Background: C1q is expressed in Alzheimer disease brain and blocks fibrillar amyloid-β neurotoxicity in vitro.
Results: C1q promotes neuroprotection by activating the transcription factor CREB and by increasing LRP1B and GPR6 expression.
Conclusion: C1q is up-regulated early in response to injury and induces protective pathways against Aβ toxicity.
Significance: C1q initiates a neuroprotective program that may be a beneficial therapeutic target in neurodegenerative disorders.
Keywords: Alzheimer disease, Amyloid, Complement, Gene Expression, Neurodegeneration, C1q, GPR6, LRP1B, Neuroprotection
Abstract
Complement protein C1q is induced in the brain in response to a variety of neuronal injuries, including Alzheimer disease (AD), and blocks fibrillar amyloid-β (fAβ) neurotoxicity in vitro. Here, we show that C1q protects immature and mature primary neurons against fAβ toxicity, and we report for the first time that C1q prevents toxicity induced by oligomeric forms of amyloid-β (Aβ). Gene expression analysis reveals C1q-activated phosphorylated cAMP-response element-binding protein and AP-1, two transcription factors associated with neuronal survival and neurite outgrowth, and increased LRP1B and G protein-coupled receptor 6(GPR6) expression in fAβ-injured neurons. Silencing of cAMP-response element-binding protein, LRP1B or GPR6 expression inhibited C1q-mediated neuroprotection from fAβ-induced injury. In addition, C1q altered the association of oligomeric Aβ and fAβ with neurons. In vivo, increased hippocampal expression of C1q, LRP1B, and GPR6 is observed as early as 2 months of age in the 3×Tg mouse model of AD, whereas no such induction of LRP1B and GPR6 was seen in C1q-deficient AD mice. In contrast, expression of C1r and C1s, proteases required to activate the classical complement pathway, and C3 showed a significant age-dependent increase only after 10–13 months of age when Aβ plaques start to accumulate in this AD model. Thus, our results identify pathways by which C1q, up-regulated in vivo early in response to injury without the coordinate induction of other complement components, can induce a program of gene expression that promotes neuroprotection and thus may provide protection against Aβ in preclinical stages of AD and other neurodegenerative processes.
Introduction
Alzheimer disease (AD),2 the most common neurodegenerative disease of the elderly, is associated with the loss of cognitive function and the presence of neuropathological changes such as synaptic and neuronal loss, neurofibrillary tangles, and plaques of aggregated amyloid β (Aβ) peptides. Complement protein C1q, the recognition component of the classical complement pathway, binds to β-sheet fibrillar Aβ (fAβ) plaques and, when associated with C1r and C1s as in the C1 complex, activates the complement cascade that can have both a protective effect by enhancing clearance of Aβ through C1q- and C3-dependent opsonization and detrimental inflammatory consequences through production of the chemotactic factor C5a and subsequent recruitment and activation of microglia to the site of injury (1).
C1q is known, however, to be a multifunctional macromolecule with some inherent functions independent of its association with C1r and C1s such as enhancing clearance of apoptotic cells by phagocytic cells (including microglia) and modulating cytokine production to limit inflammation (2–4). In addition, synthesis of the complement protein C1q has been found up-regulated in the brain in response to a variety of neuronal injuries, including models of AD (1, 5–7). The early components of the complement cascade, C1q and C3, are also involved in central nervous system (CNS) development (8), and we recently demonstrated that C1q has direct neuroprotective properties (9, 10) and suppressed fAβ-induced neuronal death in vitro (9). This neuroprotective effect of C1q against fAβ was independent of modulation of the typical apoptotic pathways such as caspase or calpain activation (9) suggesting that C1q may induce nonconventional neuroprotective mechanisms in fAβ-injured neurons.
In this study, we extend our initial observations to show that C1q can protect both immature and mature primary cortical neurons against fAβ and oligomeric Aβ-induced neurotoxicity by modulating neuronal gene expression to induce a neuroprotective response that engages low density lipoprotein receptor-related protein 1B (LRP1B) and G protein-coupled receptor 6 (GPR6). In addition, C1q prevents Aβ association with neurons. Consistent with these observations, in vivo hippocampal expression of C1q, LRP1B, and GPR6 was found to be increased at 2–4 months in the 3×Tg AD mouse but not in hippocampus of C1q-deficient AD mice. These results identify C1q as a component of the response to early injury in AD, before expression of other components of the complement system, and prior to the formation of complement-activating fAβ plaques.
EXPERIMENTAL PROCEDURES
Reagents
Serum-free neurobasal (NB), B27 supplement, and l-glutamine were obtained from Invitrogen. Poly-l-lysine hydrobromide and LRP1B, GPR6, and β-actin antibodies were from Sigma. Microtubule-associated protein (MAP)-2 antibody and anti-mouse lysosome-associated membrane protein 1 (LAMP1) were from Abcam. β-Amyloid(1–16) (6E10) monoclonal antibodies were from Covance, and anti-human C1q were from DAKO. Rabbit antibodies against phosphorylated JNK (pJNK), phosphorylated cAMP-response element-binding protein (pCREB), CREB, and phosphorylated c-Jun (pc-Jun) were from Cell Signaling. Alexa 405-, 488-, or 555-conjugated secondary antibodies were from Invitrogen. Cy3 anti-chicken and HRP-conjugated anti-mouse or rabbit secondary antibodies were from Jackson ImmunoResearch. Human C1q was isolated from plasma as described previously (11) and modified (12). Human Aβ(1–42) (Aβ), provided by Dr. Charles Glabe (University of California at Irvine), was synthesized as described previously (13). Fibrillar and oligomeric forms of Aβ were prepared as described previously (14). fAβ concentration was determined by spectrophotometry, and peptide conformation was analyzed by circular dichroism as described previously (9).
Animals, Neuron Isolation, and Culture
All animal experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee of University of California at Irvine. The 3×TgBUBC1q−/− mice were generated as described previously (15). Briefly, the 3×Tg mice (harboring the Swedish mutation (KM670/671NL), a human four repeat Tau (P301L) mutation and a knock-in mutation of presenilin1 (PS1M146V) (16)) were backcrossed for six generations to the BUB/BnJ strain (The Jackson Laboratory, Bar Harbor, ME) to generate 3×TgBUB mice. The 3×TgBUB mice were then crossed to C1q knock-out mice (C1qa−/−) (17), previously backcrossed to BUB/BnJ for six generations, generating 3×TgBUBC1q−/− mice (validated by PCR and/or qPCR and test breeding). Nontransgenic mice of the same background were used as controls. Mice were anesthetized with a mixture of ketamine/xylazine (67/27 mg/kg) and perfused with PBS. After dissection, hippocampi were immediately frozen on dry ice. Cortical neurons were isolated from day 18 Sprague-Dawley rat embryos (Charles River Laboratories, Inc., Wilmington, MA) or day 16 C57BL/6 mouse embryos as described previously (10). Neurons were grown for 4 days in vitro (immature) or 10 days in vitro (mature) before stimulation with C1q, fAβ, or oligomeric Aβ. In some experiments, neurons were transfected at 3 days in vitro with 10 nm scrambled siRNA (Ambion) or siRNA specific for CREB (Cell Signaling Technology) or GAPDH, GPR6, or LRP1B (Ambion) using the GeneSilencer siRNA transfection reagent (Genlantis, San Diego).
