Background: STAT2 is a key transcription factor that mediates the protective role of type I interferons in host defense.
Results: Type I interferons induce the phosphorylation of STAT2 at serine 287.
Conclusion: Serine 287-STAT2 is a regulatory site involved in modulating the transcriptional and cellular responses to type I interferons.
Significance: Deregulated STAT2 signaling may contribute to heightened type I interferon responses and susceptibility to many diseases.
Keywords: Chromatin, Interferon, Protein Phosphorylation, STAT Transcription Factor, Virus
Abstract
STAT2 is a positive modulator of the transcriptional response to type I interferons (IFNs). STAT2 acquires transcriptional function by becoming tyrosine phosphorylated and imported to the nucleus following type I IFN receptor activation. Although most STAT proteins become dually phosphorylated on specific tyrosine and serine residues to acquire full transcriptional activity, no serine phosphorylation site in STAT2 has been reported. To find novel phosphorylation sites, mass spectrometry of immunoprecipitated STAT2 was used to identify several phosphorylated residues. Of these, substitution of serine 287 with alanine (S287A) generated a gain-of-function mutant that enhanced the biological effects of IFN-α. S287A-STAT2 increased cell growth inhibition, prolonged protection against vesicular stomatitis virus infection and enhanced transcriptional responses following exposure of cells to IFN-α. In contrast, a phosphomimetic STAT2 mutant (S287D) produced a loss-of-function protein that weakly activated IFN-induced ISGs. Our mechanistic studies suggest that S287A-STAT2 likely mediates its gain-of-function effects by prolonging STAT2/STAT1 dimer activation and retaining it in transcriptionally active complexes with chromatin. Altogether, we have uncovered that in response to type I IFN, STAT2 is serine phosphorylated in the coiled-coil domain that when phosphorylated can negatively regulate the biological activities of type I IFNs.
Introduction
Type I interferons (IFN-α/β) are pleiotropic cytokines that induce the expression of multiple genes with antiviral, antiproliferative and immunomodulatory activities (1). It is widely recognized that the transcriptional responses to type I IFNs are triggered by activation of the canonical JAK/STAT pathway. Binding of type I IFNs to the IFNAR1-IFNAR2 heterodimeric receptor complex activates the receptor-associated tyrosine kinases JAK1 and TYK2, which, in turn, transphosphorylate the intracellular domains of IFNAR1 and IFNAR2, thus creating binding sites for the transcription factors STAT1 and STAT2 (2). JAK1 and TYK2 phosphorylate STAT1 at tyrosine (Tyr)-701 and STAT2 at Tyr-690 in human cells (3–5). The ternary IFN-stimulated gene factor 3 (ISGF3)2 transcriptional complex is then formed consisting of STAT1 and STAT2 bound as heterodimers via reciprocal phosphotyrosyl-SH2 interactions together with the IFN regulatory factor-9 (IRF9). As a result, ISGF3 translocates to the nucleus and binds to the interferon-stimulated response element (ISRE) of interferon-stimulated genes (ISGs) and initiates gene transcription (6, 7).
A common feature shared by all members of the STAT family is a requirement of ligand-induced phosphorylation of a conserved tyrosine residue, located N-terminal of the transactivation domain, for their activation (3, 5, 8–12). A second phosphorylation event occurs on a serine residue 20 amino acids or more C-terminal of the activating tyrosine site, mapped to the transactivation domain. Phosphorylation of this residue influences the transcriptional activity of STAT proteins (13–17). In the case of STAT1, phosphorylation of Tyr-701 and translocation to the nucleus are both required for its stable association with chromatin and the ensuing phosphorylation of Ser-727 (18). Unlike the other members of the STAT family, no such serine phosphorylation event has been reported in STAT2.
Tyrosine phosphorylation is not the only known IFN-inducible post-transcriptional modification in STAT2. In addition to STAT1 and IRF9, IFN-α induces the acetylation of STAT2 at lysine 390 by cAMP responsive element-binding protein (19). Acetylated Lys-390 alters the interaction between STAT2 and STAT1, thereby facilitating IFN-induced activation and antiviral activity. The transcriptional activity of STAT2 can also be modulated by a stretch of amino acid residues mapped to the SH2 domain of STAT2. Our laboratory identified a novel motif consisting of proline 630, tyrosine 631, threonine 632, and lysine 633 that we termed PYTK and is conserved in STAT1 and STAT3 (20, 21). A point mutation of Pro-630 to leucine surfaced under IFN-α selection pressure and was found to abrogate IFN-α induced apoptosis, whereas mutation of Y631 to phenylalanine had the opposite effect as it promoted the apoptotic effects of IFN-α. These observations indicate that the function of STAT2 is dictated by specific residues, with some being targeted for post-translational modifications.
Our study identifies for the first time that STAT2 is serine phosphorylated in an IFN-dependent manner at Ser-287, a residue mapped to the coiled-coil domain of STAT2. This novel post-translational event is important for regulating STAT2/ISGF3 signaling. Mutating Ser-287 to alanine (S287A) generated a gain-of-function STAT2 mutant that augmented the anti-proliferative and antiviral response to IFN-α, whereas a phosphomimetic mutation of Ser-287 dramatically impaired the transcriptional activity of STAT2. Our findings support the existence of a novel regulatory serine residue in STAT2 that when phosphorylated affects type I IFN signaling.
EXPERIMENTAL PROCEDURES
Cell Lines, Cell Culture Reagents, and Antibodies
STAT2-deficient human U6A fibrosarcoma cells, kindly obtained from Dr. Ana Costa-Pereira (Imperial College London, London, UK), and 293FT cells were cultured in DMEM (Cellgro Mediatech, Inc., Manassas, VA) supplemented with 10% fetal calf serum (Gemini Bio-Products), 100 units/ml penicillin (Cellgro), 100 μg/ml streptomycin (Cellgro), and 1× GlutaMAX (Invitrogen) at 37 °C in a humidified atmosphere containing 5% CO2. Recombinant human IFN-α-2a, (specific activity 2 × 107 units/ml), was purchased from PeproTech, Inc. (Rock Hill, NJ). Rabbit anti-STAT2 (C-20), mouse anti-STAT2 (A-7), rabbit anti-STAT1 (E-23), and rabbit anti-ISGF3γ p48 (IRF9; C-20) were purchased from Santa Cruz Biotechnology (Santa Cruz). Rabbit anti-phospho-Tyr-689-STAT2 (07-224) and rabbit anti-acetyl histone H3 (17–615) were obtained from Millipore (Bedford, MA), mouse anti-STAT1 (phospho-Tyr-701) was purchased from BD Biosciences, mouse anti-GAPDH was purchased from Proteintech Group, Inc. (Chicago, IL), and mouse anti-actin was obtained from Abcam (Cambridge, MA).
Plasmids and Mutagenesis
FLAG-tagged human STAT2 in pCDNA3 vector (provided by Dr. Curt Horvath, Northwestern University, Evanston, IL) was subcloned into pIRES2-EGFP (Clontech, Mountain View, CA) and subsequently used as DNA template to introduce single mutations with the QuikChange II site-directed mutagenesis kit (Agilent Technologies). The following primer sets synthesized by Bioneer Inc. (Alameda, CA) were used: serine 287 > alanine (S287A), 5′-GACTGAGTTGCCTGGTTGCCTATCAGGATGACCCTC-3′ and 5′-GAGGGTCATCCTGATAGGCAACCAGGCAACTCAGTC-3′; serine 287 > aspartic acid (S287D), 5′-GACTGAGTTGCCTGGTTGACTATCAGGATGACCCTC-3′ and 5′-GAGGGTCATCCTGATAGTCAACCAGGCAACTCAGTC-3′. Mutagenesis was confirmed by DNA sequencing of entire STAT2. Mutated and wild type (WT) STAT2 were also subcloned into the lentivirus expression vector pCPP to transduce U6A cells and obtain pooled cell populations with reconstituted STAT2. The pCPP vector is a modified version of the pHR′-CMV-lacZ in which lacZ was removed and replaced with a multiple cloning site (22). pCPP, the packaging plasmid pCMV-dR8.2 dvpr, and the envelope plasmid pCMV-VSV-G were kindly provided by Dr. Alexander Y. Tsygankov and Dr. Xavier Graña (Temple University, Philadelphia, PA). The use of lentiviral vectors was approved by Temple University under National Institutes of Health guidelines.
