until null absorbance at 280 nm. To exchange the detergent, the resin was further washed with eight column volumes of the same buffer containing 0.15% digitonin (Sigma) recrystallized according to Ref. 34 instead of DDM. For CIII elution, the salt concentration in the elution buffer was raised to 500 mm NaCl. The CIII obtained was stored at −80 °C in the presence of 10% glycerol.
For CIV purification the flow-through from the DEAE column was incubated in batch with 4 ml of Talon® His tag purification resin (Clontech) for 2 h. The resin was loaded into a column and washed with 20 mm Tris-HCl, pH 7.2, containing 150 mm NaCl and 0.5% DDM until null absorbance at 280 nm. The same buffer but containing 0.15% digitonin instead of DDM was used for detergent replacement by washing with 8 column volumes. Adding 75 mm imidazole to the digitonin-containing buffer eluted CIV. The purified CIV was stored at −80 °C in the presence of 10% glycerol. Protein concentrations were determined using the micro-BCA protein assay kit (Thermo Scientific) according to the manufacturer's instructions.
Measurement of Ubiquinol-Cytochrome c Reductase and Cytochrome c Oxidase Activities
Purified CIII was assayed in 50 mm potassium phosphate, pH 7.2, 0.25 m sucrose, 1 mm KCN, 0.05% DDM, 50 μm horse heart cytochrome c, and 40 μm decylubiquinol (Sigma) at room temperature for ubiquinol-dependent cytochrome c reductase activity. The reaction was started by the addition of the enzyme at a final concentration of 5–15 nm. Prior to its use, decylubiquinol was freshly prepared by reduction in the presence of a few grains of sodium borohydride followed by the addition of 0.1 m HCl to quench unreacted sodium borohydride. Reduction of cytochrome c was monitored at 550 nm, and the rate of reduction was calculated using an extinction coefficient of 21.5 mm−1 cm−1. Antimycin A-insensitive activities were subtracted from each sample.
CIV activity was measured according the cytochrome c oxidase assay kit guidelines (Sigma). Purified CIV was assayed in 50 mm Tris-HCl, pH 7.0, 120 mm KCl, and 25 μm ferrocytochrome c for cytochrome c oxidase activity. Cytochrome c reduction was done by incubation of a 1 mm solution in the presence of 2 mm dithiothreitol for 20 min. The ratio of absorbance at 550 nm to 565 nm was measured, and the ferrocytochrome c solution was only used if this value was between 10 and 20. The reaction was started after addition of 5–15 nm CIV. Oxidation of ferrocytochrome c was monitored by the decrease of absorbance at 550 nm. KCN-insensitive activities were subtracted from each sample.
Turnover numbers are expressed as mole of cytochrome c reduced or oxidized per mol of cytochrome bc1 complex monomer or CIV monomer, respectively, per s under steady-state conditions.
Determination of CL in Purified CIII and CIV by Electrospray Ionization-Mass Spectrometry (ESI-MS)
Phospholipids were extracted from samples of CIII and CIV (quadruplicate samples of 50 μg of protein each) with a mixture of methanol, chloroform, 0.1 n HCl (containing 11 mm ammonium acetate) in a proportion of 1:1:0.9 as described previously (18, 29). For quantification of CL, 0.8 nmol of CL (14:0)4 was added as an internal standard prior to extraction of lipids.
ESI-MS analysis of CL was performed as described previously (18). Samples for CL quantitation were injected into a normal phase HPLC column (Luna 5-μm silica 100 Å, 2.0 × 150 mm column; Phenomenex) coupled to an API 3200 mass spectrometer running in negative ion mode. Negative ion ESI was carried out at −4,000 V, with declustering potential of −100 V, focusing potential of −350 V, and entrance potential of −10 V. The solvent system consisted of hexane, isopropyl alcohol, 20 mm ammonium acetate in water, pH 5.5, in a ratio of 30:40:7 (v/v/v) for solvent A and hexane/isopropyl alcohol (30:40, v/v) for solvent B. In separate runs, 0.34, 0.69, or 1.37 nmol of CL (18:1)4 as a reference and 0.8 nmol of CL (14:0)4) as an internal standard were used to establish the standard curve. The nanomoles of individual molecular species of CL in analyzed samples were calculated by determining the ratio of the peak area of the CL molecular species of interest to the internal standard CL (14:0)4. This ratio, along with the slope and intercept of the standard curve made with CL (18:1)4, was used to calculate nanomoles of CL (23). Total CL was determined as the sum of the most prevalent CL species, and values in nanomoles/mg of the protein was calculated. The number of CL molecules per mol of CIII or CIV was calculated using molecular masses of 560 or 240 kDa, respectively.
Phospholipase Treatment
CIV (48 μg) was incubated for 2 h in the presence of 50 mm Tris-HCl, pH 7.2, 2 mm CaCl2, 0.15% digitonin, 0.3 m NaCl, and 100 units/ml phospholipase A2 in a final volume of 300 μl. Adding EDTA to a final concentration of 10 mm stopped the reaction. Phospholipase was removed using a centrifugal filter with a 50-kDa molecular mass cutoff (Amicon, Millipore) and buffer containing 50 mm Tris-HCl, pH 7.2, 150 mm NaCl, and 0.15% digitonin.
Preparation of Liposomes
Lipid mixtures in chloroform were evaporated under nitrogen. The lipid film was solubilized with 50 mm Tris-HCl, pH 7.2, containing 150 mm NaCl and vortexed 20 min. The final volume of buffer used for lipid solubilization was calculated taking into account the final volume of the protein mixture used in the proteoliposome reconstitution experiments (see below). The multilamellar vesicles were dispersed by sonication for 15 min with an ultrasonic cleaner from Avanti Polar Lipids or by probe sonication (Bronwill Biosonik) for the preparation of small unilamellar vesicles. Lipid amounts are expressed as mol % throughout.
Proteoliposome Reconstitution
Equimolar quantities of CIII and CIV in 0.15% digitonin were mixed in the presence of small unilamellar phospholipid vesicles (see above) at a lipid to protein weight ratio of 15:1. The lipid was dissolved in 2.5 times the volume of the protein mixture to dilute the detergent while adding the lipid. Proteoliposomes were formed as digitonin was diluted below its critical micelle concentration (≈0.06%) by incubation for 2 h at room temperature with gentle shaking.
Protein Extraction, Separation, and Band Quantification
Proteoliposomes were sedimented by ultracentrifugation at 4 °C at 90,000 × g for 15 min. The supernatant was discarded, and the sediment was resuspended in 50 mm Tris-HCl buffer, pH 7.2, containing 150 mm NaCl and 3% digitonin and incubated for 30 min. After a second centrifugation under the same conditions, the supernatant containing the extracted proteins was subjected to BN-PAGE in a 3–12% gradient gel. Typically gels were run 14 h at an initial current of 12 mA. The gels were stained for protein visualization using Bio-Safe Coomassie G-250 and scanned. The images were analyzed using ImageJ software (National Institutes of Health, imagej.nih.gov). The bands identified on Western blots made from the above gels (see below) as the supercomplexes (containing both CIII and CIV) and for free CIII (representative of the protein that was not integrated into the supercomplexes) were quantified and normalized by the sum of the quantities obtained for all the bands considered.
