Background: Neuregulin (NRG) is overexpressed in 30% of breast cancers and mediates tumor cell migration and invasion.
Results: NRG mediates its effects on tumor cell migration via inhibition of PKD1.
Conclusion: NRG is a negative regulator of PKD1 and acts through Rac1.
Significance: We provide a mechanism through which the NRG/Rac1 pathway cross-talks with PKD1 signaling pathways to facilitate directed cell migration.
Keywords: Breast Cancer, Cell Migration, Migration, Protein Kinase D (PKD), Rac1, NRG, Neuregulin
Abstract
Neuregulin (NRG; heregulin) is overexpressed in ∼30% of breast cancers and mediates various processes involved in tumor progression, including tumor cell migration and invasion. Here, we show that NRG mediates its effects on tumor cell migration via PKD1. Downstream of RhoA, PKD1 can prevent directed cell migration through phosphorylation of its substrate SSH1L. NRG exerts its inhibitory effects on PKD1 through Rac1/NADPH oxidase, leading to decreased PKD1 activation loop phosphorylation and decreased activity toward SSH1L. The consequence of PKD1 inhibition by NRG is decreased binding of 14-3-3 to SSH1L, localization of SSH1L to F-actin at the leading edge, and increased cofilin activity, resulting in increased reorganization of the actin cytoskeleton and cell motility. Our data provide a mechanism through which the Rho GTPase Rac1 cross-talks with PKD1 signaling pathways to facilitate directed cell migration.
Introduction
Neuregulin (NRG)4 is a secreted growth factor that binds to the ErbB3 and ErbB4 receptors. It is required in the morphogenesis and differentiation of the normal mammary gland but is also overexpressed in ∼30% of breast tumors (1, 2). Up-regulation of NRG expression in mammary epithelial cells can be sufficient to drive malignant transformation and the development of tumors (3–5). Additionally, members of the NRG family are detected in pre-invasive ductal breast cancer (6), and NRG has been shown to induce breast cancer cell invasion and metastasis through activation of ErbB3 (5, 7–9).
NRG increases cell motility through regulation of the actin cytoskeleton, enhancing the formation of lamellipodia, membrane ruffles, F-actin stress fibers, and filopodia (10, 11). Actin reorganization at the leading edge of migrating cells is initiated by F-actin filament severing through cofilin (12). In the absence of a chemotactic stimulus, cofilin is bound to phosphatidylinositol 4,5-bisphosphate at the plasma membrane (13–15). Activation of cofilin is initiated through receptor tyrosine kinase-activated phospholipase enzymes that cleave phosphatidylinositol 4,5-bisphosphate, allowing its release from the plasma membrane at sites of stimulation. Cofilin enzymatic activity is tightly regulated through inhibition of phosphorylation at Ser-3. Phosphorylation of this site is mediated by LIM domain kinases LIMK1 and LIMK2 and testicular protein kinases TESK1 and TESK2 (13–15). Phosphatases such as SSH1L (slingshot 1-like) and chronophin keep cofilin dephosphorylated and active or re-enter phosphorylated cofilin into the active pool (16, 17).
The MCF-7 breast cancer cell line is a bona fide cell culture model to investigate NRG-mediated signaling leading to cell adhesion, cell motility, and actin-remodeling processes (18). In MCF-7 cells, actin reorganization at the leading edge in lamellipodia is triggered by local activation of SSH1L (19). Stimulation with NRG induces translocation of SSH1L to F-actin-rich lamellipodia, correlating with cofilin dephosphorylation (19). However, the mechanisms by which NRG activates SSH1L are not well defined.
We and others have shown that SSH1L can be negatively regulated by the PKD family of serine/threonine kinases (20–22). Phosphorylation of SSH1L at Ser-978 by PKD1 leads to binding of 14-3-3 protein, resulting in its release from F-actin and sequestration in the cytosol (20, 21). In addition, PKD1 also decreases cofilin activity through the PAK4 (p21-activated kinase 4)/LIMK pathway by direct phosphorylation and activation of PAK4 (22). The activator of PKD1 that leads to such signaling is the small Rho GTPase RhoA (20). In invasive breast cancer, PKD1 is down-regulated in its expression compared with normal epithelium, whereas the two other isoforms of this kinase family, PKD2 and PKD3, remain unchanged in their expression, indicating a specific function for this PKD isoform in this cancer (23). Moreover, the knockdown of PKD1 in low-motility breast cancer cell lines (e.g. MCF-7) or the re-expression of active PKD1 in highly invasive cells (e.g. MDA-MB-231) showed that PKD1 is a key negative regulator of breast caner cell invasion (23).
Here, we show that NRG mediates its stimulatory effects on breast cancer cell migration via Rac1/NADPH oxidase-induced inhibition of PKD1, thus decreasing PKD1 activity toward SSH1L. The consequence of PKD1 inhibition by NRG is the localization of SSH1L to F-actin at the leading edge and increased cofilin activity, leading to reorganization of the actin cytoskeleton and cell motility.
EXPERIMENTAL PROCEDURES
Cell Lines, Antibodies, and Reagents
Cell lines were obtained from American Type Culture Collection and maintained in DMEM with 10% FBS. MCF-7 cells stably expressing shRNA directed against PKD1 or control virus have been described previously (23). Anti-PAK4, anti-phospho-Ser-474 PAK4, anti-cofilin, anti-phospho-Ser-3 cofilin, and anti-phospho-Ser-744/748 PKD (recognizes active PKD1–PKD3) antibodies were from Cell Signaling Technology (Danvers, MA). Anti-Rac1, anti-RhoA, anti-GST, anti-PKD1, anti-Myc (mouse monoclonal), and anti-14-3-3β antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-HA and anti-β-actin antibodies were from Sigma-Aldrich. Anti-PKD2 antibody was from Millipore (Billerica, MA). Anti-PKD3 and anti-SSH1L antibodies were from Bethyl Laboratories (Montgomery, TX). Rabbit anti-Myc polyclonal antibody was from Abcam (Cambridge, MA). Anti-active Rac1 antibody was from NewEast Biosciences (Malvern, PA). The anti-PKD1 antibody was raised against a peptide corresponding to amino acids 372–385 in human PKD1 and was characterized previously (23). A rabbit polyclonal antibody specific for human SSH1L phosphorylated at Ser-978 (anti-phospho-Ser-978 SSH1L antibody) was raised by 21 Century Biochemicals (Marlboro, MA) using COOH-aminohexanoic acid-PLKRSH(pS)LAKL-amide and acetyl-LKRSH(pS)LAKLGS-aminohexanoic acid-COOH-amide peptides as antigens. HRP-linked secondary antibodies were from Millipore. The secondary antibodies used for immunofluorescence (Alexa Fluor 488-conjugated goat anti-rabbit F(ab′)2 and Alexa Fluor 568-conjugated goat anti-mouse F(ab′)2) and Alexa Fluor 633-conjugated phalloidin were from Invitrogen. Lipofectamine 2000 (Invitrogen) was used for transient transfection. Recombinant human NRG (heregulin-β1) was from PeproTech Inc. (Rocky Hill, NJ). Rho Inhibitor I was from Cytoskeleton Inc. (Denver, CO). Diphenyleneiodonium (NADPH oxidase inhibitor) was from Sigma-Aldrich.
DNA Expression Vectors and Lentiviral shRNA Expression
Expression plasmids for HA-tagged and GFP-tagged human PKD1, PKD1(S738E/S742E) (constitutively active), and PKD1(K612W) (kinase-dead) have been described previously (20). The expression plasmids for Myc-tagged SSH1L and SSH1L(S978A) were obtained from Dr. K. Mizuno (19). The expression plasmid for dominant-negative Rac1 (Rac1.N17) was obtained from Dr. A. Mercurio (University of Massachusetts Medical School). pLKO.1-puro vectors encoding shRNA directed against PKD1 (NM_002742.x-2498s1c1) and the non-target sequence control were obtained from Sigma-Aldrich and have been described previously (20). The ViraPower lentiviral expression system (Invitrogen) was used for an optimized mixture of packaging plasmids that supply the structural and replication proteins that are required to produce lentivirus in HEK293FT cells.