Transfection and Transduction
U6A cells stably expressing human STAT2 were generated by transfecting U6A cells with pIRES2-EGFP encoding WT or mutant STAT2 using TurboFect (Fermentas, Inc., Burlington, Canada) transfection reagent according to the manufacturer's instructions. Twenty-four hours post-transfection, cells were reseeded at a lower density, and G418 (Mediatech, Inc.) was added to a final concentration of 500 μg/ml to select for STAT2-expressing cells. Individual colonies were picked, expanded, and screened for STAT2-expression by immunoblotting. Clones stably expressing STAT2 were maintained in complete DMEM supplemented with 300 μg/ml of G418. To make STAT2 lentiviral particles, 293FT-cells were co-transfected with pCPP-STAT2, pCMV-dR8.2 dvpr, and pCMV-VSV-G at a ratio of 6:4:3, respectively. Supernatants containing lentivirus were collected after 48 h, passed through a 0.45-μm filter, and subsequently added to U6A cells together with 10 μg/ml polybrene (hexadimethrine bromide; Sigma-Aldrich) for 24 h. Infected cells were placed under selection 48 h post-infection by adding 2 μg/ml puromycin (Sigma-Aldrich). Single clones and pooled populations of U6A cells reconstituted with different STAT2 constructs were both used in our studies and yielded the same responses to IFN stimulation.
Mass Spectrometry and Data Analysis
U6A cells stably expressing WT-STAT2 were left untreated or treated with 1000 units/ml IFN-α for 30 min. STAT2 was immunoprecipitated with anti-STAT2 antibody (C-20), and immune complexes were resolved on 4–12% NuPAGE Bis-Tris gels (Invitrogen) and visualized using Coomassie Blue stain. A protein band corresponding to the molecular weight of STAT2 was excised and subjected to surface enhanced laser desorption/ionization time-of-flight mass spectrometry. Mass spectrometry of STAT2 was performed by ITSI Biosciences (Johnstown, PA) using an LTQ XL mass spectrometer (Thermo Scientific, Rockford, IL). Resulting mass spectra was searched against a custom STAT2_human database from UniProt using Proteome Discoverer (version 1.2, Thermo Scientific) and SEAQUEST algorithms. Proteins were identified when two or more unique peptides had X-correlation scores greater than 1.5, 2.0, and 2.5 for respective peptide charge states of +1, +2, and +3, and Δ score was ≥ 0.1. Methylated C termini, carbamidomethyl cysteine, and methylated glutamic and aspartic acid residues were used as static modifications. Phosphorylated serine, threonine, and tyrosine along with oxidation of methionine were variable modifications. Two missed cleavages were allowed, and mass tolerance was set to 1 Da.
Cell Extract Preparation, Immunoprecipitation, and Immunoblotting
Cells were washed in ice-cold phosphate-buffered saline (PBS) and resuspended in lysis buffer (50 mm Tris pH 7.5, 150 mm NaCl, 2 mm EDTA, 0.5% Triton X-100) supplemented with 1× protease inhibitor mixture (Roche Diagnostics), 1 mm sodium orthovanadate, 1 mm PMSF, and 1× phosphatase inhibitor mixtures 1 and 2 (Sigma) for 15 min. After centrifugation, supernatants were collected, and protein concentrations were measured using Bio-Rad protein assay kit (Bio-Rad). In some experiments, cell extracts were subjected to immunoprecitation with anti-IRF9 antibodies. Immunocomplexes were precipitated using protein G Sepharose (GE Healthcare). Lysates or immunoprecipitates were separated on NuPAGE 4–12% Bis-Tris gels. Proteins were transferred to PVDF membrane (Millipore), blocked in blocker casein TBS (Thermo Scientific), and incubated with primary antibodies overnight at 4 °C in TBS+3% chicken egg albumin (Sigma-Aldrich) followed by incubation with anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies (1:10,000; Invitrogen) for 1 h in room temperature. Membranes were developed by chemiluminescence using SuperSignal West Pico (Thermo Scientific). Quantification of protein bands was performed using ΑlphaView software (ProteinSimple, CA).
cDNA Synthesis and Quantitative Real-time PCR
Total RNA was isolated from U6A cells incubated with IFN-α (1000 units/ml) for the indicated times using TRIzol reagent (Invitrogen). Contaminating DNA was removed with DNA-free kit (Ambion), and cDNA was synthesized from total RNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, CA). cDNA (50 ng) was mixed with 2x TaqMan Master Mix (Applied Bioscience) and gene-specific FAM dye-labeled TaqMan MGB probes corresponding to IFN-inducible genes and ran in triplicate wells. Real-time analysis was performed using a 7300 real time PCR system (Applied Biosystems). Actin cDNA was also measured to normalize for differences in cDNA concentration.
Virus Infections
Cells were seeded in 12-well plates and left untreated or treated with increasing concentrations of IFN-α for 16 h prior to infection with green fluorescent protein (GFP)-expressing vesicular stomatitis virus (VSV) (23). Plaque-purified VSV was added to cells at a multiplicity of infection of 0.01 under serum-free medium conditions for 1 h at 37 °C. Cells were then washed twice with PBS followed by readdition of complete DMEM. Infected cells were visualized using a Zeiss Axiovert 40 CFC (Carl Zeiss, Inc.) inverted fluorescent microscope, and progeny virion titers in culture supernatants were quantified by standard plaque assay on baby hamster kidney (BHK-21) cells.
Cell Growth Inhibition Assay
Cells were seeded in flat bottom 96-well plates at a density of 500 cells in 100 μl volume/well. Cells were stimulated with or without IFN-α (100, 400, or 1000 units/ml) and incubated for 72 h at 37 °C. Cell proliferation was determined by using CellTiter 96® AQueous One solution reagent (Promega, Madison, WI) according to the manufacturer's instructions. Absorbance was measured at 490 nm using a VICTORTMX5 Multilabel Plate Reader (PerkinElmer Life Sciences). Background values were subtracted from each well before proceeding with determination of percent cell growth inhibition. Data are shown as % of control (untreated cells).
Subcellular Fractionation and Nuclear Localization of STAT2
Cells were washed twice in cold PBS and resuspended for 15 min in cold hypotonic buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm sodium orthovanadate, 10 mm β-glycerophosphate, 1 mm DTT, 0.5 mm PMSF, 1× protease inhibitor mixture) before adding Triton X-100 to a final concentration of 0.6%. Samples were then vortexed vigorously and centrifuged at 1000 × g at 4 °C. Supernatants were collected as cytoplasmic fractions. Next, nuclear pellets were resuspended for 15 min in hypertonic buffer (20 mm HEPES, pH 7.9, 400 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm sodium orthovanadate, 10 mm β-glycerophosphate, 1 mm DTT, 1 mm PMSF, 1× protease inhibitor mixture), and subsequently centrifuged for 5 min at 15,000 × g. Resulting supernatants were collected as nuclear fractions. Protein concentration was measured by Bio-Rad Protein Assay. Localization and activation of STAT2 was analyzed in cytoplasmic and nuclear fractions by immunoblot analysis.
EMSA
Five μg of nuclear protein was mixed with 32P-labeled double-stranded oligonucleotide probe corresponding to the IFN-γ activated sequence (GAS) of the FcγRI gene or an ISRE sequence derived from the promoter of the ISG15 gene as described previously (20, 21). Rabbit anti-STAT2 (C-20) and rabbit anti-STAT1 (E-23) were used for supershifting STAT2 and STAT1, respectively. The corresponding binding buffer was added (ISRE probe (40 mm KCl, 20 mm HEPES, pH 7.0, 1 mm MgCl2, 0.5 mm DTT, 4% Ficoll, 0.02% Triton X-100); for GAS probe (10 mm Tris, pH 7.4, 5 mm MgCl2, 100 mm KCl, 1 mm DTT, 10% glycerol)), and samples were incubated at room temperature for 30 min. Loading buffer (40% glycerol, bromphenol blue, xylene cyanole FF, 250 mm Tris-HCl, pH 7.5) was added, and samples were separated on a 5% native polyacrylamide gel. Gels were vacuum-dried and developed using x-ray autoradiography film.
Chromatin Immunoprecipitation Analysis
Chromatin immunoprecipitation (ChIP) assay kit (Millipore) was used to detect protein-chromatin interactions according to manufacturer's instructions. The immunoprecipitated chromatin DNA was used as template to amplify the ISRE-responsive elements in the promoters of ISGs. DNA from cell lysates taken before immunoprecipitation was used as input control. DNA primers for quantitative ChIP analysis were purchased from Applied Biosystems. Primers sequences used for quantitative ChIP analysis were as follows: IFIT2 promoter, 5′-CTTTCCCTTTTGTAACGTCAGC-3′, 5′-TGCACTCTTCAGAAATCTTCCTC-3′ and probe, 5′-AGGTCTCTTCAGCATTTATTGGTGGCA-3′; OAS1 promoter, 5′-GTTGGCTGGAGGTTAAAATGC-3′ and 5′-TCTGCTTCCTGAAACTTACCC-3′ and probe, 5′-CAGAGTTCAGAGAAAGGCTGGGCT-3′; ISG15 promoter, 5′-CCACTTTTGCTTTTCCCTGTC-3′, 5′-AGTTTCGGTTTCCCTTTCCC-3′, and probe: 5′-ATGCCCCAGAGTGAGCGGAAG-3′. Co-immunoprecipitated promoter abundance was normalized to promoter abundance of input DNA.
