Background: Nonmuscle myosin IIB (NMIIB) is a key player in cell motility.
Results: Although the individual NMIIB molecules are not processive, NMIIB thick filaments show robust processive motion.
Conclusion: NMIIB forms processive thick filament in vitro, which is likely the functional unit in cells.
Significance: We demonstrate how processive systems can be formed from nonprocessive individual molecules.
Keywords: Actin, Kinetics, Molecular Motors, Myosin, Single Molecule Biophysics, TIRF Microscopy, Nonmuscle Filament Motility, Optical Tweezers, Single Molecule Kinetics
Abstract
Nonmuscle myosin IIB (NMIIB) is a cytoplasmic myosin, which plays an important role in cell motility by maintaining cortical tension. It forms bipolar thick filaments with ∼14 myosin molecule dimers on each side of the bare zone. Our previous studies showed that the NMIIB is a moderately high duty ratio (∼20–25%) motor. The ADP release step (∼0.35 s−1) of NMIIB is only ∼3 times faster than the rate-limiting phosphate release (0.13 ± 0.01 s−1). The aim of this study was to relate the known in vitro kinetic parameters to the results of single molecule experiments and to compare the kinetic and mechanical properties of single- and double-headed myosin fragments and nonmuscle IIB thick filaments. Examination of the kinetics of NMIIB interaction with actin at the single molecule level was accomplished using total internal reflection fluorescence (TIRF) with fluorescence imaging with 1-nm accuracy (FIONA) and dual-beam optical trapping. At a physiological ATP concentration (1 mm), the rate of detachment of the single-headed and double-headed molecules was similar (∼0.4 s−1). Using optical tweezers we found that the power stroke sizes of single- and double-headed heavy meromyosin (HMM) were each ∼6 nm. No signs of processive stepping at the single molecule level were observed in the case of NMIIB-HMM in optical tweezers or TIRF/in vitro motility experiments. In contrast, robust motility of individual fluorescently labeled thick filaments of full-length NMIIB was observed on actin filaments. Our results are in good agreement with the previous steady-state and transient kinetic studies and show that the individual nonprocessive nonmuscle myosin IIB molecules form a highly processive unit when polymerized into filaments.
Introduction
The myosin superfamily of actin-based molecular motors consists of at least 35 different classes (1, 2). The myosin II or conventional class is the largest among them; in mammals it includes skeletal, cardiac, smooth muscle, and nonmuscle myosin II (NMII)2 isoforms. NMII isoforms contribute to various cellular processes, such as cell migration (3–5), cell adhesion (6–8), and cytokinesis (9). Mammals have three different NMII heavy chain genes (MYH9, MYH10, and MYH14). The NMIIs generated by these heavy chain genes are termed NMIIA, NMIIB, and NMIIC, respectively (10, 11). These heavy chain genes homodimerize through their coiled-coil tail structures, and each heavy chain binds a regulatory (RLC) and essential (ELC) light chain when expressed in cells. This results in a myosin molecule containing two globular N-terminal motor domains harboring ATPase and actin binding activities, two neck regions with bound light chains, a long α-helical coiled-coil region responsible for filament formation, and a short C-terminal nonhelical tail region (12). All three nonmuscle myosin isoforms are activated by phosphorylation of the RLC, which is catalyzed by a number of different kinases, most commonly myosin light chain kinase (MLCK) and Rho kinase (10, 11, 13–15).
Although the NMII isoforms have similar primary and secondary structure, they have distinct enzymatic properties. Detailed in vitro studies of the kinetic parameters of actin-activated ATPase activity of NMIIA, IIB, and IIC myosins reveal subtle differences in the rate and equilibrium constants for the ATPase reaction (16–20). NMIIB is one of the slowest characterized myosins with respect to its rate of actin-activated MgATPase activity and rate of translocation of actin filaments in the in vitro motility assay (16, 18). The IIB isoform, which is the most abundant NMII isoform expressed in the central nervous system and is highly expressed in cardiac muscle (21–24), has the highest ADP affinity among the myosins documented so far, and its ADP release rate is very slow compared with other conventional myosins (16, 18). As a consequence, the duty ratio (the fraction of time the myosin head spends strongly bound to actin during an ATPase cycle) of this myosin is significantly greater than that of muscle myosin II but is lower than that of processive myosins such as myosin V (25, 26). NMIIB can be thus characterized as an intermediate duty ratio motor (10, 17, 18).
Similar to skeletal and cardiac myosins, NMIIs also form supramolecular structures. Vertebrate skeletal myosin forms highly ordered arrays of thick filaments that are ∼1.6 μm in length. The individual heads within these filaments function independently with phosphate release being the rate-limiting step in their enzymatic cycle. As a consequence they have a very low (∼0.03) duty ratio (27, 28). For a molecular motor like the skeletal myosin these enzymatic characteristics are crucial because they ensure that the individual myosin heads spend only a very short period of time attached to actin to generate force and therefore do not interfere negatively with other myosin heads during sarcomere shortening. The NMIIA filaments consist of only ∼28 heavy chains on each side of the bipolar structure (29, 30), so the duty ratio has to be higher to maintain processivity and to ensure that at least a few myosin heads are attached to an actin filament at any given moment.
The aim of this study was to systematically characterize NMIIB at different levels of structural organization and assess whether NMIIB is capable of interacting with actin in a processive manner at the same organizational levels. We compared the kinetic and mechanical behavior of single-headed (NMIIB-SH-HMM) and double-headed HMM (NMIIB-HMM) molecules to gain insight into the cooperative nature of the myosin heads at the single molecule level. To achieve our goal we utilized a single molecule motility assay using both total internal reflection fluorescence (TIRF) microscopy and optical tweezers. The results showed that at physiological ATP concentrations the rate of detachment of NMIIB-SH-HMM from actin was very similar to that of NMIIB-HMM and that these rates are consistent with the ADP release rates reported from solution kinetic studies (16, 18). This suggests that the individual myosin heads act independently, in a noncooperative manner. At this level of organization (one myosin head or two myosin heads coupled together in the form of an HMM fragment), the NMIIB acted as a low duty ratio, nonprocessive motor. In stark contrast to these results, we found that NMIIB filaments exhibit robust processive behavior, as observed both in TIRF and optical trapping experiments.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant human nonmuscle heavy meromyosin IIB (NMIIB-HMM) with a C-terminal EGFP and FLAG tag was co-expressed with bovine nonmuscle regulatory light chain (MYL9) and chicken essential (MYL6) light chains in the baculovirus/Sf9 system (Invitrogen). For TIRF microscopy experiments, the RLC was fused with a GFP on its N terminus (GFP-RLC) (31). The NMIIB-SH-HMM was obtained by co-expressing HMM with subfragment-2 (S2), GFP-RLC, and ELC. The S2 fragment contained a C-terminal His tag. The full-length NMIIB was co-expressed with GFP-RLC and ELC. The infected Sf9 cells were harvested by sedimentation after 24–72 h of growth and stored at −80 °C. The proteins were purified as described by Wang et al. (32). Briefly, the cell pellets were extracted and homogenized in a buffer containing 0.5 m NaCl, 10 mm MOPS (pH 7.3), 10 mm MgCl2, 1 mm EGTA, 3 mm NaN3, 2 mm ATP, 0.1 mm phenylmethylsulfonyl fluoride, 0.1 mm dithiothreitol, 5 μg/ml leupeptin, and proteinase inhibitor mixture (2 μg/ml chymostatin (MP Biochemicals, Santa Ana, CA), 1 μg/ml pepstatin (MP Biochemicals), 1 μg/ml Nα-tosyl-Lys-chloromethylketone-HCl (TLCK) (Sigma), 1 μg/ml N-p-tosyl-l-phenylalanine chloromethyl ketone (TPCK) (Sigma). The full-length nonmuscle myosin IIB, the NMIIB-HMM, and the NMIIB-SH-HMM were co-purified with the light chains by FLAG-affinity chromatography using M2 FLAG affinity gel (Sigma). The NMIIB-SH-HMM samples were subsequently purified using a nickel-nitrilotriacetic acid resin under native conditions following the manufacturer's instructions (Qiagen). The HMM IIB and the full-length nonmuscle myosin IIB with either wild-type RLC or GFP-RLC at 0.5–4.2 μm were phosphorylated with MLCK (1–10 nm) overnight on ice in case of full-length NMIIB myosin molecules and 15 min at room temperature in case of NMIIB-HMM molecules in a reaction mixture containing 10 mm MOPS (pH 7.3), 50 mm KCl (300 mm KCl in the case of full-length nonmuscle myosin IIB to avoid aggregation), 5 mm MgCl2, 0.2 mm CaCl2, 0.1 mm EGTA, 0.1 μm calmodulin, 1 mm DTT, and 0.2 mm ATP. The full-length rabbit smooth muscle MLCK (NP_001075775) was expressed in the baculovirus/Sf9 system and purified via an N-terminal FLAG tag. The phosphorylation of the NMIIB-HMM molecules was confirmed with acrylamide pendant Phos-tag phosphate affinity SDS-PAGE system (Wako Chemicals, Richmond, VA). The MgATPase activity was measured using an NADH-coupled assay at 25 °C in 10 mm MOPS (pH 7.0), 2 mm MgCl2, 1 mm ATP, 50 mm KCl, 0.15 mm EGTA, 40 units/ml l-lactic dehydrogenase, 200 units/ml pyruvate kinase, 200 μm NADH, and 1 mm phospho(enol)pyruvate. During the TIRF experiments the nonmuscle filaments were formed by reducing the KCl concentration by fast dilution to 150 mm in the assay buffer.