RNA Extraction, Microarray Analysis, and qRT-PCR
Total RNA from cortical neuron cultures or pulverized mouse hippocampi (5 mg) was extracted using the Illustra RNAspin mini isolation kit (GE Healthcare). Gene expression profiles were studied using the Rat Gene 1.0 ST array (Affymetrix). RNA labeling and hybridization were performed by the University of California at Irvine Genomics High Throughput Facility. Data processing and analysis were performed using JMP Genomics 5.0 software (SAS Institute Inc., Cary, NC). Significant differences in gene expression in C1q- and fAβ-treated neurons or fAβ-treated neurons compared with untreated neurons were identified by ANOVA test using the Bonferroni multiple testing method and a false-positive rate (α error) of 0.05. Functional classification of modulated genes was performed using DAVID software (david.abcc.ncifcrf.gov) (18). All data were entered in the Gene Expression Omnibus database (accession numbers GSE18860 and GSE28886). Identification of transcription factor-binding sites was performed using PAINT (19) and MatInspector (20). The cDNA synthesis was performed using the Moloney murine leukemia virus reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Quantitative RT-PCR was performed using the iCycler iQ and the iQ5 software (Bio-Rad) using the maxima SYBR/Green Master Mix (Thermo Fisher Scientific). The rat and mouse primers (supplemental Table S1) were designed using primer-blast (ncbi.nlm.nih.gov) and obtained from Operon (Huntsville, AL). The relative mRNA levels in vivo were determined as follows: mRNA levels = 2−ΔCt, ΔCt = (CtTarget − CtGAPDH). The fold-change (FC) was determined as follow: FC = 2−ΔΔCt, where ΔΔCt = (CtTarget − CtGAPDH)test − (CtTarget − CtGAPDH)untreated. Ct values represent the number of cycles for which the fluorescence signals were detected (21).
Immunocytochemistry
Neurons were fixed with 3.7% paraformaldehyde and permeabilized with 0.1% Triton X-100. Immunocytochemistry was performed according to standard procedures adapted from Glynn and McAllister (22) as described previously (10). Slides were mounted with Prolong gold antifade reagent with or without DAPI (Invitrogen). Cells were examined using the Nikon Eclipse Ti-E confocal microscope and the EZ-C1 and NIS-Element AR 3.00, sp7 software. The MAP-2 area and ratios of pCREB and pc-Jun over DAPI were quantified using ImageJ as described previously (23). The average size of Aβ aggregates was determined using the “analyze particles” function of ImageJ and the scale bar as a size reference (only aggregates >2 μm2 were analyzed). The total neurite length was determined using NeuronJ (24). Co-localization between fAβ and MAP-2 or oligomeric Aβ and LAMP1 was determined using the NIS-Element AR 3.00, sp7 software and Pearson's correlation coefficient.
Western Blot
Neurons were washed with 1 ml of cold Hanks' balanced salt solution and harvested in 200 μl of RIPA buffer containing 10 μm sodium fluoride, 2 μm EDTA, 1 mm PMSF, 200 μm activated sodium vanadate, and 1× protease inhibitor mixture (Roche Applied Science). Neurons were scraped and incubated for 1 h on ice, and the lysate was centrifuged for 15 min at 14,000 rpm at 4 °C. The protein concentration in the soluble fraction was determined by microBCA assay (Pierce) using bovine serum albumin (BSA) as standards. Equal amounts of proteins were separated by 10% SDS-PAGE and then transferred to nitrocellulose membranes (GE Healthcare). The membranes were then incubated in blocking buffer (5% BSA/Tris buffer saline (TBS)/Tween 0.1%) for 1 h at room temperature and incubated overnight at 4 °C with primary antibodies. After three washes, the membranes were incubated with HRP-conjugated anti-rabbit or anti-mouse secondary antibodies for 1 h at room temperature. The proteins were then developed using enhanced chemiluminescence plus (ECL +, GE Healthcare) and analyzed using the Nikon D700 digital SLR camera and the ImageJ software as described previously (25).
Statistical Analysis
Results were compared with two-tailed nonparametric Mann-Whitney U test, one-way ANOVA (Kruskall-Wallis test) or two-way ANOVA, followed by Bonferroni post hoc test, α error = 0.05 for all tests. Differences were considered significant when p was < 0.05.
RESULTS
C1q Protects Immature and Mature Neurons against Fibrillar and Oligomeric Aβ Toxicity
We previously showed that C1q protects immature rat neurons against fAβ-induced toxicity (9) and promotes neurite outgrowth in nutrient-stressed rat and mouse primary cortical neurons (10), suggesting that C1q is a potent neuroprotective factor. These findings are now extended with studies demonstrating that C1q protects against fAβ toxicity in mature (Fig. 1, A and B) as well as immature mouse primary cortical neurons (Fig. 1C). Specifically, although neuronal integrity (measured as total MAP-2 area normalized to percentage of untreated neurons (9)) was significantly (p < 0.05) decreased by fAβ by an average of 50% within 24 h, C1q restored neuronal integrity in fAβ-treated mature (Fig. 1, A and B, 15 μm Aβ and 20 nm C1q, p < 0.05) and immature (Fig. 1C, 5 μm Aβ and 10 nm C1q, p < 0.01) neurons to levels of untreated neurons. Although synthesis of C1q has been demonstrated in vivo in human AD brains (26), in several AD mouse models (15), in hippocampal slices stimulated with Aβ (27, 28), and at specific times during development (6, 8), it is of note that C1q has not been detected intracellularly or in the supernatants of these cultured neurons (data not shown and Ref. 9). If low levels of C1q are produced by these neurons in culture, it seems to have a limited role in this experimental setup because immature neurons isolated from C1q-deficient mice did not show exacerbated loss in neuronal integrity induced by 5 μm fAβ (Fig. 1D, 52 ± 5%, p < 0.01), and C1q (10 nm) restored neuronal integrity in fAβ-injured C1q-deficient neurons to levels of untreated neurons (Fig. 1D, p < 0.01), similarly to what was observed in C1q-sufficient neurons. We report for the first time that C1q also protects neurons against toxicity induced by Aβ oligomers (Fig. 1E), which are considered to be the principal toxic forms of Aβ (29–31). After 24 h of treatment with 1 μm oligomeric Aβ, neurons exhibited a 45% decrease in integrity (Fig. 1E, p < 0.001) that was prevented by addition of 10 nm C1q to levels of integrity of untreated neurons (Fig. 1E, p < 0.01). This neuroprotection was also validated by measuring neurite outgrowth (data not shown). Finally, C1q protected neurons and prevented fAβ toxicity when added up to 16 h after fAβ (Fig. 1F). Altogether, these results suggest that C1q is a potent neuroprotective molecule that can protect neurons against different toxic forms of Aβ.
FIGURE 1.
C1q protects immature and mature neurons against fibrillar and oligomeric Aβ toxicity. A and B, mouse primary mature neurons were treated with 15 μm fAβ and/or 20 nm C1q for 24 h. C–F, immature neurons isolated from wild-type (C, E, and F) or C1q−/− (D) mice were treated with 5 μm fAβ and/or 10 nm C1q (C and D) or 1 μm oligomeric (oligo) Aβ and/or 10 nm C1q (E) for 24 h or 5 μm fAβ for 24 h with 10 nm C1q added for the last 8 h of incubation (F). Neuronal integrity was assessed by MAP-2 immunocytochemistry (A, images representative of three independent experiments) and image analysis (B–F). Scale bar, 100 μm. Results represent mean ± S.D. (n = 3, five fields per condition) and were compared using one-way ANOVA (Kruskall-Wallis test). *, p < 0.05; **, p < 0.01. and ***, p < 0.001.
C1q Modulates Gene Expression and Induces LRP1B and GPR6 in Aβ-injured Neurons
To characterize the molecular basis of C1q neuroprotection against Aβ-induced injury, gene expression in fAβ-injured primary immature rat neurons incubated for 3 h in the presence and absence of C1q was assessed by microarray analysis. C1q modulated genes associated with plasma membrane/cytoskeleton functions, secreted/extracellular functions, cell adhesion, signal transduction, and programmed cell death in fAβ-injured neurons (Fig. 2, A and B). Of the genes modulated (supplemental Table S2,a subset of these genes are shown in Table 1, 3rd column), C1q increased the expression of two membrane-associated receptors, the low density lipoprotein receptor-related protein 1B (LRP1B, Table 1), and G protein-coupled receptor 6 (GPR6, Table 1 and Fig. 2D), while decreasing the expression of the nucleation-promoting factors Wiskott-Aldrich syndrome protein family member 2 (WASF2) and the transcription factor immediate early response 3 (IER3) compared with neurons treated with fAβ alone (Table 1). Importantly, C1q promoted a similar gene expression program in rat and mouse primary neurons as assessed by qRT-PCR of selected genes (Fig. 2C), suggesting that the neuroprotective response induced by C1q is conserved among species.