In-gel Enzyme Activity
For CIII activity, the extracted protein was subjected to colorless native-PAGE as described previously (35). The gel was then incubated in the presence of 0.05% (w/v) diaminobenzidine in 50 mm phosphate buffer, pH 7.2, for 2 h at room temperature. For CIV activity, the protein was loaded onto a BN-polyacrylamide gel. After running, the gel was incubated in the presence of 0.05% (w/v) cytochrome c and 0.05% diaminobenzidine (w/v) in 50 mm phosphate buffer, pH 7.2, for 1 h at room temperature (36).
Western Blot Analysis
Stained gels were transferred for Western blot analysis to a PVDF membrane. For CIII visualization, a monoclonal antibody or a polyclonal antibody specific for the Qcr7p subunit was used. For CIV visualization, an antibody recognizing the His tag was used. Two different techniques were used for the acquisition of the Western blot image. When the signal was captured by a Fluor®-S Max multi-imager (Bio-Rad), secondary anti-mouse or anti-rabbit (stabilized peroxidase conjugate, Thermo Scientific) antibody was used. SuperSignal Western Femto Sensitivity kit (Thermo Scientific) was used to develop the signal according the manufacturer's instructions. Alternatively, we used the Odyssey Infrared Imaging System (LI-COR Biosciences). In this case, the secondary IRDye 680RD goat anti-rabbit and IRDye 800CW donkey anti-guinea pig antibodies were used to detect CIII and CIV polyclonal antibodies or Rcf1p and CIV polyclonal antibodies, simultaneously. The guidelines suggested by the manufacturer were used. SDS (0.1%) was included in the final wash of the membrane. This step was mandatory not only to reduce the background due to the use of the IRDye 680 secondary antibody but also to wash the Coomassie R-250 stain out of the membrane.
Purification of the Reconstituted and Native Supercomplexes
Proteoliposomes containing the reconstituted supercomplex were sedimented by ultracentrifugation at 4 °C at 90,000 × g for 15 min. The supernatant was discarded, and the sediment was resuspended in 50 mm Tris-HCl buffer, pH 7.2, containing 150 mm NaCl and 3% digitonin and incubated for 30 min. After a second centrifugation, the supernatant (0.1 ml) containing the extracted proteins was loaded onto a 2-ml sucrose gradient (0.75–1.5 m sucrose in 15 mm Tris-HCl, pH 7.2, 20 mm KCl, and 0.4% digitonin) and centrifuged at 4 °C for 20 h at 30,000 rpm (TLS-55 swinging bucket rotor). Fractions (60–80 μl) from the gradient were analyzed by BN-PAGE and Western blotting. Fractions enriched in III2IV2 supercomplex were analyzed by single particle negative stain EM. Purification of natural supercomplexes from strain USY00b was performed as described previously (29).
Negative Stain Imaging by EM
A continuous thin carbon support film was prepared by depositing carbon onto a freshly cleaved mica sheet from a graphite rod using a Denton Vacuum DU-502A Evaporator. The carbon support film was floated onto a water surface and to a Quantifoil carbon grid (Quantifoil R2/2 holey carbon film on 400 mesh copper grids. West Chester, PA). The carbon-coated grid was glow discharged for 2 min prior to sample application. The negative stain grids were prepared using a droplet method. A 5-μl drop of natural supercomplex or reconstituted supercomplex was placed on the holey carbon grid. After 1 min, the excess sample was removed by using a filter paper followed by placement of a 5-μl drop of 2% ammonium molybdate (Aldrich) on the grid. After 1 min, the excess stain was removed using a filter paper. The stain deposition was repeated once. After 30 s, the grid was blotted with filter paper and air-dried. Samples were imaged at liquid nitrogen temperature using a Polara 300-kV electron microscope (FEI Co.) equipped with a 4 × 4K CCD camera (TVIPS GmbH, Gauting, Germany). Images were recorded at low dose conditions at 0.5 to 3 μm underfocus and a ×39,000 nominal magnification.
Particle picking and image processing were performed using SPARX software package (37). All the micrographs containing particles were manually selected and windowed using e2boxer.py (29). For reconstituted supercomplex, a total of 8,578 particles were selected from 529 micrographs. For natural supercomplex, a total of 3,579 particles was selected from 150 micrographs. EMAN2 software program was used for generating reference free class averages (e2refine2d) (38) from the single particle projection data sets of the reconstituted supercomplex and the natural supercomplex.
RESULTS
Analysis of Purified CIII and CIV
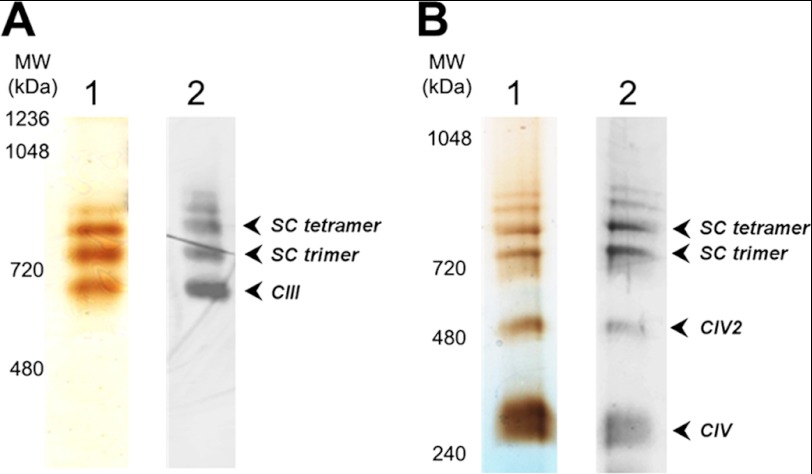
Purified CIII and CIV displayed activities of about 40 and 600 s−1, respectively, which is comparable with published results (33, 39, 40). Even though the purification of the complexes utilized detergents, tightly bound lipids were previously found associated with highly purified preparations (41). For instance, yeast CIII (as the ubiquinol:cytochrome c oxidoreductase dimer) contains 14 structural lipids in the crystal structure (42), of which four are CL, two are PI, six are PE, and two are PC molecules. In the case of the bovine CIV (as the cytochrome c oxidase monomer), the crystal structure revealed the presence of 13 lipids, including two CL, one PC, three PE, four PG, and three triacylglycerol molecules (43). Furthermore, CL was shown to be essential for the optimum activity of both CIII and CIV (27, 44–46). We confirmed the presence of tightly bound CL molecules by ESI-MS and estimated about eight (8.4 ± 0.1) or two (1.8 ± 0.1) per purified CIII (560 kDa) or CIV (240 kDa), respectively, using the CL amounts shown in Fig. 1. Interestingly, CL bound both to CIII and CIV showed the same proportion among the five major CL species (Fig. 1) as in the mitochondrial inner membrane (29). The higher amount of CL in CIII than was determined in the crystal structure is a result of the less delipidation of this enzyme because only one step of the purification procedure was used instead of the two steps as described previously (33). We chose not to use the second step, which resulted in the loss of subunit Qcr10p. The amount of CL in the purified CIV is the same as was determined in the crystal structure of bovine CIV (43).