TIRF/in Vitro Motility Assays
The TIRF/in vitro motility assay was performed in a motility buffer containing 20 mm MOPS (pH 7.4), 5 mm MgCl2, 0.1 mm EGTA, 50 mm KCl, 1 mm ATP, 25 μg/ml glucose oxidase, 45 μg/ml catalase, 2.5 mg/ml glucose, and 50 mm dithiothreitol at 25 °C. The TIRF assay was performed on an Olympus IX81 microscope equipped for objective type TIRF microscopy using a PlanApo (×60, NA 1.45) objective lens and a relay lens (PE5, ×5 or PE2.5, ×2.5; Olympus) connected to an EMCCD (iXon, Andor Technology, South Windsor, CT). Excitation light sources were 488 (diode lasers, Crystal Laser, Reno, NV and Coherent, Santa Clara, CA) and 532 nm (diode laser, IK Series He-Cd laser, Kimmon Koha, Centennial, CO). To prevent nonspecific binding of fluorescently labeled protein, the coverslips (top, 18 × 18 mm and bottom, 24 × 50 mm) comprising the flow cell were prepared as follows. Coverslips were soaked in concentrated H2SO4 overnight, boiled in ddH2O for 1 h, and dried with a stream of nitrogen. The flow cell, ∼30 μl in volume, was prepared using double-sided Scotch tape (3M). For TIRF assays, 30 μl of 2 mg/ml biotin-BSA solution was applied to the flow cell, incubated for 4 min, and washed with motility assay buffer (2 × 200 μl). Next, 60 μl of NeutrAvidin solution (1 mg/ml, Pierce) was applied, incubated for 2 min, and washed with motility assay buffer (2 × 200 μl). Filaments of rabbit actin (biotinylated to 10%, 50 nm) labeled with rhodamine phalloidin (Invitrogen) were applied to the flow cell and incubated for 2 min. Unbound actin was washed off with 1 ml of motility assay buffer. Fluorescently (GFP) labeled phosphorylated full-length NMIIB-HMM or NMIIB-SH-HMM molecules (10–50 nm final concentration) in 100 μl of motility assay buffer were introduced into the flow cell, which was then mounted onto the microscope for imaging. Data were captured and analyzed using MetaMorph (Universal Imaging, Molecular Devices LLC, Sunnyvale, CA) and Origin 8.5 (OriginLab Corp., Northampton, MA). The tracking of dwell times of the fluorescently labeled particles was performed using a customized particle-tracking program. This program was modified by Chris A. Combs (Light Microscopy Core Facility at the NHLBI, National Institutes of Health) and Ronn Kling (Ronn Kling Consulting, Warrenton, VA) from open-source particle tracking software, developed by D. Grier (New York University), J. Crocker (University of Pennsylvania), and E. R. Weeks (Emory University). The program was developed and run within IDL software programming environment (ITT Visual Information Solutions, Boulder, CO). The FIONA analysis was performed with Video Spot Tracker v.05.23 (CISMM at UNC-CH) (33, 34).
Optical Trapping
Three-bead assays (35, 36) were performed using a system similar to that described by Vanzi et al. (37) and Takagi et al. (38). Briefly, an in vitro force assay chamber (volume ∼40 μl), was constructed using two coverslips, one of which had been sparsely decorated with 2.1-μm diameter glass microspheres (Bangs Laboratories, Fishers, IN) using a 0.1% nitrocellulose-amylacetate solution, assembled using double-sided adhesive tape. Anti-GFP antibody (3–15 nm in PBS; MP Biomedicals) was allowed to bind nonspecifically inside the chamber for 5 min, and then the chamber was flushed extensively with PBS. After blocking the surface with 1 mg/ml BSA for 2 min, phosphorylated NMIIB-HMM or NMIIB-SH-HMM proteins were diluted to a concentration of 0.1–0.3 nm in 25 mm KCl, 25 mm imidazole, 4 mm MgCl2, 1 mm EGTA (pH 7.4) at 22 °C (AB buffer) (39) and allowed to bind specifically to antibodies within the chamber. AB buffer supplemented with 50 mm DTT, 1 mm ATP, 3 mg/ml glucose, 0.1 mg/ml glucose oxidase, and 0.02 mg/ml catalase (24) was used in the final chamber mixture, together with 0.2 nm rhodamine phalloidin-labeled, 10% biotinylated filamentous actin, and NeutrAvidin-coated 1-μm biotin-labeled polystyrene beads conjugated with tetramethyl-rhodamine B-isothiocyanate-BSA (39). Under fluorescence imaging, a single actin filament was attached to two 1-μm beads, via manipulation of the optical traps. These beads/actin dumbbells (length ∼ 5–7 μm) were pretensioned (to 4–5 piconewtons) and positioned above the glass microspheres attached to the surface of the chamber (functioning as a pedestal), to record transient unitary acto-myosin interactions. Only one of nine pedestals exhibited unitary acto-myosin interactions, providing statistical support for the assumption that only a single nonmuscle myosin NMIIB-HMM and NMIIB-SH-HMM was capable of interacting with the actin filament at any instance. Experiments were performed using an optical trap stiffness of 0.01–0.022 piconewton/nm per trap. Similarly, as reported by Baboolal et al. (40) data were sampled at 20 kHz whereas sine waves (frequency = 200 Hz) of amplitudes (∼200 nm, peak-to-peak) were applied to one of the optical traps (41, 42).
In the applied three-bead assay, the detached cross-bridge has no influence on the motion and position of the detector bead; it is influenced only by thermal motion or by the oscillation from acousto-optic deflector through the tethered actin filament and motor bead. If the actin filament dumbbell is not attached to the surface through a myosin molecule, then the variance of the motion of the detector bead is going to be relatively high. When the interaction occurs, the system is stiffened and the variance drops markedly by up to 10-fold. Thus, if the variance is plotted in a trap system in which no interactions are occurring, the plotted histogram can be fitted with a single gaussian curve. If interactions occur, the variance histogram will show two peaks, and it can be fitted with a double gaussian curve. The higher variance peak will represent the detached state of the acto-myosin system, and the lower variance value corresponds to the attached state. Next, the program compares the variance of each data point to the “attached” and “detached” variance values to determine the beginning and the end point of the interactions. If the variance value of the individual data point is between the attached and detached variance value, the score (attached or detached) of individual data points depends on the score of the previous data point, which greatly reduces the probability of false positive hits. A similar method was used by Laakso et al. (43) and described thoroughly in Knight et al. (44).
The recorded data were used to compute a running mean bead position and running variance computed over sliding windows of 4 ms or 40 ms. A marked decrease in the running variance of the noise level of this sine wave was used to distinguish regions of the collected data as periods of myosin attachments (40, 43, 44). Data analyzed over a 4-ms running variance allowed identification of both short and long durations, whereas use of a 40-ms running variance acts as a low pass filter and isolates the longer duration attachments from much shorter duration attachments. Power stroke sizes were measured by plotting a histogram of the displacements and fitting the data to a gaussian distribution (36). The shift in the peak position of the displacement gaussian distribution from zero is considered a measure of the power stroke size. With phosphorylated NMIIB HMM molecules the orientation of a given actin filament being studied could be ascertained by the directionality of the accumulated steps. In the case of the short lived interactions observed with the phosphorylated NMIIB fragments, the orientation of the filament was obtained by observing the directionality of the long lived events using that filament. In the case of unphosphorylated HMM, only filaments were used that had a large number (>100) of recorded events over multiple pedestals. The frequency versus displacement plots yielded gaussian distributions that were centered on 0 nm. These data were pooled and used to construct a displacement histogram. Analysis was performed using custom software written in LabView 8.0 (National Instruments), and histograms were plotted using Origin 8.5 (OriginLab Corp.).
Electron Microscopy
For negative staining, NMIIB samples were diluted to ∼100 nm with a buffer containing 10 mm MOPS (pH 7.0), 2 mm MgCl2, 0.1 mm EGTA, and 150 mm KCl in the case of full-length myosin or 25 mm KCl in the case of HMM. A drop of sample was applied to a carbon-filmed EM grid and directly stained with 1% uranyl acetate. Micrographs were recorded at × 40,000 on a JEOL 1200EX II microscope. Data were recorded on an AMT XR-60 CCD camera.
RESULTS
Protein Expression, Purification, and Sample Preparation
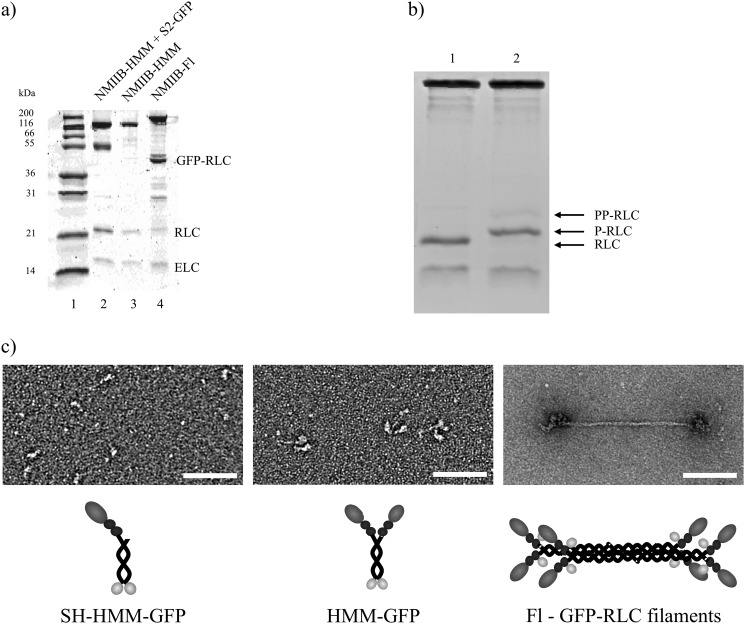
To analyze NMIIB kinetically and mechanically at the single molecule level, different fragments and the full-length molecule were expressed and purified (Fig. 1a). To facilitate the specific binding of NMIIB-HMM and NMIIB-SH-HMM molecules to a nitrocellulose surface in optical trapping experiments a GFP molecule was fused to the C terminus of these molecules. These molecules could be captured via an anti-GFP antibody bound to the coverslip surface. The GFP was also used in TIRF experiments as a fluorescent label. In the case of the full-length NMIIB, the GFP was fused to the regulatory light chain (GFP-RLC) (31) instead of the heavy chain of the molecule to avoid possible problems during nonmuscle filament formation. In all cases, except where stated, the myosin or myosin fragment was phosphorylated with MLCK prior to use in the assays. This treatment resulted in the majority of the regulatory light chains of the myosin being monophosphorylated (Fig. 1b).
FIGURE 1.
Purified proteins used in the study. a, Coomassie Blue-stained 4–12% SDS-polyacrylamide gel image of the samples. Lane 1, molecular mass standards. Lane 2, single-headed NMIIB-HMM-GFP. The ∼51-kDa fragment is the His-tagged S2-GFP. Lane 3, NMIIB-HMM-GFP. Lane 4, full-length NMIIB with GFP-RLC. The ∼43-kDa fragment is the GFP-tagged RLC. b, acrylamide pendant Phos tag phosphate affinity SDS-PAGE image of a representative preparation of NMIIB-HMM molecules before (lane 1) and after (lane 2) MLCK treatment. Densitometry analysis shows that 80% of RLCs were monophosphorylated (P-RLC), 12% were double-phosphorylated (PP-RLC), and 8% were not phosphorylated (RLC). c, rotary shadowed electron micrographs of the NMIIB constructs (upper) and simplified schematic diagrams of the molecules (lower). Scale bars: SH-HMM-GFP and HMM-GFP, 50 nm; full-length NMIIB filament with GFP-RLC, 100 nm.