Immunoblotting, Immunoprecipitation, and PAGE
Cells were washed twice with cold PBS (140 mm NaCl, 2.7 mm KCl, 8 mm Na2HPO4, and 1.5 mm KH2PO4, pH 7.2) and lysed with Triton lysis buffer (50 mm Tris-HCl, pH 7.4, 1% Triton X-100, 150 mm NaCl, and 5 mm EDTA, pH 7.4) plus protease inhibitor mixture (Sigma-Aldrich). Lysates were incubated on ice for 30 min. Following centrifugation (13,000 rpm, 15 min, 4 °C), protein concentration was determined. The proteins of interest were immunoprecipitated by a 1-h incubation with a specific antibody (2 μg), followed by a 30-min incubation with protein G-Sepharose (Amersham Biosciences). Immune complexes were washed three times with TBS (50 mm Tris-HCl, pH 7.4, and 150 mm NaCl) and resolved in 20 μl of TBS and 2× Laemmli buffer. Samples were subjected to SDS-PAGE, transferred to nitrocellulose membranes, and visualized by immunostaining.
Pulldown Assays for Active Rac and Rho
Pulldown assays for active Rho GTPases were performed using rhotekin-Rho-binding domain beads (pulldown of active Rho) or PAK-p21-binding domain beads (pulldown of active Rac) and the RhoA Activation or Rac1 Activation Assay Biochem kits purchased from Cytoskeleton Inc. All assays were performed according to the manufacturer's instructions.
Immunofluorescence Microscopy
Cells were transfected and plated on glass coverslips. The next day, cells were washed twice with PBS. Following fixation with 3.5% paraformaldehyde (15 min, 37 °C), cells were washed three times with PBS and permeabilized with 0.1% Triton X-100 in PBS for 2 min at room temperature. Samples were blocked with 3% bovine serum albumin and 0.05% Tween 20 in PBS (blocking solution) for 30 min at room temperature. The coverslips were then incubated for 2 h at room temperature with primary antibodies (diluted 1:2000 in blocking solution). Cells were washed five times with PBS and incubated with secondary antibodies (diluted 1:500 in blocking solution) for 2 h at room temperature. F-actin was stained together with secondary antibodies by incubation with Alexa Fluor 633-conjugated phalloidin in blocking solution. After extensive washes with PBS, coverslips were mounted in Fluoromount-G (Southern Biotech, Birmingham, AL). Samples were examined using an LSM 510 META confocal laser scanning microscope (Zeiss, Jena, Germany) with a Plan-Apochromat 63×/1.4 differential interference contrast oil immersion objective. Alexa Fluor 488 was detected with a 488 nm laser line, Alexa Fluor 568 with a 543 nm laser line, and Alexa Fluor 633-conjugated phalloidin with a 633 nm laser line in a Multi-track configuration, switching tracks after each line. Images shown depict single confocal sections. Images were processed using NIH ImageJ.
Impedance-based Real-time Chemotaxis Analysis
Cells were transfected as indicated and, after 24 h, seeded on Transwell CIM-Plate 16 plates (Roche Applied Science). After attachment, cells were stimulated with NRG as indicated, and migration toward NIH-3T3 cell-conditioned medium over the indicated time periods was continuously monitored in real-time using the xCELLigence RTCA dual plate instrument (Roche Applied Science). Error bars (gray) represent three experiments.
Wound Healing/Scratch Assays
Cells were transfected as indicated. At 90% confluence, cell layers were wounded by scratching using 1-mm pipette tips. Cells were stimulated with NRG as indicated, and different scratch regions (n = 30) were photographed at the indicated times. Scratch area was determined using Image-Pro Plus (Media Cybernetics Inc., Rockville, MD).
RESULTS
Neuregulin Decreases PKD1 Basal Activity
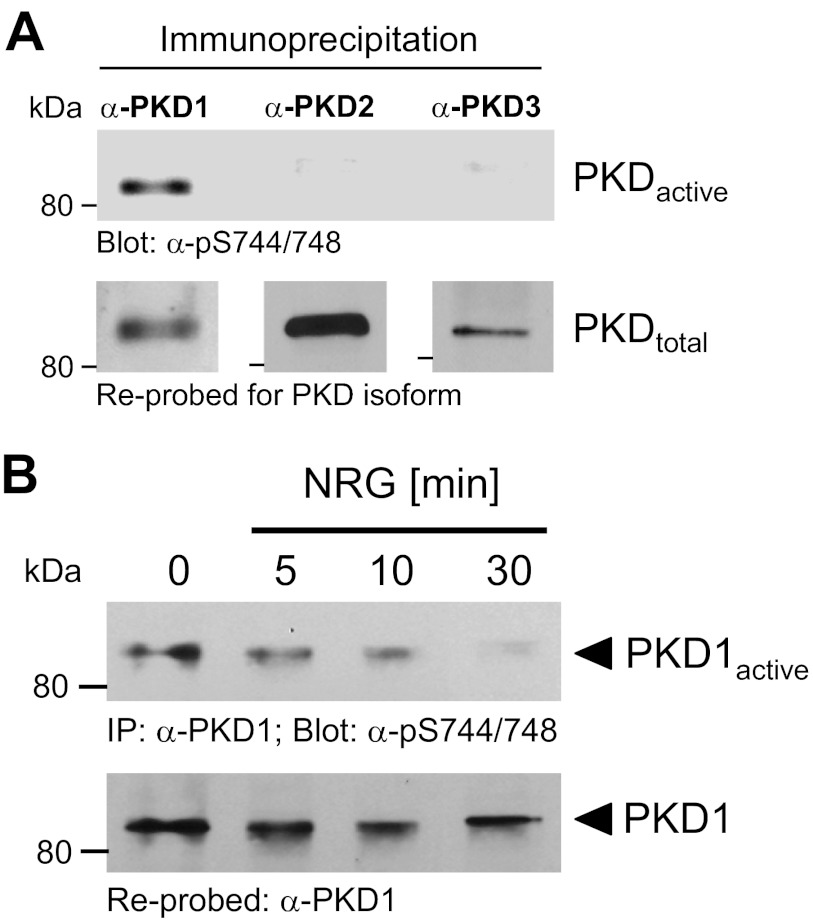
PKD1 negatively regulates cell migration by targeting substrates that control actin reorganization at the leading edge (20–22). Analysis of the breast cancer cell line MCF-7 for expression and activity of the three PKD family members (PKD1–PKD3) indicated that all are expressed, but only PKD1 showed basal activity (Fig. 1A). PKD activity was determined by probing with an antibody (anti-phospho-Ser-744/748 PKD antibody) that recognizes phosphorylation at the activation loop serines of all three PKD isoforms, an event that is required for kinase activation (24). MCF-7 cells under normal growth conditions are non- or low-motile, but treatment with NRG increases their migratory potential (19). Therefore, we tested if NRG in MCF-7 cells affects PKD1 basal activity. We found that treatment of cells with NRG led to a time-dependent down-regulation of endogenous PKD1 activity (Fig. 1B).
FIGURE 1.
NRG decreases PKD1 basal activity. A, endogenous PKD1, PKD2, or PKD3 was immunoprecipitated from MCF-7 cells under normal growth conditions using isoform-specific antibodies. Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed for PKD activity by immunoblotting for activation loop phosphorylation (anti-phospho-Ser-744/748 PKD antibody). Samples were reprobed for the respective PKD isoform. B, MCF-7 cells were serum-starved for 16 h and then treated with NRG (100 ng/ml) for the indicated times. Endogenous PKD1 was immunoprecipitated (IP; anti-PKD1 antibody), and samples were subjected to SDS-PAGE, transferred to nitrocellulose, analyzed by immunoblotting for activation loop phosphorylation (anti-phospho-Ser-744/748 PKD antibody), and reprobed for PKD1.