Immunofluorescence Microscopy
Cellular localization of STAT2 was evaluated by culturing cells on coverslips for 48 h before treatment with (1000 units/ml) IFN-α for the indicated times. Cells were washed with PBS and fixed for 20 min using 4% paraformaldehyde. The cells were washed with PBS and permeabilized with PBS + 0.1% Triton X-100 for 5 min. After incubation with blocking buffer (PBS + 4% goat serum) for 1 h, the cells were incubated with primary rabbit anti-STAT2 antibodies (C-20) in blocking buffer overnight at 4 °C. The cells were washed with PBS+0.1% Triton X-100 before and after 1 h of incubation with secondary donkey anti-rabbit DyLight 594 antibodies (BioLegend) in blocking buffer at room temperature. The coverslips were mounted on glass slides using Vectashield mounting medium with DAPI (Vector Laboratories, Inc.), and visualized using a Nikon Eclipse TE-2000U fluorescence microscope.
Statistical Analysis
Student's t test and two-way analysis of variance followed by Tukey's post hoc test were used to determine any statistically significant differences between samples.
RESULTS
Identification of Phosphorylation Sites in STAT2
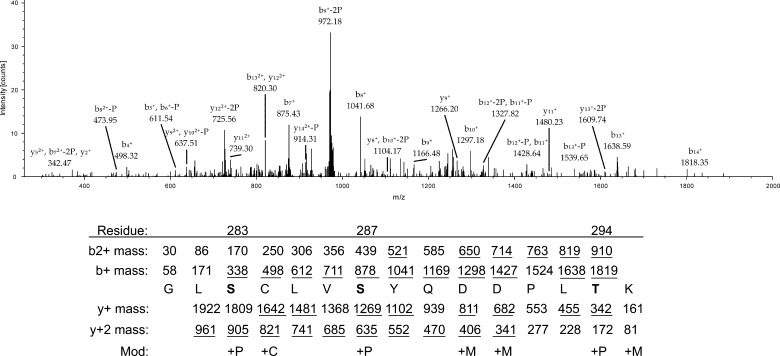
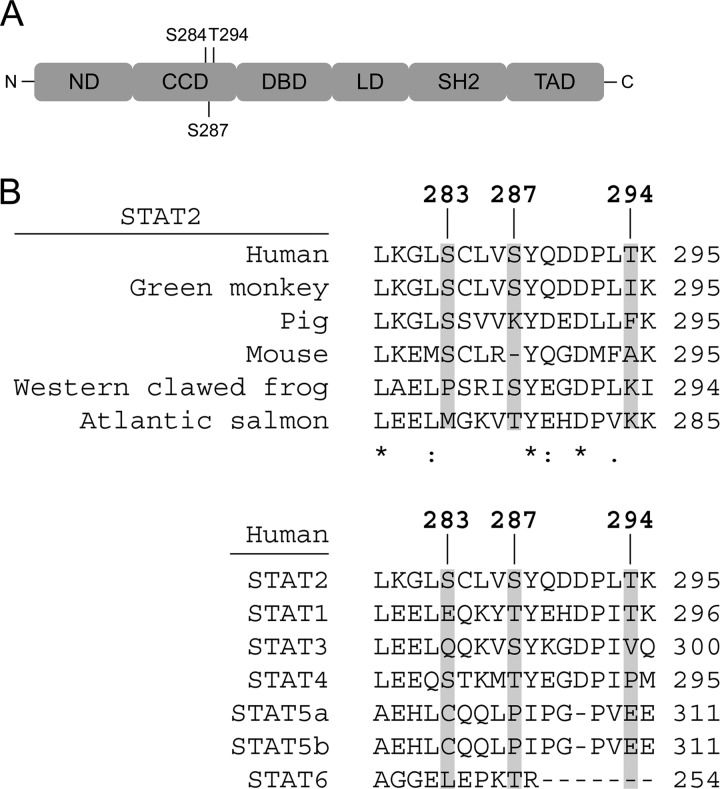
To identify novel phosphorylation sites, STAT2 immunoprecipitates prepared from STAT2-deficient U6A cells reconstituted with wild type (WT) STAT2 and stimulated with or without IFN-α were resolved by SDS-PAGE. Protein bands corresponding to STAT2 were excised from the gel and processed for mass spectrometry. The proteins in both the untreated and IFN-α-treated gel bands were positively identified as STAT2 via peptide mass fingerprinting (data not shown). Consequently, we conducted additional mass spectrometry and identified peptides corresponding to amino acids 281–295 with Ser-283, Ser-287, and Thr-294 as phosphorylated following IFN-α treatment for 30 min (Figs. 1 and 2A). To determine which phospho-residue of the three identified to most likely be important for the function of STAT2, we aligned the human STAT2 peptide sequence in question with STAT2 from several other species as well as with other members of the STAT family (Fig. 2B). Taking the functionally similar properties of threonine and serine into account, Ser-287 was found to be more evolutionarily conserved between species and within the STAT family than both Ser-283 and Thr-294. Therefore, Ser-287 was selected for further studies.
FIGURE 1.
MS/MS analysis of peptides corresponding to amino acids 281–295 of STAT2. S and T in boldface type indicate phosphorylated residues, and underlined mass indicates a match to a peak in the spectrum. P, phosphoryl group (79.97Da); C, carbamidomethyl (57.02 Da); M, methyl (14.02 Da). Not all peaks are annotated in the mass spectrum. Matches were made with a match tolerance of 3 Da.
FIGURE 2.
Structural organization of STAT2, location of detected phosphorylation sites, and conservation of the sites between species and STATs. ND, N-terminal domain; LD, Linker domain; SH2, Src homology 2 domain; TAD, transactivation domain.
Mutation of Ser-287-STAT2 to Alanine Selectively Regulates the Expression of Type I IFN-induced Genes
To evaluate the role of Ser-287 in the transcriptional function of STAT2, site-directed mutagenesis was performed in which Ser-287 was replaced with alanine. We generated STAT2-deficient U6A cells stably reconstituted with WT-STAT2 or S287A-STAT2 and evaluated the IFN-α-inducible expression of six ISGs that are representative of the antiproliferative, apoptotic, and antiviral effects of type I IFNs (MX1, TRAIL, OAS1, IFIT2, RSAD2, and ISG15) by quantitative real-time PCR (qRT-PCR) analysis. As shown in Table 1, compared with WT-STAT2, expression of S287A-STAT2 enhanced IFN-induced OAS1, IFIT2, RSAD2, TRAIL, and ISG15 transcription most notably at 18 h of IFN-α treatment. Interestingly, S287A-STAT2 failed to induce MX1 gene expression to the same levels as WT-STAT2 did at both 6 and 18 h of IFN-α treatment. Thus, our data suggest that the outcome of Ser-287 phosphorylation may be promoter-specific and to various degrees affect IFN-induced ISG expression.
TABLE 1.
Induction of ISGs in U6A cells in response to 1000 units/ml IFN-α
Values shown in the table are mRNA abundance normalized to β-actin mRNA as measured by quantitative RT-PCR. Student's t test was used to test for significant differences in ISG induction between WT-STAT2 and S287A-STAT2 expressing cells for each time point (n = 5); *, p < 0.05.
