Abstract
Context:
GH receptor (GHR) exon 3-deleted/full-length (d3/fl) polymorphism has been proposed to affect the responsiveness to GH therapy. Conventional multiplex PCR genotyping method for this polymorphism detection requires DNA samples, which may be difficult to obtain due to ethical and procedural issues and may limit further studies into the role of this polymorphism in health and disease.
Objective:
The objective of the study was to develop a simple genotyping-alternative method for GHRd3 identification by directly measuring serum levels of total GH binding protein (tGHBP) and exon 3-positive GHBP[(E3(+)GHBP] by immunoassay and thereby assess the GHRd3 status.
Design:
The GHRd3 genotype was determined by PCR, and tGHBP and E3(+)GHBP levels were measured in serum of 88 healthy adults.
Main Outcome Measures:
The GHRd3 chemotype by ELISA was compared with the genotype by conventional PCR.
Results:
The concordance rate of GHR exon 3 status identification between PCR genotyping and ELISA chemotyping was shown to be 100%. There were negligible detectable serum levels of E3(+)GHBP in d3/d3 subjects. The ratio of serum levels E3(+)GHBP vs. tGHBP in fl/fl and d3/fl subjects was (mean ± sd) 96.6 ± 5.1 and 57.1 ± 8.4%, respectively (P < 0.0001). Interestingly, we observed that d3/d3 subjects had significantly lower serum levels of tGHBP compared with fl/fl and d3/fl genotypes.
Conclusions:
This dual ELISA against tGHBP and E3(+)GHBP can be used as an alternative method for determining GHRd3 polymorphism status. The implications of differences in serum levels of tGHBP among different genotypes and responsiveness to GH therapy need to be further investigated.
GH is widely used in treatment of children with short stature and the response to GH therapy is highly variable (1). GH circulates bound to a GH binding protein (GHBP), which is a cleaved extracellular fragment of the GH receptor (GHR). A polymorphism conferring two isoforms of GHR transcripts, differing by the retention (GHRfl) or exclusion (GHRd3), of exon 3 as a consequence of an early homologous recombination event due to two retroviral-repeat sequences flanking exon3 (2). The relation of GHR exon3 deletion polymorphism with responsiveness to GH has been examined in numerous of studies. Dos Santos et al. (3) proposed that idiopathic short stature patients with the GHRd3 variant were more responsive to GH than GHRfl carriers. This finding was observed in several studies on various GH indications (4–8). However, other researchers were unable to demonstrate an effect of GHRd3 polymorphism associated with GH response (9–14). The discrepancy among the studies could be partially caused by small sample sizes or different experiment design; however, the variation among individual circulating levels of GHBP might contribute to difference in the responsiveness to GH.
The conventional PCR method by Pantel et al. (2) is widely used as to detect GHRd3/fl polymorphism. The limitations of this method are the DNA required, the time-consuming procedures, and misgenotyping. A rapid high-throughput genotyping method of GHR exon3 deletion polymorphism using a tag single-nucleotide polymorphism was recently reported by Glad et al. (15). Although this method dramatically improved efficiency and accuracy of GHRd3/fl genotyping, it provides no information about individual circulating levels of the GHBP protein. Seidel et al. (16) developed immunoassays against total GHBP (tGHBP) translated from both GHRfl and GHRd3 transcripts and exon 3-positive GHBP [E3(+)GHBP] translated from GHRfl transcript; however, GHRd3/fl polymorphisms could not be absolutely predicted due to a partial overlap in the serum levels of E3(+)GHBP between GHRfl/fl and GHRd3/f1 individuals.
In the current study, we developed in-house dual-ELISA assays against the tGHBP and E3(+)GHBP using polyclonal anti-GHBP antibodies and a specific anti-GHBP exon 3 antibody. The circulating levels of tGHBP and E3(+)GHBP were measured in the serum and compared with GHR genotypes determined by conventional PCR. The GHRd3 status was predicted according to the ratio of E3(+)GHBP and tGHBP. This approach, which we termed chemotyping, represents an alternative to a traditional DNA-based determination of a clinically relevant polymorphism.