FIGURE 1.
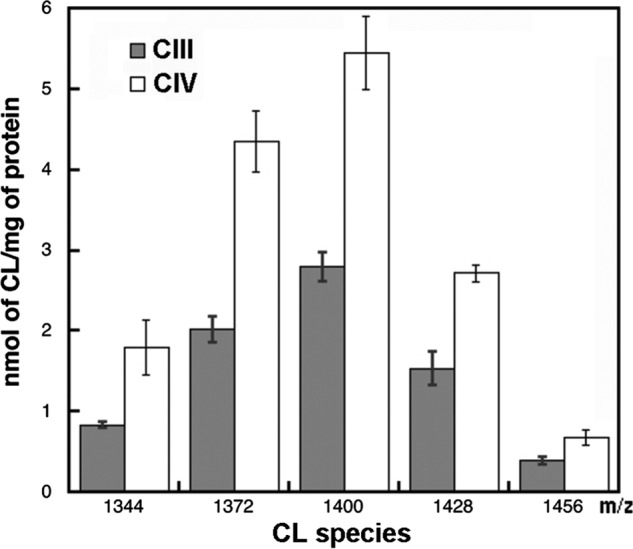
CL species and content in the purified CIII and CIV determined by ESI-MS. Total CL was determined as the sum of the five prevalent CL species (1344 (16:1)4; 1372 (16:1)3(18:1)1; 1400 (18:1)2(16:1)2; 1428 (18:1)3(16:1)1, and 1456 (18:1)4) from values of singly ionized CL. Data represent the average of four independent runs, and the bars indicate standard deviations.
Reconstitution of CIII and CIV in Proteoliposomes
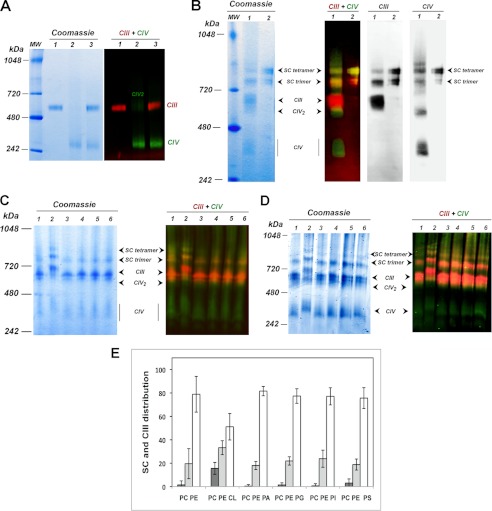
Purified CIII and CIV were mixed with accompanying dilutions of digitonin below its critical micelle concentration. BN-PAGE and Western blot analyses only showed the individual complexes with no evidence of higher order association of CIII and CIV into a supercomplex. (Fig. 2A, lanes 3 are a mixture of individual complexes shown in lanes 1 and 2). A low level of CIV dimer was present in the purified CIV, which is consistent with published results for bovine CIV (47). Therefore, the lipids tightly bound to purified CIII and CIV were not sufficient to generate supercomplex formation from these individual complexes. Thus, we decided to perform reconstitution of CIII and CIV into proteoliposome containing the major phospholipids of the mitochondrial membrane, PC, PE, and CL. When the two complexes were mixed together in the presence of an equimolar concentration of PC and PE plus 20% CL and analyzed after digitonin solubilization of the sedimented proteoliposomes by BN-PAGE, clear formation of higher order structures was observed (Fig. 2B). Two distinct bands with lower mobility than CIII and CIV dimer (CIV2) appeared (Fig. 2B, lane 1 in the polyacrylamide gel). Formation of supercomplexes composed of CIII and CIV was confirmed by recognition by individual polyclonal antibodies directed at CIII and CIV. Both slow migrating bands cross-reacted with polyclonal antibodies to the individual CIII and CIV (Fig. 2B, lanes 1, in Western blots). Molecular weight standards in BN-PAGE do not reflect actual molecular weights of the supramolecular aggregates and were used to correlate relative mobility of sample bands between gels. To evaluate CIII-CIV supercomplexes formed in PC/PE/CL liposomes, we used a mixture of the purified natural supercomplex III2IV1 (supercomplex trimer) and III2IV2 (supercomplex tetramer) as a standard (Fig. 2B, lanes 2). As seen in Fig. 2B, one of reconstituted supercomplexes corresponds to the natural trimer and another one to the natural tetramer. From hereon, we refer to these two higher order complexes as the reconstituted supercomplex trimer (III2IV1) and reconstituted supercomplex tetramer (III2IV2).
FIGURE 2.
Reconstitution of the supercomplexes from individual CIII and CIV in the presence of different lipid mixtures. A, BN-PAGE stained with Coomassie R-250 (left panel) and Western blot (right panel) with red or green fluorescent-tagged secondary antibodies detecting CIII or CIV, respectively, are shown. Samples were CIII (lanes 1), CIV (lanes 2), and CIII and CIV together (lanes 3), all in the absence of added phospholipid. B, BN-PAGE stained with Coomassie R-250 (left panel) and Western blot (2nd panel from left) with red or green fluorescent-tagged polyclonal antibodies directed against CIII or CIV, respectively, Western blot with polyclonal antibody against CIII (2nd panel from right), and Western blot with polyclonal antibody against CIV (right panel) are shown. Samples were CIII and CIV reconstituted in proteoliposomes containing an equimolar mixture of PC and PE supplemented with 20% CL (lanes 1) or native purified supercomplexes (lanes 2). The composition of the various bands is noted based on antibody cross-reactivity and mobility relative to the native trimer and tetramer. SC denotes supercomplex. C, BN-PAGE and Western blots as in B of CIII and CIV reconstituted together in liposomes containing an equimolar mixture of PC and PE (lanes 1) supplemented with 20% CL (lanes 2), PA 1% (lanes 3), PG 1% (lanes 4), PI 15% (lanes 5), and PS 2% (lanes 6) of the total phospholipid. D, same as in C, except all anionic phospholipids were supplemented to 20% of the total phospholipid. E, quantification of the different species visualized in the BN-PAGE after Coomassie R-250 staining (mean value of at least four different experiments with bars indicating standard deviation). In white is CIII; in light gray is the supercomplex trimer, and in dark gray is the supercomplex tetramer.