FIGURE 2.
C1q modulates gene expression profiles of fAβ-injured neurons. A, Pearson correlation coefficient-based heat map (complete linkage method) representation of the log2 fold-change of significantly modulated genes 3 h after treatment of rat primary immature neurons with fAβ (5 μm) and/or C1q (10 nm) over untreated neurons shown with a color gradient from blue (down-regulated) to red (up-regulated). Neurons were from the same pool of primary rat cortical neurons with each condition (untreated, Aβ, Aβ+C1q) performed in separate triplicate wells, and each replicate was run on a separate array. B, gene ontology-based functional classification of genes modulated by C1q in fAβ-treated neurons. C, C1q-dependent modulation of LRP1B, GPR6, WASF2, and IER3 mRNA levels assessed by qRT-PCR in fAβ-injured rat or mouse primary immature neurons (n = 3). Dotted lines represent a 2-fold increase and decrease by C1q over fAβ alone-treated neurons. D, GPR6 and actin protein levels in neurons stimulated with fAβ (5 μm) and/or C1q (10 nm) for 24 h by Western blot. E, heat map of significantly over-represented transcription factor-binding sites in the promoter sequences of genes up-regulated by C1q using PAINT and MatInspector. Each red square indicates the presence of the transcription factor-binding site (for example AP-1) in a gene up-regulated by C1q (gene tree indicated by lines at the bottom).
TABLE 1.
Selection of significantly modulated genes by C1q in fAβ-injured neurons
| Probeset ID | mRNA accession no. | Gene symbol | fAβ + C1qa,b | fAβa,b | fAβ+C1q − fAβc | Fold-difference fAβ+ C1q − fAβd |
|---|---|---|---|---|---|---|
| 10777788 | NM_138533 | Spon2 | 0.689 | −2.007 | 2.696 | 6.480 |
| 10940544 | NM_031806.1 | Gpr6 | −0.280 | −2.969 | 2.689 | 6.449 |
| 10845051 | NM_001107843 | Lrp1b | 1.309 | −0.418 | 1.727 | 3.310 |
| 10889919 | NM_001031823 | Gpr33 | −0.264 | −1.735 | 1.470 | 2.770 |
| 10855862 | NM_181381 | Abcg2 | 1.722 | 0.393 | 1.329 | 2.512 |
| 10858071 | NM_033441 | Rho | 2.103 | 0.782 | 1.321 | 2.498 |
| 10888662 | BC099168 | Wdr43 | −0.381 | −1.683 | 1.302 | 2.466 |
| 10702829 | NM_001113371 | Synj2 | −0.435 | −1.608 | 1.173 | 2.255 |
| 10880331 | NM_001013167 | Wasf2 | −2.134 | 0.012 | −2.146 | 0.226 |
| 10831077 | NM_212505 | Ier3 | −2.818 | −0.586 | −2.233 | 0.213 |
| 10896486 | NM_001009665 | Ebag9 | −0.118 | 2.128 | −2.246 | 0.211 |
| 10793243 | NM_001107331 | Irx1 | −2.080 | 0.641 | −2.721 | 0.152 |
a Differences in mRNA expression (microarray analysis) are from untreated neurons expressed as log2 fold-change.
b Neurons were from the same pool of primary rat cortical neurons with each condition (untreated, Aβ, Aβ+ C1q) performed in separate triplicate wells and each replicate run on a separate array.
c Column 4 (fAβ+ C1q) − column 5 (fAβ).
d Log2 values from column 6 (fAβ+C1q − fAβ) were converted to fold difference ( = 2column 6).
To start to delineate the signaling cascades stimulated by C1q, the transcription factor-binding sites present in the promoter region of the C1q-modulated genes were identified using PAINT and MatInspector. This analysis reveals that 10 transcription factors, including MyoD, v-Maf, c-Myb, Oct-1, and AP-1, can be activated by C1q (Fig. 2E). Among these transcription factors, AP-1 appears to be able to induce a large number of the genes up-regulated by C1q (Fig. 2E), which suggests that AP-1 might be a central transcription factor in C1q-induced neuroprotection.
C1q Activates the Transcription Factor Activator Protein (AP)-1 in a pCREB-dependent JNK-independent Pathway
AP-1 is a heterodimer of the basic leucine zipper proteins Fos and Jun that positively regulates both synaptic strength and synapse number (32). To confirm that C1q activates AP-1 in fAβ-injured neurons, immature mouse neurons were stimulated with fAβ in the presence or absence of C1q for 30 min and 1 and 3 h, and pc-Jun nuclear translocation was assessed by immunostaining and image quantification (Fig. 3, A and B). C1q significantly (p < 0.001) increased pc-Jun nuclear translocation in fAβ-treated neurons after 1 and 3 h of stimulation as compared with fAβ alone (Fig. 3, A and B). AP-1 can be activated in a JNK-dependent pathway or can be induced in a cAMP/pCREB-dependent pathway (32, 33). In fAβ-injured neurons, C1q did not increase phosphorylation of JNK but did phosphorylate CREB (Fig. 3, C–H). Indeed, although no differences in pJNK levels were observed after 15 or 30 min of stimulation with C1q and/or fAβ (Fig. 3, C and D), C1q significantly increased both the phosphorylation of CREB (Fig. 3, G and H, p < 0.05) and its nuclear translocation (Fig. 3, E and F, p < 0.05) after 30 min of stimulation in fAβ-injured neurons. In addition, this activation of pCREB by C1q was sustained over time because the levels of pCREB remained significantly (p < 0.05) higher after 3 h of stimulation as compared with fAβ alone (Fig. 3, G and H). Altogether these results show that AP-1 is activated by C1q probably through a pCREB-dependent JNK-independent pathway in fAβ-injured neurons.
FIGURE 3.
CREB is a central transcription factor in C1q-mediated neuroprotection. A and B, mouse primary immature neurons were stimulated with 5 μm fAβ and/or 10 nm C1q for 30, 60, and 180 min, stained for MAP-2 (red) and pc-Jun (green), mounted in DAPI (blue), and analyzed by confocal microscopy (A) and image quantification (B) to determine pc-Jun nuclear translocation (pc-Jun area over DAPI). Images are representative of three independent experiments. C and D, mouse primary immature neurons were stimulated with 5 μm fAβ and/or 10 nm C1q for 15 and 30 min, and phosphorylation of JNK was determined by Western blot (C) and band intensity analysis (D). Blots are representative of two independent experiments. E–H, mouse primary immature neurons were stimulated with 5 μm fAβ and/or 10 nm C1q for 15, 30, 60, and 180 min. E and F, neurons were stained for MAP-2 (red), pCREB (green), mounted in DAPI (blue), and analyzed by confocal microscopy (E, images representative of three independent experiments) and image quantification (F) to determine pCREB nuclear translocation (pCREB area over DAPI). G and H, phosphorylation of CREB determined by Western blot (G) and band intensity analysis (H). Blots are representative of three independent experiments. I, neurons were transfected with 10 nm scrambled siRNA (scr siRNA) or siRNA targeting CREB 24 h before treatment with 5 μm fAβ ± 10 nm C1q. CREB inhibition was determined by WB 24 h post-transfection (inset). Neuronal integrity was assessed by MAP-2 staining and quantitative image analysis after 24 h of treatment, n = 2 (5 fields per condition). All results represent means ± S.E. and are compared using two-way ANOVA test, *, p < 0.05; **, p < 0.01 and ***, p < 0.001. Scale bar, 10 μm. NT, untransfected.
