Background: UNC93B1 sorts nucleic acid-sensing TLRs to endosomes.
Results: Stimulation of HUVEC by TLR3, but not other TLR agonists, up-regulates the endogenous UNC93B1, which translocates TLR3 to plasma membrane. Poly(I:C) strongly potentiates activation of TLR9 via UNC93B1.
Conclusion: Up-regulation of UNC93B1 through TLR3 activation forms a positive feedback loop that primes cells for the augmented response against infection.
Significance: This work presents unique TLR3-UNC93B1 interrelationship.
Keywords: Glycosylation, Interferon, Toll-like Receptors (TLR), Trafficking, Transcription Regulation, Toll-like Receptor 3, Toll-like Receptor 9, Cell Surface Localization, Interferon-β
Abstract
Translocation of nucleic acid-sensing (NAS) Toll-like receptors (TLRs) to endosomes is essential for response to microbial nucleic acids as well as for prevention of the autoimmune response. The accessory protein UNC93B1 is indispensable for activation of NAS TLRs because it regulates their response through trafficking to endosomes. We observed that poly(I:C) up-regulates transcription of UNC93B1 and promotes trafficking of TLR3 to the plasma membrane in human epithelial cell line. Up-regulation of UNC93B1 is triggered through TLR3 activation by poly(I:C). Further studies revealed that expression of UNC93B1 promotes trafficking of differentially glycosylated TLR3, but not other NAS TLRs, to the plasma membrane. UNC93B1 promoter region contains binding sites for poly(I:C)- and type I interferon-inducible regulatory elements. UNC93B1 also increases the protein lifetime of TLR3 and TLR9 and augments signaling of all NAS TLRs. Furthermore, we discovered that poly(I:C) pretreatment primes B-cells to the activation by ssDNA via up-regulation of UNC93B1. Our findings identified TLR3 as the important regulator of UNC93B1 that in turn governs the responsiveness of all NAS TLRs.
Introduction
Toll-like receptors (TLRs)2 play an essential role in innate immune response by recognition of conserved molecular patterns of pathogens or endogenous danger associated signals (1, 2). TLRs differ in ligand specificity, utilization of the signaling adapter proteins, downstream signaling effects, and in cellular localization of the receptors (1, 3). TLR3 comprises a large N-terminal horseshoe-shaped ectodomain that consists of 23 conserved leucine-rich repeats, a transmembrane helix, and a C-terminal cytoplasmic Toll-interleukin-1 receptor domain. Of 15 predicted N-linked glycosylation sites on the ectodomain of TLR3, 11 were observed in the crystal structure (4, 5). TLR3 is activated by double-stranded RNA (dsRNA), which is formed during the replication process of many viruses (6, 7) and is weaker by the endogenous cellular mRNA or small interfering RNAs (siRNA) (8). TLR3 differs from other TLRs in its dependence on Toll/IL-1R domain-containing adapter-inducing interferon-β (TRIF) rather than on the myeloid differentiation primary response gene 88 (MyD88) adaptor-mediated signaling. Recognition of dsRNA leads to the TLR3 dimerization, subsequent activation of a signaling cascade, phosphorylation of interferon regulatory transcription factor-3 (IRF-3), and activation of type I interferon (IFN) response (9). TLR3 is a member of a group of TLRs along with TLR7, TLR8, TLR9, and TLR13 that recognize nucleic acids. In unstimulated cells NAS TLRs mainly reside in the endoplasmic reticulum (ER). Upon stimulation they translocate to the endosomes where they encounter the internalized nucleic acid ligands (10, 11). Endosomal maturation and acidification are essential for the TLR response. Compounds that block the endosomal maturation such as bafilomycin A prevent activation of NAS TLRs (12–14). Activation of TLR3 and binding affinity with ligand is pH and length-dependent. The strongest response to dsRNA is achieved between pH 5.7 and 6.7 (13, 14). Acidic pH is crucial for TLR3-ligand interaction in activation with oligomeric dsRNA molecules (below 40 bp), whereas TLR3 can be activated at pH 7 with large dsRNA oligomers such as poly(I:C), suggesting that binding of dsRNA is only weakened but not completely abolished at neutral pH (13).
Translocation of NAS TLRs to endosomes is driven by UNC93B1 (15), which is a 12-helical membrane spanning ER resident protein (16). It physically interacts with transmembrane domains of NAS TLRs. The UNC93B1 3d mutation (H412R) results in a complete deficiency in NAS TLRs signaling (17). Mice with 3d mutation are highly susceptible to infection with intracellular pathogens such as cytomegalovirus, Listeria monocytogenes, and Staphylococcus aureus (16, 17). A patient with a deficiency in UNC93B1 had impaired interferon-α/β and -λ antiviral response (18). Mutation (D34A), which modifies the selectivity of UNC93B1 for TLR7 versus TLR9, results in a lethal autoimmune response (19, 20).
We show that poly(I:C), but not by CpG DNA or LPS, markedly up-regulates the endogenous UNC93B1 mRNA expression as well as translocation of TLR3 to the plasma membrane in human umbilical vein endothelial cells (HUVEC). We observed two distinct effects of UNC93B1 overexpression. First, up-regulation of UNC93B1 increased the amount of TLR3, but not other NAS TLRs, at the plasma membrane. Besides that, it extended the lifetime of TLR3 and TLR9 in cells. Plasma membrane localization correlated with the appearance of differentially glycosylated form of TLR3. Up-regulated transcription of UNC93B1 gene is TLR3-dependent as bafilomycin A completely abolished UNC93B1 mRNA levels after poly(I:C) treatment. Analysis of human UNC93B1 gene promoter region revealed transcriptional factor binding sites (TFBS) for IRF-3, NF-κB, AP-1, and cJun-ATF2. Furthermore, we investigated the consequences of UNC93B1 up-regulation in cells. UNC93B1 most potently enhanced activation of TLR9. Thus, priming of B-cells by TLR3 ligand significantly augmented the cellular response to TLR9 agonist.
We propose the role of UNC93B1 in surface localization of TLR3. A low amount of the circulating endogenous agonist of TLR3 in uninfected organism, in contrast to agonists of TLR9, TLR7, or TLR8 (i.e. endogenous ssDNA and ssRNA), permits the safe localization of TLR3 to the cell surface where it can capture circulating dsRNA that results from the viral infection (21). TLR3 can potentiate activation of TLR9 through up-regulation of UNC93B1. This may link the viral infections and the development of autoimmune diseases, as the processes, which lead to the up-regulation of UNC93B1 or NAS TLRs, can initiate or aggravate the inflammatory response to endogenous nucleic acids (22).
EXPERIMENTAL PROCEDURES
Cell Cultures
Human embryonic kidney cells (HEK) 293 and HEK293T were cultivated in DMEM (Invitrogen) supplemented with 10% (v/v) FBS (Invitrogen) at 37 °C in 5% CO2. HUVEC (Lonza) were cultivated in microvascular endothelial cell growth medium-2 (EGM2 BulletKit; Lonza) according to the supplier's recommendations at 37 °C and 5% CO2. Ramos-Blue cells (InvivoGen) were cultivated in Iscove's modified Dulbecco's medium (Invitrogen) supplemented with 2–4 mm l-glutamine, 10% (v/v) FBS.
Plasmids and Reagents
Expression plasmids containing sequences of TLR3 (pUNO-hTLR3), TLR9 (pUNO-hTLR9-HA), TLR7 (pUNO-hTLR7-HA), TLR8 (pUNO-hTLR8-HA), and UNC93B1 (pUNO1-hUNC93B1) were from InvivoGen, TLR3-mCer (pcDNA3-hTLR3-mCerulean)-containing plasmid was prepared in our laboratory (7), TLR9-YFP (pcDNA3-hTLR9-YFP) and TLR7-YFP (pcDNA3-hTLR7-YFP) were from Addgene, and plasmid constitutively expressing Renilla luciferase-phRL-TK was from Promega. The following plasmids were gifts: plasmid coding for firefly luciferase under NF-κB promoter (pELAM-1-luciferase) and pMyc-hTLR4 from C. Kirschning (Institute for Medical Microbiology, University of Duisburg-Essen, Essen, Germany), the plasmid coding for firefly luciferase under IFN-β promoter (pIFN-β-luciferase) from J. Hiscott (Departments of Microbiology and Medicine, McGill University, Montreal, QC, Canada), and pmCerulean-C1 provided by D. Piston (Vanderbilt University, Nashville, TN).
Cells were treated with different TLR ligands: polyinosinic-polycytidylic acid-poly(I:C) (InvivoGen), type A CpG-oligodeoxynucleotide ODN2216 (InvivoGen), type B CpG-oligodeoxynucleotide ODN10104 (Coley Pharmaceutical Group) and smooth LPS from Salmonella abortus equi (kindly provided by K. Brandenburg-Research Center Borstel, Germany). Human recombinant interferon β1a (IFN-β) was from Biomol.
Transfection and Reporter Gene Assay
HEK293 cells were harvested from an actively growing culture and plated onto CoStar White 96-well plates (Corning) at 2.2 × 104 cells/well. After 24 h at 50% confluence, the cells were transfected with the following plasmids: pIFN-β-luciferase (40 ng DNA/well) or ELAM1-luciferase reporter plasmid (40 ng DNA/well), TLR3 (20 ng DNA/well), TLR9 (20 ng DNA/well), UNC93B1 (1 ng DNA/well), and phRL-TK (5 ng DNA/well) (unless stated otherwise). Empty vector pcDNA3 (20 ng DNA/well) was used as a negative control. Plasmids were transfected using Lipofectamine 2000 reagent according to manufacturer's instructions (Invitrogen). 24 h post transfection the culture medium was replaced with fresh DMEM with 10% FBS, and cells were stimulated with TLR ligands poly(I:C) (10 μg/ml) and ODN10104 (10 μg/ml) (unless stated otherwise). 18 h after treatment the cells were lysed in Passive Lysis Buffer (Promega). The expression of the firefly and Renilla luciferase reporter gene was analyzed using the dual luciferase assay. Luminescence was quantified using the plate reader OrionII (Berthold Technologies). The relative luciferase expression (relative luciferase units) for each sample was calculated.