Next, we examined the specificity for the CL requirement in reconstitution of the trimeric and tetrameric supercomplexes. We compared reconstitution of the supercomplexes in proteoliposomes containing CL with their reconstitution in proteoliposomes containing equimolar amounts of PE and PC without or with a series of anionic phospholipids in place of CL. When the two complexes were mixed together in the presence of PC/PE and analyzed after digitonin solubilization of the sedimented proteoliposomes by BN-PAGE, there was formation of low and diffuse staining in the region of the trimer (Fig. 2, C and D, lanes 1), but no tetramer was observed. Formation of trimer was confirmed by recognition by individual polyclonal antibodies directed at CIII and CIV as indicated by the change of the band from green or bright orange to a pale yellow-orange color in Fig. 2, C and D. Overall, any formation of higher order trimeric complexes was poorly defined and variable with only PC/PE in the liposomes (see high level of deviation in Fig. 2E). A marked difference with clear formation of both trimer and tetramer in PC/PE liposomes containing 20% CL was observed (Fig. 2, C and D, lanes 2). Interestingly, in the presence of CL, all bands were retarded in mobility relative to samples without CL (Fig. 2, C and D), which may be due to an increase in lipid content due to specific affinity of the complexes for CL. There was also material running slower than the tetramer, which, based on immune reactivity with only antibody to CIV (Fig. 2B, far right panel, and Fig. 2, C and D, lanes 2 of immunoblot), may be a higher order self-association of CIV.
The summary quantification of several experiments is shown in Fig. 2E. For PC/PE liposomes, the level of Coomassie R-250 staining in the region of the tetramer was near background and showed a wide variation for the trimer region with a diffuse appearance when probed by antibodies. Tetramer formation was definite upon addition of CL with the formation of distinct trimer bands in all cases. Specificity for CL among other anionic lipids (PA, PG, PI, and PS) for tetramer formation was confirmed either at the low levels of lipids found in mitochondria (Fig. 2C, lanes 3–6) or at 20% (Fig. 2D, lanes 3–6). Addition of anionic phospholipids other than CL did not result in a slower mobility for the trimer, but there was a distinct sharpening of the trimer band. Fig. 2E shows the summary of several experiments, which confirms little or no tetramer formation in the absence of CL, variable trimer formation without anionic phospholipids, and distinct trimer formation with the addition of anionic phospholipids.
Dependence of supercomplex formation on the level of CL showed a similar result. For the tetramer (Fig. 3A), there was little if any formation without CL and increasing amounts of tetramer at increasing CL levels that maximized at 20% CL and above. In PC/PE liposomes, there was a broad variability in trimer formation (Fig. 3B) but low variability and constant amounts at CL levels of 5% and above.
FIGURE 3.
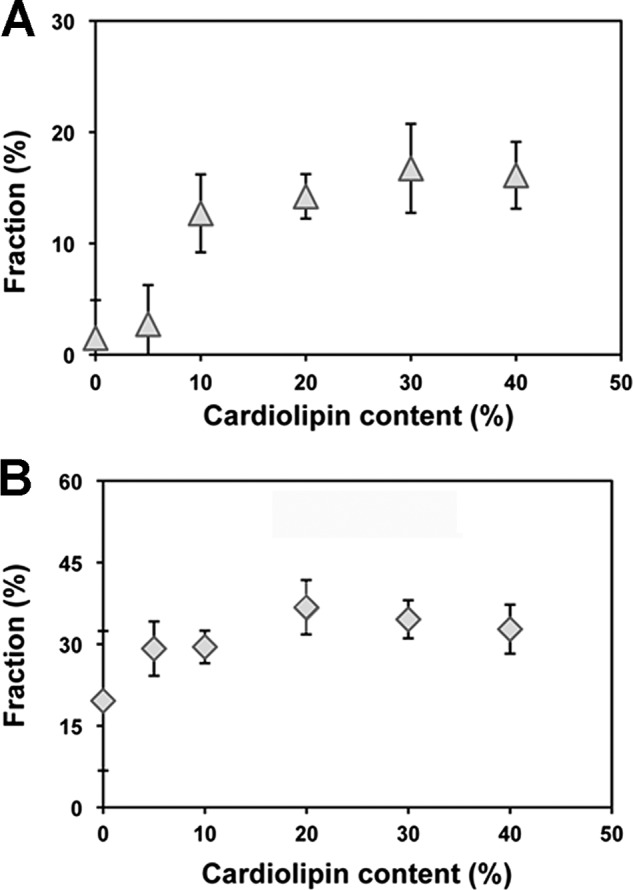
CL concentration dependence for supercomplex formation in proteoliposomes. Quantification of the amount of tetramer (A) and trimer (B) obtained after reconstitution in the presence of an equimolar mixture of PC and PE containing variable amounts of CL. The results shown are the average of four different experiments, and the bars indicate standard deviation.
Tightly Bound CL in CIV Is Required for Trimer Formation
To assess if the lipids co-purifying with CIV were required for trimer formation, CIV was treated with phospholipase A2. This treatment resulted in the loss of ∼40% of the original CIV activity. When compared with untreated CIV, phospholipase treatment reduced trimer formation to undetectable levels when reconstituted with untreated CIII in the presence of PC/PE alone (Fig. 4A). However, addition of CL restored both trimer and tetramer levels to those observed without phospholipase treatment (Fig. 4B). This result indicates that lipids co-purifying with CIV are necessary for trimer formation in the absence of added CL. Again, CL addition resulted in the most dramatic effect with its addition being absolutely required for restoration of both trimer and tetramer formation. A similar experiment was attempted with CIII. The phospholipase-treated CIII, showing 70% of the original activity, was poorly incorporated into proteoliposomes, and the formation of CIII and CIV containing oligomers was not visualized (data not shown). This was most likely due to irreversible damage to the organization of CIII subunits.
FIGURE 4.
