Background: Biosynthetic pathways to structurally complex plant medicinals are incomplete or unknown.
Results: Next generation sequencing/bioinformatics and metabolomics analysis of Podophyllum tissues gave putative unknown genes in podophyllotoxin biosynthesis.
Conclusion: Regio-specific methylenedioxy bridge-forming CyP450s were identified catalyzing pluviatolide formation.
Significance: Database of several medicinal plant transcriptome assemblies and metabolic profiling are made available for scientific community.
Keywords: Cytochrome P450, Enzyme Catalysis, Metabolism, Metabolomics, Transcriptomics, Lignans, Methylenedioxy Bridge Formation, Plant Medicinals, Podophyllotoxin
Abstract
Podophyllum species are sources of (−)-podophyllotoxin, an aryltetralin lignan used for semi-synthesis of various powerful and extensively employed cancer-treating drugs. Its biosynthetic pathway, however, remains largely unknown, with the last unequivocally demonstrated intermediate being (−)-matairesinol. Herein, massively parallel sequencing of Podophyllum hexandrum and Podophyllum peltatum transcriptomes and subsequent bioinformatics analyses of the corresponding assemblies were carried out. Validation of the assembly process was first achieved through confirmation of assembled sequences with those of various genes previously established as involved in podophyllotoxin biosynthesis as well as other candidate biosynthetic pathway genes. This contribution describes characterization of two of the latter, namely the cytochrome P450s, CYP719A23 from P. hexandrum and CYP719A24 from P. peltatum. Both enzymes were capable of converting (−)-matairesinol into (−)-pluviatolide by catalyzing methylenedioxy bridge formation and did not act on other possible substrates tested. Interestingly, the enzymes described herein were highly similar to methylenedioxy bridge-forming enzymes from alkaloid biosynthesis, whereas candidates more similar to lignan biosynthetic enzymes were catalytically inactive with the substrates employed. This overall strategy has thus enabled facile further identification of enzymes putatively involved in (−)-podophyllotoxin biosynthesis and underscores the deductive power of next generation sequencing and bioinformatics to probe and deduce medicinal plant biosynthetic pathways.
Introduction
Massively parallel sequencing technologies (1) are rapidly evolving and increasingly provide unprecedented opportunities to significantly enhance the understanding of biosynthetic processes, including those in important and yet poorly understood (non-model) medicinal plants. One main advantage is that such technologies can potentially lower the time frame for discovery of new genes and thus more rapidly improve our understanding of metabolism, e.g. when compared with more traditional approaches including labeled precursor administration, potential intermediate identification, enzyme purification and characterization, gene cloning, expressed sequence tag (EST)2 libraries, etc. In this context several recent investigations have used these massive parallel sequencing technologies to study a variety of non-model plants, with transcriptome assemblies mainly being generated from data from 454 and Illumina sequencing. Among others, these include Panax quinquefolius L (2), Panax ginseng (3), Gynostemma pentaphyllum (4), Phalaenopsis orchids (5, 6), Camellia sinensis (7), Catharanthus roseus (8), Papaver somniferum (9), Acacia auriculiformis, Acacia mangium (10), Cicer arietinum (11), and Abies balsamea (12). Although massive amounts of information can be obtained in this way, an informed analysis is required to help select candidate genes and carefully determine if they have a specific biosynthetic function of interest.
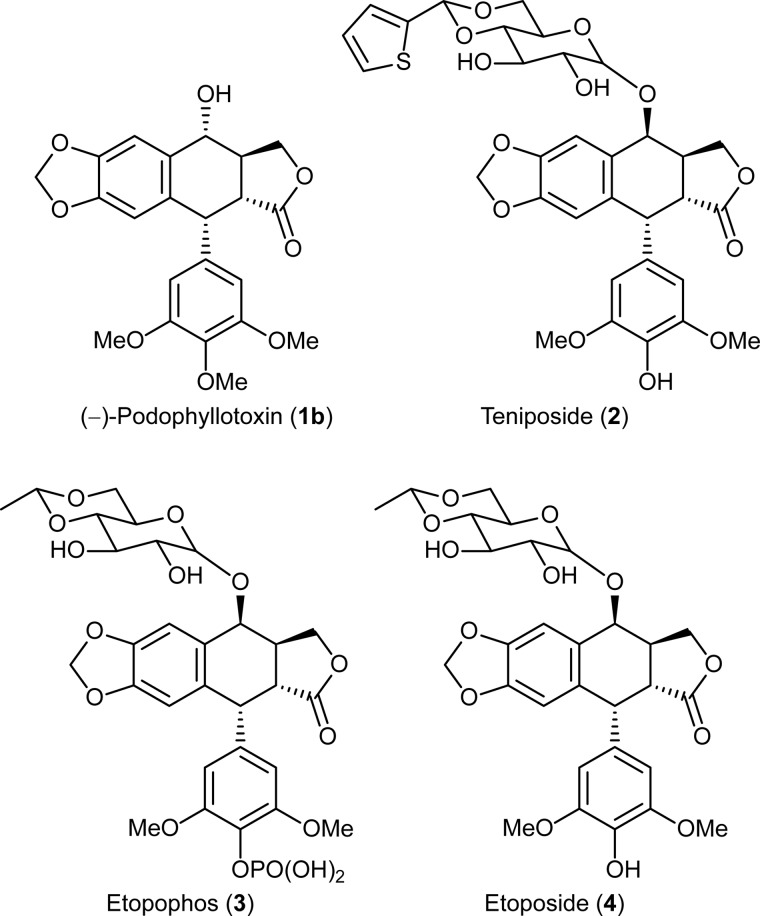
Podophyllum species produce the aryltetralin lignan, (−)-podophyllotoxin (1b), which is of great medicinal importance due to its extensive use in the semi-synthesis of the anticancer drugs, teniposide (2), etopophos® (3), and etoposide (4) (Fig. 1). The latter are topoisomerase II inhibitors that are widely used for treating several cancers, including lung and testicular cancers (13). However, as the main source of (−)-podophyllotoxin (1b), Podophyllum hexandrum is intensively collected, and some reports suggest it has become endangered due to over-harvesting (14).
FIGURE 1.
(−)-Podophyllotoxin (1b) and its derivatives teniposide (2), etopophos (3), and etoposide (4) used in cancer treatment.
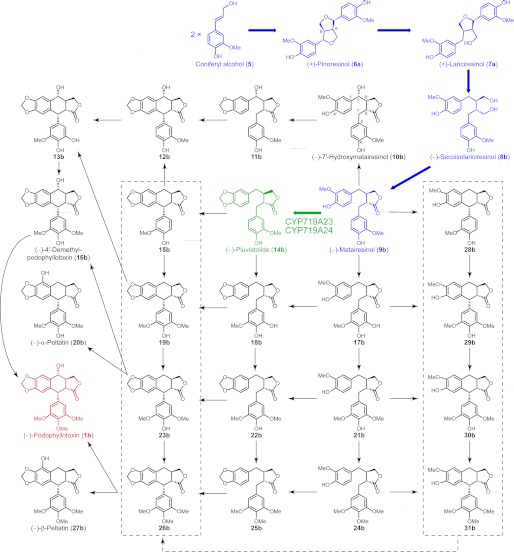
Although various synthetic chemical approaches to (−)-podophyllotoxin (1b) have been described, its production is not economical through such routes (15–17). An alternative approach that may be more productive is to obtain it in higher amounts via biotechnological manipulation whether in cell culture or in whole plants. Yet this is currently not possible as our knowledge of the (−)-podophyllotoxin (1b) biosynthetic pathway is still incomplete. Nevertheless, after monolignol formation, the entry point in its biosynthetic pathway occurs via stereoselective coupling of two E-coniferyl alcohol (5)-derived free radicals involving the participation of dirigent proteins (18–20) to afford (+)-pinoresinol (6a) (Fig. 2). The latter then undergoes enantiospecific reduction via action of pinoresinol/lariciresinol reductase to sequentially afford (+)-lariciresinol (7a) and (−)-secoisolariciresinol (8b). Stereospecific dehydrogenation next converts the latter into (−)-matairesinol (9b), the last unequivocal known step in (−)-podophyllotoxin (1b) biosynthesis (18, 21). Several of these known steps have been subsequently confirmed using Podophyllum and Linum species (20, 22). On the other hand, putative downstream steps converting (−)-matairesinol (9b) into (−)-podophyllotoxin (1b) have only been reported using crude enzymatic assays; no genes have yet been identified or the enzymes purified to homogeneity (23–27).
FIGURE 2.
Possible biosynthetic pathway and/or grid leading to (−)-podophyllotoxin (1b) and related lignans. Known biosynthetic steps are highlighted in blue, and the reaction catalyzed by CYP719A23 and CYP719A24 described in this work is in green.
The investigation herein describes the use of transcriptome sequencing using Illumina technologies and bioinformatics together with metabolomic analysis as a strategy to facilitate rapid gene discovery in (−)-podophyllotoxin (1b) biosynthesis. Specifically, this led to discovery of two new genes in P. hexandrum and Podophyllum peltatum that encode enzymes capable of catalyzing methylenedioxy bridge formation through conversion of (−)-matairesinol (9b) into (−)-pluviatolide (14b).
EXPERIMENTAL PROCEDURES
Plant Material
P. hexandrum and P. peltatum plants were obtained from Digging Dog Nursery (Albion, CA) and Companion Plants (Athens, OH), respectively, and maintained in Washington State University greenhouse facilities.