Isolation of RNA, RT-PCR, and Real-time PCR
HUVEC cells were plated onto a 12-well plate at 1.2 × 105 cells/well. 48 h later the cells were stimulated with poly(I:C) (25 μg/ml), ODN2216 (5 μm), LPS (25 ng/ml), or recombinant human IFN-β (1 nm) for 4, 8, 14, or 21 h. They were lysed with TRIzol reagent (Invitrogen). RNA was isolated using Pure Link RNA Mini Kit (Invitrogen). Reverse transcription was performed using High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Real-time PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems). The primers used for mRNA quantification were: hTLR3 (forward, 5′-GCTCCGAAGGGTGGCCCTTAAA-3′; reverse, 5′-GGTTTGCGTGTTTCCAGAGCCG-3′), hTLR9 (forward, 5′-CCCAGCATGGGTTTCTGCCGCA-3′; reverse, 5′-TGACATTGCCACGGGGTGCTG-3′), hTLR4 (forward, 5′-ATGGCTTGTCCAGTCTCGAAGT-3′; reverse, 5′-CAGGAAGGTCAAGTTTCTCAGCTC-3′), hIFN-β (forward, 5′-GGACAGGATGAACTTTGAC-3′; reverse, 5′-TGATAGACATTAGCCAGGAG-3′), hIFN-α (forward, 5′-CATCTCAGCAAGCCCAGAAGTATC-3′; reverse, 5′-AGCACCACCAGGACCATCAG-3′), or hUNC93B1 (forward, 5′-TGGTATCGCCTTGGGCTATGGC-3′; reverse, 5′-ATGAAGGTGAGCAGCAGGTGCAC-3′). Expression of human hypoxanthine guanine phosphoribosyl transferase (forward, 5′-AGATGGTCAAGGTCGCAAG-3′; reverse, 5′-TTCATTATAGTCAAGGGCATATCC-3′) mRNA was detected in each sample and used for normalization of target gene mRNA expression.
FACS Analysis
HEK293T cells were seeded onto 6-well plates at 1.0 × 106 cells/well. After 24 h at 50% confluence, they were transfected with TLR3 (3 μg of DNA/well (unless stated otherwise)), UNC93B1 (50 and 100 ng DNA/well), or control vector pcDNA3 (3 μg DNA/well). After 48 h, the cells were harvested and washed with ice-cold FACS buffer (1× PBS (pH 7.4), 0.5% BSA (Sigma)). HUVEC cells were seeded onto 12-well plates at 1.0 × 106 cells/well. At 100% confluence, the cells were stimulated with poly(I:C) (50 μg/ml), LPS (50 ng/ml), ODN2216 (50 μg/ml), or IFN-β (1 nm) for 24 h. Cells were harvested and washed twice with ice-cold FACS buffer. For intracellular staining, cells were fixed with 2% paraformaldehyde and permeabilized with 0.1% Triton X-100 (both Sigma). Cells were stained with mouse monoclonal anti-TLR3 (clone TLR3.7, InvivoGen) or a negative control mouse IgG1 control antibody (ab18443, Abcam) for 30 min on ice. Cells were washed with FACS buffer and stained with phycoerythrin-labeled goat F(ab)2 anti-mouse IgG secondary antibodies (IM0551, Beckman Coulter) for 30 min on ice in the dark. Flow cytometry analysis was performed with the CyFlow space flow cytometer (Partec). In each sample 10,000 cells were analyzed. Collected data were analyzed using FlowJo (Tree Star) software.
SDS-PAGE and Western Analysis
HEK293T cells were seeded onto 12-well plates (Techno Plastic Products) at 2.2 × 105 cells/well. After 24 h at 50% confluence they were transfected with TLR3, TLR9, TLR4 (all 900 ng DNA/well), UNC93B1 (10 or 30 ng of DNA/well), and control vector pcDNA3 (900 ng of DNA/well). 48 h after transfection, the cells were lysed using RIPA buffer (50 mm Tris (pH 7.5), 150 mm NaCl, 1% (v/v) Triton X-100, 0.1% SDS, 0.5% deoxycholate) with Complete Mini protease inhibitors (Roche Applied Science), sonicated, and centrifuged. HUVEC cells were seeded onto 6-well plates at 3 × 105 cells/well. After 24 h the cells were stimulated with poly(I:C) (50 μg/ml), LPS (50 ng/ml), IFN-β (1 nm), or ODN2216 (5 μm). 24 h after stimulation, the cells were lysed with RIPA buffer, sonicated, and centrifuged. The protein-containing supernatants were harvested, and the total protein amount was quantified using the BCA protein assay (Sigma). Cell extracts (30 μg of total proteins) were incubated at 65 °C for 5 min in sample buffer (SDS with 2-mercaptoethanol) and loaded onto a 12% SDS-PAGE gel (unless stated otherwise). After electrophoresis, proteins were transferred onto nitrocellulose membranes Hybond-ECL (GE Healthcare) and detected with the following primary antibodies: mouse monoclonal anti-TLR3 (IMG-315A, Imgenex), rabbit anti-HA (H6908, Sigma), rabbit anti-Myc (C3956, Sigma), mouse anti-β-actin (3700, Cell Signaling), rabbit anti-green fluorescent protein (A11122, Invitrogen). Used secondary antibodies were goat anti-mouse IgG-HRP (sc-2005, Santa Cruz) and goat anti-rabbit IgG-HRP (ab6721, Abcam). The blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce). Membranes were recorded with G:BOX Chemi using GeneSnap software (Syngene). Band analysis and densitometry was performed with GeneTools analysis software (Syngene).
Analysis of Human UNC93B1 Promoter Region
The analysis of the human UNC93B1 gene promoter region for potential TFBS was done using the MatInspector prediction software (Genomatix). MatInspector identifies TFBS in nucleotide sequences using a library of position weight matrices (23). The analyzed sequence was a region spanning 2800 bp upstream and 200 bp downstream the transcription starting site.
siRNA Silencing of IRF-3
HUVEC cells were seeded onto a 12-well tissue culture plate at 1.5 × 105 cells/well. 24 h later, the cells were transfected with siRNA ON-TARGETplus SMARTpool specific to human IRF-3 or negative control siRNA ON-TARGETplus Non-targeting Pool using DharmaFECT 4 transfection reagent (all Darmacon) according to the manufacturer's instructions. After 48 h the cells were treated with poly(I:C) (25 μg/ml) for 4, 8, and 14 h or left untreated. They were lysed using TRIzol reagent and subjected to RNA isolation and real-time PCR. For confirmation of successful siRNA silencing, cells were lysed in RIPA buffer and subjected to Western blotting. IRF-3 was detected with rabbit monoclonal anti-IRF-3 primary antibodies (4302, Cell Signaling) and goat anti-rabbit IgG-HRP secondary antibodies (ab6721, Abcam).
Confocal Microscopy
HEK293T cells were seeded onto 8-well tissue culture chambers (Ibidi) at 2.2 × 105 cells/well. After 24 h the cells were transfected with 150 ng of DNA/well of the TLR3-mCer and TLR9-YFP alone or cotransfected with UNC93B1 (30 ng of DNA/well). Localization was visualized 48 h post transfection. A plasma membrane was stained with either SynaptoRed (Biotioum) or Cholera Toxin Subunit B Alexa Fluor 555 (Molecular Probes, Invitrogen). Images were acquired using the Leica TCS SP5 inverted laser-scanning microscope on a Leica DMI 6000 CS module equipped with a HCX Plane-Apochromat lambda blue 63 × oil-immersion objective with NA 1.4 (Leica Microsystems). Images were processed with LAS AF software (Leica Microsystems) followed by ImageJ software (NIMH, National Institutes of Health, Bethesda, MD). Colocalization was identified with LAS AF software. Fluorescence intensity from two channels, TLR and plasma membrane, were measured in line profile (n = 9) in a distance of 3-μm crossing membrane. Florescence intensities per distance were plotted (GraphPad Prism software), and maximum fluorescence intensities of TLR were compared with those of plasma membrane.
Peptide N-Glycosidase F Treatment
HEK293T cells were seeded onto 12-well plates at 2.2 × 105 cells/well. After 24 h, at 50% confluence, they were transfected with TLR3 (900 ng of DNA/well), UNC93B1 (30 ng of DNA/well), or control vector pcDNA3 (900 ng of DNA/well). The cells were lysed 48 h after transfection using RIPA buffer with protease inhibitors, and the total protein amount was quantified. Lysates were treated with peptide N-glycosidase F according to manufacturer's instructions (New England Biosystems). Samples were subsequently subjected to Western blotting.
Biotinylation and Purification of Cell Surface Proteins
HEK293T cells plated onto a 60-mm tissue culture dishes at 8.5 × 105 cells/dish. After 24 h, at 50% confluence, they were transfected with TLR3 (5 μg DNA/well) alone or cotransfected with UNC93B1 (150 ng of DNA/well) and control vector pcDNA3 (5 μg of DNA/well). All cells were transfected with 25 ng of DNA/well of a pmCerulean-C1 expressing Cerulean fluoescent protein, which was used as a marker for cytoplasmic fraction. 48 h after transfection, the cells were washed with PBS (pH 8) and detached from the plate with cell scraper. Membrane-impermeable EZ-Link Sulfo-NHS-LC-LC-Biotin (sulfosuccinimidyl-6-[biotinamido]-6-hexanamido hexanoate; Pierce) was added to detached cells to covalently bind to primary amine-containing proteins exposed on the cell surface. Incubation for 30 min on 4 °C (to prevent internalization) with gentle shaking was performed. Subsequently, cells were washed with PBS with 100 mm glycine (pH 8) to quench and remove excess biotin reagent and lysed with lysis buffer (50 mm Tris-HCl (pH 8), 150 mm NaCl, 1.25% Triton X-100, 0.25% SDS, 5 mm EDTA, 5 mg/ml iodoacetamide, 0.5% deoxycholate, Complete Mini protease inhibitors) and sonicated. The amount of total cargo proteins was determined with BCA assay. Dynabeads MyOne Streptavidin C1 superparamagnetic beads (Invitrogen) were used to capture the biotinylated proteins from lysates. Beads were separated with a magnet and washed several times with PBS (pH 7.4). The biotin-streptavidin bond was broken by boiling the sample for 5 min in sample buffer (SDS with 2-mercaptoethanol). Each sample obtained was considered to contain an equal amount of proteins. Samples were subsequently subjected to Western blotting.