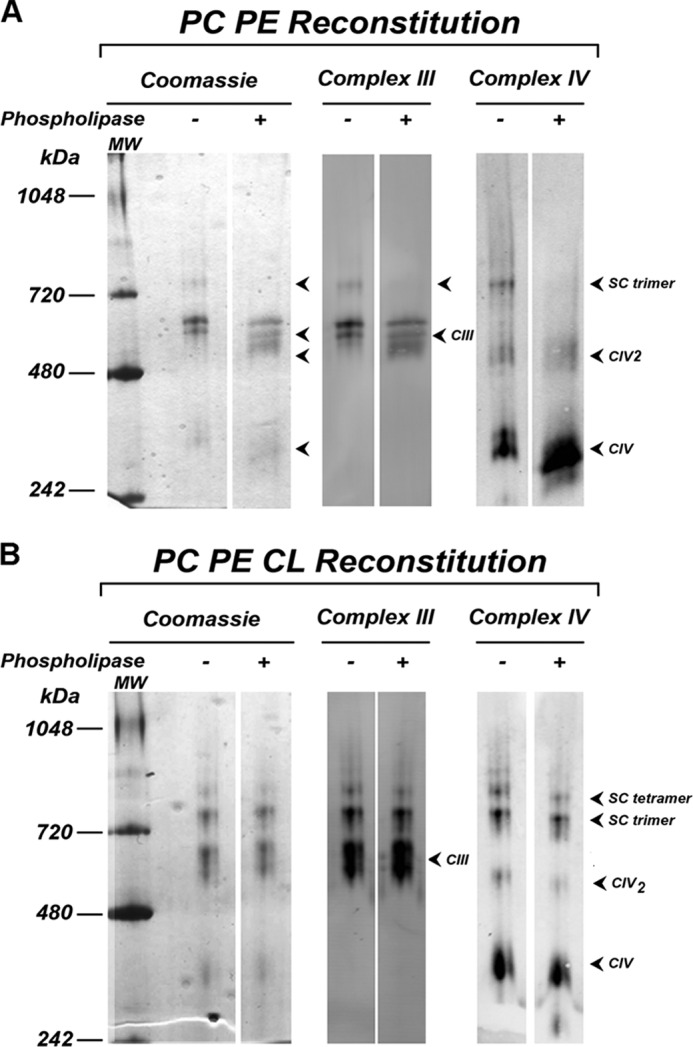
Reconstitution of supercomplexes after phospholipase treatment of CIV. A, BN-PAGE stained with Coomassie R-250 and Western blot with CIII and CIV polyclonal antibodies of CIII and CIV with (+) or without (−) phospholipase treatment followed by reconstitution in either PC/PE liposomes (A) or PC/PE/CL liposomes (B). SC denotes supercomplex.
Reconstituted Supercomplexes Are Formed by Active CIII and CIV
To further characterize the reconstituted supercomplexes formed in the presence of CL, the reconstituted material was subjected to colorless native PAGE and BN-PAGE to assess the in-gel CIII and CIV activity, respectively. As shown in Fig. 5, the reconstituted material remains active, indicating that the interaction between CIII and CIV does not preclude the activity of either of the individual complexes, as was described for the native supercomplex (26, 29, 48). Interestingly, in-gel CIII activity of the band containing individual CIII is significantly lower than activity of the bands represented by tetramer or trimer (Fig. 5). This may mean that CIII was stabilized and/or activated by interaction with CIV and suggests an important physiological role for supercomplex formation. A similar stabilization of complex I by CIII in mammals (49) was observed.
FIGURE 5.
In-gel activity of reconstituted CIII and CIV. CIII and CIV were reconstituted in the presence of PC, PE, and CL (CL 40%). For visualization of CIII activity, the digitonin-solubilized proteoliposome sample was subjected to colorless native-PAGE (A). For CIV activity, a BN-PAGE (B) was used. In-gel activities were performed as described under “Experimental Procedures” (lanes 1), or protein identification of the bands by Western blots (lanes 2) using antibody against CIII (A) or CIV (B) was used. SC denotes supercomplex.
Structural Organization of Reconstituted Tetramer
Both activity and Western blots (Fig. 5) showed low but variable levels of species larger than the tetrameric supercomplex. Such species were observed in some reconstitution experiments. As noted earlier, the trimer also showed a diffuse band pattern in the absence of CL. Therefore, we decided to analyze the structural organization of the tetramer by EM to determine whether a native organization of individual complexes was achieved. The purified S. cerevisiae supercomplex was previously studied by using negative stain EM (50). Analysis of two-dimensional class averages resulted in a proposed three-dimensional arrangement of the individual complexes in which CIII is flanked on each side by a monomer of CIV (supercomplex IV1-III2-IV1). The most important information was extracted from the projection map of the side view of the supercomplex. Later, our detailed studies of the three-dimensional structure of this supercomplex by single particle cryo-EM confirmed this arrangement (29). Thus, we concluded that the two-dimensional class averages of the side view of the reconstituted supercomplex tetramer obtained by negative stain EM could be used as a fast and convenient method for identification and confirmation of the native structure of the reconstituted supercomplex.
The reconstituted material extracted from proteoliposomes by digitonin was subjected to sucrose gradient ultracentrifugation (Fig. 6A), similar to that described for the purification of mitochondrial supercomplexes (29). Fractions 2 and 3, containing a mixture of the trimer and tetramer and enriched in the latter, were used for negative stain grid preparation in the presence of ammonium molybdate. A total of 529 micrographs were obtained, and 8,578 single particles were selected and subjected for further analysis. Some of the obtained class averages representing a side view of the reconstituted supercomplex tetramer (classified as in Ref. 50) are shown in Fig. 6B. These projection averages were compared with the projection averages representing the corresponding view of the natural purified supercomplex (Fig. 6C). The supercomplex tetramer obtained from yeast mitochondria has a distinctive, symmetrical organization (Fig. 6C) and (50). The CIII, recognizable by its large mitochondrial matrix domain, is located in the center of the hetero-oligomer and is flanked by the two CIV monomers. The class averages projections obtained for the reconstituted supercomplex side view (Fig. 6B) closely resemble the side view projections obtained for the natural supercomplex (Fig. 6C and Ref. 50). Importantly, there were no side view projections of the reconstituted supercomplex tetramer displaying a different organization than in the natural supercomplex. Specifically, because CIV can form a dimer, one might expect supercomplex tetramer organized of CIII with a dimer of CIV rather than CIV monomers flanking CIII.
FIGURE 6.
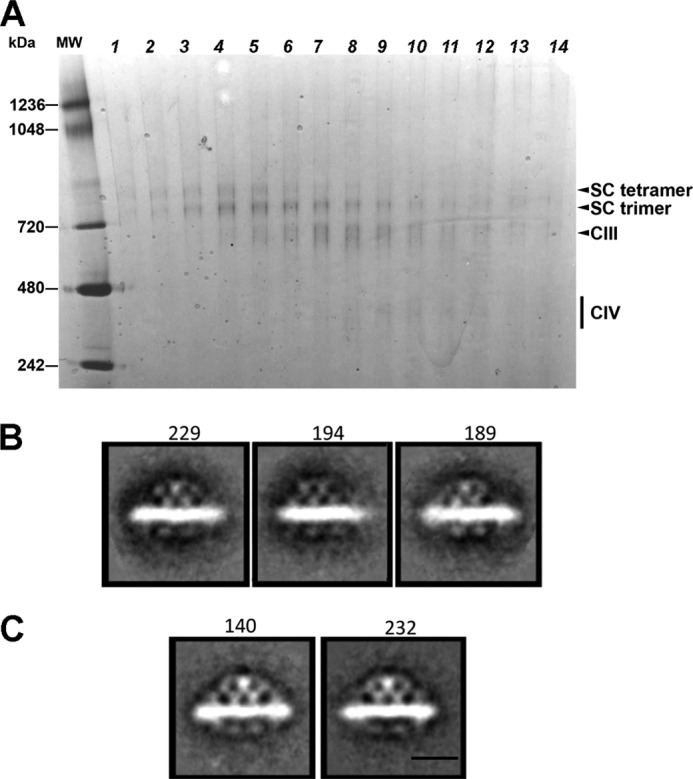
Single particle EM of reconstituted supercomplex and native III2IV2. After reconstitution of CIII and CIV in PC/PE/CL (CL 20%) liposomes, the III2IV2 supercomplex was purified by sucrose gradient centrifugation after solubilization with digitonin. BN-PAGE (A) of aliquots taken from the lower half of the sucrose gradient is shown. Fractions 2 and 3 near the bottom of the sucrose gradient were enriched in III2IV2 supercomplex and were used for the single particle EM analysis. Projection maps of the side view of the isolated reconstituted supercomplex tetramer stained with ammonium molybdate (B) and the purified natural supercomplex stained with ammonium molybdate (C) are shown. The number of particles used for each average is indicated on the top of each projection. Scale bar, 10 nm. SC denotes supercomplex.