| IFN-α | WT-STAT2 |
S287A-STAT2 |
||||
|---|---|---|---|---|---|---|
| 0 h | 6 h | 18 h | 0 h | 6 h | 18 h | |
| MX1 | 0.1 | 1.5 | 1.1 | 0.04 | 0.22* | 24* |
| OAS1 | 0.4 | 16.9 | 8.9 | 0.4 | 11.9 | 13.6* |
| IFIT2 | 0.04 | 9.58 | 0.34 | 0.06 | 12.61 | 0.91* |
| RSAD2 | 0.002 | 6.155 | 0.784 | 0.004 | 4.012 | 2.284* |
| TRAIL | 0.001 | 0.299 | 0.018 | 0.002 | 0.324 | 0.082* |
| ISG15 | 0.3 | 13.4 | 16.7 | 0.3 | 10.5 | 27.0* |
Biological Consequences of S287A-STAT2
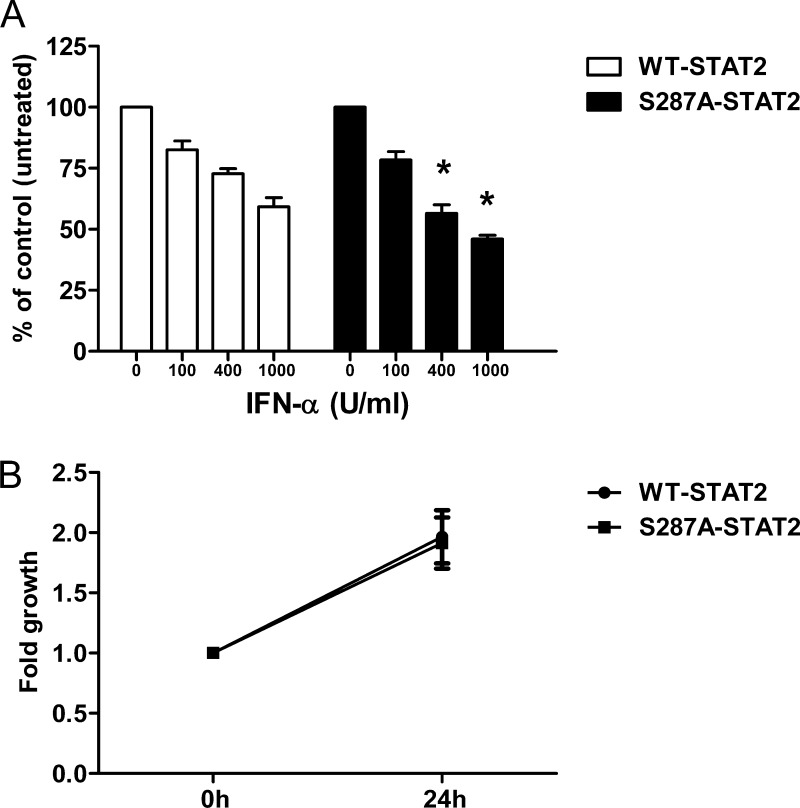
Cells exposed to type I IFNs can exhibit decreased cell proliferation, induction of apoptosis, and protection against viral infection (24). Our qRT-PCR data shown in Table 1 was restricted in that we only evaluated a small group of ISGs out of the ∼2000 IFN target genes (as listed in the Interferome database: www.interferome.org; (25)). The expression of these select genes, with the exception of Mx1, was enhanced by S287A-STAT2. This observation suggested that the cell growth inhibitory and antiviral capacities of IFN-α could be different when signaling via WT-STAT2 and S287A-STAT2. To test this, we first compared cell growth inhibition following IFN-α treatment for 72 h. As expected, in the presence of WT-STAT2, IFN-α decreased cell proliferation in a dose-dependent manner (Fig. 3A). However, the anti-proliferative effect of IFN-α was more pronounced in the presence of S287A-STAT2 when compared with WT-STAT2 at doses higher than 100 units/ml IFN-α (Fig. 3A). We also demonstrate that expression of S287A-STAT2 did not influence cell growth rate, indicating that a reduction in cell proliferation was directly linked to IFN-α stimulation (Fig. 3B). It is worth mentioning that the enhanced cell growth inhibitory effect observed with S287A-STAT2 was cytostatic and not due to activation of cell death.
FIGURE 3.
S287A-STAT2 enhances the antiproliferative effects of IFN-α. A, U6A cells stably expressing WT-STAT2 or S287A-STAT2 were treated with IFN-α for 72 h or left untreated. Cell viability was measured by MTS assay. Results are presented as the percentage of viable cells treated with IFN-α compared with untreated cells (100%). Data were obtained from four individual experiments and are shown as mean ± S.E. An asterisk signifies a statistical difference (p < 0.05) compared with cells expressing WT-STAT2 and treated with the same concentration of IFN-α. B, growth rate was determined by MTS assay at two time points (referred to as 0 and 24 h in figure). Data presented are from five independent experiments and shown as mean ± S.E. U, units.
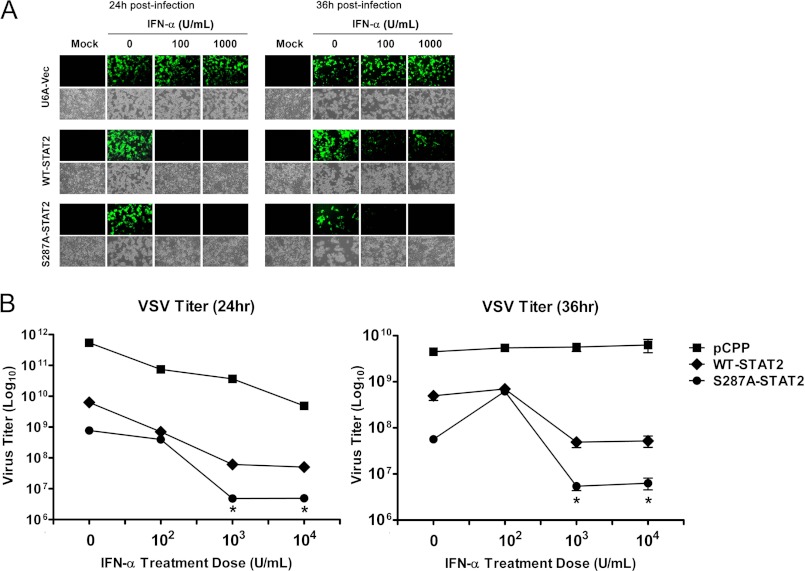
To determine whether the observed differences in gene expression we detected between WT- and S287A-STAT2 also translated into altered cellular resistance to viral infection, U6A cells stably reconstituted with empty vector (U6A-Vec), WT-STAT2 or S287A-STAT2 were infected with GFP-expressing VSV (multiplicity of infection of 0.01), a negative strand RNA virus with well established sensitivity to the antiviral effects of IFN (Fig. 4A). Without IFN-α pretreatment, all cells were visibly infected by VSV, as seen by the expression of GFP and appearance of cytopathic effects; regardless of STAT2 expression. As expected, in the absence of STAT2, IFN-α pretreatment did not prevent viral infection, whereas reconstitution of WT-STAT2 conferred protection at the various doses of IFN-α tested at 24 h post-infection. This protection, however, was limited as these cells succumbed to viral infection by 36 h even when higher doses of IFN-α were used. In contrast, S287A-STAT2 conferred continued protection against VSV even at 36 h post-infection with IFN-α pretreatment. We also measured progeny virion yield in the culture supernatants of VSV-infected cells lacking STAT2 or carrying WT-STAT2 or S287A-STAT2. Less of the virus was produced in IFN-α pretreated cells expressing S287A-STAT2 than in similarly treated control cells expressing WT-STAT2 (Fig. 4B). Together, these results show that the S287A-STAT2 mutant can functionally enhance type I IFN antiviral responses.
FIGURE 4.
STAT2-S287A confers prolonged protection against VSV infection. A, U6A cells stably expressing empty vector, WT-STAT2, or S287A-STAT2 were pretreated for 16 h with different concentrations of IFN-α and then infected with 0.01 multiplicity of infection GFP-expressing VSV. Mock controls are uninfected cells. Infection with VSV was detected by the presence of GFP expression. Bright field images show cells with cytopathic effects associated with virus infection. This is a representative experiment of two that were performed. B, same as in A, except viral titers were determined and compared between the supernatants of VSV infected U6A-cells expressing empty vector, WT-STAT2 or S287A-STAT2. Viral titers are shown as the mean ± S.E. from two independent experiments. *, p < 0.05. U, units.
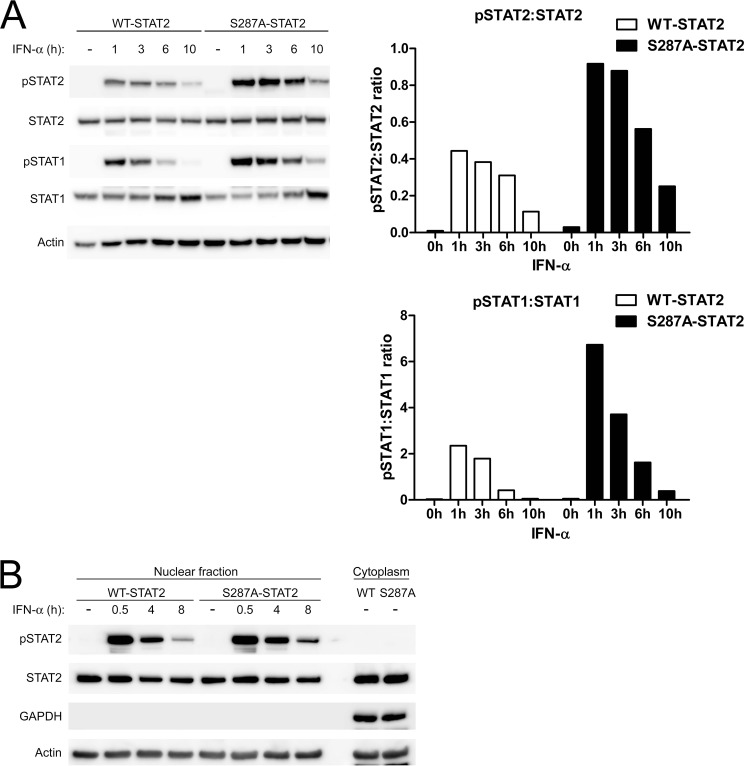
S287A-STAT2 Prolongs STAT2 and STAT1 Tyrosine Phosphorylation
To study whether the gain of function S287A altered STAT activation, U6A cells expressing WT-STAT2 or S287A-STAT2 were stimulated with IFN-α for various times. Immunoblot analysis revealed that the S287A mutation caused an increase in the level of phosphorylated Tyr-690 of STAT2 and Tyr-701 of STAT1 when compared with WT-STAT2 (Fig. 5A; and quantitative analysis). Furthermore, S287A-STAT2 extended the duration of tyrosine phosphorylation of STAT1 and STAT2. Although cells expressing WT-STAT2 showed little to no activation of STAT1 and STAT2 between 6 h and 10 h of IFN-α stimulation, cells expressing S287A-STAT2 continued to show detectable levels of activated STAT1 and STAT2. Of note, S287A-STAT2 alone did not cause activation of either STAT, suggesting that its effect remained IFN-α dependent.