The negatively stained EM images confirmed the expected shape and size of NMIIB-SH-HMM, NMIIB-HMM, and full-length molecules (Fig. 1c). The actin-activated MgATPase activities of the samples were found to be 85–115% of the steady-state MgATPase rates reported in earlier solution kinetic studies (16, 18).
Single Molecule TIRF Experiments
TIRF microscopy is a powerful method to observe and characterize the behavior of myosin molecules at the single molecule level. GFP-labeled myosin molecules or fragments were added to flow chambers in which fluorescently (rhodamine phalloidin) labeled, biotinylated actin filaments were bound via NeutrAvidin and biotinylated BSA to a cleaned glass coverslip. The NMIIB-SH-HMM and NMIIB-HMM molecules appeared as diffraction-limited spots. We recorded the binding and the unbinding of the GFP-labeled myosin molecules to determine their attachment lifetimes and whether or not they moved processively as single molecules. Each myosin molecule was labeled at the C terminus of the heavy chain, and on the N terminus of RLC molecules, so the fragments contained either 3 (NMIIB-SH-HMM-GFP) or 4 (NMIIB-HMM-GFP) GFP molecules. Thus, the disappearance of a fluorescent spot is most likely a dissociation event of that molecule from actin rather than loss of signal through photobleaching or blinking. The binding and the unbinding of the GFP-labeled NMIIB-SH-HMM-GFP and NMIIB-HMM-GFP to rhodamine phalloidin-labeled actin were recorded at 1 mm ATP with 67-ms temporal resolution (50-ms acquisition time plus the 17-ms readout time of the CCD chip) over a time span of 1 min or more (supplemental movies 1 and 2).
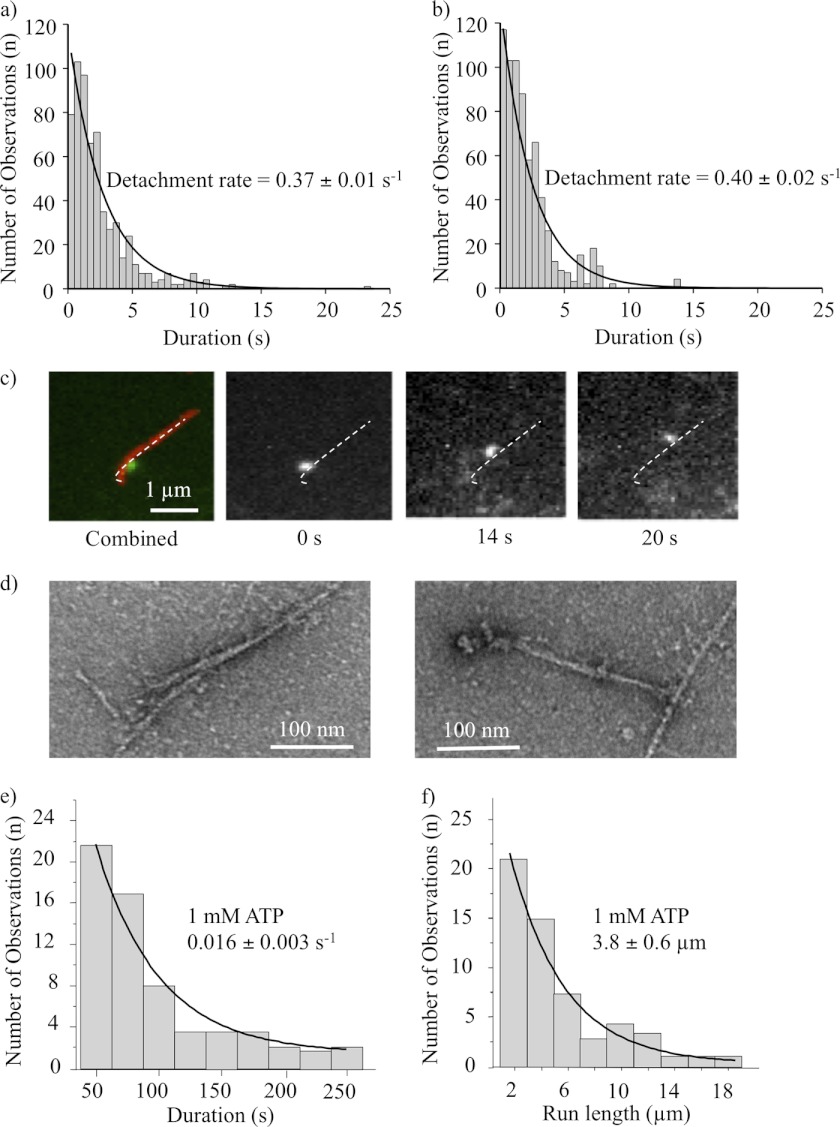
The dissociation kinetics of the NMIIB-SH-HMM-GFP and NMIIB-HMM-GFP were determined by monitoring the dwell times of individual molecules on actin filaments and were plotted as histograms (Fig. 2, a and b). For both NMIIB-SH-HMM-GFP and NMIIB-HMM-GFP the histograms of dwell times were well fitted by single exponential decay functions. The average detachment rates determined from these histograms were 0.37 ± 0.01 s−1 for NMIIB-HMM and 0.4 ± 0.02 s−1 for NMIIB-SH-HMM molecules (Fig. 2, a and b). No obvious processive movements were detected using this assay with either of these fragments.
FIGURE 2.
TIRF microscopy analysis of actin-myosin interactions. a, histogram of dwell times of NMIIB-HMM-actin interactions at 1 mm ATP. Solid line is the single exponential fit to the data which gave a detachment rate constant of 0.37 ± 0.01 s−1, n = 611. b, histogram of dwell times of NMIIB-SH-HMM-actin interactions at 1 mm ATP. Solid line is the single exponential fit to the data which gave a detachment rate constant of 0.40 ± 0.02 s−1, n = 683. c, filament of full-length NMIIB molecule with GFP-RLC (green) interactions with rhodamine phalloidin-labeled actin (red), at 1 mm ATP (left panels). Right panels, GFP-labeled nonmuscle filament moving processively along a single actin filament. Scale bar, 1 μm. d, electron micrographs of NMIIB nonmuscle filaments interacting with actin filaments in the presence of ATP. Scale bars, 100 nm. e, histogram of dwell times of NMIIB filament-actin interactions at 1 mm ATP. Solid line is the single exponential fit to the data which gave a detachment rate constant of 0.016 ± 0.003 s−1, n = 62. f, distribution of run lengths of NMIIB filaments on actin filaments at 1 mm ATP. Solid line is the single exponential fit to the data which gave a run length of 3.8 ± 0.6 μm, n = 56.
In contrast, NMIIB polymerized into filaments showed strikingly different behavior in the TIRF in vitro motility assays (Fig. 2c). These bipolar filaments were readily imaged in the TIRF assay as objects that are slightly larger than diffraction limited spots, and in some cases, a long and short axis could be distinguished. At 1 mm ATP the nonmuscle myosin filaments moved along F-actin as processive units (Fig. 2c) with an average run length of 3.8 ± 0.6 μm (Fig. 2f) and an average velocity of 48 ± 2 nm/s. This behavior produced a much slower filament detachment rate constant (0.016 ± 0.003 s−1) (Fig. 2e) than was observed for the single HMM molecules reported above (Fig. 2, a and b). Filaments that reached the end of the actin filaments and dissociated have been excluded from the run length analysis. In some cases the NMIIB filaments moved with their long axis parallel to the actin filaments, and in others they moved with their long axis perpendicular to the actin filament (supplemental Movie 3). NMIIB filaments could be observed to switch back and forth between these orientations while moving.
Electron micrographic images of NMIIB filaments interacting with F-actin in the presence of ATP revealed additional insight into the nature of this interaction (Fig. 2d). It is apparent from the EM images that the myosin heads of the nonmuscle filament are in close association with the actin filament. Similar to the behavior described above in the TIRF assay, we observed that in some cases heads on both sides of the bare zone of NMIIB filaments were bound to actin (Fig. 2d, left), whereas in other cases only the heads at one side of the NMIIB filament interacted with actin (Fig. 2d, right) (supplemental Movie 3).
Optical Trapping Experiments
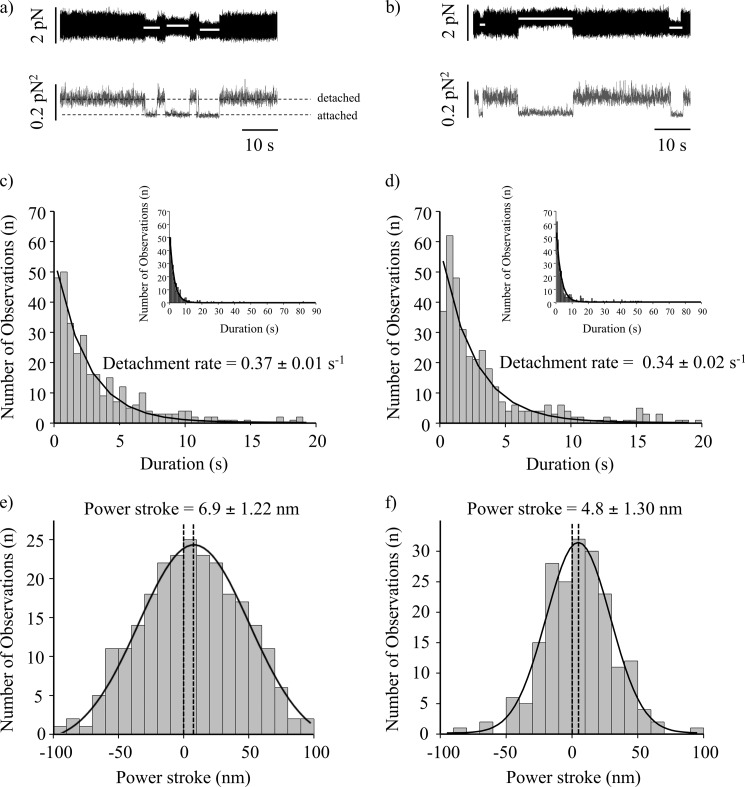
The interaction of various constructs of NMIIB with actin was examined at the single molecule level using optical tweezers. We applied the three-bead assay, in which the myosin molecule of interest was bound to a nitrocellulose-coated surface, and an actin filament (tethered between two optically trapped beads) was brought into close proximity. The NMIIB-SH-HMM and NMIIB-HMM molecules were tagged at their C termini with a GFP molecule to facilitate attachment via an anti-GFP antibody bound to the nitrocellulose surface. In the case of optical trapping experiments we used myosin fragments expressed with wild-type RLC, so the single-headed and double-headed molecules were tethered to the surface only by their C termini. The optically trapped beads were oscillated with 200 Hz and 200-nm amplitude, which increased the signal-to-noise ratio of the attachment events (Fig. 3, a and b). Data were sampled at 20,000 Hz and were sometimes converted (filtered) into 2000 Hz by averaging 10 consecutive data points (Figs. 3, a and b, and 4, a and b). Two types of attachment events were observed in the unfiltered data (Fig. 4, a and b). There were numerous short lived (duration <50 ms) events as well as less frequent longer lived events that were more consistent with the known kinetics of NMIIB as well as with the lifetimes observed in the TIRF experiments presented earlier. For both NMIIB-HMM and NMIIB-SH-HMM the short lived events account for >80% of the observed interactions (Table 1).