Rcf1p Presence
Rcf1p is a small protein that was recently proposed to act as a bridge between CIII and CIV to support supercomplex organization (51–53). To test the presence of this protein in our reconstituted supercomplexes, we employed Western blot analysis with Rcf1p-specific antibody. Purified CIV reconstituted with CIII in either PC/PE without (Fig. 7, lanes 1) or with CL (Fig. 7, lanes 2) contained Rcf1p-positive material in individual CIV as well as in the trimer and tetramer. Because in the absence of CL no tetramer was detected, the presence of Rcf1p was not sufficient for the tetramer formation. However, CL was absolutely required.
FIGURE 7.
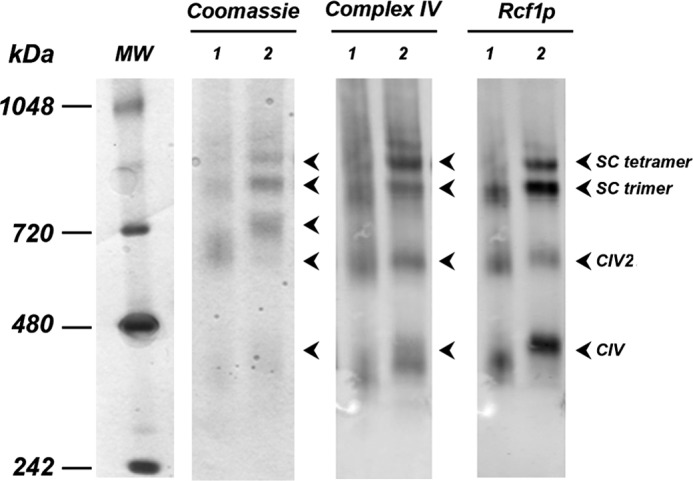
Presence of Rcf1p in the reconstituted supercomplexes. BN-PAGE stained with Coomassie R-250 and Western blots using CIV or Rcf1p polyclonal antibodies (as indicated) of the digitonin extracts containing supercomplexes reconstituted from CIII and CIV in the presence of PC and PE (lanes 1) or PC, PE, and CL (20% CL) (lanes 2) are shown. SC denotes supercomplex.
DISCUSSION
The trimeric and tetrameric structures observed after reconstitution dependent on incorporation into liposomes retain enzymatic activity, and in the case of CIII reconstitution may further stimulate the CIII activity. Single particle EM analysis of the reconstituted tetramer showed a strikingly similar IV1-III2-IV1 organization of the reconstituted supercomplex when compared with the native supercomplex. Here, we present for the first time the reconstitution of yeast respiratory chain supercomplexes from individual complexes in the presence of specific phospholipids. Exposure of individual complexes to phospholipids was required for higher order complex formation, and the reconstitution of the tetrameric form was absolutely dependent on the addition of CL. No other anionic phospholipid was as effective as CL in tetramer formation. In addition, we established that CL is directly required for formation of the tetramer and not just as an integral component of CIII and CIV. As shown in vivo using a Δcrd mutant (24, 26, 45), the presence of CL in the membrane is essential for the formation and stabilization of these supercomplexes. In the mutant, PG levels are elevated to the level of CL and presumably substitute for CL in stabilizing the individual complexes as shown for the mammalian complexes (44). However, both in vivo and in vitro, the presence of PG is not sufficient to support formation of the stable tetramer organization of the respiratory chain components. Results presented here with several anionic phospholipids and results from the Δcrd mutant (54) where PG levels are elevated indicate that some stabilization of the trimer may be supported by anionic phospholipids either present in the individual complexes or supplied in excess over this amount.
Recent structural studies of the mammalian and yeast respiratory supercomplexes by single particle cryo-EM demonstrated that individual complexes have few contact points, which are located at the lipid-water interface, and larger spaces between the membrane-embedded surfaces of these complexes (28–30). This implies that individual complexes are held together by phospholipids that fill the gaps between complexes and are in excess over the integral phospholipids observed in the crystal structures of these complexes. We previously identified (29) about 50 molecules of CL in the purified III2IV2 natural supercomplex, which is quite in excess of the eight CL molecules suggested from crystal structures. It is tempting to speculate that the additional CL molecules fill these gaps in the reconstituted and native III2IV2 supercomplexes in addition to the tightly bound CL molecules that are important for formation and stabilization of the supercomplex tetramer.
Some trimer was formed even in the absence of added CL and only required the incorporation of the purified CIII and CIV into liposomes containing PE and PC. As discussed above, the crystal structures for both of these complexes contain structurally important lipids, including CL, for stabilization. However, phospholipase treatment of CIV abolished trimer formation and required addition of CL to restore trimer formation. Whether CL alone is sufficient to restore trimer formation was difficult to determine because CL alone did not form suitable liposomes. However, in our experiments, the addition of extra CL was required for the formation of the tetrameric structure, indicating that the lipids present in the isolated complexes, and specifically tightly bound CL molecules were not sufficient to support formation of III2IV2 species.
In the case of the trimer, our experiments show that CL tightly bound to CIV is important for association of CIV and CIII into a trimer. However, according to our cryo-three-dimensional structure of the yeast tetramer III2IV2, the surface of CIV, which contains tightly bound CL, does not face CIII (29). This implies that CL tightly bound to CIV does not interact directly with CIII. Therefore, the CLs tightly bound to CIV may be important for the correct conformation of CIV required for higher order assembly. This is consistent with a strong decrease in CIV activity after phospholipase treatment. It also might allow CIV to interact with some additional proteins, for example Rcf1p, which may be involved in supercomplex formation. Another interesting possibility is that in the trimer III2IV1, CIII and CIV have a different orientation with respect to each other than in the tetramer thereby interacting with each other through different surfaces than in the tetramer. The difference in the III-IV arrangement in the tetramer and trimer could stem from the fact that these supercomplexes interact in the mitochondrial membrane with different dehydrogenases and are formed under different growth conditions and might be sensitive to CL levels in the membrane. To test these possibilities, future experiments will include EM single particle analyses of the trimers reconstituted in the absence of CL and in the presence of CL, as well as of the natural trimers obtained under various growth conditions.