Chemicals
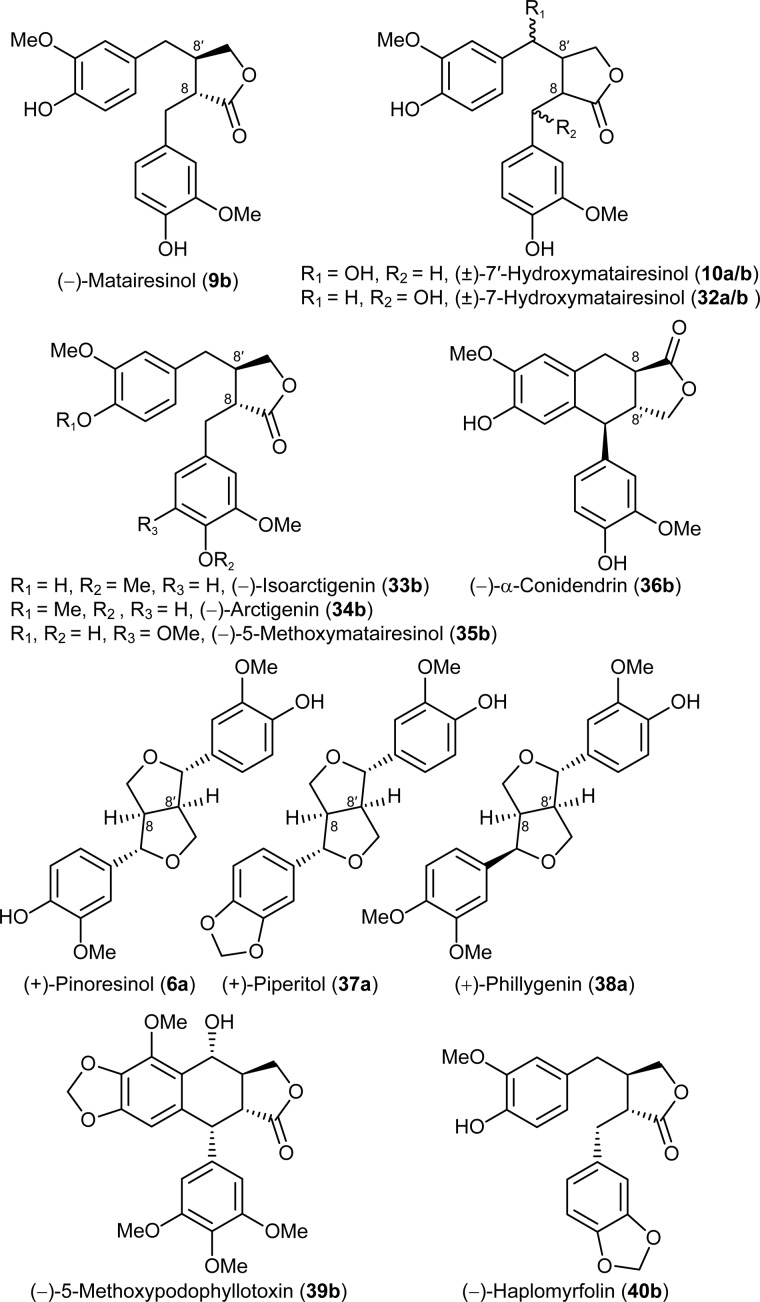
(−)-Matairesinol (9b) (28), (−)-arctigenin (34b, Fig. 3), and (+)-phillygenin (38a) (29) were isolated from Forsythia intermedia. (±)-Pinoresinols (6a/b) (22), (±)-7′-hydroxymatairesinols (10a/b) (18), (±)-7-hydroxymatairesinols (32a/b), (±)-isoarctigenins (33a/b) (29), and (±)-piperitols (37a/b) (30) were synthesized as described. (−)-α-Conidendrin (36b) and (−)-5-methoxymatairesinol (35b) were gifts from Dr. Eric P. Swan (Forentek), whereas (−)-α- and (−)-β-peltatins (20b and 27b) were obtained from Dr. Paul M. Dewick (University of Nottingham, UK). (−)-Podophyllotoxin (1b) was purchased from Sigma.
FIGURE 3.
Lignans tested as putative substrates in assays for CYP719A23 and CYP719A24 methylenedioxy bridge formation and (−)-haplomyrfolin (40b).
Metabolite Extraction and Analysis
Rhizomes, stems, and leaves (2 g, fresh weight) were individually harvested, immediately frozen in liquid nitrogen, ground to a fine powder, and subsequently lyophilized. Each tissue was then successively sized via passage through a 150-μm sieve and extracted with 10 μl/mg methanol-water (7:3, v/v) with the corresponding extracts maintained at −80 °C until analysis. Samples were analyzed by liquid chromatography using a Waters Acquity ultra performance liquid chromatography system equipped with a Waters BEH C18 column (1.7-μm particles, 2.1 × 50 mm), with detection at 280 nm and by electrospray ionization mass spectrometry in the positive mode (Table 1). The gradient program was as follows: flow rate of 0.3 ml/min and a linear gradient of water with 0.1% formic acid and acetonitrile with 0.1% formic acid from 95:5 to 75:25 in 11 min, to 60:40 in 5 min, and to 0:100 in 4 min followed by 1.5 min at 0:100. The column temperature was held at 25 °C, and sample injection volume was 5 μl. Masses were determined using a Waters Xevo G2 Q-TOF mass spectrometer and using leucine-enkephalin as a lock-mass standard.
TABLE 1.
Characteristic ions from detected lignans in positive mode mass spectral analyses
Base peaks are in bold.
| Lignan | Molecular mass | ESI-MS m/z |
|---|---|---|
| Daltons | ||
| (−)-Podophyllotoxin (1b) | 414 | 437, 432, 415, 397, 247 |
| (−)-α-Peltatin (20b) | 400 | 423, 418, 401, 247 |
| (−)-ß-Peltatin (27b) | 414 | 415, 247, 203 |
| Podophyllotoxin-glucoside (41) | 576 | 599, 594, 397 |
| α-Peltatin-glucoside (42) | 562 | 580, 563, 409, 247 |
| ß-Peltatin-glucoside (43) | 576 | 594, 577, 415, 409, 247 |
| 4′-Demethylpodophyllotoxin (16) | 400 | 423, 418, 401, 383, 247 |
(−)-Podophyllotoxin (1b)
m/z 437.1215 ([M + Na]+, 81%), calculated 437.1207; 432.1652 ([M + NH4]+, 25%), calculated 432.1653; 415.1391 ([M + H]+, 36%), calculated 415.1387; 397.1285 ([M + H − H2O]+, 100%), calculated 397.1282; 247.0605 (19%), calculated 247.0601.
(−)-α-Peltatin (20b)
m/z 423.1059 ([M + Na]+, 60%), calculated 423.1050; 418.1503 ([M + NH4]+, 45%), calculated 418.1496; 401.1236 ([M + H]+, 71%), calculated 401.1231; 247.0608 (100%), calculated 247.0601.
(−)-β-Peltatin (27b)
m/z 415.1397 ([M + H]+, 44%), calculated 415.1387; 247.0605 (100%), calculated 247.0601; 203.0708 (1%), calculated 203.0703.
Podophyllotoxin-glucoside (41)
m/z 599.1739 ([M + Na]+, 61%), calculated 599.1735; 594.2181 ([M + NH4]+, 11%), calculated 594.2182; 397.1289 ([M + H − H2O − Glc]+, 100%), calculated 397.1282.
α-Peltatin-glucoside (42)
m/z 580.2030 ([M + NH4]+, 3%), calculated 580.2025; 563.1763 ([M + H]+, 3%), calculated 563.1759; m/z 409.1134 (33%), calculated 409.1129; 247.0603 (100%), calculated 247.0601.
β-Peltatin-glucoside (43)
m/z 594.2183 ([M + NH4]+, 1%), calculated 594.2182; 577.1926 ([M + H]+, 11%), calculated 577.1916; 415.1393 ([M + H − Glc]+, 22%), calculated 415.1387; 409.1140 (10%), calculated 409.1129; 247.0599 (100%), calculated 247.0601.
4′-Demethylpodophyllotoxin (16)
m/z 423.1057 ([M + Na]+, 53%), calculated 423.1050; 418.1499 ([M + NH4]+, 37%), calculated 418.1496; 401.1229 ([M + H]+, 14%), calculated 401.1231; 383.1134 ([M + H − H2O]+, 100%), calculated 383.1125; 247.0606 (21%), calculated 247.0601.
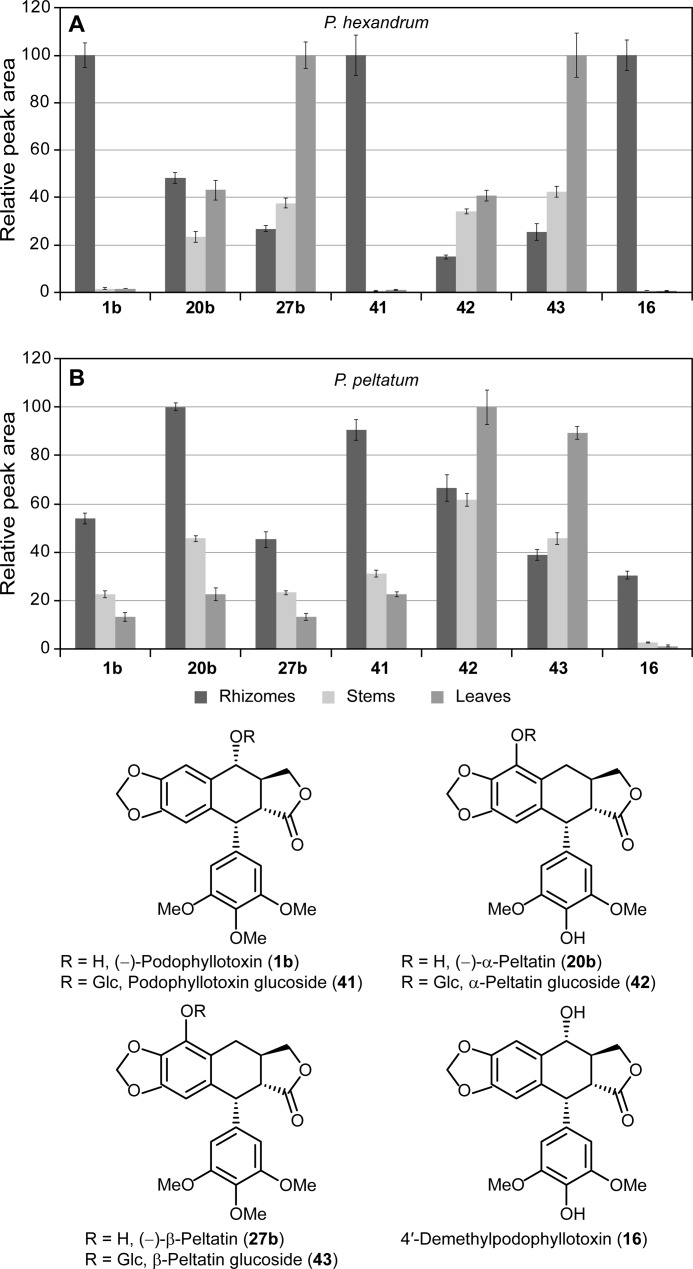
For relative abundance assessment of metabolites, integration was performed using extracted specific ion chromatogram for each compound: m/z 397.128 for (−)-podophyllotoxin (1b), m/z 247.060 for (−)-α- and (−)-β-peltatin (20b and 27b), m/z 594.218 for podophyllotoxin-glucoside (41), m/z 409.113 for α-peltatin glucoside (42), m/z 577.193 for β-peltatin glucoside (43), and m/z 383.113 for 4′-demethylpodophyllotoxin (16) (see Fig. 4 for structures).
FIGURE 4.
Relative lignan contents in different tissues of P. hexandrum and P. peltatum. Lignans identified included (−)-podophyllotoxin (1b), (−)-α-peltatin (20b), (−)-β-peltatin (27b), podophyllotoxin-glucoside (41), α-peltatin-glucoside (42), β-peltatin-glucoside (43), and 4′-demethylpodophyllotoxin (16) in both P. hexandrum (A) and P. peltatum (B) tissues. The individual lignan amounts are presented as relative peak areas (280 nm) with each given compound having a relative peak area of 100% for the most abundant amount, and the others are reported as a percentage of that value.