Determination of Protein Half-life
HEK293T cells were seeded onto 12-well plates (Techno Plastic Products) at 2.2 × 105 cells/well. After 24 h, at 50% confluence, they were transfected with TLR3 or TLR9 (900 ng DNA/well) and UNC93B1 (30 ng DNA/well) or with control vector pcDNA3 (900 ng DNA/well). After 48 h, the cells were treated with 80 μg/ml cycloheximide, inhibitor of protein synthesis (Sigma) for 1, 3, 5, and 7 h. The cells were lysed with RIPA buffer. The protein-containing supernatants were harvested, and the total protein amount was quantified. Lysates were subsequently subjected to Western blotting. Band analysis and densitometry were performed with GeneTools analysis software (Syngene). The one phase exponential nonlinear fit was performed with GraphPad Prism 5 (GraphPad Prism Software) to calculate protein half-life. Plateau was constrained to a constant value of 0.5, and Y value at time 0 was defined as 1.
Experiments on Ramos-Blue Cells
Ramos-Blue are a B lymphocyte cell line that stably expresses an NF-κB/AP-1-inducible secreted embryonic alkaline phosphatase reporter gene. Cells were seeded onto 12-well plates at 2 × 106 cells/well. Immediately, the cells were stimulated with poly(I:C) (20 μg/ml), ODN2216 (20 μg/ml), ODN10104 (20 μg/ml), or R-848 (20 μg/ml) for 4, 8, 16, or 24 h. Supernatants were collected and transferred onto a 96-well plate in triplicates. Because secreted embryonic alkaline phosphatase is secreted in medium, it can be readily monitored using QUANTI-Blue reagent (InvivoGen) at 630 nm with a spectrophotometer. Residual cells were lysed with TRIzol reagent and subjected to RNA isolation and real-time PCR. To confirm that UNC93B1 mRNA transcription is TLR3-dependent, we stimulated cells with poly(I:C) (20 μg/ml) alone or together with 0.2 μm bafilomycin A (LC Laboratories). RNA was isolated and subjected to real-time PCR.
For priming experiments cells were seeded onto 96-well plates at 2 × 105 cells/well. After 24 h the cells were incubated for 4, 8, or 12 h with medium or first TLR agonist and then treated with second agonist for additional 12 h. At time 0 h, both agonists were added simultaneously. Agonists used were poly(I:C) (0.5 μg/ml) and ODN2216 (0.5 μg/ml). Agonist concentrations were much lower, because at higher concentrations responses reached plateau. After 12 h cells were centrifuged, supernatants were transferred in another 96-well plate, and levels of secreted embryonic alkaline phosphatase were measured.
Statistical Analysis
Error bars represent mean S.D. of triplicate samples. Data were compared for significance using the unpaired t test and were considered significant with p value of 0.05.
RESULTS
Stimulation by poly(I:C) Up-regulates Transcription of UNC93B1 and Intracellular Translocation of TLR3 to the Cell Surface in HUVEC Cells
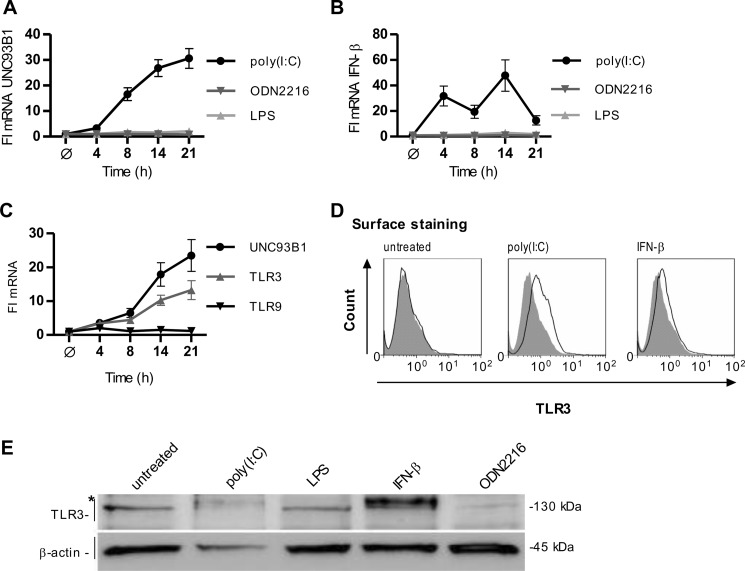
TLR3 is localized in the ER in unstimulated cells, whereas it translocates to endosomes upon stimulation with dsRNA (11). However, TLR3 has been also observed at the cell surface of several cell types (24–26). We analyzed how activation of TLRs affects the expression of UNC93B1 HUVEC cells. The cells were stimulated by poly(I:C), ODN2216, and LPS as agonists of TLR3, TLR9, and TLR4, respectively. Only stimulation by poly(I:C) induced strong up-regulation of UNC93B1 mRNA (Fig. 1A). In addition, poly(I:C) stimulation led to the up-regulated transcription of TLR3 and TLR9 (supplemental Fig. S1A). Only poly(I:C) induced the rapid and robust increase in the IFN-β mRNA transcription (Fig. 1B), whereas IFN-α mRNA transcription was induced upon LPS and ODN2216 stimulation but not by poly(I:C) (supplemental Fig. S1B). Poly(I:C) induced robust IFN-β response already after 4 h, and subsequently the increased transcription of UNC93B1 was detected after 8 h. Treatment of HUVEC cells with IFN-β increased UNC93B1 and TLR3 but not TLR9 mRNA transcripts (Fig. 1C). Stimulation by poly(I:C) and IFN-β (Fig. 1D), but not by ODN2216 (supplemental Fig. S1C), also increased the amount of cell surface TLR3 in HUVEC cells. Localization of TLR3 to the cell surface correlates with the appearance of an additional form of TLR3. This form with increased molecular weight was observed in cells stimulated with poly(I:C) and IFN-β but not in cells stimulated with LPS and ODN, which therefore did not direct TLR3 toward the cell membrane (Fig. 1E).
FIGURE 1.
Activation of human umbilical vein endothelial cells by poly(I:C) up-regulates transcription of UNC93B1 and surface localization of TLR3. A and B, HUVEC cells were incubated with poly(I:C) (25 μg/ml), ODN2216 (5 μm), or LPS (25 ng/ml) for the indicated times. UNC93B1 (A) and IFN-β (B) mRNA transcripts were determined by real-time PCR. C, HUVEC cells were incubated with IFN-β (1 nm) for the indicated times. Induction of UNC93B1, TLR3, and TLR9 mRNA transcripts was measured by real-time PCR. A–C, the results are represented by mean values with S.D. from triplicate wells. The representative data from three experiments are shown. D, expression of cell surface-associated TLR3 is shown. HUVEC cells were either untreated or stimulated with poly(I:C) (50 μg/ml) or IFN-β (1 nm) for 24 h. Histograms of unstained cells (filled areas) or cells stained with anti-TLR3 antibodies (solid lines) are shown. Data are representative of two experiments. E, HUVEC cells were stimulated with poly(I:C) (50 μg/ml), LPS (50 ng/ml), IFN-β (1 nm), or ODN2216 (5 μm) for 24 h, and cell lysates were separated by 5% SDS-PAGE. A Western blot was performed with anti-TLR3 antibody and anti-β-actin antibodies. * indicates the differentially glycosylated form of TLR3. The representative image from two experiments is shown.
These results show that transcription of UNC93B1 gene is up-regulated by stimulation with poly(I:C) and IFN-β. Increased expression of UNC93B1 correlates with localization of TLR3 on the plasma membrane.
Analysis of the UNC93B1 Promoter Region
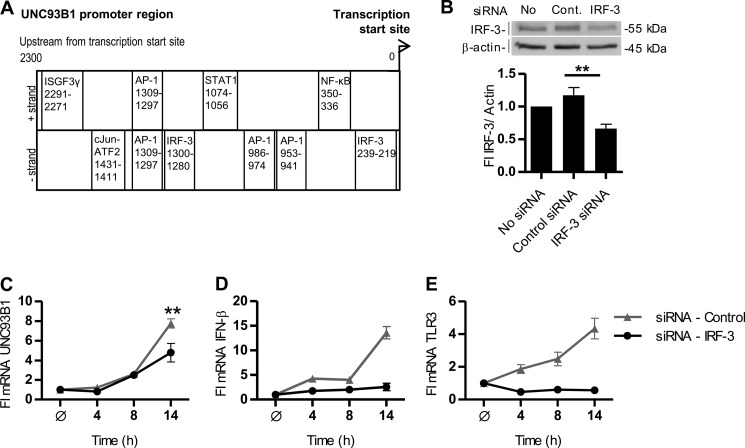
Because we showed that the UNC93B1 mRNA transcription is up-regulated by treatment with poly(I:C) and IFN-β, we set to investigate the transcriptional regulation of the UNC93B1 gene. We examined its promoter region 2800 bp upstream and 200 bp downstream of the transcription site start for TFBS using the TFBS prediction software MatInspector (23). Binding sites for IRF-3, NF-κB, AP-1, and cJun-ATF2 (Fig. 2A) were identified. Those transcription factors can be activated by TRIF- and MyD88-mediated signaling (1, 27–29) and are also responsible for the transcriptional induction of the ifn-β gene (9). IFN-β binds to the IFNAR1 and IFNAR2 receptor complex leading to the activation of Tyk2 and Jak1 with subsequent formation of STAT1-STAT2 heterodimers (30). In the nucleus STAT1-STAT2 heterodimer associates with the DNA-binding protein ISGF3γ (31) to form a heterotrimeric complex ISGF3, which binds the interferon-sensitive response element of IFN-α/β-responsive genes (32). Interferon-sensitive response element was not found in the UNC93B1 promoter region; however, STAT1 and ISGF3γ binding sites were predicted by MatInspector. Silencing of the IRF-3 transcription factor by 50% (Fig. 2B) partially inhibited the UNC93B1 mRNA transcription (Fig. 2C), whereas IFN-β and TLR3 transcripts were strongly impaired (Fig. 2, D and E). These data suggest that UNC93B1 mRNA transcription is regulated by the coordinated action of poly(I:C)-induced transcriptional factors with contribution of the type I interferon signaling.