Inhibition of PKD1 Activity by NRG Leads to Increased Cell Motility
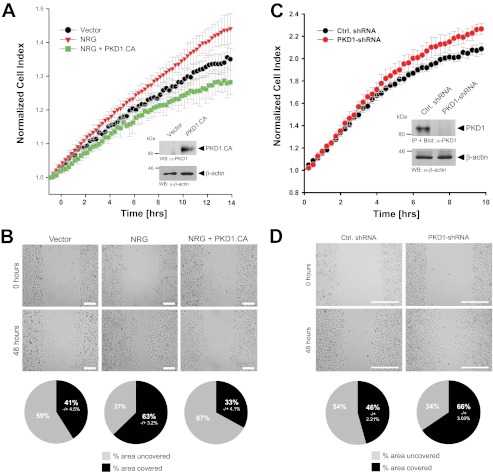
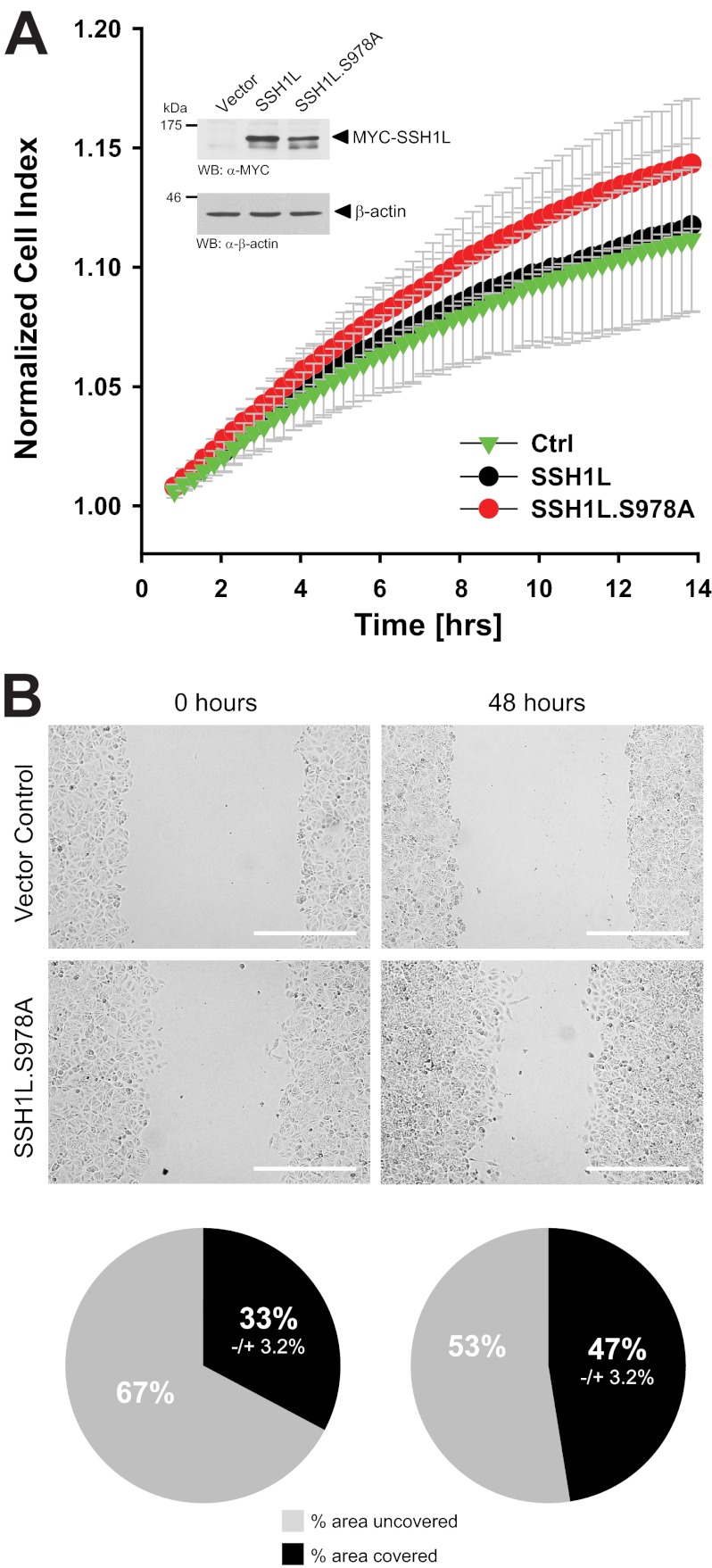
To test if NRG-induced inhibition of basal PKD1 activity contributes to directed cell migration, we next investigated whether a constitutively active PKD1 allele with activation-mimicking phosphorylations (PKD1(S738E/S742E)) is capable of blocking or reducing NRG-induced directed cell migration. Although treatment of cells with NRG increased directed cell migration compared with control cells (Fig. 2A, red versus black), additional ectopic expression of an active allele of PKD1 blocked NRG-induced directed cell migration (Fig. 2A, green versus red). Similar effects were observed when cells were subjected to wound healing (scratch) assays (Fig. 2B). We then tested if the basal activity of PKD1 can contribute to the non-motile phenotype of MCF-7 cells. We found that depletion of PKD1 from cells increased their ability to migrate. This was found in impedance-based Transwell assays toward a chemotactic stimulus (Fig. 2C) and in wound healing assays (Fig. 2D). Our data suggest that inhibition of endogenous basal PKD1 activity is a mechanism through which NRG induces cell migration.
FIGURE 2.
Inhibition of PKD1 activity by NRG leads to increased cell motility. A and B, MCF-7 cells were transfected with a vector control or constitutively active PKD1 (PKD1.CA). Cells were seeded on Transwell CIM-Plate 16 plates and subjected to real-time migration assays (xCELLigence) over a time period of 14 h (A). Error bars (gray) represent three experiments. Control Western blotting (WB) was performed to determine overexpression of constitutively active PKD1. Equal loading was controlled by probing for β-actin (anti-β-actin antibody). Cells were also subjected to wound healing assays over a time period of 48 h. The area closure of the scratch wound was quantified (Image-Pro Plus), and the pie graphs show the percent area covered or uncovered (B). C and D, MCF-7 cells stably expressing shRNA directed against PKD1 or control (Ctrl.) virus were seeded in Transwell CIM-Plate 16 plates and, after adhesion, subjected to real-time migration assays (xCELLigence) over a time period of 10 h. Error bars (gray) represent three experiments. Additionally, lysates of cells were analyzed for efficient knockdown of PKD1. PKD1 was immunoprecipitated (IP; anti-PKD1 antibody), subjected to SDS-PAGE, and transferred to nitrocellulose, and samples were stained with PKD1-specific antibody. Western blot staining for β-actin served to show equal loading. Cells were also subjected to wound healing assays over a time period of 48 h. The area closure of the scratch wound was quantified (Image-Pro Plus), and the pie graphs show the percent area covered or uncovered (D).
NRG Regulates Cofilin Activity via Slingshot and Not PAK4
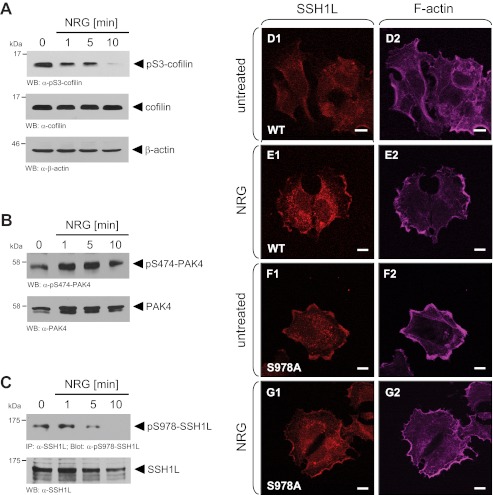
One key pathway by which NRG induces directed cell migration is by increasing the activity of cofilin (19). Time-dependent treatment of MCF-7 cells with NRG decreased cofilin phosphorylation at Ser-3, indicating its activation (Fig. 3A). PKD1 can regulate cofilin phosphorylation at Ser-3 via both activation of PAK4 and inactivation of SSH1L (direct phosphorylation at Ser-978). The net result of such signaling is accumulation of phosphorylated inactive cofilin (22, 25–27). Therefore, we tested if NRG acts through both enzymes by probing for PKD1-mediated phosphorylations. In MCF-7 cells, PAK4 is constitutively active as judged by its phosphorylation at the activation loop (anti-phospho-Ser-474 PAK4), and treatment with NRG did not have any inhibitory effect (Fig. 3B). Correlating with the observed basal PKD1 activity, SSH1L was basally phosphorylated at Ser-978 (Fig. 3C). Phosphorylation at this site was shown previously to retain SSH1L in the cytosol and render it inactive toward cofilin at the leading edge (20). Treatment with NRG decreased SSH1L phosphorylation at this site over time (Fig. 3C). This resulted in increased localization of SSH1L to the leading edge, where it co-localized with F-actin structures (Fig. 3, D and E). In contrast, an SSH1L(S978A) mutant was constitutively co-localized with F-actin at the leading edge independent of NRG treatment (Fig. 3, F and G). Taken together, these results indicate that, in response to NRG, SSH1L phosphorylation at Ser-978 is decreased, and active SSH1L is located at the leading edge, where it increases cofilin activity.