FIGURE 3.
Myosin-actin interactions at 1 mm ATP sampled at 20,000 Hz and filtered to 2000 Hz. a, NMIIB-HMM-actin interactions at 1 mm ATP. Top trace, bead position record. Bottom trace, variance. The white lines in the top trace represent the unitary actin-myosin interactions. b, NMIIB-SH-HMM-actin interactions at 1 mm ATP. Top trace, bead position record. Bottom trace, variance. The white lines in the top trace represent unitary actin-myosin interactions. c, histogram of dwell times of NMIIB-HMM-actin interactions at 1 mm ATP (0.37 ± 0.01 s−1, n = 327). Solid line shows the single exponential fit of the data. Inset shows data collected over a longer time frame. d, histogram of dwell times of NMIIB-SH-HMM-actin interactions at 1 mm ATP (0.34 ± 0.2 s−1, n = 371). Solid line shows the single exponential fit of the data. Inset shows data collected over a longer time frame. e, power stroke size of NMIIB-HMM. Fitting the displacement data collected from the optical trap during each acto-myosin interaction to a gaussian distribution (solid line) yielded histograms centered at 6.9 ± 1.2 nm, n = 257. f, power stroke size of NMIIB-SH-HMM) molecules. Fitting the displacement data collected from the optical trap during each acto-myosin interaction to a gaussian distribution yielded a histogram centered at 4.8 ± 1.3, n = 197.
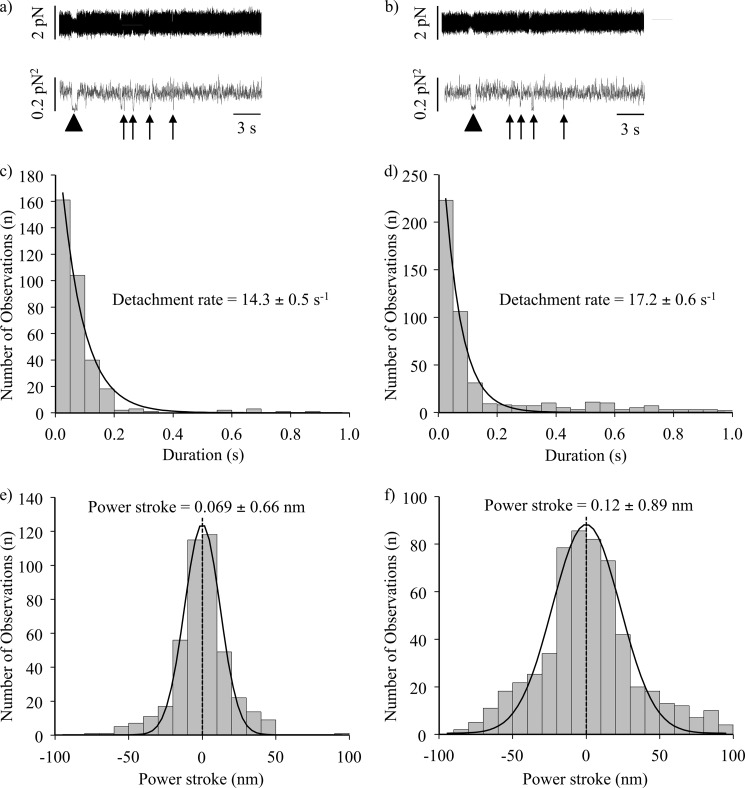
FIGURE 4.
Short lived acto-myosin interactions are observed in the raw bead position traces. a, NMIIB-HMM-actin interactions at 1 mm ATP. Top trace, bead position record. Bottom trace, variance. b, NMIIB-SH-HMM-actin interactions at 1 mm ATP. Top trace, bead position record. Bottom trace, variance. Arrows underneath the variance traces for a and b show short lived interactions; the arrowhead marks the position of a single longer lived interaction in these traces such as observed in Fig. 3, a and b. Note the difference in time scale for these data compared with those shown in Fig. 3. c, histogram of dwell times of fast actin-myosin interactions for NMIIB-HMM-actin interactions at 1 mm ATP. Solid line is a single exponential fit of the data which gave a detachment rate constant of 14.3 ± 0.5 s−1, n = 336. d, histogram of dwell times of fast actin-myosin interactions for NMIIB-SH-HMM-actin interactions at 1 mm ATP. Solid line is a single exponential fit of the data which gave a detachment rate constant of 17.2 ± 0.6 s−1, n = 459. e, power stroke sizes of NMIIB-HMM. f, power stroke sizes of SH-NMIIB-HMM. Fitting the displacement data collected from the optical trap during each acto-myosin interaction to gaussian distributions yielded histograms centered at ∼0 nm for both NMIIB-HMM (n = 440) and SH-HMM (n = 579) molecules.
TABLE 1.
Effect of attachment mode on percentage of short lived events
| Attachment mode | NMIIB-HMMa | NMIIB-SH-HMMa |
|---|---|---|
| % of short events | % of short events | |
| No anti-GFP antibodies, no HMM | 0 | NDb |
| Anti-GFP antibodies alone | 100 | ND |
| HMM bound directly to surface | 89 ± 2.7 | ND |
| HMM bound via anti-GFP antibody | 83.9 ± 1.6 | 87.6 ± 1.3 |
| HMM bound via anti-GFP antibody at 150 mm KCl | 84.6 ± 2.5 | ND |
a No. of short lived events/(no. of short lived events + no. of long lived events) × 100 ± S.E.
b ND, not determined.
For the analysis of dwell times of the longer lived events the filtered data were used which largely eliminated the short lived events. The histograms of dwell times were fitted to a single exponential decay function (Fig. 3, c and d) to determine the detachment rate constant. These values were 0.37 ± 0.01 s−1 for NMIIB-HMM, and 0.34 ± 0.02 s−1 for NMIIB-SH-HMM at 1 mm ATP, which are similar to the detachment rates observed in the TIRF experiments. The records from both the single-headed and double-headed fragments were virtually identical, and no evidence for processive stepping events was observed.
To determine whether the number of myosin motor domains on the molecule affects the size of the power stroke, we plotted the attachment positions of the long lived interactions and fitted the data to a gaussian distribution. The shift in peak of this distribution represents the power stroke size (44). Power strokes of 6.9 ± 1.2 nm (NMIIB-HMM, Fig. 3e) and 4.8 ± 1.3 nm (NMIIB-SH-HMM, Fig. 3f) were measured for the two molecules, which are within the ranges typically observed for myosin II molecules (for review, see Refs. 45, 46). The unpaired Student's t test showed that the difference between the power stroke size values is not statistically significant (the two-tailed p value equals 0.8141).
We next investigated the nature of the numerous very short lived (<100 ms) interactions that were clearly visible as brief sharp drops in variance of the motion of the beads when the data are displayed at 20,000 Hz without low pass filtering (Fig. 4, a and b). Arrows underneath the variance traces point out these short lived interactions compared with the longer lived interactions (marked by an arrowhead) that were scored in Fig. 3, a and b. For this analysis events with lifetimes longer than 1 s were excluded. When only these short lived interactions were examined, the detachment rate constants were 14.3 ± 0.5 s−1 (NMIIB-HMM, Fig. 4c) and 17.2 ± 0.6 s−1 (NMIIB-SH-HMM, Fig. 4d) at 1 mm ATP which are approximately 50 times faster than those measured for the long lived events. We hypothesize that these short lived events most likely represent the very transient, nonproductive, weak binding interactions between NMIIB and actin.