Acknowledgments
We thank Dr. Irina Serysheva for use of the Odyssey Infrared Imaging System. We also acknowledge the generous gift of antibodies by Drs. Rosemary Stuart and Bernard Trumpower. We thank Dr. Robert Murphy for the use of the LIPID MAPS mass spectrometry facility at the University of Colorado at Denver, which is supported by National Institutes of Health Large Scale Collaborative Grant GM-069338.
This work was supported, in whole or in part, by National Institutes of Health Grant GM R01 56389 (to W. D.). This work was also supported by the John Dunn Research Foundation (to W. D.).
Complex III in eukaryotes is a structural and functional dimer of two ubiqunol:cytochrome c oxidoreductase multisubunit enzymes (cytochrome bc1 complex), and when in a supercomplex it is referred to as III2.
- CIII
- complex III
- CIV
- complex IV
- BN-PAGE
- blue native-PAGE
- CL
- cardiolipin
- PE
- phosphatidylethanolamine
- PC
- phosphatidylcholine
- PG
- phosphatidylglycerol
- PA
- phosphatidic acid
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- cryo-EM
- cryo-electron microscopy
- DDM
- dodecylmaltoside.
REFERENCES
- 1. Chance B., Williams G. R. (1955) A method for the localization of sites for oxidative phosphorylation. Nature 176, 250–254 [DOI] [PubMed] [Google Scholar]
- 2. Dudkina N. V., Kouril R., Peters K., Braun H. P., Boekema E. J. (2010) Structure and function of mitochondrial supercomplexes. Biochim. Biophys. Acta 1797, 664–670 [DOI] [PubMed] [Google Scholar]
- 3. Lenaz G., Genova M. L. (2012) Supramolecular organisation of the mitochondrial respiratory chain. A new challenge for the mechanism and control of oxidative phosphorylation. Adv. Exp. Med. Biol. 748, 107–144 [DOI] [PubMed] [Google Scholar]
- 4. Schägger H. (2001) Respiratory chain supercomplexes. IUBMB Life 52, 119–128 [DOI] [PubMed] [Google Scholar]
- 5. Stuart R. A. (2008) Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J. Bioenerg. Biomembr. 40, 411–417 [DOI] [PubMed] [Google Scholar]
- 6. Wittig I., Schägger H. (2009) Supramolecular organization of ATP synthase and respiratory chain in mitochondrial membranes. Biochim. Biophys. Acta 1787, 672–680 [DOI] [PubMed] [Google Scholar]
- 7. Acín-Pérez R., Fernández-Silva P., Peleato M. L., Pérez-Martos A., Enriquez J. A. (2008) Respiratory active mitochondrial supercomplexes. Mol. Cell 32, 529–539 [DOI] [PubMed] [Google Scholar]
- 8. Piccoli C., Scrima R., Boffoli D., Capitanio N. (2006) Control by cytochrome c oxidase of the cellular oxidative phosphorylation system depends on the mitochondrial energy state. Biochem. J. 396, 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamashita T., Nakamaru-Ogiso E., Miyoshi H., Matsuno-Yagi A., Yagi T. (2007) Roles of bound quinone in the single subunit NADH-quinone oxidoreductase (Ndi1) from Saccharomyces cerevisiae. J. Biol. Chem. 282, 6012–6020 [DOI] [PubMed] [Google Scholar]
- 10. Mileykovskaya E., Dowhan W. (2009) Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta 1788, 2084–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gómez L. A., Hagen T. M. (2012) Age-related decline in mitochondrial bioenergetics. Does supercomplex destabilization determine lower oxidative capacity and higher superoxide production? Semin. Cell Dev. Biol. 23, 758–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paradies G., Petrosillo G., Paradies V., Ruggiero F. M. (2011) Mitochondrial dysfunction in brain aging. Role of oxidative stress and cardiolipin. Neurochem. Int. 58, 447–457 [DOI] [PubMed] [Google Scholar]
- 13. McMillin J. B., Dowhan W. (2002) Cardiolipin and apoptosis. Bioch. Biophys. Acta 1585, 95–107 [DOI] [PubMed] [Google Scholar]
- 14. Ostrander D. B., Sparagna G. C., Amoscato A. A., McMillin J. B., Dowhan W. (2001) Decreased cardiolipin synthesis corresponds with cytochrome c release in palmitate-induced cardiomyocyte apoptosis. J. Biol. Chem. 276, 38061–38067 [DOI] [PubMed] [Google Scholar]
- 15. Huang Z., Jiang J., Tyurin V. A., Zhao Q., Mnuskin A., Ren J., Belikova N. A., Feng W., Kurnikov I. V., Kagan V. E. (2008) Cardiolipin deficiency leads to decreased cardiolipin peroxidation and increased resistance of cells to apoptosis. Free Radic. Biol. Med. 44, 1935–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saini-Chohan H. K., Holmes M. G., Chicco A. J., Taylor W. A., Moore R. L., McCune S. A., Hickson-Bick D. L., Hatch G. M., Sparagna G. C. (2009) Cardiolipin biosynthesis and remodeling enzymes are altered during development of heart failure. J. Lipid Res. 50, 1600–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sparagna G. C., Chicco A. J., Murphy R. C., Bristow M. R., Johnson C. A., Rees M. L., Maxey M. L., McCune S. A., Moore R. L. (2007) Loss of cardiac tetralinoleoyl cardiolipin in human and experimental heart failure. J. Lipid Res. 48, 1559–1570 [DOI] [PubMed] [Google Scholar]
- 18. Sparagna G. C., Johnson C. A., McCune S. A., Moore R. L., Murphy R. C. (2005) Quantitation of cardiolipin molecular species in spontaneously hypertensive heart failure rats using electrospray ionization mass spectrometry. J. Lipid Res. 46, 1196–1204 [DOI] [PubMed] [Google Scholar]
- 19. Dumas J. F., Peyta L., Couet C., Servais S. (2012) Implication of liver cardiolipins in mitochondrial energy metabolism disorder in cancer cachexia. Biochimie, in press [DOI] [PubMed] [Google Scholar]
- 20. McKenzie M., Lazarou M., Thorburn D. R., Ryan M. T. (2006) Mitochondrial respiratory chain supercomplexes are destabilized in Barth syndrome patients. J. Mol. Biol. 361, 462–469 [DOI] [PubMed] [Google Scholar]
- 21. Joshi A. S., Zhou J., Gohil V. M., Chen S., Greenberg M. L. (2009) Cellular functions of cardiolipin in yeast. Biochim. Biophys. Acta 1793, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schlame M., Ren M. (2009) The role of cardiolipin in the structural organization of mitochondrial membranes. Biochim. Biophys. Acta 1788, 2080–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bogdanov M., Mileykovskaya E., Dowhan W. (2008) Lipids in the assembly of membrane proteins and organization of protein supercomplexes. Implications for lipid-linked disorders. Subcell. Biochem. 49, 197–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang M., Mileykovskaya E., Dowhan W. (2002) Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 277, 43553–43556 [DOI] [PubMed] [Google Scholar]