Materials and Methods
Study design
The study was approved by the institutional review boards of the University of California, Los Angeles (UCLA), and Albert Einstein University. Subjects were selected from a case-control human longevity study (17). PCR was conducted as previously described (2) to detect GHR exon 3 retention/exclusion. Eighty-eight serum samples were further analyzed by chemotyping, including 37 subjects with GHR fl/fl genotype, 33 subjects with GHR d3/fl genotype, and 18 subjects with GHR d3/d3 genotype. The investigator team conducting the ELISA assays at UCLA was blinded to the genotyping results performed at Albert Einstein University.
Total human GHBP immunoassay
Serum levels of total human GHBP were measured by a UCLA in-house ELISA, and results were verified by the DSL-10-48100 human GHBP (hGHBP) ELISA kit (Diagnostic Systems Laboratories, Inc., Webster, TX). For the in-house hGHBP ELISA, each well of microtiter plates was coated with 0.3 μg hGHBP monoclonal antibody (Diagnostic Systems Laboratories) in 100 μl 50 mm PBS (pH 7.2) at room temperature for 4 h. The plates were then blocked with SuperBlock T20 (PBS) blocking buffer (Thermo Scientific, Rockford, IL). The recombinant human GHBP (R&D Systems, Minneapolis, MN) used as a standard was prepared with assay buffer (50 mm PBS, 0.1% Tween 20, 0.25% BSA) in the range 9–1126 pm (lower detectable limit < 2 pm). The serum samples were prediluted 5 times with assay buffer. Fifty-microliter standard, control, or sample was added followed by addition of 50 ng biotin-labeled goat anti-hGHBP polyclonal antibody (pAb) (R&D Systems) in an assay buffer with 2% normal goat serum and incubated overnight. Streptavidin-horseradish peroxidase conjugate was added and incubated on a shaker for 30 min at room temperature after washes. After that procedure, 100 μl of o-phenylenediamine dihydrochloride solution was added and incubated for 10–20 min. The reaction was stopped by the addition of 2N H2SO4, and absorbance was determined on a plate spectrophotometer (Molecular Designs, Sunnyvale, CA) at 490 nm. The intra- and interassay coefficient variations (CVs) were 4.5–8.3 and 6.5–9.7%, respectively. The lower detectable limit of the DSL hGHBP ELISA, defined as the double sd of the zero standard, was 1.7 pm. The intra- and interassay CVs were 4.9–5.6 and 5.1–8.4%, respectively.
E3(+) human GHBP in-house immunoassay
The UCLA in-house E3(+) human GHBP ELISA was developed according to the method of Kratzsch et al. (18). Anti-E3(+)GHBP IgG isolated from the anti-E3(+)GHBP rabbit serum was biotinylated by EZ-Link Sulfo-NHS-LC-Biotin (Thermo Scientific) and used as detection antibody. Briefly, goat anti-hGHBP pAb 0.2 μg (R&D Systems) in 100 μl sodium carbonate-bicarbonate (50 mm, pH 9.5) was added to each well and incubated for 3 h at room temperature. Standard was prepared as mentioned in total hGHBP assay. After blocking and washing, 50 μl standard, control, or 1:5 diluted samples were added followed by the addition of 0.2 μg biotin-anti-E3(+)GHBP pAb in assay buffer with 2% normal goat serum and was incubated overnight. Absorbance was determined by a spectrophotometer followed by streptavidin-horseradish peroxidase and o-phenylenediamine dihydrochloride steps. The measurement range of the E3(+)GHBP assay was between 7 and 1745 pm. The intra- and interassay CVs were below 10%. Total hGHBP and E3(+)GHBP immunoassay was performed simultaneously.
Statistics
The ELISA standard curves were analyzed using a four-parameter logistic curve fit. Results are reported as the mean ± sd. A correlation analysis was performed to compare in-house tGHBP assay with the Diagnostic Systems Laboratories kit. A two-tailed Student t test was used to compare the difference of total GHBP and E3(+)GHBP among the subjects with GHR fl/fl, d3/fl, or d3/d3 genotypes. P < 0.05 was considered significantly different. All statistics were performed with Prism GraphPad (La Jolla, CA).