FIGURE 3.
NRG regulates cofilin activity via slingshot and not PAK4. A–C, MCF-7 cells were serum-starved for 24 h and then treated with NRG (100 ng/ml) for the indicated times. Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed for phosphorylation of endogenous cofilin (A), PAK4 (B), or SSH1L (C) using phospho-specific antibodies (anti-phospho-Ser-3 cofilin, anti-phospho-Ser-474 PAK4, or anti-phospho-Ser-978 SSH1L). Samples were restained for total protein using anti-cofilin, anti-PAK4, or anti-SSH1L antibody. WB, Western blot; IP, immunoprecipitation. D–G, MCF-7 cells were transfected with Myc-tagged wild-type SSH1L or SSH1L(S978A), serum-starved, and then treated with NRG (100 ng/ml). Samples were subjected to immunofluorescence staining using anti-Myc antibodies for visualization of SSH1L as well as phalloidin for staining of F-actin structures.
PKD1 Activity Determines SSH1L Localization to the Leading Edge
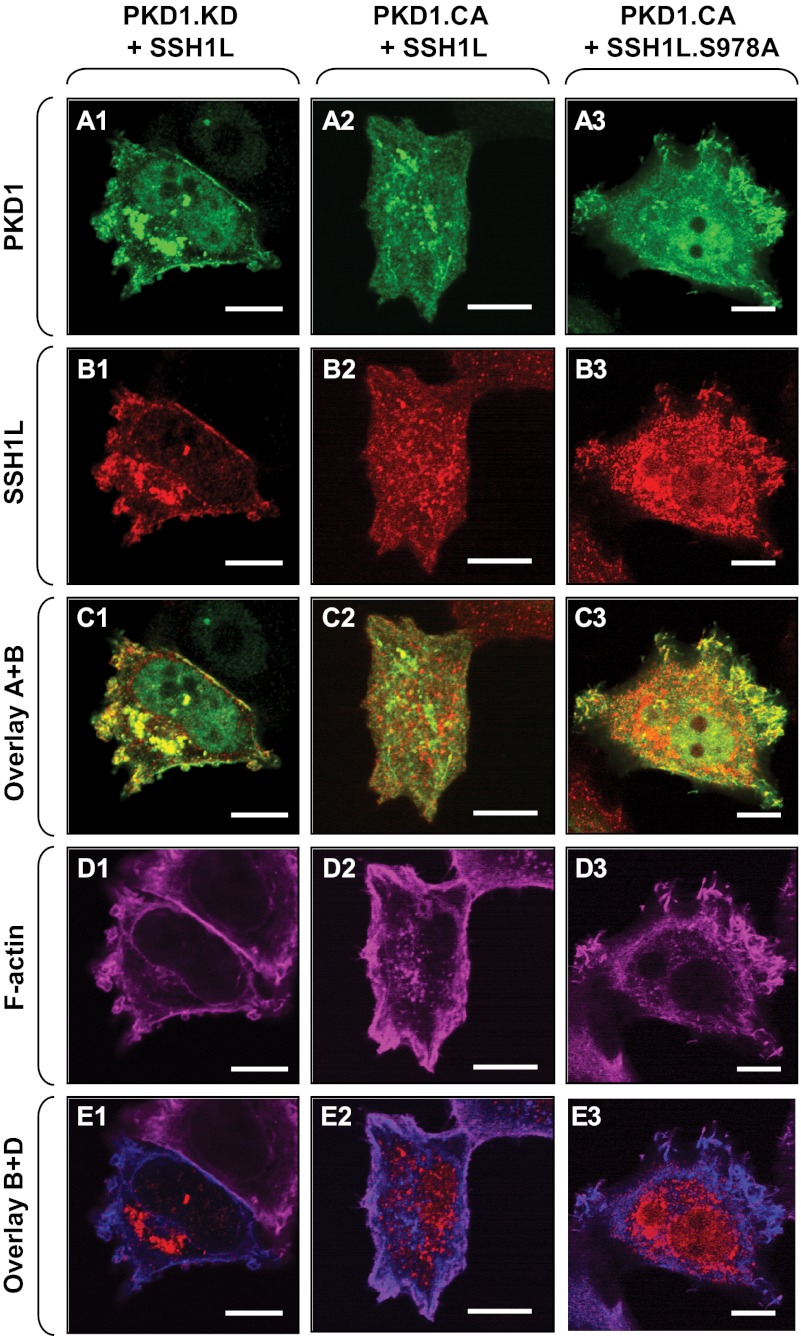
Localization of SSH1L to areas of actin reorganization is prevented by PKD-mediated phosphorylation at Ser-978, which retains it in the cytosol (20, 21). In MCF-7 cells, endogenous SSH1L showed basal phosphorylation at Ser-978 (Fig. 3C), and consequently, overexpressed wild-type SSH1L was also mostly cytosolic (Fig. 3D). Expression of a dominant-negative PKD1 allele (kinase-dead PKD1) led to localization of wild-type SSH1L to F-actin structures at the leading edge, thus emulating NRG effects (Fig. 4, A1–E1). Expression of constitutively active PKD1 resulted in the localization of SSH1L almost exclusively to the cytosol and prevented co-localization with F-actin (Fig. 4, compare E2 with E1). This is dependent on PKD1-mediated phosphorylation of SSH1L at Ser-978 because a slingshot mutant harboring a mutation in the PKD phosphorylation site (SSH1L(S978A)) remained localized at membrane ruffles at the leading edge in the presence of active PKD1 (Fig. 4, A3–E3).
FIGURE 4.
PKD1 activity determines SSH1L localization to the leading edge. A–E, MCF-7 cells were cotransfected with either Myc-tagged wild-type SSH1L or SSH1L(S978A) and kinase-dead (PKD1.KD) or constitutively active (PKD1.CA) PKD1 as indicated. Samples were fixed, and cells were subjected to immunofluorescence analysis. A, PKD1, stained with anti-PKD1 antibody. B, SSH1L, stained with anti-Myc antibody. D, F-actin structures, stained with phalloidin. C shows an overlay of PKD1 and SSH1L signals, and E shows an overlay of SSH1L and F-actin structures.
SSH1L Phosphorylation at Ser-978 Affects Directed MCF-7 Cell Migration
The data obtained so far suggested that NRG regulates directed cell migration via inhibition of PKD1, leading to decreased phosphorylation of SSH1L at Ser-978. Therefore, we next tested if the SSH1L(S978A) mutant, which cannot be regulated by PKD1, has similar effects on directed cell migration as treatment of cells with NRG (Fig. 2A) or knockdown of endogenous PKD1 (Fig. 2B). Ectopic expression of the SSH1L(S978A) mutant increased directed cell migration, whereas expression of wild-type SSH1L, which can be regulated by endogenous basal PKD1 activity, had no effect (Fig. 5A). Similar effects on cell motility were observed using wound healing assays as a readout for cell motility (Fig. 5B). This suggests that the motility of MCF-7 cells is indeed mediated through a NRG/PKD1/SSH1L pathway.
FIGURE 5.
SSH1L phosphorylation at Ser-978 affects directed MCF-7 cell migration. A and B, MCF-7 cells were transfected with vector control or Myc-tagged SSH1L or SSH1L(S978A). 16 h after transfection, cells were seeded in Transwell CIM-Plate 16 plates and, after adhesion, subjected to real-time migration assays (xCELLigence) over a time period of 14 h. Error bars (gray) represent three experiments. Control Western blotting (WB) was performed to determine overexpression of SSH1L or its mutant by probing with anti-Myc antibodies. Probing for β-actin (anti-β-actin antibody) served to show equal loading. Cells were also subjected to wound healing assays over a time period of 48 h. The area closure of the scratch wound was quantified (Image-Pro Plus), and the pie graphs show the percent area covered or uncovered (B).