FIGURE 5.
S287A-STAT2 increases IFN-α induced tyrosine phosphorylation of STAT2 and STAT1 and is retained in the nucleus. U6A cells stably expressing WT-STAT2 or S287A-STAT2 were treated with IFN-α (1000 units/ml) for the indicated times. A, tyrosine phosphorylation of STAT2 (pSTAT2) and STAT1 (pSTAT1) were evaluated by immunoblot analysis. Quantification of A is depicted as bar graphs to the right. B, nuclear extracts were analyzed for the presence of activated STAT2 in the nucleus. GAPDH served as a negative control to check the purity of our extracts. Immunoblots shown are one representative experiment of three performed.
To determine whether tyrosine phosphorylated S287A-STAT2 detected at 6 to 10 h of IFN-α stimulation was found in the nucleus, an indication that active STAT2 was available for gene transcription, nuclear fractions of IFN-α treated U6A WT-STAT2 and S287A STAT2 cells were prepared. Immunoblot analysis confirmed an increased amount of tyrosine phosphorylated S287A-STAT2 at 8 h of IFN-α treatment compared with WT-STAT2 (Fig. 5B). These results showed that the observed enhanced ISG expression is likely a consequence of prolonged ISGF3 activation.
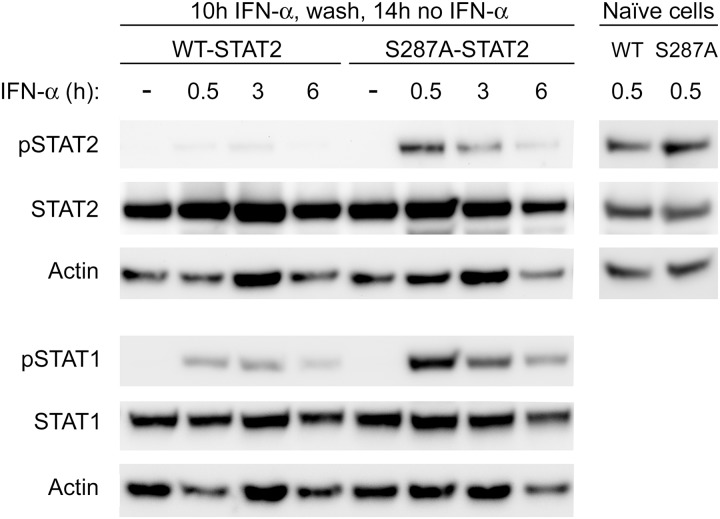
S287A-STAT2 Is Less Susceptible to IFN-α-mediated Desensitization
Continuous exposure to type I IFN is known to temporarily desensitize cells to further IFN stimulation (26). This unresponsive phase can last up to 24 h from the initial IFN exposure. After the continued presence of IFN, desensitized cells remain unresponsive to newly added IFN as further induction of ISGs transcription is severely diminished. Desensitized cells (after 16 h of IFN exposure) subjected to a recovery period by removal of IFN for 6 h only show modest restoration of STAT2 activation with little to no STAT1 activation (27). Because we first found that S287A-STAT2 provided longer protection against VSV infection after an overnight incubation with IFN-α, and second, tyrosine phosphorylation of STAT1 and STAT2 was prolonged, we next determined whether this heightened response could be attributed to resistance to IFN-α induced desensitization. Naïve U6A cells expressing WT-STAT2 or S287A-STAT2 were subjected to continuous exposure to IFN-α for 10 h; this is the time point at which STAT1 and STAT2 are weakly phosphorylated in the presence of WT-STAT2 (Fig. 6). This was followed by a recovery period of no IFN-α for 14 h before readdition of fresh IFN-α for various time points. No tyrosine phosphorylated STAT1 and STAT2 were detected in cells carrying WT-STAT2 or S287A-STAT2 prior to IFN-α re-exposure, indicating that STAT1 and STAT2 had returned to their initial inactive state. As expected, cells expressing WT-STAT2 were refractory to further IFN-α stimulation as STAT1 and STAT2 were found to be weakly activated by IFN-α. In contrast, cells expressing S287A-STAT2 were resensitized to IFN-α as tyrosine phosphorylation of STAT1 and STAT2 was reinduced and markedly different from cells expressing WT-STAT2 (Fig. 6). Indeed, in the presence of S287A-STAT2, the level of tyrosine phosphorylated STAT2 in previously desensitized cells after a 30 min re-exposure to IFN-α was similar to that observed in naïve cells expressing either WT-STAT2 or S287A-STAT2 that have been exposed for the same time to IFN-α. These results clearly indicate that S287A-STAT2 is more resistant to IFN-α-induced desensitization.
FIGURE 6.
S287A-STAT2 is protected from IFN-α desensitization. U6A cells stably expressing WT-STAT2 or S287A-STAT2 were treated with IFN-α (1000 units/ml) for 10 h, followed by two washes with PBS, and addition of regular DMEM for 14 h. Cells were then restimulated with IFN-α for the indicated times. Naïve cells indicate U6A WT-STAT2 or S287A-STAT2 that have been treated with IFN-α for 0.5 h without prior desensitization. Tyrosine phosphorylation of STAT2 (pSTAT2) and STAT1 (pSTAT1) were evaluated by immunoblot analysis. Immunoblots shown are one representative experiment of three performed.
Prolonged activation of STAT1 and STAT2 with S287A-STAT2 could be attributed to several reasons, including 1) a delay in the rate of STAT tyrosine dephosphorylation, and 2) increased association with IRF-9. To examine the first possibility, pulse-chase experiments were conducted in the presence of the protein kinase inhibitor staurosporine. No obvious differences were seen in the rate of STAT1 and STAT2 tyrosine dephosphorylation between cells expressing WT-STAT2 and S287A-STAT2 (supplemental Fig. S1).
Next, we focused on the interaction between STAT2 and IRF-9. The crystal structure of STAT2 remains unsolved; yet threading analysis using the protein structure of STAT1 hints Ser-287 to be part of the loop between the third and fourth α helix of the coiled-coil domain (CCD) and, therefore, accessible for phosphorylation by kinases or direct protein-protein interactions (28). The CCD of STAT2, which encompasses amino acids 137–316, is known to bind IRF9 (29). STAT2-IRF9 complexes can mediate the expression of ISRE driven target genes independently of STAT1 (30). Consequently, we wanted to examine whether Ser-287-STAT2 altered the binding of STAT2 to IRF9. Co-immunoprecipitation assay using anti-IRF9 antibodies showed no differences in the amount of protein interactions between IRF9 and S287A-STAT2 compared with WT-STAT2 following treatment with IFN-α (supplemental Fig. S2). This result indicates that S287A-STAT2 does not appear to influence IRF9-STAT2 complex formation.