RNA Extraction and cDNA Preparation
Total RNA was individually isolated from 100 mg of flash-frozen rhizome using Invitrogen Plant RNA Purification Reagent, and from stems, and leaves using a Qiagen (Valencia, CA) RNeasy Mini kit according to the manufacturer's instructions including the additional Qiagen cleanup protocol. An aliquot (1 μg) of each was subsequently used for cDNA preparation. After DNase I (Invitrogen) treatment, cDNA was prepared using SuperScript® III First-Strand Synthesis System (Invitrogen) according to the manufacturer's instructions and used for candidate gene amplification.
Transcriptome Sequencing and Library Assembly
Total RNA samples (∼25 μg) dissolved in water were evaluated for integrity using the Bioanalyzer 2100 (Agilent, CA) with samples having an RNA integrity number (31) (RIN) >5.0 processed further. RNA sample concentrations were estimated using a RiboGreen assay in a Qubit fluorometer, and each was then processed using either Illumina RNA-Seq or Illumina TruSeq RNA sample preparation kits, according to Illumina protocols. Briefly, poly-A+ RNA was isolated from total RNA samples using oligo-d(T)25 magnetic beads (Dynabeads; Invitrogen), then fragmented with the supplied reagents. First-strand cDNA synthesis followed using random hexamers and the enzyme master mix provided. After second strand cDNA synthesis, the cDNA fragments were end-repaired by treatment with T4 DNA polymerase and a Klenow fragment of Escherichia coli DNA polymerase I followed by the addition of a single deoxyadenosine to the 3′ end of blunt-ended phosphorylated fragments. Sequencing adapters were attached with bacteriophage T4 ligase followed by agarose gel electrophoresis with excision of fragments circa 500 bp in length. Each library DNA was purified from agarose using Qiagen QiaQuick gel extraction reagents and subjected to 15 cycles of PCR. Amplified libraries were evaluated for quality and quantity using the Bioanalyzer 2100 and Nanodrop ND-1000 (Thermo Scientific), respectively. Based on Nanodrop concentrations, libraries were normalized to 10 nm, and accurate concentrations of sequenceable molecules were determined by quantitative PCR using a reference library of known concentration as a standard. Flow cells were prepared on the Illumina Cluster Station, and paired-end 54-bp sequence reads were obtained using an Illumina Genome Analyzer IIx instrument.
Initial read sets were examined for gross anomalies (e.g. over-represented reads due to library preparation issues) and for the presence of known sequencing artifacts, specifically φX and known Illumina adapters. Read sets were then partitioned into collections of roughly one million reads, with these being quality-rimmed using the FASTX Toolkit at the level of phrap Q = 10.
Cleaned paired-end read data were next subjected to multiple assemblies using ABySS (32, 33) run in parallel mode over a range of kmers (k) with 24 ≤ k ≤ 54. Contigs thus generated were named synthetic ESTs, which in turn were created by performing multiple assemblies using different kmer sizes, pooling the resulting synthetic ESTs, and as described below, performing a subsequent assembly with a standard EST assembler. Typically this led to the production of at least 20 sets of synthetic ESTs.
Contigs resulting from each ABySS assembly were scaffolded using the ABySS scaffolder taking advantage of read pairing constraints. The NNN gap spacers inserted through scaffolding were resolved using GapCloser from the SOAPdenovo suite (34), and synthetic EST sets, per kmer, were constructed from the resulting scaffolds of at least 80 nucleotides. All read data were incorporated in the assembly producing an overall transcript reference.
Final Assembly
Pooled synthetic EST sets were assembled using MIRA in EST assembly mode (35). To control redundancy explicitly (at 98% sequence identity), the assembly results were processed with cd-hit (36). Resulting contigs of at least 100 nucleotides were reported as the final contig set for the build. Manual assessments of the longest contigs and contigs with anomalously low or high read counts were performed by inspection of the pileup-view using Tablet (37). Final assembly for all tissues and species investigated can be accessed in the Medplants website.
Bioinformatic Analysis
Amino acid sequences of reference genes for shikimate, phenylpropanoid, and lignan biosynthetic pathways were obtained from the NCBI database. Most genes were from Arabidopsis thaliana, except Petunia × hybrida for prephenate aminotransferase, Nicotiana tabacum for hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase, Forsythia × intermedia for dirigent protein, and pinoresinol/lariciresinol reductase and P. peltatum for secoisolariciresinol dehydrogenase and dirigent protein, respectively (Table 2 and supplemental Table S1). Amino acid sequences of each reference gene were applied to tblastn in BioEdit (Version 7.0.5.3, Oct. 28, 2005) (38) to search homologous genes against either P. hexandrum or P. peltatum contig databases. Some contigs with high homology (with identity ≥ 30% and E ≤ 5 × 10−23) against each reference gene were selected, and the respective ORFs for each contig were determined using ORF Finder (NCBI). ORFs were individually translated into amino acid sequence using EMBOSS Transeq (European Bioinformatics Institute), and each amino acid sequence translation was applied to tblastn (NCBI) to confirm whether it corresponded to each target gene. The procedures mentioned above were carried out to avoid ambiguity of chimeric contigs (39, 40). In addition, E-values and identity (%) of each candidate gene were calculated using blastp (NCBI) against the amino acid sequence of the corresponding reference genes. Unknown candidate genes for (−)-podophyllotoxin (1b) biosynthesis were selected using the same approach based on known sequences for cinnamate 4-hydroxylase (CYP73A1), p-coumaroyl CoA 3-hydroxylase (CYP98A44), ferulate 5-hydroxylase (CYP84A3), flavonoid 6-hydroxylase (CYP71D9), flavonoid 3′-hydroxylase (CYP75A1), and corytuberine synthase (CYP80G2) as well as the methylenedioxy bridge-forming enzymes piperitol/sesamin synthase CYP81Q1 (41) and (S)-canadine synthase CYP719 (42) (supplemental Table S2).
TABLE 2.
BLAST result for known protein sequences from P. peltatum and P. hexandrum
Pp, P. peltatum; Ph, P. hexandrum.
| Protein | Query accession number/species of origin | Species/transcript | Identity | E value |
|---|---|---|---|---|
| % | ||||
| Dirigent protein | AAK38666.1/P. peltatum | PpDir1_Pp27246 | 99.0 | 2 E−115 |
| PhDir1_Ph08051 | 92.0 | 4 E−93 | ||
| Pinoresinol/lariciresinol reductase | ACF71492.1/P. hexandrum | PhPLR2_Ph140193 | 99.4 | 0 |
| PpPLR1_Pp37193 | 95.5 | 0 | ||
| Secoisolariciresinol dehydrogenase | AAK38664.1/P. peltatum | PpSDH1_Pp12640 | 99.3 | 3 E−163 |
| PhSDH1_Ph12248 | 97.5 | 2 E−152 |
Gene Cloning and Yeast Expression
Candidates for CyP450 genes were amplified from P. hexandrum cDNA using the primers described in supplemental Table S2. Amplification was performed using PfuTurbo DNA polymerase (Agilent PCR) in a thermocycler with 35 cycles of 94 °C denaturing for 30 s, 55 °C annealing for 30 s, and 70 °C extension for 3 min, and a final extension for 10 min. PCR products were resolved in 1% agarose gels, where single bands of ∼1500 bp were obtained. Sequences were deposited in the GenBankTM database under accession numbers KC110988–KC110998 (supplemental Table S2).
Products were cloned into pENTR/d-TOPO (Invitrogen) and subsequently transferred to a yeast expression vector pYES-DEST52 (Invitrogen) according to the manufacturer's instructions. From P. hexandrum, yeast expression clones pYES-DEST52::CYP719A23 and pYES-DEST52::CYP73A107 were obtained and from P. peltatum, pYES-DEST52::CYP719A24. Each was subsequently individually introduced in the Saccharomyces cerevisiae strain WAT11 (43) using the lithium acetate procedure according to the vector manufacturer's instructions. An empty vector pYES-DEST52 was also introduced into WAT11 and used as a negative control.
Transformed yeasts were selected using synthetic complete media lacking uracil (SC-U) plates with 2% agar and 2% glucose. Single colonies of transformed yeast were spiked in liquid SC-U (10 ml) containing 2% glucose, then grown overnight until A600 reached 3–4 and subsequently inoculated in Erlenmeyer flasks (1 liter) containing induction media (200 ml SC-U with 1% raffinose and 2% galactose) to a final A600 of 0.05. Inductions were carried out for 24 h at 30 °C in an orbital shaker at 300 rpm until cells were harvested for immediate microsome preparation.
Microsome Preparations
After induction, cells for each candidate recombinant enzyme were harvested by centrifugation at 3900 × g for 10 min then resuspended in 3 ml/g of cell weight Tris-HCl buffer (50 mm, pH 7.4) containing EDTA (1 mm), sorbitol (600 mm), DTT (0.1 mm), and PMSF (0.4 mm). Cells were disrupted using 0.5-mm glass beads (Biospec Products, Inc.) with approximately half of the total volume added to cell suspensions in Falcon tubes (50 ml) by vortexing for 10 × 30 s at full speed with 30 s intervals on ice. Cell lysates were individually separated from glass beads by decantation, and glass beads were washed twice with half the total volume of buffer used initially to resuspend the cells. Cell debris was removed by centrifugation at 15,000 × g for 10 min, and microsomal preparations were individually obtained as gelatinous pellets after ultracentrifugation of the supernatant at 91,000 × g for 75 min. Each microsomal fraction was resuspended in Tris-HCl buffer (50 mm, pH 7.4) containing EDTA (1 mm) and 30% glycerol (500 μl/g of fresh weight of cell harvested) and homogenized using a Dounce homogenizer. Microsomal preparations were kept at −80 °C for up to 8 weeks with no detectable loss in activity.
Enzymatic Assays
Assays were performed in sodium phosphate buffer (200 μl, 100 mm, and pH 7.5) with the addition of a methanol solution of the substrates (10 μl) at the desired concentration (from 0.1 to 25 mm) followed by NADPH (50 mm, 10 μl) in sodium phosphate buffer (100 mm, pH 7.5) and finally the microsomal preparation (30 μl) with a protein concentration of ∼60 μg/μl. Upon the addition of each microsomal preparation reaction, the mixtures were vortexed and incubated at 25 °C for 5 min with constant shaking. Reactions were individually terminated by the addition of glacial acetic acid (10 μl) and then centrifuged at 16,000 × g for 30 min. Aliquots of supernatant were then directly analyzed by ultra performance liquid chromatography using the same methodology described for metabolite analysis. For all kinetic data, assays were performed in three independent experiments. Kinetic parameters (Km and kcat) were estimated by nonlinear least-squares data fitting (44), and cytochrome P450 (CyP450) content in microsomes was determined by the reduced CO difference spectrum (45) using a Lambda 20 UV-visible spectrophotometer (PerkinElmer Life Sciences).