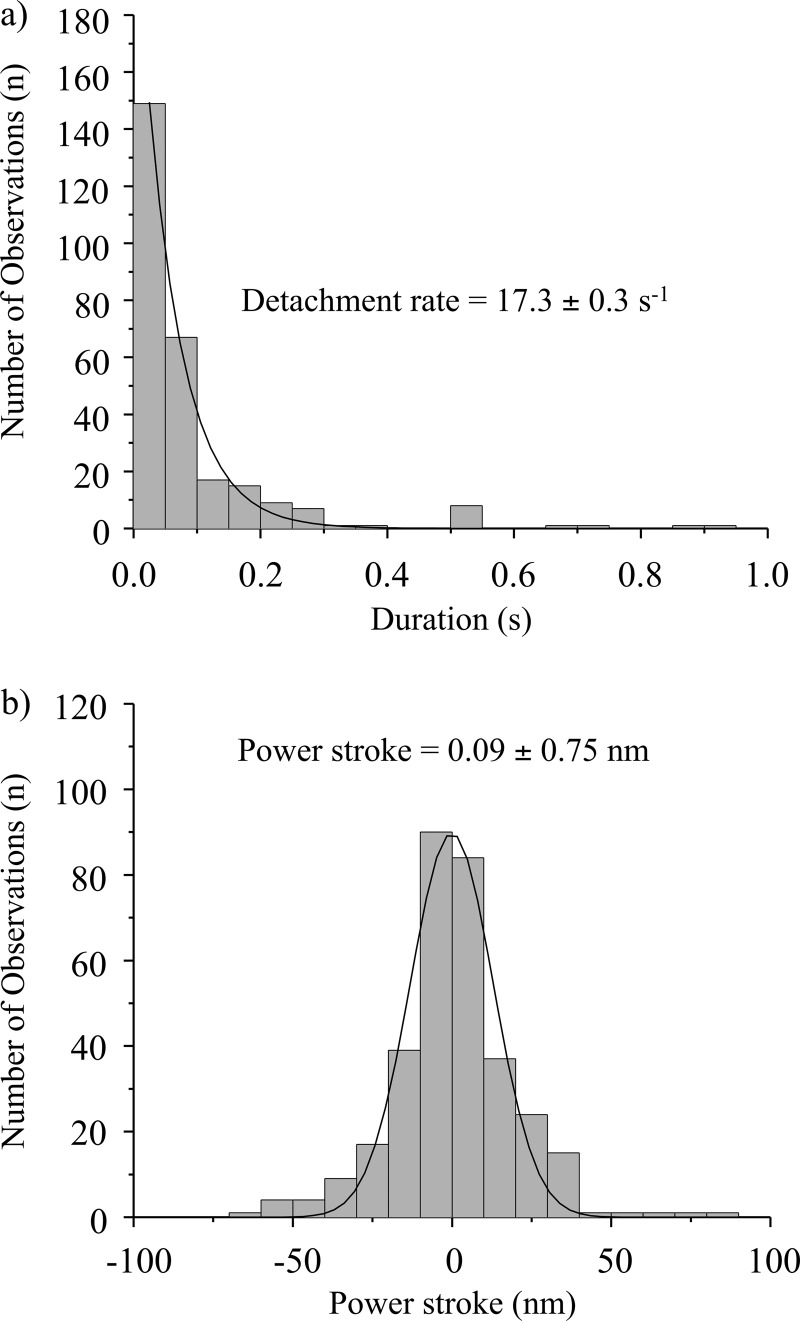
To test this hypothesis we repeated the optical trapping experiments using unphosphorylated NMIIB-HMM molecules. It has been shown that the unphosphorylated NMIIB binds to actin but does not support net movement, most likely because under these conditions the myosin cross-bridges are in a weakly bound state. The analysis of unfiltered data revealed that the detachment rate constant of the unphosphorylated NMIIB-HMM (17.3 ± 0.3 s−1) is identical to those of the fast interactions observed in the cases of phosphorylated NMIIB-HMM and NMIIB-SH-HMM (Fig. 5a). No long lived events were detected for unphosphorylated NMIIB-HMM.
FIGURE 5.
Unphosphorylated NMIIB-HMM shows short lived interactions with actin. a, histogram of dwell times. Solid line shows a single exponential fit to the data which yield a detachment rate constant of 17.3 ± 0.3 s−1, n = 326. b, power stroke size distribution. Solid line shows a gaussian fit to the data which had a peak at 0.09 ± 0.75 nm, n = 338.
Weak binding cross-bridges should not undergo a power stroke. We used the same method as above to measure the “power stroke” of the short lived interactions seen in the raw traces when trapping the phosphorylated NMIIB fragments. Analysis of the displacement sizes show a gaussian distribution with a peak close to 0 nm (0.07 ± 0.66 nm for NMIIB-HMM, Fig. 4e and 0.12 ± 0.89 nm for NMIIB-SH-HMM, Fig. 4f). Similar 0-nm displacement curves have been seen for Drosophila melanogaster myosin-18, which does not hydrolyze ATP (47). To further support the hypothesis that the short lived interactions are weak actin-myosin interactions, we measured the power stroke size of the short lived interactions observed when using unphosphorylated NMIIB-HMM molecules. The peak of the gaussian distribution was also close to 0 nm (Fig. 5b). Thus, these short lived interactions occurred on a time scale faster than what would be expected from the measured solution kinetic values of NMIIB and did not result in a power stroke.
Further evidence that the short lived interactions represent transient, nonproductive attachments of myosin comes from analysis of the frequency of the events is given in Table 2. Essentially no short lived interactions were observed if there was no myosin or no anti-GFP antibody added to the observation chambers. If pedestal beads coated with anti-GFP antibody are probed, approximately 3.2 events/min are detected compared with values of 13.6 and 16/min when NMIIB-HMM or NMII-SH-HMM is probed. The binding constant for actin for many myosins is strikingly weakened by increasing the ionic strength, and one would predict that increasing ionic strength would decrease the frequency of short lived events. However, the affinity on NMIIB-HMM for actin is not dramatically weakened by increasing the ionic strength (Table 3), and thus, this test cannot be easily applied (Table 1).
TABLE 2.
Effect of attachment mode on frequency of short lived events
| Attachment mode | Frequency of short events |
|---|---|
| events/min ± S.E. | |
| No anti-GFP antibodies, no HMM | ∼0 |
| Anti-GFP antibodies alone | 3.2 ± 0.46 |
| HMMIIB-HMM + anti-GFP antibodies | 13.6 ± 2.1 |
| HMMIIB-D+SH-HMM + anti-GFP antibodies | 16.0 ± 2.3 |
TABLE 3.
Effect of ionic strength on actin-activated ATPase activity of NMIIB-HMM
| KCl concentration | Vmax | KATPase |
|---|---|---|
| mm | s−1 | μm |
| 50 | 0.15 | 7.9 |
| 100 | 0.16 | 9.5 |
| 150 | 0.15 | 17.9 |
No evidence for processive movements on single- or double-headed HMM fragments was obtained from either the TIRF single molecule motility assay or the optical trapping. The optical trap imposed a small load onto the myosin, which may be preventing the myosin from stepping processively. To probe for possible processive movements in finer detail and under conditions where there is no load, we employed the FIONA (48) technique to localize the fluorescently labeled HMM molecules in the xy plane (Fig. 6). This superresolution light microscopy technique is capable of detecting potential movements of the fluorescently labeled myosin fragments with high spatial resolution. During the experiments the fluorescence intensity of the HMM particles remained constant, and the detailed analysis revealed no directional movement or stepping behavior. No steps of the expected 5–7 nm in size were detected during a 1–10 s observation period, and the largest “displacements” observed were ∼2 nm, which is within the error of our detection (1.4 nm).
FIGURE 6.
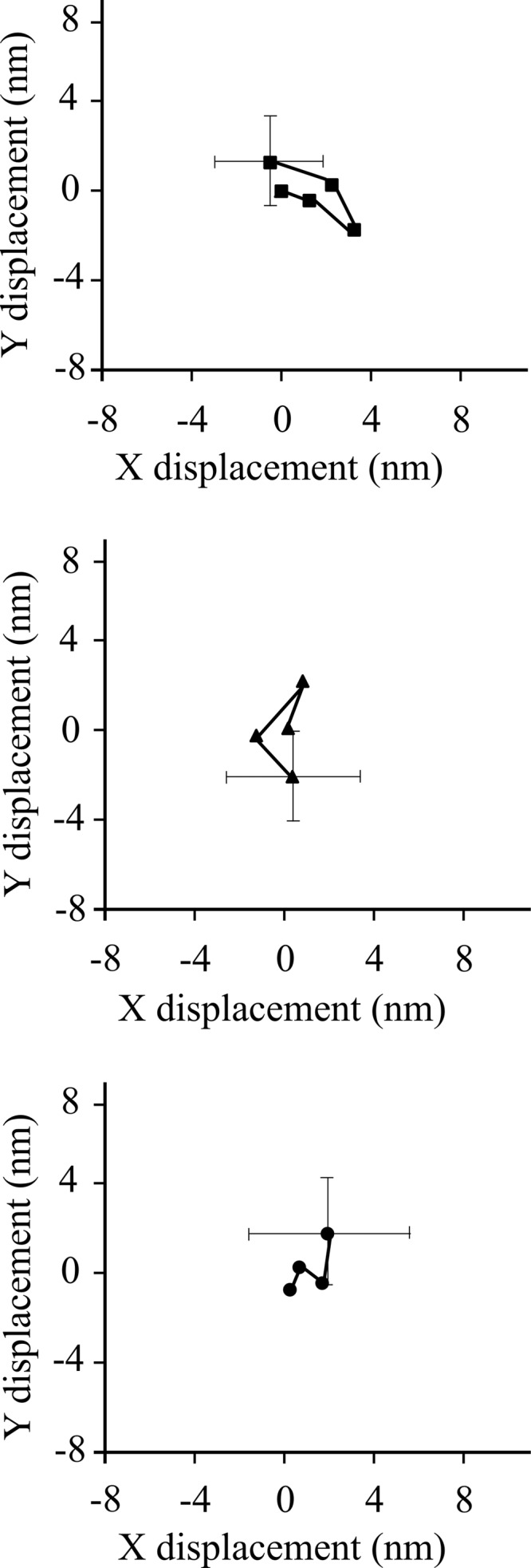
Displacement of HMM molecules in TIRF assay analyzed by FIONA. The positions of three molecules at 2-s intervals are shown. Error bars show the uncertainty of the measurements.
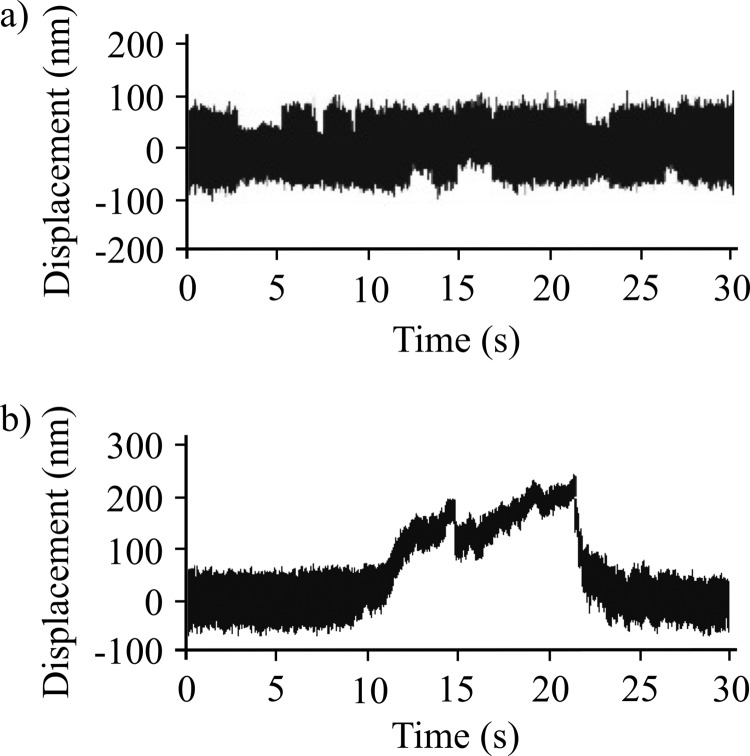
The mechanical properties of full-length NMIIB were also investigated at the single molecule level using optical tweezers. First, the myosin sample was deposited on nitrocellulose in the AB buffer containing 300 mm KCl. EM studies showed that at this ionic concentration single, two-headed myosins bind to the surface. Similar to behavior of NMIIB-SH-HMM and NMIIB-HMM molecules, the individual full-length NMIIB dimers showed unitary interactions with the actin molecule, and no signs of processive movement were observed (Fig. 7a). In a second set of experiments, we lowered the KCl concentration in the myosin solution to 150 mm to form bipolar filaments prior to sample deposition (Fig. 1c). We were able to visualize the nonmuscle filaments in our optical trapping microscope by exciting the GFP-RLC molecules bound to the myosin via epifluorescence imaging using a standard mercury lamp. We did not use anti-GFP antibodies to attach myosin dimer molecules or nonmuscle filaments to the surface during these experiments. The nonmuscle myosin filaments appeared as bright, diffraction-limited spots. This allowed us to determine which pedestal was coated with a myosin filament for targeting during the optical trapping experiments. In contrast to the nonprocessive behavior observed with individual myosin molecules, we observed numerous processive runs with myosin filaments which lasted for several hundred nanometers, typically until one of the actin-polystyrene beads of the dumbbell was pulled out from the trap or the actin filament broke (Fig. 7b).