Results
Comparison of total GHBP measured by in-house ELISA and Diagnostic Systems Laboratories kit
As shown in Supplemental Fig. 1, published on The Endocrine Society's Journals Online web site at http://jcem.endojournals.org, the serum levels of tGHBP measured by the in-house ELISA were highly correlated with the tGHBP levels measured by the Diagnostic Systems Laboratories kit (r = 0.99, P < 0.0001).
Comparison of GHR exon 3 genotype between PCR and ELISA
To address the question whether ELISA chemotyping can be used as an alternative to the genotyping method for GHR exon3 status identification, we compared the result of genotype by PCR and chemotype by ELISA. As seen from Table 1, the genotypes of 83 of 88 subjects determined by PCR and ELISA matched each other (concordance rate 94.3%). The ELISA studies on these five subjects were repeated with nearly identical results. These five subjects as well as the other 83 subjects were regenotyped by the same PCR method, and the results of this second run indicated that the five subjects were miscategorized. The final results shown in Table 1 demonstrate that GHR exon 3 chemotype determined by the ELISA method was 100% accurate and suggest that the PCR genotyping method is far more error prone than the ELISA method.
Table 1.
Concordance rate of GHR exon 3 genotype between RCR and ELISA
| PCR | ELISA |
Concordance ratea | ||
|---|---|---|---|---|
| fl/fl | d3/fl | d3/d3 | ||
| First | ||||
| fl/fl | 35 | 0 | 0 | 94.3% (83/88) |
| d3/fl | 2 | 30 | 0 | |
| d3/d3 | 0 | 3 | 18 | |
| Secondb | ||||
| fl/fl | 37 | 0 | 0 | 100% (88/88) |
| d3/fl | 0 | 33 | 0 | |
| d3/d3 | 0 | 0 | 18 | |
The concordance rate was calculated by total number of genotype agreed by both PCR and ELISA methods divided by total subjects.
The second PCR was performed on all initial samples.
Serum levels of total GHBP and E3(+)GHBP
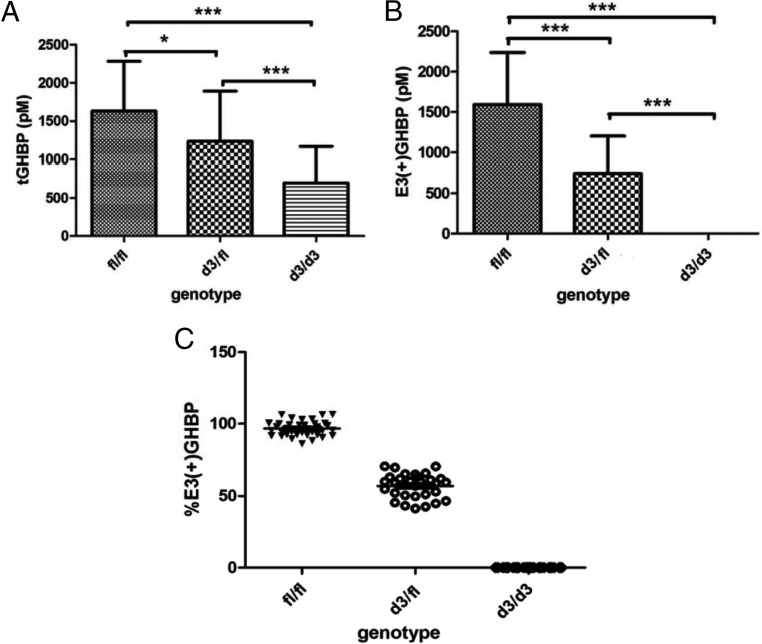
The serum level of tGHBP in subject with the d3/d3 genotype was significantly lower than that in subjects with the d3/fl genotype (P < 0.001) or that in subjects with the fl/fl genotype (P < 0.0001) (Fig. 1A). The tGHBP levels in the fl/fl group and the d3/fl group were also significantly different (P < 0.05), and the subjects with the fl/fl genotype had higher levels of tGHBP in serum. The serum levels of tGHBP in the fl/fl group, the d3/fl group, and the d3/d3 group were 1628 ± 652, 1233 ± 657, and 697 ± 471 pm, respectively. There was negligible detectable E3(+) GHBP in the serum of subjects with the d3/d3 genotype at the ELISA sensitivity of 1.7 pm (Fig. 1B). The serum levels of E3(+)GHBP in subjects with the fl/fl genotype and the d3/fl genotype were 1590 ± 645 and 742 ± 458 pm, respectively. The mean ratio of E3(+)GHBP and tGHBP is 96.6 ± 5.1% in the fl/fl group and 57.1 ± 8.4% in the d3/fl group. Distribution of the individual ratios within the three genotypes did not overlap between genotypes, indicating reliable and accurate genotype classification by the levels of E3(+)GHBP and tGHBP and their ratio (Fig. 1C). The levels of both tGHBP and E3(+)GHBP levels were observed to be reduced by the ex vivo addition of recombinant GH in a dose-response manner; however, their ratio did not significantly change (see Supplemental Table 1).