Isolation of Enzymatic Product
150 enzymatic assays using microsome preparations of yeast expressing CYP719A23 and with (−)-matairesinol (9b) as substrate were pooled together, and the whole (∼37.5 ml) preparation was extracted 3 times with chloroform (40 ml). The combined organic solubles were evaporated to dryness in vacuo and resuspended in methanol (1 ml), and the enzymatically formed (−)-pluviatolide (14b) was next purified by HPLC using a SymmetryShield RP18 column (Waters, 5 μm particle size, 3.9 × 150 mm) eluted as follows: flow rate of 1 ml/min and linear gradient of water and acetonitrile from 9:1 to 4:6 in 25 min and to 1:0 in 2.5 min followed by 4.5 min at 1:0. Fractions containing (−)-pluviatolide (14b) were pooled, freeze-dried, and subjected to 1H, 13C, and 13C,1H heteronuclear single quantum coherence NMR spectroscopic analyses using deuterated chloroform as solvent and tetramethylsilane as the internal standard in a Varian VNMRS 600 MHz spectrometer (supplemental Table S3 and Figs. S1–S3).
(−)-Pluviatolide (14b)
δH (CDCl3): 2.45–2.62 (4H, m); 2.89 (1H, dd, J = 7.0 and 14.1); 2.96 (1H, dd, J = 5.2 and 14.0); 3.85 (3H, s); 3.86 (1H, dd, J = 7.4 and 9.1); 4.11 (1H, dd, J = 7.1 and 9.2); 5.93 (1H, d, J = 1.4); 5.94 (1H, d, J = 1.4); 6.44–6.47 (2H, m); 6.63 (1H, dd, J = 1.8 and 7.9); 6.67 (1H, d, J = 1.8); 6.69 (1H, d, J = 7.7); 6.84 (1H, d, J = 8). δC (CDCl3) 178.64, 147.85, 146.65, 146.32, 144.52, 131.59, 129.43, 122.07, 121.55, 114.22, 111.48, 108.79, 108.31, 101.04, 71.19, 55.87, 46.59, 41.00, 38.30, 34.62. MS: m/z 379.1155 ([M + Na]+, calculated 379.1157), 357.1337 ([M + H]+, calculated 357.1338), 339.1230 ([M + H − H2O]+, calculated 339.1232), 161.0604 (calculated 161.0603), 137.0604 (calculated 137.0603) and 135.0445 (calculated 135.0446).
RESULTS AND DISCUSSION
Metabolite Profiling
First, metabolite profiling was carried out to ensure that target and biochemically related metabolites were present in the various tissues of the Podophyllum species investigated. Thus, utilizing ultra performance liquid chromatography-electrospray ionization-mass spectrometry, the extracts of rhizome, stem, and leaves were examined. Based on metabolite UV, retention time, and mass spectra, it was readily possible to detect and confirm the presence of the target metabolite, (−)-podophyllotoxin (1b). In P. hexandrum, it accumulates in higher amounts in the rhizome, but was barely detectable in leaves and stem (Fig. 4A). In P. peltatum, the same trend was observed, with a higher accumulation in the rhizomes, although the stem and leaves also had (−)-podophyllotoxin (1b) contents closer to that in the rhizomes (Fig. 4B). Its identification was performed by comparison with an authentic standard, having the same retention time as well as mass spectrum (Table 1 and “Experimental Procedures”). Other two related lignans, (−)-α- and (−)-β-peltatins (20b and 27b), were detected in different tissues of both species and also identified using authentic standards (Table 1 and “Experimental Procedures”). Interestingly, in P. hexandrum, the accumulation pattern of these two lignans was quite different from that observed for (−)-podophyllotoxin (1b), with (−)-α-peltatin (20b) being detected throughout all the different tissues. Conversely, there was a higher accumulation of (−)-β-peltatin (27b) in the aerial tissues, especially in leaves (Fig. 4A). In P. peltatum, on the other hand, both lignans accumulated in a somewhat similar pattern to that of (−)-podophyllotoxin (1b), with increasing amounts from leaves to stems and with the highest abundance in the rhizomes (Fig. 4B).
Based on the mass spectroscopic analyses (Table 1 and “Experimental Procedures”), it was also possible to identify the known (46–48) glycosylated forms of the aforementioned lignans: podophyllotoxin-glucoside (41), α-peltatin-glucoside (42), and β-peltatin-glucoside (43). It was also possible to detect 4′-demethylpodophyllotoxin (16) with mass identical to the isobaric (−)-α-peltatin (20b) but with the additional base peak resulting from loss of water [M + H − H2O]+ of m/z 383.1134 (calculated 383.1125). The accumulation patterns of podophyllotoxin-glucoside (41) and 4′-demethylpodophyllotoxin (16) were similar to that of podophyllotoxin (1), with higher amounts in the underground tissues in both species, especially in P. hexandrum. As for (−)-α- and (−)-β-peltatin glucosides (42b and 43b), they accumulated in all tissues, with slightly higher levels in the leaves (Fig. 4).
Based on the metabolite profiles observed for (−)-podophyllotoxin (1b) and the other related lignans described above, it could be considered that the overall functional biosynthetic pathway leading to these lignans might be present in all tissues in both species; on the other hand, specific hydroxylation enzymes regiospecifically placing hydroxyl groups leading either to (−)-podophyllotoxin (1b) or to (−)-α- and (−)-β-peltatin (20b and 27b) might be tissue-specific.
Transcriptome Assembly and Analysis
Upon confirmation and analysis of metabolites as compared with authentic standards, RNA was extracted from tissues, and transcriptome data were generated as described under “Experimental Procedure.” From the crude RNA, poly-A+ RNA was isolated, fragmented, and converted into cDNA using random hexamers and end-paired to increase data quality for later assembly. After the addition of adapters and quality control of the products, sequencing was performed using the Illumina Genome Analyzer IIX generating the read sets. These first read sets were then examined for known anomalies like significant sequence similarity to either the φX genome or the Illumina adapter reference set, and those anomalies were discarded; the initial data were thus slightly reduced, typically by 1–5%. The reads were then partitioned in paired data blocks to take advantage of parallel processing in the workflow and subsequently assembled using ABySS, which provides the ability to associate input files of read data with their source library. In this step, kmer size tended to be the most sensitive factor in the construction and analysis of the de Bruijn graph, and different choices often led to similar, yet different, assemblies. Contigs from these assemblies tended to be 100–500 bp long and were treated as “synthetic ESTs,” and these were then assembled into the final contig set. It is important to stress that working with large quantities of data in this way presents a number of challenges, and for this reason each of the stages above includes a number of integrity checks (e.g. incomplete processing caused by system failures), basic data quality measurements (e.g. abundance of sequencing contaminants), and biological significance. The transcriptome database assembled for P. hexandrum and P. peltatum produced final databases of 227,885 and 147,960 contigs, respectively, including several complete and incomplete ORFs. The transcriptome data obtained is available alongside that of other important medicinal plants and can be accessed in the Medplants website.
Next, comparative analysis of metabolite profiles and transcriptome assembly data from each of the various tissues (rhizomes, stems and leaves) was carried out to (a) verify whether known genes previously cloned from P. peltatum (18, 22) were correctly assembled; (b) identify candidate genes in the shikimate-chorismate pathway to phenylalanine, the entry point into the phenylpropanoid pathway and in the core phenylpropanoid pathway leading to monolignols, such as coniferyl alcohol (5) (the latter is also the entry point metabolite into the (−)-podophyllotoxin (1b) biosynthetic pathway); (c) conduct a bioinformatics analysis to identify potential candidate genes encoding steps beyond (−)-matairesinol (9b) and leading to the target compound, (−)-podophyllotoxin (1b).
Thus, assemblies were first interrogated to assess the validity of the presence of contigs corresponding to previously described genes and to obtain a first measure of the assembly quality. In our earlier studies of (−)-podophyllotoxin (1b) biosynthesis in P. peltatum, we had cloned and characterized a dirigent protein responsible for mediating the stereoselective coupling of E-coniferyl alcohol (5) leading to (+)-pinoresinol (6a) (18) and a secoisolariciresinol dehydrogenase, responsible for the conversion of (−)-secoisolariciresinol (8b) into (−)-matairesinol (9b) (22). When the assemblies obtained for P. peltatum were interrogated using these known sequences, contigs with very high identity (>98%) and low E value (<10−110) to the previously described genes were observed (Table 2). This suggested that the assembly process was providing high quality data related to known genes; however, whether the sequences were fully correct or not (e.g. if they had point mutations, etc.) was not explored further, i.e. by confirmation through cloning, protein expression, etc.
The search for homologs in the shikimate/phenylpropanoid and monolignol-forming pathways was also successful, with several homologs to each gene in both species identified. This included 10 enzymes in the initial shikimate/chorismate pathway and 9 enzymes from the core phenylpropanoid pathway (supplemental Table S1) with identity ≥30% and E ≤ 5 × 10−23, as stated under “Experimental Procedure.” Overall, this successful search for homologs for all known genes from the shikimate/phenylpropanoid pathway that lead to the target lignans and several other important compounds (e.g. flavonoids, lignin, etc.) indicated that the assemblies obtained had a satisfactory coverage of the species transcriptomes. On the other hand, cloning and confirmation of the absolute accuracy of the assembly and the actual function of the corresponding gene was not carried out for non-CyP450s as this was outside the scope of the current study.
Even considering the highly encouraging results from this initial assessment of the assembled datasets, the potential limitations of this overarching approach need to be considered. This is because the assembly protocol is a tradeoff of the stringency of the assembly and the length of the transcripts obtained. It is thus always possible to find chimeric contigs, especially in cases of highly similar transcripts (close homologs) or in those cases where repeating elements are present. On the other hand, if the stringency of the assembly procedure is increased to minimize the presence of chimeric contigs, the result can be an increase in the number of incomplete and redundant (more than one contig for the same transcript) contigs. During the assembly process, several parameters were thus optimized to reach a balance, but in the final assembly examples of both could still be observed.
CyP450s in Podophyllotoxin Biosynthesis
Once the quality of the assemblies was evaluated and validated in silico, a search for unknown putative genes involved in the last steps in (−)-podophyllotoxin (1b) biosynthesis was then undertaken. The possible biochemical modifications required for conversion of (−)-matairesinol (9b) into (−)-podophyllotoxin (1b) are readily deduced (Fig. 2), but the order of these reactions, on the other hand, has several possible permutations. Indeed, these could possibly either occur in parallel in a biosynthetic “grid” or through a specific sequence of conversions (Fig. 2). To verify the role of possible substrates, it would be necessary to test a variety of compounds, most of which are not commercially available. From (−)-matairesinol (9b), the biosynthetic pathway leading to (−)-podophyllotoxin (1b) can, however, be expected to include several CyP450s involving two hydroxylations, one carbon-carbon (C-C) coupling/cyclization and methylenedioxy bridge formation, respectively (Fig. 2). Additionally, although the C-C coupling and hydroxylation on the 7 position could be putatively performed by enzymes other than CyP450 (e.g. by a laccase in the first case and/or an oxoglutarate dependent dioxygenase for the latter), the formation of a methylenedioxy bridge functionality should be catalyzed by CyP450s. Accordingly, the first genes chosen for investigation were CyP450s.