FIGURE 7.
Optical trap analysis of NMIIB nonmuscle filament-actin interactions. a, bead position record of full-length NMIIB-actin interaction. In this case the myosin molecules were deposited on nitrocellulose surface in high salt (300 mm KCl AB buffer) buffer, and as a result single, two-headed myosin molecules are probed. No signs of processive runs were observed. b, bead position record of a NMIIB filament-actin interaction. Prior to sample deposition the ionic strength of the buffer was lowered to 150 mm, and as a result the myosin molecules formed filaments. The fluorescent GFP-RLC-labeled NMIIB filaments were clearly visible on top of the pedestals, and during the measurements these pedestals were targeted. The NMIIB filaments show robust motility.
DISCUSSION
NMIIB is an unusual conventional myosin in that solution kinetic studies show that it has an intermediate duty ratio (16, 18). The duty ratio of a myosin is an important determinant of its mechanical properties. Two-headed myosins with high duty ratios such as myosin V have the ability to move processively along actin as single molecules, a very useful feature for a myosin designed to transport cargo in cells (49). Very low duty ratio myosins such as mammalian skeletal muscle myosin are designed to work in ensemble with the hundreds of other myosins polymerized into thick filaments. The low duty ratio allows for rapid shortening of muscle fibers because myosin motors do not remain attached for long after completing a power stroke. The intermediate duty ratio of NMIIB raises questions of how this affects its mechanical properties. Based strictly on calculations using the duty ratio determined in solution kinetic studies, a two-headed NMIIB would not be predicted to be capable of moving processively along actin as a single molecule, but a bipolar filament containing approximately 50 NMIIB motor domains should be capable of processively moving on actin filament.
The aim of the current study was 2-fold. First, we wanted to probe the mechanical properties of NMIIB at a single molecule level. We asked the question how the relatively slow ADP off rate (0.35 s−1) (16) translates to dwell times in single molecule fluorescence and optical trapping experiments. Second, we wanted to characterize the nonmuscle myosin IIB “machinery” at different levels of organization and complexity. We compared the kinetic and mechanical properties of a single- and double-headed myosin fragments and characterized the behavior of full-length NMIIB when it was polymerized into bipolar filaments, which probably represents the in vivo functional unit of NMIIB. We found that single- and double-headed HMM molecules behave very similarly with regard to their power stroke size and dwell time on actin filaments. Instead of using subfragment-1 (S1), which is truncated immediately after the neck region, for a comparison we expressed a single-headed HMM molecule because the attachment of S1 molecules to a specimen surface might compromise the mechanical performance of the molecule. The detachment rates at 1 mm ATP of the two molecule species were very similar and were consistent with the ADP release rates of NMIIB S1 and HMM measured in vitro with transient kinetic techniques (18).
Using optical tweezers, Tyska et al. (50) showed that in case of smooth and skeletal myosins the dwell times of the single- and double-headed molecules were very similar to each other in both unloaded and loaded conditions, which is consistent with our findings for NMIIB HMM. These authors also demonstrated that the stroke sizes of the double-headed molecule smooth and skeletal muscle myosins were approximately double that of the single-headed fragment. They invoked the concept that “two heads are better than one” and suggested that the second, not actively attached myosin head, might serve to orient the active attached head to bring about maximum efficiency of that head. In our case the stroke sizes of the NMIIB-SH-HMM and HMM molecules were similar. The Tyska et al. study created the single-headed myosins by limited proteolytic digestion of intact myosins followed by chromatographic separation of the two species. The proteolytic treatment of myosin often results in cleavage of important surface loops near the nucleotide binding site and the actin binding site. We used a recombinant NMIIB-SH-HMM molecule in which a FLAG-tagged HMM heavy chain was co-expressed with a His-tagged S2 fragment which dimerizes with itself and with the S2 portion of the HMM. Single-headed HMM is obtained by purification on an anti-FLAG-affinity column to eliminate S2 dimers followed by a Ni-column which binds the single-headed-HMM but not the double-headed HMM. By using this construct we avoided a harsh protease treatment, leaving the myosin motor surface loops intact, and could control for an attachment mechanism via the surface-attached anti-GFP antibody which specifically binds the C terminus of the HMM constructs. Another possibility for the difference in our results and those of Tyska et al. might be due to differences in the flexibility of the S2 domains of the various molecules.
There is a wide range (3.5–15 nm) of values reported in the literature for power stroke sizes of various myosin II molecules or their subfragments (29, 38, 42, 43). Some of this variability likely arises from different methods of measuring the power stroke size, but some are probably attributable to the type of surface attachment employed. Because the NMIIB-SH-HMM molecule contains a coiled-coil segment, it is physically bigger than the S1 proteolytic fragment and is more likely to attach to the surface in an orientation that allows the full-length of the lever arm to make the power stroke. Thus, we were able to compare the kinetics and mechanics of attachment of single- and double-headed constructs where both molecules had identical attachment geometries to the surface and identical motor domains presented to the actin filaments. Our results suggest that, at least for NMIIB, the number of heads the molecule contains does not affect the size of the power stroke.
This, in turn, supports the notion that only one of the two heads of HMM is likely to be interacting with actin for the majority of the attachment time in the presence of ATP (51, 52). Support for this comes from the identity of the attachment lifetimes observed in the optical trap and in the TIRF assay for single- and double-headed HMM constructs and in the fact that we saw only one level of variance during attachment events in the trap. A two-headed attachment should give a lower variance than a one-headed attachment because its stiffness should be higher. In fact, such behavior was observed with myosin V, where at stall forces two levels of stiffnesses were apparent (26). However, we cannot rule out a case where initially two heads are attached and the trail head detaches rapidly due to the strain exerted on it by the attached lead head on a time scale too fast for the apparatus to detect.
The observation of two classes of attachment events occurring on separable time scales has not been previously described in optical trapping experiments of myosins. The numerous short lived interactions could have two sources. They may be the result of interactions of the actin filament with the anti-GFP antibody. Nonspecific actin-antibody interactions have been observed previously in our laboratory when trying to use antibodies for specifically attaching myosins in the sliding actin in vitro actin gliding motility assay, and we see less frequent short lived interactions when probing surfaces coated with anti-GFP antibodies in the absence of HMM (Table 2). However, in the current study the short lived interactions were also seen even when anti-GFP antibody was not used as an attachment strategy for NMIIB (Tables 1 and 2) and were the only type of interaction seen when unphosphorylated NMIIB-HMM was probed. More likely the short lived interactions represented the weakly bound, nonproductive interactions of NMIIB with actin filaments (i.e. a high stiffness, no power stroke state). Such interactions were first detected in relaxed skinned muscle fibers which were oscillated at high frequency (53) and were later proposed to arise from nonspecific ionic interactions between loop II of myosin with actin (54). Such interactions were predicted to be the basis for the ability of unphosphorylated smooth muscle myosin to retard the movement of actin filaments by phosphorylated smooth muscle myosin in in vitro actin gliding motility assays (55). The fact that the short lived interactions detected in the present study have no net displacement suggests that they are nonproductive and are probably not associated with the hydrolysis of ATP. These weak binding interactions could function in the cell to help tether and align the myosin filament with the actin filament without expending further ATP.
Norstrom et al. (56) reported that single molecules of NMIIB HMM take multiple forward and backward steps along the actin filament. This result was very intriguing, and we have made a great effort to investigate the possibility that NMIIB-HMM is a processive myosin, but we have failed to demonstrate such behavior utilizing TIRF/in vitro single molecule motility assays and single molecule optical trapping. In TIRF/in vitro single molecule motility experiments the actin filament was attached to the surface, which might interfere with the motion of the NMIIB-HMM molecules because these molecules would have to spiral around the actin filament given their short power strokes. However, we used the same method for actin filament attachment as described by Sakamoto et al. (57). These authors demonstrated that a mutant myosin V HMM molecule with a short neck region, which binds only two calmodulin molecules per chimeric heavy chain and was thus similar in length to the neck of NMIIB HMM, was able to move processively along the surface attached actin filament even though its step size was only 11 nm (57). Thus, this geometry of actin filament attachment should not limit the movement of myosin.
We observed no obvious processive movements in the optical trap utilizing a similar configuration to the assay used by Norstrom et al. Most of our data were collected while oscillating the actin-attached beads to facilitate detection of events, but even in the absence of oscillations, no processive movements were observed. Similarly, varying the trap stiffness values (0.01–0.022 piconewton/nm) and ATP concentrations (1 μm and 1 mm) did not result in processive movements.
The myosin molecule is subject to low but finite loading forces in the optical trap, and these forces may hinder the ability of the myosin to move processively along actin. In contrast, there is no external load in the single molecule TIRF motility assay, and yet we did not observe processive runs in this assay either. One could argue that if the NMIIB-HMM were taking 7-nm steps, then a large number of steps would be required for movement of the fluorescent spot to be observed in this assay. However, two lines of evidence argue against this. First, the lifetimes of attachment in the single molecule TIRF assay system were virtually identical for single-headed and double-headed HMM molecules, and the values for each of these were very similar to the values obtained in the optical trap where it could be reliably determined that only single power strokes were being made. Second, we used a superresolution light microscopy technique, FIONA (48), which can detect 7-nm steps if they were occurring. No individual movements >2 nm were observed, which is smaller than the positional error in the measurement system. Thus, we find no evidence by two different single molecule assays that NMIIB HMM molecules are processive. It should be noted that our studies used human NMIIB molecules whereas Norstrom et al. (56) used chicken NMIIB molecules.