Neuregulin Blocks 14-3-3 Binding-induced Inhibition of SSH1L
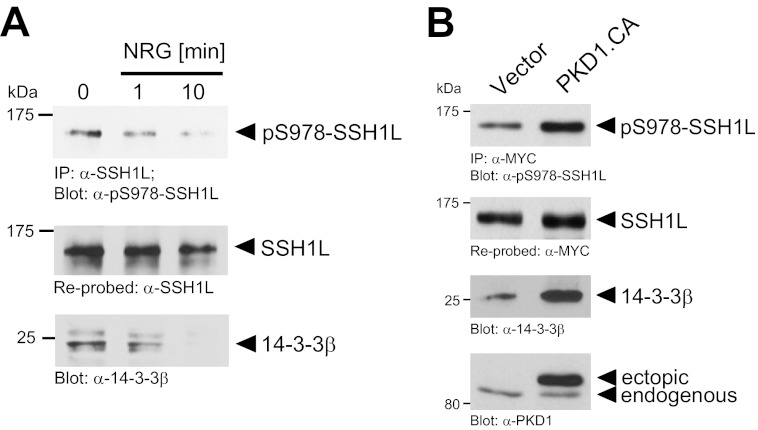
One mechanism by which SSH1L can be retained in the cytosol is through binding to proteins of the 14-3-3 family (19). Binding of 14-3-3 to SSH1L is regulated by its phosphorylation at Ser-978 (20). In MCF-7 cells, stimulation with NRG led to a time-dependent decrease in basal SSH1L phosphorylation at Ser-978, correlating with a decrease in 14-3-3 binding (Fig. 6A). However, the ectopic expression of a constitutively active allele of PKD1 increased both SSH1L phosphorylation at Ser-978 and binding to 14-3-3 (Fig. 6B). This suggests that loss of 14-3-3 binding is the mechanism by which NRG induces cofilin dephosphorylation in MCF-7 cells.
FIGURE 6.
NRG blocks 14-3-3 binding-induced inhibition of SSH1L. A, MCF-7 cells were serum-starved for 16 h and then treated with NRG (100 ng/ml) for the indicated times. Endogenous SSH1L was immunoprecipitated (IP; anti-SSH1L antibody), and samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting for binding to 14-3-3 (anti-14-3-3β antibody). Samples were also control-stained for SSH1L phosphorylation (anti-phospho-Ser-978 SSH1L antibody) and reprobed for SSH1L. B, MCF-7 cells were cotransfected with Myc-tagged SSH1L and vector control or constitutively active PKD1 (PKD1.CA) as indicated. SSH1L was immunoprecipitated (anti-Myc antibody), and samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed for binding of 14-3-3β (anti-14-3-3β antibody). For controls, samples were analyzed for phosphorylation of SSH1L at Ser-978 (anti-phospho-Ser-978 SSH1L antibody) and then reprobed for SSH1L (anti-Myc antibody). PKD1 expression was detected by Western blotting of lysates with anti-PKD1 antibody.
Neuregulin Negatively Regulates PKD1 via Rac1/NADPH Oxidase
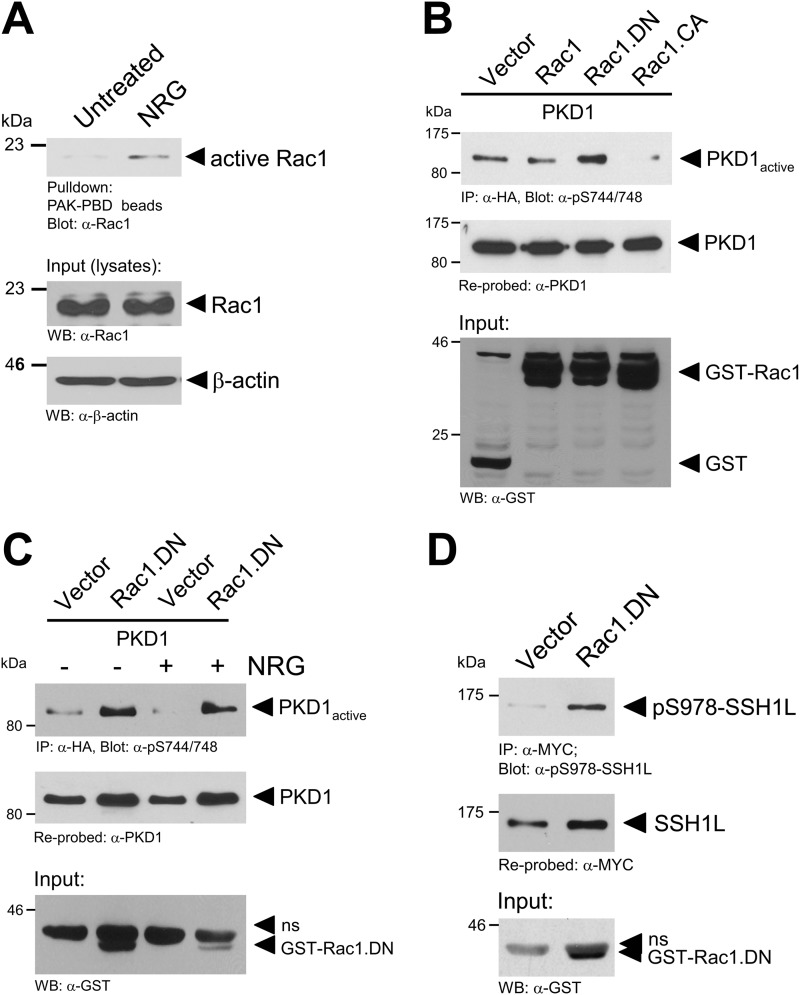
We next investigated how PKD1 activity is negatively regulated by NRG. NRG was shown previously to mediate its action on cell migration via activation of Rac1 (19). In MCF-7 cells, NRG led to an increase in Rac1 activity (Fig. 7A). We next tested if NRG negatively affects PKD1 activity through Rac1. Using a set of constitutively active (Rac1.V12 mutant) or dominant-negative (Rac1.N17 mutant) expression constructs, we first tested if Rac1 activity can impact PKD1 activity. Constitutively active Rac1 inhibited PKD1 activity (Fig. 7B), indicating that Rac1 may act as a suppressor of PKD1 activity in MCF-7 cells. In support of this, increased PKD1 activity was detected when a dominant-negative allele of Rac1 was expressed (Fig. 7B). In addition, ectopic expression of dominant-negative Rac1 effectively blocked the inhibitory effects of NRG on PKD1 (Fig. 7C), and increased PKD1 activity induced by dominant-negative Rac1 correlated with an increase in SSH1L phosphorylation at Ser-978 (Fig. 7D).
FIGURE 7.
NRG negatively regulates PKD1 via Rac1. A, MCF-7 cells were serum-starved for 24 h and then stimulated with NRG (100 ng/ml). Cells were lysed, and active Rac1 was pulled down. Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed for Rac1 by immunoblotting with anti-Rac1 (total Rac1) antibody. Cell lysates were also stained with anti-Rac1 antibody to determine total Rac1 (input control). WB, Western blot. B, cells were transfected with HA-tagged PKD1 and GST-tagged Rac1, dominant-negative Rac1 (Rac1.DN), or constitutively active Rac1 (Rac1.CA) as indicated. 24 h after transfection, cells were lysed, and PKD1 was immunoprecipitated (IP; anti-HA antibody). Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed for PKD1 activity by immunoblotting with anti-phospho-Ser-744/748 PKD antibody (which recognizes PKD1 phosphorylated at Ser-738 and Ser-742). Immunoblots were restained for PKD1 (anti-PKD1 antibody). Cell lysates were stained with anti-GST antibody for overexpression of GST-Rac1. C, MCF-7 cells were transfected with vector control, PKD1, or dominant-negative Rac1 as indicated. After transfection, cells were serum-starved for 16 h and then stimulated with NRG (100 ng/ml) as indicated. Cells were lysed, and PKD1 was immunoprecipitated (anti-HA antibody). Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed for PKD1 activity by immunoblotting with phospho-Ser-744/748 PKD antibody. For controls, immunoblots were restained for PKD1 (anti-PKD1 antibody). Cell lysates were stained with anti-GST antibody for overexpression of GST-Rac1. D, MCF-7 cells were transfected with Myc-tagged SSH1L and GST-tagged dominant-negative Rac1 as indicated. 24 h after transfection, cells were lysed, and SSH1L was immunoprecipitated (anti-Myc antibody). Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed for phosphorylation of SSH1L at Ser-978 using anti-Ser-978 SSH1L antibody. For controls, immunoblots were restained for ectopically expressed SSH1L (anti-Myc antibody). Cell lysates were stained with anti-GST antibody for overexpression of GST-Rac1.