The CyP450s are a very large (e.g. more than 200 genes in A. thaliana) and diverse family of pivotal importance in plant secondary metabolism (49–51). Many are involved in phenylpropanoid metabolism and have been identified, cloned, and characterized from several plant species. More specifically, they can be employed in monolignol biosynthesis, where cinnamate-4-hydroxylase, p-coumaroyl CoA 3-hydroxylase, and ferulate 5-hydroxylase are responsible for the successive hydroxylation of the C6C3 core (52, 53) as well as in the biosynthetic pathway leading to many downstream products, e.g. in flavonoid (54) and lignan (41, 55) biosynthesis.
The prediction of CyP450 physiological function is, however, frequently a difficult endeavor. This is because members of distinct gene families can perform similar functions, and in some cases, members of the same families can catalyze different reactions (51). To increase the probability of obtaining the presumed CyP450s responsible for methylenedioxy bridge formation, several different known CyP450s were used as templates for the search of homologs that could be involved in (−)-podophyllotoxin (1b) and related phenylpropanoid biosynthesis. Initially focusing on P. hexandrum, its transcriptome was mined as described under “Experimental Procedure” for homologs to cinnamate 4-hydroxylase (CYP73A1), p-coumaroyl CoA 3-hydroxylase (CYP98A), ferulate 5-hydroxylase (CYP84A3), flavonoid 6-hydroxylase (CYP71D9), flavonoid 3′-hydroxylase (CYP75A1), corytuberine synthase (CYP80G2), (+)-δ-canadinene 8′-hydroxylase (CYP706B1), (S)-canadine synthase (CYP719), and piperitol/sesamin synthase (CYP81Q). The highest homology contigs in the assembled transcriptome were then selected for cloning and further analysis (supplemental Table S2). All genes cloned matched transcriptome contig sequences perfectly (100%), therefore, again validating the sequencing and assembly procedures employed.
Each of the above-cloned candidates was heterologously expressed as described under “Experimental Procedure” for methylenedioxy bridge-forming enzymes and assayed against a range of possible substrates. The first genes to be tested encoded homologs of well established enzymes CYP73A107 (cinnamate 4-hydroxylase homolog), CYP98A68 (p-coumaroyl CoA 3-hydroxylase homolog), CYP84A52 (ferulate 5-hydroxylase homolog), and CYP71BE30 (flavonoid 6-hydroxylase homolog); these CyP450s were assayed and found to carry out the anticipated enzymatic conversion (data not shown). CYP73A107 showed the highest activity and was thus used as a positive control in all assays. We then proceeded to investigate the methylenedioxy bridge-forming steps of (−)-podophyllotoxin (1b) and related lignans.
Methylenedioxy Bridge Formation in Podophyllotoxin Biosynthesis
Methylenedioxy bridge formation by CyP450s has been described in isoflavonoid (56), lignan (41, 55), and alkaloid (42, 57–59) biosynthesis, with encoding genes cloned and characterized. Transcriptome data were interrogated looking for sequences similar to either CyP450 families, and we were able to identify putative methylenedioxy bridge-forming enzyme homologs to both lignan (CYP81Q1 and CYP81Q2) (41) and alkaloid (CYP719A1 and AY610513) (42, 57) biosynthesis. In the assembled P. hexandrum transcriptome, there was one full-length transcript with ∼50% identity to the former, coded CYP81B57 (supplemental Table S2), and another full-length candidate with ∼68% identity to the latter, coded CYP719A23 (Fig. 5). Later, the P. peltatum assembled transcriptome was also interrogated, and the transcript coded CYP719A24 was selected showing ∼68% identity to CYP719A1 and ∼96% identity to CYP719A23 (Fig. 5).
FIGURE 5.
Sequence alignment of methylenedioxy bridge-forming cytochrome P450s. CYP719A23 and CYP719A24, cloned from P. hexandrum and P. peltatum, respectively, show ∼68% identity to Coptis japonica (S)-canadine synthase (CYP719A1).
These three candidates were cloned and their sequences confirmed by traditional Sanger sequencing (60). Subsequently, each was individually expressed in yeast, and the corresponding recombinant proteins were assayed for methylenedioxy bridge formation using a range of potential substrates and analogs thereof including (−)-matairesinol (9b), the last confirmed intermediate in (−)-podophyllotoxin (1b) biosynthesis in Podophyllum species, (±)-7′-hydroxymatairesinols (10a/b), (±)-7-hydroxymatairesinols (32a/b), (−)-5-methoxymatairesinol (35b), (±)-isoarctigenins (33a/b), (−)-arctigenin (34b), (±)-pinoresinols (6a/b), (±)-piperitols (37a/b), (+)-phillygenin (38a), and (−)-α-conidendrin (36b) (Fig. 3). Of these, (−)-7′-hydroxymatairesinol (10b) has been shown to be an intermediate in (−)-5-methoxypodophyllotoxin (39b) formation in Linum flavum (18), and isoarctigenin (33) could also be an intermediate in (−)-podophyllotoxin (1b) biosynthesis if (−)-matairesinol (9b) underwent methylation in the 4′ position before other modifications. The other analogs, (±)-pinoresinols (6a/b), (−)-arctigenin (34b), (−)-α-conidendrin (36b), (±)-piperitols (37 a/b), and (+)-phillygenin (38a) have similar overall structure to possible intermediates and were tested to assess the enzyme substrate versatility.
The first candidate evaluated was the homolog to the piperitol/sesamin synthase, namely CYP81B57. In our hands no catalytic activity could be observed, including toward piperitol/sesamin synthase natural substrates, (+)-pinoresinol (6a) and (+)-piperitol (37a) (data not shown).
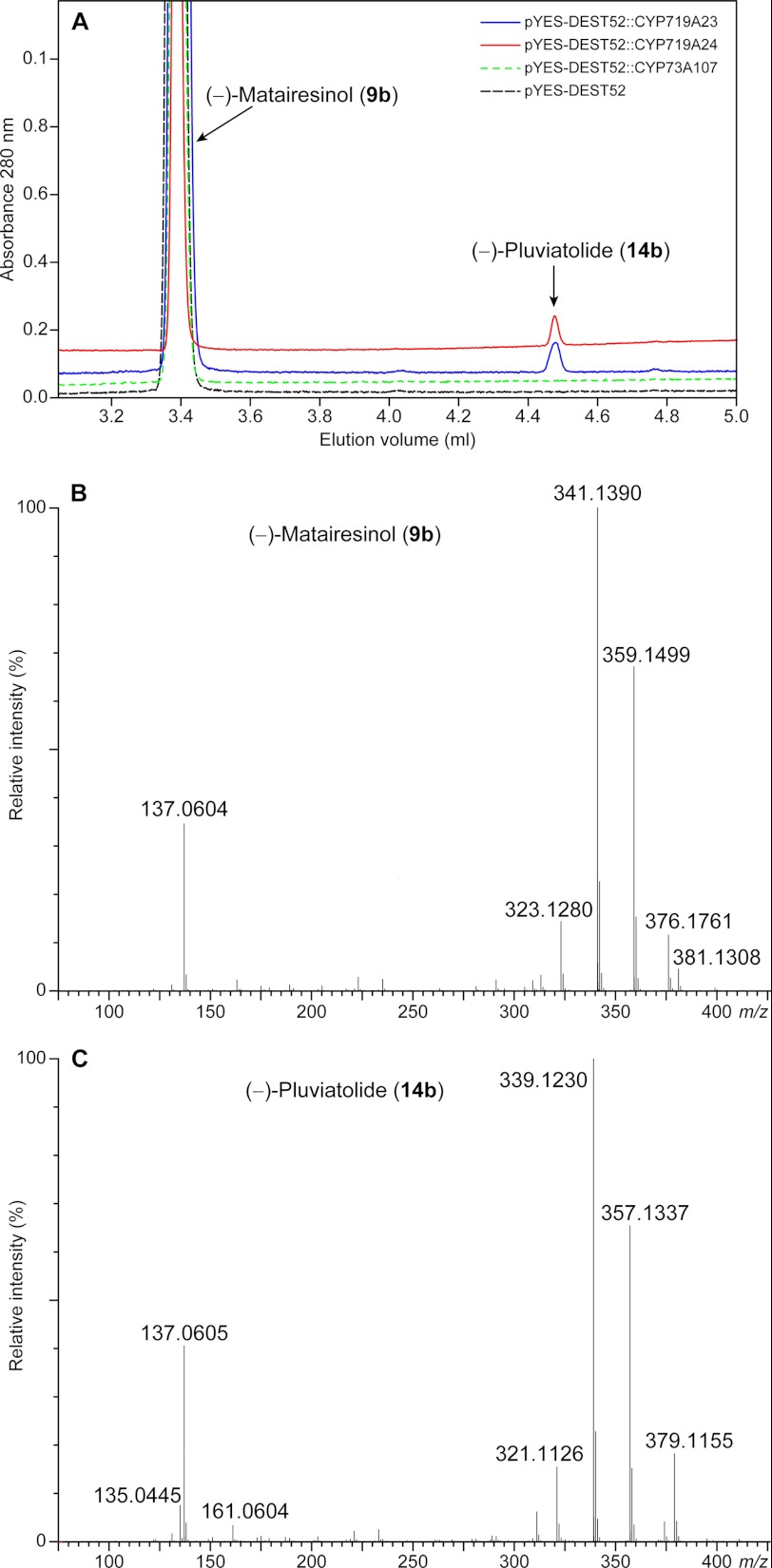
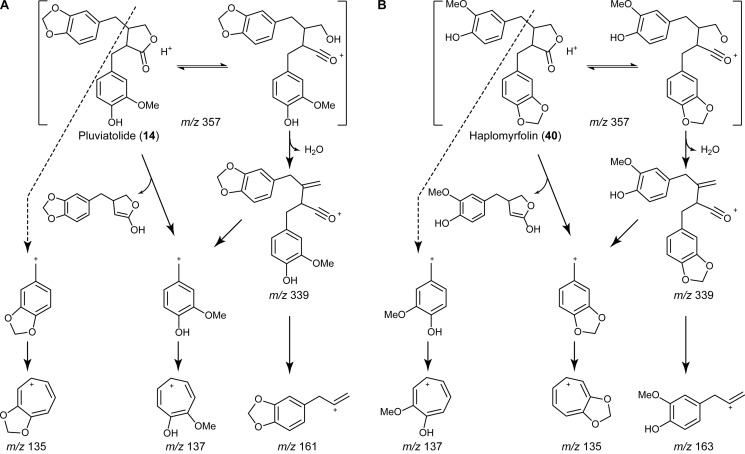
We then proceeded to evaluate the methylenedioxy bridge-forming candidate homologs to the enzymes putatively annotated as involved in alkaloid biosynthesis. These (S)-canadine synthase homologs were found to act on (−)-matairesinol (9b) in the presence of NADPH, leading to the formation of a product with a longer retention time in ultra performance liquid chromatography analysis (Fig. 6A), whose protonated molecular ion was at m/z 357 (Fig. 6C), corresponding to the loss of two hydrogens as compared with (−)-matairesinol (9b) (Fig. 6B). It was not possible to detect any activity and/or product formation, however, with the other putative substrates tested (data not shown), indicating that the (S)-canadine synthase homologs had a considerable degree of specificity toward (−)-matairesinol (9b). From the three possible products formed by a methylenedioxy bridge-forming enzyme (either with formation of one methylenedioxy bridge in either one of the aromatic rings or in both), the expected biosynthetic pathway reaction product to (−)-podophyllotoxin (1b) would be (−)-pluviatolide (14b) rather than (−)-haplomyrfolin (40b). In this respect, the fragmentation pattern of the enzymatic product was consistent to that described in the literature (61, 62) for (−)-pluviatolide (14b) (Fig. 7). The product had a base peak of m/z 355.1183, corresponding to [M − H]− (calculated 355.1182) in the negative ion mode (data not shown). The positive ion mode spectrum was more informative (Fig. 6C), showing a base peak of m/z 339.1230 corresponding to [M + H − H2O]+ (calculated 339.1232) and further peaks of m/z 357.1337 and m/z 379.1155 corresponding to [M + H]+ (calculated 357.1338) and [M + Na]+ (calculated 379.1157), respectively. More importantly, it was possible to observe the predicted fragmentation that unequivocally indicated formation of the methylenedioxy bridge; that is, the characteristic substituted tropylium cation corresponding to the methylenedioxy group with m/z 135.0445 (calculated 135.0446) as well as the hydroxymethoxy-substituted fragment with m/z 137.0604 (calculated 137.0603). Finally, a fragment of m/z 161.0605 was also observed that allowed the distinction between the two potential isobaric products, (−)-pluviatolide (14b) and (−)-haplomyrfolin (40b) (Fig. 7); this corresponded to the allylbenzodioxole fragment (calculated 161.0603) and shows that the methylenedioxy bridge was formed in the expected ring forming (−)-pluviatolide (14b), as the alternative product (−)-haplomyrfolin (40b) would produce a 4-allyl-2-methoxyphenol fragment of m/z 163.0754 (Fig. 7). Therefore, the product obtained had the methylenedioxy bridge introduced into the correct aromatic ring. Analysis of the 1H, 13C, and heteronuclear single quantum correlation NMR spectra of the enzymatically obtained product also clearly supports the formation of the product, in accordance with literature values (63), e.g. with the methylenedioxy bridge characteristic peaks at 5.9 and 101 ppm in 1H and 13C NMR spectra, respectively (supplemental Table S3 and Figs. S1–S3).