FIGURE 2.
Transcriptional regulation of UNC93B1. A, shown is a schematic map of the 2300 bp of the human UNC93B1 promoter region. TFBS for transcriptional factors promoting antiviral response are distributed according to the distance from the transcription start site. B–E, HUVEC cells were transfected with an siRNA pool specific to human IRF-3 or negative control siRNA non-target pool. B, HUVEC cells were lysed and subjected to Western blot. IRF-3 was detected with anti-IRF-3 antibodies. Anti-β-actin antibodies were used as a loading control. IRF-3 expression was normalized to the loading control and calculated as a percentage of the intensity of the untreated control. Densitometric analysis is shown below the blot. This experiment was repeated twice, and the representative data are shown. Statistical significance is indicated by **, p ≤ 0.05. C–E, the effect of IRF-3 knockdown on transcription of UNC93B1, IFN-β, and TLR3 is shown. 48 h after siRNA transfection cells were treated with poly(I:C) (25 μg/ml) for 4–21 h or left untreated. Induction of UNC93B1 (C), IFN-β (D), and TLR3 (E) mRNA was subsequently measured by real-time PCR. The results are represented by mean values with S.D. from the triplicate wells showing the representative data from three experiments. Statistical significance is indicated by **, p ≤ 0.05.
UNC93B1 Directs Localization of Differentially Glycosylated TLR3 to the Plasma Membrane
When UNC93B1 was up-regulated, we observed translocation of TLR3 toward the cell surface of HUVEC cells. We set to investigate the mechanism of this phenomenon.
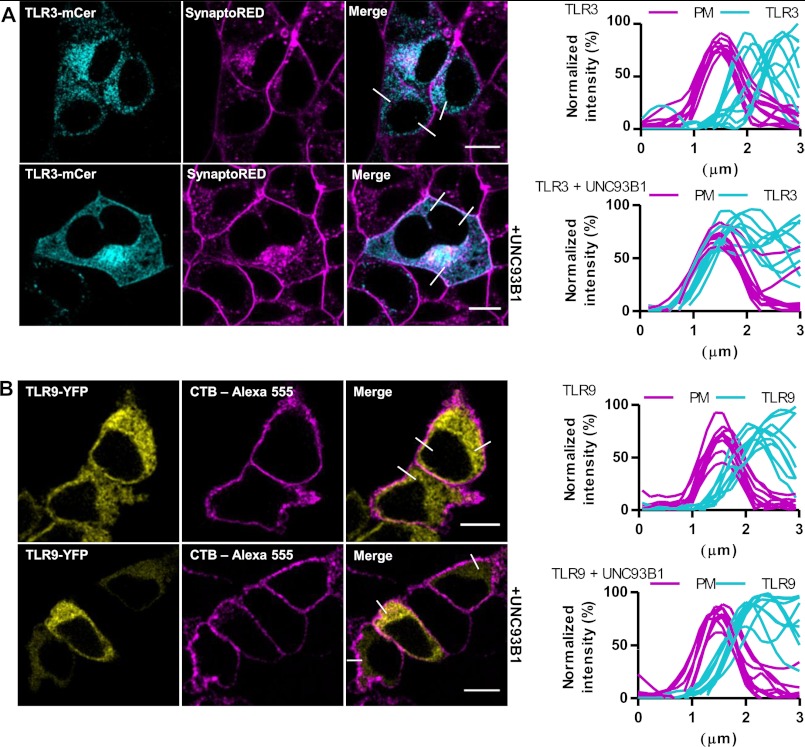
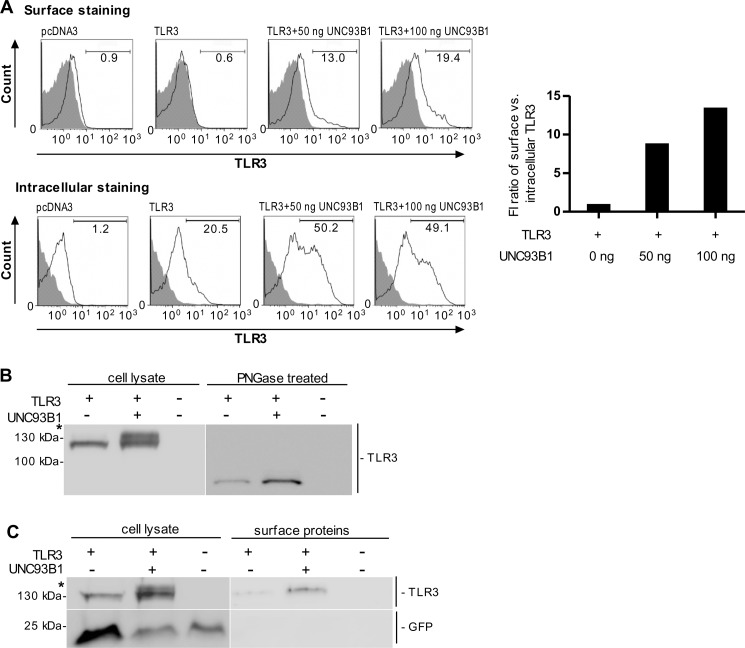
Human embryonic kidney cells 293T (HEK293T) were transfected with TLR3-mCer alone or together with plasmid encoding UNC93B1. Confocal microscopy revealed that overexpression of UNC93B1 induced translocation of TLR3 to the cell surface (Fig. 3A). However, no translocation of TLR9 (Fig. 3B), TLR7 (supplemental Fig. S2A) or TLR8 (supplemental Fig. S2B) toward the plasma membrane was observed upon UNC93B1 overexpression. Analysis of colocalization revealed that in cells overexpressing TLR3 and only endogenous UNC93B1, traces from plasma membrane and TLR3 do not colocalize, whereas in cells overexpressing UNC93B1 all curves overlap (Fig. 3A, right). Those results were confirmed by flow cytometry where coexpression of UNC93B1 led to a distinct increase (about 13-fold) of the cell population expressing cell surface-associated TLR3 (Fig. 4A, right). However, surface localization of TLR3 was not caused by the increased protein expression as the overexpression of TLR3 (supplemental Fig. S3) without up-regulated UNC93B1 failed to increase the population of cells with surface-localized TLR3 (supplemental Fig. S3, right).
FIGURE 3.
UNC93B1 augments surface localization of TLR3 in HEK293T cells. A and B, HEK293T cells were transfected with a TLR3-mCer (cyan) (A) or TLR9-YFP (yellow) (B) alone (top) or with UNC93B1 encoding plasmid (bottom). Plasma membrane markers SynaptoRed (A) and cholera toxin subunit B Alexa Fluor 555 (CTB-Alexa) (B) are shown in magenta. A and B, TLR membrane localization was evaluated from plots of normalized fluorescence intensities of TLR and plasma membrane (PM) within 3-μm line profiles (n = 9). Three representative lines are marked on merged images. Images are selected from five independent experiments. Scale bars, 10 μm.
FIGURE 4.
UNC93B1 translocates the differentially glycosylated TLR3 to the plasma membrane. A, HEK293T cells were transfected with TLR3 alone or with UNC93B1. Cell surface and intracellular expression of TLR3 was determined using flow cytometry. Histograms of mock-transfected cells (pcDNA3, filled areas) or cells transfected with TLR3 (solid lines) are shown. The representative data from three experiments are shown. Increase of cell population expressing plasma membrane-localized TLR3 normalized by the number of cells expressing intracellular TLR3 is shown on the right. B, HEK293T cells were transiently transfected with plasmid encoding TLR3 alone or with UNC93B1. 48 h post-transfection cell lysates were loaded onto a 5% SDS-PAGE gel. TLR3 was detected on a Western blot using anti-TLR3 antibody (left). * indicates the differentially glycosylated form of TLR3. Cell lysates were treated with peptide N-glycosidase F (PNGase; right). The representative image from three experiments is shown. C, HEK293T cells were transiently transfected with TLR3 alone or with UNC93B1. 48 h post-transfection surface proteins were biotinylated. Cells were lysed, and the biotinylated proteins were isolated, blotted, and detected using anti-TLR3 antibody. Cytoplasmic mCerulean was detected using anti-GFP antibody. * indicates differentially glycosylated form of TLR3. The representative image from three experiments is shown.
When UNC93B1 was overexpressed along with TLR3, an additional band of TLR3 higher than 130 kDa was observed. This was similar to what we showed in HUVEC cells and as reported before (33). This increase was due to glycosylation of TLR3 as it is peptide N-glycosidase F-sensitive (Fig. 4B). To test if a differentially glycosylated TLR3 is restricted to the cell surface, we biotinylated the surface proteins of HEK293T cells expressing TLR3 in the absence or presence of UNC93B1 overexpression. Biotinylated TLR3 was detected in the lysate of cells overexpressing UNC93B1, whereas the size of the band corresponded only to a differentially glycosylated form (Fig. 4C).
Similar reports were recently published by Qi et al. (33) where they also showed a differently modified form of TLR3 on the cell surface upon UNC93B1 coexpression. Our results demonstrate that UNC93B1 directs TLR3 to the cell compartment where it is differentially glycosylated and targeted toward the cell membrane, whereas UNC93B1 itself does not reach the cell surface (Ref. 15); data now shown). This phenomenon, however, is unique to TLR3 as no cell surface localization or modified glycosylation pattern due to the up-regulated UNC93B1 was observed for TLR9, TLR7, or TLR8 (supplemental Fig. S4A).