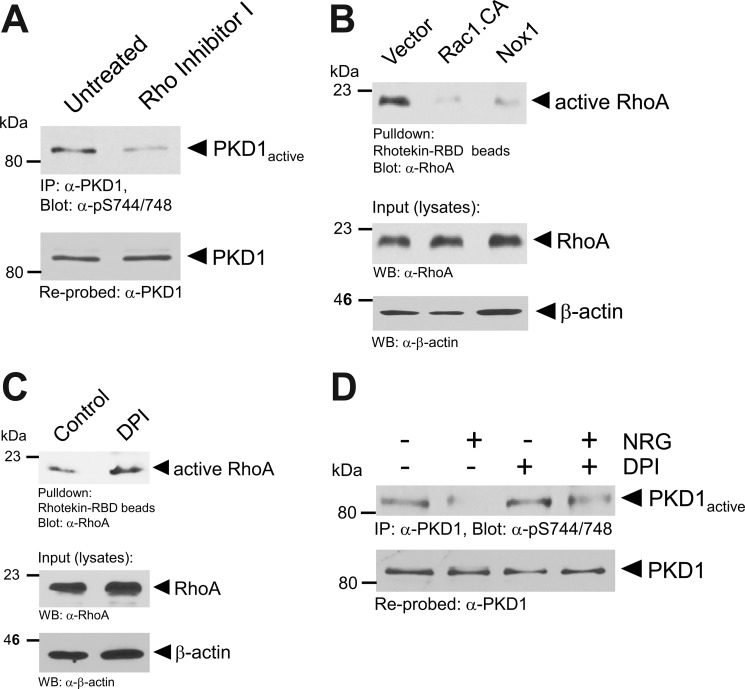
We next investigated the mechanism Rac1 uses to negatively affect basal PKD1 activity in MCF-7 cells. Treatment of cells with C3 transferase-based RhoA Inhibitor I decreased basal PKD1 activity (Fig. 8A), similar to the results obtained with constitutively active Rac1 (Fig. 7B). This indicates that basal PKD1 activity in MCF-7 cells is mediated through RhoA activity. Basal RhoA activity was inhibited when constitutively active Rac1 or Nox1 was expressed (Fig. 8B). Additionally, inhibition of NADPH oxidase with diphenyleneiodonium increased the levels of active RhoA in cells (Fig. 8C). Taken together, the data obtained indicate a negative regulatory function of the Rac1/NADPH oxidase pathway in RhoA/PKD1 signaling. Because Rac1 acts downstream of NRG in MCF-7 cells, we next tested if inhibition of NADPH oxidase can block the inhibitory effects of NRG on PKD1. We found that inhibition of NADPH oxidase restored PKD1 basal activity in the presence of NRG (Fig. 8D).
FIGURE 8.
Rac1 inhibits RhoA/PKD1 signaling through NADPH oxidase. A, MCF-7 cells were treated with Rho Inhibitor I (2 μg/ml, 4 h). Cells were lysed, and PKD1 was immunoprecipitated (IP; anti-PKD1 antibody). Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting for PKD1 activation loop phosphorylation (anti-phospho-Ser-744/748 PKD). Immunoblots were restained for PKD1 (anti-PKD1 antibody). B, MCF-7 cells were transfected as indicated. Cells were lysed and subjected to rhotekin-Rho-binding domain pulldown assays for active RhoA. Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting for RhoA (anti-RhoA antibody). For input and loading controls, lysates of samples were stained for total RhoA (anti-RhoA antibody) and β-actin (anti-β-actin antibody). Rac1.CA, constitutively active Rac1; WB, Western blot. C, MCF-7 cells were treated with diphenyleneiodonium (DPI; 20 μm, 1 h) as indicated. Cells were lysed and subjected to rhotekin-Rho-binding domain pulldown assays for active RhoA. Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting for RhoA (anti-RhoA antibody). For input and loading controls, lysates of samples were stained for total RhoA (anti-RhoA antibody) and β-actin (anti-β-actin antibody). D, MCF-7 cells were treated with NRG (100 ng/ml, 10 min) and diphenyleneiodonium (20 μm, 1 h) as indicated. Cells were lysed, and PKD1 was immunoprecipitated (anti-PKD1 antibody). Samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblotting for PKD1 activation loop phosphorylation (anti-phospho-Ser-744/748 PKD antibody). Immunoblots were restained for PKD1 (anti-PKD1 antibody).
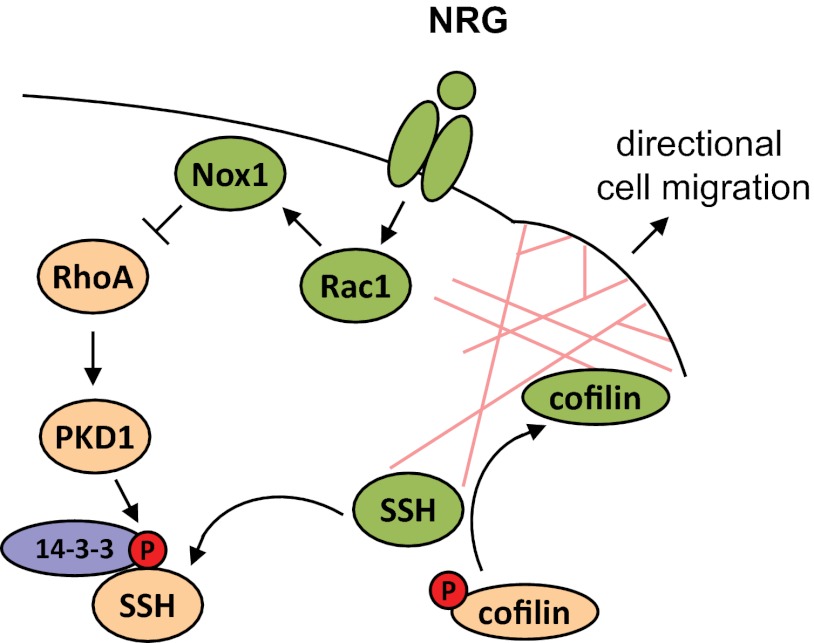
In summary, our data indicate that NRG inhibits PKD1 through Rac1/NADPH oxidase. This results in activation of SSH1L and dephosphorylation of cofilin, allowing increased directed cell migration (Fig. 9).
FIGURE 9.
Schematic showing how PKD1 is involved in NRG-mediated cell motility. NRG regulates cofilin activity, actin reorganization, and directed cell migration at the leading edge of MCF-7 cells by inhibiting PKD1 through Rac1/NADPH oxidase. When activated (i.e. via RhoA as shown previously (20)), PKD1 phosphorylates SSH1L at Ser-978. This leads to binding of SSH1L to 14-3-3 and retention in the cytosol. Inhibition of PKD1 in response to NRG signaling leads to loss of this phosphorylation and results in localization of SSH1L to the leading edge, where it dephosphorylates and activates cofilin to contribute to directed cell migration. Our data provide a mechanism by which both Rho GTPases may cross-talk to facilitate directed cell migration.
DISCUSSION
Neuregulins are cell-cell signaling proteins that play essential roles in the nervous system, heart, and breast (28). In the normal breast, NRG is involved in development during pregnancy and lactation (29). NRG also has a role in the pathogenesis of breast cancer (28), where it contributes to tumor formation and progression to a more aggressive phenotype (30, 31). For example, expression of NRG can lead to acquisition of hormone independence (7) and anti-estrogen resistance (32, 33). Furthermore, NRG mediates the spread and motility of neoplastic cells (30, 31) and is sufficient for the development of mammary tumors and metastasis (5). NRG promotes breast cancer cell invasion by increasing cell motility through induction of actin reorganization (11). In this study, we have described the functional relationship between NRG and PKD1, a kinase that was shown to inhibit directed cell migration and invasion in breast, prostate, and gastric cancers (23, 34, 35).