Fig. 1.
Serum levels of tGHBP (A) and E3(+)GHBP (B) in subjects with the GHR fl/fl, d3/fl, and d3/d3 genotypes (n = 37 for fl/fl, n = 33 for d3/d3, and n = 18 for d3/d3) and ratios of E3(+)GHBP to tGHBP (C). tGHBP and E3(+)GHBP were measured by an in-house ELISA, and the genotype was determined by conventional PCR (confirmed by chemotyping). *, P < 0.05; ***, P < 0.0001.
Discussion
The conventional GHR d3/fl genotyping method developed by Pantel et al. (2) was widely used in studies of growth disorders, acromegaly, and other states. The disadvantages of this method include the DNA required, laborious and time-consuming technique, and as shown here as well as in the other studies, genotyping failure or genotyping error due to multiplex PCR inefficiency, DNA quality, and sample contamination (15, 19). Genotyping error was observed in our study at a rate of 5% (involving d3/d3). A second round of PCR for samples initially genotyped as d3/d3 using the primer specific to the GHRfl isoform is recommended to avoid false homozygous GHRd3 genotyping. This method limits a large-scale cohort study. Seidel et al. (16) developed immunoassays against tGHBP and E3(+)GHBP; however, GHRd3/fl polymorphisms could not be absolutely predicted due to a partial overlap in the levels of E3(+)GHBP in plasma between GHRfl/fl and GHRf1/d3 individuals. In this study, we purified the anti-E3(+)GHBP IgG from E3(+)GHBP antiserum and biotinylated it. The biotinylated anti-E3(+)GHBP IgG was used as a detection antibody instead of the anti-E3(+)GHBP serum used by Seidel et al., thus improving the assay's sensitivity and specificity. Our results indicate that chemotyping by ELISA can be used as an alternative to the PCR genotyping method of GHR d3/fl due to 100% concordance rate between PCR and ELISA.
Acromegalic patients carrying the GHRd3 allele were shown to have lower levels of circulating GH (20). In our study, we demonstrated the d3/d3 group had dramatically lower serum tGHBP levels compared with the d3/fl or fl/fl group. This could translate into higher GH turnover (not measured here) that could be associated with different GH responsiveness. The tGHBP levels of the d3/fl group were also significantly lower compared with the fl/fl group. Association of the GHR exon3 polymorphism with responsiveness to GH in children and adults has been intensively investigated, but the association remains controversial. Several studies showed growth prediction models based on clinical and laboratory data can explain approximately 50% of the observed variability in short-term end points and are less accurate in predicting final height after long-term GH treatment. Hence, more anthropometric variables, biological parameters, and molecular genetics should be recruited to improve the current models. The variation in individual tGHBP levels may also play an important role in differences in responsiveness to GH and need to be investigated further in the future.
This chemotyping assay developed here to determine the GHR/GHBP polymorphism status of individuals by directly assaying serum should prove valuable in clinical research on GH and potentially have useful clinical applications due to limitations of the PCR-based genotyping method.
Acknowledgments
We thank Christian Strasburger for his help and advice on writing the manuscript.
This work was supported by grants from Eli Lilly and Co. and from National Institutes of Health Grants P01AG027734 and 2P30DK063491.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- CV
- Coefficient variation
- E3(+)GHBP
- exon 3-positive GHBP
- GHBP
- GH binding protein
- GHR
- GH receptor
- GHRd3
- transcripts with exclusion of exon 3
- GHRfl
- GHR transcripts with the retention of exon 3
- hGHBP
- human GHBP
- pAb
- polyclonal antibody
- tGHBP
- total GHBP
- UCLA
- University of California, Los Angeles.