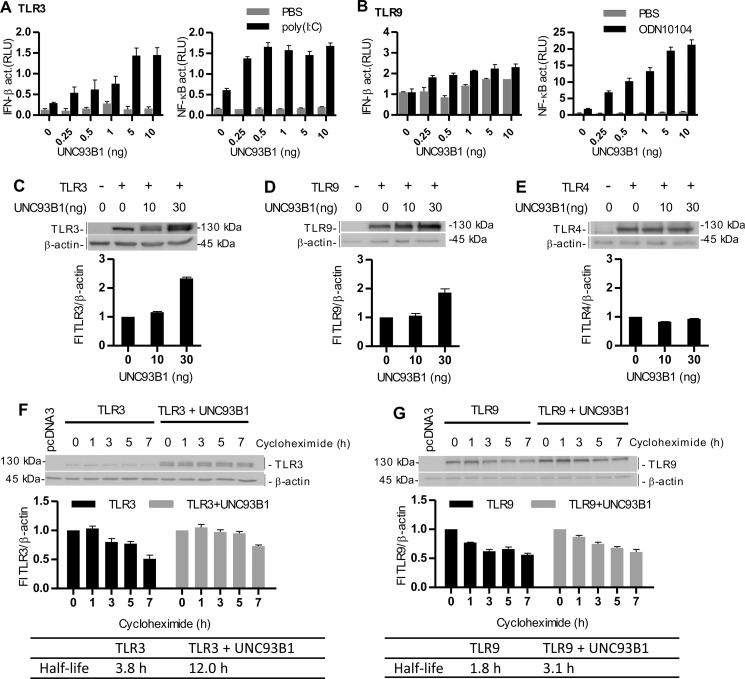
UNC93B1 Augments Signaling and Stability of TLR3 and TLR9
Furthermore, we investigated the effect of UNC93B1 up-regulation on the responsiveness of different TLRs. Overexpression of UNC93B1 led to the amplified responsiveness to TLR3 and TLR9 (Fig. 5, A and B, respectively) and TLR7 and TLR8 agonists (supplemental Fig. S4, B and C). The increased activation was most pronounced for TLR9. Additionally, overexpression of UNC93B1 markedly increased the protein levels of TLR3 (Fig. 5C) and TLR9 (Fig. 5D), whereas it had no effect on TLR4 (Fig. 5E), which does not interact with UNC93B1 (17). In this experimental setting UNC93B1 could not influence the transcription of TLRs as they are introduced into HEK293 cells without their endogenous gene regulatory context. The increased protein level is, therefore, most likely a consequence of the increased lifetime of TLR proteins associated with UNC93B1. Overexpression of UNC93B1 decreased the degradation rate of TLR3 upon the inhibition of protein synthesis by cycloheximide (3.8-h versus 12-h half-life). Similar results were obtained with TLR9 with an increase of a protein half-life from 1.8 to 3.1 h. Those data suggest that UNC93B1 increases the half-life of associated proteins in cells probably by protection against the proteolytic degradation (Fig. 5, F and G). Results are in agreement with a report of Qi et al. (33) where they showed that UNC93B1 coexpression increased a TLR3 protein half-life from 3 to 6 h.
FIGURE 5.
UNC93B1 augments signaling and protein lifetime of TLR3 and TLR9. A and B, HEK293 cells were transfected with plasmid encoding TLR3 (A) or TLR9 (B) with increasing amounts of UNC93B1. Cells were cotransfected with IFN-β- or NF-κB-responsive reporter plasmids and Renilla normalization reporter plasmid. 18 h after stimulation with poly(I:C) (10 μg/ml) (A) or ODN10104 (10 μg/ml) (B), luciferase activity (relative luciferase units (RLU)) was measured in the cell lysates. The results are represented by mean values with S.D. from the triplicate wells. The representative data from three experiments are shown. C–E, HEK293T cells were transfected with TLR3 (C), TLR9 (D), and TLR4 (E) alone or with UNC93B1. A Western blot was performed using anti-TLR3 (C), anti-HA (D), or anti-Myc (E) antibodies. Anti-β-actin antibodies were used as a loading control. TLR3, TLR9, or TLR4 protein expression was normalized to the loading control and calculated as a percentage of the intensity of the proteins in cells not overexpressing UNC93B1. Densitometric analyses are shown below each blot. The representative data from three experiments are shown. F and G, HEK293T cells were transfected with TLR3 (F) or TLR9 (G) alone or with UNC93B1. 48 h post-transfection, the cells were incubated with cycloheximide for the indicated times. Cell lysates were prepared, and a Western blot was performed using anti-TLR3 (F) or anti-HA (G) antibodies. Anti-β-actin antibodies were used as a loading control. TLR3 and TLR9 protein expression was normalized to the loading control and calculated as a percentage of the intensity of the proteins in the untreated cells. Densitometric analyses and calculated half-lives of proteins are shown below each blot. The representative data from three experiments are shown.
Priming of B-cells by poly(I:C) Potentiates the TLR9 Response via UNC93B1
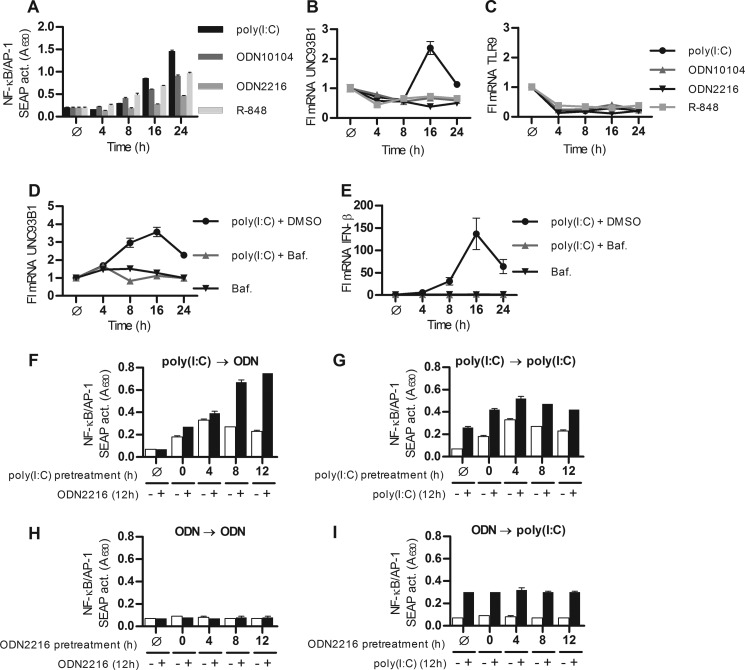
Up-regulation of TLRs and UNC93B1 is characteristic for several autoimmune conditions (34–36). Because the stimulation of TLR3 up-regulates UNC93B1, which most effectively potentiates activation of TLR9, we reasoned that the prior cell stimulation by a TLR3 agonist could increase the responsiveness of cells to a subsequent stimulation by a TLR9 agonist. Human B-lymphocytes (Ramos-Blue cells) express several pattern recognition receptors and respond to TLR3, TLR9, and TLR7 agonists such as poly(I:C), ODN10104 (B type CpG ODN), ODN2216 (A type CpG ODN), and R-848 (Fig. 6A). Stimulation of B-cells by poly(I:C), but not by other TLR agonists, was able to up-regulate the transcription of UNC93B1 (Fig. 6B), as we previously demonstrated in HUVEC cells. None of the agonists increased the transcription of TLR9 (Fig. 6C). Elevated transcription of UNC93B1 mRNA was abolished after pretreatment with bafilomycin A (Fig. 6D). Bafilomycin A also abolished poly(I:C)-induced IFN-β mRNA transcription (Fig. 6E). These data demonstrate that UNC93B1 gene transcription is triggered by TLR3 and not by cytosolic dsRNA sensors.
FIGURE 6.
Poly(I:C) pretreatment primes B-cells to the activation by ssDNA via up-regulation of UNC93B1. A–C, Ramos-Blue cells were stimulated with poly(I:C) (20 μg/ml), ODN2216 (20 μg/ml), ODN10104 (20 μg/ml), and R-848 (20 μg/ml) for the indicated times. A, NF-κB/AP-1-dependent secreted embryonic alkaline phosphatase (SEAP) activity (A630) was measured in supernatants. UNC93B1 (B) and TLR9 (C) mRNA were subsequently measured by real-time PCR. D and E, cells were stimulated with poly(I:C) (20 μg/ml) + DMSO alone or pretreated with bafilomycin A (Baf.) for the indicated times. UNC93B1 (D) and IFN-β (E) mRNA were measured by real-time PCR. The results are represented by mean values with S.D. from triplicate wells. The representative data from three experiments are shown. F–I, Ramos-Blue B-cells were pretreated with medium (Ø) or first agonist for 0–12 h and subsequently treated with second agonist for additional 12 h. Agonist used were poly(I:C) (0.5 μg/ml) and ODN2216 (0.5 μg/ml). F, the first agonist was poly(I:C), and the second agonist was ODN2216. G, the first agonist was poly(I:C), and the second agonist was also poly(I:C). H, the first agonist was ODN2216, and the second agonist was also ODN2216. I, the first agonist was ODN2216, and the second agonist was poly(I:C). Results shown in F–I were all preformed in one experiment setting. The results are represented by mean values with S.D. from triplicate wells. The representative data from three experiments are shown.
For the priming experiments we compared different sequential combinations of TLR3 and TLR9 agonists. Cells were pretreated with the first agonist for 0–12 h and then treated with the second agonist for additional 12 h. A strongly augmented response took place only when cells were pretreated with poly(I:C) and subsequently stimulated with ODN2216 (Fig. 6F). Pretreatment and treatment with poly(I:C) resulted in an additive response (Fig. 6G). Although stimulation with ODN alone or pretreatment with the same agonist resulted in only weak activation of B-cells (Fig. 6H). Finally, no priming effect was observed when cells were pretreated with ODN and subsequently stimulated with poly(I:C) (Fig. 6I). It should be noted that agonist concentrations in priming experiments were much lower, as at higher concentrations responses reached plateau before we could observe any priming effect.