MCF-7 cells are a bona fide cell model to study the mechanisms by which NRG regulates cell migration (19). They are non- or low-motile and endogenously express basally active PKD1 and active RhoA (Figs. 1A and Fig. 8, B and C). Basal RhoA/PKD1 activity most likely is caused by serum components because it was shown previously that it can be decreased with serum deprivation (36). Previously, we have shown that knockdown of PKD1 in MCF-7 cells significantly increases their capacity to migrate and invade the extracellular matrix in two- and three-dimensional assay systems (23). However, in these cells, PKD1 has also been described to increase cell proliferation through accelerating the G0/G1-to-S phase transition via the MEK/Erk1/Erk2 signaling pathway (37). Although NRG can induce a motile phenotype, it was also shown to have antiproliferative effects on breast cancer cells (1, 38, 39). Thus, the basal activity of PKD1 may maintain proliferation, and its negative regulation by NRG may be a mechanism for cancer cells to switch from a proliferative to a migratory phenotype.
NRG induces directed MCF-7 cell migration via activation of SSH1L (19). Localization of SSH1L to areas of actin reorganization is dependent on its dephosphorylation at Ser-978 and release from binding to 14-3-3 proteins (19, 20). We have shown here that, in MCF-7 cells, the basal activity of PKD1 is sufficient to promote phosphorylation of SSH1L at Ser-978, leading to its binding to 14-3-3 proteins (Fig. 6). When stimulated with NRG, PKD1 is inhibited, and SSH1L phosphorylation at Ser-978 is decreased (Figs. 1 and 3). This allows SSH1L to localize to the leading edge and fulfill its function in mediating cofilin dephosphorylation and directed cell migration (Figs. 3–5).
In breast cancer cells, Rac1 is a downstream effector of NRG/ErbB3 signaling and mediates migratory responses (40). Downstream of NRG, Rac1 mediates SSH1L activation and its effects on actin reorganization at the leading edge (19). It was shown in keratinocytes that the absence of Rac1 activity leads to inhibition of slingshot through binding of 14-3-3 proteins; moreover, Rac1 activation by α6β4 integrin leads to slingshot-mediated dephosphorylation of cofilin to increase their migration (41). Our data show that a similar mechanism is in place in MCF-7 cells. In these cells, basal PKD1 activity is decreased when Rac1 is activated by NRG, and this translates to decreased SSH1L phosphorylation at Ser-978 (Fig. 7).
The mechanism by which PKD1 is inhibited is most likely through cross-talk between Rho GTPases. PKD1 has been shown to be activated downstream of RhoA (20, 42, 43), and basal PKD1 activity in MCF-7 cells is also due to RhoA activity (Fig. 8A). The balance between Rac1 and RhoA activities determines cell morphology and migratory behavior (44). For example, in NIH-3T3 fibroblasts, growth factor receptor-mediated activation of Rac1 or expression of constitutively active Rac1 can down-regulate RhoA activity. A bona fide target for Rac1 is NADPH oxidase (45). It was shown that Rac1 activates this superoxide-generating enzyme complex and that superoxide can contribute to directed cell migration by activating the SSH1L/cofilin pathway (46). We have shown here that the RhoA/PKD1 signaling pathway is negatively regulated by Rac1/NADPH oxidase signaling (Fig. 8). On the other hand, PKD1 has been shown to be activated by the superoxide breakdown product hydrogen peroxide (47). This implicates that the balance of superoxide and hydrogen peroxide at the leading edge of migrating cells may provide a switch to regulate actin reorganization processes. This will be the focus of future investigations.
Our data provide a mechanism through which the Rho GTPases Rac1 and RhoA cross-talk at the level of PKD1 to facilitate directed cell migration. It can be speculated that PKD1 is active in all areas where RhoA signaling occurs and contributes to focal adhesion formation, cell adhesion, and stress fiber formation, whereas Rac1-mediated branching of actin at the leading edge requires inactivation of PKD1. However, there are also other mechanisms by which the NRG/PKD1 cross-talk could contribute to cell invasiveness. For example, PKD1 negatively affects extracellular matrix degradation and cell invasion by regulating the expression of many matrix metalloproteinases, including MMP-9 (23). It was shown that NRG increases expression of MMP-9 (5), suggesting that PKD inhibition through NRG may be a mechanism by how MMP-9 levels are regulated. In summary, our data indicate that, in MCF-7 cells, regulation of PKD1 activity by the NRG/Rac1/NADPH oxidase pathway provides a molecular switch with SSH1L as an effector for inducing actin reorganization processes that promote cell migration (Fig. 9).
Acknowledgments
We thank our colleagues in the Storz laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM086435 and CA140182. This work was also supported by Bankhead-Coley Program Grant 10BG11 from the Florida Department of Health (to P. S.).
- NRG
- neuregulin.
REFERENCES
- 1. Breuleux M. (2007) Role of heregulin in human cancer. Cell. Mol. Life Sci. 64, 2358–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang Y., Spitzer E., Meyer D., Sachs M., Niemann C., Hartmann G., Weidner K. M., Birchmeier C., Birchmeier W. (1995) Sequential requirement of hepatocyte growth factor and neuregulin in the morphogenesis and differentiation of the mammary gland. J. Cell Biol. 131, 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li Q., Ahmed S., Loeb J. A. (2004) Development of an autocrine neuregulin signaling loop with malignant transformation of human breast epithelial cells. Cancer Res. 64, 7078–7085 [DOI] [PubMed] [Google Scholar]
- 4. Schmitt M., Walker M. P., Richards R. G., Bocchinfuso W. P., Fukuda T., Medina D., Kittrell F. S., Korach K. S., DiAugustine R. P. (2006) Expression of heregulin by mouse mammary tumor cells: role in activation of ErbB receptors. Mol. Carcinog. 45, 490–505 [DOI] [PubMed] [Google Scholar]
- 5. Atlas E., Cardillo M., Mehmi I., Zahedkargaran H., Tang C., Lupu R. (2003) Heregulin is sufficient for the promotion of tumorigenicity and metastasis of breast cancer cells in vivo. Mol. Cancer Res. 1, 165–175 [PubMed] [Google Scholar]
- 6. Marshall C., Blackburn E., Clark M., Humphreys S., Gullick W. J. (2006) Neuregulins 1–4 are expressed in the cytoplasm or nuclei of ductal carcinoma (in situ) of the human breast. Breast Cancer Res. Treat. 96, 163–168 [DOI] [PubMed] [Google Scholar]
- 7. Tang C. K., Perez C., Grunt T., Waibel C., Cho C., Lupu R. (1996) Involvement of heregulin-β2 in the acquisition of the hormone-independent phenotype of breast cancer cells. Cancer Res. 56, 3350–3358 [PubMed] [Google Scholar]
- 8. Tsai M. S., Shamon-Taylor L. A., Mehmi I., Tang C. K., Lupu R. (2003) Blockage of heregulin expression inhibits tumorigenicity and metastasis of breast cancer. Oncogene 22, 761–768 [DOI] [PubMed] [Google Scholar]
- 9. Yao J., Xiong S., Klos K., Nguyen N., Grijalva R., Li P., Yu D. (2001) Multiple signaling pathways involved in activation of matrix metalloproteinase-9 (MMP-9) by heregulin-β1 in human breast cancer cells. Oncogene 20, 8066–8074 [DOI] [PubMed] [Google Scholar]
- 10. Adam L., Vadlamudi R., Kondapaka S. B., Chernoff J., Mendelsohn J., Kumar R. (1998) Heregulin regulates cytoskeletal reorganization and cell migration through the p21-activated kinase-1 via phosphatidylinositol-3 kinase. J. Biol. Chem. 273, 28238–28246 [DOI] [PubMed] [Google Scholar]
- 11. Hijazi M. M., Thompson E. W., Tang C., Coopman P., Torri J. A., Yang D., Mueller S. C., Lupu R. (2000) Heregulin regulates the actin cytoskeleton and promotes invasive properties in breast cancer cell lines. Int. J. Oncol. 17, 629–641 [DOI] [PubMed] [Google Scholar]
- 12. Bamburg J. R., McGough A., Ono S. (1999) Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 9, 364–370 [DOI] [PubMed] [Google Scholar]
- 13. Mouneimne G., DesMarais V., Sidani M., Scemes E., Wang W., Song X., Eddy R., Condeelis J. (2006) Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 16, 2193–2205 [DOI] [PubMed] [Google Scholar]