FIGURE 6.
Ultra performance liquid chromatography-mass spectrometry analysis of enzymatic assays. A, ultra performance liquid chromatography chromatogram shows product formation in CYP719A24 and CYP719A23 assays in comparison to negative controls (empty vector and cinnamate-4-hydroxylase, CYP73A107). Positive ion mass spectra of substrate (−)-matairesinol (9b, B) and product (−)-pluviatolide (14b, C) show loss of two mass units.
FIGURE 7.
Fragmentation pattern of (−)-pluviatolide (14b) and (−)-haplomyrfolin (40b). The expected fragments generated by the two isobaric compounds 14b (A) and 40b (B) during LC-ESI-MS analyses are shown (adapted from Schmidt et al. (61)). The fragment at m/z 161 and the absence of a fragment at m/z 163 point to (−)-pluviatolide (14b) as the products of CYP719A23 and CYP719A24.
Kinetic Data of Putative Pluviatolide Synthase
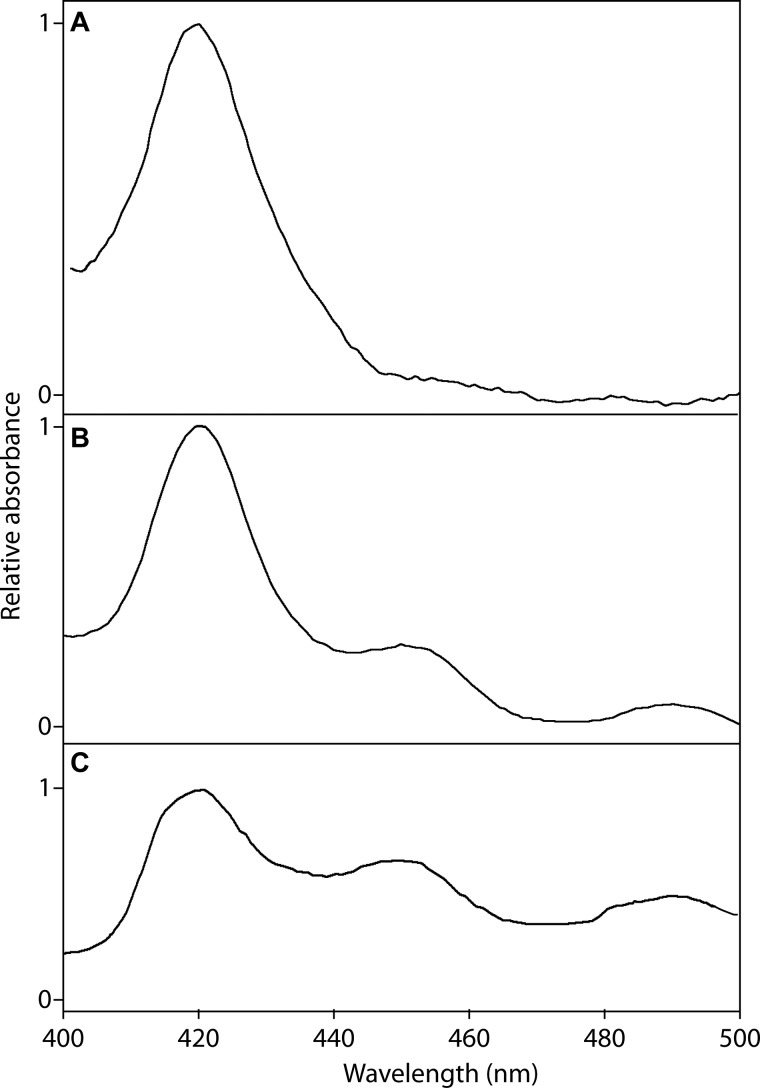
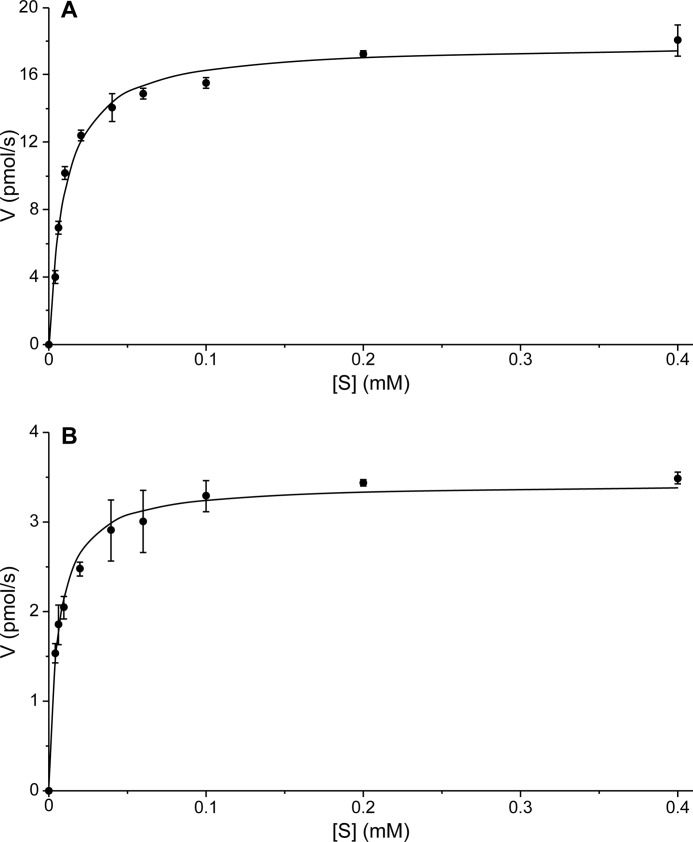
Using (−)-matairesinol (9b) as a substrate, microsomes from S. cerevisiae expressing the P. hexandrum CYP719A23, with a concentration of 72 pmol as determined by analysis of the reduced CO binding spectrum (Fig. 8B), showed a saturation curve consistent with Michaelis-Menten kinetics with a Km of 9.7 ± 2.2 μm (Fig. 9A) and kcat of 14.9 ± 1.0 min−1. In addition, the homolog cloned from P. peltatum, CYP719A24, with a concentration of 26 pmol as determined by reduced CO binding spectrum (Fig. 8C), displayed an apparent Km of 5.8 ± 1.4 μm (Fig. 9 B) and kcat of 7.8 ± 0.4 min−1. This tight binding toward (−)-matairesinol (9b) indicated that these CyP450s from both species can be provisionally presumed to be those involved in the methylenedioxy bridge formation leading to (−)-podophyllotoxin (1b) and pathway related lignans. Still, to determine if (−)-pluviatolide (14b) is the true biosynthetic intermediate will ultimately require in vivo verification, e.g. by down-regulation or knock-out of this gene and identification of corresponding changes in metabolite profile. However, to date there is no system in place to transform Podophyllum species.
FIGURE 8.
Reduced CO binding spectra for (−)-pluviatolide synthases. Spectra were obtained with microsomes isolated from S. cerevisiae (strain WAT11) transformed with pYES-DEST52 (empty vector as negative control) (A), pYES-DEST52::CYP719A23 (B), and pYES-DEST52::CYP719A24 (C).
FIGURE 9.
Kinetic parameters for CYP719A23 and CYP719A24. Steady state Michaelis-Menten kinetics derived from initial rates of CYP719A23 (A) and CYP719A24 (B) enriched microsomes with (−)-matairesinol (9b) as substrate. Assays were performed in triplicate.