Elevated response upon pretreatment with poly(I:C) could be a consequence of the enhanced expression of TLR9, as we observed the up-regulation of TLR9 mRNA (supplemental Fig. S1A) in HUVEC cells. However, no up-regulation of TLR9 was observed in RAMOS-Blue cells with either poly(I:C) or ODN (Fig. 6C). From our data we can conclude that the mechanism of enhanced response to TLR9 ligand upon poly(I:C) pretreatment is probably a consequence of the UNC93B1 up-regulation.
DISCUSSION
TLR3 differs from other NAS TLRs in several aspects. It depends on the TRIF adaptor in contrast to MyD88-mediated signaling of other TLRs, and its signaling cascade activates the IRF-3 transcription factor rather than IRF-7 and NF-κB, which are induced by other NAS TLRs (6, 9). TLR3 also represents a separate evolutionary group distinct from single-stranded polynucleotide recognizing TLR7, TLR8, and TLR9 (37, 38). We found out that UNC93B1 induces translocation of differentially glycosylated TLR3, but not TLR7, TLR8, or TLR9, to the plasma membrane. This is another unique characteristic of TLR3. In this report we showed that UNC93B1 expression is up-regulated by poly(I:C) stimulation in HUVEC and RAMOS-Blue B-cell line. We described two distinct effects of UNC93B1 up-regulation in cells. It promotes the translocation of differentially glycosylated form of TLR3, but not other NAS TLRs, to the cell surface. In addition to the translocation, it also augments the responsiveness of NAS TLRs by increasing their lifetime.
Activation by poly(I:C) and IFN-β triggered the up-regulation of UNC93B1 and TLR3 in HUVEC cells. UNC93B1 mRNA levels started to increase only after the IFN-β mRNA peak, suggesting that UNC93B1 is IFN-β-inducible. However, neutralizing antibodies to IFN-β only partially decreased the UNC93B1 mRNA (data not shown). Analysis of UNC93B1 promoter region suggests that its transcription is regulated by multiple, probably synergistic TLR3-induced transcription factors as silencing of IRF-3 resulted in only partial suppression of UNC93B1.
TLR3 and TLR4 both trigger TRIF-TBK1-dependent IRF-3 activation (39). However; no induction of UNC93B1 mRNA was observed in HUVEC cells after stimulation with LPS. Koehn et al. (40) reported minor induction of UNC93B1 mRNA with LPS in monocyte-derived dendritic cells but not in monocyte-derived macrophages, suggesting cell type-specific regulation.
It has been reported that knockdown of UNC93B1 reduced both the full-length TLR3 at the plasma membrane as well as the secretion of TLR3 ectodomain (41), but it has not been known that TLR3 stimulation up-regulates UNC93B1, forming a positive feedback loop. dsRNA can be detected by cytoplasmic receptors such as MDA5 and RIG-1, which also lead to antiviral interferon response (42). However, the sensitivity to bafilomycin A demonstrates that UNC93B1 up-regulation triggered by poly(I:C) is caused by the endosomal and not by cytosolic dsRNA sensors.
Qi et al. (33) also showed the presence of differentially glycosylated form of TLR3 in cells coexpressing UNC93B1. UNC93B1 probably translocates TLR3 to the compartment where additional glycosylation takes place. This different glycosylation could be due to the addition of N-linked glycans or to the introduction of complex type N-linked glycans (43). TLR3 glycosylation is important for its bioactivity as the inhibition of glycosylation by tunicamycin prevents TLR3-induced signaling (13, 44). However, it is unlikely that glycosylation affects ligand binding as glycosylation sites are positioned on the surface of TLR3 ectodomain that does not interfere with the ligand (5). Qi et al. (33) reported that a portion of TLR3 in cells coexpressing UNC93B1 is endoglycosidase H-resistant, suggesting that the resistant protein portion traffics through Golgi complex where additional glycosylation process takes place. We demonstrated that differentially glycosylated TLR3 is limited to the cell surface, as only TLR3 with a higher molecular weight was detected in cell surface protein fraction. Different glycosylation probably exerts its effect through trafficking and potentially through protein stability (43). In general, glycans are important for direct targeting of proteins to specific locations within the cell (43).
Endosomal localization with acidification is crucial for robust signaling of NAS TLRs (10, 12–14). It prevents activation with endogenous nucleic acids (45), which can lead to the development of autoimmune diseases such as psoriasis and systemic lupus erythematous (46, 47). It was reported that chimeric TLR9 receptor, in which the ectodomain of murine TLR9 was fused to the transmembrane and cytosolic domains of murine TLR4, localized to the cell surface and was able to recognize the endogenous DNA (48). Chimeric murine TLR9 comprising a transmembrane segment and part of the ectodomain of murine TLR3 also localized to the cell surface and no longer required proteolysis for the activation by the endogenous ssDNA, resulting in the development of lethal autoinflammation in mice (49). In contrast to TLR3, those chimeric receptors were not sensitive to bafilomycin A (48, 49). However, similar TLR9-TLR4 chimera in another study failed to localize to the membrane (50). We argue that the inability of TLR9, TLR7, and TLR8 to be translocated by UNC93B1 toward the cell surface can be considered as an evolutionary adaptation to prevent autoimmune response to self-DNA and ssRNA. In contrast, TLR3 is present at the cell surface of MRC-5 fibroblasts (51), human corneal epithelial cells (24), human lung microvascular endothelial cell (26), and HUVEC (25) without pathological consequences. In the airway epithelial cells A549, TLR3 translocation to the cell surface was observed upon the respiratory syncytial virus infection. Respiratory syncytial virus infection sensitized the airway epithelial cells to subsequent stimulation by dsRNA by the up-regulation of TLR3 (21). Hewson et al. (52) demonstrated that rhinovirus infection increased the expression of TLR3 mRNA and TLR3 protein at the surface of human bronchial epithelial cells BEAS-2B, which is, as we showed in this report, probably a result of the up-regulated UNC93B1. Furthermore, it was reported that the exogenous dsRNA also increased the surface TLR3 expression (21, 52). We observed the increased surface localization of TLR3 by stimulation of HUVEC cells with poly(I:C) and IFN-β. In addition, cell surface localization coincided with the differentially glycosylated form of TLR3. We conclude that this effect is a consequence of up-regulation of UNC93B1 by those two agents.
The functional role of the surface-localized TLR3 is still an open question. Although some reports claim that activation with dsRNA occurs entirely in acidic environment (13, 14), several studies reported that surface-expressed TLR3 participates in the recognition of dsRNA and triggers signaling pathway (21, 51, 52). De Bouteiller et al. (13) demonstrated that in contrast to dsRNA, activating antibodies against TLR3 ectodomain activated TLR3-CD32 chimeric protein from the cell surface in a pH-independent manner. We cannot completely rule out the possibility that recognition of dsRNA also occurs at the plasma membrane. Our results (data not shown) and others (51) on the inhibition of signaling by inhibitory monoclonal antibodies led us to propose that TLR3 may have a role for detecting dsRNA at the cell surface, but the largest contribution to signaling occurs in the acidic endosomes. Endocytosis of TLR3-dsRNA complex and endosomal acidification should enhance binding of dsRNA to TLR3 and receptor dimerization with augmented signaling.
In the eukaryotic organisms dsRNA is mainly limited to complementary segments of ssRNA, mainly localized in the cytosol (53). Therefore, the exposure of TLR3 ectodomain at the cell surface in contrast to TLR9 and TLR7/8 does not pose the threat of initiating the autoimmune response.
We showed that UNC93B1 extends the lifetime of associated NAS TLRs in cells and potentiates the response to their respective agonists, probably due to the increased endosomal translocation. In vivo, during infection, multiple PRR receptors are typically activated as the pathogens harbor various distinct molecular patterns. Priming and tolerance between different TLR agonists have been described (54); however, the mechanisms and direct mediators of cross-priming have not been characterized. We investigated the role of UNC93B1 as the contributor to these effects. Based on the important contribution of the level of UNC93B1 expression on the TLR9 responsiveness, we anticipated the priming effect. Pretreatment of Ramos B-cells with poly(I:C) indeed resulted in the augmented response to TLR9 agonist. Our data are in agreement with a study on C3H/HeJ bone marrow-derived macrophages where previous exposure to poly(I:C) resulted in augmented TNF-α production after the TLR9 stimulation by ODN. It was proposed that the TRIF-dependent response primes the MyD88-dependent signaling (54); however, we found that the mechanism of priming is more specific than the general activation of a TRIF or MyD88-dependent pathway. In B-cells poly(I:C) did not up-regulate TLR9. Therefore, we conclude that the priming effect of TLR3 activation is probably a consequence of the increased UNC93B1 expression as no priming was observed in cells pretreated with ODN, which did not up-regulate the UNC93B1. A minor priming effect had been observed after pretreatment with ODN and subsequent stimulation with poly(I:C) (54), but we did not observe any such effect on Ramos B-cells; therefore, this difference could be due to cell- or assay-specific characteristics.