- 14. Wang W., Mouneimne G., Sidani M., Wyckoff J., Chen X., Makris A., Goswami S., Bresnick A. R., Condeelis J. S. (2006) The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell Biol. 173, 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Toshima J., Toshima J. Y., Amano T., Yang N., Narumiya S., Mizuno K. (2001) Cofilin phosphorylation by protein kinase testicular protein kinase 1 and its role in integrin-mediated actin reorganization and focal adhesion formation. Mol. Biol. Cell 12, 1131–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang T. Y., DerMardirossian C., Bokoch G. M. (2006) Cofilin phosphatases and regulation of actin dynamics. Curr. Opin. Cell Biol. 18, 26–31 [DOI] [PubMed] [Google Scholar]
- 17. Gohla A., Birkenfeld J., Bokoch G. M. (2005) Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat. Cell Biol. 7, 21–29 [DOI] [PubMed] [Google Scholar]
- 18. Tan M., Grijalva R., Yu D. (1999) Heregulin β1-activated phosphatidylinositol 3-kinase enhances aggregation of MCF-7 breast cancer cells independent of extracellular signal-regulated kinase. Cancer Res. 59, 1620–1625 [PubMed] [Google Scholar]
- 19. Nagata-Ohashi K., Ohta Y., Goto K., Chiba S., Mori R., Nishita M., Ohashi K., Kousaka K., Iwamatsu A., Niwa R., Uemura T., Mizuno K. (2004) A pathway of neuregulin-induced activation of cofilin-phosphatase Slingshot and cofilin in lamellipodia. J. Cell Biol. 165, 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eiseler T., Döppler H., Yan I. K., Kitatani K., Mizuno K., Storz P. (2009) Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat. Cell Biol. 11, 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peterburs P., Heering J., Link G., Pfizenmaier K., Olayioye M. A., Hausser A. (2009) Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res. 69, 5634–5638 [DOI] [PubMed] [Google Scholar]
- 22. Spratley S. J., Bastea L. I., Döppler H., Mizuno K., Storz P. (2011) Protein kinase D regulates cofilin activity through p21-activated kinase 4. J. Biol. Chem. 286, 34254–34261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eiseler T., Döppler H., Yan I. K., Goodison S., Storz P. (2009) Protein kinase D1 regulates matrix metalloproteinase expression and inhibits breast cancer cell invasion. Breast Cancer Res. 11, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rozengurt E., Rey O., Waldron R. T. (2005) Protein kinase D signaling. J. Biol. Chem. 280, 13205–13208 [DOI] [PubMed] [Google Scholar]
- 25. Dan C., Kelly A., Bernard O., Minden A. (2001) Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J. Biol. Chem. 276, 32115–32121 [DOI] [PubMed] [Google Scholar]
- 26. Soosairajah J., Maiti S., Wiggan O., Sarmiere P., Moussi N., Sarcevic B., Sampath R., Bamburg J. R., Bernard O. (2005) Interplay between components of a novel LIM kinase-slingshot phosphatase complex regulates cofilin. EMBO J. 24, 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang W., Eddy R., Condeelis J. (2007) The cofilin pathway in breast cancer invasion and metastasis. Nat. Rev. Cancer 7, 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Falls D. L. (2003) Neuregulins: functions, forms, and signaling strategies. Exp. Cell Res. 284, 14–30 [DOI] [PubMed] [Google Scholar]
- 29. Li L., Cleary S., Mandarano M. A., Long W., Birchmeier C., Jones F. E. (2002) The breast proto-oncogene, HRGα regulates epithelial proliferation and lobuloalveolar development in the mouse mammary gland. Oncogene 21, 4900–4907 [DOI] [PubMed] [Google Scholar]
- 30. Krane I. M., Leder P. (1996) NDF/heregulin induces persistence of terminal end buds and adenocarcinomas in the mammary glands of transgenic mice. Oncogene 12, 1781–1788 [PubMed] [Google Scholar]
- 31. Schelfhout V. R., Coene E. D., Delaey B., Thys S., Page D. L., De Potter C. R. (2000) Pathogenesis of Paget's disease: epidermal heregulin-α, motility factor, and the HER receptor family. J. Natl. Cancer Inst. 92, 622–628 [DOI] [PubMed] [Google Scholar]
- 32. Grunt T. W., Saceda M., Martin M. B., Lupu R., Dittrich E., Krupitza G., Harant H., Huber H., Dittrich C. (1995) Bidirectional interactions between the estrogen receptor and the c-erbB2 signaling pathways: heregulin inhibits estrogenic effects in breast cancer cells. Int. J. Cancer 63, 560–567 [DOI] [PubMed] [Google Scholar]
- 33. Pietras R. J., Arboleda J., Reese D. M., Wongvipat N., Pegram M. D., Ramos L., Gorman C. M., Parker M. G., Sliwkowski M. X., Slamon D. J. (1995) HER-2 tyrosine kinase pathway targets estrogen receptor and promotes hormone-independent growth in human breast cancer cells. Oncogene 10, 2435–2446 [PubMed] [Google Scholar]
- 34. Jaggi M., Rao P. S., Smith D. J., Hemstreet G. P., Balaji K. C. (2003) Protein kinase Cμ is down-regulated in androgen-independent prostate cancer. Biochem. Biophys. Res. Commun. 307, 254–260 [DOI] [PubMed] [Google Scholar]
- 35. Kim M., Jang H. R., Kim J. H., Noh S. M., Song K. S., Cho J. S., Jeong H. Y., Norman J. C., Caswell P. T., Kang G. H., Kim S. Y., Yoo H. S., Kim Y. S. (2008) Epigenetic inactivation of protein kinase D1 in gastric cancer and its role in gastric cancer cell migration and invasion. Carcinogenesis 29, 629–637 [DOI] [PubMed] [Google Scholar]
- 36. Bastea L. I., Döppler H., Balogun B., Storz P. (2012) Protein kinase D1 maintains the epithelial phenotype by inducing a DNA-bound, inactive SNAI1 transcriptional repressor complex. PLoS ONE 7, e30459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karam M., Legay C., Auclair C., Ricort J. M. (2012) Protein kinase D1 stimulates proliferation and enhances tumorigenesis of MCF-7 human breast cancer cells through a MEK/ERK-dependent signaling pathway. Exp. Cell Res. 318, 558–569 [DOI] [PubMed] [Google Scholar]
- 38. Daly J. M., Jannot C. B., Beerli R. R., Graus-Porta D., Maurer F. G., Hynes N. E. (1997) Neu differentiation factor induces ErbB2 down-regulation and apoptosis of ErbB2-overexpressing breast tumor cells. Cancer Res. 57, 3804–3811 [PubMed] [Google Scholar]
- 39. Guerra-Vladusic F. K., Scott G., Weaver V., Vladusic E. A., Tsai M. S., Benz C. C., Lupu R. (1999) Constitutive expression of heregulin induces apoptosis in an erbB2-overexpressing breast cancer cell line SKBr-3. Int. J. Oncol. 15, 883–892 [DOI] [PubMed] [Google Scholar]
- 40. Wertheimer E., Gutierrez-Uzquiza A., Rosemblit C., Lopez-Haber C., Sosa M. S., Kazanietz M. G. (2012) Rac signaling in breast cancer: a tale of GEFs and GAPs. Cell. Signal. 24, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kligys K., Yao J., Yu D., Jones J. C. (2009) 14-3-3ζ/τ heterodimers regulate Slingshot activity in migrating keratinocytes. Biochem. Biophys. Res. Commun. 383, 450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cowell C. F., Yan I. K., Eiseler T., Leightner A. C., Döppler H., Storz P. (2009) Loss of cell-cell contacts induces NF-κB via RhoA-mediated activation of protein kinase D1. J. Cell Biochem. 106, 714–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yuan J., Slice L. W., Rozengurt E. (2001) Activation of protein kinase D by signaling through Rho and the α subunit of the heterotrimeric G protein G13. J. Biol. Chem. 276, 38619–38627 [DOI] [PubMed] [Google Scholar]
- 44. Sander E. E., ten Klooster J. P., van Delft S., van der Kammen R. A., Collard J. G. (1999) Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147, 1009–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heyworth P. G., Knaus U. G., Settleman J., Curnutte J. T., Bokoch G. M. (1993) Regulation of NADPH oxidase activity by Rac GTPase-activating protein(s). Mol. Biol. Cell 4, 1217–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim J. S., Huang T. Y., Bokoch G. M. (2009) Reactive oxygen species regulate a slingshot-cofilin activation pathway. Mol. Biol. Cell 20, 2650–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Storz P., Toker A. (2003) Protein kinase D mediates a stress-induced NF-κB activation and survival pathway. EMBO J. 22, 109–120 [DOI] [PMC free article] [PubMed] [Google Scholar]