To determine whether CREB was required for C1q-mediated neuroprotection, CREB expression was silenced using siRNA 24 h before stimulation with fAβ and C1q (Fig. 3I, inset). Transfection with CREB siRNA affected neuronal survival because untreated neurons exhibited a 40% decrease in neuronal integrity compared with untransfected or scrambled (scr) transfected neurons (Fig. 3I), results in agreement with previous studies showing a central role of CREB in neuronal survival (34–37). Nevertheless, whereas C1q protected neurons against fAβ toxicity in untransfected or neurons transfected with scr siRNA, CREB silencing abolished the protective effect of C1q resulting in significant (p < 0.05) loss in neuronal integrity (Fig. 3I) to levels similar to fAβ-treated neurons in the absence of C1q. These results demonstrate that CREB is a central transcription factor activated by C1q and required for neuroprotection.
LRP1B and GPR6 Are Central Mediators of C1q-induced Neuroprotection against Aβ
LRP1B and GPR6 expression were repressed by fAβ, and C1q restored their expression in fAβ-treated neurons (Table 1). These membrane receptors may represent central effectors in the C1q-induced neuroprotective pathway because GPR6 has been shown to increase intracellular cAMP production and to promote neurite extension (38), whereas LRP1B has been shown to modulate Aβ production and uptake (39, 40). To determine whether the C1q-induced expression of LRP1B and GPR6 functionally contributes to the neuroprotective response against fAβ-induced injury, their expression was silenced in mouse primary immature neurons by siRNA 24 h before stimulation with fAβ and C1q (Fig. 4). LRP1B expression was transiently reduced to less than 40% of base line by 24 h but returned to 80% expression by 48 h (Fig. 4A), whereas GPR6 expression was reduced by ∼50% 24–48 h post-transfection (Fig. 4A). Knockdown efficiencies were determined by qRT-PCR in accordance with other studies using similar transfection methods in primary neurons (41, 42). It is of note that transfection with scr siRNA or siRNA specific for LRP1B or GPR6 did not affect neuronal integrity in the absence of any stimulation (Fig. 4B, untreated) and did not increase susceptibility to Aβ toxicity after 24 h of treatment with fAβ (Fig. 4B). Although C1q protected neurons against fAβ toxicity in neurons transfected with scr siRNA, the inhibition of LRP1B and GPR6 expression abolished the protective effect of C1q resulting in significant (p < 0.001) loss in neuronal integrity (Fig. 4B) to levels similar to fAβ-treated neurons in the absence of C1q. In addition, the induction of GPR6 seems to be dependent on the expression of LRP1B because silencing LRP1B also prevents GPR6 induction by C1q in fAβ-treated neurons (Fig. 4C). These results suggest that the neuroprotective response induced by C1q requires at least LRP1B and GPR6 and that LRP1B acts upstream of GPR6.
FIGURE 4.

LRP1B and GPR6 are central effectors of C1q-induced neuroprotection in fAβ-injured neurons. Mouse primary immature neurons were transfected with 10 nm scr siRNA or siRNA targeting LRP1B, GPR6, or GAPDH 24 h before treatment with 5 μm fAβ ± 10 nm C1q. A, LRP1B, GAPDH, and GPR6 mRNA levels in neurons transfected with LRP1B, GPR6, or GAPDH siRNA normalized to levels in neurons transfected with scr siRNA were assessed by qRT-PCR at 24 and 48 h post-transfection. B, neuronal integrity was assessed by MAP-2 staining and quantitative image analysis after 24 h of treatment, n = 3 (five fields per condition). Results represent means ± S.E. and are compared using two-way ANOVA test; *, p < 0.05; **, p < 0.01, and ***, p < 0.001. C, GPR6 and actin protein levels identified by Western blot in neurons transfected with scr, LRP1B, or GPR6 siRNA for 24 h and then stimulated with 5 μm fAβ and/or 10 nm C1q for 24 h. Blots are representative of two independent experiments. UT, untreated.
C1q-dependent Expression of LRP1B and GPR6 in AD Mice
To determine whether C1q modulates the expression of LRP1B and GPR6 in vivo, we analyzed the hippocampal expression of LRP1B and GPR6 in the 3×TgBUB transgenic mouse model of AD with and without a genetic deficiency of C1q (Fig. 5, solid lines and symbols). Hippocampal LRP1B (Fig. 5A) and GPR6 (Fig. 5B) mRNA levels were significantly reduced in C1q-deficient mice compared with C1q-sufficient mice at 2 and 4 months of age, and after the fAβ plaques start to accumulate in the brain (between 10 and 13 months of age in this cohort of animals (15)), these levels were decreased, and no significant differences were observed between C1q-sufficient and -deficient mice (Fig. 5, A and B). Interestingly, C1q mRNA levels were significantly (p < 0.05) increased as early as 2 months of age in the 3×TgBUB mice compared with the nontransgenic BUB C1q-sufficient mouse (Fig. 5C, no detection of C1q mRNA was observed in any C1q-deficient mouse as expected). In contrast, C1r (Fig. 5D), C1s (Fig. 5E), and C3 (Fig. 5F) showed a significant age-dependent increase only after 10–13 months of age, i.e. when the fAβ plaques started to form (15), compared with the nontransgenic BUB C1q-sufficient and -deficient mice. The absolute levels of mRNA expression of C1q, C1r, and C1s differ by 3 orders of magnitude (C1q > C1r > C1s), although the physiological significance of these differences has to be determined. Nevertheless, these results suggest that C1q increased very early in the progression to AD in response to primary neuronal injury, perhaps due to low but chronic oligomeric Aβ production that occurs very early on before neuronal loss or in the presence of transgenic APP and/or mutated Tau or PS1. This regulated C1q expression then contributes to the regulation of LRP1B and GPR6 in the injured brain.
FIGURE 5.
Increased hippocampal expression of C1q, LRP1B, and GPR6 at early ages in AD mice. LRP1B (A), GPR6 (B), C1q (C), C1r (D), C1s (E), and C3 (F) mRNA levels were assessed by qRT-PCR in the hippocampus of 3×TgBUB (● and ■, solid lines) or nontransgenic BUB (○ and □, dotted lines) mice sufficient (● and ○) or deficient (■ and□) for C1q at 2 (n = 4–5), 4 (n = 8), 10 (n = 6), 13 (n = 6), and 18 (n = 6) months of age (performed in duplicate). Results represent means ± S.E. of mRNA levels relative to GAPDH (2−ΔCt, ΔCt = (CtTarget − CtGAPDH)) and are compared using two-way ANOVA test. *, p < 0.05; **, p < 0.01, and ***, p < 0.001.
In addition, levels of LRP1B and GPR6 expression decreased concomitantly with the increased expression of C1r, C1s, and C3, even though C1q expression was further up-regulated (Fig. 5). This demonstrates a consequential switch in the functional activities of C1q due to the induced presence of, and C1q association with, C1r and C1s, resulting in suppression of protective functions and induction of potentially detrimental consequences due to the activation of the complement cascade via interaction of the fAβ plaques with the induced intact C1, the classical complement pathway initiating complex.
C1q Alters Aβ Association with Neurons through Enhanced Aβ Aggregation
In addition to directly interacting with neurons to stimulate a neuroprotective program as described above, C1q has been shown to bind Aβ and enhance Aβ aggregation (43). To assess whether C1q may also protect neurons by affecting the association of Aβ with neurons, primary mouse immature neurons were incubated with fAβ, and association of Aβ was assessed by evaluating the co-localization between fAβ and MAP-2 (Fig. 6, A and B). In the absence of C1q, fAβ closely associated with neurons (Fig. 6, A and B), probably through binding to neuronal surfaces. In the presence of C1q, the amount of fAβ associated with neurons was significantly (p = 0.0004) decreased over a 24-h time period compared with fAβ alone (Fig. 6, A and B).