The priming effect could have an important physiological relevance in response to dsRNA originating from the viral infection in combination with bacterial infection or autoimmune activation by endogenous nucleic acids. This mechanism can account for the observed effect of the respiratory syncytial virus infection, which activates TLR3 and primes cells for the response to subsequent bacterial or viral infection (55). Activation of TLR3 by respiratory syncytial virus infection induces TLR3 production (21), whereas the addition of TLR9 agonist increases the disease severity (56). Enhanced expression of TLR3 has been observed in the dermis of patients with scleroderma in contrast to healthy control skin biopsies (35). The synovial fluid of patients with rheumatoid arthritis expression of TLR3 and TLR7 were increased as well, although the expression of UNC93B1 has rarely been investigated. Viral components as well as the endogenous ligands were found in the synovium, suggesting the possible involvement of viral infection and TLR signaling in the rheumatoid arthritis pathogenesis (34). It has been demonstrated that TLR signaling by the endogenous nucleic acids is central to the production of autoantibodies as UNC93B1 3d mutation abolished IgG anti-nuclear antibodies and ameliorated the disease in the lupus-prone mice strains (57). A recent study demonstrated significantly higher expressions of UNC93B1 mRNA and intracellular level of UNC93B1 protein in peripheral blood mononuclear cells and B cells from active systemic lupus erythematous patients. Expression of UNC93B1 also correlated with high titers of anti-dsDNA antibody (36). In vivo, any processes that up-regulate UNC93B1 expression could lead toward the enhanced response to agonists of NAS TLRs and pathogenesis of systemic lupus erythematous and other autoimmune conditions. Additionally, this effect may also explain the effect of poly(I:C) as the vaccine adjuvant (58). Therefore, tight regulation of UNC93B1 gene transcription and transcription of receptor itself prevents the excessive response of NAS TLRs.
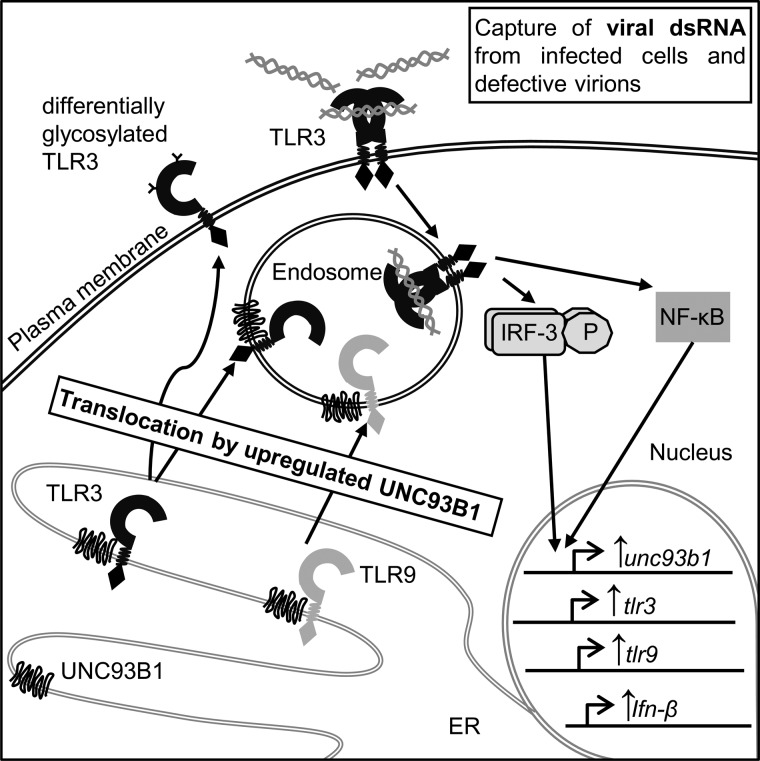
Our findings identified TLR3 as the regulator of UNC93B1 that in turn governs the responsiveness of all NAS TLRs. Activation of TLR3 increases both its own transcription as well as its localization to the cell membrane where it can capture viral dsRNA from infected cells or defective virions (21). Because acidification is important for TLR3 response, it is less likely that signaling would occur from the plasma membrane. UNC93B1 also increases stability of associated TLRs, thus amplifying response to nucleic acids (Fig. 7).
FIGURE 7.
Model of the TLR3-responsive positive feedback loop mediating the response to nucleic acids through UNC93B1 up-regulation. Activation of TLR3 increases the transcription of UNC93B1 mRNA. UNC93B1 interacts with TLR3 and other NAS TLRs in the ER. UNC93B1 induces differential glycosylation and translocation of TLR3 to the plasma membrane where it can be activated by viral dsRNA from infected cells and defective virions and primes the responsiveness of TLR9.
This work was supported by the Slovenian Research Agency, International Centre for Genetic Engineering and Biotechnology (CRP-ICGEB Research Grant Project CRP/SLO08–01) and EN-FIST Centre of Excellence.

This article contains supplemental Figs. S1–S4.
- TLR
- Toll-like receptor
- NAS
- nucleic acid sensing
- TRIF
- Toll/IL-1R domain-containing adapter-inducing interferon-β
- ER
- endoplasmic reticulum
- HUVEC
- human umbilical vein endothelial cells
- IRF-3
- interferon regulatory transcription factor-3
- MyD88
- myeloid differentiation primary response gene 88
- ODN
- oligodeoxynucleotide
- poly(I:C)
- polyinosinic-polycytidylic acid
- TFBS
- transcriptional factor binding site
- RIPA
- radioimmune precipitation assay buffer.
REFERENCES
- 1. Akira S., Takeda K. (2004) Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 2. Beutler B. (2004) Innate immunity. An overview. Mol. Immunol. 40, 845–859 [DOI] [PubMed] [Google Scholar]
- 3. McGettrick A. F., O'Neill L. A. (2010) Localisation and trafficking of Toll-like receptors. An important mode of regulation. Curr. Opin. Immunol. 22, 20–27 [DOI] [PubMed] [Google Scholar]
- 4. Choe J., Kelker M. S., Wilson I. A. (2005) Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science 309, 581–585 [DOI] [PubMed] [Google Scholar]
- 5. Bell J. K., Botos I., Hall P. R., Askins J., Shiloach J., Segal D. M., Davies D. R. (2005) The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proc. Natl. Acad. Sci. U.S.A. 102, 10976–10980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 7. Pirher N., Ivicak K., Pohar J., Bencina M., Jerala R. (2008) A second binding site for double-stranded RNA in TLR3 and consequences for interferon activation. Nat. Struct. Mol. Biol. 15, 761–763 [DOI] [PubMed] [Google Scholar]
- 8. Karikó K., Bhuyan P., Capodici J., Weissman D. (2004) Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J. Immunol. 172, 6545–6549 [DOI] [PubMed] [Google Scholar]
- 9. Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. (2003) Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301, 640–643 [DOI] [PubMed] [Google Scholar]
- 10. Latz E., Schoenemeyer A., Visintin A., Fitzgerald K. A., Monks B. G., Knetter C. F., Lien E., Nilsen N. J., Espevik T., Golenbock D. T. (2004) TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5, 190–198 [DOI] [PubMed] [Google Scholar]
- 11. Johnsen I. B., Nguyen T. T., Ringdal M., Tryggestad A. M., Bakke O., Lien E., Espevik T., Anthonsen M. W. (2006) Toll-like receptor 3 associates with c-Src tyrosine kinase on endosomes to initiate antiviral signaling. EMBO J. 25, 3335–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Häcker H., Mischak H., Miethke T., Liptay S., Schmid R., Sparwasser T., Heeg K., Lipford G. B., Wagner H. (1998) CpG-DNA-specific activation of antigen-presenting cells requires stress kinase activity and is preceded by non-specific endocytosis and endosomal maturation. EMBO J. 17, 6230–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Bouteiller O., Merck E., Hasan U. A., Hubac S., Benguigui B., Trinchieri G., Bates E. E., Caux C. (2005) Recognition of double-stranded RNA by human toll-like receptor 3 and downstream receptor signaling requires multimerization and an acidic pH. J. Biol. Chem. 280, 38133–38145 [DOI] [PubMed] [Google Scholar]
- 14. Leonard J. N., Ghirlando R., Askins J., Bell J. K., Margulies D. H., Davies D. R., Segal D. M. (2008) The TLR3 signaling complex forms by cooperative receptor dimerization. Proc. Natl. Acad. Sci. U.S.A. 105, 258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim Y. M., Brinkmann M. M., Paquet M. E., Ploegh H. L. (2008) UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 452, 234–238 [DOI] [PubMed] [Google Scholar]
- 16. Tabeta K., Hoebe K., Janssen E. M., Du X., Georgel P., Crozat K., Mudd S., Mann N., Sovath S., Goode J., Shamel L., Herskovits A. A., Portnoy D. A., Cooke M., Tarantino L. M., Wiltshire T., Steinberg B. E., Grinstein S., Beutler B. (2006) The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7, and 9. Nat. Immunol. 7, 156–164 [DOI] [PubMed] [Google Scholar]
- 17. Brinkmann M. M., Spooner E., Hoebe K., Beutler B., Ploegh H. L., Kim Y. M. (2007) The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J. Cell Biol. 177, 265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Casrouge A., Zhang S. Y., Eidenschenk C., Jouanguy E., Puel A., Yang K., Alcais A., Picard C., Mahfoufi N., Nicolas N., Lorenzo L., Plancoulaine S., Sénéchal B., Geissmann F., Tabeta K., Hoebe K., Du X., Miller R. L., Héron B., Mignot C., de Villemeur T. B., Lebon P., Dulac O., Rozenberg F., Beutler B., Tardieu M., Abel L., Casanova J. L. (2006) Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314, 308–312 [DOI] [PubMed] [Google Scholar]
- 19. Fukui R., Saitoh S., Matsumoto F., Kozuka-Hata H., Oyama M., Tabeta K., Beutler B., Miyake K. (2009) Unc93B1 biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA- but against RNA-sensing. J. Exp. Med. 206, 1339–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fukui R., Saitoh S., Kanno A., Onji M., Shibata T., Ito A., Onji M., Matsumoto M., Akira S., Yoshida N., Miyake K. (2011) Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity 35, 69–81 [DOI] [PubMed] [Google Scholar]
- 21. Groskreutz D. J., Monick M. M., Powers L. S., Yarovinsky T. O., Look D. C., Hunninghake G. W. (2006) Respiratory syncytial virus induces TLR3 protein and protein kinase R, leading to increased double-stranded RNA responsiveness in airway epithelial cells. J. Immunol. 176, 1733–1740 [DOI] [PubMed] [Google Scholar]
- 22. Marshak-Rothstein A. (2006) Toll-like receptors in systemic autoimmune disease. Nat. Rev. Immunol. 6, 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005) MatInspector and beyond. Promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 24. Ueta M., Hamuro J., Kiyono H., Kinoshita S. (2005) Triggering of TLR3 by polyI:C in human corneal epithelial cells to induce inflammatory cytokines. Biochem. Biophys. Res. Commun. 331, 285–294 [DOI] [PubMed] [Google Scholar]