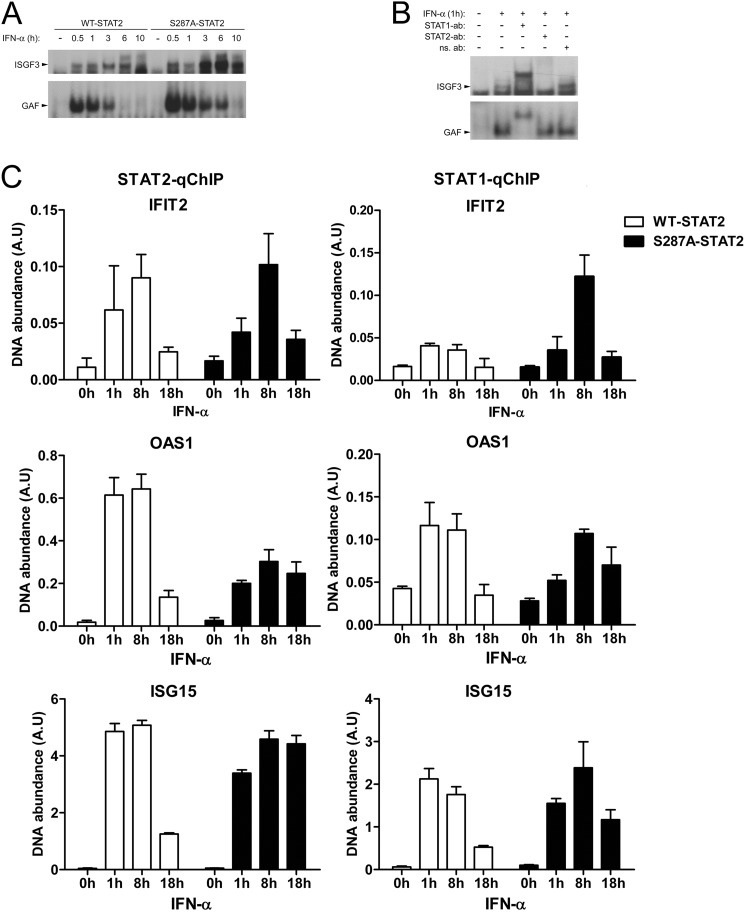
S287A-STAT2 Binding to DNA
To distinguish whether the difference we observed in the kinetics of STAT2 and STAT1 activation between WT STAT2 and S287A-STAT2 correlated with alterations in STAT binding to DNA, we tested this possibility first by EMSA using an ISRE or a GAS DNA probe. No obvious differences were observed in ISGF3 binding to ISRE between WT or S287A-STAT2 at 30 min and 1 h of IFN-α treatment (Fig. 7A). At later time points, however, S287A-STAT2 caused an increase in the amount ISGF3 bound to the ISRE probe; which was detectable even at 10 h. Similarly, S287A-STAT2 also increased the amount of γ-associated factors (GAF; STAT1 homodimers) to the GAS probe when compared with WT-STAT2. However, this increase in STAT1 homodimers binding to DNA detected in the presence of S287A STAT2 did not augment the mRNA expression of the STAT1 controlled gene IRF1 (supplemental Fig. S3). We also demonstrate by supershift EMSA that in U6A cells expressing S287A STAT2, the identity of the protein-DNA complex we found bound to the ISRE is indeed ISGF3 containing both STAT1 and STAT2, whereas only STAT1 homodimers were found bound to GAS (Fig. 7B).
FIGURE 7.
Differential recruitment of WT-STAT2 and S287A-STAT2 to chromatin. A, nuclear extracts prepared from U6A cells stably expressing WT-STAT2 or S287A-STAT2 were treated with or without IFN-α (1000 units/ml) at the indicated times and assayed for binding of ISGF3 to the ISRE probe (top) and binding of γ-associated factors (GAF; STAT1 homodimers) to the GAS probe (bottom) by EMSA. Shown is a representative experiment of three that were performed. B, similar to A, except STAT1 and STAT2 were supershifted with specific antibodies. Nonspecific antibody (ns) served as a negative control. C, qChIP assay was performed with antibodies against STAT2 (left column) and STAT1 (right column) using cell extracts treated with or without IFN-α (1000 units/ml) for the indicated time points. Specific qPCR primers were used to amplify the ISRE-containing promoter region of IFIT2, OAS1, and ISG15.
To confirm our EMSA results and more importantly, to assess in vivo the dynamic interaction of the ISGF3 complex binding to chromatin, we employed quantitative chromatin immunoprecipitation (qChIP) assay using STAT2 or STAT1 antibodies to pull down STAT-containing complexes. We evaluated binding of STAT1 and STAT2 to the ISRE motifs in the promoters of IFIT2, OAS1, and ISG15 as these were the genes whose expressions we showed earlier (Table 1) to be increased by S287A-STAT2 after 18 h of IFN-α treatment. Both STAT2 and STAT1 were recruited to the promoters of all genes analyzed (Fig. 7C). When compared with WT-STAT2, enhanced recruitment of S287A-STAT2 to the ISG promoters was observed at 18 h of IFN-α stimulation. This observation agrees with our EMSA result in that cells expressing S287A-STAT2 showed more ISGF3 bound to DNA at later time points, including 10 h. Our qChIP data are also in agreement with our previous observation in that IFIT2, OAS1, and ISG15 transcription at 18 h is higher in cells expressing S287A-STAT2 and less in cells expressing WT-STAT2 (Table 1). Acetylation of histone H3 (AcH3) at the ISRE site of the promoters of IFIT2, OAS1, and ISG15 was also assessed in cells expressing either WT-STAT2 or S287A-STAT2. Without stimulation, a basal level of AcH3 was detected in both cells lines, which was further increased with IFN-α stimulation over time (supplemental Fig. S4). A modest but not significant increase in the amount of AcH3 was detected in the presence of S287A-STAT2 at 18 h of IFN-α treatment. These results indicate that S287A-STAT2 is present in the nucleus much longer as an active transcriptional complex with chromatin.
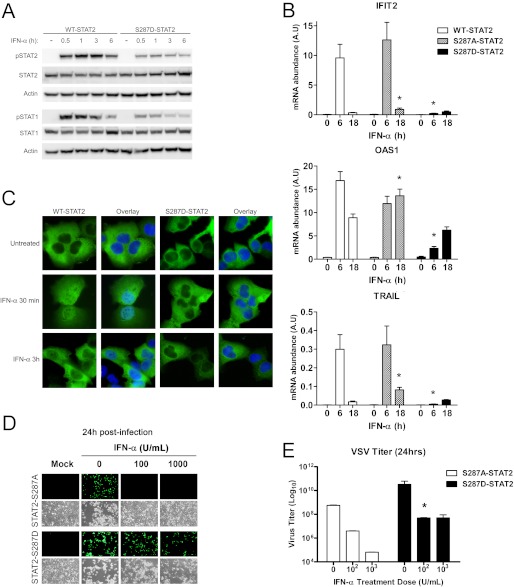
A Phosphomimetic Mutation at Ser-287 Impairs STAT2 Activation and ISG Induction
Our mass spectrometry revealed Ser-287 to be phosphorylated in response to 30 min of exposure to IFN-α (Fig. 1). Because our above findings indicated that S287A-STAT2 is a gain-of-function mutant, we next tested whether a phosphomimetic mutant of Ser-287, by changing this site to aspartate to mimic serine phosphorylation, will have the opposite effect of S287A-STAT2 and behave as a loss-of-function mutant. Indeed, in response to IFN-α, U6A cells stably expressing S287D-STAT2 showed a marked decrease in both STAT2 and STAT1 tyrosine phosphorylation when compared with WT-STAT2 expressing cells (Fig. 8A). Next, we measured IFN-α induction of ISGs and found this response to be dramatically affected, as seen by the poor mRNA expression of OAS1, IFIT2, and TRAIL (Fig. 8B). As suspected, S287D-STAT2 displayed a dramatic reduction in nuclear STAT2 (Fig. 8C). Furthermore, in contrast to S287A-STAT2, expression of S287D-STAT2 failed to protect cells against VSV infection after pretreatment with IFN-α (Fig, 8, D and E). These data strongly indicate that the phosphomimetic S287D-STAT2 mutant impairs IFN signaling via defects in STAT2 nuclear translocation and consequently, STAT2 transcriptional activity, thus suggesting that Ser-287 is located in a critical regulatory region important for the activation and retention of the ISGF3 complex bound to chromatin.
FIGURE 8.
Phosphomimetic S287D-STAT2 affects IFN-induced transcriptional and cellular responses. U6A cells stably expressing WT-STAT2, S287A-, or S287D-STAT2 were treated with IFN-α (1000 units/ml) for the indicated times. A, tyrosine phosphorylation of STAT2 (pSTAT2) and STAT1 (pSTAT1) was evaluated by immunoblot analysis. B, expression of ISGs was quantified by qRT-PCR and is shown as mRNA abundance. The bars represent the mean ± S.E.; n = 5; *, p < 0.05. C, cellular localization of STAT2 (green) by immunofluorescence staining. The nuclear stain DAPI (blue) was used in the overlay to discriminate between nucleus and cytoplasm. D, S287A-STAT2 and S287D-STAT2 cells untreated or pretreated with various concentrations of IFN-α for 16 h were infected with 0.01 multiplicity of infection GFP-expressing VSV. Mock controls are uninfected cells. E, viral titers from supernatants in D were determined and compared using Student's t test. Data presented are from two independent experiments and shown as mean ± S.E. *, p < 0.05.