There is no evidence in cells that NMIIB functions as single molecules. Instead, it is more likely that the functional unit is the short bipolar filaments such as seen in vitro by electron microscopy in the current study and from images of cells in culture (30). It is important that this functional unit behaves processively, and indeed, this behavior is seen both in the ability of single NMIIB myosin filaments to move processively along actin and in its ability to translocate actin in the optical trap when the myosin filament is fixed to the surface. In this regard, it is interesting to note that the much larger muscle myosin filaments have also been shown to move actin filaments processively in vitro (58–60).
The rate of movement of NMIIB myosin filaments, 48 nm/s, in this assay appears to be too fast to be explained by the kinetics of the single molecules measured herein. In the simplest case the expected velocity would be given by the stroke size times the cycle time or 7 nm × 0.4 s−1 = ∼3 nm/s. However, two factors complicate this simple assumption. First, Kovacs et al. (61) showed that positive strain increases the rate of ADP release from NMIIB heads by a factor of approximately 4. The magnitude of this increase might increase further if larger positive strains are applied to an attached ADP-bound. A second possible explanation arises for the architecture of the filament itself. EM images show that the myosin heads at the end of a bipolar filament can be spread out over a large (based on EM images at least 1 order of magnitude greater than the power stroke size) distance and should be able to explore a relatively large distance along a single actin filament.3 We propose as the nonmuscle myosin filament moves on the actin filament, the attached heads at the front bias the diffusion of the trailing heads. After detachment these heads might rebind to the actin filament far from their original point of attachment and hence give a much larger apparent step size than would be possible on a single molecule level. This could translate into a faster velocity.
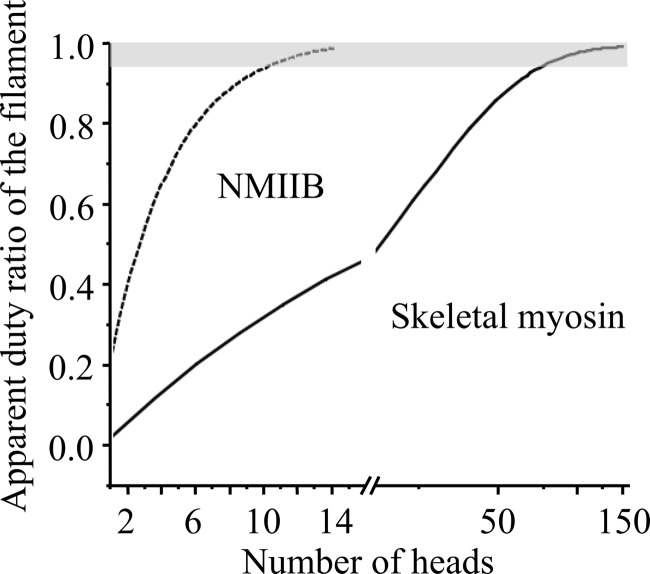
The duty ratio of the myosin V molecules is high (25, 26), and a theoretical model suggests that the “cumulative” or apparent duty ratio of the skeletal muscle thick filament is also close to unity (62). In a simple model, a myosin head is bound to actin with probability r (technically r is the duty ratio), and consequently, the probability of the detached state is (1−r). In this overly simplified model the strain has no apparent effect on attachment or detachment rates. In case of an HMM molecule (two independently acting heads), the probability that none of the heads is bound to actin is (1−r)2. As we increase the number of heads (n), the apparent duty ratio of the system will increase, and the probability of the detached state (1−r)n, will exponentially decrease. In Fig. 8 this apparent duty ratio of skeletal (r = 0.04) (63) and NMIIB (r = 0.22) (61) is plotted as a function of the number of the myosin heads. Even though this overly simple model operates solely with the numerical values of the duty ratios of myosin motors and number of heads, it shows that NMIIB reaches the high duty ratio regime when the number of heads is approximately a dozen, whereas the skeletal myosin turns into a processive system when the number of heads reaches approximately 120. These numbers are in a surprisingly good agreement with the numbers of individual heads of NMIIB molecules in nonmuscle filaments and skeletal myosin molecules in thick filaments. Our experimental findings and the above calculations clearly show that processive molecular systems can be built from nonprocessive building blocks, and the minimal number of required building blocks, in our case myosin molecules, is determined by the duty ratio of individual molecules.
FIGURE 8.
Dependence of the duty ratio on number of myosin heads for skeletal muscle and NMIIB. In case of NMIIB (duty ratio is ∼0.22) the system reaches the processive zone (depicted in gray) when the number of available heads is ∼12–13. In case of skeletal myosin (duty ratio is 0.04) this number is 1 order of magnitude higher.
Future experiments are required to reveal the effect of load on the duty ratio of NMIIB. Kovacs et al. used transient kinetic techniques to demonstrate that load modulates the rate of ADP release from actin-bound myosin heads (61). Laakso et al. (43) showed the kinetic parameters and the duty ratio of myosin Ib motor strongly depend on tension. Such a behavior would not be unexpected in case of a myosin motor which is responsible for maintaining cortical tension (65). Load has also been shown to affect the kinetics of other myosins in the optical trap (38, 64, 66, 67). Norstrom et al. (56) predicted that the backward load would increase the detachment rate, but our initial experiments do not show this effect. Also, it is not known yet how many myosin heads interact with the actin molecule at any given moment or what is the force output of the individual nonmuscle filaments. The combination of optical tweezers with TIRF microscopy might be the right tool to answer these questions.
Acknowledgments
We thank Dr. Fang Zhang for expert technical assistance, the members of the Laboratory of Molecular Physiology for support and advice, and Dr. Zenon Grabarek (Boston Biomedical Research Institute) for the clone of myosin light chain kinase.
This work was supported, in whole or part, by National Institutes of Health intramural funds from the NHLBI.

This article contains supplemental Movies 1–3.
N. Billington, A. Wang, R. S. Adelstein, and J. R. Sellers, unpublished data.
- NMIIB
- nonmuscle myosin IIB
- ELC
- essential light chain
- FIONA
- fluorescence imaging with 1-nm accuracy
- HMM
- heavy meromyosin
- MLCK
- myosin light chain kinase
- RLC
- regulatory light chain
- S1 and S2
- subfragment-1 and -2, respectively
- SH
- single-headed
- TIRF
- total internal reflection fluorescence.
REFERENCES
- 1. Odronitz F., Kollmar M. (2007) Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8, R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de la Fuente H., Mittelbrunn M., Sánchez-Martín L., Vicente-Manzanares M., Lamana A., Pardi R., Cabañas C., Sánchez-Madrid F. (2005) Synaptic clusters of MHC class II molecules induced on DCs by adhesion molecule-mediated initial T-cell scanning. Mol. Biol. Cell 16, 3314–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lo C. M., Buxton D. B., Chua G. C., Dembo M., Adelstein R. S., Wang Y. L. (2004) Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol. Biol. Cell 15, 982–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Betapudi V., Licate L. S., Egelhoff T. T. (2006) Distinct roles of nonmuscle myosin II isoforms in the regulation of MDA-MB-231 breast cancer cell spreading and migration. Cancer Res. 66, 4725–4733 [DOI] [PubMed] [Google Scholar]
- 5. Vicente-Manzanares M., Zareno J., Whitmore L., Choi C. K., Horwitz A. F. (2007) Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 176, 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shewan A. M., Maddugoda M., Kraemer A., Stehbens S. J., Verma S., Kovacs E. M., Yap A. S. (2005) Myosin 2 is a key Rho kinase target necessary for the local concentration of E-cadherin at cell-cell contacts. Mol. Biol. Cell 16, 4531–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ivanov A. I., Samarin S. N., Bachar M., Parkos C. A., Nusrat A. (2009) Protein kinase C activation disrupts epithelial apical junctions via ROCK-II dependent stimulation of actomyosin contractility. BMC Cell Biol. 10, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conti M. A., Even-Ram S., Liu C., Yamada K. M., Adelstein R. S. (2004) Defects in cell adhesion and the visceral endoderm following ablation of nonmuscle myosin heavy chain II-A in mice. J. Biol. Chem. 279, 41263–41266 [DOI] [PubMed] [Google Scholar]
- 9. Pollard T. D. (2010) Mechanics of cytokinesis in eukaryotes. Curr. Opin. Cell Biol. 22, 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heissler S. M., Manstein D. J. (2012) Nonmuscle myosin-2: mix and match. Cell Mol. Life Sci., DOI 10.1007/s00018-012-1002-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vicente-Manzanares M., Ma X., Adelstein R. S., Horwitz A. R. (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conti M. A., Adelstein R. S. (2008) Nonmuscle myosin II moves in new directions. J. Cell Sci. 121, 11–18 [DOI] [PubMed] [Google Scholar]
- 13. Kamm K. E., Stull J. T. (2001) Dedicated myosin light chain kinases with diverse cellular functions. J. Biol. Chem. 276, 4527–4530 [DOI] [PubMed] [Google Scholar]
- 14. Matsumura F. (2005) Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 15, 371–377 [DOI] [PubMed] [Google Scholar]
- 15. Vicente-Manzanares M., Rey M., Pérez-Martinez M., Yáñez-Mó M., Sancho D., Cabrero J. R., Barreiro O., de la Fuente H., Itoh K., Sánchez-Madrid F. (2003) The RhoA effector mDia is induced during T cell activation and regulates actin polymerization and cell migration in T lymphocytes. J. Immunol. 171, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 16. Wang F., Kovacs M., Hu A., Limouze J., Harvey E. V., Sellers J. R. (2003) Kinetic mechanism of non-muscle myosin IIB: functional adaptations for tension generation and maintenance. J. Biol. Chem. 278, 27439–27448 [DOI] [PubMed] [Google Scholar]
- 17. Kovacs M., Wang F., Hu A., Zhang Y., Sellers. J. R. (2003) Functional divergence of human cytoplasmic myosin II: kinetic characterization of the non-muscle IIA isoform. J. Biol. Chem. 278, 38132–38140 [DOI] [PubMed] [Google Scholar]
- 18. Rosenfeld S. S., Xing J., Chen L. Q., Sweeney H. L. (2003) Myosin IIb is unconventionally conventional. J. Biol. Chem. 278, 27449–27455 [DOI] [PubMed] [Google Scholar]