- 25. Lundberg A. M., Drexler S. K., Monaco C., Williams L. M., Sacre S. M., Feldmann M., Foxwell B. M. (2007) Key differences in TLR3/poly I:C signaling and cytokine induction by human primary cells. A phenomenon absent from murine cell systems. Blood 110, 3245–3252 [DOI] [PubMed] [Google Scholar]
- 26. Pegu A., Qin S., Fallert Junecko B. A., Nisato R. E., Pepper M. S., Reinhart T. A. (2008) Human lymphatic endothelial cells express multiple functional TLRs. J. Immunol. 180, 3399–3405 [DOI] [PubMed] [Google Scholar]
- 27. Fitzgerald K. A., Rowe D. C., Barnes B. J., Caffrey D. R., Visintin A., Latz E., Monks B., Pitha P. M., Golenbock D. T. (2003) LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the toll adapters TRAM and TRIF. J. Exp. Med. 198, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goodbourn S., Didcock L., Randall R. E. (2000) Interferons. Cell signalling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81, 2341–2364 [DOI] [PubMed] [Google Scholar]
- 29. Jiang Z., Mak T. W., Sen G., Li X. (2004) Toll-like receptor 3-mediated activation of NF-κB and IRF3 diverges at Toll-IL-1 receptor domain-containing adapter inducing IFN-β. Proc. Natl. Acad. Sci. U.S.A. 101, 3533–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Novick D., Cohen B., Rubinstein M. (1994) The human interferon α/β receptor. Characterization and molecular cloning. Cell 77, 391–400 [DOI] [PubMed] [Google Scholar]
- 31. Veals S. A., Schindler C., Leonard D., Fu X. Y., Aebersold R., Darnell J. E., Jr., Levy D. E. (1992) Subunit of an α-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 12, 3315–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Levy D. E., Kessler D. S., Pine R., Reich N., Darnell J. E. (1988) Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 2, 383–393 [DOI] [PubMed] [Google Scholar]
- 33. Qi R., Singh D., Kao C. C. (2012) Proteolytic processing regulates Toll-like receptor 3 stability and endosomal localization. J. Biol. Chem. 287, 32617–32629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roelofs M. F., Joosten L. A., Abdollahi-Roodsaz S., van Lieshout A. W., Sprong T., van den Hoogen F. H., van den Berg W. B., Radstake T. R. (2005) The expression of toll-like receptors 3 and 7 in rheumatoid arthritis synovium is increased and costimulation of toll-like receptors 3, 4, and 7/8 results in synergistic cytokine production by dendritic cells. Arthritis Rheum. 52, 2313–2322 [DOI] [PubMed] [Google Scholar]
- 35. Agarwal S. K., Wu M., Livingston C. K., Parks D. H., Mayes M. D., Arnett F. C., Tan F. K. (2011) Toll-like receptor 3 up-regulation by type I interferon in healthy and scleroderma dermal fibroblasts. Arthritis Res. Ther. 13, R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakano S., Morimoto S., Suzuki S., Watanabe T., Amano H., Takasaki Y. (2010) Up-regulation of the endoplasmic reticulum transmembrane protein UNC93B in the B cells of patients with active systemic lupus erythematosus. Rheumatology 49, 876–881 [DOI] [PubMed] [Google Scholar]
- 37. Chuang T. H., Ulevitch R. J. (2001) Identification of hTLR10. A novel human Toll-like receptor preferentially expressed in immune cells. Biochim. Biophys. Acta 1518, 157–161 [DOI] [PubMed] [Google Scholar]
- 38. Heil F., Ahmad-Nejad P., Hemmi H., Hochrein H., Ampenberger F., Gellert T., Dietrich H., Lipford G., Takeda K., Akira S., Wagner H., Bauer S. (2003) The Toll-like receptor 7 (TLR7)-specific stimulus loxoribine uncovers a strong relationship within the TLR7, -8, and -9 subfamily. Eur. J. Immunol. 33, 2987–2997 [DOI] [PubMed] [Google Scholar]
- 39. Takeda K., Akira S. (2004) TLR signaling pathways. Semin. Immunol. 16, 3–9 [DOI] [PubMed] [Google Scholar]
- 40. Koehn J., Huesken D., Jaritz M., Rot A., Zurini M., Dwertmann A., Beutler B., Korthäuer U. (2007) Assessing the function of human UNC-93B in Toll-like receptor signaling and major histocompatibility complex II response. Hum. Immunol. 68, 871–878 [DOI] [PubMed] [Google Scholar]
- 41. Qi R., Hoose S., Schreiter J., Sawant K. V., Lamb R., Ranjith-Kumar C. T., Mills J., San Mateo L., Jordan J. L., Kao C. C. (2010) Secretion of the human Toll-like receptor 3 ectodomain is affected by single nucleotide polymorphisms and regulated by Unc93b1. J. Biol. Chem. 285, 36635–36644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Randall R. E., Goodbourn S. (2008) Interferons and viruses. An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89, 1–47 [DOI] [PubMed] [Google Scholar]
- 43. Bhatia P. K., Mukhopadhyay A. (1999) in Advances in Biochemical Engineering/Biotechnology (Bhatia P. K., Danielsson B., Gemeiner P., Grabley S., Lammers F., Mukhopadhyay A., Ramanathan K., Saleemuddin M., Scheper T., Stefuca V., Thiericke R., Xie B., eds) pp. 155–201, Springer Berlin/Heidelberg [Google Scholar]
- 44. Sun J., Duffy K. E., Ranjith-Kumar C. T., Xiong J., Lamb R. J., Santos J., Masarapu H., Cunningham M., Holzenburg A., Sarisky R. T., Mbow M. L., Kao C. (2006) Structural and functional analyses of the human Toll-like receptor 3. Role of glycosylation. J. Biol. Chem. 281, 11144–11151 [DOI] [PubMed] [Google Scholar]
- 45. Barbalat R., Ewald S. E., Mouchess M. L., Barton G. M. (2011) Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol. 29, 185–214 [DOI] [PubMed] [Google Scholar]
- 46. Lande R., Gregorio J., Facchinetti V., Chatterjee B., Wang Y. H., Homey B., Cao W., Wang Y. H., Su B., Nestle F. O., Zal T., Mellman I., Schröder J. M., Liu Y. J., Gilliet M. (2007) Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature 449, 564–569 [DOI] [PubMed] [Google Scholar]
- 47. Christensen S. R., Kashgarian M., Alexopoulou L., Flavell R. A., Akira S., Shlomchik M. J. (2005) Toll-like receptor 9 controls anti-DNA autoantibody production in murine lupus. J. Exp. Med. 202, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barton G. M., Kagan J. C., Medzhitov R. (2006) Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat. Immunol. 7, 49–56 [DOI] [PubMed] [Google Scholar]
- 49. Mouchess M. L., Arpaia N., Souza G., Barbalat R., Ewald S. E., Lau L., Barton G. M. (2011) Transmembrane mutations in Toll-like receptor 9 bypass the requirement for ectodomain proteolysis and induce fatal inflammation. Immunity 35, 721–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leifer C. A., Brooks J. C., Hoelzer K., Lopez J., Kennedy M. N., Mazzoni A., Segal D. M. (2006) Cytoplasmic targeting motifs control localization of toll-like receptor 9. J. Biol. Chem. 281, 35585–35592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Matsumoto M., Kikkawa S., Kohase M., Miyake K., Seya T. (2002) Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling. Biochem. Biophys. Res. Commun. 293, 1364–1369 [DOI] [PubMed] [Google Scholar]
- 52. Hewson C. A., Jardine A., Edwards M. R., Laza-Stanca V., Johnston S. L. (2005) Toll-like receptor 3 is induced by and mediates antiviral activity against rhinovirus infection of human bronchial epithelial cells. J. Virol. 79, 12273–12279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kramerov D. A., Grigoryan A. A., Ryskov A. P., Georgiev G. P. (1979) Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences. Evidence from DNA cloning experiments. Nucleic Acids Res. 6, 697–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bagchi A., Herrup E. A., Warren H. S., Trigilio J., Shin H. S., Valentine C., Hellman J. (2007) MyD88-dependent and MyD88-independent pathways in synergy, priming, and tolerance between TLR agonists. J. Immunol. 178, 1164–1171 [DOI] [PubMed] [Google Scholar]
- 55. Rudd B. D., Burstein E., Duckett C. S., Li X., Lukacs N. W. (2005) Differential role for TLR3 in respiratory syncytial virus-induced chemokine expression. J. Virol. 79, 3350–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Johnson T. R., Rao S., Seder R. A., Chen M., Graham B. S. (2009) TLR9 agonist, but not TLR7/8, functions as an adjuvant to diminish FI-RSV vaccine-enhanced disease, while either agonist used as therapy during primary RSV infection increases disease severity. Vaccine 27, 3045–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kono D. H., Haraldsson M. K., Lawson B. R., Pollard K. M., Koh Y. T., Du X., Arnold C. N., Baccala R., Silverman G. J., Beutler B. A., Theofilopoulos A. N. (2009) Endosomal TLR signaling is required for anti-nucleic acid and rheumatoid factor autoantibodies in lupus. Proc. Natl. Acad. Sci. U.S.A. 106, 12061–12066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jelinek I., Leonard J. N., Price G. E., Brown K. N., Meyer-Manlapat A., Goldsmith P. K., Wang Y., Venzon D., Epstein S. L., Segal D. M. (2011) TLR3-specific double-stranded RNA oligonucleotide adjuvants induce dendritic cell cross-presentation, CTL responses, and antiviral protection. J. Immunol. 186, 2422–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]