DISCUSSION
Our objective was to identify novel phosphorylation sites in STAT2. All other STATs have been confirmed to be tyrosine and serine phosphorylated to acquire transcriptional activity and become fully active (13–15, 31), with the exception of STAT6, in which serine phosphorylation inactivates its transcriptional function (16, 17). Although the transcriptional activity of STAT2 is driven by tyrosine phosphorylation and nuclear translocation, it is less clear whether the transcriptional response to type I IFNs also requires STAT2 to be serine phosphorylated. We used mass spectrometry and identified Ser-287 as a bona fide phosphorylation site. This amino acid residue is mapped to the CCD of STAT2 that when mutated to alanine (S287A) generated a gain-of-function mutant that increased the biological effects of type I IFNs over that of WT-STAT2. In contrast, substitution of Ser-287 with aspartic acid (S287D), to mimic serine phosphorylation, had the expected opposite behavior of S287A-STAT2. The dramatic negative effect S287D-STAT2 had on IFN-α signaling indicates that the role of phosphorylated Ser-287 is to limit type I IFN signaling.
Recently, the analogous site to Ser-287 in STAT1, threonine 288, was found to be mutated to alanine in several patients with chronic mucocutaneous candidiasis (32, 33). The T288A substitution was one of 12 mutations clustered spatially around the same region in the CCD. The resulting phenotype for 11 of the 12 mutations, including T288A, was resistance to phospho-Tyr-701 dephosphorylation after activation with IFN-γ, IFN-α or IL-27, which led to increased GAS binding (EMSA) and GAS-dependent but not ISRE-dependent ISG induction. This observation parallels to what we have observed with S287A in STAT2. Although the exact mechanism of STAT2 inactivation is not clear, the most feasible model of STAT1 inactivation has been proposed by the group of James Darnell. As phosphorylated STAT1 homodimers disassociate from DNA, the STAT1 dimer now interacts via their N-terminal domains (28, 34). This facilitates reorientation of the STAT1 dimer from a parallel (active) to an anti-parallel (inactive) conformation (35, 36). Instead of phospho-tyrosyl-SH2 interactions, the new anti-parallel dimer is stabilized by reciprocal interaction between the CCD of one monomer and the DBD of the other monomer. The final step of this reorientation process is the dephosphorylation of the now exposed phospho-tyrosines. The critical residues for the STAT1 CCD-DBD interactions have been mapped to Phe-172 in the CCD and Gln-340, Gly-384, and Gln-408 of the DBD. Mutation of any of these amino acids leads to prolonged STAT1-phosphorylation due to impaired dephosphorylation, similarly to what was seen in Liu et al. (32). It is, therefore, plausible that the mutations identified in chronic mucocutaneous candidiasis patients, all localized near the CCD-DBD interaction surface, disrupt the formation of the anti-parallel STAT1 dimer necessary for proper dephosphorylation of phospho-Tyr-701-STAT1. Curiously, only Phe-172 is conserved in STAT2, whereas the other three critical residues conserved in the STAT1 DBD are not, indicating that the exact mechanism of STAT2/STAT1 anti-parallel binding might be different from that of STAT1 homodimers. Therefore, until the structure of STAT2-containing complexes is solved, mutational studies are warranted to identify regulatory residues that are critical in the regulation of STAT2 function.
Protein modeling of STAT2 using the crystal structure of STAT1 as a protein template infers that Ser-287 is exposed and available for post-translational modifications and/or direct protein interaction with a second protein. Furthermore, according to the solved structures of other STATs (28, 37), this site is located on the side of the CCD that is facing away from the DBD and SH2 domain as the STAT-dimer is associated with DNA. Therefore, it makes it less likely that the S287A mutation directly affects STAT functions that are linked to DNA binding and/or SH2-mediated STAT dimerization.
Type I IFNs activate the transcriptional activity of STAT2. Our qRT-PCR results showed that S287A-STAT2 augmented the expression of several IFN target genes; which correlated with sustained activation of STAT1 and STAT2. Our qChIP experiments confirmed increased binding of S287A-STAT2 to chromatin at later time points compared with WT-STAT2. This helps explain why ISG expression was more pronounced at later times in the presence of S287A-STAT2 as detected in our initial qRT-PCR experiment. Although we did not detect significant alterations in the level of acetylated histone H3 at the ISRE region in the ISG promoters when cells express either WT-STAT2 or S287A-STAT2, it is most probable that acetylation and/or methylation of specific lysine residues in histone H3 may take place with S287A-STAT2 (38). Alternatively, binding of acetylated histone H3 to other regions distal from the promoter site may be preferred in the presence of S287A-STAT2 following IFN-α stimulation (39). It is also possible that S287A-STAT2 may slowly increase the binding affinity for adaptor molecules/coactivators as it has been reported to occur with the INHAT component, pp32 (40). pp32 interacts with tyrosine phosphorylated STAT2 and STAT1 and potentiates the expression of ISGs. Furthermore, IFN-α-induced acetylation of STAT2 Lys-390 is presumed to facilitate the reorientation of the antiparallel STAT1/STAT2 heterodimer to the parallel conformation and promote the antiviral effects of type I IFNs (19). This leads to speculation that perhaps S287A-STAT2 augments and/or extends the duration of acetylated STAT2 during which STAT2/STAT1 heterodimers are kept in the active parallel orientation leading to enhanced gene transcription. Alternatively, the S287A mutation may turn STAT2 into a more flexible molecule in which reorientation of antiparallel STAT1/STAT2 heterodimers to the parallel conformation in IFN naïve and/or IFN desensitized cells is more efficient and stable, resulting in prolonged STAT activation. Overall, our study reports the first observation that STAT2 is serine-phosphorylated in response to type I IFNs. Unlike serine phosphorylation of other STATs, Ser-287-STAT2 phosphorylation evokes a negative regulatory role in the antiproliferative and antiviral effects of type I IFNs. This information could serve as the platform in the development of therapeutics to disable resistance and improve the clinical efficacy of type I IFNs by targeting the responsible kinase.
This work was supported by National Cancer Institute Grants RO1CA140499 and K22CA095326 (to A. M. G.). This work was also supported by an ACS Research Scholar Grant (RSG-09-195-01-MPC) with additional funding provided by the Fox Chase Cancer Center institutional support via the Kidney Cancer Keystone Program.

This article contains supplemental Figs. S1–S4.
- ISGF3
- IFN-stimulated gene factor 3
- CCD
- coiled-coil domain
- IRF9
- IFN regulatory factor 9
- ISG
- interferon-stimulated gene
- ISRE
- interferon stimulated response element
- GAS
- gamma activated sequence
- VSV
- vesicular stomatitis virus
- qRT-PCR
- quantitative real-time PCR
- qChIP
- quantitative ChIP
- DBD
- DNA-binding domain.