FIGURE 6.
C1q alters fAβ association with neurons in an LRP1B-dependent manner. Mouse primary immature neurons were treated with 5 μm fAβ and/or 10 nm C1q for 24 h (A and B) or transfected with 10 nm scr siRNA or siRNA targeting LRP1B or GPR6 24 h before treatment with 5 μm fAβ ± 10 nm C1q (C), stained for MAP-2 (red), C1q (blue), and Aβ (green), and analyzed by confocal microscopy. A, images representative of three different experiments. Scale bar, 10 μm. B and C, scatter dot plot of Pearson's correlation coefficient for co-localization between Aβ and MAP-2 (n = 3, 5 fields per condition); red line, mean values. Results were compared using two-tailed non parametric Mann-Whitney U test (B) and one-way ANOVA (Kruskall-Wallis test) (C).
Members of the LDL receptor family such as LRP1B have been shown to modulate Aβ production and uptake (39, 40). Because C1q modulates association of Aβ with neurons and enhances the expression of LRP1B and GPR6, we next investigated whether LRP1B and/or GPR6 plays a role in the C1q-dependent modulation of fAβ association with neurons. Aβ association with neurons after siRNA knockdown of LRP1B or GPR6 in neurons was determined as described above. Although neurons transfected with scr siRNA or GPR6 siRNA showed less fAβ association with neurons when incubated with C1q over a 24-h time period (Fig. 6C, p < 0.05) similar to that observed for untransfected fAβ + C1q-treated neurons (Fig. 6B), addition of LRP1B siRNA prevented the effect of C1q. In fact, after inhibition of LRP1B expression, the association of fAβ with neurons, determined using the Pearson's correlation coefficient for co-localization between MAP-2 and Aβ, in the presence of C1q was similar to fAβ-treated neurons in the absence of C1q (Fig. 6C).
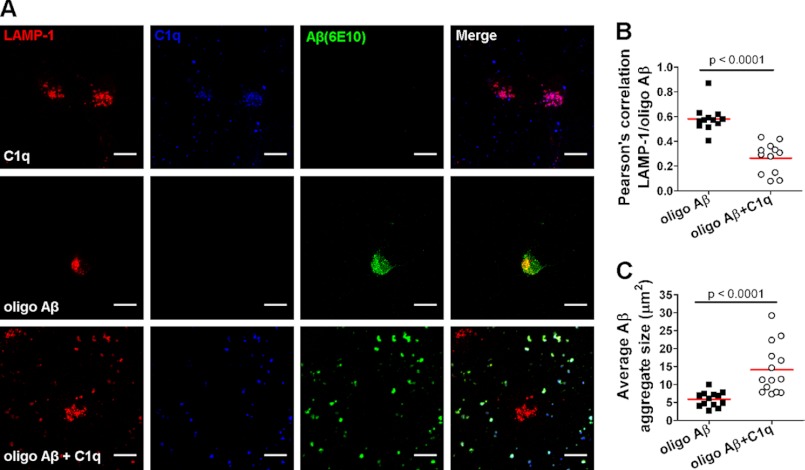
To then evaluate the effect of C1q on the neuronal uptake of other Aβ assembly states, primary mouse immature neurons were incubated with oligomeric Aβ, and uptake of Aβ was assessed by evaluating its co-localization with the lysosomal marker LAMP1 (Fig. 7, A and B). The amount of Aβ that co-localizes with intraneuronal LAMP1-positive vacuoles was significantly (p < 0.0001) decreased by C1q when compared with oligomeric Aβ alone (Fig. 7, A and B), suggesting that C1q may prevent accumulation of Aβ in neurons. In agreement with the known capacity of C1q to bind Aβ (43), C1q strongly co-localized with Aβ aggregates (Fig. 7A) and enhanced Aβ aggregation (Fig. 7C). Indeed, the average size of Aβ aggregates increased from 5.9 ± 2.0 μm2 in absence of C1q to 14.1 ± 6.9 μm2 in presence of C1q (Fig. 7C). Previous studies have shown that extracellular sequestration of Aβ oligomers, which are considered to be the principal toxic forms of Aβ (29–31), into larger aggregates decreases neurotoxicity (44, 45). Altogether, these data suggest that C1q prevents Aβ neurotoxicity by modulating neuronal gene expression to induce a neuroprotective response that requires LRP1B and GPR6 and that LRP1B, but not GPR6, may also prevent Aβ association with neurons (Fig. 8).
FIGURE 7.
C1q decreases internalization of Aβ oligomers through enhanced Aβ aggregation. Mouse primary immature neurons were treated with 1 μm oligomeric Aβ and/or 10 nm C1q for 8 h, stained for LAMP1 (red), C1q (blue), and Aβ (green), and analyzed by confocal microscopy. A, images representative of three different experiments. Scale bar, 10 μm. B and C, scatter dot plot of Pearson's correlation coefficient for co-localization between LAMP1 and Aβ (B) and average Aβ aggregate size in μm2 (C) (n = 3, 5 fields per condition); red line, mean values. Results were compared using two-tailed nonparametric Mann-Whitney U test.
FIGURE 8.

Model of C1q-induced neuroprotective pathways against Aβ. C1q induces CREB phosphorylation and AP-1 activation followed by up-regulation of the expression of LRP1B, which is involved in C1q-dependent GPR6 expression and C1q-mediated neuroprotection. LRP1B can also bind Aβ and reduces its internalization. In addition, C1q directly binds Aβ and increases its aggregation that sequesters Aβ away from the neurons and thus limits its toxic association with neurons.
DISCUSSION
Our study describes a distinct induction of C1q expression in the brains of AD mice at very early ages, identifies previously undescribed molecular mechanisms induced by C1q in neurons that protect against both oligomeric and fAβ neurotoxicity in vitro, and suggests that C1q and its downstream effectors LRP1B and GPR6, which appear to be induced in a C1q-dependent fashion in vivo, are part of a neuroprotective response that is triggered early on in the progression to AD before the accumulation of fAβ plaques and subsequent complement activation and ensuing inflammation.
While studying the molecular basis for the C1q-induced neuroprotection, we found that C1q profoundly affected the transcriptional response of fAβ-injured neurons. Recently, we published that C1q triggers a complex program of gene expression in nutrient-stressed primary neurons that enhance neurite outgrowth and limit neuronal stress and inflammation (10). Interestingly, the transcriptional programs stimulated by C1q in fAβ-injured neurons, while sharing some pathways with nutrient-stressed neurons (such as pathways associated with plasma membrane function and signal transduction), also show differences, suggesting that the environment and the stress stimuli influence the cellular state and thus the specific pathways induced by C1q that coalesce to a neuroprotective response. It is of note that the signaling cascade seems to involve common effectors. Here, the addition of C1q to fAβ-injured neurons resulted in increased phosphorylation and nuclear translocation of CREB, similar to what was observed in nutrient-stressed neurons (10) and in monocytes (46). CREB is a transcription factor vital for long term memory and synaptic plasticity (34), neurogenesis (47–49), and induction of neurotrophic factors in the CNS (35). In addition, pCREB is a central transcription factor in brain-derived neurotrophic factor signaling (50) and also induces brain-derived neurotrophic factor upon activation (51). These results suggest that pCREB is a central transcription factor activated very early on by C1q in neurons and that C1q may induce similar signaling cascades as brain-derived neurotrophic factor.
C1q increased the expression of GPR6 in fAβ-injured neurons, and inhibition of GPR6 prevented the C1q-neuroprotective effect against Aβ. GPR6 is a constitutively active G protein-coupled receptor that is highly expressed in the CNS. In primary rat cerebellar neurons, overexpression of GPR6 increases intracellular cAMP production and promotes neurite extension (38, 52). The induced expression of GPR6 by C1q may thus stimulate a positive feedback to sustain pCREB activation through increased cAMP levels and subsequent neuronal survival.