References
- 1. Cohen P, Germak J, Rogol AD, Weng W, Kappelgaard AM, Rosenfeld RG; American Norditropin Study Group 2010. Variable degree of growth hormone (GH) and insulin-like growth factor (IGF) sensitivity in children with idiopathic short stature compared with GH-deficient patients: evidence from an IGF-based dosing study of short children. J Clin Endocrinol Metab 95:2089–2098 [DOI] [PubMed] [Google Scholar]
- 2. Pantel J, Machinis K, Sobrier ML, Duquesnoy P, Goossens M, Amselem S. 2000. Species-specific alternative splice mimicry at the growth hormone receptor locus revealed by the lineage of retroelements during primate evolution. J Biol Chem 275:18664–18669 [DOI] [PubMed] [Google Scholar]
- 3. Dos Santos C, Essioux L, Teinturier C, Tauber M, Goffin V, Bougnères P. 2004. A common polymorphism of the growth hormone receptor is associated with increased responsiveness to growth hormone. Nat Genet 36:720–724 [DOI] [PubMed] [Google Scholar]
- 4. Binder G, Baur F, Schweizer R, Ranke MB. 2006. The d3-growth hormone (GH) receptor polymorphism is associated with increased responsiveness to GH in Turner syndrome and short small-for-gestational-age children. J Clin Endocrinol Metab 91:659–664 [DOI] [PubMed] [Google Scholar]
- 5. Jorge AA, Marchisotti FG, Montenegro LR, Carvalho LR, Mendonca BB, Arnhold IJ. 2006. Growth hormone (GH) pharmacogenetics: influence of GH receptor exon 3 retention or deletion on first-year growth response and final height in patients with severe GH deficiency. J Clin Endocrinol Metab 91:1076–1080 [DOI] [PubMed] [Google Scholar]
- 6. Räz B, Janner M, Petkovic V, Lochmatter D, Eblé A, Dattani MT, Hindmarsh PC, Flück CE, Mullis PE. 2008. Influence of growth hormone (GH) receptor deletion of exon 3 and full-length isoforms on GH response and final height in patients with severe GH deficiency. J Clin Endocrinol Metab 93:974–980 [DOI] [PubMed] [Google Scholar]
- 7. Wassenaar MJ, Dekkers OM, Pereira AM, Wit JM, Smit JW, Biermasz NR, Romijn JA. 2009. Impact of the exon 3-deleted growth hormone (GH) receptor polymorphism on baseline height and the growth response to recombinant human GH therapy in GH-deficient (GHD) and non-GHD children with short stature: a systematic review and meta-analysis. J Clin Endocrinol Metab 94:3721–3730 [DOI] [PubMed] [Google Scholar]
- 8. Dörr HG, Bettendorf M, Hauffa BP, Mehls O, Rohrer T, Stahnke N, Pfäffle R, Ranke MB; German KIGS Group 2011. Different relationships between the first 2 years on growth hormone treatment and the d3-growth hormone receptor polymorphism in short small-for-gestational-age (SGA) children. Clin Endocrinol (Oxf) 75:656–660 [DOI] [PubMed] [Google Scholar]
- 9. Audí L, Carrascosa A, Esteban C, Fernández-Cancio M, Andaluz P, Yeste D, Espadero R, Granada ML, Wollmann H, Fryklund L; Spanish SGA Study Group 2008. The exon 3-deleted/full-length growth hormone receptor polymorphism does not influence the effect of puberty or growth hormone therapy on glucose homeostasis in short non-growth hormone-deficient small-for-gestational-age children: results from a two-year controlled prospective study. J Clin Endocrinol Metab 93:2709–2715 [DOI] [PubMed] [Google Scholar]
- 10. Blum WF, Machinis K, Shavrikova EP, Keller A, Stobbe H, Pfaeffle RW, Amselem S. 2006. The growth response to growth hormone (GH) treatment in children with isolated GH deficiency is independent of the presence of the exon 3-minus isoform of the GH receptor. J Clin Endocrinol Metab 91:4171–4174 [DOI] [PubMed] [Google Scholar]