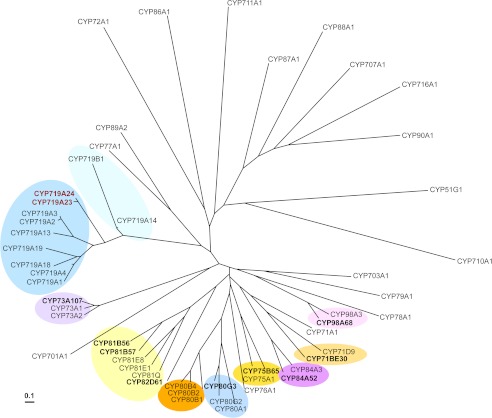
It is very interesting that the candidates with the highest homology to enzymes from alkaloid biosynthesis, the CYP719A1 (S)-canadine synthase (Fig. 10), were found to be capable of performing the same reaction in lignan metabolism, whereas the candidate CYP81B57, closely related to the lignan piperitol/sesamin synthase from the Pedaliaceae, Sesamum indicum, had no activity detected. Podophyllum species, however, belong to the Berberidaceae family and Ranunculales order, the only family with CYP719 genes described thus far (64). The Podophyllum species also form a closely related phylogenetic group (65) with other species known for biosynthesizing similar aryltetralin lignans but with no alkaloids in them reported so far, such as Dysosma (66, 67) and Diphylleia (68) species. This clade forms a monophyletic group with many benzylisoquinoline alkaloid producing species (69), including those from which other CYP719s have been described. It can thus be tentatively postulated that this Podophyllum group has either no or strongly reduced alkaloid level of biosynthesis while “recruiting” some of its genes for the podophyllotoxin pathway.
FIGURE 10.
Phylogenetic analysis of cloned (bold) and known cytochrome P450 enzymes. The phylogenetic tree was generated by sequence alignment using ClustalW (Version 1.4), constructed with neighboring joining clustering algorithm (Version 3.5c), and visualized with Tree view (Version 1.6.6). Amino acid sequences were obtained from UniProtKB, SwissProt, or GenBankTM with the following accession numbers: AB014459, CYP51G1, A. thaliana; AF212990, CYP701A1, Cucurbita maxima; AB006790, CYP703A1, Petunia × hybrida; NM_202845, CYP707A1, A. thaliana; M32885, CYP71A1, Persea americana; O81971, CYP71D9, Glycine max; NM_129002, CYP710A1, A. thaliana; NP_850074, CYP711A1, A. thaliana; NM_123002, CYP716A1, A. thaliana; Q948Y1, CYP719A1, Coptis japonica; EU882969, CYP719A2 and AB126256, CYP719A3, Eschscholzia californica; EU883001, CYP719A4, Thalictrum flavum; EF451151, CYP719A13, Argemone mexicana; EF451152, CYP719A14, A. mexicana; AB374407, CYP719A18, C. japonica; AB374408, CYP719A19, C. japonica; EF451150, CYP719B1, P. somniferum; L10081, CYP72A1, Catharanthus roseus; Z17369, CYP73A1, Helianthus tuberosus; NP_180607, CYP73A2, A. thaliana; Z22545, CYP75A1, Petunia × hybrida; X71658, CYP76A1, Solanum melongena; X71656, CYP77A1, S. melongena; P48420, CYP78A1, Zea mays; U32624, CYP79A1, Sorghum bicolor; U09610, CYP80A1, Berberis stolonifera; AF014801, CYP80B1, E. californica; AB025030, CYP80B2, C. japonica; AY610509, CYP80B4, T. flavum; AB288053, CYP80G2, C. japonica; P93147, CYP81E1, Glycyrrhiza echinata; AY278229, CYP81E8, Medicago truncatula; BAE48234, CYP81Q, Sesamum indicum; NP_195345, CYP84A3, A. thaliana; P48422, CYP86A1, A. thaliana; AF216313, CYP87A1, H. annuus; U32579, CYP88A1, Z. mays; U61231, CYP89A2, A. thaliana; Q42569, CYP90A1, A. thaliana; NP_850337, CYP98A3, A. thaliana. A 10% change is indicated by the scale bar.
Conclusions
In this work the utility of massively parallel sequencing was demonstrated for the study of non-model Podophyllum medicinal plants. The Illumina-based technology produced abundant high quality data, with 100% agreement with sequences obtained from the cloned transcripts obtained using traditional Sanger sequencing. In addition to verifying the validity of known sequences in the lignan pathway leading to (−)-podophyllotoxin (1b), the two CyP450s from P. hexandrum and P. peltatum studied herein are putative pluviatolide synthases, i.e. capable of catalyzing methylenedioxy bridge formation for (−)-podophyllotoxin (1b) biosynthesis. As an extension of this work and as a resource for the scientific community, the transcriptome data and metabolic profiling of the species investigated herein and in several other important medicinal plants are available for the community in the open websites MedPlTranscriptome and Medplants.
Acknowledgments
We thank Prof. David Nelson (University of Tennessee) for assigning standard nomenclature to the CyP450 described herein and Amy Hetrick (Institute of Biological Chemistry) for growing the Podophyllum and Linum plants. We also gratefully acknowledge the assistance of Robin Kramer (National Center for Genomic Resources) in the production of transcript assemblies used in this project.
This work was supported, in whole or in part, by National Institutes of Health Grant 1RC2GM092561(NIGMS). This work was also supported by National Science Foundation Grant MCB-1052557 and by the G. Thomas and Anita Hargrove Center for Plant Genomic Research.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) KC110988–KC110998.

This article contains supplemental Tables S1 and S2 and Figs. S1–S3.
- EST
- expressed sequence tag
- CyP450
- cytochrome P450.
REFERENCES
- 1. Ansorge W. J. (2009) Next-generation DNA sequencing techniques. N. Biotechnol. 25, 195–203 [DOI] [PubMed] [Google Scholar]
- 2. Sun C., Li Y., Wu Q., Luo H., Sun Y., Song J., Lui E. M., Chen S. (2010) De novo sequencing and analysis of the American ginseng root transcriptome using a GS FLX titanium platform to discover putative genes involved in ginsenoside biosynthesis. BMC Genomics 11, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen S., Luo H., Li Y., Sun Y., Wu Q., Niu Y., Song J., Lv A., Zhu Y., Sun C., Steinmetz A., Qian Z. (2011) 454 EST analysis detects genes putatively involved in ginsenoside biosynthesis in Panax ginseng. Plant Cell Rep. 30, 1593–1601 [DOI] [PubMed] [Google Scholar]
- 4. Subramaniyam S., Mathiyalagan R., Jun Gyo I., Bum-Soo L., Sungyoung L., Deok Chun Y. (2011) Transcriptome profiling and in silico analysis of Gynostemma pentaphyllum using a next generation sequencer. Plant Cell Rep. 30, 2075–2083 [DOI] [PubMed] [Google Scholar]
- 5. Hsiao Y.-Y., Chen Y.-W., Huang S.-C., Pan Z.-J., Fu C.-H., Chen W.-H., Tsai W.-C., Chen H.-H. (2011) Gene discovery using next-generation pyrosequencing to develop ESTs for Phalaenopsis orchids. BMC Genomics 12, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Su C.-L., Chao Y.-T., Alex Chang Y.-C., Chen W.-C., Chen C.-Y., Lee A.-Y., Hwa K. T., Shih M.-C. (2011) De novo assembly of expressed transcripts and global analysis of the Phalaenopsis aphrodite transcriptome. Plant Cell Physiol. 52, 1501–1514 [DOI] [PubMed] [Google Scholar]
- 7. Shi C.-Y., Yang H., Wei C.-L., Yu O., Zhang Z.-Z., Jiang C.-J., Sun J., Li Y.-Y., Chen Q., Xia T., Wan X.-C. (2011) Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics 12, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giddings L.-A., Liscombe D. K., Hamilton J. P., Childs K. L., DellaPenna D., Buell C. R., O'Connor S. E. (2011) A stereoselective hydroxylation step of alkaloid biosynthesis by a unique cytochrome P450 in Catharanthus roseus. J. Biol. Chem. 286, 16751–16757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Desgagné-Penix I., Khan M. F., Schriemer D. C., Cram D., Nowak J., Facchini P. J. (2010) Integration of deep transcriptome and proteome analyses reveals the components of alkaloid metabolism in opium poppy cell cultures. BMC Plant Biol. 10, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wong M. M., Cannon C. H., Wickneswari R. (2011) Identification of lignin genes and regulatory sequences involved in secondary cell wall formation in Acacia auriculiformis and Acacia mangium via de novo transcriptome sequencing. BMC Genomics 12, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hiremath P. J., Farmer A., Cannon S. B., Woodward J., Kudapa H., Tuteja R., Kumar A., Bhanuprakash A., Mulaosmanovic B., Gujaria N., Krishnamurthy L., Gaur P. M., Kavikishor P. B., Shah T., Srinivasan R., Lohse M., Xiao Y., Town C. D., Cook D. R., May G. D., Varshney R. K. (2011) Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotechnol. J. 9, 922–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zerbe P., Chiang A., Yuen M., Hamberger B., Hamberger B., Draper J. A., Britton R., Bohlmann J. (2012) Bifunctional cis-abienol synthase from Abies balsamea discovered by transcriptome sequencing and its implications for diterpenoid fragrance production. J. Biol. Chem. 287, 12121–12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Canel C., Moraes R. M., Dayan F. E., Ferreira D. (2000) Podophyllotoxin. Phytochemistry 54, 115–120 [DOI] [PubMed] [Google Scholar]
- 14. Nadeem M., Palni L. M. S., Purohit A. N., Pandey H., Nandi S. K. (2000) Propagation and conservation of Podophyllum hexandrum Royle. An important medicinal herb. Biol. Conserv. 92, 121–129 [Google Scholar]
- 15. Gordaliza M., García P. A., del Corral J. M., Castro M. A., Gómez-Zurita M. A. (2004) Podophyllotoxin. Distribution, sources, applications and new cytotoxic derivatives. Toxicon 44, 441–459 [DOI] [PubMed] [Google Scholar]
- 16. Wu Y., Zhang H., Zhao Y., Zhao J., Chen J., Li L. (2007) A new and efficient strategy for the synthesis of podophyllotoxin and its analogues. Org. Lett. 9, 1199–1202 [DOI] [PubMed] [Google Scholar]
- 17. Reynolds A. J., Scott A. J., Turner C. I., Sherburn M. S. (2003) The intramolecular carboxyarylation approach to podophyllotoxin. J. Am. Chem. Soc. 125, 12108–12109 [DOI] [PubMed] [Google Scholar]
- 18. Xia Z.-Q., Costa M. A., Proctor J., Davin L. B., Lewis N. G. (2000) Dirigent-mediated podophyllotoxin biosynthesis in Linum flavum and Podophyllum peltatum. Phytochemistry 55, 537–549 [DOI] [PubMed] [Google Scholar]
- 19. Davin L. B., Wang H.-B., Crowell A. L., Bedgar D. L., Martin D. M., Sarkanen S., Lewis N. G. (1997) Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 275, 362–366 [DOI] [PubMed] [Google Scholar]