REFERENCES
- 1. de Veer M. J., Holko M., Frevel M., Walker E., Der S., Paranjape J. M., Silverman R. H., Williams B. R. (2001) Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc Biol. 69, 912–920 [PubMed] [Google Scholar]
- 2. Yan H., Krishnan K., Greenlund A. C., Gupta S., Lim J. T., Schreiber R. D., Schindler C. W., Krolewski J. J. (1996) Phosphorylated interferon-α receptor 1 subunit (IFNaR1) acts as a docking site for the latent form of the 113-kDa STAT2 protein. EMBO J. 15, 1064–1074 [PMC free article] [PubMed] [Google Scholar]
- 3. Fu X. Y. (1992) A transcription factor with SH2 and SH3 domains is directly activated by an interferon α-induced cytoplasmic protein tyrosine kinase(s). Cell 70, 323–335 [DOI] [PubMed] [Google Scholar]
- 4. Improta T., Schindler C., Horvath C. M., Kerr I. M., Stark G. R., Darnell J. E., Jr. (1994) Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc. Natl. Acad. Sci. U.S.A. 91, 4776–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shuai K., Stark G. R., Kerr I. M., Darnell J. E., Jr. (1993) A single phosphotyrosine residue of Stat91 required for gene activation by interferon-γ. Science 261, 1744–1746 [DOI] [PubMed] [Google Scholar]
- 6. Kessler D. S., Veals S. A., Fu X. Y., Levy D. E. (1990) Interferon-α regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 4, 1753–1765 [DOI] [PubMed] [Google Scholar]
- 7. Veals S. A., Santa Maria T., Levy D. E. (1993) Two domains of ISGF3 γ that mediate protein-DNA and protein-protein interactions during transcription factor assembly contribute to DNA-binding specificity. Mol. Cell Biol. 13, 196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schindler C., Shuai K., Prezioso V. R., Darnell J. E., Jr. (1992) Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257, 809–813 [DOI] [PubMed] [Google Scholar]
- 9. Zhong Z., Wen Z., Darnell J. E., Jr. (1994) Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264, 95–98 [DOI] [PubMed] [Google Scholar]
- 10. Jacobson N. G., Szabo S. J., Weber-Nordt R. M., Zhong Z., Schreiber R. D., Darnell J. E., Jr., Murphy K. M. (1995) Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J. Exp. Med. 181, 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gouilleux F., Wakao H., Mundt M., Groner B. (1994) Prolactin induces phosphorylation of Tyr-694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J. 13, 4361–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hou J., Schindler U., Henzel W. J., Ho T. C., Brasseur M., McKnight S. L. (1994) An interleukin-4-induced transcription factor: IL-4 Stat. Science 265, 1701–1706 [DOI] [PubMed] [Google Scholar]
- 13. Wen Z., Zhong Z., Darnell J. E., Jr. (1995) Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82, 241–250 [DOI] [PubMed] [Google Scholar]
- 14. Cho S. S., Bacon C. M., Sudarshan C., Rees R. C., Finbloom D., Pine R., O'Shea J. J. (1996) Activation of STAT4 by IL-12 and IFN-α: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J. Immunol. 157, 4781–4789 [PubMed] [Google Scholar]
- 15. Friedbichler K., Kerenyi M. A., Kovacic B., Li G., Hoelbl A., Yahiaoui S., Sexl V., Müllner E. W., Fajmann S., Cerny-Reiterer S., Valent P., Beug H., Gouilleux F., Bunting K. D., Moriggl R. (2010) Stat5a serine 725 and 779 phosphorylation is a prerequisite for hematopoietic transformation. Blood 116, 1548–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maiti N. R., Sharma P., Harbor P. C., Haque S. J. (2005) Serine phosphorylation of Stat6 negatively controls its DNA-binding function. J. Interferon Cytokine Res. 25, 553–563 [DOI] [PubMed] [Google Scholar]
- 17. Shirakawa T., Kawazoe Y., Tsujikawa T., Jung D., Sato S., Uesugi M. (2011) Deactivation of STAT6 through serine 707 phosphorylation by JNK. J. Biol. Chem. 286, 4003–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sadzak I., Schiff M., Gattermeier I., Glinitzer R., Sauer I., Saalmüller A., Yang E., Schaljo B., Kovarik P. (2008) Recruitment of Stat1 to chromatin is required for interferon-induced serine phosphorylation of Stat1 transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 105, 8944–8949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang X., Gao J. S., Guan Y. J., McLane K. E., Yuan Z. L., Ramratnam B., Chin Y. E. (2007) Acetylation-dependent signal transduction for type I interferon receptor. Cell 131, 93–105 [DOI] [PubMed] [Google Scholar]
- 20. Gamero A. M., Sakamoto S., Montenegro J., Larner A. C. (2004) Identification of a novel conserved motif in the STAT family that is required for tyrosine phosphorylation. J. Biol. Chem. 279, 12379–12385 [DOI] [PubMed] [Google Scholar]
- 21. Scarzello A. J., Romero-Weaver A. L., Maher S. G., Veenstra T. D., Zhou M., Qin A., Donnelly R. P., Sheikh F., Gamero A. M. (2007) A mutation in the SH2 domain of STAT2 prolongs tyrosine phosphorylation of STAT1 and promotes type I IFN-induced apoptosis. Mol. Biol. Cell 18, 2455–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hasham M. G., Tsygankov A. Y. (2004) Tip, an Lck-interacting protein of Herpesvirus saimiri, causes Fas- and Lck-dependent apoptosis of T lymphocytes. Virology 320, 313–329 [DOI] [PubMed] [Google Scholar]
- 23. Fernandez M., Porosnicu M., Markovic D., Barber G. N. (2002) Genetically engineered vesicular stomatitis virus in gene therapy: application for treatment of malignant disease. J. Virol. 76, 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maher S. G., Romero-Weaver A. L., Scarzello A. J., Gamero A. M. (2007) Interferon: cellular executioner or white knight? Curr. Med. Chem. 14, 1279–1289 [DOI] [PubMed] [Google Scholar]
- 25. Samarajiwa S. A., Forster S., Auchettl K., Hertzog P. J. (2009) INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res. 37, D852–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larner A. C., Chaudhuri A., Darnell J. E., Jr. (1986) Transcriptional induction by interferon. New protein(s) determine the extent and length of the induction. J. Biol. Chem. 261, 453–459 [PubMed] [Google Scholar]
- 27. Sakamoto S., Qin J., Navarro A., Gamero A., Potla R., Yi T., Zhu W., Baker D. P., Feldman G., Larner A. C. (2004) Cells previously desensitized to type 1 interferons display different mechanisms of activation of stat-dependent gene expression from naïve cells. J. Biol. Chem. 279, 3245–3253 [DOI] [PubMed] [Google Scholar]
- 28. Chen X., Vinkemeier U., Zhao Y., Jeruzalmi D., Darnell J. E., Jr., Kuriyan J. (1998) Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93, 827–839 [DOI] [PubMed] [Google Scholar]
- 29. Martinez-Moczygemba M., Gutch M. J., French D. L., Reich N. C. (1997) Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-α-stimulated transcription factor ISGF3. J. Biol. Chem. 272, 20070–20076 [DOI] [PubMed] [Google Scholar]
- 30. Bluyssen H. A., Levy D. E. (1997) Stat2 is a transcriptional activator that requires sequence-specific contacts provided by stat1 and p48 for stable interaction with DNA. J. Biol. Chem. 272, 4600–4605 [DOI] [PubMed] [Google Scholar]
- 31. Yamashita H., Xu J., Erwin R. A., Farrar W. L., Kirken R. A., Rui H. (1998) Differential control of the phosphorylation state of proline-juxtaposed serine residues Ser725 of Stat5a and Ser730 of Stat5b in prolactin-sensitive cells. J. Biol. Chem. 273, 30218–30224 [DOI] [PubMed] [Google Scholar]
- 32. Liu L., Okada S., Kong X. F., Kreins A. Y., Cypowyj S., Abhyankar A., Toubiana J., Itan Y., Audry M., Nitschke P., Masson C., Toth B., Flatot J., Migaud M., Chrabieh M., Kochetkov T., Bolze A., Borghesi A., Toulon A., Hiller J., Eyerich S., Eyerich K., Gulácsy V., Chernyshova L., Chernyshov V., Bondarenko A., Grimaldo R. M., Blancas-Galicia L., Madrigal Beas I. M., Roesler J., Magdorf K., Engelhard D., Thumerelle C., Burgel P. R., Hoernes M., Drexel B., Seger R., Kusuma T., Jansson A. F., Sawalle-Belohradsky J., Belohradsky B., Jouanguy E., Bustamante J., Bué M., Karin N., Wildbaum G., Bodemer C., Lortholary O., Fischer A., Blanche S., Al-Muhsen S., Reichenbach J., Kobayashi M., Rosales F. E., Lozano C. T., Kilic S. S., Oleastro M., Etzioni A., Traidl-Hoffmann C., Renner E. D., Abel L., Picard C., Maródi L., Boisson-Dupuis S., Puel A., Casanova J. L. (2011) Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J. Exp. Med. 208, 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van de Veerdonk F. L., Plantinga T. S., Hoischen A., Smeekens S. P., Joosten L. A., Gilissen C., Arts P., Rosentul D. C., Carmichael A. J., Smits-van der Graaf C. A., Kullberg B. J., van der Meer J. W., Lilic D., Veltman J. A., Netea M. G. (2011) STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N. Engl. J. Med. 365, 54–61 [DOI] [PubMed] [Google Scholar]
- 34. Mao X., Ren Z., Parker G. N., Sondermann H., Pastorello M. A., Wang W., McMurray J. S., Demeler B., Darnell J. E., Jr., Chen X. (2005) Structural bases of unphosphorylated STAT1 association and receptor binding. Mol. Cell 17, 761–771 [DOI] [PubMed] [Google Scholar]
- 35. Mertens C., Zhong M., Krishnaraj R., Zou W., Chen X., Darnell J. E. (2006) Dephosphorylation of phosphotyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain. Genes Dev. 20, 3372–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhong M., Henriksen M. A., Takeuchi K., Schaefer O., Liu B., ten Hoeve J., Ren Z., Mao X., Chen X., Shuai K., Darnell J. E., Jr. (2005) Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc. Natl. Acad. Sci. U.S.A. 102, 3966–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Becker S., Groner B., Müller C. W. (1998) Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature 394, 145–151 [DOI] [PubMed] [Google Scholar]
- 38. Testoni B., Völlenkle C., Guerrieri F., Gerbal-Chaloin S., Blandino G., Levrero M. (2011) Chromatin dynamics of gene activation and repression in response to interferon α (IFN(α)) reveal new roles for phosphorylated and unphosphorylated forms of the transcription factor STAT2. J. Biol. Chem. 286, 20217–20227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tamura T., Smith M., Kanno T., Dasenbrock H., Nishiyama A., Ozato K. (2009) Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J. Biol. Chem. 284, 12217–12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kadota S., Nagata K. (2011) pp32, an INHAT component, is a transcription machinery recruiter for maximal induction of IFN-stimulated genes. J. Cell Sci. 124, 892–899 [DOI] [PubMed] [Google Scholar]