In addition to inducing a specific transcriptional profile in neurons that promotes survival, C1q altered Aβ association with neurons and enhanced Aβ aggregation, especially that of oligomeric Aβ forms. These results are in accordance with previous data showing that C1q strongly binds Aβ and induces its aggregation (43, 53). These data also suggest that neuronal production of molecules capable of modulating Aβ aggregation (and perhaps aggregation of other misfolded protein) is part of the early response to neuronal injury. Indeed, it has been reported that Aβ increases neuronal collagen VI expression that blocks the association of Aβ oligomers with neurons, enhances Aβ aggregation, and prevents neurotoxicity (44). It is of note that C1q has a collagen-like domain, and Aβ also induces C1q expression in neurons (28). In Aβ-injured neurons and AD mice, the presence of C1q increased the expression of LRP1B. Members of the LDL receptor family have been shown to modulate Aβ production and uptake (39, 40). Specifically, LRP1 binds to APP and Aβ and enhances neuronal internalization and delivery to the lysosomal degradation pathway (54). However, continuous Aβ uptake through LRP1 may overload the degradation pathway leading to Aβ intraneuronal accumulation (54). Moreover, the rapid APP endocytosis through LRP1 may favor the amyloidogenic pathway leading to enhanced Aβ production (55, 56). LRP1B is highly expressed in the brain, including the cortex, hippocampus (dentate gyrus), and cerebellum (57–59). LRP1B interacts with APP and reduces the processing of Aβ (57) probably through a reduced internalization of APP by neurons due to the slower endocytosis rate of LRP1B than LRP1 (60). The increase of LRP1B by C1q may result in a decrease in intraneuronal accumulation of Aβ through slower endocytosis of extracellular Aβ, greater degradation through the lysosomal pathway over time, and/or slower endocytosis of APP that would limit the amyloidogenic pathway. In addition, LRP1B appears to be a prerequisite for GPR6 expression (Fig. 8), suggesting that LRP1B might indirectly promote neurite outgrowth through induction of GPR6 and cAMP (38). It is also of note that haplotypes in the LRP1B gene are significant/protective for successful aging without cognitive decline (61) suggesting that the C1q/LRP1B pathway might be a promising therapeutic target to slow down neurodegeneration.
In addition to demonstrating the early induction of C1q in the brain of 3×Tg AD mice months prior to the detection of plaque pathology, these data demonstrate that the expression of components of the complement C1 complex can be discordantly regulated, with C1q synthesized in the absence of the C1 proteases, C1r and C1s, which are induced only after the Aβ plaques start to accumulate in the brain (Fig. 5). In the Tg2576 AD mouse model, the absence of C1q resulted in decreased inflammation and neuronal loss after accumulation of fAβ plaques, probably through impaired classical complement activation due to the lack of C1q in the later stages of the disease when complement activation can contribute to detrimental enhancement of inflammation (62). One possible explanation for the apparent contrasting dichotomy of effects of C1q in AD is that once C1r and C1s are produced, most of the C1q is now in the C1 complex and can no longer interact with neurons as “free” C1q to promote neuroprotection (or at least the neuroprotective effect is likely counteracted by the inflammatory components of the cascade). In line with this hypothesis, the neuroprotective induction of LRP1B and GPR6 decreased with age concomitantly with the increased expression of the downstream components (C1r, C1s, and C3) of the complement cascade, which can lead to a proinflammatory environment with the production of C5a.
Although beneficial induction of C1q in the brain is observed during development, where in conjunction with other early complement cascade proteins it participates in the pruning of inappropriate synapses (8), the C1q effects described here are independent of the other complement components and thus identify another function of this innate recognition protein. The induction of C1q expression in several brain injury models, such as kainic acid treatment, virus infection, or ischemia/reperfusion (1), is consistent with a role for C1q and its downstream effectors in the neuronal response to injury. This potential protective effect is particularly intriguing in the developing mouse in which C1q synthesis is induced when the cochlea removal is performed after P14 but not before P11 and correlates with neuronal survival (6). In the context of AD, our results further suggest that C1q, which is detected in the hippocampus of 3×Tg mice as early as 2 months of age here and has been detected in the CNS at 3 months of age in another AD transgenic mouse (also prior to induction of other complement proteins) (63), regulates the expression of LRP1B and GPR6 early in the progression to AD.
The findings presented in this study on the neuroprotection induced by C1q, the recent report of the activation of the inflammasome by aggregated C1q in drusen (64), and our finding that soluble unaggregated C1q suppresses inflammasome activation and production of mature IL-1β in macrophages (3) suggest that this protein may have opposing functions depending on its presentation to or within the cell and the environment of the cell. Thus, C1q may be a biosensor of inflammation and injury, resulting in neuroprotection or neurodegeneration depending on the conditions in the local environment.
In summary, the selective up-regulation of C1q in the injured brain and the identification of C1q-induced genes critical for the observed neuroprotection in Aβ-injured neurons described here (Fig. 8) suggest that induction of C1q as a response to injury can initiate a potent neuroprotective program that is likely activated in a broad range of different injury models. The ability to therapeutically engage such neuroprotective pathways may be particularly beneficial in AD and potentially other neurodegenerative disorders or ischemic injury.
Acknowledgments
We thank Drs. R. Ager and M. Fonseca for generous excellent assistance on neuron culture preparation and immunostaining; Sophie Chu and Lindsey Weiner for excellent technical support; Dr. Frank LaFerla (University of California, Irvine) for the 3×Tg mice; Dr. Michael Buchmeier (University of California, Irvine) for use of confocal microscope, and Drs. Suhail Rasool and Charles Glabe (University of California, Irvine) for Aβ peptides. RNA labeling and hybridization were performed by the University of California at Irvine Genomics High Throughput Facility.
This work was supported, in whole or in part, by National Institutes of Health Grants AI 41090, NS35144, and AG 00538.

This article contains supplemental Tables S1 and S2.
- AD
- Alzheimer disease
- fAβ
- fibrillar amyloid-β
- pCREB
- phosphorylated cAMP-response element-binding protein
- AP-1
- activator protein-1
- LRP1B
- low density lipoprotein receptor-related protein 1B
- GPR6
- G protein-coupled receptor 6
- MAP-2
- microtubule-associated protein-2
- LAMP1
- lysosome-associated membrane protein 1
- CREB
- cAMP-response element-binding protein
- qRT
- quantitative RT
- ANOVA
- analysis of variance
- Aβ
- amyloid-β
- APP
- amyloid precursor protein
- pc-Jun
- phosphorylated c-Jun
- scr
- scrambled.