- 20. Davin L. B., Lewis N. G. (2000) Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol. 123, 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Broomhead A. J., Rahman M. M. A., Dewick P. M., Jackson D. E., Lucas J. A. (1991) Matairesinol as precursor of Podophyllum lignans. Phytochemistry 30, 1489–1492 [Google Scholar]
- 22. Xia Z.-Q., Costa M. A., Pelissier H. C., Davin L. B., Lewis N. G. (2001) Secoisolariciresinol dehydrogenase purification, cloning, and functional expression. Implications for human health protection. J. Biol. Chem. 276, 12614–12623 [DOI] [PubMed] [Google Scholar]
- 23. Federolf K., Alfermann A. W., Fuss E. (2007) Aryltetralin-lignan formation in two different cell suspension cultures of Linum album. Deoxypodophyllotoxin 6-hydroxylase, a key enzyme for the formation of 6-methoxypodophyllotoxin. Phytochemistry 68, 1397–1406 [DOI] [PubMed] [Google Scholar]
- 24. Kranz K., Petersen M. (2003) β-Peltatin 6-O-methyltransferase from suspension cultures of Linum nodiflorum. Phytochemistry 64, 453–458 [DOI] [PubMed] [Google Scholar]
- 25. Molog G. A., Empt U., Kuhlmann S., van Uden W., Pras N., Alfermann A. W., Petersen M. (2001) Deoxypodophyllotoxin 6-hydroxylase, a cytochrome P450 monooxygenase from cell cultures of Linum flavum involved in the biosynthesis of cytotoxic lignans. Planta 214, 288–294 [DOI] [PubMed] [Google Scholar]
- 26. van Uden W., Bouma A. S., Bracht Waker J. F., Middel O., Wichers H. J., de Waard P., Woerdenbag H. J., Kellogg R. M., Pras N. (1995) The production of podophyllotoxin and its 5-methoxy derivative through bioconversion of cyclodextrin-complexed desoxypodophyllotoxin by plant cell cultures. Plant Cell Tissue Organ Cult. 42, 73–79 [Google Scholar]
- 27. Seidel V., Windhövel J., Eaton G., Alfermann A. W., Arroo R. R., Medarde M., Petersen M., Woolley J. G. (2002) Biosynthesis of podophyllotoxin in Linum album cell cultures. Planta 215, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 28. Umezawa T., Davin L. B., Lewis N. G. (1991) Formation of lignans (−)-secoisolariciresinol and (−)-matairesinol with Forsythia intermedia cell-free extracts. J. Biol. Chem. 266, 10210–10217 [PubMed] [Google Scholar]
- 29. Ozawa S., Davin L. B., Lewis N. G. (1993) Formation of (−)-arctigenin in Forsythia intermedia. Phytochemistry 32, 643–652 [Google Scholar]
- 30. Kato M. J., Chu A., Davin L. B., Lewis N. G. (1998) Biosynthesis of antioxidant lignans in Sesamum indicum seeds. Phytochemistry 47, 583–591 [Google Scholar]
- 31. Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M., Lightfoot S., Menzel W., Granzow M., Ragg T. (2006) The RIN. An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J., Birol I. (2009) ABySS. A parallel assembler for short read sequence data. Genome Res. 19, 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Birol I., Jackman S. D., Nielsen C. B., Qian J. Q., Varhol R., Stazyk G., Morin R. D., Zhao Y., Hirst M., Schein J. E., Horsman D. E., Connors J. M., Gascoyne R. D., Marra M. A., Jones S. J. (2009) De novo transcriptome assembly with ABySS. Bioinformatics 25, 2872–2877 [DOI] [PubMed] [Google Scholar]
- 34. Li R., Zhu H., Ruan J., Qian W., Fang X., Shi Z., Li Y., Li S., Shan G., Kristiansen K., Li S., Yang H., Wang J., Wang J. (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chevreux B. (2005) MIRA: An Automated Genome and EST Assembler, Ph.D. thesis, German Cancer Research Center, Heidelberg, Germany [Google Scholar]
- 36. Li W., Godzik A. (2006) Cd-hit. A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 [DOI] [PubMed] [Google Scholar]
- 37. Milne I., Bayer M., Cardle L., Shaw P., Stephen G., Wright F., Marshall D. (2010) Tablet. Next generation sequence assembly visualization. Bioinformatics 26, 401–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hall T. A. (1999) BioEdit. A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 [Google Scholar]
- 39. Rodrigue S., Malmstrom R. R., Berlin A. M., Birren B. W., Henn M. R., Chisholm S. W. (2009) Whole genome amplification and de novo assembly of single bacterial cells. PLoS ONE 4, e6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pignatelli M., Moya A. (2011) Evaluating the fidelity of de novo short read metagenomic assembly using simulated data. PLoS ONE 6, e19984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ono E., Nakai M., Fukui Y., Tomimori N., Fukuchi-Mizutani M., Saito M., Satake H., Tanaka T., Katsuta M., Umezawa T., Tanaka Y. (2006) Formation of two methylenedioxy bridges by a Sesamum CYP81Q protein yielding a furofuran lignan, (+)-sesamin. Proc. Natl. Acad. Sci. U.S.A. 103, 10116–10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ikezawa N., Tanaka M., Nagayoshi M., Shinkyo R., Sakaki T., Inouye K., Sato F. (2003) Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J. Biol. Chem. 278, 38557–38565 [DOI] [PubMed] [Google Scholar]
- 43. Pompon D., Louerat B., Bronine A., Urban P. (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 272, 51–64 [DOI] [PubMed] [Google Scholar]
- 44. Kemmer G., Keller S. (2010) Nonlinear least-squares data fitting in Excel spreadsheets. Nat. Protoc. 5, 267–281 [DOI] [PubMed] [Google Scholar]
- 45. Omura T., Sato R. (1964) The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J. Biol. Chem. 239, 2379–2385 [PubMed] [Google Scholar]
- 46. von Wartburg A., Angliker E., Renz J. (1957) Lignanglucoside aus Podophyllum peltatum L. 7. Mitteilung über mitosehemmende Naturstoffe. Helv. Chim. Acta 40, 1331–1357 [Google Scholar]
- 47. Stähelin H. F., von Wartburg A. (1991) The chemical and biological route from podophyllotoxin glucoside to etoposide. Ninth Cain Memorial Award Lecture. Cancer Res. 51, 5–15 [PubMed] [Google Scholar]
- 48. Lim C. K. (1996) Analysis of aryltetrahydronaphthalene lignans and their glucoside conjugates in podophyllin resin by high-performance liquid chromatography. J. Chromatogr. A 722, 267–271 [Google Scholar]
- 49. Bak S., Beisson F., Bishop G., Hamberger B., Höfer R., Paquette S., Werck-Reichhart D. (2011) The Arabidopsis Book, e0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ehlting J., Hamberger B., Million-Rousseau R., Werck-Reichhart D. (2006) Cytochromes P450 in phenolic metabolism. Phytochem. Rev. 5, 239–270 [Google Scholar]
- 51. Nelson D. R. (2006) Plant cytochrome P450s from moss to poplar. Phytochem. Rev. 5, 193–204 [Google Scholar]
- 52. Umezawa T. (2010) The cinnamate/monolignol pathway. Phytochem. Rev. 9, 1–17 [Google Scholar]
- 53. Humphreys J. M., Hemm M. R., Chapple C. (1999) New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 96, 10045–10050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Winkel-Shirley B. (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jiao Y., Davin L. B., Lewis N. G. (1998) Furanofuran lignan metabolism as a function of seed maturation in Sesamum indicum. Methylenedioxy bridge formation. Phytochemistry 49, 387–394 [Google Scholar]
- 56. Clemens S., Barz W. (1996) Cytochrome P450-dependent methylenedioxy bridge formation in Cicer arietinum. Phytochemistry 41, 457–460 [Google Scholar]
- 57. Samanani N., Park S.-U., Facchini P. J. (2005) Cell type-specific localization of transcripts encoding nine consecutive enzymes involved in protoberberine alkaloid biosynthesis. Plant Cell 17, 915–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Díaz Chávez M. L., Rolf M., Gesell A., Kutchan T. M. (2011) Characterization of two methylenedioxy bridge-forming cytochrome P450-dependent enzymes of alkaloid formation in the Mexican prickly poppy Argemone mexicana. Arch. Biochem. Biophys. 507, 186–193 [DOI] [PubMed] [Google Scholar]
- 59. Ikezawa N., Iwasa K., Sato F. (2007) Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J. 274, 1019–1035 [DOI] [PubMed] [Google Scholar]
- 60. Sanger F., Coulson A. R. (1975) A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 94, 441–448 [DOI] [PubMed] [Google Scholar]
- 61. Schmidt T. J., Hemmati S., Fuss E., Alfermann A. W. (2006) A combined HPLC-UV and HPLC-MS method for the identification of lignans and its application to the lignans of Linum usitatissimum L. and L. bienne Mill. Phytochem. Anal. 17, 299–311 [DOI] [PubMed] [Google Scholar]
- 62. Schmidt T. J., Alfermann A. W., Fuss E. (2008) High-performance liquid chromatography/mass spectrometric identification of dibenzylbutyrolactone-type lignans. Insights into electrospray ionization tandem mass spectrometric fragmentation of lign-7-eno-9,9′-lactones and application to the lignans of Linum usitatissimum L. (common flax). Rapid Commun. Mass Spectrom. 22, 3642–3650 [DOI] [PubMed] [Google Scholar]
- 63. Takaku N., Choi D.-H., Mikame K., Okunishi T., Suzuki S., Ohashi H., Umezawa T., Shimada M. (2001) Lignans of Chamaecyparis obtusa. J. Wood Sci. 47, 476–482 [Google Scholar]
- 64. Nelson D., Werck-Reichhart D. (2011) A P450-centric view of plant evolution. Plant J. 66, 194–211 [DOI] [PubMed] [Google Scholar]
- 65. Wang W., Chen Z.-D., Liu Y., Li R.-Q., Li J.-H. (2007) Phylogenetic and biogeographic diversification of Berberidaceae in the northern hemisphere. Syst. Bot. 32, 731–742 [Google Scholar]
- 66. Yu P.-Z., Wang L.-P., Chen Z.-N. (1991) A new podophyllotoxin-type lignan from Dysosma versipellis var. tomentosa. J. Nat. Prod. 54, 1422–1424 [Google Scholar]
- 67. Jiang R.-W., Zhou J.-R., Hon P.-M., Li S.-L., Zhou Y., Li L.-L., Ye W.-C., Xu H.-X., Shaw P.-C., But P. P. (2007) Lignans from Dysosma versipellis with inhibitory effects on prostate cancer cell lines. J. Nat. Prod. 70, 283–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Broomhead A. J., Dewick P. M. (1990) Tumour-inhibitory aryltetralin lignans in Podophyllum versipelle, Diphylleia cymosa, and Diphylleia grayi. Phytochemistry 29, 3831–3837 [Google Scholar]
- 69. Liscombe D. K., Macleod B. P., Loukanina N., Nandi O. I., Facchini P. J. (2005) Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry 66, 1374–1393 [DOI] [PubMed] [Google Scholar]