- 11. Carrascosa A, Esteban C, Espadero R, Fernández-Cancio M, Andaluz P, Clemente M, Audí L, Wollmann H, Fryklund L, Parodi L; Spanish SGA Study Group 2006. The d3/fl-growth hormone (GH) receptor polymorphism does not influence the effect of GH treatment (66 microg/kg per day) or the spontaneous growth in short non-GH-deficient small-for-gestational-age children: results from a two-year controlled prospective study in 170 Spanish patients. J Clin Endocrinol Metab 91:3281–3286 [DOI] [PubMed] [Google Scholar]
- 12. Carrascosa A, Audí L, Esteban C, Fernández-Cancio M, Andaluz P, Gussinyé M, Clemente M, Yeste D, Albisu MA. 2008. Growth hormone (GH) dose, but not exon 3-deleted/full-length GH receptor polymorphism genotypes, influences growth response to two-year GH therapy in short small-for-gestational-age children. J Clin Endocrinol Metab 93:147–153 [DOI] [PubMed] [Google Scholar]
- 13. Carrascosa A, Audí L, Fernández-Cancio M, Esteban C, Andaluz P, Vilaró E, Clemente M, Yeste D, Albisu MA, Gussinyé M. 2008. The exon 3-deleted/full-length growth hormone receptor polymorphism did not influence growth response to growth hormone therapy over two years in prepubertal short children born at term with adequate weight and length for gestational age. J Clin Endocrinol Metab 93:764–770 [DOI] [PubMed] [Google Scholar]
- 14. Marchisotti FG, Jorge AA, Montenegro LR, Berger K, de Carvalho LR, Mendonca BB, Arnhold IJ. 2009. Comparison between weight-based and IGF-I-based growth hormone (GH) dosing in the treatment of children with GH deficiency and influence of exon 3 deleted GH receptor variant. Growth Horm IGF Res 19:179–186 [DOI] [PubMed] [Google Scholar]
- 15. Glad CA, Johannsson G, Carlsson LM, Svensson PA. 2010. Rapid and high throughput genotyping of the growth hormone receptor exon 3 deleted/full-length polymorphism using a tagSNP. Growth Horm IGF Res 20:270–273 [DOI] [PubMed] [Google Scholar]
- 16. Seidel B, Glasow A, Schutt M, Kiess W, Wu Z, Strasburger CJ, Kratzsch J. 2003. Association between the GH receptor/exon 3 genotype and the level of exon 3-positive GH-binding protein in human serum. Eur J Endocrinol 148:317–324 [DOI] [PubMed] [Google Scholar]
- 17. Barzilai N, Atzmon G, Schechter C, Schaefer EJ, Cupples AL, Lipton R, Cheng S, Shuldiner AR. 2003. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA 290:2030–2040 [DOI] [PubMed] [Google Scholar]
- 18. Kratzsch J, Wu Z, Kiess W, Dehmel B, Bosse-Henck A, Reuter W, Pflaum CD, Strasburger CJ. 2001. The exon 3-retaining and the exon 3-deleted forms of the growth hormone-binding protein (GHBP) in human serum are regulated differently. Clin Endocrinol (Oxf) 2001:61–68 [DOI] [PubMed] [Google Scholar]
- 19. Audí L, Esteban C, Carrascosa A, Espadero R, Pérez-Arroyo A, Arjona R, Clemente M, Wollmann H, Fryklund L, Parodi LA; Spanish SGA Study Group 2006. Exon 3-deleted/full-length growth hormone receptor polymorphism genotype frequencies in Spanish short small-for-gestational-age (SGA) children and adolescents (n = 247) and in an adult control population (n = 289) show increased fl/fl in short SGA. J Clin Endocrinol Metab 91:5038–5043 [DOI] [PubMed] [Google Scholar]
- 20. Schmid C, Krayenbuehl PA, Bernays RL, Zwimpfer C, Maly FE, Wiesli P. 2007. Growth hormone (GH) receptor isoform in acromegaly: lower concentrations of GH but not insulin-like growth factor-1 in patients with a genomic deletion of exon 3 in the GH receptor gene. Clin Chem 53:1484–1488 [DOI] [PubMed] [Google Scholar]