REFERENCES
- 1. Alexander J. J., Anderson A. J., Barnum S. R., Stevens B., Tenner A. J. (2008) The complement cascade. Yin-Yang in neuroinflammation–neuro-protection and -degeneration. J. Neurochem. 107, 1169–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bohlson S. S., Fraser D. A., Tenner A. J. (2007) Complement proteins C1q and MBL are pattern recognition molecules that signal immediate and long-term protective immune functions. Mol. Immunol. 44, 33–43 [DOI] [PubMed] [Google Scholar]
- 3. Benoit M. E., Clarke E. V., Morgado P., Fraser D. A., Tenner A. J. (2012) Complement protein C1q directs macrophage polarization and limits inflammasome activity during the uptake of apoptotic cells. J. Immunol. 188, 5682–5693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fraser D. A., Pisalyaput K., Tenner A. J. (2010) C1q enhances microglial clearance of apoptotic neurons and neuronal blebs, and modulates subsequent inflammatory cytokine production. J. Neurochem. 112, 733–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Melchior B., Garcia A. E., Hsiung B. K., Lo K. M., Doose J. M., Thrash J. C., Stalder A. K., Staufenbiel M., Neumann H., Carson M. J. (2010) Dual induction of TREM2 and tolerance-related transcript, Tmem176b, in amyloid transgenic mice. Implications for vaccine-based therapies for Alzheimer's disease. ASN Neuro. 2, e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harris J. A., Iguchi F., Seidl A. H., Lurie D. I., Rubel E. W. (2008) Afferent deprivation elicits a transcriptional response associated with neuronal survival after a critical period in the mouse cochlear nucleus. J. Neurosci. 28, 10990–11002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Howell G. R., Macalinao D. G., Sousa G. L., Walden M., Soto I., Kneeland S. C., Barbay J. M., King B. L., Marchant J. K., Hibbs M., Stevens B., Barres B. A., Clark A. F., Libby R. T., John S. W. (2011) Molecular clustering identifies complement and endothelin induction as early events in a mouse model of glaucoma. J. Clin. Invest. 121, 1429–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stevens B., Allen N. J., Vazquez L. E., Howell G. R., Christopherson K. S., Nouri N., Micheva K. D., Mehalow A. K., Huberman A. D., Stafford B., Sher A., Litke A. M., Lambris J. D., Smith S. J., John S. W., Barres B. A. (2007) The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178 [DOI] [PubMed] [Google Scholar]
- 9. Pisalyaput K., Tenner A. J. (2008) Complement component C1q inhibits β-amyloid- and serum amyloid P-induced neurotoxicity via caspase- and calpain-independent mechanisms. J. Neurochem. 104, 696–707 [DOI] [PubMed] [Google Scholar]
- 10. Benoit M. E., Tenner A. J. (2011) Complement protein C1q-mediated neuroprotection is correlated with regulation of neuronal gene and microRNA expression. J. Neurosci. 31, 3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tenner A. J., Lesavre P. H., Cooper N. R. (1981) Purification and radiolabeling of human C1q. J. Immunol. 127, 648–653 [PubMed] [Google Scholar]
- 12. Young K. R., Jr., Ambrus J. L., Jr., Malbran A., Fauci A. S., Tenner A. J. (1991) Complement subcomponent C1q stimulates Ig production by human B lymphocytes. J. Immunol. 146, 3356–3364 [PubMed] [Google Scholar]
- 13. Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. (2003) Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 14. Kayed R., Head E., Sarsoza F., Saing T., Cotman C. W., Necula M., Margol L., Wu J., Breydo L., Thompson J. L., Rasool S., Gurlo T., Butler P., Glabe C. G. (2007) Fibril-specific, conformation-dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fonseca M. I., Chu S. H., Berci A. M., Benoit M. E., Peters D. G., Kimura Y., Tenner A. J. (2011) Contribution of complement activation pathways to neuropathology differs among mouse models of Alzheimer's disease. J. Neuroinflammation 8, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oddo S., Caccamo A., Shepherd J. D., Murphy M. P., Golde T. E., Kayed R., Metherate R., Mattson M. P., Akbari Y., LaFerla F. M. (2003) Triple-transgenic model of Alzheimer's disease with plaques and tangles. Intracellular Aβ and synaptic dysfunction. Neuron 39, 409–421 [DOI] [PubMed] [Google Scholar]
- 17. Botto M., Dell'Agnola C., Bygrave A. E., Thompson E. M., Cook H. T., Petry F., Loos M., Pandolfi P. P., Walport M. J. (1998) Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat. Genet. 19, 56–59 [DOI] [PubMed] [Google Scholar]
- 18. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 19. Vadigepalli R., Chakravarthula P., Zak D. E., Schwaber J. S., Gonye G. E. (2003) PAINT. A promoter analysis and interaction network generation tool for gene regulatory network identification. OMICS 7, 235–252 [DOI] [PubMed] [Google Scholar]
- 20. Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005) MatInspector and beyond: promoter analysis based on transcription factor-binding sites. Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 21. Schmittgen T. D., Livak K. J. (2008) Analyzing real time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 22. Glynn M. W., McAllister A. K. (2006) Immunocytochemistry and quantification of protein colocalization in cultured neurons. Nat. Protoc. 1, 1287–1296 [DOI] [PubMed] [Google Scholar]
- 23. Noursadeghi M., Tsang J., Haustein T., Miller R. F., Chain B. M., Katz D. R. (2008) Quantitative imaging assay for NF-κB nuclear translocation in primary human macrophages. J. Immunol. Methods 329, 194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meijering E., Jacob M., Sarria J. C., Steiner P., Hirling H., Unser M. (2004) Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58, 167–176 [DOI] [PubMed] [Google Scholar]
- 25. Khoury M. K., Parker I., Aswad D. W. (2010) Acquisition of chemiluminescent signals from immunoblots with a digital single-lens reflex camera. Anal. Biochem. 397, 129–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fonseca M. I., Kawas C. H., Troncoso J. C., Tenner A. J. (2004) Neuronal localization of C1q in preclinical Alzheimer's disease. Neurobiol. Dis. 15, 40–46 [DOI] [PubMed] [Google Scholar]
- 27. Fan R., Tenner A. J. (2004) Complement C1q expression induced by Aβ in rat hippocampal organotypic slice cultures. Exp. Neurol. 185, 241–253 [DOI] [PubMed] [Google Scholar]
- 28. Fan R., Tenner A. J. (2005) Differential regulation of Aβ42-induced neuronal C1q synthesis and microglial activation. J. Neuroinflammation 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Glabe C. G. (2006) Common mechanisms of amyloid oligomer pathogenesis in degenerative disease. Neurobiol. Aging 27, 570–575 [DOI] [PubMed] [Google Scholar]
- 30. Stroud J. C., Liu C., Teng P. K., Eisenberg D. (2012) Toxic fibrillar oligomers of amyloid-β have cross-β structure. Proc. Natl. Acad. Sci. U.S.A. 109, 7717–7722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glabe C. G., Kayed R. (2006) Common structure and toxic function of amyloid oligomers implies a common mechanism of pathogenesis. Neurology 66, S74–S78 [DOI] [PubMed] [Google Scholar]
- 32. Sanyal S., Sandstrom D. J., Hoeffer C. A., Ramaswami M. (2002) AP-1 functions upstream of CREB to control synaptic plasticity in Drosophila. Nature 416, 870–874 [DOI] [PubMed] [Google Scholar]
- 33. Bowden G. T. (2004) Prevention of non-melanoma skin cancer by targeting ultraviolet-B-light signaling. Nat. Rev. Cancer 4, 23–35 [DOI] [PubMed] [Google Scholar]
- 34. Josselyn S. A., Nguyen P. V. (2005) CREB, synapses, and memory disorders. Past progress and future challenges. Curr. Drug Targets CNS Neurol. Disord. 4, 481–497 [DOI] [PubMed] [Google Scholar]
- 35. McCauslin C. S., Heath V., Colangelo A. M., Malik R., Lee S., Mallei A., Mocchetti I., Johnson P. F. (2006) CAAT/enhancer-binding protein δ and cAMP-response element-binding protein mediate inducible expression of the nerve growth factor gene in the central nervous system. J. Biol. Chem. 281, 17681–17688 [DOI] [PubMed] [Google Scholar]