- 25. Boumans H., Grivell L. A., Berden J. A. (1998) The respiratory chain in yeast behaves as a single functional unit. J. Biol. Chem. 273, 4872–4877 [DOI] [PubMed] [Google Scholar]
- 26. Zhang M., Mileykovskaya E., Dowhan W. (2005) Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J. Biol. Chem. 280, 29403–29408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wenz T., Hielscher R., Hellwig P., Schägger H., Richers S., Hunte C. (2009) Role of phospholipids in respiratory cytochrome bc1 complex catalysis and supercomplex formation. Biochim. Biophys. Acta 1787, 609–616 [DOI] [PubMed] [Google Scholar]
- 28. Althoff T., Mills D. J., Popot J. L., Kühlbrandt W. (2011) Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 30, 4652–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mileykovskaya E., Penczek P. A., Fang J., Mallampalli V. K., Sparagna G. C., Dowhan W. (2012) Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J. Biol. Chem. 287, 23095–23103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dudkina N. V., Kudryashev M., Stahlberg H., Boekema E. J. (2011) Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc. Natl. Acad. Sci. U.S.A. 108, 15196–15200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dowhan W., Bibus C. R., Schatz G. (1985) The cytoplasmically made subunit IV is necessary for assembly of cytochrome c oxidase in yeast. EMBO J. 4, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoshikawa S., Shinzawa-Itoh K., Tsukihara T. (1998) Crystal structure of bovine heart cytochrome c oxidase at 2.8 Å resolution. J. Bioenerg. Biomembr. 30, 7–14 [DOI] [PubMed] [Google Scholar]
- 33. Palsdottir H., Hunte C. (2003) in Membrane Protein Purification and Crystallization: A Practical Guide, pp. 191–203, Academic Press, San Diego [Google Scholar]
- 34. Stuart R. A. (2009) Supercomplex organization of the yeast respiratory chain complexes and the ADP/ATP carrier proteins. Methods Enzymol. 456, 191–208 [DOI] [PubMed] [Google Scholar]
- 35. Wittig I., Karas M., Schägger H. (2007) High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteomics 6, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 36. Sabar M., Balk J., Leaver C. J. (2005) Histochemical staining and quantification of plant mitochondrial respiratory chain complexes using blue-native PAGE. Plant J. 44, 893–901 [DOI] [PubMed] [Google Scholar]
- 37. Hohn M., Tang G., Goodyear G., Baldwin P. R., Huang Z., Penczek P. A., Yang C., Glaeser R. M., Adams P. D., Ludtke S. J. (2007) SPARX, a new environment for cryo-EM image processing. J. Struct. Biol. 157, 47–55 [DOI] [PubMed] [Google Scholar]
- 38. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN. Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 39. Meunier B., Maréchal A., Rich P. R. (2012) Construction of histidine-tagged yeast mitochondrial cytochrome c oxidase for facile purification of mutant forms. Biochem. J. 444, 199–204 [DOI] [PubMed] [Google Scholar]
- 40. Taanman J. W., Capaldi R. A. (1992) Purification of yeast cytochrome c oxidase with a subunit composition resembling the mammalian enzyme. J. Biol. Chem. 267, 22481–22485 [PubMed] [Google Scholar]
- 41. Hunte C., Richers S. (2008) Lipids and membrane protein structures. Curr. Opin. Struct. Biol. 18, 406–411 [DOI] [PubMed] [Google Scholar]
- 42. Hunte C. (2005) Specific protein-lipid interactions in membrane proteins. Biochem. Soc. Trans. 33, 938–942 [DOI] [PubMed] [Google Scholar]
- 43. Shinzawa-Itoh K., Aoyama H., Muramoto K., Terada H., Kurauchi T., Tadehara Y., Yamasaki A., Sugimura T., Kurono S., Tsujimoto K., Mizushima T., Yamashita E., Tsukihara T., Yoshikawa S. (2007) Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 26, 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gomez B., Jr., Robinson N. C. (1999) Phospholipase digestion of bound cardiolipin reversibly inactivates bovine cytochrome bc1. Biochemistry 38, 9031–9038 [DOI] [PubMed] [Google Scholar]
- 45. Pfeiffer K., Gohil V., Stuart R. A., Hunte C., Brandt U., Greenberg M. L., Schägger H. (2003) Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880 [DOI] [PubMed] [Google Scholar]
- 46. Sedlák E., Panda M., Dale M. P., Weintraub S. T., Robinson N. C. (2006) Photolabeling of cardiolipin binding subunits within bovine heart cytochrome c oxidase. Biochemistry 45, 746–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Musatov A., Ortega-Lopez J., Robinson N. C. (2000) Detergent-solubilized bovine cytochrome c oxidase: dimerization depends on the amphiphilic environment. Biochemistry 39, 12996–13004 [DOI] [PubMed] [Google Scholar]
- 48. Schägger H., Pfeiffer K. (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Acín-Pérez R., Bayona-Bafaluy M. P., Fernández-Silva P., Moreno-Loshuertos R., Pérez-Martos A., Bruno C., Moraes C. T., Enríquez J. A. (2004) Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13, 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Heinemeyer J., Braun H. P., Boekema E. J., Kouril R. (2007) A structural model of the cytochrome c reductase/oxidase supercomplex from yeast mitochondria. J. Biol. Chem. 282, 12240–12248 [DOI] [PubMed] [Google Scholar]
- 51. Chen Y. C., Taylor E. B., Dephoure N., Heo J. M., Tonhato A., Papandreou I., Nath N., Denko N. C., Gygi S. P., Rutter J. (2012) Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 15, 348–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Strogolova V., Furness A., Robb-McGrath M., Garlich J., Stuart R. A. (2012) Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol. Cell. Biol. 32, 1363–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vukotic M., Oeljeklaus S., Wiese S., Vögtle F. N., Meisinger C., Meyer H. E., Zieseniss A., Katschinski D. M., Jans D. C., Jakobs S., Warscheid B., Rehling P., Deckers M. (2012) Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 15, 336–347 [DOI] [PubMed] [Google Scholar]
- 54. Zhang M., Su X., Mileykovskaya E., Amoscato A. A., Dowhan W. (2003) Cardiolipin is not required to maintain mitochondrial DNA stability or cell viability for Saccharomyces cerevisiae grown at elevated temperatures. J. Biol. Chem. 278, 35204–35210 [DOI] [PubMed] [Google Scholar]