- 19. Heissler S. M., Manstein D. J. (2011) Comparative kinetic and functional characterization of the motor domains of human nonmuscle myosin-2C isoforms. J. Biol. Chem. 286, 21191–21202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Margossian S. S., Lowey S. (1973) Substructure of the myosin molecule. 3. Preparation of single-headed derivatives of myosin. J. Mol. Biol. 74, 301–311 [DOI] [PubMed] [Google Scholar]
- 21. Kawamoto S., Adelstein R. S. (1991) Chicken nonmuscle myosin heavy chains: differential expression of two mRNAs and evidence for two different polypeptides. J. Cell Biol. 112, 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maupin P., Phillips C. L., Adelstein R. S., Pollard T. D. (1994) Differential localization of myosin-II isozymes in human cultured cells and blood cells. J. Cell Sci. 107, 3077–3090 [DOI] [PubMed] [Google Scholar]
- 23. Rochlin M. W., Itoh K., Adelstein R. S., Bridgman P. C. (1995) Localization of myosin II A and B isoforms in cultured neurons. J. Cell Sci. 108, 3661–3670 [DOI] [PubMed] [Google Scholar]
- 24. Kelley C. A., Sellers J. R., Gard D. L., Bui D., Adelstein R. S., Baines I. C. (1996) Xenopus nonmuscle myosin heavy chain isoforms have different subcellular localizations and enzymatic activities. J. Cell Biol. 134, 675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De La Cruz E. M., Wells A. L., Rosenfeld S. S., Ostap E. M., Sweeney H. L. (1999) The kinetic mechanism of myosin V. Proc. Natl. Acad. Sci. U.S.A. 96, 13726–13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Veigel C., Wang F., Bartoo M. L., Sellers J. R., Molloy J. E. (2002) The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 4, 59–65 [DOI] [PubMed] [Google Scholar]
- 27. Geeves M. A., Holmes K. C. (1999) Structural mechanism of muscle contraction. Annu. Rev. Biochem. 68, 687–728 [DOI] [PubMed] [Google Scholar]
- 28. Holmes K. C., Geeves M. A. (2000) The structural basis of muscle contraction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niederman R., Pollard T. D. (1975) Human platelet myosin. II. In vitro assembly and structure of myosin filaments. J. Cell Biol. 67, 72–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Verkhovsky A. B., Svitkina T. M., Borisy G. G. (1995) Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J. Cell Biol. 131, 989–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kengyel A., Wolf W. A., Chisholm R. L., Sellers J. R. (2010) Nonmuscle myosin IIA with a GFP fused to the N terminus of the regulatory light chain is regulated normally. J. Muscle Res. Cell Motil. 31, 163–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang F., Harvey E. V., Conti M. A., Wei D., Sellers J. R. (2000) A conserved negatively charged amino acid modulates function in human nonmuscle myosin IIA. Biochemistry 39, 5555–5560 [DOI] [PubMed] [Google Scholar]
- 33. Kural C., Balci H., Selvin P. R. (2005) Molecular motors one at a time: FIONA to the rescue. J. Phys. Condens. Matter 17, S3979–3995 [DOI] [PubMed] [Google Scholar]
- 34. Snyder G. E., Sakamoto T., Hammer J. A., 3rd, Sellers J. R., Selvin P. R. (2004) Nanometer localization of single green fluorescent proteins: evidence that myosin V walks hand-over-hand via telemark configuration. Biophys. J. 87, 1776–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Finer J. T., Simmons R. M., Spudich J. A. (1994) Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature 368, 113–119 [DOI] [PubMed] [Google Scholar]
- 36. Molloy J. E., Burns J. E., Kendrick-Jones J., Tregear R. T., White D. C. (1995) Movement and force produced by a single myosin head. Nature 378, 209–212 [DOI] [PubMed] [Google Scholar]
- 37. Vanzi F., Takagi Y., Shuman H., Cooperman B. S., Goldman Y. E. (2005) Mechanical studies of single ribosome/mRNA complexes. Biophys. J. 89, 1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takagi Y., Homsher E. E., Goldman Y. E., Shuman H. (2006) Force generation in single conventional actomyosin complexes under high dynamic load. Biophys. J. 90, 1295–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kron S. J., Spudich J. A. (1986) Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. U.S.A. 83, 6272–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baboolal T. G., Sakamoto T., Forgacs E., White H. D., Jackson S. M., Takagi Y., Farrow R. E., Molloy J. E., Knight P. J., Sellers J. R., Peckham M. (2009) The SAH domain extends the functional length of the myosin lever. Proc. Natl. Acad. Sci. U.S.A. 106, 22193–22198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Veigel C., Bartoo M. L., White D. C., Sparrow J. C., Molloy J. E. (1998) The stiffness of rabbit skeletal actomyosin cross-bridges determined with an optical tweezers transducer. Biophys. J. 75, 1424–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Batters C., Wallace M. I., Coluccio L. M., Molloy J. E. (2004) A model of stereocilia adaptation based on single molecule mechanical studies of myosin I. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 1895–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Laakso J. M., Lewis J. H., Shuman H., Ostap E. M. (2008) Myosin I can act as a molecular force sensor. Science 321, 133–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Knight A. E., Veigel C., Chambers C., Molloy J. E. (2001) Analysis of single-molecule mechanical recordings: application to acto-myosin interactions. Prog. Biophys. Mol. Biol. 77, 45–72 [DOI] [PubMed] [Google Scholar]
- 45. Tyska M. J., Warshaw D. M. (2002) The myosin power stroke. Cell Motil. Cytoskeleton 51, 1–15 [DOI] [PubMed] [Google Scholar]
- 46. Yanagida T., Kitamura K., Tanaka H., Hikikoshi Iwane A., Esaki S. (2000) Single molecule analysis of the actomyosin motor. Curr. Opin. Cell Biol. 12, 20–25 [DOI] [PubMed] [Google Scholar]
- 47. Guzik-Lendrum S., Nagy A., Takagi Y., Houdusse A., Sellers J. R. (2011) Drosophila melanogaster myosin-18 represents a highly divergent motor with actin tetering properties. J. Biol. Chem. 286, 21755–21766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yildiz A., Selvin P. R. (2005) Fluorescence imaging with one nanometer accuracy: application to molecular motors. Acc. Chem. Res. 38, 574–582 [DOI] [PubMed] [Google Scholar]
- 49. Hammer J. A., 3rd, Sellers J. R. (2012) Walking to work: roles for class V myosins as cargo transporters. Nat. Rev. Mol. Cell Biol. 13, 13–26 [DOI] [PubMed] [Google Scholar]
- 50. Tyska M. J., Dupuis D. E., Guilford W. H., Patlak J. B., Waller G. S., Trybus K. M., Warshaw D. M., Lowey S. (1999) Two heads of myosin are better than one for generating force and motion. Proc. Natl. Acad. Sci. U.S.A. 96, 4402–4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Greene L. E. (1981) Comparison of the binding of heavy meromyosin and myosin subfragment 1 in F-actin. Biochemistry 20, 2120–2126 [DOI] [PubMed] [Google Scholar]
- 52. Greene L. E., Sellers J. R. (1987) Effect of phosphorylation on the binding of smooth muscle heavy meromyosin X ADP to actin. J. Biol. Chem. 262, 4177–4181 [PubMed] [Google Scholar]
- 53. Brenner B., Schoenberg M., Chalovich J. M., Greene L. E., Eisenberg E. (1982) Evidence for cross-bridge attachment in relaxed muscle at low ionic strength. Proc. Natl. Acad. Sci. U.S.A. 79, 7288–7291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Furch M., Remmel B., Geeves M. A., Manstein D. J. (2000) Stabilization of the actomyosin complex by negative charges on myosin. Biochemistry 39, 11602–11608 [DOI] [PubMed] [Google Scholar]
- 55. Cuda G., Pate E., Cooke R., Sellers J. R. (1997) In vitro actin filament sliding velocities produced by mixtures of different types of myosin. Biophys. J. 72, 1767–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Norstrom M. F., Smithback P. A., Rock R. S. (2010) Unconventional processive mechanics of non-muscle myosin IIB. J. Biol. Chem. 285, 26326–26334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sakamoto T., Yildez A., Selvin P. R., Sellers J. R. (2005) Step-size is determined by neck length in myosin V. Biochemistry 44, 16203–16210 [DOI] [PubMed] [Google Scholar]
- 58. Sellers J. R., Kachar B. (1990) Polarity and velocity of sliding filaments: control of direction by actin and of speed by myosin. Science 249, 406–408 [DOI] [PubMed] [Google Scholar]
- 59. West J. M., Higuchi H., Ishijima A., Yanagida T. (1996) Modification of the bi-directional sliding movement of actin filaments along native thick filaments isolated from a clam. J. Muscle Res. Cell Motil. 17, 637–646 [DOI] [PubMed] [Google Scholar]
- 60. Previs M. J., Beck Previs S., Gulick J., Robbins J., Warshaw D. M. (2012) Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science 337, 1215–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kovács M., Thirumurugan K., Knight P. J., Sellers J. R. (2007) Load-dependent mechanism of nonmuscle myosin 2. Proc. Natl. Acad. Sci. U.S.A. 104, 9994–9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Smith D. A. (2004) How processive is the myosin-V motor? J. Muscle Res. Cell Motil. 25, 215–217 [DOI] [PubMed] [Google Scholar]
- 63. Harris D. E., Warshaw D. M. (1993) Smooth and skeletal muscle myosin both exhibit low duty cycles at zero load in vitro. J. Biol. Chem. 268, 14764–14768 [PubMed] [Google Scholar]
- 64. Veigel C., Molloy J. E., Schmitz S., Kendrick-Jones J. (2003) Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat. Cell Biol. 5, 980–986 [DOI] [PubMed] [Google Scholar]
- 65. Bridgman P. C., Dave S., Asnes C. F., Tullio A. N., Adelstein R. S. (2001) Myosin IIB is required for growth cone motility. J. Neurosci. 21, 6159–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Veigel C., Schmitz S., Wang F., Sellers J. R. (2005) Load-dependent kinetics of myosin-V can explain its high processivity. Nat. Cell Biol. 7, 861–869 [DOI] [PubMed] [Google Scholar]
- 67. Greenberg M. J., Lin T., Goldman Y. E., Shuman H., Ostap E. M. (2012) Myosin IC generates power over a range of loads via a new tension-sensing mechanism. Proc. Natl. Acad. Sci. U.S.A. 109, E2433–E2440 [DOI] [PMC free article] [PubMed] [Google Scholar]