- 36. Lee B., Cao R., Choi Y. S., Cho H. Y., Rhee A. D., Hah C. K., Hoyt K. R., Obrietan K. (2009) The CREB/CRE transcriptional pathway. Protection against oxidative stress-mediated neuronal cell death. J. Neurochem. 108, 1251–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Caccamo A., Maldonado M. A., Bokov A. F., Majumder S., Oddo S. (2010) CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 107, 22687–22692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tanaka S., Ishii K., Kasai K., Yoon S. O., Saeki Y. (2007) Neural expression of G protein-coupled receptors GPR3, GPR6, and GPR12 up-regulates cyclic AMP levels and promotes neurite outgrowth. J. Biol. Chem. 282, 10506–10515 [DOI] [PubMed] [Google Scholar]
- 39. Jaeger S., Pietrzik C. U. (2008) Functional role of lipoprotein receptors in Alzheimer's disease. Curr. Alzheimer Res. 5, 15–25 [DOI] [PubMed] [Google Scholar]
- 40. Kim J., Castellano J. M., Jiang H., Basak J. M., Parsadanian M., Pham V., Mason S. M., Paul S. M., Holtzman D. M. (2009) Overexpression of low density lipoprotein receptor in the brain markedly inhibits amyloid deposition and increases extracellular Aβ clearance. Neuron 64, 632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cui H., Hayashi A., Sun H. S., Belmares M. P., Cobey C., Phan T., Schweizer J., Salter M. W., Wang Y. T., Tasker R. A., Garman D., Rabinowitz J., Lu P. S., Tymianski M. (2007) PDZ protein interactions underlying NMDA receptor-mediated excitotoxicity and neuroprotection by PSD-95 inhibitors. J. Neurosci. 27, 9901–9915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Supnet C., Grant J., Kong H., Westaway D., Mayne M. (2006) Amyloid-β-(1–42) increases ryanodine receptor-3 expression and function in neurons of TgCRND8 mice. J. Biol. Chem. 281, 38440–38447 [DOI] [PubMed] [Google Scholar]
- 43. Webster S., O'Barr S., Rogers J. (1994) Enhanced aggregation and β structure of amyloid β peptide after coincubation with C1q. J. Neurosci. Res. 39, 448–456 [DOI] [PubMed] [Google Scholar]
- 44. Cheng J. S., Dubal D. B., Kim D. H., Legleiter J., Cheng I. H., Yu G. Q., Tesseur I., Wyss-Coray T., Bonaldo P., Mucke L. (2009) Collagen VI protects neurons against Aβ toxicity. Nat. Neurosci. 12, 119–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ojha J., Masilamoni G., Dunlap D., Udoff R. A., Cashikar A. G. (2011) Sequestration of toxic oligomers by HspB1 as a cytoprotective mechanism. Mol. Cell. Biol. 31, 3146–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fraser D. A., Arora M., Bohlson S. S., Lozano E., Tenner A. J. (2007) Generation of inhibitory NFκB complexes and phosphorylated cAMP response element-binding protein correlates with the anti-inflammatory activity of complement protein C1q in human monocytes. J. Biol. Chem. 282, 7360–7367 [DOI] [PubMed] [Google Scholar]
- 47. Tchantchou F., Lacor P. N., Cao Z., Lao L., Hou Y., Cui C., Klein W. L., Luo Y. (2009) Stimulation of neurogenesis and synaptogenesis by bilobalide and quercetin via common final pathway in hippocampal neurons. J. Alzheimers Dis. 18, 787–798 [DOI] [PubMed] [Google Scholar]
- 48. Li Y. F., Cheng Y. F., Huang Y., Conti M., Wilson S. P., O'Donnell J. M., Zhang H. T. (2011) Phosphodiesterase-4D knock-out and RNA interference-mediated knockdown enhance memory and increase hippocampal neurogenesis via increased cAMP signaling. J. Neurosci. 31, 172–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nakagawa S., Kim J. E., Lee R., Chen J., Fujioka T., Malberg J., Tsuji S., Duman R. S. (2002) Localization of phosphorylated cAMP response element-binding protein in immature neurons of adult hippocampus. J. Neurosci. 22, 9868–9876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tong L., Balazs R., Thornton P. L., Cotman C. W. (2004) β-Amyloid peptide at sublethal concentrations down-regulates brain-derived neurotrophic factor functions in cultured cortical neurons. J. Neurosci. 24, 6799–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kokubo M., Nishio M., Ribar T. J., Anderson K. A., West A. E., Means A. R. (2009) BDNF-mediated cerebellar granule cell development is impaired in mice null for CaMKK2 or CaMKIV. J. Neurosci. 29, 8901–8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heiber M., Docherty J. M., Shah G., Nguyen T., Cheng R., Heng H. H., Marchese A., Tsui L. C., Shi X., George S. R., (1995) Isolation of three novel human genes encoding G protein-coupled receptors. DNA Cell Biol. 14, 25–35 [DOI] [PubMed] [Google Scholar]
- 53. Webster S., Rogers J. (1996) Relative efficacies of amyloid β peptide (Aβ)-binding proteins in Aβ aggregation. J. Neurosci. Res. 46, 58–66 [DOI] [PubMed] [Google Scholar]
- 54. Fuentealba R. A., Liu Q., Zhang J., Kanekiyo T., Hu X., Lee J. M., LaDu M. J., Bu G. (2010) Low-density lipoprotein receptor-related protein 1 (LRP1) mediates neuronal Aβ42 uptake and lysosomal trafficking. PLoS ONE 5, e11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ulery P. G., Beers J., Mikhailenko I., Tanzi R. E., Rebeck G. W., Hyman B. T., Strickland D. K. (2000) Modulation of β-amyloid precursor protein processing by the low density lipoprotein receptor-related protein (LRP). Evidence that LRP contributes to the pathogenesis of Alzheimer disease. J. Biol. Chem. 275, 7410–7415 [DOI] [PubMed] [Google Scholar]
- 56. Cam J. A., Zerbinatti C. V., Li Y., Bu G. (2005) Rapid endocytosis of the low density lipoprotein receptor-related protein modulates cell surface distribution and processing of the β-amyloid precursor protein. J. Biol. Chem. 280, 15464–15470 [DOI] [PubMed] [Google Scholar]
- 57. Cam J. A., Zerbinatti C. V., Knisely J. M., Hecimovic S., Li Y., Bu G. (2004) The low density lipoprotein receptor-related protein 1B retains β-amyloid precursor protein at the cell surface and reduces amyloid-β peptide production. J. Biol. Chem. 279, 29639–29646 [DOI] [PubMed] [Google Scholar]
- 58. Marschang P., Brich J., Weeber E. J., Sweatt J. D., Shelton J. M., Richardson J. A., Hammer R. E., Herz J. (2004) Normal development and fertility of knockout mice lacking the tumor suppressor gene LRP1b suggest functional compensation by LRP1. Mol. Cell. Biol. 24, 3782–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu C. X., Li Y., Obermoeller-McCormick L. M., Schwartz A. L., Bu G. (2001) The putative tumor suppressor LRP1B, a novel member of the low density lipoprotein (LDL) receptor family, exhibits both overlapping and distinct properties with the LDL receptor-related protein. J. Biol. Chem. 276, 28889–28896 [DOI] [PubMed] [Google Scholar]
- 60. Bu G., Cam J., Zerbinatti C. (2006) LRP in amyloid-β production and metabolism. Ann. N.Y. Acad. Sci. 1086, 35–53 [DOI] [PubMed] [Google Scholar]
- 61. Poduslo S. E., Huang R., Spiro A., 3rd (2010) A genome screen of successful aging without cognitive decline identifies LRP1B by haplotype analysis. Am. J. Med. Genet. Neuropsychiatr. Genet. 153B, 114–119 [DOI] [PubMed] [Google Scholar]
- 62. Fonseca M. I., Zhou J., Botto M., Tenner A. J. (2004) Absence of C1q leads to less neuropathology in transgenic mouse models of Alzheimer's disease. J. Neurosci. 24, 6457–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Reichwald J., Danner S., Wiederhold K. H., Staufenbiel M. (2009) Expression of complement system components during aging and amyloid deposition in APP transgenic mice. J. Neuroinflammation 6, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Doyle S. L., Campbell M., Ozaki E., Salomon R. G., Mori A., Kenna P. F., Farrar G. J., Kiang A. S., Humphries M. M., Lavelle E. C., O'Neill L. A., Hollyfield J. G., Humphries P. (2012) NLRP3 has a protective role in age-related macular degeneration through the induction of IL-18 by drusen components. Nat. Med. 18, 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]